Abstract
Major histocompatibility complex class II mRNAs encode heterodimeric proteins involved in the presentation of exogenous antigens during an immune response. Their 3′UTRs bind a protein complex in which we identified two factors: EBP1, an ErbB3 receptor-binding protein and DRBP76, a double-stranded RNA binding nuclear protein, also known as nuclear factor 90 (NF90). Both are well-characterized regulatory factors of several mRNA molecules processing. Using either EBP1 or DRBP76/NF90-specific knockdown experiments, we established that the two proteins play a role in regulating the expression of HLA-DRA, HLA-DRB1 and HLA-DQA1 mRNAs levels. Our study represents the first indication of the existence of a functional unit that includes different transcripts involved in the adaptive immune response. We propose that the concept of ‘RNA operon’ may be suitable for our system in which MHCII mRNAs are modulated via interaction of their 3′UTR with same proteins.
INTRODUCTION
Major histocompatibility complex class II molecules (MHCII) are cell-surface glycoproteins that have a central role in the adaptive immune response because they present peptides, mainly derived from extracellular proteins, to the antigen receptor of CD4+ T cells (1). The MHCII molecules are expressed by professional antigen-presenting cells (APCs) that are cells of haematopoietic origin, such as dendritic cells (DCs), B lymphocytes and cells of the monocyte/macrophage lineage. These professional APCs are able to induce both primary and secondary immune responses because of the constitutive expression of MHCII molecules, as well as of potent costimulatory molecules, differently than non-professional APCs. The latter definition includes non-bone marrow-derived cells that do not express a complete range of costimulatory molecules, constitutively expressing MHCII molecules, such as thymic epithelial cells and endothelial cells in various organs, as well as cell types that do not express basal levels of MHCII molecules but can be induced to express MHCII molecules in response to IFNγ and some tumour cells from several neoplastic tissues (2,3)
MHCII molecules are composed of two non-covalently associated polypeptides, α- and β-chains, encoded by different genes located on the short arm of chromosome 6. MHCII is a multigenic system including different isotypes (HLA-DR, HLA-DP and HLA-DQ), whose expression is cell-type regulated at the transcription level by a highly conserved MHCII enhanceosome (4). The functioning of the immune response in infection, transplantation and autoimmunity is strictly dependent on the level of expression of MHCII molecules on the APC surface (5), which regulate the efficacy of antigen presentation. Moreover, MHCII expression level in cancer cells seems to correlate with increased tumourigenicity, metastatic potential and reduced immunogenicity (3,6). One of the elements that influences the density of these molecules is certainly the difference in the regulation of transcription demonstrated for DQB1 (7), DRB1 (8) and DQA1 (9,10) genes. It has been suggested that this variability is caused by an allelic polymorphism in the 5′ regulatory region and contributes to the difference in susceptibility to autoimmune disease in different individuals (11). Other evidence indicates that the post-transcriptional regulation is yet another level of control of MHCII expression (12,13). It is possible that variability of cis-sequences located in the 5′ and 3′UTR of MHCII mRNAs encoded by different alleles by binding trans-acting factors could also influence their expression and play a role in the antigen presentation.
It has been shown that the inhibition of protein synthesis results in a significant reduction of the half-life of all MHCII mRNAs studied (14) by a mechanism in which ongoing translation per se is required for the stabilization of the transcripts (15). A sequence of HLA-DRA mRNA able to bind nuclear and cytoplasmic protein factors has been previously identified in the 3′UTR region (16). Its ability to cross-compete with HLA-DQA1 mRNA for complex formation suggests that different 3′UTR of MHCII mRNAs share consensus sequences or secondary structures able to interact with ribonucleoproteins (RNPs). The 3′UTRs and the 5′UTRs are the transcript target sequences involved in the RNPs binding, inside the ‘RNA operon’ (17–19). This is a functional unit in which multiple physiologically related transcripts can be co-ordinately regulated during splicing, export, stability, localization and translation. These subpopulations of mRNAs bind the same RNP complex in a dynamic manner because the protein components could change during the different steps of the RNA processing and each mRNA can join different RNA operons.
Many studies have confirmed that the expression of proteins involved in immune-mediated inflammation is modulated by a ‘post-transcriptional operon’ (20,21). In this framework, the stabilities (22,23) of distinct groups of mRNAs change in a co-ordinate way in order to efficiently produce functional protein groups according to the cell needs. For example, Human antigen R (HuR) (24,25) and tristetraproline (TTP) (26), in activated T lymphocytes and in cancer cells, respectively, are able to modulate the half-lives of many common mRNA targets including cytokines and chemokines that must be synchronously regulated during the immune response.
In the current study, we present data relative to the identification and characterization of two proteins participating in the complex that interacts with the 3′UTR of MHCII mRNAs. Our data suggest that these factors play a role in the RNA expression, consistent with the ‘RNA operon model’ in which functionally related transcripts such as MHCII mRNAs are regulated by the same RNPs.
MATERIALS AND METHODS
Cell lines
The M14 cell line was established from specimens obtained by primary tumours from melanoma patients (27) and was cultured in RPMI medium 1640 with 10% FCS (GIBCO) and 1% penicillin/streptomycin (SIGMA). Raji is a B lymphoma cell line and was cultured in RPMI with 10% FCS (GIBCO) and 1% penicillin/streptomycin (SIGMA).
Flow cytometry analysis
Determination of cell surface expression of MHCII antigens was performed by cytofluorimetric analysis using the FACS ARIA cell-sorting system and analysed by the DIVA software (BD Biosciences). FITC mouse anti-human HLA-DR, along with the appropriate FITC mouse IgG isotype controls were purchased from BD Biosciences. Cell cycle analysis was performed following propidium iodide staining and flow cytometry analysis.
In Vitro transcription
All DNA fragments used for the riboprobe synthesis were obtained by PCR, using full length cDNAs as template and the primers indicated in Table 1. The transcription reactions were performed to obtain cold and [32P] UTP labelled probes using T7 in vitro transcription system MAXIScript T7 (Ambion).
Table 1.
Sequences of primers used for riboprobes template synthesis, target sequences of siRNA and sequences of primers used for qRT–PCR, respectively
| Probe | Primers | Sequence |
|---|---|---|
| Primers used for PCR of riboprobes templates | ||
| 3-DRA | 3DRAT7 | TAATACGACTCACTATAGGACATGGAGGTGATGGTGTTTC |
| 3DRAR | TTCCACCCAAGATCATCAAA | |
| 3-DQA1 | 3DQA1T7 | TAATACGACTCACTATAGGCCATCCTGGAAGGGAAGTG |
| 3DQA1R | TCAGGAGGTCAGGGAAAGAA | |
| 3-DRB1 | 3DRB1T7 | TAATACGACTCACTATAGGAATGCAGATGACCACATTCAAG |
| 3DRB1R | TGAGAAACATTTAATAATGTAATG | |
| 3-DQB1 | 3DQB1T7 | TAATACGACTCACTATAGGGCACTGACTCCTGAGACTATT |
| 3DQB1R | TGCTTCTCTTGAGCAGTCTGAG | |
| GFP | GFPT7 | TAATACGACTCACTATAGGAAGAGTGCCATGCCCGAAGGT |
| GFPR | GCTGCTGGGATTACACATGGC | |
| siRNA target sequences | ||
| siNF90 | 5′-GCCCACCTTTGCTTTTTAT-3′ | |
| siEBP1 | 5′-TCCCACCAGCATTTCGGTAAA-3′ | |
| siDRA | 5′-CAGGAATCATGGGCTATCAAA-3′ | |
| siDRB1 | 5′-CCTGGACAGATACTTCTATAA-3′ | |
| Primers used for qRT–PCR | ||
| β-Actin | ACT-F | TCATGAAGTGTGACGTTGACA |
| ACT-R | CCTAGAAGCATTTGCGGTGCAC | |
| GAPDH | G-F | AACGGATTTGGTCGTATTGGGC |
| G-R | TCGCTCCTGGAAGATGGTGATG | |
| HLA-DRA | DRA-F | GGACAAAGCCAACCTGGAAA |
| DRA-R | AGGACGTTGGGCTCTCTCAG | |
| HLA-DRA 3′UTR | 3DRA-F | ACATGGAGGTGATGGTGTTT |
| 3DRA-R | AGCCAGCTAGATGTTAGAGTACG | |
| HLA-DRB1 | DRB1-F | CTCAGCATCTTGCTCTTGTGCAG |
| DRB1-R | CAGCATTAAAGTCAGGTGGTTCC | |
| HLA-DQA1 | DQA1-F | GGTGTAAACTTGTACCAGT |
| DQA1-R | GGAGACTTGGAAAACACT | |
| HLA-A-B-C | MHCI_F | CTCAGCATCTTGCTCTTGTGCAG |
| MHCI_R | ATGTAATCCTTGCCGTCGTA | |
| EBP1 | EBP1-F | CTGCACGCCAATAGAAGG |
| EBP1-R | AGTAAACGGCATGGCATCA | |
| DRBP76/NF90 | NF90-F | CTGGTGCTGCTGTGTAAGGA |
| NF90-R | AGGGACAATGGAGGCTCTTT | |
The underlined sequence represents the T7 promoter.
