Abstract
DNA double-strand breaks (DSBs) and other lesions occur frequently during cell growth and in meiosis. These are often repaired by homologous recombination (HR). HR may result in the formation of DNA structures called Holliday junctions (HJs), which need to be resolved to allow chromosome segregation. Whereas HJs are present in most HR events in meiosis, it has been proposed that in vegetative cells most HR events occur through intermediates lacking HJs. A recent screen in yeast has shown HJ resolution activity for a protein called Yen1, in addition to the previously known Mus81/Mms4 complex. Yeast strains deleted for both YEN1 and MMS4 show a reduction in growth rate, and are very sensitive to DNA-damaging agents. In addition, we investigate the genetic interaction of yen1 and mms4 with mutants defective in different repair pathways. We find that in the absence of Yen1 and Mms4 deletion of RAD1 or RAD52 have no further effect, whereas additional sensitivity is seen if RAD51 is deleted. Finally, we show that yeast cells are unable to carry out meiosis in the absence of both resolvases. Our results show that both Yen1 and Mms4/Mus81 play important (although not identical) roles during vegetative growth and in meiosis.
INTRODUCTION
During the life cycle of a living cell, double-strand breaks (DSBs) form constantly due to both internal and external insults. Throughout the course of evolution different repair mechanisms have evolved to repair these extremely dangerous lesions. Commonly, DSB repair mechanisms are divided into either non-homologous end joining (NHEJ), which uses little or no homology, and Homologous Recombination-based mechanisms (1), which rely on sequence similarity to achieve repair.
In the early 1960s Robin Holliday proposed a recombination model that can account for the formation of gene conversion [non-crossover (NCO)] and crossover (CO) events and their association during meiosis in fungi (2). Briefly, the Holliday model suggests that during meiosis a nick is formed in the two heterozygous alleles allowing annealing to occur between complementary sequences in the two different DNA strands. The cross-molecule DNA structures formed in this process are now termed Holliday junctions (HJs) (3). Due to the symmetrical nature of these structures their resolution can result in either a CO or a NCO, depending on the orientation of the cleavage.
Subsequently, work in several labs showed that in fact, the initiating lesions in meiosis are DSBs created by the nuclease Spo11 (4,5). Thus, later recombination models suggested DSBs as initiating events, but kept HJs as a central feature; the currently accepted ones are modifications of the DSB repair model proposed by Szostak and co-workers (6). In the current models (1), recombination is initiated by a DSB, followed by strand invasion of the homologous sequence (Figure 1). If both broken arms engage in strand invasion, a double HJ (dHJ) is created (Figure 1C). The dHJ requires resolution; this can occur in both planes at each junction, generating either CO or NCO products. If both junctions are resolved independently they should result in an equal amount of CO and NCO events.
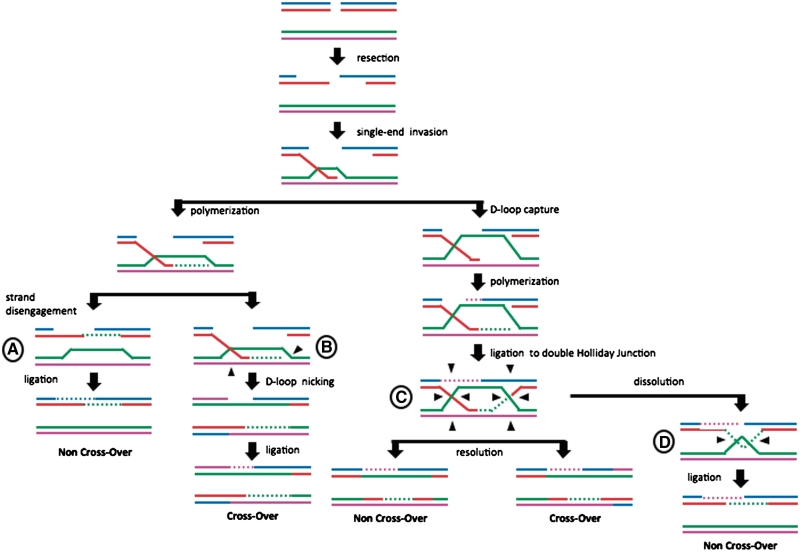
Figure 1.
Schematic representation of DSB repair by homologous recombination and its products. Following the formation of a DSB there is single-strand resection to form a 3′ overhang, which invades a homologous sequence. Single end invasion can be resolved through: (A) SDSA—strand disengagement, ligation to form a NCO product, or (B) The D-loop can be nicked and ligation may lead to a CO product. When there is also a second end capture by the D-loop, polymerization can lead to the formation of a double HJ, which can either be (C) resolved by HJ resolvases to be ligated to form NCO and CO products, or (D) undergo dissolution by the activity of a helicase and a topoisomerase to form a NCO product.
