Abstract
MicroRNAs (miRNAs), a class of post-transcriptional gene expression regulators, have recently been detected in human body fluids, including peripheral blood plasma as extracellular nuclease resistant entities. However, the origin and function of extracellular circulating miRNA remain essentially unknown. Here, we confirmed that circulating mature miRNA in contrast to mRNA or snRNA is strikingly stable in blood plasma and cell culture media. Furthermore, we found that most miRNA in plasma and cell culture media completely passed through 0.22 µm filters but remained in the supernatant after ultracentrifugation at 110 000g indicating the non-vesicular origin of the extracellular miRNA. Furthermore, western blot immunoassay revealed that extracellular miRNA ultrafiltrated together with the 96 kDa Ago2 protein, a part of RNA-induced silencing complex. Moreover, miRNAs in both blood plasma and cell culture media co-immunoprecipited with anti-Ago2 antibody in a detergent free environment. This is the first study to show that extracellular miRNAs are predominantly exosomes/microvesicles free and are associated with Ago proteins. We hypothesize that extracellular miRNAs are in the most part by-products of dead cells that remain in extracellular space due to the high stability of the Ago2 protein and Ago2-miRNA complex. Nevertheless, our data does not reject the possibility that some miRNAs can be associated with exosomes.
INTRODUCTION
MicroRNAs (miRNAs) are a class of 19–23 nt long non-coding RNAs that negatively regulate the expression of mRNAs containing respective miRNAs target sequences (1). MiRNAs have been shown to regulate genes involved in differentiation, proliferation, apoptosis and to be implicated in several diseases including cancer (2,3). Recently, significant amounts of miRNA have been found in extracellular human body fluids including blood plasma, urine, saliva and semen (4–7). Moreover, some circulating miRNAs in the blood have been successfully revealed as biomarkers for several cancers, cardiovascular disease, brain injury and liver injury (4,8–11). Mammalian cells in culture have been also reported to export miRNAs into the extracellular environment (12–15); however, the mechanism of origin and the function of extracellular miRNA remain essentially unclear. Extracellular miRNAs have recently been detected in exosomes isolated from peripheral blood and culture media of several cell lines (12,16). Although, exosomal miRNA has been hypothesized to be involved in intercellular communication (12,15) it remains unclear whether all extracellular miRNAs are associated with exosomes and whether extracellular miRNA are present in physiologically relevant amounts for cell-to-cell signaling. Another study has indicated that cells in culture predominantly exported miRNA in exosome-independent form (13).
In this study, we show that most of the extracellular miRNA in blood plasma and cell culture is independent from exosomes and is bound to Ago2 protein—a part of RNA-induced silencing complex. Further, we found some indications that also other Ago proteins such as Ago1, Ago3 and Ago4 might be associated with extracellular miRNA. This and further presented results indicate that large parts of circulating miRNAs might be by-products of dead/dying cells which persist due to the stability of the miRNA/Ago2 complex. If and to what extent miRNA/Ago2 complexes can be purposively released from cells and act in paracrine manner remain to be investigated.
MATERIALS AND METHODS
Cell culture
All cells were obtained from the American Type Culture Collection (ATCC). Human HEK293T, MCF7 and MDAMB231 cells were grown in Dulbecco’s modified Eagle’s medium (DMEM) (Invitrogen, cat. 11960-044) supplemented with 10% heat-inactivated fetal bovine serum (FBS), 4 mM l-Glutamine and penicillin/streptomycin at 37°C in 5% CO2. Human T47D and HCC1954 cells were grown in RPMI supplemented with 10% FBS, 1% non-essential amino acids and penicillin/streptomycin at 37°C in 5% CO2. For RNA isolation, cells were plated at a density of 105 cells per well in a 24-well plate, the media was renewed ∼12 h after plating and the total RNA isolated 3 days after plating (along with the collection of conditioned media). For overexpression of hsa-miR-34b, MCF7 cells were transfected with 400 ng (per well) of plasmid DNA encoding pri-miR-34b (System Biosciences, cat. PMIRH34B PA-1) using Turbofect transfection reagent (Fermentas) according to the manufacturer’s protocol. For the overexpression of Ago proteins HEK293T cells were transiently transfected with pIRESneo-FLAF-HA-Ago1, pIRESneo-FLAF-HA-Ago2, pIRESneo-FLAF-HA-Ago3 and pIRESneo-FLAF-HA-Ago4 using Turbofect transfection reagent (Fermentas) according to the manufacturer’s protocol. Next day after transfection the media was renewed. Conditioned media was collected 72 h post-transfection, concentrated 10 times using Vivaspin2 10 kDa MWCO columns (Sartorius Stedim GmbH, Heidelberg, Germany) and used in co-immunoprecipitation experiments.
Collection of human blood and plasma preparation
EDTA-Blood was collected from healthy donors and processed for plasma isolation immediately after collection. Blood plasma was centrifuged at 14 000g for 10 min and diluted 1/10 in 1×PBS (w/o Ca, Mg) before filtration through 0.22 μm filters (Millex®GS, Millipore), ulftrafiltration, ultracentrifugation and RNA isolation. This study has been approved by the ethics committee, Heidelberg, Germany.
Preparation of conditioned media, ultracentrifugation and ultrafiltration
To obtain conditioned media for RNA isolation, ultracentrifugation and ultrafiltration, MCF7 cells were seeded in culture flasks and the media was renewed ∼12 h after plating. After 3 days in culture, media were collected and centrifuged at 1200g for 3 min at RT to remove living cells and subsequently centrifuged at 14 000g for 10 min at RT to remove cell debris. For the ultracentrifugation, 6 ml of conditioned media were combined with 6 ml of DMEM and the resulting 12 ml were loaded into the ultracentrifugation tube. Ultracentrifugation of blood plasma (diluted 1/10 in PBS) and conditioned media was performed at 110 000g for 2 h at 4°C. The 110 000 g pellets were washed twice with 1 ×PBS and then dissolved in 700 μl of Qiazol reagent for total RNA isolation (Qiagen). Ultrafiltration of blood plasma (diluted 1/10 in PBS) and conditioned media was performed using Vivaspin20 columns (Sartorius Stedim GmbH) with 300, 100 and 50 kDa MWCO according to the manufacturer’s instructions.
