Abstract
The barley stem rust resistance gene Reaction to Puccinia graminis 1 (Rpg1), encoding a receptor-like kinase, confers durable resistance to the stem rust pathogen Puccinia graminis f. sp. tritici. The fungal urediniospores form adhesion structures with the leaf epidermal cells within 1 h of inoculation, followed by hyphae and haustorium formation. The RPG1 protein is constitutively expressed and not phosphorylated. On inoculation with avirulent urediniospores, it is phosphorylated in vivo within 5 min and subsequently degraded. Application of arginine-glycine-aspartic acid peptide loops prevented the formation of adhesion structures for spore attachment, the phosphorylation of RPG1, and germination of the viable spores. Arginine-glycine-aspartic acid affinity chromatography of proteins from the ungerminated avirulent rust spores led to the purification and identification of a protein with fibronectin type III and breast cancer type 1 susceptibility protein domains and a vacuolar protein sorting-associated protein 9 with a coupling of ubiquitin to endoplasmic reticulum degradation domain. Both proteins are required to induce in vivo phosphorylation and degradation of RPG1. Combined application of both proteins caused hypersensitive reaction on the stem rust-resistant cultivar Morex but not on the susceptible cultivar Steptoe. Expression studies indicated that mRNA of both genes are present in ungerminated urediniospores and are constitutively transcribed in sporelings, infected leaves, and haustoria in the investigated avirulent races. Evidence is presented that RPG1, in yeast, interacts with the two protein effectors from the urediniospores that activate cooperatively the stem rust resistance protein RPG1 long before haustoria formation.
Disease resistance in many plant pathogen interactions results from the expression of a resistance gene (R) in the plant and its cognate Avr gene in the pathogen and is often associated with rapid localized cell death or hypersensitive response (HR). Several R genes that respond to specific bacterial, fungal, viral, or oomycete pathogens have been isolated from a variety of plant species. The encoded R proteins are mostly cytoplasmic and are classified by their domain structure (1). Unlike the R genes, the Avr genes are diverse and do not share structural similarities. Several Avr genes have been cloned, and the location of their encoded proteins in the infected plant has been investigated. Molecular recognition of the pathogen effectors or avirulence proteins has been demonstrated to take place within the plant cell in some biotrophic pathogens, such as the rust, downy mildew, and powdery mildew fungi (2, 3). However, recognition of some effectors from necrotrophic fungi, such as Cladosporium fulvum (4), Rhynchosporium secalis (5), and Fusarium oxysporum (6), does not occur inside the plant cell. Recent evidence demonstrates that pathogen effectors are delivered into the host cells by several different mechanisms. The Gram-negative bacteria use the type III secretory system (7, 8), whereas certain fungi exploit haustoria for synthesis and translocation of the effectors into the host cytoplasm as is well documented with the flax rust Melampsora lini (9–11). The effector ToxA from the fungus Pyrenophora tritici repentis does not require haustoria for its delivery. Tox A is translocated into the plant cells by binding an unknown receptor via an arginine-glycine-aspartic acid (RGD) peptide loop (12, 13). Oomycetes, on the other hand, exploit both haustoria and apoplast as means for delivering the effectors (14, 15). Certain oomycete effector proteins have a unique recognizable amino acid motif (RXLR followed by an E/D-rich domain) that is assumed to function as a host-targeting signal (16). In contrast, the rice blast fungus Magnaporthe oryzae delivers the effectors secreted from the invasive hyphae through the bitrophic interfacial complex (17). Thus, the mode of effector delivery is diverse, as is its recognition and action. For instance, deviations from the classic gene-for-gene hypothesis, which supported the “one effector-one target theory,” have now become very common. Resistance pathways are activated by either a direct or indirect interaction between the R and Avr gene products. Direct physical interaction has been demonstrated for the tomato Pto R protein and the AvrPto protein (18, 19). Direct interaction has also been established for the rice PITA gene and the corresponding Avr proteins of the blast fungus Magnaporthe grisea (20) and for Arabidopsis RRS1 R and Ralstonia solanacearum PopP2 (21). The L567 flax rust avirulence genes of M. lini are expressed in haustoria, and their protein products are secreted into the plant cells (9, 10). Failure of certain Avr gene products to interact directly with corresponding R gene products formed the basis for the “guard hypothesis” (22), according to which R proteins “guard” certain host proteins (guardees) that are manipulated by pathogen effectors (23). One well-known guardee is RIN4, which is targeted by at least three bacterial effector proteins (AvrRpm1, AvrB, and AvrRpt2) and guarded by two R proteins (RPM1 and RPS2) (24–26). Another guardee is PBS1, which is targeted by the bacterial effector protein AvrPphB and guarded by RPS5 (27). Although the guard hypothesis supports the indirect recognition of the pathogen by the NBS-LRR proteins, it fails to address the virulence activity (28, 29). This failure gave rise to another concept called the “decoy model” (30). The decoy model describes the effectors to act as a molecular sensor of the virulence activity of pathogens. Support for the decoy model comes from the Pto gene, which was originally thought to be a Pseudomonas syringae virulence target, with its partner, Prf, acting as a guard monitoring the activity of AvrPto and AvrPtoB (22). However, the virulence targets of AvrPto and AvrPtoB now appear to be the kinase domains of the receptor-like kinases CERK1, BAK1, EFR1, and FLS2 (31–34). These observations suggest that Pto is being used as bait by Prf to interact with effector proteins that normally target other kinases. In summary, different effectors may target a single R gene or a single effector may target several R genes. These R and Avr gene products may interact either directly or indirectly with each other, but no two Avr genes or effectors have so far been shown to interact with each other.