One of the main enigmas over the years has been how the resolution of HJs takes place. The discovery of resolvases (enzymes capable of resolving the HJ structure) in the bacteriophage T4 (7) and in Escherichia coli (8,9) suggested that resolvases may be universal features of HR. In budding yeast the first enzyme which showed resolvase activity was the mitochondrial enzyme, Cruciform Cutting endonuclease (Cce1) (10). The first nuclear enzyme identified to have a resolvase activity was Mus81 (together with its partner Eme1), discovered in fission yeast and in human cells (11,12). Mus81 belongs to the RAD1/XPF family of 3′-flap endonucleases that play a role in removal of DNA lesions formed by different cross-linking agents (13). The Mus81/Eme1 [Mus81/Mms4 complex in budding yeast (14)], was shown to be a non-classical resolvase, which cleaves the HJ structures asymmetrically (13). More importantly, it was found to have a significant preference for nicked structures. In contrast to fission yeast, which shows <1% viable spores in the absence of either mus81 or eme1, in budding yeast deletion of MUS81 reduces spore viability to only 40% (15,16). This led to the understanding that there must be additional enzymes that can resolve HJ structures in budding yeast.
Using an in vitro HJ resolution activity, West and co-workers have recently found another protein, Yen1, able to cleave HJ structures. Its human ortholog, GEN1, has the same HJ resolvase activity (17). These two proteins are members of the Rad2/XPG structure-specific endonucleases (18). West and colleagues (19) also examined the interaction between MUS81 and YEN1 and found that cells lacking both show severe sensitivity to a variety of DNA-damaging agents. The two genes were also shown to play redundant roles in the resolution of joint plasmids in vivo (20,21).
If we summarize our knowledge about the resolution of HJs we can come up with the following possible strategies for the formation of CO or NCO events (Figure 1). First, there is the possibility of DSB repair without HJ formation: following the invasion of the donor sequences, the recently extended ssDNA strand can disengage and ligate to the other broken DNA arm. This process is known as synthesis-dependent strand annealing [SDSA; (22) (Figure 1—marked as A)]. A second possibility comes from the fact that Mus81/Mms4 was shown to have higher resolvase activity on nicked HJ structures (13). If invasion occurs but instead of disengagement, the D-loop is cleaved by the Mus81/Mms4 (Figure 1—marked as B), ligation of this structure will create a CO product. Thirdly, resolution can occur as proposed in the DSB repair model (6,23): following end invasion and capture of the second end by the D-loop, a double HJ structure is formed that can be resolved in two planes (horizontal or vertical). If the two junctions are resolved in the same plane a NCO product is obtained, whereas if they are resolved in different planes the product will be a crossover (Figure 1—marked as C). This resolution can be performed by either a classical HJ resolvase, such as Yen1, or by non-classical HJ resolvases, such as Mus81/Mms4, with or without the assistance of flap endonucleases (17,24). In addition, there can also be dissolution of the double HJ structures (Figure 1—marked as D). This process involves the activity of helicases such as the BLM and Sgs1 proteins, with the help of type III topoisomerase (24–26).
In order to decipher the role that each resolution pathway plays in DSB repair we have examined the genetic interactions between YEN1 and MMS4, and their interaction with genes known to play central roles in different repair mechanism. Our results suggest that Yen1 and Mus81/Mms4 have both overlapping as well as separate activities in resolving homologous recombination intermediates. In addition, we show that Yen1 and Mms4 play a redundant role in meiosis in budding yeast.
MATERIALS AND METHODS
Yeast strains
All of the yeast strains used in the present study are isogenic derivatives of strain MK202 (MATa-inc ura3-HOcs lys2:: 5.6 kb ura3::HOcs-incRB ade3::GALHO ade2-1 leu2-3,112 his3-11,15 trp1-1 can1-100) (27). The ura3-HOcs allele on chromosome V is a 39-bp oligonuclotide insertion at the NcoI site. A 5.6-kb URA3 fragment containing the Hocs-inc sequence carrying BamHI and EcoRI polymorphisms was inserted at a HpaI site within LYS2 sequences, as described previously (27).
Deletion of YEN1 was obtained by transformation of MK202 with a PCR product produced on the appropriate yen1::KanMX strain from the Saccharomyces Genome Deletion Project array. The Mms4::TRP1 allele was introduced by transformation of MK202 with PCR product produced on the appropriate strain, provided by K.J. Myung (28). Deletions of the RAD52, RAD51, RAD1 and RAD18 genes were created by one- or two-step transplacement by using plasmids pSM20 (29), pAM28 (30), pRR46 (31) and plasmid K211 (gift from F. Fabre), respectively. All chromosomal configurations were verified by Southern blot analysis after transformation.