Isolation of miRNAs
Isolation of extracellular miRNAs from liquid samples was performed using Tri-Reagent LS (Sigma) together with miRNeasy kit (Qiagen). Briefly, 1.2 ml of Tri-Reagent LS were added to 0.4 ml of blood plasma or conditioned media, mixed by vigorous shaking for 10 s and then incubated for 10 min at RT to ensure complete dissociation of nucleoprotein complexes. Then (i) 5 pg of synthetic miRNA-39 from Caenorhabditis elegans (cel-miRNA-39) were added as a spike-in control for purification efficiency; (ii) 1.2 μl of glycogen (10 mg/ml) were added to enhance the efficiency of RNA column binding. After supplying 320 μl of chloroform, the mixture was vigorously shaken for 45 s and allowed to stand for 5 min at RT. Following the centrifugation at 14 000g for 20 min total RNA was precipitated from the upper (aqueous) phase by adding 1.5 V of 100% ethanol. Purification of extracted total RNA was performed with miRNeasy columns (Qiagen) according to the manufacturer’s instructions. From the Qiagen columns, RNA was eluted in 30 μl of RNase-free water. Total RNA from cells monolayers grown on 24-well plates was isolated using Qiazol reagent (Qiagen) in combination with miRNAeasy kit (Qiagen) according to the manufacturer’s protocol. To monitor the extraction efficacy, 5 pg of cel-miR-39 was added to the cell/Qiazol lysates. RNA was eluted in 30 μl of RNase-free water.
Quantitative real-time PCR
A fixed volume of 5 μl of total RNA from 30 μl eluates obtained after RNA isolation of a given sample was used as an input into a reverse transcription (RT) reaction. The levels of α-tubulin and β-actin mRNAs were measured using corresponding TaqMan Gene Expression Assays (Applied Biosystems) according to the manufacturer’s instructions. The levels of mature hsa-miR-16, hsa-miR-21, hsa-miR-24, hsa-miR-34b, cel-miR-39 mRNAs were measured using individual TaqMan microRNA Assays (Applied Biosystems) according to the manufacturer’s instructions with minor modifications. Cel-miR-39 was used for the normalization of both extracellular and intracellular miRNAs levels. Since we compared intracellular and extracellular expression of miRNAs in the same cell line it was possible to use only cel-mir-39 as spiked-in control for the normalization of miRNAs in the cells as well as in the media. Reverse transcriptase reactions contained 5 μl of purified total RNA, 50/N nM stem–loop RT primers (where N is a number of analysed miRNAs), 1 ×RT buffer, 0.25 mM each of dNTPs, 3.33 U/µl MultiScribe reverse transcriptase and 0.25 U/µl RNase inhibitor. RT reaction had final volume of 15 µl were incubated for 30 min at 16°C, 30 min at 42°C, 5 min at 85°C and then held at 4°C. RT product was further diluted six times with RNAse-free water. Real-time PCR was performed using Taqman Universal PCR Master Mix. A 10 µl PCR reaction included 2 µl of diluted RT product, 1×TaqMan Universal PCR Master Mix (Applied Biosystems) and 1 ×of the corresponding miRNA assay primers. The reactions were incubated in 384-well plates at 95°C for 10 min, followed by 50 cycles of 95°C for 15 s and 60°C for 1 min. All reactions were run in duplicate. Real-time PCR was performed using the LightCycler 480 Real-Time PCR System (Roche). Data was analyzed with the LightCycler 480 software (Roche), determining the threshold cycle (Ct) by the second derivative max method. Relative quantities of miRNA were calculated using the ΔΔCt method after normalization to the cel-miR-39 spiked-in control.
miRNA profiling
The expression of miRNA in the blood plasma, MCF7 cells, MCF7 cells conditioned media, control DMEM+ 10% FBS and 110 000 g pellets was profiled using stem–loop RT–PCR-based TaqMan Human MicroRNA Arrays (Applied Biosystems). The arrays represented 667 mature miRNAs and miRNAs* present in the Sanger miRBase v12 in a two-card set of arrays (Array A v2.1 and Array B v2.0). RT-PCR reactions were performed according to the manufacturer’s instructions. Briefly, total RNA (3 μl per reaction) was reverse transcribed using TaqMan miRNA Reverse Transcription Kit (Applied Biosystems) in combination with the stem–loop Megaplex primers pool sets A and B in total volume of 7.5 μl. The RT cycling conditions were: 40 cycles of 16°C for 2 min, 42°C for 1 min and 50°C for 1 s. Megaplex RT products were further pre-amplified using TaqMan PreAmp Master Mix and Megaplex PreAmp primers (Applied Biosystems). Briefly, 2.5 μl of the megaplex RT products were mixed with 2.5 μl of Megaplex PreAmp Primers (pool A or B) and 12.5 μl TaqMan PreAmp Master Mix in a 25 μl PCR reaction. The pre-amplification cycling conditions were 95°C for 10 min, 55°C for 2 min and 75°C for 2 min followed by 12 cycles of 95°C for 15 s and 60°C for 4 min. The pre-amplified cDNA was diluted with 0.1 × TE (pH 8.0) to 100 μl and 10 μl of diluted cDNA was used in each plate for real-time PCR reactions. The expression profiles of miRNA were acquired using TaqMan Low-Density Arrays. Briefly, quantitative real-time PCR was carried out on Applied BioSystems 7900HT thermocycler using the manufacturer’s recommended cycling conditions. Cycle threshold (Ct) values were calculated with the SDS software v2.3 using automatic baseline settings with assigned minimum threshold of 0.2. Because of the fact that the Ct value of 35 represents single template detection, Ct value > 35 were considered to be below the detection level of the assays (17). Therefore, all miRNAs with Ct > 35 were included in the analysis as ‘undetected’ and assigned the Ct values of 40. Array experiments were performed only once using samples from one biological experiment. Those miRNAs that appeared on the DMEM+10% FBS (negative control sample) array were removed from the analysis (for all miRNAs that were more than 1.5 Δ Ct, as compared to conditioned media samples).
Immunoprecipitation
Immunoprecipitation of extracellular miRNAs with anti-Ago2 antibody (Abnova, clone 2E12-1C9; cat. H00027161-M01) was performed using Pierce Classic IP kit (Thermo Scientific). Binding with anti-Ago2 was carried out in a detergent-free solution. The maximal efficiency of immunoprecipitation was achieved when 5 μg of anti-Ago2 antibody was combined with 5 μl of sample. Briefly, 10 μl of 1× PBS were added to 5 μl of 2× concentrated human blood plasma (or 10× concentrated conditioned media) to form 15 μl of miRNA binding solution. The concentration of the samples was performed using Vivaspin2 columns with 10 kDa MWCO (Sartorius Stedim GmbH). To form Ago2/anti-Ago2 complexes, 15 μl of corresponding miRNA binding solutions were combined with 5 μg (10 μl) of anti-Ago2 antibody and incubated overnight at +4°C. To capture the immune complexes, 20 μl Pierce Protein A/G Agarose suspended in 200 μl of 1×PBS was added to the antibody/lysate samples and incubated for 2 h on a shaker at RT. The A/G Agarose resin was washed two times with 1 ml of IP Lysis/Wash buffer. Afterwards, 700 μl of Qiazol reagent was added to the resin and total RNA was isolated with the miRNeasy kit (Qiagen) as described above. Cel-miR-39 (5 pg) was spiked into the samples after addition of Qiazol reagent and used as normalization control. Immunoprecipitation of exogenously expressed FLAG-fused Ago proteins was approached similarly, except that 100 μl of 10× concentrated conditioned media was combined with 200 μl of Pierce IP Lysis buffer (0.025 M Tris, 0.15 M NaCl, 0.001 EDTA, 1% NP-40, 5% glycerol, pH 7.4) before incubating with 10 μg of mouse anti-FLAG antibody (Sigma, clone M2).