RNA electrophoretic mobility shift assay (REMSA) and supershift
S100 protein extracts were obtained from M14 melanoma cells, lysed with five cell volumes of hypotonic buffer: 10 mM HEPES pH 7.9, 1.5 mM MgCl2, 10 mM KCl, 0.5 mM DTT, 0.2 mM PMSF, 1xPI (protease inhibitors cocktail). The extracts were centrifuged at 1000g and the supernatant was mixed with 0.11 volumes of S100 extraction buffer (0.3 M HEPES pH 7.9; 30 mM MgCl2; 1.4 M KCl; 1xPI). After centrifugation at 100 000g, the samples were dialysed against 50 volumes of dialysis buffer: 20 mM HEPES pH 7.9, 20% glycerol, 100 mM KCl, 0.5 mM DTT, 0.2 mM EDTA. REMSAs were performed as described previously (16). In brief, 30 µg protein was incubated for 30 min at room temperature with 5 × 105 cpm of 32P-UTP-labelled 3-DRA or 3-DQA1 riboprobes. Differently 3-DRB1 and 3-DQB1 riboprobes were refolded by heating to 70°C for 5 min and slow-cooling to room temperature over 1 h before use in binding experiments (28). After incubation, the reaction mixtures were treated with 10 U RNase T1 to digest the RNA sequences unprotected by the complex binding and with heparin at a final concentration of 5 mg/ml, at 22°C. Competition assays were performed by pre-incubating 0.5 and 5 µg of cold probe and 0.5 and 5 µg of poly(U) or poly(G) or poly(C) or poly(A) homopolymers (SIGMA) with the protein extract prior to the incubation with the radio-labelled riboprobes. For antibody supershift assays 0.5, 2 and 6 µg of specific polyclonal rabbit anti-EBP1 (UPSTATE Cell Signalling) and monoclonal mouse anti-DRBP76 (Biosciences Pharmigen) or 6 µg control IgG were pre-incubated with cell extracts prior to the addition of probes. The binding reactions with 3-DRA or 3-DQA1 and 3-DRB1 or 3-DQB1 riboprobes were separated on native 6% or 5% PAGE, respectively. The gels were subsequently dried and radioactive image acquired by Typhoon analysis (Amersham Bioscience).
RNA interference-mediated gene silencing
Transfections of siRNA for gene silencing were performed using HiPerFect Transfection Reagent (QIAGEN). The siRNAs used in this study were all purchased from QIAGEN and were those showing the highest and more specific interference potential against the intended targets, chosen in the list of pre-designed siRNAs. The target sequences of siRNA used are listed in Table 1. Briefly, cells were plated at 3 × 105 cells/well in six-well plates and incubated for 24 h before performing transfection with 50 nM siRNAs concentration. Cells were collected by trypsinization at 0, 24, 48 and 72 h after the transfection for both flow cytometry and cell cycle analysis and for RNA isolation.
Quantitative measurement of specific transcripts by qRT–PCR assay
For the isolation of total RNA, after the cells were lysed in the QIAzol Lysis Reagent (QIAGEN), the RNA was purified using phenol–chloroform extraction. All reverse transcriptase reactions were performed using the QuantiTect RT Kit (QIAGEN). The accumulation of specific transcripts was measured by qRT–PCR, using the DNA Engine Opticon Real-Time PCR Detection System (BIORAD). The qRT–PCR assays were performed using the amount of cDNA obtained retro-transcribing 0.5 µg of total RNA. The QuantiTect SYBR Green PCR Kit (QIAGEN) was used to perform all the reactions in presence of 0.2 μM primers (Table 1), synthesized by PRIMM; each assay was run in triplicate. The relative amount of specific transcripts was calculated by the comparative cycle threshold method given by Livak and Schmittgen (29). To correct for sample to sample variations in qRT–PCR efficiency and errors in quantization, the level of GAPDH transcripts was tested for use in normalization of specific RNA levels. External standards were used to establish standard PCR curves for quantifying copies of those transcripts that required an absolute, comparative quantization. In the graphs, we report the mRNA fold variation as the ratio of the measured value for an experimental sample to the value for the control sample, when the control is equal to 1.
Northwestern blot (NW) and western blot (WB) assays
Extracts were separated on 12.5% SDS–PAGE and electrotransferred to PVDF membrane (Amersham). For NW membrane-bound proteins were sequentially immersed at 22°C in binding buffer 1 (BB1; 50 mM Tris–HCl, pH 7.5, 50 mM NaCl, 1 mM EDTA, 1 mM DTT) containing decreasing concentrations of guanidine–HCl (5 min each in 6 M, 3 M, 1.5 M, 0.75 M, 0.37 M and 0.18 M). Thereafter the membranes were rinsed in BB1 and blocked for 1 h in 2.5% nonfat milk powder. RNA binding was carried out in BB1 containing 150 000 cpm/ml of the corresponding labelled RNA probe (50–100 ng/ml) and 10 µg/ml each of yeast RNA for 2 h at 22°C. After three washes in BB1, bound labelled RNA was detected by Typhoon analysis (Amersham Bioscience) (30). The RNA binding protein used as negative control was hnRNPH1 (31).
In western blot analysis, we used mouse monoclonal anti-DRBP76 (Biosciences Pharmigen) and rabbit polyclonal anti-EBP1 antibodies. After incubation with goat anti-rabbit secondary antibody, all membranes were developed using ECL kit (Amersham Bioscience) and exposed to X-ray film. EBP1 expression plasmid encoding the larger p48 isoform of the protein was kindly provided by Professor S. Curry and recombinant protein was prepared according to the published protocol (28).
Protein biochemical purification
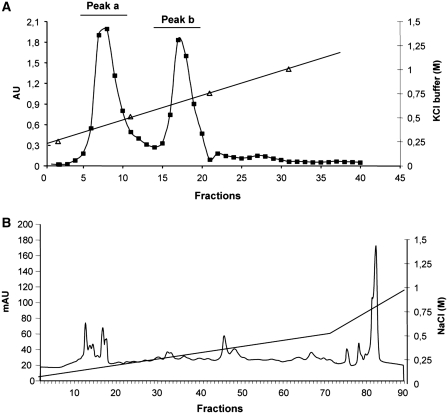
Heparin-sepharose CL-6B (GE-Healthcare) was swelled in distilled water and then equilibrated in buffer A: 10 mM HEPES pH 7.5, 40 mM KCl, 3 mM MgCl2 and 5% glycerol. About 80 mg of Raji S100 extract was incubated in batch with heparin-sepharose resin. Thereafter the bound resin was packed in a small column. Non-adsorbed proteins were washed out with Buffer A and bound proteins were eluted with four 10 ml steps of KCl in Buffer A (0.25 M; 0.5 M; 0.75 M and 1.0 M). For each step, 10 fractions (500 µl) were recovered. The absorbance reading at 280 nm allowed us to see two peaks (a and b) as shown in Figure 3, panel A. The peaks obtained from the columns were pooled separately and dialysed against 50 volumes of Buffer A (20 mM Tris–HCl pH 8.0, 0.1 mM EDTA, 0.1 mM DTT). About 30 µg of these heparin binding fractions were checked for binding activity by REMSA using 3-DRA labelled probe (data not shown).
Figure 3.
Chromatograms of two-step purification of MHCII binding proteins. (A) Elution profile of the heparin-sepharose chromatography. The column was loaded with S100 Raji extract, washed and eluted with KCl discontinuous gradient. (B) Elution profile of the Mono Q chromatography. The column was loaded with the pooled fractions of peak b of the heparin-sepharose column, washed and eluted with a NaCl linear gradient. On the x-axis is indicated the fraction number, on the primary y-axis the absorbance at 280 nm, on the secondary y-axis the concentration of elution buffers, for both panels.