Media and growth conditions
Saccharomyces cerevisiae strains were grown at 30°C, unless specified otherwise. Standard YEP medium (1% yeast extract, 2% Bacto Peptone) supplemented with 3% glycerol (YEP-Gly), 2% galactose (YEP-Gal) or 2% dextrose (YEPD) was used for non-selective growth. We added 1.8% Bacto Agar for solid media.
Growth rate calculation
Cultures were grown to stationary phase in liquid YEPD culture, and diluted (1:5) into fresh YEPD media in a 96-well plate. All cultures were then grown and measured every 20 min using the TEKAN liquid handling robotic system for at least 12 h. The OD595 values were plotted as a function of time and the slope was obtained using Microsoft Excel. Doubling time was measured by dividing ln2 by the slope.
Repair efficiency measurement
Each strain was streaked onto YEP-Gly plates. Individual colonies were resuspended in water, appropriately diluted and plated on YEPD and YEP-Gal plates. Colonies were counted after 3–5 days of incubation at 30°C. In order to analyze, colony-formation cells were photographed following 2 days on YEPD and 3–5 days on YEP-Gal plates.
Induction experiment
Single colonies were grown to logarithmic phase in rich YEP-Gly medium, centrifuged and resuspended in YEP-Gal. DNA was extracted from samples at timely intervals (0, 2, 10 and 24 h) and subjected to Southern blot analysis using a 1.2-kb BamHI URA3 sequence as probes. The blots were quantified with the ImageMaster 1D Image Analysis Software.
Southern blot analysis
Southern blotting was carried out as described previously (32).
Sporulation efficiency experiment
The appropriate strains were mated to create the different diploid strains (wild-type, double heterozygous, homozygous yen1Δ, homozygous mms4Δ and double homozygous yen1Δ mms4Δ). Cells were confirmed to be diploids by a mating test. Mating cells were micromanipulated in order to select diploid cells. Following 2 days growth in YEPD plates, three diploid colonies from each strain were transferred to liquid YEPD media for over-night growth. The cultures were centrifuged and re-suspended in SM sporulation medium (2% potassium acetate). Cultures were left to sporulate for 5 days at 25°C. To quantify sporulation efficiency more than 1000 cells were counted under the microscope. The percentage of asci and spore-containing cells out of the total cells counted is shown.
RESULTS
mms4Δ and yen1Δ show a synthetic genetic interaction
In vitro experiments have recently shown that both Mms4 and Yen1 have resolvase activity (17). In order to study the role that these two proteins play in DSB repair, we created a double mutant yen1Δ mms4Δ. Tetrad analysis of a diploid strain heterozygous for each deletion showed that while each single mutant displayed a colony size similar to that of the wild-type haploids, the double mutant has a significantly smaller colony size (Figure 2), implying a synthetic sick interaction between the two mutants. The fitness reduction in the double mutant is also apparent when following growth in liquid cultures: whereas each single mutant has only a slight effect on fitness, the double mutant grows very slowly (Figure 2B). The fitness effect indicates a synergistic genetic interaction (33). These results suggest that Mms4 and Yen1 carry out alternative, redundant activities, which are needed quite frequently during vegetative growth.
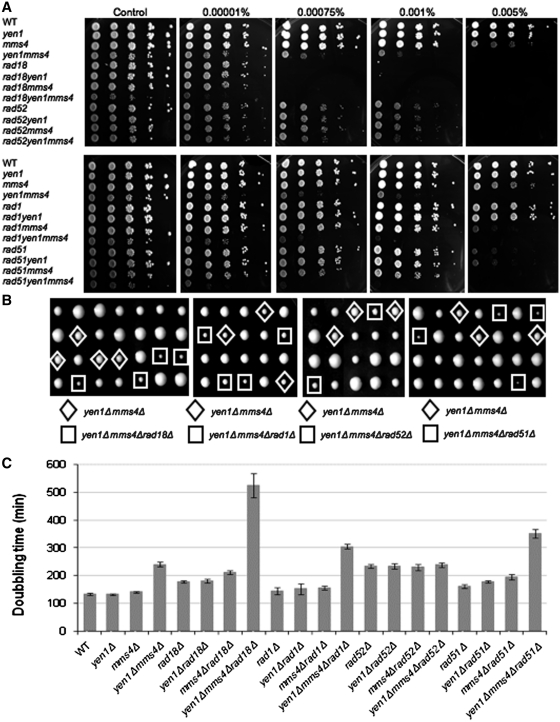
Figure 2.
yen1Δ mms4Δ cells show growth defects. (A) Tetrad analysis of a double heterozygous diploid. Colony size indicates that yen1Δ mms4Δ colonies have a slow growth phenotype. yen1Δ single deletion (circle), mms4Δ single deletion (square) and yen1Δ mms4Δ double deletion (diamond) are marked. (B) Calculated doubling time of wild-type cells compared to single (yen1Δ and mms4Δ) and double mutant yen1Δ mms4Δ. Quantification of growth rate shows a prolonged doubling time for the double deletion yen1Δ mms4Δ.