Western blot analysis
Retinates or pellets collected after ultrafiltration or ultracentrifugation, respectively, were dissolved in 100 μl of Laemmli loading buffer (250 mM Tris–HCl pH 6.4; 2% SDS; 100 nM b-MeEtOH), separated by 8% or 12% SDS-PAGE and transferred to nitrocellulose membranes (Amersham Pharmacia, Freiburg, Germany). Membranes were blocked with 5% non-fat milk powder (Roche, Mannheim, Germany) in 1×TBS containing 0.1% Tween-20. Incubation with primary monoclonal mouse anti-Ago2 antibodies (Abnova, clone 2E12-1C9; cat. H00027161-M01), rabbit polyclonal anti-NPM1 (Sigma, clone AB1) or mouse anti-CD9 (R&D systems GmbH) was performed at 4°C overnight in a 1:1000 dilution. After incubation with the primary antibody, membranes were washed twice in 1×TBS with 0.1% Tween-20 for 10 min and incubated for 1 h at room temperature with horseradish peroxidase (HRP) conjugated goat anti-mouse IgG (Santa Cruz, sc-2005) or goat anti-rabbit IgG (Santa Cruz, sc-2004) diluted (1:10 000) in blocking solution. Membranes were washed four times for 10 min in TBS–Tween20 and finally developed with Pierce ECL Western Blotting Substrate (Thermo Scientific) according to the instructions of the manufacturer.
RESULTS
Extracellular versus intracellular miRNA expression pattern
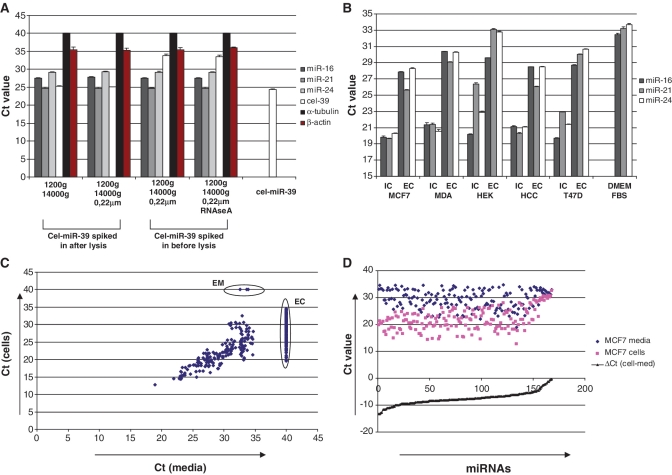
We first confirmed the presence of the nuclease-resistant miRNA species in an extracellular environment. For that purpose, total RNA was isolated from the conditioned media of MCF7 cells and subjected to TaqMan quantitative real-time PCR (qRT–PCR) mature miRNA assays (Applied Biosystems) using pre-designed primers. Species of hsa-miR-16, hsa-miR-21 and hsa-miR-24 as well as α-tubulin and β-actin mRNAs were detected as described in ‘Materials and Methods’ section (Figure 1A). Centrifugation of the cell-free conditioned media at 14 000 g for 10 min and subsequent filtration through 0.22 μm filters did not decrease the concentration of extracellular miRNAs in the samples. This indicated that these miRNAs species were neither associated with cells debris nor micro-vesicles. Extracellular miRNAs were resistant to nuclease degradation as synthetic cel-miR-39 was immediately degraded when spiked into the media 1 min before the addition of denaturing Tri-Reagent LS (Figure 1A). Furthermore, incubation of conditioned media with 100 ng/ml of RNAseA for 15 min did not affect the levels of endogenous miRNAs (Figure 1A). On the contrary, no α-tubulin mRNA and only trace amounts of β-actin mRNA was detected in the conditioned media.
Figure 1.
(A) miRNAs but not detectable mRNA are present in conditioned media in nuclease resistant form. Conditioned media was collected from MCF7 cells and further treated as indicated on the graph. Cel-miR-39 (5 pg) was added into the media either before or after addition of denaturing Trizol LS reagent (Sigma). As a control for miRNA recovery, 5 pg of cel-miR-39 was dissolved in 30 μl of nuclease-free water. Total RNA from each sample was eluted in 30 μl of nuclease-free water and the content of miRNAs, β-actin and α-tubulin mRNAs were analyzed using quantitative real-time PCR as described in the ‘Materials and Methods’ section. Data presented as raw Ct values, each bar represents mean (SD) (n = 3). (B) Extracellular (EC) and intracellular (IC) levels of three miRNA species in five different cancer cell lines were measured by quantitative real-time PCR. Data presented as Ct values normalized on spiked in cel-miR-39 control; each bar represents mean (SD) of three independent RNA isolations. (n = 3). (C and D). Intra- and extra-cellular miRNA spectra for MCF-7 cells were measured by TaqMan miRNA Low density array (Applied Biosystems). (C). Correlation plot shows Ct values of miRNAs obtained from total RNA extracted from 400 μl of conditioned media and corresponding MCF7 cells monolayer. Those miRNAs that were not detected on the array (Ct > 35) were assigned Ct values of 40. EC—miRNAs presented exclusively in the cells; EM—miRNAs presented exclusively in the media (lists of exclusively expressed miRNAs Cp value <35 in Supplementary Table S1). (D) Those miRNAs that were detected in both cells and the media demonstrated a dramatic difference in extracellular/intracellular amount. Note, miRNAs content in the media was much lower [average ΔCt (cell-media) = −7.5].
