Abstract
The posttranslational addition of palmitate to cysteines occurs ubiquitously in eukaryotic cells, where it functions in anchoring target proteins to membranes and in vesicular trafficking. Here we show that the Saccharomyces cerevisiae palmitoyltransferase Pfa4 enhanced heterochromatin formation at the cryptic mating-type loci HMR and HML via Rif1, a telomere regulatory protein. Acylated Rif1 was detected in extracts from wild-type but not pfa4Δ mutant cells. In a pfa4Δ mutant, Rif1-GFP dispersed away from foci positioned at the nuclear periphery into the nucleoplasm. Sir3-GFP distribution was also perturbed, indicating a change in the nuclear dynamics of heterochromatin proteins. Genetic analyses indicated that PFA4 functioned upstream of RIF1. Surprisingly, the pfa4Δ mutation had only mild effects on telomeric regulation, suggesting Rif1's roles at HM loci and telomeres were more complexly related than previously thought. These data supported a model in which Pfa4-dependent palmitoylation of Rif1 anchored it to the inner nuclear membrane, influencing its role in heterochromatin dynamics.
Keywords: transcriptional silencing, chromosome architecture
Transcriptional silencing in Saccharomyces cerevisiae, a form of gene repression that occurs at the HM loci and telomeres, requires formation of distinctive chromatin structures that span extended chromosomal domains forming the budding-yeast version of heterochromatin (1–3). Silencing is established by direct recruitment of Sir2/3/4 proteins (Sir complex) by sequence-specific DNA binding proteins bound to DNA elements called “silencers.” The Sir2 NAD-dependent deacetylase positioned at silencers removes acetyl groups from an adjacent nucleosome, creating another binding site for an additional Sir complex. Iterative cycles of histone deacetylation and Sir complex binding expand heterochromatin (“silent” chromatin) over several kilobases of DNA. Yeast heterochromatin is dispensable for cell viability but critical for controlling gene expression at the HM loci and telomeres, both easily measured with phenotypic assays. Thus, yeast silencing has served as a powerful tool for defining mechanisms of heterochromatin formation.
The SIR genes were isolated in forward genetic screens focused on HM silencing and encode nonessential proteins that are structural components of yeast heterochromatin (4). However, several modified genetic screens have identified proteins with impacts in chromosome biology that extend beyond heterochromatin. For example, mutant versions of the origin recognition complex (ORC), a silencer binding protein that also functions as the eukaryotic initiator of DNA replication, were identified in silencing screens (5–7). A common feature of these modified screens is that silencing is genetically compromised by mutation to sensitize it to further exacerbating defects; alleles of ORC were identified in screens focused on HMR silencing in which the HMR silencer had been compromised by cis-mutation (6, 7). Variations of sensitized silencing screens have uncovered roles in chromosome biology for other conserved factors that are essential for viability, highly redundant, or otherwise difficult to define (8–10).
We exploited a genetically sensitized form of HMR silencing and identified PFA4, a gene that encodes a DHHC (asp-his-his-cys) domain-containing, integral membrane S-palmitoyltransferase of the endoplasmic reticulum (ER) (11, 12). The data supported a model in which Pfa4 modulated HM silencing, Sir-complex dynamics, and macromolecular aspects of telomere architecture by palmitoylating the telomere binding protein Rif1.
Results and Discussion
PFA4 Contributed to HMR Silencing.
To uncover new regulators of chromatin, several genetic modifications of HMR silencing were exploited. First, SIR1 was deleted. Sir1, one of four Sir proteins, contributes to the initial recruitment of the Sir complex to the HM loci (13). In its absence, silencing is weakened but not abolished, indicating that Sir1-independent mechanisms for Sir complex recruitment exist (14). Second, the parent strain was further sensitized by use of a synthetic silencer (HMR-SS), making HMR more sensitive to SIR1 loss (15). Third, modest overexpression of the cell-cycle regulator FKH1 (FKH1hc) was used to partially restore silencing crippled by these modifications. FKH1hc recreates bona fide Sir complex silencing at HMR in sir1Δ HMR-SS cells (16). The strain was subjected to transposon-based mutagenesis and screened for mutants that reduced silencing (17). LEU2 insertion mutations in YKU80 and PFA4 were identified and cosegregated with the silencing defect. YKU80 was discussed previously (17).
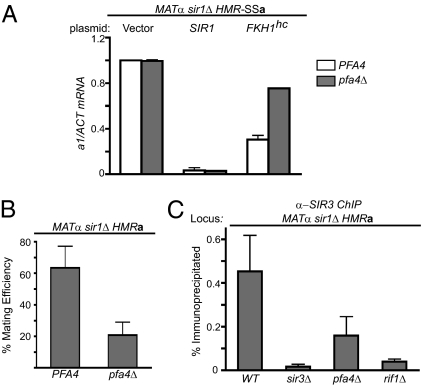
To confirm a role for PFA4 in HMR silencing, the effect of a complete deletion of the gene on transcription of the a1-gene was determined by RT-PCR (Fig. 1A). Transcription of a1 is the direct consequence of reduced silencing at HMR. In SIR1 cells, a1 mRNA was not detected regardless of PFA4 genotype. In contrast, FKH1-assisted silencing was sensitive to PFA4 as a1 mRNA levels were twofold greater in the pfa4Δ mutant compared with wild-type cells. These data solidified the conclusion that PFA4 contributed to FKH1-assisted silencing and that SIR1 masked PFA4's contribution.
Fig. 1.