Double mutants mms4Δ yen1Δ show high sensitivity to DNA damage
We next examined the sensitivity of the double mutant to DNA-damaging agents. MMS4 was first discovered due to its sensitivity to the alkylating agent Methyl methanesulfonate (MMS) (34). The Mms4 protein works in a complex with Mus81; cells deleted for MUS81 also show sensitivity to a variety of DNA-damaging agents (35). Mus81 was previously shown to exhibit enhanced sensitivity to DNA-damaging agents when combined with a YEN1 deletion (19,20). We performed a drop assay to examine the MMS sensitivity of the double mutant compared to the each single mutant and the wild-type (Figure 3A). The single mutant yen1Δ shows the same lack of MMS sensitivity as the wild-type strain. As expected, the single mutant mms4Δ exhibits a mild MMS sensitivity apparent only at high MMS concentrations (0.005%). In contrast, the double mutant yen1Δ mms4Δ shows a significantly higher sensitivity to MMS compared to both single mutants (Figure 3A).
Figure 3.
Genetic interactions of yen1Δ and mms4Δ with different repair enzymes. (A) Drop assay to analyze MMS sensitivity of yen1Δ, mms4Δ and double deletion yen1Δ mms4Δ with repair enzymes (rad18Δ, rad52Δ, rad1Δ or rad51Δ). (B) Tetrad analysis of double deletion yen1Δ mms4Δ combined with mutations in additional repair enzymes. The yen1Δ mms4Δ strain (diamond) and the triple deletion (square) are marked. (C) Doubling time of yen1Δ, mms4Δ and yen1Δ mms4Δ combined with mutations in additional repair enzymes.
These results indicate that Mms4/Mus81 and Yen1 play redundant roles in DNA damage repair. Thus, in the absence of one activity the other can compensate to some degree. It is important to note that the results show that in the absence of Mms4, Yen1 can only partially compensate, resulting in a mild MMS sensitivity in the single mms4Δ cells.
Genetic interactions of mms4Δ and yen1Δ
In order to investigate in what genetic pathways MMS4 and YEN1 participate, we examined their genetic interactions with mutations affecting various DNA repair pathways. First, we analyzed the interaction between these genes and the post-replication repair (PRR) pathway. As a representative of this pathway, we chose the E3 ubiquitin ligase Rad18 (36). Mutations affecting this protein inactivate both the PRR’s error-free and the error-prone sub-pathways (37,38). Tetrad analysis of a diploid strain heterozygous for mms4Δ, yen1Δ and rad18Δ indicates that the triple mutant yen1Δ mms4Δ rad18Δ has a severe growth defect (Figure 3B). This observation was confirmed by measuring generation time (Figure 3C) and imply a synergistic effect of deletion of rad18Δ on the growth of the double mutant mms4Δ yen1Δ (33). We next examined the MMS sensitivity of the single mutant rad18Δ, the double mutant with mms4Δ or yen1Δ as well as the triple mutant yen1Δ mms4Δ rad18Δ. We found that each of the double mutants is as sensitive as the single rad18Δ mutant (Figure 3A); however, the triple mutant shows severe sensitivity to MMS even in the most diluted concentration (0.00001% MMS). Together, these results indicate a synergistic relationship between RAD18 and MMS4 and YEN1 both in fitness and in sensitivity to DNA damage, suggesting that the PRR pathway acts as an alternative repair mechanism to the two HJ resolution pathways (the Mms4/Mus81 dependent and Yen1 dependent) but that the three pathways may partially overlap (see ‘Discussion’ section).
The second interaction we examined was with Rad1, an excision repair protein that works in a complex with Rad10 (39). The Rad1/Rad10 complex plays a role in flap removal in different DNA repair processes, including DSB repair (40–43). It was previously suggested that this complex might also play a role in the resolution of HJs (44) [however, see also (45)]. Later experiments found additional support for this idea, including an in vitro interaction between Rad1 and the HJ-interacting proteins Slx4 and Mms4/Mus81 (46) and the stimulation of Rad1 activity by Slx4 phosphorylation (47).