We further investigated if cells in culture secreted miRNA specifically. For that purpose we compared extracellular and intracellular levels of hsa-miR-16, hsa-miR-21 and hsa-miR-24 species in several breast cancer cell lines (Figure 1B). Interestingly, bovine miRNAs were detected in DMEM medium supplemented with 10% bovine serum, furthermore indicating enormous stability of the extracellular miRNAs. However, in the conditioned media (following incubation with the cells) the analyzed miRNAs were present in significantly higher amounts (several magnitudes) than in DMEM+ 10% bovine serum alone (Figure 1B). Therefore, the presence of bovine miRNA had no substantial effect on the analysis. We further observed that in all cell lines intracellular and extracellular patterns of miRNA-16 and miRNA-24 were well correlated (Figure 1B). Overexpression of randomly selected miRNA-27a with pri-miR-27a encoding plasmid in both HEK293 and MCF7 cells led to the proportional increase of miRNA-27a level in the conditioned media (data not shown), so favoring the hypothesis of non-specific export of miRNAs. However, in all cell lines analyzed, the amount of miRNAs exported into the media over 3-day period was dramatically lower as compared to the miRNAs amount in the monolayers of the parental cells (Figure 1B). The mean ΔCt values of extracellular to intracellular miRNAs signals in five analyzed cell lines were as follows: 8.5 ± 0.84 for miR-16; 6.6 ± 0.78 for miR-21; 8.8 ± 1.08 for miR-24. Surprisingly, higher concentration of miR-21 in the media was found as compared to extracellular miR-16 and miR-24, indicating that intracellular and extracellular miRNA levels were not parallel for all miRNAs. The observed difference between extracellular/intracellular levels of miRNA-21 and miR-16/miR-24 cannot be attributed to the difference in primers’ efficacy as they were near equal for these miRNA species (data not shown). Second, we evaluated miRNA expression profile of MCF7 breast cancer cells versus miRNA content in their media. Considering miRNAs with Ct values of <35 as ‘detected’ 150 miRNA species were exclusively detected in the cells monolayers while 168 miRNAs were detected both in the cells and in the media (Figure 1C). Only three miRNA species were found exclusively in the media (Figure 1C, EM; Supplementary Table S1). However, the Ct values of these miRNAs ranged from 32.6 to 33.8 and were, therefore, close to the undetectable levels. The profiles of extracellular and intracellular miRNAs (for miRNAs detected in both cells and media) were strongly correlated (Figure 1C and D). The average ΔCt of cellular to extracellular miRNAs was 7.5 indicating that the amount of miRNA was approximately 180 times higher in the cell monolayers than in 3-day conditioned media. Small RNA species of non-miRNA origin including U6, RNU24, RNU48, RNU6B, RNU43 and RNU44 were presented in high amounts (Ct 12.9–24.8) within the cells but were either totally absent (RNU43, RNU44, RNU6B) or had significantly lower Ct values (ΔCt > 12) in the media (Table 1). On the whole-miRNA profile level, we did not perform any quantitative calculations of extracellular to intracellular miRNA amounts but merely aimed to qualitatively demonstrate that: (i) none of the miRNAs was present in higher concentrations outside the cells and (ii) the extracellular profile of miRNAs closely but not completely relates to the intracellular miRNA expression pattern.
Table 1.
Non-miRNA species in cells and conditioned media
| Non-miRNA species | AvCt [cells] ± SD | AvCt [media] ± SD |
|---|---|---|
| MammU6 | 12.9 ± 0.7 | 24.6 ± 0.5 |
| RNU24 | 16.9 ± 0.04 | 33.5 ± 0.5 |
| RNU43 | 23.1 ± 0.2 | Undetermined |
| RNU44 | 15.1 ± 0.4 | Undetermined |
| RNU48 | 13.2 ± 0.4 | 29.0 ± 0.4 |
| RNU6B | 24.8 ± 0.2 | Undetermined |
Most of the extracellular miRNAs are not associated with exosomes
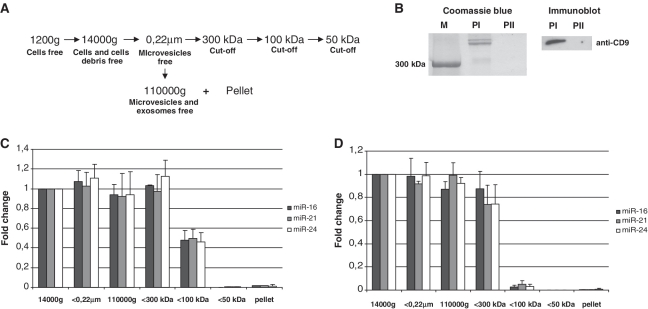
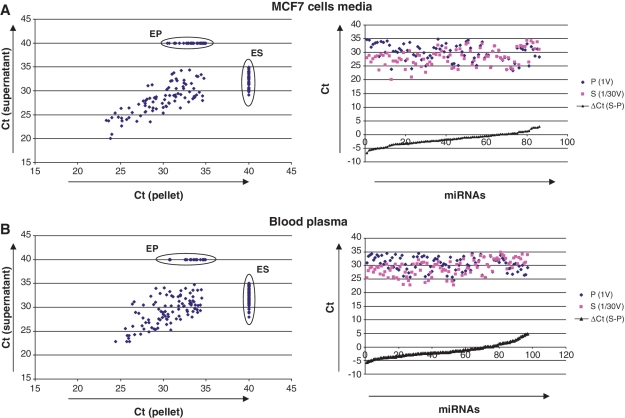
Extracellular miRNAs have been previously detected in cell-exported exosomes, small membrane vesicles of 50–100 nm in diameter (12,14–16). However, to our knowledge, no experiments demonstrating that extracellular miRNA is associated exclusively with exosomes have been described. We have noticed that incubation of either blood plasma or conditioned media with the 1% NP-40 detergent did not restore the nuclease sensitivity of extracellular miRNAs (data not shown). To further confirm the hypothesis of exosome-free extracellular miRNA, we fractionated blood plasma and conditioned media of MCF7 cells according to well-developed methods for exosomes isolation (12,18). Thus, cell debris and microvesicles were removed from the samples by filtration through 0.22 μm filters; the exosome-free fractions were obtained by further ultracentrifuging the samples at 110 000g for 2 h at +4°C (Figure 2A and B). The levels of three miRNA species (hsa-miR-16, hsa-miR-21 and hsa-miR-24) were quantitatively assayed in the samples after each fractionation step with individual TaqMan miRNA Assays. To our surprise, ultracentrifugation did not significantly decrease the level of any of these miRNAs in both blood plasma and conditioned media (Figure 2C and D). Accordingly, 110 000g pellets contained only trace amounts of the respective miRNAs (blood plasma: miR-16: 0.34 ± 0.07%; miRNA-21: 0.21 ± 0.14%; miRNA-24: 0.94 ± 0.68%; MCF7 media: miR-16: 1.57 ± 0.5%; miRNA-21: 1.9 ± 1.18%; miRNA-24: 1.19 ± 0.80%). The presence of exosomal marker protein CD9 in 110 000g pellets was confirmed by western immunoblot analysis (Figure 2B). Furthermore, TaqMan Low Density Array (Applied Biosystems) of 707 different miRNAs and miRNAs* performed on total RNAs isolated from 400 μl (1/30 V) of 110 000g supernatants and total 110 000g pellets revealed that only a small proportion of all miRNAs was specifically associated with the pellet fractions in both plasma and conditioned media (Figure 3). Those miRNAs that appeared on the profile with Ct value of <35 were considered as ‘detected’. In the conditioned media the average ΔCt of miRNAs detected in 1/30 V supernatant and 1 V pellet was −1.6 indicating ∼90 times higher miRNA amounts in the supernatant compared to pellet (Figure 3A). Extracellular miRNAs that were found exclusively in the exosomes-free supernatant and in the exosomes-containing pellet are listed in Supplementary Tables S2 and S3. Out of 188 miRNA species detected in the blood plasma, 97 miRNAs were detected both in the pellet and supernatant (Figure 3B) and 69 miRNA species were found exclusively in the supernatant (Figure 3B, ES; Supplementary Table S4). Only 22 different miRNAs were detected exclusively in the pellet fraction of blood plasma (Figure 3B, EP; Supplementary Table S5), however, the Ct values of these miRNAs ranged from 30.8 to 34.9. The average ΔCt for miRNAs between 1/30 V supernatant and 1 V pellet was −1.4 indicating that the amount of miRNA was ∼80 times higher in the plasma supernatant compared to the pellet. Therefore, our data strongly indicates that the level of miRNAs that can be associated with exosomes is dramatically lower compared to the amount of exosomes-free miRNAs per volume of blood plasma or conditioned media.