The emergence of the deadly wheat stem rust race TTKSK (aka Ug99) has renewed interest in understanding how plants perceive these pathogens and activate resistance pathways. The barley stem rust R gene, Reaction to Puccinia graminis 1 (Rpg1), confers durable resistance against Puccinia graminis f. sp. tritici (Pgt) (35). Rpg1 was cloned (36), and transgenic plants of the susceptible barley cultivar (cv.) Golden Promise expressing Rpg1 were completely resistant against stem rust (37). Rpg1 encodes a receptor-like kinase with dual kinase domains (36). The protein kinase 2 (pK2) domain is catalytically active, whereas the pK1 is a pseudokinase, but both domains are required for stem rust resistance (38). Rpg1 is constitutively expressed (39) and is mostly cytoplasmic, with a small fraction associated with the plasma membrane (38). On inoculation with stem rust urediniospores, RPG1 is phosphorylated in vivo within 5 min (40) and is subsequently degraded (41). Both actions are required for resistance.
Function and adhesion of bean rust (Uromyces appendiculatus) urediniospores can be blocked by a synthetic RGD peptide loop, which masks the extracellular domain of an integrin-like receptor and prevents the formation of appressoria, which is required for successful infection (42). Schindler et al. (43) used RGD affinity chromatography to purify a 70- to 72-kDa polyprotein that cross-reacted with antivitronectin antisera from soybean cells. This led us to use RGD affinity chromatography to purify proteins that bind to RGD peptide loops from the ungerminated avirulent stem rust urediniospores of barley. It led to the isolation and identification of two discrete proteins, a hypothetical protein [P. graminis f. sp. tritici genome (PGTG_10537.2)] with fibronectin type III and breast cancer type 1 susceptibility protein (BRCA1) C-terminal domains and a vacuolar protein sorting-associated protein 9 (VPS9) with a coupling of ubiquitin to endoplasmic reticulum degradation (CUE) domain (PGTG_16791). We suggest that the fibronectin domain of the hypothetical protein binds to the RGD column and the VPS9 protein coelutes with the RGD-binding protein, because VPS9 does not bind RGD and they probably exist as a complex in vivo.
Results
Adhesion Structures of Pgt Urediniospores to Barley Leaf Epidermal Cells Appear Within a Single Hour of Inoculation, and Their Formation Is Prevented by Synthetic RGD Peptide Loops.
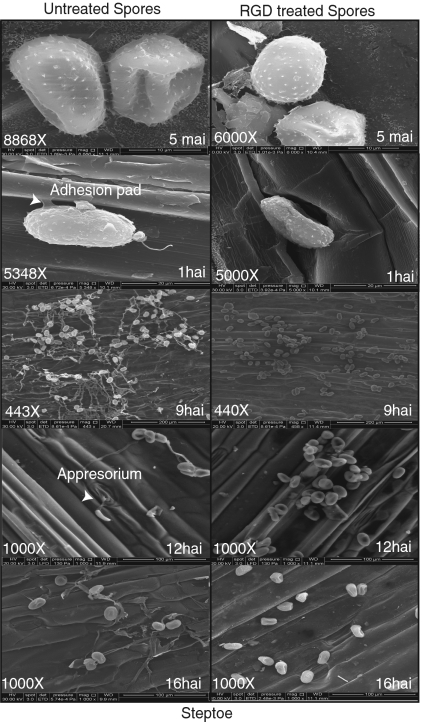
We incubated 100 mg of MCCF urediniospores with 750 μL (10 μg/μL) of RGD peptides for 15 min and dried them for 2 h. After rehydration for 4 h, they were inoculated onto the leaves. The spores treated with the RGD peptides failed to induce phosphorylation of RPG1 despite being viable based on their ability to grow on 2% (wt/vol) water agar. They also did not cause disease or sporulation on the susceptible cv. Steptoe. Analyses by scanning EM yielded the following results. Although we could not find any morphological differences in the first 5 min, within which time RPG1 is phosphorylated, the RGD-treated spores did not germinate or grow on the leaf surfaces (Fig. 1, Fig. S1, and Table S1). On the other hand, the untreated spores germinated and formed adhesion pads and germtubes within 1 h after inoculation when studied on an angle to the epidermis (Fig. 1). The observed adhesion pads may provide a means for transfer of avirulence proteins from the urediniospores to the host.
Fig. 1.
Inhibition of adhesion pad formation and germination by treatment of Pgt race MCCF urediniospores with RGD peptide loops. Untreated or RGD peptide-treated spores were inoculated on the leaf surface of 10-d-old susceptible cv. Steptoe seedlings. Leaf samples were observed at 5 min to 16 h after inoculation by scanning EM. Untreated spores formed adhesion connections between the epidermal cell and the spore within 1 h after inoculation, followed by germination. The RGD-treated spores failed to form adhesion pads and germinate. hai, hours after inoculation; mai, minutes after inoculation.
Isolation and Characterization of RGD-Binding and VPS9-Like Proteins.
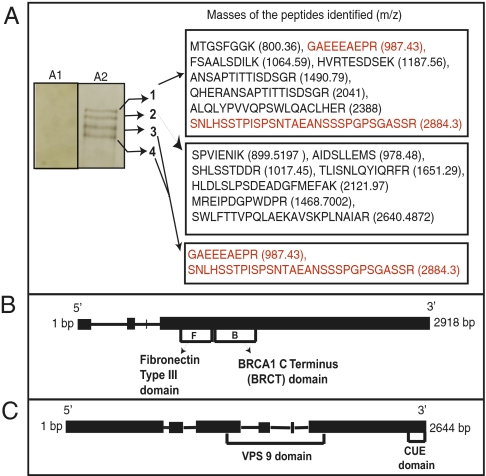
Urediniospores treated with the synthetic RGD peptide failed to induce RPG1 phosphorylation (Fig. S2). Such spores were able to germinate on 2% (wt/vol) water agar, indicating that they are viable, but did not cause disease on susceptible cultivars. This prompted us to isolate and purify proteins that bind to the RGD tripeptide using RGD-coupled Sepharose. The unbound Sepharose by itself did not bind or purify any protein from the rust spore extract, but RGD affinity chromatography of the spore extract yielded a product that caused RPG1 phosphorylation, degradation, and HR when applied to barley leaves (Fig. S3 A and B). Gel fractionation of this product yielded four distinct bands (Fig. 2A). MS and comparison with the Puccinia database (Genebank NCBI Genome Project Identification #18535 Puccinia graminis f. sp. tritici CRL 75-36-700-3) identified band 1 protein (Fig. 2A and Fig. S4A) as a hypothetical protein (PGTG_10537.2) with fibronectin type III and BRCA1 C-terminal domains (Fig. 2B), henceforth referred to as the RGD-binding protein. The RGD-binding gene, with four exons and three introns, codes for a predicted protein of 818 aa. Band 2 protein is a VPS9 with a CUE domain (PGTG_16791) (Fig. 2 A and C and Fig. S4B). The VPS9 gene, with six exons and five introns, codes for a predicted protein of 744 aa. The VPS9 protein coeluted with the RGD-binding protein. Bands 3 and 4 are truncated fragments of the RGD-binding protein.