PFA4 in HMR silencing. (A) A pfa4Δ mutation reduced FKH1-assisted silencing but not SIR1-mediated silencing. MATα sir1Δ HMR-SSa cells that were either PFA4 or pfa4Δ were transformed with a 2-μ URA3-plasmid that was empty (Vector) or contained either SIR1 or FKH1 (FKH1 “high copy”; FKH1hc) and a1 mRNA levels were measured by RT-PCR (17). (B) PFA4's role in silencing natural HMR was examined by quantitative mating assays in which the precise number of cells in a population that can mate is determined by serial dilutions into an excess of MATa cells, as in A, except all of the cells are counted (17). Mating efficiency is indicated as the percentage of cells that could form diploid colonies on selective agar plates. The significance of the difference between PFA4 and pfa4Δ cells was assessed with a t test (P = 0.0006). (C) Sir3 binding to HMR was examined by ChIP using monoclonal Sir3 antibodies (16). The significance of the differences between PFA4 and the mutant cells indicated in the figure were assessed with t tests and the P values were: sir3Δ, P = 0.010; pfa4Δ, P = 0.052; rif1Δ, P = 0.012.
FKH1-assisted silencing was used as a tool to sensitize silencing to SIR1-independent mechanisms that are likely to be more conserved in eukaryotes (17). However, if PFA4 had a fundamental role in chromosome biology that affected HMR, its contribution should not be limited to this specialized condition. Therefore, quantitative mating experiments with sir1Δ cells containing a native HMR locus and chromosomal FKH1 were performed (Fig. 1B). Loss of PFA4 caused a threefold reduction in HMR silencing in these cells. Silent chromatin in these cells was also monitored by ChIP with monoclonal antibodies against Sir3 (16). Sir3 levels at HMR were reduced more than twofold in the pfa4Δ mutant cells (Fig. 1C). Thus, PFA4 could modulate heterochromatin formation at the native HMR locus.
Because PFA4 encodes a palmitoyltransferase, we tested whether its catalytic activity was required for HMR silencing. A pfa4C108A allele produces a catalytically inactive version of Pfa4 that fails to direct the trafficking of the chiten synthase, a Pfa4 substrate (12). Importantly, in contrast to a wild-type PFA4-HA allele, a pfa4C108A-HA allele expressed on either a CEN or 2-μm plasmid failed to complement the HMR silencing defect of a pfa4Δ mutant (Fig. S1). Thus, the catalytic activity of Pfa4 was required for HMR silencing in sir1Δ cells.
PFA4 Was Required for S-Acylation of Rif1.
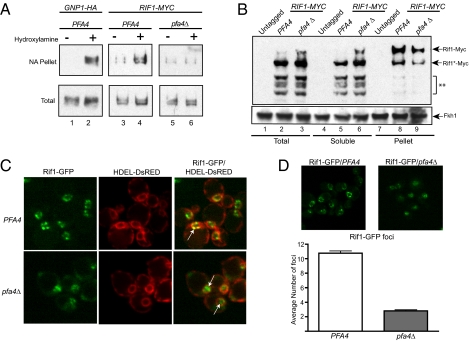
Palmitate is a 16-carbon fatty acid, and as a protein modification functions as a membrane anchor, tethering a protein domain to cellular membranes. Although palmitoylation is most recognized for its role in vesicular trafficking, it seemed significant that Pfa4 resides within the ER membrane, as this organelle's membrane is contiguous with the nuclear membranes (11, 18). The catalytic site of S-palmitoyltranferases face the cytoplasm. Thus, Pfa4 could palmitoylate a nuclear protein that would anchor it to the ER membrane, allowing it to enter the nucleus via translocation through nuclear pores. Thus, an attractive hypothesis was that Pfa4 directly palmitoylated a nuclear protein relevant to silencing. Rif1, a protein with roles in telomere structure and silencing, was recovered in a proteomic survey of palmitoylated proteins in yeast (19). Thus, the modification status of Rif1 was examined more closely with an acyl-biotin exchange (ABE) protocol (20) (Fig. S2A) in which free thiols are first protected by treatment with N-ethylmaleimide (NEM). Hydroxylamine is then used to cleave thiol ester bonds of any existing protein-acyl linkages. Finally a thiol-specific biotinylation reagent (EZ-Link; Thermo Scientific) covalently links biotin to the newly liberated thiols, and biotinylated proteins are purified by neutravidin-affinity chromatography. GNP1-3xHA, a positive control, was acylated in a PFA4-dependent manner, whereas Orc1 and Fkh1 were not (19) (Fig. 2A and Fig. S2B). Rif1-9xMyc was also enriched in the acylated fraction, and this enrichment was reduced by omission of the hydroxylamine step. Most significantly, this enrichment was reduced to background levels by deletion of PFA4 (Fig. 2A and Fig. S2B). NEM protection was performed under highly denaturing conditions (6 M Urea), making it unlikely that Rif1 was purified on the basis of its interaction with a secondary modified protein. Therefore, one or more cysteines of Rif1 were acylated. Given the dependence on PFA4, these data provided evidence that Rif1 was palmitoylated.
Fig. 2.