Figure 2.
(A) The procedure for the fractionation of human blood plasma and conditioned media. Cell-debris free (1200g following 14 000g centrifugation) blood plasma diluted 12 times in PBS and MCF7 cells conditioned media were used as starting samples. Microvesicles were removed from the samples by filtration through 0.22 μm filters; exosome-free fractions were obtained by ultracentrifugation at 110 000g. Furthermore, fractionation of 0.22 μm filtered samples was performed using 300, 100 and 50 kDa MWCO nanomembranes. (B) The efficacy of ultracentrifugation to remove exosomes and large proteins from plasma was assessed by SDS–PAGE gel of the 110 000g pellets. Coomassie blue (left) staining and western immunoblot (right) had shown that the first ultracentrifugation (PI pellet) removed large proteins (>300 kDa) and exosomes (as judged by exosomal marker protein CD9) from the plasma supernatant. While the pellet obtained after second ultracentrifugation of the same supernatant (PII) at the same force did not contain any detectable amounts of proteins including exosomal marker. (C and D) The level of three miRNA species miR-16, miR-21 and miR-24 was quantitatively assayed in the conditioned media (C) and blood plasma (D) samples after each fractionation step with individual TaqMan miRNA Assays. Spiked in cel-miR-39 was used as normalization control for all samples. Data presented as relative miRNA fold change. Each bar represents mean (SD) of three independent experiments (n = 3).
Figure 3.
miRNA spectra of the supernatants and the pellets obtained after ultracentrifugation of MCF7 cells conditioned media (A) and human blood plasma (B) as measured by TaqMan miRNA Low Density arrays (Applied Biosystems). Blood plasma (1 ml) was combined with 11 ml of 1×PBS before ultracentrifugation. ‘Left’: Correlation plots shows Ct values of miRNA signals obtained from total RNA extracted from 400 μl (1/30 volume) of supernatants and total pellets (pellets were obtained after ultracentrifugation of total 12 ml of samples). Note that only 1/30 of total supernatants were taken for the RNA isolation and subsequently for the TaqMan Low Density Arrays. MiRNAs which were undetected on the array (Ct > 35) were assigned Ct values of 40. ES—miRNAs presented exclusively in the supernatant; EP—miRNAs presented exclusively in the pellets (exclusively detected miRNAs are listed in Supplementary Data). ‘Right’: Those miRNAs that were detected in both supernatants and pellets demonstrated a dramatic difference in the content. Note: amount of miRNAs in the pellets is much lower than in supernatants.
Extracellular miRNA is associated with Ago2 protein
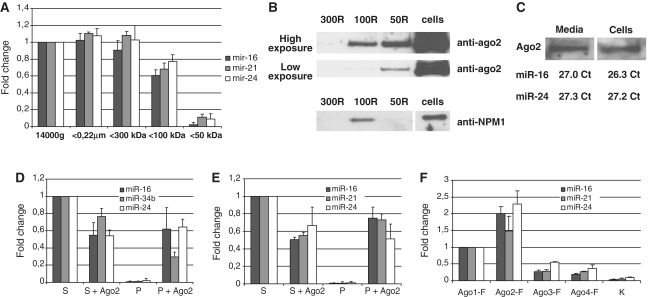
We further hypothesized that exosome-free extracellular miRNA could be protected from nuclease digestion by associated proteins. To estimate the approximate molecular weight of ‘extracellular miRNA binding protein’, we applied a sequential ultrafiltration procedure (Figure 2A and B). For this purpose 0.22 μm filtered blood plasma and conditioned media were further fractionated by centrifugal ultrafiltration using nanomembrane concentrators of 300, 100 and 50 kDa MWCO. Individual TaqMan miRNA Assays were used to quantify the species of hsa-miR-16, hsa-miR-21 and hsa-miR-24 which remained in the samples after each ultrafiltration step. In conditioned media, extracellular miRNA completely passed through 300 kDa MWCO filters, ∼50% of the investigated miRNAs passed through 100 kDa MWCO filter and only a negligible amount of miRNAs were found in the 50 kDa filtrate (Figure 2C). The fact that 300 kDa MWCO nanomembranes were permeable for extracellular miRNAs provided an additional evidence of exosome-free presence of extracellular miRNA. Similarly to the conditioned media, most of the miRNA was still found in the 300 kDa filtrates in blood plasma (Figure 2D). However, roughly 90% of miRNA did not pass through 100 kDa filters and 50 kDa filtrates were almost totally depleted of miRNAs (Figure 2D). When serum-free 1-day conditioned media from MCF7 cells was sequentially fractionated, the percent of miRNA that passed through 100 kDa MWCO nanomembranes was about 70% while near 5% of the original miRNA amount was still detected in 50 kDa filtrate (Figure 4A). This data suggest that extracellular miRNA in blood plasma and conditioned media is associated with a protein of MW between 300 and 50 kDa. It is well-known that in the cell mature miRNAs are directly bound to Ago2, a 96 kDa protein responsible for the endonuclease cleavage of targeted mRNA (1). Accordingly, anti-Ago2 monoclonal antibody has previously been shown to be effective in capturing cellular miRNAs (19). To determine if the extracellular miRNA is also associated with Ago2 protein, the retinates (remnants that did not pass through the nanofilters) collected after ultrafiltration of serum-free conditional media (Figure 4A) were subjected to western immunoblot analysis with anti-Ago2 monoclonal antibody (Figure 4B). As we expected, Ago2 specific bands were observed in both 100 and 50 kDa retinates, while no Ago2 was detected in 300 kDa retinates. The relative intensities of Ago2 bands on the western blot corresponded to the amounts of miRNAs depleted from the filtrates (Figure 4A). More importantly, the amount of Ago2 in MCF7 conditioned media and in MCF7 cell lysates was proportional to their miRNAs content that was estimated by visual comparison of Ago2 signals on western blot and normalized Ct values of miR-16 and miR-24 (Figure 4C). To further confirm the binding of extracellular miRNAs to Ago2 protein in blood plasma and conditioned media, anti-Ago2 and control immunoprecipitates were subjected to TaqMan miRNA Assays (Figure 4D and E). In consistency with the western blot data, miRNAs were detected in anti-Ago2 immunoprecipitates while control immunoprecipitates (containing no anti-Ago2) were almost totally free of miRNAs. Moreover, incubation of both blood plasma and cells media with anti-Ago2 antibody caused the depletion of miRNA from the samples. Ultrafiltration and immunoprecipitation experiments were performed in a detergent-free environment, therefore, targeting a native form of extracellular miRNA/Ago2. Figure 4D and E included only the co-immunoprecipitation performed when 5 μg of anti-Ago2 was used (when the maximal recovery of miRNAs was observed). Further increase of the anti-Ago2 antibody per reaction (from 5 to 10 μg) did not significantly increase miRNA content in the pellet. The second immunoprecipitation with anti-Ago2 yielded significantly smaller amount of miRNA as compared to first immunoprecipitation (data not shown). The remaining miRNA in the supernatant is, therefore, unlikely to be attributed to larger parts to the efficiency of co-immunoprecipitation but could be associated with other Ago proteins, specifically Ago1, Ago3 and Ago4. Unfortunately, there are no well described antibodies against Ago1, Ago3 and Ago4 for the immunoprecipitation available. To test the hypothesis that other Ago proteins loaded with miRNA can also be present in the extracellular environment we have tested the ability of anti-FLAG antibody to precipitate miRNAs from 3-days conditional media of cells transfected with constructs encoding FLAG-fused human Ago1, Ago2, Ago3 and Ago4 (Figure 4F). The highest amount of miRNA in the media was still associated with Ago2; however, the media of the cells transfected with Ago1 also contained significant amounts of miRNA. It is therefore, feasible that the remaining miRNA in the blood plasma and conditioned media can be bound to other members of the Ago family.