Fig. 2.
Isolation and characterization of RGD-binding and VPS9 proteins. (A1) Control sample from unmodified Sepharose column. (A2) Proteins isolated by Sepharose RGD affinity chromatography, separated by SDS/PAGE on a 10–20% gradient gel, and visualized by silver staining. Band 1 shows the amino acid sequence and mass of protein with a BRCA1 C-terminal domain and a fibronectin type III domain (red letters). Band 2 shows VPS9 with a CUE domain. Bands 3 and 4 show truncated versions of protein in band 1. (B) Gene structure of PGTG_10537.2 encoding RGD-binding protein with 818 amino acids in four exons. (C) Gene structure of PGTG_16791 encoding VPS9 with 744 amino acids.
RGD-Binding and VPS9 Proteins Together Induce in Vivo Phosphorylation and Degradation of RPG1.
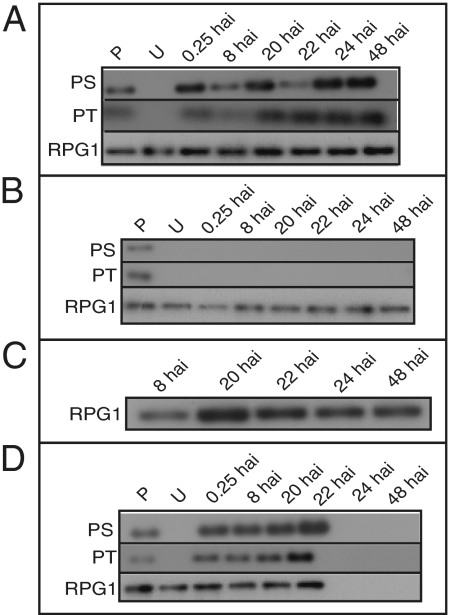
The two genes were cloned, and the proteins were purified from Pichia pastoris transformants. Application of the RGD-binding protein onto leaves of barley cv. Morex containing a functional Rpg1 gene induced RPG1 protein phosphorylation (Fig. 3A). However, the phosphorylated RPG1 protein persisted for at least 48 h. Application of the VPS9 protein did not induce RPG1 phosphorylation (Fig. 3B) or degradation (Fig. 3C). Application of both proteins together in equal quantities induced RPG1 phosphorylation and degradation after 22 h, as observed after inoculation with avirulent urediniospores (Fig. 3D).
Fig. 3.
RGD-binding and VPS9 proteins together induce in vivo phosphorylation and degradation of RPG1. (A) Infiltration of leaf with purified RGD-binding protein induced phosphorylation of RPG1 but failed to degrade RPG1 after 22 h. Infiltration with purified VPS9 failed to induce phosphorylation of RPG1 (B) and RPG1 degradation up to 48 h (C). (D) Infiltration of both proteins together induced RPG1 phosphorylation within 0.25 h and degradation before 24 h. RPG1 was immunoprecipitated with specific antibodies, and phosphorylation was detected with either phosphoserine or phosphothreonine antibody. The RPG1 row is a loading control. hai, hours after inoculation with stem rust urediniospores; P, in vitro phosphorylated RPG1 protein l; PS, phosphoserine; PT, phosphothreonine; U, uninoculated control.
RGD-Binding and VPS9 Proteins Together Induce HR in Barley Harboring a Functional Rpg1 Gene.
We tested the ability of the RGD-binding and VPS9 proteins individually or in combination to elicit HR in plants. HR was observed only when both proteins were applied together and only on resistant (with Rpg1) cv. Morex and not on susceptible (without Rpg1) cv. Steptoe (Fig. 4A). To rule out the possibility that HR may be nonspecific because of infiltration of increased protein concentration in the combined protein sample, we tested the ability of BSA to cause HR by itself or in a 1:1 ratio with either the RGD-binding protein or the VPS9 protein. HR is a specific reaction attributable to the infiltration of the VPS9 and the RGD-binding proteins only and did not manifest in the presence of BSA (Fig. S5A). BSA by itself did not cause HR. Further, the infiltration of a 1:1:1 ratio of BSA/RGD-binding protein/VPS9 protein retained its ability to cause HR only in the resistant cv. Morex, indicating that BSA did not have an inhibitory effect on HR (Fig. S5A). Because Morex and Steptoe differed for this clear and easily scored HR, we mapped the gene controlling the phenotype on a Steptoe × Morex mapping population (44). The HR cosegregated with the Rpg1 gene in 33 selected lines. An additional segregant from a different population, ASM170, was tested because it has a crossover event within the Rpg1 gene rendering it nonfunctional (36). This line failed to show HR (Fig. 4B).
Fig. 4.