PFA4 was required for S-acylation of Rif1 and formation of Rif1-GFP foci. (A) ABE chemistry (20) was used to test whether Gnp1, a protein known to be palmitoylated by PFA4 (19), and Rif1 were acylated (also see Fig. S1). Biotinylated proteins should be present in the neutravidin-agarose (NA) pellet when the extracts are treated with hydroxylamine (+). Protein immunoblotting was used to detect Gnp1-3xHA and Rif1-9xMyc (22). (B) Wild-type yeast cells (Untagged) or cells containing RIF1-9xMYC that were either PFA4 or pfa4Δ were fractionated by centrifugation into membrane-containing and soluble fractions (21). Rif1-9xMyc was detected by protein immunoblotting. Rif1-9xMyc contains the Myc epitopes at its C terminus. Full-length Rif1-9xMyc is indicated (Rif1-Myc). Rif1-Myc* is a truncated form of the protein that lacks about 400 amino acids of the N terminus but retains the C-terminal Myc epitopes. The shorter fragments (**) are further N-terminally truncated Rif1-9xMyc fragments. Fkh1 was detected with an anti-Fkh1 antibody (16). (C) Serial sectioning and confocal microscopy was used to visualize Rif1-GFP. The ER was detected with HDEL-DsRED (26) that stains the ER, which is contiguous with the nucleus (small red circles), as well as the cortical ER that leads to signal intensity near the plasma membrane. White arrows in the merged data panel indicate selected Rif1-GFP foci. (D) The number of Rif1-GFP foci in wild type (PFA4) and pfa4Δ mutant cells was determined by counting distinct foci from 97 PFA4 cells and 98 pfa4Δ mutant cells. Confocal sections were acquired at 300-nm intervals through the nucleus. The significance of the difference was determined by a t test (P = 1.76 × 10−48). Images were acquired on a swept field confocal microscope (Nikon Ti-E) equipped with a Roper CoolSnap HQ2 CCD camera using a Nikon 60×, 1.4NA Planapo oil objective lens, and a 1.5× Optivar auxillary magnifier.
Palmitoylation of Rif1 would be expected to modulate Rif1's association with membranes and thus its solubility in cellular extracts. Therefore, the solubility of Rif1-9xMyc in PFA4 and pfa4Δ mutant cell extracts was compared by differential centrifugation (21) (Fig. 2B). Immunoblotting revealed two bands that corresponded to full-length Rif1 with epitope (Rif1-9xMyc) and an N-terminal truncated form that lacked about 400 amino acids but retained the C-terminal Myc epitope (Rif1*-9xMyc). Rif1-9xMyc and Rif1*-9xMyc were enriched in the insoluble pellet of PFA4 relative to pfa4Δ extracts. Correspondingly, these proteins were diminished in the soluble fraction of PFA4 relative to pfa4Δ extracts. In addition, several smaller Rif1-9xMyc protein fragments were detected (**) that associated primarily with the soluble fraction of both the PFA4 and pfa4Δ extracts. Finally, Fkh1, which is not palmitoylated (Fig. S1B), distributed similarly between the pellet and soluble fractions regardless of PFA4 genotype (Fig. 2B). Thus, PFA4 altered the solubility of Rif1 in yeast extracts, consistent with palmitoylation of Rif1 helping anchor it to the nuclear membrane.
PFA4 Was Required for Rif1-GFP Foci at the Nuclear Periphery.
Rif1 associates with telomere clusters that are themselves anchored to the inner nuclear membrane, forming distinct foci (22–25). To test whether Rif1's localization to these foci was affected by PFA4, a Rif1-GFP fusion protein was coexpressed in a strain expressing an HDEL-DsRed fusion protein, which is retained in the ER to allow visualization of the contiguous nuclear and ER membranes (26). Confocal sectioning of wild-type cells revealed 10 to 12 discrete Rif1-GFP foci per nucleus, most of which abutted the nuclear membrane (Fig. 2C). This pattern was similar to that observed by immunoflorecense of Rif1 and is reminiscent of other telomere-binding proteins (22, 25). In contrast, in pfa4Δ mutant cells, Rif1-GFP fluorescence was more broadly distributed across the nucleus, and only two to four larger, more diffuse foci per nucleus could be identified (Fig. 2D). Importantly, a pfa4Δ mutant did not affect levels of Rif1-GFP as measured by protein immunoblotting, and a catalytically inactive pfa4(C108A) mutant produced similar dispersion of Rif1-GFP foci to a pfa4Δ mutant (Fig. S3). Therefore, enrichment of Rif1-GFP in distinct foci at the nuclear periphery required PFA4-dependent palmitoylation.
PFA4 and RIF1 Functioned in a Common Pathway in HM Silencing.
The data presented above indicated that Rif1's nuclear distribution was affected by PFA4, suggesting that this regulation was relevant to PFA4's role in silencing. In the current view, Rif1 acts as a direct negative regulator of telomeric silencing and an indirect positive regulator of HM silencing (22). Specifically, Rif1 and the Sirs interact exclusively with Rap1; multiple Rap1 sites exist at telomeres serving as the telomeric version of a silencer. Thus, Rif1 attenuates telomeric silencing by competing with Sir complexes for binding to Rap1. Because Sir complexes are limiting, increases in Sir complexes at telomeres are balanced by reductions in Sir complexes at the HM loci (27). Therefore, deletion of RIF1 enhances telomeric silencing and simultaneously reduces HM silencing. This model might account for HMR silencing defects caused by deletion of PFA4. However, HM silencers contain single Rap1 binding sites important for silencing (28, 29), and genome-wide analysis of Rif1 binding provides evidence that it binds the HM loci (25). Thus, it is possible that Rif1 also has more direct negative roles in HM silencing. Regardless, if palmitoylation regulates Rif1's role in HM silencing, then defects caused by a rif1Δ mutation should not be further exacerbated by loss of PFA4. Therefore, we asked whether RIF1 and PFA4 functioned in a common pathway in HM silencing (Fig. 3 A and B).
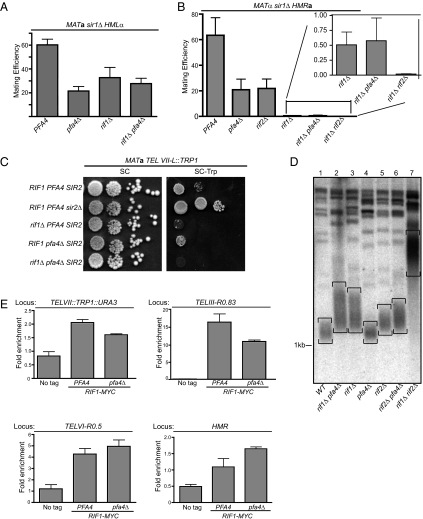
Fig. 3.