Pooled fractions of peak b were subsequently separated by FPLC Mono Q anion-exchange chromatography. The column was washed with Mono Q buffer A (20 mM Tris–HCl pH 8.0, 0.1 mM EDTA, 0.1 mM DTT) and eluted with a first linear gradient from 0% to 50% of Mono Q buffer B (1 M NaCl in Mono Q buffer A) and a second linear gradient from 50% to 100% of Mono Q buffer B (Figure 3, panel B). Each fraction was analysed by northwestern blot.
Poly(U)-Sepharose 4-B (GE-Healthcare) was swelled in distilled water for 15 min. The resin was packed in a 2 ml column and then equilibrated in Buffer 1 containing 20 mM Tris–HCl pH 7.5, 0.1 mM EDTA pH 8.0, 0.1 mM DTT and 50 mM NaCl. A quantity of 6 mg of Raji S100 extract was loaded into the column and non-adsorbed proteins were washed out with 30 volumes of Buffer 1. Bound proteins were eluted with Buffer 2 (20 mM Tris–HCl pH 7.5, 0.1 mM EDTA pH 8.0, 0.1 mM DTT and 500 mM NaCl recovering 10 fractions that were subsequently separated by FPLC Mono Q anion-exchange chromatography. Each fraction was analysed by northwestern blot using 3-DRA probes and the bands of binding analysed by nano-Electrospray Ionization Tandem Mass Spectrometry (nano-ESI-MS/MS) (data not shown).
Nano-ESI–MS/MS
The bands resulting from the purification procedure and identified by RNA binding activity assay as containing protein of interest were excised and subjected to in situ digestion with trypsin by using the procedure as described (in Protein Chip Technical note). Extracted peptides were separated by nano-HPLC (Dionex Ultimate) and sequenced online by nano-ESI MS/MS experiments with a Q-TOF mass spectrometer (Q-Star Elite; Applied Biosystems). Sequences were searched with the Mascot algorithm in the NCBInr database (human). Only peptides with individual ion scores >41 indicating identity or extensive homology (P < 0.05) were taken into account (Table 2).
Table 2.
Sequences of peptides obtained by nano-ESI-MS/MS of proteins extracted from polyacrylamide gel after in situ trypsin digestion
| Peptides identified | Mass | Amino acid position |
|---|---|---|
| EBP1 identification | ||
| SDQDYILK | 9.804.815 | 93–100 |
| LVKPGNQNTQVTEAWNK | 19.259.959 | 155–171 |
| TIIQNPTDQQK | 12.846.674 | 199–209 |
| AFFSEVER | 9.834.712 | 263–270 |
| FDAMoxPFTLR | 11.125.325 | 271–280 |
| RFDAMoxPFTLR | 12.686.336 | 270–280 |
| HELLQPFNVLYEKEGEFVAQFK | 26.823.693 | 298–319 |
| HELLQPFNVLYEK | 16.288.562 | 298–310 |
| EGEFVAQFK | 10.535.131 | 311–319 |
| FTVLLMPNGPMR | 14.067.050 | 320–331 |
| FTVLLMoxPNGPMoxR | 14.067.050 | 320–331 |
| ITSGPFEPDLYK | 13.656.816 | 332–343 |
| SEMoxEVQDAELK | 12.935.758 | 344–354 |
| ALLQSSASRK | 9.315.087 | 355–364 |
| DRBP76/NF90 identification | ||
| LFPDTPLALDANK | 14.137.504 | 49–66 |
| SSELEQYLQR | 12.516.095 | 427–436 |
| VLAGETLSVNDPPDVLDR | 19.089.793 | 458–470 |
RESULTS
Identification of cytoplasmic proteins binding to the 3′UTR of MHCII mRNAs in a non-professional APC
There is evidence that in Raji, a B lymphoma cell line acting as a professional APC, the 3′UTR of HLA-DRA and HLA-DQA1-specific mRNAs bind to factors exhibiting different subcellular locations (16). To better appreciate any variation in the expression of MHCII genes due to the manipulation of these interactions, we chose a cell model system showing a low constitutive expression of MHCII molecules and representing a non-professional APC, the M14 melanoma cell line derived from primary human melanoma (27).
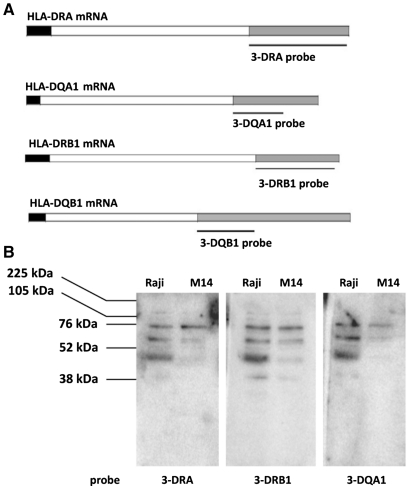
Four different probes, including sequences downstream the stop codon, were designed to analyse the cytoplasmic protein binding with 3′UTR of MHCII mRNAs: 3-DRA corresponding to a 403 bp fragment of HLA-DRA cDNA; 3-DRB1 equivalent to a 308 bp fragment of HLA-DRB1 cDNA; 3-DQA1, corresponding to a 202 bp sequences of HLA-DQA1 cDNA; and 3-DQB1, equivalent to a 295 bp sequences of HLA-DQB1 cDNA (Figure 1, panel A). 3-DRA and 3-DRB1 are the full-length 3′UTRs (except for few bases), 3-DQB1 is the region upstream the first poly(A) signal and 3-DQA1 is the minimum binding site common to all splicing variants (32).
Figure 1.
3′UTR of MHC II mRNAs-binding by northwestern. (A) Schematic representation of HLA-DRA, HLA-DRB1, HLA-DQA1 and HLA-DQB1 mRNAs with 5′UTR, coding region and 3′UTR indicated respectively as black, white and grey bars. The probes used are indicated. (B) Northwestern blot analysis of S100 extracts prepared from M14 and Raji cell lines carried out with 3-DRA, 3-DRB1 and 3-DQA1 riboprobes. The molecular weights are indicated.
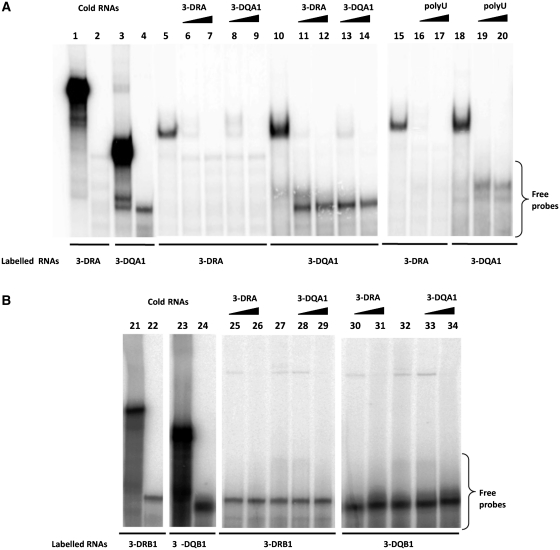
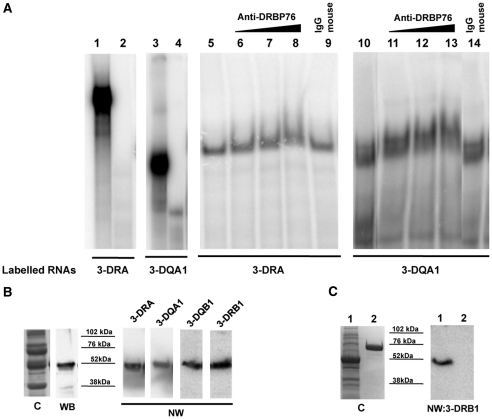
Binding assays were carried out using S100 extracts from both M14 and Raji cell lines to investigate also on the possible tissue specificity of the interactions. First, we performed northwestern blot analysis using 3-DRA and 3-DQA1 riboprobes and observed a similar pattern of binding with proteins from two cell lines immobilized on the membrane (Figure 1, panel B). The negative control was performed by using a GFP labelled riboprobe that showed no interaction band (data not shown). Then we carried out REMSA experiments that clearly confirmed the interaction between proteins present in the S100 cytoplasmic extracts from M14 and either 3-DRA (lane 5) or 3-DQA1 (lane 10) riboprobes (Figure 2, panel A). Specificity of the binding was assessed by competition experiments using unlabelled probes (lanes 6 and 7 for 3-DRA; lanes 13 and 14 for 3-DQA1). In addition, 0.5 and 5 µg of cold 3-DQA1 completely displaced the binding activity to 3-DRA probe (lanes 8 and 9) and the same amount of cold 3-DRA was able to compete totally with the binding to 3-DQA1 (lanes 11 and 12). The binding activity with 3-DRB1 and 3-DQB1was not observed with the protocol used for the previous experiment. However, it became apparent after riboprobes heating to 70°C for 5 min and slow-cooling to room temperature (Figure 2, panel B, lanes 27 and 32). About 0.5 and 5 µg of both cold 3-DRA (lanes 25 and 26) and 3-DQA1 (lanes 28 and 29) completely displaced the binding activity to 3-DRB1 probe. The same amount of cold 3-DRA (lanes 30 and 31) or 3-DQA1 was able to totally compete with the binding to 3-DQB1 (lanes 33 and 34). These results suggest that the same protein/s interact with similar 3′UTR nucleotide sequences.