RGD-binding and VPS9 proteins together induce HR in barley harboring a functional Rpg1 gene. (A) Infiltration of both purified proteins together elicited HR in the resistant cv. Morex (with Rpg1) but not in the susceptible cv. Steptoe (without Rpg1). HR was not elicited when the two proteins were infiltrated individually. (B) Combined infiltration of the two proteins failed to elicit HR in the susceptible line ASM170 with a recombined defective Rpg1. (C) Combined infiltration of RGD-binding and VPS9 proteins elicited HR in the resistant transgenic line GP/Rpg1 T1 but not in its host cv. Golden Promise, lacking Rpg1. HR also failed to manifest in the susceptible Rpg1 mutant transgenes GP/Rpg1-pK1 (KK152/153NQ) and GP/Rpg1-pK2 (KK461/462NQ). HR was photographed on the third day after protein infiltration.
Tests were conducted with three transgenic lines carrying either the Rpg1 gene (37) or defective Rpg1 genes with mutations in the pK1 (KK152/153NQ) or pK2 (KK461/462NQ) domain (38). The stem rust-susceptible parent cv. Golden Promise (without Rpg1) and the transformants with defective Rpg1 genes failed to show HR, whereas the functional transgenic GP-Rpg1/T1 in the Golden Promise genetic background (37) showed HR (Fig.4 C). These data implicate Rpg1 as the prime candidate responsible for HR.
RGD-Binding and VPS9 Proteins Interact with RPG1 and with Each Other in Yeast.
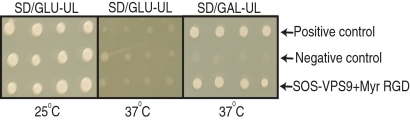
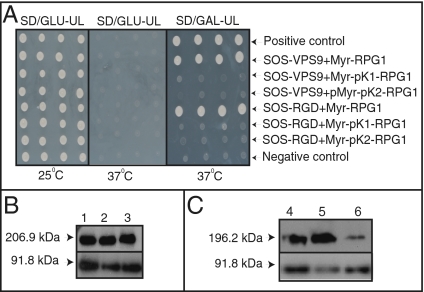
Yeast two-hybrid assays confirmed that the RGD-binding and VPS9 proteins interact with each other and with RPG1 (Figs. 5 and 6). In addition, the RPG1-pK1 (KK152/153NQ) and RPG1-pK2 (KK461/462NQ) mutants failed to interact with the RGD-binding or VPS9 protein (Fig. 6). The RPG1-pK1 mutant has a mutation in the inactive pK1 domain, in which asparagine and glutamine are substituted for lysines 152 and 153. The RPG1-pK2 mutant has a mutation in the catalytically active pK2 domain, in which asparagine and glutamine are substituted for lysines 461 and 462. These two mutants result in a susceptible phenotype (38) and fail to cause HR. The pK2 domain plays an active role in phosphorylation, and hence in resistance, whereas the role played by the pK1 domain is unclear. With these two mutants, we investigated the role of the pK1 domain in protein-protein interactions with the fungal effectors and its effect on phosphorylation and HR. Protein interactions in the interacting yeast clones were verified by Western blot analysis. RPG1 was detected by RPG1-specific antibody, whereas the RGD-binding protein and VPS9 protein were confirmed using a commercially available Son of Sevenless (SOS) antibody. All three proteins are expressed, as evidenced by Western blot analysis (Fig. 6 B and C and Fig. S5B).
Fig. 5.
RGD-binding and VPS9 proteins interact with each other in yeast. The VPS9 gene cloned in the pSOS vector was cotransformed with the RGD-binding gene cloned in the pMyr vector into the cdc25Ha yeast strain. The cotransformants were plated on Synthetic Drop out/Glucose-Uracil/Leucine medium and allowed to grow at 25 °C. Colonies growing at 25 °C were tested for protein interaction on Synthetic Drop out/Glucose-Uracil/Leucine and Synthetic Drop out/Galactose-Uracil/Leucine medium at 37 °C. The clones growing on Synthetic Drop out/Galactose-Uracil/Leucine at 37 °C and not growing on Synthetic Drop out/Glucose-Uracil/Leucine at 37 °C were scored as positive, exhibiting protein-protein interaction. Synthetic Drop out/Glucose-Uracil/Leucine is a medium that allows the growth of all clones at 25 °C and demonstrates that live yeast cells are present. SOS-MAFB + Myr-MAFB served as a positive control, and SOS-MAFB + Myr Lamin C served as a negative control. MAFB, V-maf masculoaboneurotic fibrosarcoma oncogene homolog B; SD/GAL-UL, Synthetic Drop out/Galactose-Uracil/Leucine; SD/Glu-UL, Synthetic Drop out/Glucose-Uracil/Leucine.
Fig. 6.
RGD-binding and the VPS9 proteins interact with RPG1. (A) RGD-binding and VPS9 proteins interact with RPG1, whereas the RPG1-pK1 and RPG1-pK2 mutant proteins fail to interact with either the RGD-binding or VPS9 protein. The VPS9 and RGD-binding genes were cloned in pSOS vector as the bait. RPG1 and its mutant derivatives, RPG1-pK1 (KK152/153NQ) and RPG1-pK2 (KK461/462NQ), were cloned in the pMyr vector as prey. The prey and bait constructs were cotransformed into the cdc25Ha yeast strain in different combinations as indicated in the figure. The cotransformants were plated on Synthetic Drop out/Glucose-Uracil/Leucine medium and allowed to grow at 25 °C. The colonies growing at 25 °C were checked for interaction on Synthetic Drop out/Glucose-Uracil/Leucin and Synthetic Drop out/Galactose-Uracil/Leucine medium at 37 °C. The clones growing on Synthetic Drop out/Galactose-Uracil/Leucine medium at 37 °C and not growing on Synthetic Drop out/Glucose-Uracil/Leucin medium at 37 °C were scored as positive, exhibiting protein-protein interaction. A description of the media and growing temperature functions is included in the legend for Fig. 5. SD/Gal-UL, Synthetic Drop out/Galactose-Uracil/Leucine; SD/Glu-UL, Synthetic Drop out/Glucose-Uracil/Leucine. (B and C) Western blot showing that the Myr-RPG1, Myr-pK1-RPG1, Myr-pK2-RPG1, SOS-RGD, and SOS-VPS9 proteins are expressed in the yeast clones used for the protein interaction assay. SOS-RGD and SOS-VPS9 were detected by SOS antibody, whereas Myr-RPG1 and its mutant derivatives were detected by RPG1 antibody. SOS-MAFB + Myr-MAFB served as a positive control, and SOS-MAFB + Myr Lamin C served as a negative control. The positive and negative controls were provided by the suppliers of the Cytotrap kit (Stratagene). MAFB, V-maf masculoaboneurotic fibrosarcoma oncogene homolog B.