We therefore examined the relationship between Rad1 and the pathway that involves Mms4 and Yen1. We created a diploid strain heterozygous for yen1Δ, mms4Δ and rad1Δ. Tetrad analysis as well as growth rate measurements showed no effect for rad1Δ in either mms4Δ or yen1Δ backgrounds (Figure 3B and C). However, there is a decrease in growth rate in the triple mutant yen1Δ mms4Δ rad1Δ compared to the double mutant yen1Δ mms4Δ (Figure 3B and C). Examination of the sensitivity to MMS shows that deletion of RAD1 in the yen1Δ background has no effect (Figure 3A). However, both in an mms4Δ background (compare the sensitivity to 0.005% MMS of the double mutant rad1Δ mms4Δ to that of the mms4Δ strain) and in a yen1Δ mms4Δ background (compare the sensitivity to 0.001% MMS of the triple mutant yen1Δ mms4Δ rad1Δ strain to that of the double mutant yen1Δ mms4Δ) deletion of RAD1 confers increased sensitivity to MMS.
In order to examine the interaction of yen1Δ and mms4Δ with the homologous recombination repair pathway, we crossed yen1Δ mms4Δ strains to either rad51Δ or rad52Δ strains. Tetrad analysis, as well as growth rate measurements, show that rad52Δ exhibits an epistatic relation with the yen1Δ mms4Δ double mutant: the triple mutant yen1Δ mms4Δ rad52Δ grows as slowly as the double yen1Δ mms4Δ or the single rad52 mutant (Figure 3B and C). Similar relationships are observed when sensitivity to MMS is measured (Figure 3A). Surprisingly, deletion of RAD51 in a yen1Δ mms4Δ double mutant leads to lower doubling time and increased sensitivity to MMS (Figure 3). The additivity in DNA damage sensitivity is particularly apparent under very low MMS concentrations (e.g. 0.00001% MMS, Figure 3A). Thus, in the absence of Yen1 and Mms4 deletion of RAD51 creates a phenotype that is not shared by yen1Δ mms4Δ rad52Δ strains (see ‘Discussion’ section).
Double mutants yen1Δ mms4Δ show a specific reduction in crossover events
We next examined the role of Mms4 and Yen1 in the repair of DSBs. We utilized a system previously used in our lab to examine the repair of a single DSB at a specific site in the genome (27). The haploid strain used (MK202) bears two copies of the URA3 gene; one of them, located on chromosome V, carries the recognition site for the yeast HO site-specific endonuclease (Figure 4). The second copy, a 5.6 kb fragment located on chromosome II, carries a similar site containing a single-base pair mutation that prevents recognition by the endonuclease (ura3-HOcs-inc). In addition, the ura3 alleles differ at two restriction sites, located to the left (BamHI) and to the right (EcoRI) of the HOcs-inc insertion; these polymorphisms are used to monitor the transfer of information between the chromosomes. In these strains, the HO gene is under the transcriptional control of the GAL1 promoter (27). Upon transfer of the cells to galactose-containing medium, the HO endonuclease is produced at high levels. The enzyme creates a DSB in essentially the whole-cell population (27,32). The broken chromosomes are then repaired by a mechanism that copies the HOcs-inc information, together with the flanking markers, resulting in a NCO event (gene conversion), that may be accompanied by a crossing over between the ectopic site, resulting in a detectable translocation event (32) (Figure 4A). There is no genetic selection for recombinational products; instead, repair is monitored in the entire cell population. During the course of the experiment, cell viability remains high in the wild-type strain, MK202, which constitutes our standard.
Figure 4.
Role of resolvases in the repair of DSB. (A) Schematic representation of the system used to examine DSB repair of a single-induced DSB. The endogenous URA3 gene on Ch. V carries an HO endonuclease recognition site. In the LYS2 locus on Ch. II, there is an insertion of 5.6 kb sequence homologous to the URA3 sequence, with a mutated HOcs (HOcs-inc) and two polymorphisms of BamHI (B) and EcoRI (R) sites. The cells also contain the HO endonuclease under a galactose-inducible GAL1 promoter. Following the transfer of the cells to galactose, a single DSB is formed on Ch. V. The repair of the break can lead to either non-crossover or crossover products. Restriction enzyme sites (arrowhead) and fragment sizes expected in a Southern blot are indicated. (B) Graphic representation of the repair efficiency of wild-type, yen1Δ, mms4Δ and yen1Δ mms4Δ cells. Repair efficiency is calculated by comparing the number of cells grown on glucose compared to colonies formed on galactose-containing medium. (C) Southern blot of DNA from cells taken 0, 2, 10 or 24 h after transfer to galactose-containing medium. The DNA was digested with PvuII and ApaLI and probed with a fragment of Ch. V carrying the URA3 gene. The percentage of crossover product was calculated in the 24 h time point by densitometer quantification of the autoradiogram. The percentage of the crossover bands was divided by the total DNA content (Ch. II, Ch. V and cross-over). The numbers in parentheses are the standard deviation (SD) values for three independent experiments.