RIF1 and PFA4 in silencing, telomere length and Rif1 binding. Quantitative mating-assays were used to measure the epistatic relationship between RIF1 and PFA4 at (A) HML, and (B) HMR. Quantitative mating assays were performed as previously described (17). (C) Telomeric silencing was assessed by measuring expression of the TRP1 reporter gene engineered at the left telomere on chromosome VII (TEL VII-L::TRP1) by comparing growth on rich medium to that on synthetic medium lacking tryptophan. (D) Average length of bulk telomeres for a set of congenic strains was determined by DNA blot hybridization (45). (E) The effect of PFA4 on Rif1-9xMyc binding to several loci was measured by ChIP.
Data for HML were straightforward. Quantitative mating showed that HML silencing was reduced similarly when either PFA4 or RIF1 was deleted, and that the effect was no greater when the two deletions were combined (Fig. 3A). The data for HMR were more complex, but nevertheless consistent (Fig. 3B). Quantitative mating showed that HMR silencing was reduced when either PFA4 or RIF1 was deleted, but the defect caused by RIF1 deletion was more severe. This different quantitative contribution was also revealed by Sir3-ChIP experiments (Fig. 1C). Significantly, however, cells lacking both PFA4 and RIF1 exhibited an HMR-silencing defect that was the same as that of cells lacking only RIF1 (Fig. 3B, Inset). The absence of an additive defect was not a result of assay limitations because cells lacking both RIF1 and RIF2 [RIF1 and RIF2 have overlapping functions (30)] showed greater silencing defects. Thus, RIF1 and PFA4 acted in a common genetic pathway to silence HML and HMR.
PFA4 Had Mild Effects on Telomeric Silencing and Length Control.
As discussed above, Rif1 acts as a negative regulator of telomeric silencing by binding to telomere-bound Rap1 and blocking recruitment of the Sir complex. Thus, in a rif1Δ mutant, telomeric silencing is enhanced. This effect is also thought to be the basis of RIF1's positive role in HM silencing. Thus, based on the HM silencing data as well as delocalization of Rif1-GFP, we expected that a pfa4Δ mutation would enhance telomeric silencing. Silencing of the left telomere of chromosome VII was assessed using a TRP1 reporter gene (Fig. 3C). In wild-type cells, TRP1 at telomere VII-L is silenced, hence cells grow poorly on medium lacking tryptophan (31). The Sir complex is essential for telomeric silencing, hence cells lacking SIR2 express TRP1 and grow on medium lacking tryptophan (32). As expected, deletion of RIF1 enhanced telomeric silencing (33). However, a pfa4Δ mutation had only a slight effect on silencing of telomere VII-L.
Rif1 also negatively regulates telomere length (34). We therefore tested whether PFA4 regulated telomere length by measuring the average length of bulk telomeres (Fig. 3D). DNA was cleaved with XhoI at a location near yeast chromosomal ends, and telomeres were detected by DNA blot hybridization with a probe complementary to yeast telomeres. Telomeres appear as a smear in these experiments (Fig. 3D, telomeres are bracketed). As expected, a deletion of RIF1 led to an increase in telomere length. A deletion of PFA4, however, had no effect on telomere length. Telomeres in cells lacking both PFA4 and RIF1 were similar in length to those in cells lacking only RIF1. As in silencing (Fig. 3B), RIF1 and RIF2 have additive roles in telomere length, such that deletion of both genes leads to substantially longer telomeres (30). Thus, a rif2Δ mutation might sensitize telomere length to defects in RIF1 caused by a pfa4Δ mutation. A pfa4Δ mutation did lead to slightly longer telomeres in rif2Δ mutant cells (compare rif2Δ to rif2Δ pfa4Δ) but the effect was small. Thus, PFA4 had only a very modest role in controlling telomere length. Therefore, Rif1's molecular roles at telomeres remained largely intact in pfa4Δ mutant cells.
To reconcile dispersion of Rif1-GFP with retention of telomere functions in a pfa4Δ mutant, ChIPs in cells expressing Rif1-9xMyc were performed (Fig. 3E). Deletion of PFA4 reduced Rif1 binding to the right telomere of chromosome III (TEL III-R) by ∼30%. A similar reduction occurred at TEL VII-L. Rif1-9xMyc association with a third telomere TEL VI-R was not reduced. Thus, Rif1's average occupancy at telomeres did not change substantially in a pfa4Δ mutant, providing an explanation for the mild effects that the pfa4Δ mutant had on telomere silencing and length control. Considering the substantial dispersion of Rif1-GFP in pfa4Δ mutants, these data suggest that Rif1 is normally present in excess concentration at telomeres for its roles in telomeric silencing and length regulation.
Because a genome-wide analysis detected Rif1 binding to HMR (25), Rif1-9xMyc binding to HMR was examined (Fig. 3E). At HMR, Rif1-9xMyc occupancy was actually enhanced by ∼30% by deletion of PFA4. One explanation was that in a pfa4Δ mutant, Rif1, now released from pools normally tightly sequestered into foci at the nuclear periphery, was free to sample Rap1 proteins bound to more internal sites, such as HM silencers, more efficiently. Thus, perhaps reduced HMR silencing in a pfa4Δ mutant was caused in part by direct negative regulation of this locus by Rif1, and in part by some sequestration of Sir proteins at telomeres, as in the current model. These two mechanisms might act additively to cause reductions in HM silencing, and could explain how PFA4-dependent regulation of Rif1 could reduce HM silencing measurably without simultaneously affecting telomeric silencing or length control substantially.