Figure 2.
3′UTR of MHC II mRNAs binding by REMSA. (A) REMSAs experiments performed using 3-DRA (lane 1) and 3-DQA1 (lane 3) riboprobes; lanes 2 and 4 show the digestion of riboprobes with T1 RNase. Lanes 5 and 15 show bands of interaction of M14 extract with 3-DRA, lanes 10 and 18 with 3-DQA1; competition experiments of 3-DRA binding were performed using cold 3-DRA (lanes 6 and 7), cold 3-DQA1 (lanes 8 and 9) and poly(U) homopolymers (lanes 16 and 17). Competition experiments of 3-DQA1 binding were performed using cold 3-DRA (lanes 11 and 12), cold 3-DQA1 (13 and 14) and poly(U) homopolymers (lanes 19 and 20). (B) REMSAs experiments carried out using 3-DRB1 (lane 21) and 3-DQB1 (lane 22) riboprobes. Lanes 22 and 24 show the digestion of riboprobes with T1 RNase; lanes 27 and 32 show bands of interaction of M14 extract with 3-DRB1 and 3-DQB1.Competition experiments of 3-DRB1 binding were performed using cold 3-DRA (lane 25 and 26) and cold 3-DQA1 (28 and 29). Competition experiments of 3-DQB1 binding were performed using cold 3-DRA (lanes 30 and 31) and cold 3-DQA1 (33 and 34).
Finally, we assessed that poly(A), poly(C) and poly(G) homopolymers were unable to compete with the binding (data not shown), while poly(U) homopolymers were able to successfully inhibit the formation of both 3-DRA and 3-DQA1protein complexes (Figure 2A, lanes 16, 17, 19 and 20), indicating that the interacting proteins belong to the class of U-rich binding proteins.
Two known RNA binding proteins, DRBP76/NF90 and EBP1, are included in the RNP complex interacting with 3′UTR of MHCII mRNAs
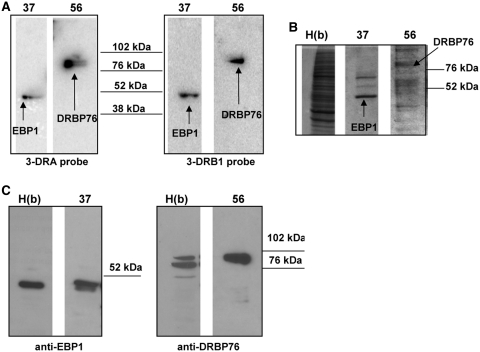
We first purified the proteins included in the complex between S100 Raji extract and 3′UTR of MHCII mRNAs by sequential fractionation using affinity and anionic exchange chromatography. Protein identification was carried out by functional binding assay and mass spectrometry analysis of the proteins separated through SDS–PAGE. In detail, the S100 fraction from Raji cells was processed by affinity with a column of heparin-sepharose. Retained proteins were eluted by a KCl discontinuous gradient and fractions collected were analysed by REMSA. All the samples tested, except the flow-through, showed RNA binding activity (data not shown). Two major protein peaks (indicated as a and b in Figure 3, panel A) were evident in the profile. After dialysis, pooled fractions from either a or b peaks were further purified using FPLC Mono-Q sepharose anion-exchange chromatography. Proteins were eluted using a linear gradient of NaCl (Figure 3, panel B) and their binding activity was tested by northwestern blot. This assay shows that two fractions from peak b (fractions 37 and 56) exhibited significant binding activity (Figure 4, panel A) with both 3-DRA and 3-DRB1 riboprobes. There were no fractions from peak a that showed any binding activity with either riboprobes (data not shown). Both fractions 37 and 56 are composed of several proteins, but 3-DRA and 3-DRB1 probes identified the same proteins in the pool (see coomassie gel in panel B of Figure 4). In fraction 37, one binding protein with an apparent molecular weight of 50 KDa is detected by both probes, and in fraction 56 one protein of 76 KDa mass. These two proteins were trypsin digested, extracted from the polyacrylamide gel and analysed by nano-ESI-MS/MS for identification. As shown in Table 2, the 50 KDa protein was identified as EBP1 through the overall sequencing of 14 peptides, while the 76 KDa protein turned out to be DRBP76/NF90 (three peptides, Table 2). No other peptides were identified from these bands, except for those deriving from the trypsin used for digestion. After probe stripping, northwestern membranes were used for western blot with anti-EBP1- and anti-DRBP76-specific antibodies and the identity of both proteins was confirmed (Figure 4, panel C).
Figure 4.
Identification of RNA binding proteins. (A) Northwestern blot analysis performed with 3-DRA probe and 3-DRB1 of Raji cell extracts after Mono Q chromatography. The lanes indicated with numbers 37 and 56 represent the fractions showing the binding; the identified proteins are indicated. The molecular weight markers are shown in the middle. (B) SDS–PAGE analysis of heparin-sepharose pool b, indicated as H(b) and of fractions 37 and 56. The gel was stained with coomassie blue. (C) western blot analysis with anti-EBP1 and anti-DRBP76 antibodies, after stripping the membranes analysed by northwestern blot.
Likewise, the 50 KDa protein was also obtained after sequential purification using a first step of poly(U)-sepharose affinity chromatography followed by Mono Q anion-exchange chromatography. The protein showed a clear RNA binding activity using northwestern assay and was identified as EBP1 by nano-ESI-MS/MS analysis (data not shown).
To further confirm that EBP1 and DRBP76/NF90 are components of RNP complex binding to MHCII mRNAs, we carried out a supershift assay on non-denaturing gel using antibody against EBP1 or DRBP76. Both 3-DRA (lanes 6–8, Figure 5, panel A) and 3-DQA1 (lanes 11–13) riboprobes showed a slower electrophoretic mobility band when anti-DRBP76 antibody was added to the reaction, compared to the effect of IgG control antibody (lanes 9 and 14). However, retardation of complex mobility was not detected using anti-EBP1 antibody, probably due to RNA masking of the EBP1 interaction with specific antibody. To sort out this problem and to further validate the involvement of EBP1 protein in the RNP complex, we produced recombinant EBP1 protein (28) whose identity was confirmed by western blot with anti-EBP1 antibody. The purified EBP1 protein (data not shown) or total Escherichia coli extract expressing the recombinant protein was used in a northwestern binding assay that further demonstrated the interaction between the EBP1 and the four riboprobes. The NW performed with 3-DRB1 demonstrate absence of binding with a non-related RNA binding protein (31) (Figure 5, panel C).
Figure 5.
Analysis of RNPs. (A) Supershift of complex with anti-DRBP76 in REMSA performed with 3-DRA (lane 1) and 3-DQA1 probes (lane 3). Lanes 2 and 4 show the digestion of riboprobes with T1 RNase. Lane 5 shows the interaction of M14 extract with 3-DRA probe while lanes 6–8 clearly show the gel mobility retardation of the complex, in binding reaction containing 0.5, 2 and 6 µg of anti-DRBP76 antibody. No supershift was found in the presence of 6 µg of control IgG (lane 9). In the same way, the binding of 3-DQA1 probe was performed in the absence of antibody (lane 10) or in the presence of 0.5, 2 and 6 µg of anti-DRBP76 antibody (lanes 11–13). No supershift was found in the presence of 6 µg of control IgG (lane 14). (B) Analysis of E. coli protein extract expressing recombinant EBP1. C represents the coomassie of SDS–PAGE, WB is the western blot performed with anti-EBP1, NW are the northwestern blot carried out with indicated riboprobes. (C) Analysis of E. coli protein extract expressing recombinant EBP1 (Lane 1). C represents the coomassie of SDS–PAGE, NW is the northwestern blot carried out with 3-DRB1 riboprobe. In lane 2 is loaded hnRNPH1 protein.