To validate the interactions further, we tested the ability of RPG1, RPG1-pK1, RPG1-pK2, RGD-binding protein, and VPS9 protein to interact with a known noninteractor of RPG1, the Arabidopsis Sleepy 1 gene product (AtSLY1) (45). RPG1, RPG1-pK1, and RPG1-pK2 failed to interact with AtSLY1 (Fig. S6 A and B). All six proteins were expressed, as evidenced by Western blot analysis (Figs. S6B and S7 B and C). An advantage of using AtSLY1 is that it is an F-box protein known to recruit protein complexes and to mediate protein-protein interactions (45). Therefore, failure of RPG1 and its mutant derivatives, RGD-binding and VPS9 proteins, to interact with AtSLY1 further strengthens our yeast two-hybrid data.
Expression Studies and Sequencing of the RGD-Binding and VPS9 Genes from the Avirulent Race MCCF.
To compare the RGD-binding and VPS9 gene sequences from the avirulent race MCCF used in this study with the avirulent race SCCLc7a, for which a genome sequence is available, we amplified and resequenced the mRNA from both genes. Sequence analysis indicated that both the RGD-binding and VPS9 genes appear to be functional in the avirulent race MCCF, with a few SNPs that did not alter the amino acid sequence compared with the SCCLc7a sequence. Expression studies indicated that mRNA of both targeted genes is present in ungerminated urediniospores and is constitutively transcribed in sporelings, infected leaves, and haustoria in the avirulent races MCCF and SCCLc7a.
Discussion
Fibronectins are globular proteins with an RGD-binding motif and act as adhesive ligands connecting the cytoskeleton and the ECM (46). They mediate cellular interactions between the ECM and plasma membrane (47). Proteins containing potentially active RGD motifs attach to mammalian cells by binding via integrin transmembrane proteins with one domain outside the plasma membrane and the other inside (48, 49). They mediate bidirectional transmembrane signaling events involved in pathogenesis and defense (50). BRCA1 C-terminal domain-containing proteins are involved in cell cycle regulation, DNA metabolism, phosphopeptide binding, and protein-protein interactions (51, 52). VPS9 is a vacuolar sorting protein with a CUE domain that catalyzes the nucleotide exchange of Rab5 through G proteins, activating the GDP-GTP exchange factors, which serve an essential function in intracellular protein trafficking and cell signaling events (53). The CUE domain of VPS9 forms a high-affinity ubiquitin-binding pocket required for vacuolar or endocytotic transport (53) of surface-attenuated receptors, thereby promoting intramolecular ubiquitination (54). Shan et al. (55) have shown that Avr1b protein synthesized in the yeast P. pastoris could trigger an interaction with Rps1b when infiltrated in soybean leaves, suggesting that the protein could enter the plant cells unaided by any other pathogen-encoded molecules. In our case, the two proteins were infiltrated together to cause HR by a yet to be identified mechanism. RPG1 degradation could be related to HR and could possibly act to stop runaway HR. Another hypothesis for the effect of RPG1 degradation is that it initiates a disease resistance signaling complex, which may be negatively regulated by RPG1.
Conclusion
Here, we present evidence that RPG1 interacts in yeast, with the RGD-binding and VPS9 proteins from stem rust pathogens triggering rapid phosphorylation and eventual degradation of RPG1. The process initiates HR and resistance to stem rust. A possible hypothesis is that RPG1 behaves as a receptor recognizing the pathogen and transducing the transmembrane signals. The RPG1 protein does not have a RGD motif. Therefore, it is possible that the pathogen RGD-binding protein recognizes some other receptor in a complex with RPG1 initiating the signaling process. We show that two protein effectors associated with the stem rust urediniospore surface work cooperatively to activate the stem rust R protein RPG1 long before haustoria formation. Based on these observations, we speculate that a unique mechanism for pathogen recognition and signaling in the host cell is functioning here.
Materials and Methods
Rust Inoculation.
Seedlings were grown in a growth chambers at 24 °C and 80% relative humidity for 1 wk before inoculation. Plants were inoculated with the rust fungus (at a rate of 0.025 mg of spores per plant) as previously described (40). Tissue samples were collected as indicated in the figures.
Treatment of Spores with RGD Peptides.
About 100 mg of MCCF urediniospores was mixed with 750 μL (10 μg/μL) of the RGD peptide (Sigma–Aldrich) and incubated for 15 min at room temperature. The urediniospore suspension was spun at 13,000 × g for 5 min and dried for 2 h at room temperature. The urediniospores were rehydrated in a Petri dish for 4 h at room temperature and applied onto the leaves. The plants were sampled 15 and 30 min after application and assayed for RPG1 phosphorylation as previously described (40).
Isolation and Analysis of RGD-Binding Protein from Pgt Race MCCF Spores by RGD Affinity Chromatography.