Figure 4.
(A–C) The levels of extracellular miRNAs correlate with the level of extracellular Ago2 but not NPM1 protein. One-day serum-free conditioned media from MCF7 cells was fractionated using nanomembrane concentrators of 300, 100 and 50 kDa MWCO. (A) Individual TaqMan miRNA assays were used to quantify the species of miR-16, miR-21 and miR-24 which remained in the solution after each ultrafiltration step. (B) Concentrated fractions (retinates) were subjected to western immunoblot analysis using anti-Ago2 and anti-NPM1 antibody. Note: 300R = retinate from 300 kDa MWCO filter; 100R = 100 kDa retinate from 100 kDa MWCO filter and 50R = retinate from 50 kDa MWCO filter. (C) The amount of Ago2/miRNA in conditioned media was proportional to the amount of Ago2/miRNA in the cells. MCF7 cells lysate and concentrated serum-free conditioned media were prepared and miRNA content was assessed using TaqMan miRNA assays. After that, cell lysate was diluted so that the miRNA content in it equaled approximately the miRNA content in the concentrated serum-free media. Finally, western immunoblot was performed on concentrated media and diluted cell lysates and the amounts of Ago2 were visually compared. (D and E) Extracellular miRNA co-immunoprecipitated with anti-Ago2 antibody in a detergent-free environment. Species of miR-16, miR-21, miR-24 and exogenously expressed miR-34b were measured both in anti-Ago2 co-immunoprecipitates (P and P+Ago2) and in the remained supernatants (S and S+Ago2) obtained from MCF7 conditioned media (D) and blood plasma (E). Data presented as a fold change compared to control supernatants (assigned value 1), each bar represents mean (SD) of three independent experiments. Cel-miR-39 spiked in during RNA isolation step was used as normalization control. Note, the anti-Ago2 antibody induced a depletion of the miRNAs from conditioned media and blood plasma. (F) The levels of miR-16, miR-21 and miR-24 were measured in the anti-FLAG pellets obtained from HEK293T cells conditioned medias 72 h after transfection with plasmids-encoding FLAG-Ago1, FLAG-Ago2, FLAG-Ago3, FLAG-Ago4 or empty vector (K).
In one recent report it was hypothesized that miRNA in serum-free conditioned media can be bound to nucleophosmin (NPM1) (13). This assumption was based on the fact that NPM1 was able to protect miRNA from RNAses when mixed with synthetic miRNA in vitro and was present in serum-free conditioned media in relatively high amounts. We indeed detected high amounts of NPM1 in 1-day conditioned serum-free media from MCF7 cells. However, the ultrafiltration patterns of NPM1 and extracellular miRNA were significantly different (Figure 4A and B). It remains to be elucidated, why 33 kDa NPM1 was found exclusively in 100 kDa retinate fraction (did not pass through 100 kDa filters). However, in the cell NPM1 forms complexes with some other proteins (like nucleolin and ribonucleoproteins) and undergo oligomerization (20).
miRNA remains stable for the prolonged periods after the cell death
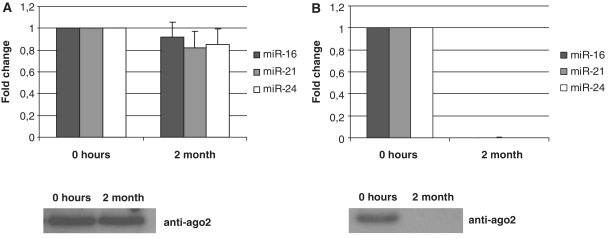
We have also noticed that extracellular miRNA levels in conditioned media increased with higher rate of cell death (data not shown). This observation was the basis for our hypothesis that extracellular miRNAs might be by-products of dead cells that persist in the extracellular space due to their unusual stability. We further evaluated the stability of the intracellular miRNAs in the lysates of MCF-7 cells. For that reason, 5 × 106 MCF7 cells were lysed in 1 ml of buffer containing mild detergent (1% NP-40) and no protease or phosphatase inhibitors. The lysate was then diluted 10 times with PBS to reduce the concentration of the detergent, passed through 0.22 μm filters and split in two parts. One part of the lysate was immediately frozen and the other part was stored at room temperature. Two months later, total RNA was isolated from both lysates. Species of hsa-miR-16, hsa-miR-21 and hsa-miR-24 were analyzed by TaqMan miRNA qRT–PCR (Figure 5A). To our surprise, both the levels of miRNAs and the amount of Ago2 protein in two lysates were not significantly different. On the contrary, no miRNA or Ago2 have been detected in conditioned serum-free media from MCF7 cells which was stored for 2 months at RT (Figure 5B). The sensitivity of both miRNA and Ago2 to the overtime degradation in serum-free media can be explained by the fact that it contains significantly less contaminating proteins as compared to serum-containing conditioned media, blood plasma or cell lysate. It is known that miRNA processing machinery in the cell leads to the production of mature miRNA strand attached to one of the Ago proteins and so far there were no indications that intracellular mature miRNAs can be directly associated with other proteins. Given a remarkable stability of miRNA/Ago2 complexes it can be logically anticipated that miRNA are found in significant amounts in human body fluids.
Figure 5.
Stability of miRNA and Ago2. (A) MCF7 cells were lysed in a buffer containing no protease or phosphatase inhibitors and either frozen or left at room temperature for 2 months. (B) Serum-free conditioned media obtained after incubation with MCF7 cells monolayer for 24 h was either frozen or left at RT for 2 months. The species of miRNAs and Ago2 content were analyzed by TaqMan miRNA qRT–PCR assays and western immunoblot, respectively, in both lysates and conditioned medias. Data presented as a fold change at 2 month point versus 0 point (frozen samples), each bar represents mean (SD) of three independent RNA isolations (n = 3). Cel-miR-39 was used as normalization control.