Knockdown of EBP1 and DRBP76/NF90 indicates that they have a role in the accumulation of MHCII mRNAs
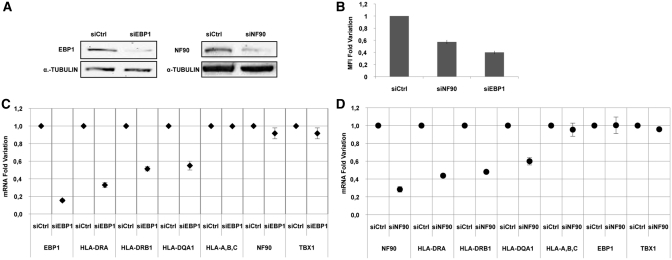
We used siRNAs specifically interfering with the expression of EBP1 and DRBP76/NF90 (Table 1) in M14 cells to establish whether or not the silencing of these two proteins affects the amount of MHCII molecules. The knockdown of proteins was checked after 48 h by western blot with specific antibodies (Figure 6, panel A). We measured the cell surface HLA-DR expression by flow cytometry after 72 h of transfection and observed a reduction of 60% mean fluorescence intensity (MFI) after EBP1 silencing and of 45% MFI following DRBP76/NF90 silencing (Figure 6, panel B).
Figure 6.
Modulation of MHCII expression following knockdown of EBP1 and DRBP76/NF90 proteins in M14 cells. (A) Western blot analysis of cell extracts after silencing of two proteins using siEBP1 and siNF90, performed with anti-EBP1, anti-DRBP76 and anti-α tubulin antibodies. (B) Flow cytometry analysis of cells transfected with siEBP1, siNF90 and siCtrl and stained with HLA-DR specific antibodies. Results are plotted as fold change of MFI (mean fluorescence intensity) value. (C) qRT–PCR analysis of total mRNAs after silencing with either siCtrl or siEBP1. The graph illustrates the mRNA fold variation of the transcripts indicated on the x-axis. (D) qRT–PCR analysis of total mRNAs following silencing with either siCtrl or siNF90. The graph illustrates the mRNA fold variation of the transcripts indicated on the x-axis. The standard deviations are shown in all graphs.
Next, we looked at MHCII-specific mRNA accumulation by qRT–PCR to establish at which level EBP1 and DRBP76/NF90 silencing interferes with the cell surface expression of these molecules. We measured the copy number of HLA-DRA, HLA-DRB1, HLA-DQA1 mRNAs at 24, 48 and 72 h, respectively, after transfection of cells with specific siEBP1 and with control siRNA (siCtrl); the results at 48 h are shown in Figure 6, panel C, as fold of mRNA variation. Following the EBP1 knockdown, we observed a fold change of 0.67 ± 0.03, 0.49 ± 0.02 and 0.45 ± 0.04 for DRA, DRB1 and total DQA1, respectively. As control for the specificity of silencing, we examined the expression of a group of genes related to our system, the HLA-A-B-C genes (33) and one totally unrelated gene, Tbx1 (34). In both cases, we observed no change in the accumulation of their transcripts. This result clearly indicates that the EBP1 silencing induces a decrease in the accumulation of total MHCII mRNAs. We then looked at MHCII-specific mRNA accumulation following siNF90 transfection. The oligo used (35) targets a sequence at exon 18 of the gene and selectively reduced only two isoforms of RNA coding for NF90 proteins without affecting the other isoforms (including those coding for NF110). We measured the copy number of MHCII mRNAs and control messengers; the results obtained after 48 h of transfection are shown in Figure 6, panel D, as fold of mRNA decrease. We obtained a fold change of 0.57 ± 0.008, 0.52 ± 0.01 and 0.4 ± 0.04 for DRA, DRB1 and DQA1, respectively, after NF90 silencing. Similarly to EBP1, this result indicates that the NF90 reduction induces a decrease in the accumulation of all MHCII mRNAs. To eliminate the possibility that the effect observed on MHCII expression consequent to the knockdown of the two proteins was due to a more general effect on the cell cycle progression, we analysed M14 cells after transfection with either siEbp1 and siNF90 by PI labelling and flow cytometry. We did not observe any changes in the cell cycle profile (data not shown).
Cytoplasmic co-regulation of α- and β-mRNAs
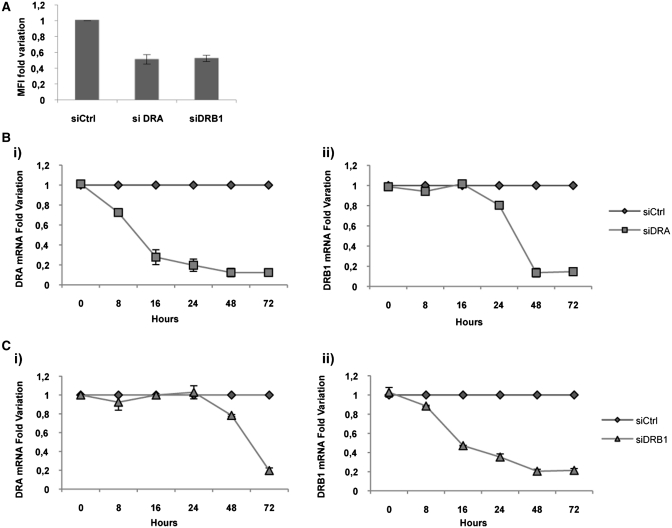
Having demonstrated that EBP1 and NF90 interact with similar binding sites on different MHCII messengers, our aim was to investigate the possibility of a co-regulation of HLA-DRA and HLA-DRB1-specific mRNAs in the cytoplasm. We used siRNAs targeting HLA-DRA and HLA-DRB1 mRNAs (siDRA and siDRB1, respectively, Table 1) to individually reduce each messenger level in M14. In cells transfected with siDRA we observed, after 72 h, a decrease of 50% of HLA-DR surface expression compared to the untreated cells, as measured by MFI (Figure 7, panel A). When we carried out a time course of DRA mRNA variation, by qRT–PCR, we observed a progressive decrease with a fold change of 0.87 ± 0.017 at 72 h after the transfection (Figure 7, panel Bi), compared to the untreated cells. Surprisingly, after siDRA transfection, we also observed a significant delayed decrease of DRB1 mRNA, already apparent at 24 h with a fold variation of 0.2 ± 0.005 and culminating at 72 h with a fold change of 0.86 ± 0.01 (Figure 7, panel Bii).
Figure 7.
Downregulation of MHCII expression following knockdown of DRA or DRB1 mRNAs in M14 cells. (A) Flow cytometry analysis of the cell line transfected with and siCtrl, siDRA and siDRB1 and stained with HLA-DR-specific antibodies. Results are plotted as fold change of MFI (mean fluorescence intensity) value. (B) qRT–PCR analysis of total mRNAs following silencing with siDRA and siCtrl, analysed at different times after transfection. The B(i) panel shows HLA-DRA mRNA fold variation and the B(ii) shows HLA-DRB1 mRNA fold variation. (C) qRT–PCR analysis of total mRNAs following silencing with siDRB1 and siCtrl, analysed at different times after transfection. The C(i) panel shows HLA-DRA mRNA fold variation and C(ii) HLA-DRB1 mRNA fold variation. The standard deviations are shown in all graphs.
Likewise, following siDRB1 transfection, we observed a decrease of 50% of HLA-DR heterodimer expression (Figure 7, panel A) corresponding to a fold change of HLA-DRB1-specific mRNA of 0.79 ± 0.01, after 72 h (Figure 7, panel Cii). When we looked at HLA-DRA mRNA accumulation, we observed, also in this experiment, an important decrease of transcript already apparent at 48 h with a fold change of 0.22 ± 0.01 and reaching 0.81 ± 0.02 at 72 h compared to the expression in untreated cells (Figure 7, panel Ci). Our data confirm that the specific silencing of the mRNA encoding one chain of the heterodimer induces the diminution of the other mRNA, with a delay of 24 h. This result indicates that the amount of one mRNA is related to the quantity of the other messenger and that they could be co-regulated in their cytoplasmic processing.