The efficiency of DSB repair can be estimated by comparing the ability of the cells to form colonies on galactose-containing medium (in which the HO endonuclease is constitutively expressed) to that seen on glucose (no HO induction) (27). We examined the repair efficiency of strains deleted for YEN1 or MMS4, as well as that of the double mutant yen1Δ mms4Δ. As seen in Figure 4B, the two single deletion strains show similar repair efficiencies, which are indistinguishable from that of the wild-type strain. In contrast, the double mutant shows a significant reduction in repair efficiency compared to the other strains. This reduction, although significant, is not dramatic, indicating that most of the repair is not compromised in the absence of both Yen1 and Mms4.
In order to better understand the repair events that take place in the absence of Yen1 and Mms4, we examined the repair products by Southern blot analysis (see ‘Materials and Methods’ section). Cells were grown to mid-logarithmic phase in medium containing glycerol (YEP-Gly), and a DSB was induced by transferring the cells to galactose-containing medium (YEP-Gal). Cells were harvested at different times and subjected to Southern blot analysis. As depicted in Figure 4A, repair of the DSB can create either NCO or CO products. The latter can be detected in Southern blots due to the creation of novel restriction fragments as a consequence of the resulting translocation (Figure 4A). Cleavage by the restriction enzymes PvuII and ApaLI shows two bands corresponding to the CO products (Figure 4). We compared the hybridization pattern of the wild-type strain at the time of transfer to galactose (0 h), 2 h, 10 h and 24 h later, to the same time points in the single mutants yen1Δ, mms4Δ and mms4Δ yen1Δ double deletion strains (Figure 4C). Quantification of CO bands in all strains shows that while wild-type and yen1Δ strains have similar percentage of CO products (16 and 15%, respectively), in the single mutant mms4Δ there is only a slight reduction in CO products (12%). In the double mutant, yen1Δmms4Δ there is a significant reduction in CO product after 24 h in galactose (7%). Note that in the double mutant, there is a low DSB signal 2 h following transfer to galactose. This is due to a delay in DSB formation in this strain as previously described (20). For this reason, we chose to quantify CO products 24 h after the transfer to galactose to allow all of the cells to undergo break and repair.
Differential requirement for Mms4 and Yen1 in meiosis
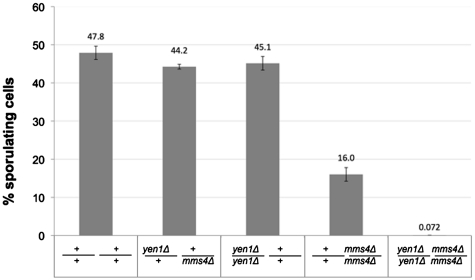
One of the main indications for the existence of more than one resolvase in S. cerevisiae is the fact that, in contrast to S. pombe, in the absence of Mus81/Mms4 cells are still able to undergo meiosis (sporulation), albeit at a significantly lower level than the wild-type strain (15,16). We thus decided to examine the possibility that Yen1 might play a role in the resolution of meiotic CO. We created a series of isogenic diploid strains carrying different combinations of YEN1 and MMS4 alleles, and we measured their sporulation efficiency. Figure 5 shows that wild-type, double heterozygotes and strains homozygous for the YEN1 deletion, all showed similar high levels of sporulation (∼50%). As expected, diploids homozygous for mms4Δ exhibit a significantly lower efficiency of meiosis (16%). In contrast, almost no sporulation could be observed for the diploid double homozygous yen1Δ/ yen1Δ mms4Δ/ mms4Δ (Figure 5). The few cells (out of thousands examined) that exhibited indications of sporulation, carried a single spore, instead of the four spores seen in most of the asci of the other strains. We therefore conclude that Yen1 and Mms4/Mus81 act as alternative resolvases for recombination events in meiosis, and the activity of at least one of them is essential to ensure proper meiosis and sporulation.
Figure 5.
Meiotic defects in the absence of Yen1 and Mms4. Graphic representation of the percentage of sporulating cells in wild-type, double heterozygous yen1Δ mms4Δ, homozygous yen1Δ, homozygous mms4Δ and double homozygous yen1Δ mms4Δ. Diploid cells were incubated at 25°C for 5 days in sporulation medium and were analyzed under the microscope for the presence of meiotic spores. For each strain, three independent samples were examined. The wild-type strain, the double heterozygous and the homozygous yen1Δ strain show high sporulation (∼50%); the homozygous mms4Δ strain shows reduced sporulation (16%) and almost no spores (0.072%) were observed in the double homozygous yen1Δmms4Δ strain.