RGD peptides were synthesized and coupled to Sepharose by Alpha Diagnostics. The RGD-binding protein was purified according to a modified protocol of Schindler et al. (43). The rust urediniospores (100 mg) were hydrated at room temperature and grown in 500 mL of germination solution containing 1 mL of 500× stock germination solution [72 μL of nonanol, 0.5 μL of Tween 20, 10 mL of ethanol, and 10 mL of Milli Q water (Barnstead International, Dubuque, IA)] and 499 mL of Milli Q water for 3 h. The germlings were filtered through a 0.4-μm muslin cloth, and associated proteins were solubilized by the addition of 20 mL of 200 mM octyl β-d-glucoside in PBS (pH 7.2) containing 1 mM CaCl2, 1 mM MgCl2, and 1 mM PMSF. The mix was vigorously vortexed for 5 min, and the extraction was allowed to take place for 30 min at 4 °C, after which the insoluble cell debris was pelleted at 15,000 × g. The supernatant was applied to a 10-mL column containing RGD peptides coupled to Sepharose (20 mg of peptide per 1 mL of Sepharose) and allowed to bind overnight at 4 °C. The column was washed with 50 column volumes of PBS containing 50 mM octyl β-d-glucoside and eluted with the same buffer containing 0.1 mg/mL RGD. The eluate was concentrated 50-fold by centrifugation with a Centricon membrane filter (30-kDa cutoff; Millipore) and examined by running on a 10–20% SDS/PAGE gel and silver staining.
The crude extract was applied to the leaves and investigated for phosphorylation and degradation of RPG1. The separated proteins were cut from the silver-stained SDS/PAGE gels and subjected to MALDI-MS and MS/MS analysis, and proteins were identified by blasting against the Puccinia group (GenBank NCBI Genome Project Identification #18535 Puccinia graminis f. sp. tritici CRL 75-36-703) database. Primers were designed based on the race SCCLc7a sequence, and the genes were amplified by PCR and sequenced from the avirulent race MCCF. Because of the inability to amplify the full-length RGD-binding gene by RT-PCR directly, it was synthesized by Genscript (Genscript USA Inc. Piscathaway, NJ) based on the sequence derived from the avirulent race MCCF, cloned into a pUC vector, and used as a template for amplifying and cloning into the expression vector and the yeast two hybrid vectors as described below. The full-length VPS9 gene was cloned by RT-PCR, using the Advantage 2 polymerase from Clontech, and was cloned into pGEMT vector (Promega) for further cloning into the expression vectors as described in SI Materials and Methods.
Bait and Prey Construction.
Full-length Rpg1 cDNA was cloned in-frame with human SOS (h-SOS) at the NotI site or in the pMyr vector, and orientation was determined by sequencing. The RPG1-pK1 and RPG1-pK2 mutants were generated by site-specific mutagenesis using the Quik Change system (Stratagene) with primers specified in Table S2. The site-directed mutations, as well as the absence of undesired mutations, were confirmed by DNA sequencing. The full-length RGD-binding and VPS9 genes were cloned at either the SalI (SalI RGD pMyr forward and reverse primers) or EcoRI/Xho1 (EcoRI VPS9 pMyr forward and XhoI VPS9 pMyr reverse primers) site of the pMyr vector or the NotI site of the pSOS vector and tested for interaction with RPG1. The full-length RGD-binding gene was cloned in frame with h-SOS at the Not1 site to test for interaction with VPS9 using the NotI RGD SOS forward and reverse primers (Table S2).
Supplementary Material
Acknowledgments
Technical assistance provided by Dr. Valerie Lynch of the Vince Franceschi Imaging and Microscopy Center is highly acknowledged. Research was supported by the National Research Initiative of the United States Department of Agriculture Cooperative State Research, Education, and Extension Service (Grant 2007-35301-18205), National Institute of Food and Agriculture (Grant 2010-65108-20568), and National Institutes of Health (Grant 1RO1GM080749-01A1).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1111771108/-/DCSupplemental.
References
- 1.Martin GB, Bogdanove AJ, Sessa G. Understanding the functions of plant disease resistance proteins. Annu Rev Plant Biol. 2003;54:23–61. doi: 10.1146/annurev.arplant.54.031902.135035. [DOI] [PubMed] [Google Scholar]
- 2.Ellis JG, Dodds PN, Lawrence GJ. Flax rust resistance gene specificity is based on direct resistance-avirulence protein interactions. Annu Rev Phytopathol. 2007;45:289–306. doi: 10.1146/annurev.phyto.45.062806.094331. [DOI] [PubMed] [Google Scholar]
- 3.Ridout CJ, et al. Multiple avirulence paralogues in cereal powdery mildew fungi may contribute to parasite fitness and defeat of plant resistance. Plant Cell. 2006;18:2402–2414. doi: 10.1105/tpc.106.043307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Luderer R, Takken FL, de Wit PJ, Joosten MH. Cladosporium fulvum overcomes Cf-2-mediated resistance by producing truncated AVR2 elicitor proteins. Mol Microbiol. 2002;45:875–884. doi: 10.1046/j.1365-2958.2002.03060.x. [DOI] [PubMed] [Google Scholar]
- 5.Rohe M, et al. The race-specific elicitor, NIP1, from the barley pathogen, Rhynchosporium secalis, determines avirulence on host plants of the Rrs1 resistance genotype. EMBO J. 1995;14:4168–4177. doi: 10.1002/j.1460-2075.1995.tb00090.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rep M, et al. A small, cysteine-rich protein secreted by Fusarium oxysporum during colonization of xylem vessels is required for I-3-mediated resistance in tomato. Mol Microbiol. 2004;53:1373–1383. doi: 10.1111/j.1365-2958.2004.04177.x. [DOI] [PubMed] [Google Scholar]