PFA4 and RIF1 Acted in a Common Pathway to Control Sir3 Dynamics.
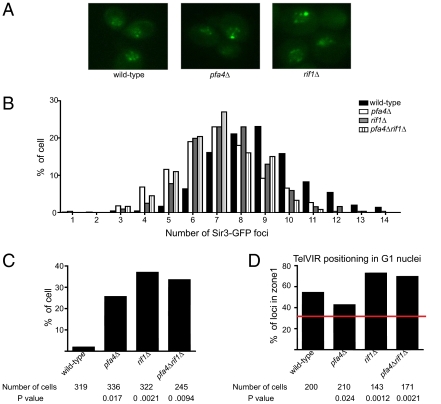
The HM data supported the idea that PFA4 regulated RIF1, but the modest effects of PFA4 on telomeres precluded epistasis analysis. Therefore, more macromolecular aspects of telomere behavior were used to examine the relationship between PFA4 and RIF1. Because most telomeres contain neighboring domains of heterochromatin, a Sir3-GFP fusion protein was used as a marker for telomere clusters. Serial sectioning by fluorescence microscopy revealed an average of 8.8 Sir3-GFP foci in nuclei of wild-type cells, and this number dropped to 7.0 and 7.4 in pfa4Δ and rif1Δ mutants, respectively (Fig. 4 A and B). Cells with both mutations (rif1Δ pfa4Δ) produced a similar number of Sir3-GFP foci as cells with either mutation alone (average = 7 per nucleus). These data suggest that both RIF1 and PFA4 acted in a common pathway to modulate telomere clustering. However, RIF1 and PFA4 had relatively small effects on telomere clustering; although the number of Sir3-GFP clusters was reduced, most appeared to remain intact. Importantly, Sir3-GFP levels were similar in wild-type and pfa4Δ mutant cells (Fig. S3D). Thus, the striking loss of Rif1-GFP foci in pfa4Δ mutants was likely not caused by a large-scale breakdown of telomere clusters but rather a dispersion of Rif1 away from these, and possibly other structures, at the nuclear periphery.
Fig. 4.
PFA4 modulated Sir3-GFP distribution and the perinuclear association of telomere VIR. (A) Examples of Sir3-GFP foci observed in wild-type, pfa4Δ, and rif1Δ cells. (B) Sir3-GFP foci were counted and the percentage of cells (y axis) containing a given number of foci (x axis) was determined. (C) The percentage of cells containing SBF formed by Sir3-GFP was determined. (D) The percentage of cells containing TelVIR in zone I of the nucleus (region closest to the nuclear periphery) was determined. Statistical data included in the figure.
An additional phenomenon was noted. Although the intensity of individual Sir3-GFP foci in wild-type cells did not vary dramatically, one or two of the foci within pfa4Δ or rif1Δ mutant cells were more intense than other foci. We therefore counted the number of exceptionally intense foci by setting an arbitrary cutoff to define the most intense spots as single bright foci (SBFs). Only 1.9% of wild-type cells contained a SBF (Fig. 4C). In contrast, the mutants contained 15- to 20-fold more (pfa4Δ = 25.6%; rif1Δ = 37%). Deletions of both genes produced a similar number of SBFs as either deletion alone (rif1Δ pfa4Δ = 33%). These data suggest that RIF1 and PFA4 acted in a common pathway to prevent formation of foci that contained abnormally high quantities of Sir3-GFP.
Because telomeres form clusters that associate with the nuclear periphery, we also examined whether PFA4 and RIF1 influenced the anchoring of a telomere to the nuclear membrane by measuring distances between a GFP-tagged lac operator array at the right end of telomere VI and the rim of fluorescence created by a GFP-tagged nuclear pore protein, Nup49-GFP (24). Measurements were normalized to the nuclear diameter and binned into three concentric zones of equal surface area. This analysis was restricted to G1 cells. In wild-type cells, TelVIR was found in zone I, the zone closest to the nuclear periphery, in 58% of the cells examined (Fig. 4D). A pfa4Δ mutation reduced the fraction in zone I to 43%, suggesting that palmitoylation helped promote telomere anchoring to the nuclear membrane. However, in contrast to HM silencing and Sir3-GFP behavior, deletion of RIF1 caused a phenotype opposite to that of pfa4Δ and actually increased the fraction of cells in zone I to 71%. Deletion of both RIF1 and PFA4 gave rise to 70% of the cells with TelVIR in zone I, similar to RIF1deletion alone. A possible explanation for these data were that palmitoylation of Rif1 by Pfa4 favored telomere anchoring by one mechanism and revealed Rif1's contribution to this anchoring, but the presence of Rif1 restricted perinuclear anchoring by another mechanism that was able to dominate only when RIF1 was completely absent. For example, absence of Rif1 would be expected to enhance the presence of Sir complexes at telomeres substantially, promoting Sir-mediated mechanisms of telomere anchoring.