DISCUSSION
The life cycle of an mRNA is thoroughly regulated by its dynamic association with mRNA binding proteins (RBPs). In the nucleus, transcripts are capped, spliced, cleaved and polyadenylated and are subject to a quality control so that only properly processed molecules are exported to the cytoplasm, where mRNAs are exposed to multiple fates including subcellular localization, translation and degradation (36). These steps were traditionally thought to occur independently, but several studies have revealed that many functionally related transcripts undergo a coupling between transcription and RNA processing (37).
In this article, we identified two proteins, EBP1 and DRP76/NF90, which bind the 3′UTR of MHCII mRNAs modulating their quantities and consequently cell surface expression of HLA class II molecules. We demonstrate that some MHCII mRNAs are subjected to a co-regulation and probably are connected inside an ‘RNA operon’, including several protein factors that take part in all transcriptional and post-transcriptional events of mRNA life.
Previous data from our lab showed a modulation of the MHC II gene expression at the stability level (14,15), while the nucleo-cytoplasm export of transcripts was correlated with the binding of 3′UTRs of HLA-DRA and HLA-DQA1 mRNAs to Raji cytoplasmic factors (16). In this article, we show that the same sequences also interact with proteins present in the cytoplasmic extracts from M14, an MHCII-positive melanoma cell line, indicating that the binding activity is not tissue specific. The nature of the binding activity with 3-DRB1 and 3-DQB1 probes is somewhat different and the interaction of these riboprobes to factors from M14 cytoplasm became apparent in REMSA experiments when RNA sequences are folded in the correct structure. In addition our data show that 3-DRA, 3-DQA, 3-DRB1 and 3-DQB1 riboprobes cross-compete for the same protein interaction, suggesting the presence of two or more similar binding sites. To unravel the identity of the proteins, we carried out two subsequent steps of biochemical purification using S100 Raji cytoplasmic extracts, since this is a B cell line derived from a professional APC. Following northwestern blot, that allows us to select the bands interacting with probes, we performed a mass-spectrometry analysis that identified two proteins corresponding to 50 and 90 KDa as EBP1 and DRBP76/NF90, respectively.
EBP1 is the human homologue of the mouse cell cycle-regulated protein p38-2G4 (38), initially identified as an ErbB3 binding protein. Many papers have suggested that the shorter form of EBP1, p42 acts as a potent tumour suppressor in various human cancers, whereas the longer form p48 might function as an oncogene, promoting cell survival and proliferation (39,40). In order to study the function of its interaction with 3′UTR of MHCII mRNAs, we decided to specifically knock down EBP1. Our results showed a significant decrease of HLA-DRA, HLA-DRB1 and HLA-DQA1 messengers while other non-correlated transcripts were unaffected. This result suggests that EBP1 may play a function at the transcriptional and/or post-transcriptional level, considering that literature data seem to indicate a double role for EBP1. This protein, indeed, is able to recruit a transcriptional repressor complex including the histone deacetilase 2 (HDAC2), the retinoblastoma protein (Rb) and Sin3A corepressor (41–43). Since other evidence has indicated that, in some tumour cells, the MHCII transcriptional repression occurs by the inhibition of the CIITA transactivator function, through the recruitment of HDAC1/2 and Sin3A in presence or not of Rb protein (44), we speculate that EBP1 could bind the repressor complex (Rb/HDAC/Sin3A), reducing its negative transcriptional effect on MHCII genes. This mechanism could regulate the conversion of cancer cells to antigen presenting cells and consequently could induce tumour antigen presentation (45).
On the other hand, the down-regulation of MHCII mRNAs, following EBP1 depletion, can also be explained by a mechanism modulating the messenger stability, as already demonstrated for BCL2 mRNA (46). Zhou et al. (47) have instead confirmed the capability of EBP1 to promote the androgen receptor (AR), mRNA decay through the interaction with a UC-rich motif within the 3′UTR. The crystal structure (28) and the other literature data (48) indicate that EBP1 may bind structured RNAs as IREs (49) and ribosomal RNA, thereby enhancing translation initiation, as well as ssRNA, dsRNA and dsDNA (28). Finally, in the cytoplasm, EBP1 has been found in association with 80S ribosomes and polysomes, consistent with a role in the control of the translation process (28,50).
The other protein interacting with MHCII mRNAs identified in our study is DRBP76 (51), a member of the dsRNA binding proteins (DRBP) family capable of binding both dsRNAs and highly structured single-stranded RNA molecules (52) through its C-terminus region. Differential splicing gives rise to two different molecular weight protein isoforms and the proteins homologous to DRBP76 with 90 KDa molecular weight are named NF90 and NFAR1(53).
Literature data indicate that NF90 recognizes the antigen receptor response element (ARRE)/nuclear factor of activated T cell (NFAT) motifs on DNA and mediates the activation of IL-2 transcription in response to T cell receptor stimulation (54). In THP1 and HeLa cells, NF90 binds to the DNase I Hypersensitive site II (DHS-II) in the first intron of the HLA-DRA gene (55); nevertheless the interaction and the consequent modulation of transcription seems to be strictly correlated to the activation status of the cells. Through its association with the 3′UTRs of different mRNAs, DRBP76/NF90 can influence the nucleus–cytoplasm export (56) or the stability of many messengers (54,57,58). Finally, the interaction of DRBP76/NF90 leads to a subsequent inhibition of translation rates (35) that may also constitute an innate immune translational surveillance mechanism in host defence against virus infection (56,59).
In this article, we show that the specific silencing of DRBP76/NF90 in the M14 cell line induces a decrease in MHCII mRNA accumulation. Similarly to EBP1, DRBP76/NF90 could down-regulate the MHCII transcription or affect the stability of the MHCII messengers. We can speculate a co-transcriptional recruitment of the mRNAs by DRBP76/NF90 that can interact with the first intron of unspliced mRNA or with its 3′UTR determining, either a transcriptional regulation or, a post-transcriptional modulation, in terms of export or stability messenger. The balance between the two mechanisms may be a consequence of the cell's need to express MHCII and to present antigen during the immune response, both in professional or non-professional APC.
Our data indicate that all MHCII mRNAs encoding α- and β-chains of HLA-DR and HLA-DQ isotypes bind both EBP1 and DRBP76/NF90 proteins (probably by a dsRNA secondary structure, such as a stem–loop) common to different transcripts, carrying an U-rich sequence. Moreover, since the specific knockdown of DRA or DRB1 mRNAs influences the accumulation of the other messenger, we propose that both messengers could be associated inside a RNP complex with each other and co-regulated during the cytoplasmic processing. It appears that EBP1 knockdown does not affect DRBP76/NF90 protein expression, nor does DRBP76/NF90 silence EBP1; the depletion of each protein is not able to completely annul the MHCII mRNA accumulation, suggesting a cooperative role. One mechanism could be the interaction of RNA binding proteins to different but similar binding sites, the depletion of one protein being partially compensated by the other.
We speculate that both proteins are joined to the same RNP complex, as occurs for the complex binding BCL2 mRNA (46) probably in association with other factors, to regulate MHCII expression in relationship to different biological pathways and in this framework it is not surprising that EBP1 and DRBP76/NF90 seem to achieve the same effect on MHCII mRNAs. Both proteins could function in cooperation to couple transcriptional and post-transcriptional events and to guarantee a co-regulated expression of all mRNAs inside the functional unit of the ‘MHC class II operon’, during the adaptive immune response.
FUNDING
Funding for open access charge: The Italian Ministry of Research (FIRB project n. RBLA033WJX).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank Prof. S. Curry for providing the plasmid-encoding EBP1 recombinant protein, Dr L. Mandrich for helping in its purification, Prof. Russo for providing RNA binding protein negative control. The IGB FACS facility is also acknowledged.