The fact that intracellular miRNAs can be preserved in the extracellular milieu for at least 2 months after cell lysis, supports our hypothesis that extracellular miRNAs probably originate as by-products from dead cells and remain stable for prolonged time in the extracellular space due to the high stability of Ago2/miRNA complex. Accordingly, non-miRNA species such as U6 RNA, RNU24, RNU43, RNU44, RNU48, RNU6B and mRNAs which are not associated with Ago proteins are either totally absent in the extracellular environment or detected in dramatically lower amounts (as compared to intracellular) (Figure 1A and Table1).
DISCUSSION
In this work, we addressed questions regarding stability, origin and possible biological function of circulating extracellular miRNA in human peripheral blood and cell culture media. As a major tool for the detection of mature miRNAs, we utilized individual TaqMan miRNA assays for three common miRNAs (hsa-miR-16, hsa-miR-21 and hsa-miR-24). These miRNAs were all readily detected in human blood plasma and conditioned media of several cancer cell lines. However, all three miRNAs were present in significantly higher levels in the cells monolayers compared to 3-day conditioned media. Furthermore, array analysis showed that intracellular and extracellular miRNA content was well correlated for most of the miRNAs. However, also some specific—especially intracellular specific—miRNAs were detected. Generally, miRNAs were present in much lower concentrations in conditioned media than in cell monolayers. Nevertheless, some miRNAs were found exclusively in the media but only in trace amounts (Ct ranging from 32.6 to 33.8). A number of recent studies demonstrating that exosomes derived from several cell lines contain miRNAs forced us to test the hypothesis of exosomes-mediated nuclease protection of extracellular miRNA (12,14–16). However, quantification of three individual miRNAs in different fractions obtained after ultrafiltration and ultracentrifugation of blood plasma and cells media revealed that >97% of these miRNA amounts are exosome free. Furthermore, array analysis of miRNA distribution in the supernatants and pellets obtained after ultracentrifugation revealed that only small amounts of miRNAs were associated with pellets (exosomes) fractions.
Previously, Wang et al. (13) reported a phenomenon of active miRNAs export from cells after serum deprivation. In accordance with our data, they also observed that significant fraction of extracellular miRNA were not associated with cell-derived vesicles. The authors, however, suggested the existence of a specific mechanism of miRNA transportation outside the cells and that the extracellular miRNA was associated with 33 kDa RNA binding protein nucleophosmin (NPM1). Our data, however, suggest that extracellular miRNAs are bound to 96 kDa Ago proteins and Ago2 in particular. These are the same proteins with which miRNAs are naturally associated within the cell as being a part of RNA-induced silencing complex (1,19). Ago2/miRNAs complexes are highly nuclease/protease resistant as we had shown that miRNA remained stable in the cell lysates for at least 2 months. The unusual stability of the intracellular miRNA provides a firm grounding for the proposition that the large portion of extracellular miRNA might be by-products of dead cells. According to our hypothesis, the miRNA/Ago2 complexes are released from the cells upon necrosis or apoptosis. Unlike other RNA species, miRNA remains stable in the extracellular space for long periods due to the significant resistance of Ago2 exonuclease to degradation. Further our array analyses point to the direction that some miRNA/Ago2 complexes might be directly released by cells. It remains to be shown if and to what extent miRNA/Ago complexes can be released from cells in a directed manner in order to fulfill a paracrine function. Nevertheless, our results are in agreement with the observation that some miRNAs detected in plasma or conditioned media can indeed be associated with exosomes. Thus, TaqMan Low Density Array analysis of 110 000g pellets and supernatants of both blood plasma and conditioned media revealed the presence of several pellet-specific miRNAs (however, these miRNA had Ct > 30.7). Furthermore, when studying exosome-specific miRNAs investigators have to take into account the possible contamination of exosome fractions with Ago2 loaded miRNAs.
Recently, it has been suggested that extracellular miRNA entrapped in exosomes to be involved in intercellular communication (12,14,15). Our data, however, show that only a small minority of extracellular miRNA can be associated with exosomes. It has to be shown, if Ago/miRNA complexes can be released from cells and mediate cell–cell communication. It is questionable whether the extracellular miRNAs associated with exosomes are present outside the cells in physiologically relevant concentrations (necessary to induce functional impact in other cells). However, a paradigm of autocrine and paracrine miRNA signaling through exosomes cannot be excluded.
Within recent years, there has been an enormous interest in investigating circulating miRNAs in blood plasma/serum as potential biomarkers for early cancer diagnoses. The detection of cancer-specific miRNAs in the blood plasma/serum of cancer patients has been shown in multiple reports (4,8,16). Our study is in agreement with and strongly supports the possibility of using miRNAs for cancer diagnosis since cancer-specific Ago2-associated miRNAs should remain in the blood in stable form as any other miRNAs. One has to be aware, however, that most of peripheral plasma miRNA are likely to derive from blood cells, liver, lungs, kidney and other organs where extensive contact between cells and the blood plasma occurs. Therefore, cancer-specific miRNAs can be masked by miRNAs from other sources. However, it could be anticipated that the level of cancer-specific miRNAs to be increased in the blood plasma. In a similar way, increase in particular ‘tissue specific’ or ‘organ specific’ miRNAs in blood plasma (or other body fluids) could serve as an indication of toxicity of a particular tissue/organ (10,11).
The recent discovery of virally encoded miRNAs in several pathogenic human viruses suggests that miRNAs may function in interspecies regulation involving viral miRNAs and host genes (21,22). It is thus conceivable that circulating viral miRNAs might be exploited for the diagnostics of viral infections since viral miRNA should be preserved in the extracellular space after the infected host cells die. More importantly viral miRNAs are different from host miRNAs, and therefore can serve as a specific marker for the diagnosis, especially for those viruses for which antibody-based detection systems are not yet established. Finally, we hypothesize that extracellular miRNAs may contribute to the investigation of miRNAs—regulators of cell death. If Ago2-associated miRNAs releases after the cells die, the profiles of extracellular miRNAs should represent the miRNA profile of the cells in the very late stage of apoptosis. Therefore, quantitative comparison of miRNA signatures in given cells and the media could contribute to the identification of miRNA circuit involved in apoptosis.
Our findings also suggest that restricting the analysis of biomarkers to only exosomal miRNA ignores vast majority of the circulating miRNAs in blood plasma. Moreover, our data shed light on the problem of normalization of miRNA expression applying non-miRNA species (like U6, etc.) due to their divergent stability of circulating miRNA. Further, between miRNAs themselves normalization can be difficult if miRNAs are originating from different extracellular traveling systems (ago-bound versus exosomal) as here—and maybe even between different ago-carriers—the stability of miRNAs can be different. The best way to overcome these problems is the implementation of exactly the same standardized processing protocols of samples of different comparison groups and within study cohorts.