In summary this study provided evidence that palmitoylation controlled the behavior of Rif1, a protein that regulates both telomere dynamics and silencing. PFA4 was important for Rif1-GFP's localization to discrete foci at the nuclear periphery. Several studies provide evidence that these foci correspond to telomeric clusters (22, 25, 35). Rif1 binds telomeres via direct interactions with the telomere-binding protein Rap1, and this interaction is relevant to Rif1's molecular roles at telomeres (25, 33). However, our data demonstrat that Rap1-Rif1 interactions were insufficient to explain the discrete Rif1-GFP foci that form at the nuclear periphery, and instead suggest that palmitoylation of Rif1 promoted formation of these foci. Perhaps concentrating Rif1 at the inner nuclear membrane via palmitoylation favors its association with telomere clusters that are anchored to this membrane by several other mechanisms, thus forming concentrated pockets of Rif1 revealed by the Rif1-GFP foci (36). Simultaneously, this mechanism could sequester Rif1 from many internal Rap1-associated loci, including HMR and HML, where it might be detrimental. Regardless of the precise mechanisms that explain PFA4’s role in HM silencing, the genetic data suggest that PFA4 and RIF1 acted in a common pathway, as would be predicted if palmitoylation of Rif1 by Pfa4 regulated Rif1. Although several studies establish functional links between the inner nuclear membrane and yeast silencing (37–39), palmitoylation of Rif1 is unique as an example in which a known silencing protein's association with the inner nuclear membrane is controlled by a fatty acid modification. A recent study shows that carbohydrate modification of Sir2 can regulate silencing (40). Perhaps these unexpected modifications represent an emerging theme for how cells use nonhistone proteins to make chromatin structures more responsive to a cell physiology. S-acylation catalyzed by palmitoyltransfereases is particularly interesting in that it is reversible, providing a way to make Rif1 nuclear distribution highly responsive and dynamic. Further studies will be needed to test this idea, and reveal whether the conservation of PFA4 and RIF1 results in the use of this unique pathway in metazoans. Moreover, it is likely that other nuclear proteins are controlled by palmitoylation because it provides a dynamic means to control protein distribution and chromosome architecture.
Materials and Methods
Yeast Genetics and Molecular Biology.
Yeast strains (Table S1) were constructed by standard procedures. Mating assays were performed as previously described (17). Primers are listed in Table S2.
Quantitative PCR to determine a1 mRNA levels was performed as previously described (17).
ABE assay used the modified version of ABE chemistry (41) that used the three steps previously described (42) (Fig. S2A), except that proteins were denatured by 6 M urea and free cysteines blocked by NEM (Fluka) under these conditions. Extracts were prepared from cells growing in log-phase (A600 ∼1.0).
Rif1-9xMyc Solubility.
Extracts made from cells grown in YPD and harvested at A600∼1.0 were fractionated as previously described (21) and examined for Rif1-9xMyc (25) by protein immunoblotting.
ChIP was performed as described (16, 43, 44) with anti-Sir3 to monitor Sir3 binding or anti-Myc to monitor Rif1-9xMyc binding. HMR and telomeres were detected with specific primers (Table S2). Rif1-Myc ChIPs were performed in MATa SIR1 cells that contained natural versions of the HM loci. Signals were normalized to the ADH4 locus (17).
Telomere length measurements were performed as previously described (45).
Rif1-GFP Imaging.
Cells were grown in liquid synthetic media, diluted to an A600 = ∼0.1 to 0.2, and grown to an additional 5 h to A600 = ∼0.7. One milliliter of culture was pelleted, washed with ∼1 mL water, and resuspended in 100 μL water. Rif1-GFP and HDEL-DsRED were visualized by serial sectioning and confocal microscopy (Nikon Ti-E, equipped with a Roper CoolSnap HQ2 CCD camera). Confocal sections were acquired at 300-nm intervals through the nucleus. Foci were counted manually in images of ∼100 live cells.
Sir3-GFP Imaging and Telomere VIR Positioning.
Cells were grown in liquid YPD, diluted, and regrown to A600 = 0.2 to 0.3, pelleted, and washed with water before mounting on slides with 1.4% agarose plugs. Fields of 15 to 50 cells were imaged with a Zeiss Axioplan II fluorescence microscope (100× Plan-Apochromat objective, NA = 1.4) and Axiocam HR camera. Additional z-stacks were composed of 17 elevations, each separated by 250 nm and an acquisition time of 250 ms. Foci were counted manually in images of live cells. Datasets for each strain consisted of three trials of 50 to 150 cells and were pooled after the average values of spots per nucleus were found to lie within the same 95% confidence interval. Pooled data were evaluated relative to wild-type by t test. For foci intensity, Zeiss Axiovision software was used to obtain a unitless intensity value I = (Sf − Bf)/Bf, where Sf equals signal intensity of an area encompassing the bright spot and Bf equals intensity of a comparable nuclear area lacking a fluorescent spot. An arbitrary cutoff of I > 1.1 was chosen to define an SBF. To measure perinuclear position of TelVIR, GFP positions were determined as previously described (24).
Supplementary Material
Acknowledgments
We thank Elizabeth Chybowski and Antoinette Dummer for help with the telomere experiments; David Shore and Elizabeth Conibear for yeast strains and plasmids; and Elizabeth Craig for critiquing the manuscript. This study was initiated with support from the American Cancer Society (RSG-02-164-02-GMC) (to C.A.F.) and continued with support from the National Institutes of Health (R01 GM56890) (to C.A.F.) and an American Recovery and Reinvestment Act of 2009 supplement GM56890; a National Institutes of Health Predoctoral Training Program in Genetics (5 T32 GM07133) and a predoctoral fellowship from the American Heart Association (0615552Z) helped support E.E.P; National Institutes of Health Grant NIH GM51402 (to M.R.G.) and Ruth L. Kirschstein Predoctoral Fellowship GM078746 (to J.C.) also helped support this study.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1105262108/-/DCSupplemental.