DISCUSSION
Most recombination models (2,6) propose the existence of a HJ intermediate to account for the fact that meiotic products in most organisms show, for each gene conversion event scored, similar frequencies of CO and NCO events. In contrast, vegetative cells show a much lower association between NCO and CO [∼10–15%, (32,48,49)]. It was, therefore, suggested that mitotic recombination takes place mainly by an SDSA mechanism that does not form HJs (22). Indeed, recent work found a much lower incidence of joint molecules in mitotic cells, compared to meiosis (50). Our results, however, show that resolvases play important roles in repairing spontaneous and induced DNA damage in vegetative, as well as in meiotic cells.
We have examined the genetic relationship between mutations in two genes shown to have HJ resolvase activity in vitro, MMS4 and YEN1 (17). We have shown that a double mutant yen1Δ mms4Δ exhibits a dramatic reduction in fitness, indicating a synthetic sick interaction. This underscores the important and overlapping role played by the two resolvases during vegetative growth (Figure 2). Our results thus show that despite the lower level of HJs identified by currently available methods [(50); Figure 4C], HJ resolvases do play a role in the repair of spontaneous DNA damage in mitotic cells. We also show that the repair of an induced DSB is impaired in the yen1Δmms4Δ double deletion compared to the wild-type and to each of the single mutants (Figure 4). As mentioned above, only ∼10–15% of the DSB repair events in mitotic cells result in a crossover [(32,48–50), Figure 4C]. In the absence of both Mms4 and Yen1, however, we see a reduction of about 40% in repair efficiency (Figure 4B). These results indicate that NCO events might also be affected by the absence of these two HJ resolvases. Importantly, although there is a reduction in CO events in the double mutant yen1Δ mms4Δ, there is still a significant amount of CO events (7%) detected by Southern blot. Thus, additional mechanisms may exist, which can solve the HJ structure. Unfortunately, we could not investigate this point as deletion of SGS1 in the absence of either Yen1, Mms4 or both, is inviable [data not shown and (19)]. In addition, we also see a decrease in CO events in the absence of Mms4; however, this was not accompanied by a significant reduction in repair efficiency (Figure 4). These results might suggest that in the absence of Mms4, although CO events are reduced, repair is compensated by NCO events, presumably by a Yen1-dependent pathway.
After this work was submitted for publication, Ho et al. (20) published data on the role of Yen1 and Mus81 in diploid cells. Their results are in agreement with our conclusions. Importantly, in double homozygous yen1 mus81 diploid cells an increase in break-induced replication (BIR) compensates for the reduction of NCO and CO events (20). BIR could also be responsible for the low level of CO events observed (Figure 4C). In our study, carried out in haploid cells, BIR events are likely to cause loss of essential genes and thus lethality. Note that our Southern blot analysis does not distinguish between CO events observed in living cells from those present in cells unable to proliferate.
Our meiotic results are even more dramatic than those observed in vegetative cells: deletion of both resolvases completely abolishes meiosis (0.072%, Figure 5). One of the reasons for the initial search for additional resolvases is the fact that in contrast to fission yeast, where deletion of mus81 causes complete elimination of sporulation (16), in budding yeast deletion of either mus81 or mms4 causes only a relatively slight reduction in sporulation efficiency [(15) and Figure 5]. Interestingly, deletion of YEN1 had no visible effects on the efficiency of meiosis. This asymmetrical effect demonstrates that whereas Mus81/Mms4 can compensate completely for the absence of Yen1, Yen1 cannot fully replace Mus81/Mms4. Our results show that despite the differences in importance, at least one resolvase seems to be completely essential for proper meiosis. The simplest interpretation is that in the absence of these two enzymes there is no alternative mechanism able to resolve the HJs in meiotic cells. However, we cannot rule out the possibility that the reduction in sporulation is due to additional roles played by Yen1 and Mms4 which are not related to their HJ resolution activity.
The MMS sensitivity of mms4Δ mutants is strongly increased in the absence of Yen1 (Figure 3). This result places these two enzymes in two parallel repair pathways. In contrast to mms4Δ, the yen1Δ single mutant shows wild-type MMS sensitivity, indicating that Mms4 can fully compensate for the lack of Yen1. However, similarly to what is observed in meiosis, the MMS sensitivity of the mms4Δ single deletion indicates that Yen1 cannot fully compensate for the absence of Mus81/Mms4 in order to repair the damage caused by MMS. A possible explanation for this result may come from previous findings which showed that the Mus81/Mms4 complex has a preference for cleaving nicked structures (13), such as the D-loop resolution depicted in Figure 1B. We suggest that Yen1 can compensate for the absence of Mms4 in the resolution of canonical HJ (Figure 1C), but cannot replace Mms4 when it is necessary to solve different structures. This can also explain the differential sensitivity to MMS in the mms4Δ cells.