To summarize, in this study, we have shown that the overwhelming majority of the nuclease-resistant extracellular miRNA in plasma and cell culture media is floating outside exosomes and is bound to Ago2 protein—a part of the RNA-induced silencing complex. We also found indications that extracellular miRNA can be bound to other Ago proteins (especially Ago1). It is likely that large parts of extracellular circulating miRNAs are by-products of dead or dying cells that persist due to the high stability of the miRNA/Ago 2 complex. Whether and to what extent miRNA/Ago complexes can specifically be released from cells and function in a paracrine manner needs to be investigated.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Dietmar-Hopp Foundation, the Helmholtz Society and the German Cancer Research Center. Funding for open access charge: German Cancer Research Center.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
A.T. designed and carried out experimental part and wrote this manuscript; L.W. performed western blotting experiments and revised the manuscript; A.L. helped to establish miRNA isolation protocol and participated in qPCR experiments, B.B. conceived of the study, participated in its design and coordination and revised the manuscript.
REFERENCES
- 1.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 2.Croce CM, Calin GA. miRNAs, cancer, and stem cell division. Cell. 2005;122:6–7. doi: 10.1016/j.cell.2005.06.036. [DOI] [PubMed] [Google Scholar]
- 3.Zhang B, Pan X, Cobb GP, Anderson TA. Plant microRNA: a small regulatory molecule with big impact. Dev. Biol. 2006;289:3–16. doi: 10.1016/j.ydbio.2005.10.036. [DOI] [PubMed] [Google Scholar]
- 4.Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O'Briant KC, Allen A, et al. Circulating microRNAs as stable blood-based markers for cancer detection. Proc. Natl Acad. Sci. USA. 2008;105:10513–10518. doi: 10.1073/pnas.0804549105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hanke M, Hoefig K, Merz H, Feller AC, Kausch I, Jocham D, Warnecke JM, Sczakiel G. A robust methodology to study urine microRNA as tumor marker: microRNA-126 and microRNA-182 are related to urinary bladder cancer. Urol. Oncol. 2009;28:655–661. doi: 10.1016/j.urolonc.2009.01.027. [DOI] [PubMed] [Google Scholar]
- 6.Park NJ, Zhou H, Elashoff D, Henson BS, Kastratovic DA, Abemayor E, Wong DT. Salivary microRNA: discovery, characterization, and clinical utility for oral cancer detection. Clin. Cancer Res. 2009;15:5473–5477. doi: 10.1158/1078-0432.CCR-09-0736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zubakov D, Boersma AW, Choi Y, van Kuijk PF, Wiemer EA, Kayser M. MicroRNA markers for forensic body fluid identification obtained from microarray screening and quantitative RT-PCR confirmation. Int. J. Legal Med. 2010;124:217–226. doi: 10.1007/s00414-009-0402-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Roth C, Rack B, Muller V, Janni W, Pantel K, Schwarzenbach H. Circulating microRNAs as blood-based markers for patients with primary and metastatic breast cancer. Breast Cancer Res. 2010;12:R90. doi: 10.1186/bcr2766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gupta SK, Bang C, Thum T. Circulating microRNAs as biomarkers and potential paracrine mediators of cardiovascular disease. Circ. Cardiovasc. Genet. 2010;3:484–488. doi: 10.1161/CIRCGENETICS.110.958363. [DOI] [PubMed] [Google Scholar]
- 10.Wang K, Zhang S, Marzolf B, Troisch P, Brightman A, Hu Z, Hood LE, Galas DJ. Circulating microRNAs, potential biomarkers for drug-induced liver injury. Proc. Natl Acad. Sci. USA. 2009;106:4402–4407. doi: 10.1073/pnas.0813371106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Redell JB, Moore AN, Ward Iii NH, Hergenroeder GW, Dash PK. Human traumatic brain injury alters plasma microRNA levels. J. Neurotrauma. 27:2147–2156. doi: 10.1089/neu.2010.1481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat. Cell Biol. 2007;9:654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 13.Wang K, Zhang S, Weber J, Baxter D, Galas DJ. Export of microRNAs and microRNA-protective protein by mammalian cells. Nucleic Acids Res. 38:7248–7259. doi: 10.1093/nar/gkq601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Pegtel DM, Cosmopoulos K, Thorley-Lawson DA, van Eijndhoven MA, Hopmans ES, Lindenberg JL, de Gruijl TD, Wurdinger T, Middeldorp JM. Functional delivery of viral miRNAs via exosomes. Proc. Natl Acad. Sci. USA. 2010;107:6328–6333. doi: 10.1073/pnas.0914843107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kosaka N, Iguchi H, Yoshioka Y, Takeshita F, Matsuki Y, Ochiya T. Secretory mechanisms and intercellular transfer of microRNAs in living cells. J. Biol. Chem. 2010;285:17442–17452. doi: 10.1074/jbc.M110.107821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hunter MP, Ismail N, Zhang X, Aguda BD, Lee EJ, Yu L, Xiao T, Schafer J, Lee ML, Schmittgen TD, et al. Detection of microRNA expression in human peripheral blood microvesicles. PLoS One. 2008;3:e3694. doi: 10.1371/journal.pone.0003694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guthrie JL, Seah C, Brown S, Tang P, Jamieson F, Drews SJ. Use of Bordetella pertussis BP3385 to establish a cutoff value for an IS481-targeted real-time PCR assay. J. Clin. Microbiol. 2008;46:3798–3799. doi: 10.1128/JCM.01551-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gonzales PA, Zhou H, Pisitkun T, Wang NS, Star RA, Knepper MA, Yuen PS. Isolation and purification of exosomes in urine. Methods Mol. Biol. 641:89–99. doi: 10.1007/978-1-60761-711-2_6. [DOI] [PubMed] [Google Scholar]
- 19.Hassan MQ, Gordon JA, Lian JB, van Wijnen AJ, Stein JL, Stein GS. Ribonucleoprotein immunoprecipitation (RNP-IP): a direct in vivo analysis of microRNA-targets. J. Cell Biochem. 2010;110:817–822. doi: 10.1002/jcb.22562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jian Y, Gao Z, Sun J, Shen Q, Feng F, Jing Y, Yang C. RNA aptamers interfering with nucleophosmin oligomerization induce apoptosis of cancer cells. Oncogene. 2009;28:4201–4211. doi: 10.1038/onc.2009.275. [DOI] [PubMed] [Google Scholar]
- 21.Cullen BR. Viruses and microRNAs. Nat Genet. 2006;38(Suppl):S25–S30. doi: 10.1038/ng1793. [DOI] [PubMed] [Google Scholar]
- 22.Stern-Ginossar N, Elefant N, Zimmermann A, Wolf DG, Saleh N, Biton M, Horwitz E, Prokocimer Z, Prichard M, Hahn G, et al. Host immune system gene targeting by a viral miRNA. Science. 2007;317:376–381. doi: 10.1126/science.1140956. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.