References
- 1.Grunstein M. Molecular model for telomeric heterochromatin in yeast. Curr Opin Cell Biol. 1997;9:383–387. doi: 10.1016/s0955-0674(97)80011-7. [DOI] [PubMed] [Google Scholar]
- 2.Moazed D, Rudner AD, Huang J, Hoppe GJ, Tanny JC. A model for step-wise assembly of heterochromatin in yeast. Novartis Found Symp. 2004;259:48–56. [PubMed] [Google Scholar]
- 3.Rusche LN, Kirchmaier AL, Rine J. The establishment, inheritance, and function of silenced chromatin in Saccharomyces cerevisiae. Annu Rev Biochem. 2003;72:481–516. doi: 10.1146/annurev.biochem.72.121801.161547. [DOI] [PubMed] [Google Scholar]
- 4.Rine J, Herskowitz I. Four genes responsible for a position effect on expression from HML and HMR in Saccharomyces cerevisiae. Genetics. 1987;116:9–22. doi: 10.1093/genetics/116.1.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bell SP. The origin recognition complex: From simple origins to complex functions. Genes Dev. 2002;16:659–672. doi: 10.1101/gad.969602. [DOI] [PubMed] [Google Scholar]
- 6.Micklem G, Rowley A, Harwood J, Nasmyth K, Diffley JF. Yeast origin recognition complex is involved in DNA replication and transcriptional silencing. Nature. 1993;366:87–89. doi: 10.1038/366087a0. [DOI] [PubMed] [Google Scholar]
- 7.Foss M, McNally FJ, Laurenson P, Rine J. Origin recognition complex (ORC) in transcriptional silencing and DNA replication in S. cerevisiae. Science. 1993;262:1838–1844. doi: 10.1126/science.8266071. [DOI] [PubMed] [Google Scholar]
- 8.Tyler JK, et al. The RCAF complex mediates chromatin assembly during DNA replication and repair. Nature. 1999;402:555–560. doi: 10.1038/990147. [DOI] [PubMed] [Google Scholar]
- 9.Sharp JA, Fouts ET, Krawitz DC, Kaufman PD. Yeast histone deposition protein Asf1p requires Hir proteins and PCNA for heterochromatic silencing. Curr Biol. 2001;11:463–473. doi: 10.1016/s0960-9822(01)00140-3. [DOI] [PubMed] [Google Scholar]
- 10.Kaufman PD, Cohen JL, Osley MA. Hir proteins are required for position-dependent gene silencing in Saccharomyces cerevisiae in the absence of chromatin assembly factor I. Mol Cell Biol. 1998;18:4793–4806. doi: 10.1128/mcb.18.8.4793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Ohno Y, Kihara A, Sano T, Igarashi Y. Intracellular localization and tissue-specific distribution of human and yeast DHHC cysteine-rich domain-containing proteins. Biochim Biophys Acta. 2006;1761:474–483. doi: 10.1016/j.bbalip.2006.03.010. [DOI] [PubMed] [Google Scholar]
- 12.Lam KK, et al. Palmitoylation by the DHHC protein Pfa4 regulates the ER exit of Chs3. J Cell Biol. 2006;174:19–25. doi: 10.1083/jcb.200602049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fox CA, McConnell KH. Toward biochemical understanding of a transcriptionally silenced chromosomal domain in Saccharomyces cerevisiae. J Biol Chem. 2005;280:8629–8632. doi: 10.1074/jbc.R400033200. [DOI] [PubMed] [Google Scholar]
- 14.Pillus L, Rine J. SIR1 and the origin of epigenetic states in Saccharomyces cerevisiae. Cold Spring Harb Symp Quant Biol. 2004;69:259–265. doi: 10.1101/sqb.2004.69.259. [DOI] [PubMed] [Google Scholar]
- 15.McNally FJ, Rine J. A synthetic silencer mediates SIR-dependent functions in Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:5648–5659. doi: 10.1128/mcb.11.11.5648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Casey L, Patterson EE, Müller U, Fox CA. Conversion of a replication origin to a silencer through a pathway shared by a Forkhead transcription factor and an S phase cyclin. Mol Biol Cell. 2008;19:608–622. doi: 10.1091/mbc.E07-04-0323. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Patterson EE, Fox CA. The Ku complex in silencing the cryptic mating-type loci of Saccharomyces cerevisiae. Genetics. 2008;180:771–783. doi: 10.1534/genetics.108.091710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mitchell DA, Vasudevan A, Linder ME, Deschenes RJ. Protein palmitoylation by a family of DHHC protein S-acyltransferases. J Lipid Res. 2006;47:1118–1127. doi: 10.1194/jlr.R600007-JLR200. [DOI] [PubMed] [Google Scholar]
- 19.Roth AF, et al. Global analysis of protein palmitoylation in yeast. Cell. 2006;125:1003–1013. doi: 10.1016/j.cell.2006.03.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Drisdel RC, Alexander JK, Sayeed A, Green WN. Assays of protein palmitoylation. Methods. 2006;40:127–134. doi: 10.1016/j.ymeth.2006.04.015. [DOI] [PubMed] [Google Scholar]
- 21.Liang C, Stillman B. Persistent initiation of DNA replication and chromatin-bound MCM proteins during the cell cycle in cdc6 mutants. Genes Dev. 1997;11:3375–3386. doi: 10.1101/gad.11.24.3375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mishra K, Shore D. Yeast Ku protein plays a direct role in telomeric silencing and counteracts inhibition by rif proteins. Curr Biol. 1999;9:1123–1126. doi: 10.1016/s0960-9822(99)80483-7. [DOI] [PubMed] [Google Scholar]