- 7.Abramovitch RB, Anderson JC, Martin GB. Bacterial elicitation and evasion of plant innate immunity. Nat Rev Mol Cell Biol. 2006;7:601–611. doi: 10.1038/nrm1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Block A, Li G, Fu ZQ, Alfano JR. Phytopathogen type III effector weaponry and their plant targets. Curr Opin Plant Biol. 2008;11:396–403. doi: 10.1016/j.jbi.2008.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Catanzariti AM, Dodds PN, Lawrence GJ, Ayliffe MA, Ellis JG. Haustorially expressed secreted proteins from flax rust are highly enriched for avirulence elicitors. Plant Cell. 2006;18:243–256. doi: 10.1105/tpc.105.035980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dodds PN, Lawrence GJ, Catanzariti AM, Ayliffe MA, Ellis JG. The Melampsora lini AvrL567 avirulence genes are expressed in haustoria and their products are recognized inside plant cells. Plant Cell. 2004;16:755–768. doi: 10.1105/tpc.020040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kemen E, et al. Identification of a protein from rust fungi transferred from haustoria into infected plant cells. Mol Plant Microbe Interact. 2005;18:1130–1139. doi: 10.1094/MPMI-18-1130. [DOI] [PubMed] [Google Scholar]
- 12.Manning VA, Ciuffetti LM. Localization of Ptr ToxA produced by Pyrenophora tritici-repentis reveals protein import into mesophyll cells. Plant Cell. 2005;17:3203–3212. doi: 10.1105/tpc.105.035063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sarma GN, Manning VA, Ciuffetti LM, Karplus PA. Structure of Ptr ToxA: An RGD-containing host-selective toxin from Pyrenophora tritici-repentis. Plant Cell. 2005;17:3190–3202. doi: 10.1105/tpc.105.034918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Whisson SC, et al. A translocation signal for delivery of oomycete effector proteins into host plant cells. Nature. 2007;450:115–118. doi: 10.1038/nature06203. [DOI] [PubMed] [Google Scholar]
- 15.Thomma BPHJ, VAN Esse HP, Crous PW, DE Wit PJGM. Cladosporium fulvum (syn. Passalora fulva), a highly specialized plant pathogen as a model for functional studies on plant pathogenic Mycosphaerellaceae. Mol Plant Pathol. 2005;6:379–393. doi: 10.1111/j.1364-3703.2005.00292.x. [DOI] [PubMed] [Google Scholar]
- 16.Birch PRJ, et al. Towards understanding the virulence functions of RXLR effectors of the oomycete plant pathogen Phytophthora infestans. J Exp Bot. 2009;60:1133–1140. doi: 10.1093/jxb/ern353. [DOI] [PubMed] [Google Scholar]
- 17.Khang CH, et al. Translocation of Magnaporthe oryzae effectors into rice cells and their subsequent cell-to-cell movement. Plant Cell. 2010;22:1388–1403. doi: 10.1105/tpc.109.069666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tang X, et al. Initiation of plant disease resistance by physical interaction of AvrPto and Pto kinase. Science. 1996;274:2060–2063. doi: 10.1126/science.274.5295.2060. [DOI] [PubMed] [Google Scholar]
- 19.Scofield SR, et al. Molecular basis of gene-for gene specificity in bacterial speck disease of tomato. Science. 1996;274:2063–2065. doi: 10.1126/science.274.5295.2063. [DOI] [PubMed] [Google Scholar]
- 20.Jia Y, McAdams SA, Bryan GT, Hershey HP, Valent B. Direct interaction of resistance gene and avirulence gene products confers rice blast resistance. EMBO J. 2000;19:4004–4014. doi: 10.1093/emboj/19.15.4004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Deslandes L, et al. Physical interaction between RRS1-R, a protein conferring resistance to bacterial wilt, and PopP2, a type III effector targeted to the plant nucleus. Proc Natl Acad Sci USA. 2003;100:8024–8029. doi: 10.1073/pnas.1230660100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Van der Biezen EA, Jones JD. Plant disease-resistance proteins and the gene-for-gene concept. Trends Biochem Sci. 1998;23:454–456. doi: 10.1016/s0968-0004(98)01311-5. [DOI] [PubMed] [Google Scholar]
- 23.Jones JDG, Dangl JL. The plant immune system. Nature. 2006;444:323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 24.Axtell MJ, Staskawicz BJ. Initiation of RPS2-specified disease resistance in Arabidopsis is coupled to the AvrRpt2-directed elimination of RIN4. Cell. 2003;112:369–377. doi: 10.1016/s0092-8674(03)00036-9. [DOI] [PubMed] [Google Scholar]
- 25.Mackey D, Holt BF, 3rd, Wiig A, Dangl JL. RIN4 interacts with Pseudomonas syringae type III effector molecules and is required for RPM1-mediated resistance in Arabidopsis. Cell. 2002;108:743–754. doi: 10.1016/s0092-8674(02)00661-x. [DOI] [PubMed] [Google Scholar]
- 26.Mackey D, Belkhadir Y, Alonso JM, Ecker JR, Dangl JL. Arabidopsis RIN4 is a target of the type III virulence effector AvrRpt2 and modulates RPS2-mediated resistance. Cell. 2003;112:379–389. doi: 10.1016/s0092-8674(03)00040-0. [DOI] [PubMed] [Google Scholar]
- 27.Shao F, et al. Cleavage of Arabidopsis PBS1 by a bacterial type III effector. Science. 2003;301:1230–1233. doi: 10.1126/science.1085671. [DOI] [PubMed] [Google Scholar]
- 28.Gassmann W, Hinsch ME, Staskawicz BJ. The Arabidopsis RPS4 bacterial-resistance gene is a member of the TIR-NBS-LRR family of disease-resistance genes. Plant J. 1999;20:265–277. doi: 10.1046/j.1365-313x.1999.t01-1-00600.x. [DOI] [PubMed] [Google Scholar]
- 29.Harrison BD. Virus variation in relation to resistance breaking in plants. Euphytica. 2002;124:181–192. [Google Scholar]