REFERENCES
- 1.Cresswell P. Assembly, transport, and function of MHC class II molecules. Annu. Rev. Immunol. 1994;12:259–293. doi: 10.1146/annurev.iy.12.040194.001355. [DOI] [PubMed] [Google Scholar]
- 2.Rodriguez T, Mendez R, Del Campo A, Aptsiauri N, Martin J, Orozco G, Pawelec G, Schadendorf D, Ruiz-Cabello F, Garrido F. Patterns of constitutive and IFN-gamma inducible expression of HLA class II molecules in human melanoma cell lines. Immunogenetics. 2007;59:123–133. doi: 10.1007/s00251-006-0171-9. [DOI] [PubMed] [Google Scholar]
- 3.Guardiola J, Maffei A. Control of MHC class II gene expression in autoimmune, infectious, and neoplastic diseases. Crit. Rev. Immunol. 1993;13:247–268. [PubMed] [Google Scholar]
- 4.Reith W, LeibundGut-Landmann S, Waldburger JM. Regulation of MHC class II gene expression by the class II transactivator. Nat. Rev. Immunol. 2005;5:793–806. doi: 10.1038/nri1708. [DOI] [PubMed] [Google Scholar]
- 5.Tite JP, Janeway CA., Jr Antigen-dependent selection of B lymphoma cells varying in Ia density by cloned antigen-specific L3T4a+ T cells: a possible in vitro model for B cell adaptive differentiation. J. Mol. Cell Immunol. 1984;1:253–265. [PubMed] [Google Scholar]
- 6.Campoli M, Ferrone S. HLA antigen changes in malignant cells: epigenetic mechanisms and biologic significance. Oncogene. 2008;27:5869–5885. doi: 10.1038/onc.2008.273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Beaty JS, West KA, Nepom GT. Functional effects of a natural polymorphism in the transcriptional regulatory sequence of HLA-DQB1. Mol. Cell. Biol. 1995;15:4771–4782. doi: 10.1128/mcb.15.9.4771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Perfetto C, Zacheis M, McDaid D, Meador JW, 3rd, Schwartz BD. Polymorphism in the promoter region of HLA-DRB genes. Hum. Immunol. 1993;36:27–33. doi: 10.1016/0198-8859(93)90005-l. [DOI] [PubMed] [Google Scholar]
- 9.Maffei A, Harris PE, Reed EF, Del Pozzo G, Ciullo M, Suciu-Foca N, Guardiola J. Differential expression of insulin-dependent diabetes mellitus-associated HLA-DQA1 alleles in vivo. Eur. J. Immunol. 1997;27:1549–1556. doi: 10.1002/eji.1830270634. [DOI] [PubMed] [Google Scholar]
- 10.Donner H, Seidl C, Rau H, Herwig J, Seifried E, Usadel KH, Badenhoop K. Unbalanced amounts of HLA-DQA1 allele mRNA: DQA1*03 shows high and DQA1*0501 low amounts of mRNA in heterozygous individuals. Eur. J. Immunogenet. 2002;29:321–330. doi: 10.1046/j.1365-2370.2002.00321.x. [DOI] [PubMed] [Google Scholar]
- 11.Guardiola J, Maffei A, Lauster R, Mitchison NA, Accolla RS, Sartoris S. Functional significance of polymorphism among MHC class II gene promoters. Tissue Antigens. 1996;48:615–625. doi: 10.1111/j.1399-0039.1996.tb02684.x. [DOI] [PubMed] [Google Scholar]
- 12.Meissner M, Whiteside TL, van Kuik-Romein P, Valesky EM, van den Elsen PJ, Kaufmann R, Seliger B. Loss of interferon-gamma inducibility of the MHC class II antigen processing pathway in head and neck cancer: evidence for post-transcriptional as well as epigenetic regulation. Br. J. Dermatol. 2008;158:930–940. doi: 10.1111/j.1365-2133.2008.08465.x. [DOI] [PubMed] [Google Scholar]
- 13.Radosevich M, Song Z, Gorga JC, Ksander B, Ono SJ. Epigenetic silencing of the CIITA gene and posttranscriptional regulation of class II MHC genes in ocular melanoma cells. Invest. Ophthalmol. Vis. Sci. 2004;45:3185–3195. doi: 10.1167/iovs.04-0111. [DOI] [PubMed] [Google Scholar]
- 14.Maffei A, Perfetto C, Ombra N, Del Pozzo G, Guardiola J. Transcriptional and post-transcriptional regulation of human MHC class II genes require the synthesis of short-lived proteins. J. Immunol. 1989;142:3657–3661. [PubMed] [Google Scholar]
- 15.Del Pozzo G, Guardiola J. The regulation mechanism of HLA class II gene expression at the level of mRNA stability. Immunogenetics. 1996;44:453–458. doi: 10.1007/BF02602807. [DOI] [PubMed] [Google Scholar]
- 16.Del Pozzo G, Ciullo M, Autiero M, Guardiola J. Control of nucleo-cytoplasmic HLA-DRA mRNA partitioning by interaction of a retention signal with compartmentalized proteins. J. Mol. Biol. 1994;240:193–204. doi: 10.1006/jmbi.1994.1435. [DOI] [PubMed] [Google Scholar]
- 17.Keene JD. RNA regulons: coordination of post-transcriptional events. Nat. Rev. Genet. 2007;8:533–543. doi: 10.1038/nrg2111. [DOI] [PubMed] [Google Scholar]
- 18.Keene JD. Minireview: global regulation and dynamics of ribonucleic Acid. Endocrinology. 2010;151:1391–1397. doi: 10.1210/en.2009-1250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Keene JD, Lager PJ. Post-transcriptional operons and regulons co-ordinating gene expression. Chromosome Res. 2005;13:327–337. doi: 10.1007/s10577-005-0848-1. [DOI] [PubMed] [Google Scholar]
- 20.Mazumder B, Li X, Barik S. Translation control: a multifaceted regulator of inflammatory response. J. Immunol. 2010;184:3311–3319. doi: 10.4049/jimmunol.0903778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Anderson P. Post-transcriptional regulons coordinate the initiation and resolution of inflammation. Nat. Rev. Immunol. 2010;10:24–35. doi: 10.1038/nri2685. [DOI] [PubMed] [Google Scholar]
- 22.Hao S, Baltimore D. The stability of mRNA influences the temporal order of the induction of genes encoding inflammatory molecules. Nat. Immunol. 2009;10:281–288. doi: 10.1038/ni.1699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Elkon R, Zlotorynski E, Zeller KI, Agami R. Major role for mRNA stability in shaping the kinetics of gene induction. BMC Genomics. 2010;11:259. doi: 10.1186/1471-2164-11-259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Papadaki O, Milatos S, Grammenoudi S, Mukherjee N, Keene JD, Kontoyiannis DL. Control of thymic T cell maturation, deletion and egress by the RNA-binding protein HuR. J. Immunol. 2009;182:6779–6788. doi: 10.4049/jimmunol.0900377. [DOI] [PubMed] [Google Scholar]
- 25.Mukherjee N, Lager PJ, Friedersdorf MB, Thompson MA, Keene JD. Coordinated posttranscriptional mRNA population dynamics during T-cell activation. Mol. Syst. Biol. 2009;5:288. doi: 10.1038/msb.2009.44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Al-Souhibani N, Al-Ahmadi W, Hesketh JE, Blackshear PJ, Khabar KS. The RNA-binding zinc-finger protein tristetraprolin regulates AU-rich mRNAs involved in breast cancer-related processes. Oncogene. 2010;29:4205–4215. doi: 10.1038/onc.2010.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Brady MS, Lee F, Petrie H, Eckels DD, Lee JS. CD4(+) T cells kill HLA-class-II-antigen-positive melanoma cells presenting peptide in vitro. Cancer Immunol. Immunother. 2000;48:621–626. doi: 10.1007/s002620050010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Monie TP, Perrin AJ, Birtley JR, Sweeney TR, Karakasiliotis I, Chaudhry Y, Roberts LO, Matthews S, Goodfellow IG, Curry S. Structural insights into the transcriptional and translational roles of Ebp1. EMBO J. 2007;26:3936–3944. doi: 10.1038/sj.emboj.7601817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 30.Kreft SG, Nassal M. hRUL138, a novel human RNA-binding RING-H2 ubiquitin-protein ligase. J. Cell Sci. 2003;116:605–616. doi: 10.1242/jcs.00261. [DOI] [PubMed] [Google Scholar]
- 31.Russo A, Siciliano G, Catillo M, Giangrande C, Amoresano A, Pucci P, Pietropaolo C, Russo G. hnRNP H1 and intronic G runs in the splicing control of the human rpL3 gene. Biochim. Biophys. Acta. 2010;1799:419–428. doi: 10.1016/j.bbagrm.2010.01.008. [DOI] [PubMed] [Google Scholar]