- 23.Gotta M, et al. The clustering of telomeres and colocalization with Rap1, Sir3, and Sir4 proteins in wild-type Saccharomyces cerevisiae. J Cell Biol. 1996;134:1349–1363. doi: 10.1083/jcb.134.6.1349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hediger F, Neumann FR, Van Houwe G, Dubrana K, Gasser SM. Live imaging of telomeres: yKu and Sir proteins define redundant telomere-anchoring pathways in yeast. Curr Biol. 2002;12:2076–2089. doi: 10.1016/s0960-9822(02)01338-6. [DOI] [PubMed] [Google Scholar]
- 25.Smith CD, Smith DL, DeRisi JL, Blackburn EH. Telomeric protein distributions and remodeling through the cell cycle in Saccharomyces cerevisiae. Mol Biol Cell. 2003;14:556–570. doi: 10.1091/mbc.E02-08-0457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Audhya A, Emr SD. Regulation of PI4,5P2 synthesis by nuclear-cytoplasmic shuttling of the Mss4 lipid kinase. EMBO J. 2003;22:4223–4236. doi: 10.1093/emboj/cdg397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Buck SW, Shore D. Action of a RAP1 carboxy-terminal silencing domain reveals an underlying competition between HMR and telomeres in yeast. Genes Dev. 1995;9:370–384. doi: 10.1101/gad.9.3.370. [DOI] [PubMed] [Google Scholar]
- 28.Moretti P, Freeman K, Coodly L, Shore D. Evidence that a complex of SIR proteins interacts with the silencer and telomere-binding protein RAP1. Genes Dev. 1994;8:2257–2269. doi: 10.1101/gad.8.19.2257. [DOI] [PubMed] [Google Scholar]
- 29.Moretti P, Shore D. Multiple interactions in Sir protein recruitment by Rap1p at silencers and telomeres in yeast. Mol Cell Biol. 2001;21:8082–8094. doi: 10.1128/MCB.21.23.8082-8094.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wotton D, Shore D. A novel Rap1p-interacting factor, Rif2p, cooperates with Rif1p to regulate telomere length in Saccharomyces cerevisiae. Genes Dev. 1997;11:748–760. doi: 10.1101/gad.11.6.748. [DOI] [PubMed] [Google Scholar]
- 31.Gottschling DE, Aparicio OM, Billington BL, Zakian VA. Position effect at S. cerevisiae telomeres: Reversible repression of Pol II transcription. Cell. 1990;63:751–762. doi: 10.1016/0092-8674(90)90141-z. [DOI] [PubMed] [Google Scholar]
- 32.Aparicio OM, Billington BL, Gottschling DE. Modifiers of position effect are shared between telomeric and silent mating-type loci in S. cerevisiae. Cell. 1991;66:1279–1287. doi: 10.1016/0092-8674(91)90049-5. [DOI] [PubMed] [Google Scholar]
- 33.Kyrion G, Liu K, Liu C, Lustig AJ. RAP1 and telomere structure regulate telomere position effects in Saccharomyces cerevisiae. Genes Dev. 1993;7(7A):1146–1159. doi: 10.1101/gad.7.7a.1146. [DOI] [PubMed] [Google Scholar]
- 34.Hardy CF, Sussel L, Shore D. A RAP1-interacting protein involved in transcriptional silencing and telomere length regulation. Genes Dev. 1992;6:801–814. doi: 10.1101/gad.6.5.801. [DOI] [PubMed] [Google Scholar]
- 35.Levy DL, Blackburn EH. Counting of Rif1p and Rif2p on Saccharomyces cerevisiae telomeres regulates telomere length. Mol Cell Biol. 2004;24:10857–10867. doi: 10.1128/MCB.24.24.10857-10867.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mekhail K, Moazed D. The nuclear envelope in genome organization, expression and stability. Nat Rev Mol Cell Biol. 2010;11:317–328. doi: 10.1038/nrm2894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Andrulis ED, Neiman AM, Zappulla DC, Sternglanz R. Perinuclear localization of chromatin facilitates transcriptional silencing. Nature. 1998;394:592–595. doi: 10.1038/29100. [DOI] [PubMed] [Google Scholar]
- 38.Taddei A, et al. The functional importance of telomere clustering: Global changes in gene expression result from SIR factor dispersion. Genome Res. 2009;19:611–625. doi: 10.1101/gr.083881.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Towbin BD, Meister P, Gasser SM. The nuclear envelope—A scaffold for silencing? Curr Opin Genet Dev. 2009;19:180–186. doi: 10.1016/j.gde.2009.01.006. [DOI] [PubMed] [Google Scholar]
- 40.Koch MR, Pillus L. The glucanosyltransferase Gas1 functions in transcriptional silencing. Proc Natl Acad Sci USA. 2009;106:11224–11229. doi: 10.1073/pnas.0900809106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Drisdel RC, Green WN. Labeling and quantifying sites of protein palmitoylation. Biotechniques. 2004;36:276–285. doi: 10.2144/04362RR02. [DOI] [PubMed] [Google Scholar]
- 42.Meiringer CT, Ungermann C. Probing protein palmitoylation at the yeast vacuole. Methods. 2006;40:171–176. doi: 10.1016/j.ymeth.2006.06.020. [DOI] [PubMed] [Google Scholar]
- 43.Bose ME, et al. The origin recognition complex and Sir4 protein recruit Sir1p to yeast silent chromatin through independent interactions requiring a common Sir1p domain. Mol Cell Biol. 2004;24:774–786. doi: 10.1128/MCB.24.2.774-786.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Müller P, et al. The conserved bromo-adjacent homology domain of yeast Orc1 functions in the selection of DNA replication origins within chromatin. Genes Dev. 2010;24:1418–1433. doi: 10.1101/gad.1906410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Askree SH, et al. A genome-wide screen for Saccharomyces cerevisiae deletion mutants that affect telomere length. Proc Natl Acad Sci USA. 2004;101:8658–8663. doi: 10.1073/pnas.0401263101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.