- 30.vander Hoorn RAL, Kamoun S. From Guard to Decoy: A new model for perception of plant pathogen effectors. Plant Cell. 2008;20:2009–2017. doi: 10.1105/tpc.108.060194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gimenez-Ibanez S, et al. AvrPtoB targets the LysM receptor kinase CERK1 to promote bacterial virulence on plants. Curr Biol. 2009;19:423–429. doi: 10.1016/j.cub.2009.01.054. [DOI] [PubMed] [Google Scholar]
- 32.Göhre V, Robatzek S. Breaking the barriers: Microbial effector molecules subvert plant immunity. Annu Rev Phytopathol. 2008;46:189–215. doi: 10.1146/annurev.phyto.46.120407.110050. [DOI] [PubMed] [Google Scholar]
- 33.Shan L, et al. Bacterial effectors target the common signaling partner BAK1 to disrupt multiple MAMP receptor-signaling complexes and impede plant immunity. Cell Host Microbe. 2008;4:17–27. doi: 10.1016/j.chom.2008.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Xiang T, et al. Pseudomonas syringae effector AvrPto blocks innate immunity by targeting receptor kinases. Curr Biol. 2008;18:74–80. doi: 10.1016/j.cub.2007.12.020. [DOI] [PubMed] [Google Scholar]
- 35.Steffenson BJ. Analysis of durable resistance to stem rust in barley. Euphytica. 1992;63:153–167. [Google Scholar]
- 36.Brueggeman R, et al. The barley stem rust-resistance gene Rpg1 is a novel disease-resistance gene with homology to receptor kinases. Proc Natl Acad Sci USA. 2002;99:9328–9333. doi: 10.1073/pnas.142284999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Horvath H, et al. Genetically engineered stem rust resistance in barley using the Rpg1 gene. Proc Natl Acad Sci USA. 2003;100:364–369. doi: 10.1073/pnas.0136911100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nirmala J, et al. Subcellular localization and functions of the barley stem rust resistance receptor-like serine threonine specific protein kinase receptor. Proc Natl Acad Sci USA. 2006;103:7518–7523. doi: 10.1073/pnas.0602379103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rostoks N, Steffenson BJ, Kleinhofs A. Structure and expression of the barley stem rust resistance gene Rpg1. Physiol Mol Plant Pathol. 2004;64:91–101. [Google Scholar]
- 40.Nirmala J, Drader T, Chen X, Steffenson BJ, Kleinhofs A. Stem rust spores elicit rapid RPG1 phosphorylation. Mol Plant Microbe Interact. 2010;23:1635–1642. doi: 10.1094/MPMI-06-10-0136. [DOI] [PubMed] [Google Scholar]
- 41.Nirmala J, et al. Proteolysis of the barley receptor-like protein kinase RPG1 by a proteasome pathway is correlated with Rpg1-mediated stem rust resistance. Proc Natl Acad Sci USA. 2007;104:10276–10281. doi: 10.1073/pnas.0703758104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Correa JA, Staples RC, Hoch HC. Inhibition of thigmostimulated cell differentiation with RGD-peptides in Uromyces germlings. Protoplasma. 1996;194:91–102. [Google Scholar]
- 43.Schindler M, Meiners S, Cheresh DA. RGD-dependent linkage between plant cell wall and plasma membrane: Consequences for growth. J Cell Biol. 1989;108:1955–1965. doi: 10.1083/jcb.108.5.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kleinhofs A, Graner A. In: DNA Based Markers in Plants. Phillips RL, Vasil IK, editors. Dordrecht: Kluwers; 2001. pp. 402–420. [Google Scholar]
- 45.Dill A, Thomas SG, Hu J, Steber CM, Sun TP. The Arabidopsis F-box protein SLEEPY1 targets gibberellin signaling repressors for gibberellin-induced degradation. Plant Cell. 2004;16:1392–1405. doi: 10.1105/tpc.020958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Pierschbacher MD, Ruoslahti E. Cell attachment activity of fibronectin can be duplicated by small synthetic fragments of the molecule. Nature. 1984;309:30–33. doi: 10.1038/309030a0. [DOI] [PubMed] [Google Scholar]
- 47.Hynes RO, Yamada KM. Fibronectins: Multifunctional modular glycoproteins. J Cell Biol. 1982;95:369–377. doi: 10.1083/jcb.95.2.369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Isberg RR, Tran Van Nhieu G. Binding and internalization of microorganisms by integrin receptors. Trends Microbiol. 1994;2:10–14. doi: 10.1016/0966-842x(94)90338-7. [DOI] [PubMed] [Google Scholar]
- 49.Hynes RO. Integrins: A family of cell surface receptors. Cell. 1987;48:549–554. doi: 10.1016/0092-8674(87)90233-9. [DOI] [PubMed] [Google Scholar]
- 50.Dedhar S, Hannigan GE. Integrin cytoplasmic interactions and bidirectional transmembrane signalling. Curr Opin Cell Biol. 1996;8:657–669. doi: 10.1016/s0955-0674(96)80107-4. [DOI] [PubMed] [Google Scholar]
- 51.Miyake T, Hu YF, Yu DS, Li R. A functional comparison of BRCA1 C-terminal domains in transcription activation and chromatin remodeling. J Biol Chem. 2000;275:40169–40173. doi: 10.1074/jbc.M007138200. [DOI] [PubMed] [Google Scholar]
- 52.Kobayashi M, Ab E, Bonvin AM, Siegal G. Structure of the DNA-bound BRCA1 C-terminal region from human replication factor C p140 and model of the protein-DNA complex. J Biol Chem. 2010;285:10087–10097. doi: 10.1074/jbc.M109.054106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Carney DS, Davies BA, Horazdovsky BF. Vps9 domain-containing proteins: Activators of Rab5 GTPases from yeast to neurons. Trends Cell Biol. 2006;16:27–35. doi: 10.1016/j.tcb.2005.11.001. [DOI] [PubMed] [Google Scholar]
- 54.Prag G, et al. Mechanism of ubiquitin recognition by the CUE domain of Vps9p. Cell. 2003;113:609–620. doi: 10.1016/s0092-8674(03)00364-7. [DOI] [PubMed] [Google Scholar]
- 55.Shan W, Cao M, Leung D, Tyler BM. The Avr1b locus of Phytophthora sojae encodes an elicitor and a regulator required for avirulence on soybean plants carrying resistance gene Rps1b. Mol Plant Microbe Interact. 2004;17:394–403. doi: 10.1094/MPMI.2004.17.4.394. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.