- 32.Hoarau JJ, Cesari M, Caillens H, Cadet F, Pabion M. HLA DQA1 genes generate multiple transcripts by alternative splicing and polyadenylation of the 3′ untranslated region. Tissue Antigens. 2004;63:58–71. doi: 10.1111/j.1399-0039.2004.00140.x. [DOI] [PubMed] [Google Scholar]
- 33.Marsh SG, Albert ED, Bodmer WF, Bontrop RE, Dupont B, Erlich HA, Geraghty DE, Hansen JA, Hurley CK, Mach B, et al. Nomenclature for factors of the HLA system, 2004. Tissue Antigens. 2005;65:301–369. doi: 10.1111/j.1399-0039.2005.00379.x. [DOI] [PubMed] [Google Scholar]
- 34.Fulcoli FG, Huynh T, Scambler PJ, Baldini A. Tbx1 regulates the BMP-Smad1 pathway in a transcription independent manner. PLoS ONE. 2009;4:e6049. doi: 10.1371/journal.pone.0006049. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 35.Kuwano Y, Pullmann R, Jr, Marasa BS, Abdelmohsen K, Lee EK, Yang X, Martindale JL, Zhan M, Gorospe M. NF90 selectively represses the translation of target mRNAs bearing an AU-rich signature motif. Nucleic Acids Res. 2010;38:225–238. doi: 10.1093/nar/gkp861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Moore MJ, Proudfoot NJ. Pre-mRNA processing reaches back to transcription and ahead to translation. Cell. 2009;136:688–700. doi: 10.1016/j.cell.2009.02.001. [DOI] [PubMed] [Google Scholar]
- 37.Komili S, Silver PA. Coupling and coordination in gene expression processes: a systems biology view. Nat. Rev. Genet. 2008;9:38–48. doi: 10.1038/nrg2223. [DOI] [PubMed] [Google Scholar]
- 38.Radomski N, Jost E. Molecular cloning of a murine cDNA encoding a novel protein, p38-2G4, which varies with the cell cycle. Exp. Cell Res. 1995;220:434–445. doi: 10.1006/excr.1995.1335. [DOI] [PubMed] [Google Scholar]
- 39.Liu Z, Ahn JY, Liu X, Ye K. Ebp1 isoforms distinctively regulate cell survival and differentiation. Proc. Natl Acad. Sci. USA. 2006;103:10917–10922. doi: 10.1073/pnas.0602923103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu Z, Oh SM, Okada M, Liu X, Cheng D, Peng J, Brat DJ, Sun SY, Zhou W, Gu W, et al. Human BRE1 is an E3 ubiquitin ligase for Ebp1 tumor suppressor. Mol. Biol. Cell. 2009;20:757–768. doi: 10.1091/mbc.E08-09-0983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Xia X, Cheng A, Lessor T, Zhang Y, Hamburger AW. Ebp1, an ErbB-3 binding protein, interacts with Rb and affects Rb transcriptional regulation. J. Cell Physiol. 2001;187:209–217. doi: 10.1002/jcp.1075. [DOI] [PubMed] [Google Scholar]
- 42.Zhang Y, Wang XW, Jelovac D, Nakanishi T, Yu MH, Akinmade D, Goloubeva O, Ross DD, Brodie A, Hamburger AW. The ErbB3-binding protein Ebp1 suppresses androgen receptor-mediated gene transcription and tumorigenesis of prostate cancer cells. Proc. Natl Acad. Sci. USA. 2005;102:9890–9895. doi: 10.1073/pnas.0503829102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhang Y, Woodford N, Xia X, Hamburger AW. Repression of E2F1-mediated transcription by the ErbB3 binding protein Ebp1 involves histone deacetylases. Nucleic Acids Res. 2003;31:2168–2177. doi: 10.1093/nar/gkg318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Zika E, Greer SF, Zhu XS, Ting JP. Histone deacetylase 1/mSin3A disrupts gamma interferon-induced CIITA function and major histocompatibility complex class II enhanceosome formation. Mol. Cell Biol. 2003;23:3091–3102. doi: 10.1128/MCB.23.9.3091-3102.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Khan AN, Magner WJ, Tomasi TB. An epigenetic vaccine model active in the prevention and treatment of melanoma. J. Transl. Med. 2007;5:64. doi: 10.1186/1479-5876-5-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bose SK, Sengupta TK, Bandyopadhyay S, Spicer EK. Identification of Ebp1 as a component of cytoplasmic bcl-2 mRNP (messenger ribonucleoprotein particle) complexes. Biochem. J. 2006;396:99–107. doi: 10.1042/BJ20051548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhou H, Mazan-Mamczarz K, Martindale JL, Barker A, Liu Z, Gorospe M, Leedman PJ, Gartenhaus RB, Hamburger AW, Zhang Y. Post-transcriptional regulation of androgen receptor mRNA by an ErbB3 binding protein 1 in prostate cancer. Nucleic Acids Res. 2010;38:3619–3631. doi: 10.1093/nar/gkq084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Squatrito M, Mancino M, Donzelli M, Areces LB, Draetta GF. EBP1 is a nucleolar growth-regulating protein that is part of pre-ribosomal ribonucleoprotein complexes. Oncogene. 2004;23:4454–4465. doi: 10.1038/sj.onc.1207579. [DOI] [PubMed] [Google Scholar]
- 49.Pilipenko EV, Pestova TV, Kolupaeva VG, Khitrina EV, Poperechnaya AN, Agol VI, Hellen CU. A cell cycle-dependent protein serves as a template-specific translation initiation factor. Genes Dev. 2000;14:2028–2045. [PMC free article] [PubMed] [Google Scholar]
- 50.Squatrito M, Mancino M, Sala L, Draetta GF. Ebp1 is a dsRNA-binding protein associated with ribosomes that modulates eIF2alpha phosphorylation. Biochem. Biophys. Res. Commun. 2006;344:859–868. doi: 10.1016/j.bbrc.2006.03.205. [DOI] [PubMed] [Google Scholar]
- 51.Patel RC, Vestal DJ, Xu Z, Bandyopadhyay S, Guo W, Erme SM, Williams BR, Sen GC. DRBP76, a double-stranded RNA-binding nuclear protein, is phosphorylated by the interferon-induced protein kinase, PKR. J. Biol. Chem. 1999;274:20432–20437. doi: 10.1074/jbc.274.29.20432. [DOI] [PubMed] [Google Scholar]
- 52.Barber GN. The NFAR's (nuclear factors associated with dsRNA): evolutionarily conserved members of the dsRNA binding protein family. RNA Biol. 2009;6:35–39. doi: 10.4161/rna.6.1.7565. [DOI] [PubMed] [Google Scholar]
- 53.Saunders LR, Perkins DJ, Balachandran S, Michaels R, Ford R, Mayeda A, Barber GN. Characterization of two evolutionarily conserved, alternatively spliced nuclear phosphoproteins, NFAR-1 and -2, that function in mRNA processing and interact with the double-stranded RNA-dependent protein kinase, PKR. J. Biol. Chem. 2001;276:32300–32312. doi: 10.1074/jbc.M104207200. [DOI] [PubMed] [Google Scholar]
- 54.Shi L, Godfrey WR, Lin J, Zhao G, Kao PN. NF90 regulates inducible IL-2 gene expression in T cells. J. Exp. Med. 2007;204:971–977. doi: 10.1084/jem.20052078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sakamoto S, Morisawa K, Ota K, Nie J, Taniguchi T. A binding protein to the DNase I hypersensitive site II in HLA-DR alpha gene was identified as NF90. Biochemistry. 1999;38:3355–3361. doi: 10.1021/bi982099g. [DOI] [PubMed] [Google Scholar]
- 56.Pfeifer I, Elsby R, Fernandez M, Faria PA, Nussenzveig DR, Lossos IS, Fontoura BM, Martin WD, Barber GN. NFAR-1 and -2 modulate translation and are required for efficient host defense. Proc. Natl Acad. Sci. USA. 2008;105:4173–4178. doi: 10.1073/pnas.0711222105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Vumbaca F, Phoenix KN, Rodriguez-Pinto D, Han DK, Claffey KP. Double-stranded RNA-binding protein regulates vascular endothelial growth factor mRNA stability, translation, and breast cancer angiogenesis. Mol. Cell Biol. 2008;28:772–783. doi: 10.1128/MCB.02078-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kuwano Y, Kim HH, Abdelmohsen K, Pullmann R, Jr, Martindale JL, Yang X, Gorospe M. MKP-1 mRNA stabilization and translational control by RNA-binding proteins HuR and NF90. Mol. Cell Biol. 2008;28:4562–4575. doi: 10.1128/MCB.00165-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wang P, Song W, Mok BW, Zhao P, Qin K, Lai A, Smith GJ, Zhang J, Lin T, Guan Y, et al. Nuclear factor 90 negatively regulates influenza virus replication by interacting with viral nucleoprotein. J. Virol. 2009;83:7850–7861. doi: 10.1128/JVI.00735-09. [DOI] [PMC free article] [PubMed] [Google Scholar]