There has been extensive work seeking to understand the exact nature of the spontaneous DNA damage created during replication and how it is repaired. When a replication fork encounters a lesion numerous proteins are recruited in order to either repair or bypass the damage. Generally a stalled replication fork can bypass the damage either by recruiting polymerases that are less stringent and thus can polymerize over the damage (Trans-lesion synthesis polymerases), or by using the already replicated sister strand as a template to bypass the damage site. This post-replication repair pathway is error free, and much effort is being made to elucidate its mechanism of action (51,52). It has been suggested that fork reversal may create a structure topologically similar to a HJ [the ‘chicken foot’, (53,54)]. Alternative mechanisms in which the recently replicated sister chromatid is invaded by a Rad51-dependent mechanism have also been proposed [summarized in (55)]. Either of these mechanisms may require a resolvase in order to resolve the HJ and re-start DNA replication.
An important way to understand the interface between the different repair/bypass mechanisms is by analyzing the interaction between key players in each mechanism. We thus examined the genetic interaction of yen1Δ and mms4Δ with several enzymes involved in repair and bypass mechanisms. First, we investigated the effect of deleting the RAD18 gene, which controls the post-replication repair pathways (56), in a yen1Δ mms4Δ background. There was a clear synergistic genetic interaction and the triple mutant mms4Δ yen1Δ rad18Δ showed a very severe growth defect as well as extreme sensitivity to MMS (Figure 3). These results indicate that Rad18 must participate in a compensating mechanism that repairs part of the damage left unrepaired in the absence of both Mms4 and Yen1. This compensating pathway could be the error-free branch of the post-replication repair mechanism, which has been recently shown to require strand invasion, and presumably HJ resolution (51,52). Rad18 also controls an additional repair pathway that probably does not involve Yen1 and Mms4: the trans-lesion synthesis (TLS) pathway involving error prone DNA polymerases (37,38).
Next, we examined the genetic interaction with Rad1. The Rad1/10 endonuclease has been shown to play a role in trimming non-homologous ends during homologous recombination (41) and it has also been suggested that it may play a role in processing HJs (44). Our results indicate that rad1 is completely epistatic to either yen1Δ or mms4Δ single mutants with respect to growth (Figure 3). However, when examining the sensitivity to MMS of each double mutant (yen1Δ rad1Δ versus mms4Δ rad1Δ), there is an apparent and reproducible difference. While yen1Δ rad1Δ mutants show an MMS sensitivity similar to that seen in the single mutants, mms4Δ rad1Δ shows enhanced MMS sensitivity compared to the single mms4 deletion strain. This result indicates that Rad1 activity plays a role in the absence of Mms4. Thus, the Mms4/Mus81 and the Rad1/10 nucleases share possible substrates, which may be different from canonical HJs (45). In addition, deletion of RAD1 in a yen1Δ mms4Δ background shows reduced fitness as well as increased MMS sensitivity compared to the double mutant yen1Δ mms4Δ (Figure 3). These results indicate that Rad1 plays also a role in the repair of DNA damage that is independent of the activity of the HJ resolvases.
The third interaction, we examined was with rad52. As expected, yen1Δ mms4Δ show epistatic genetic interaction to rad52. Interestingly, the phenotype of the triple mutant as well as that of the double mutants (yen1Δrad52Δ and mms4Δrad52Δ), was as severe as the single rad52Δ (and similar to the double yen1Δ mms4Δ) both in growth rate and in MMS sensitivity (Figure 3). These results indicate that Rad52 and Yen1/Mms4 work in the same pathway to solve problems that arise during vegetative growth as well as in the presence of DNA damage. As indicated above, we believe that the resolvase activity of Yen1 and Mms4 is needed during replication to resolve the structures formed when the sister strand is used for damage bypass. Rad52 plays a role in promoting DNA annealing, and also acts as a mediator for Rad51 recruitment to the resected ssDNA ends [summarized in (57)]. Mutations in RAD52 prevent almost all types of recombination events in yeast. Thus, it is to be expected that without Rad52 no further effect will be observed if the resolvases are deleted. Following this same train of thought, the final enzyme we chose to examine was Rad51. Surprisingly, rad51 mutants showed synergistic interactions with yen1 mms4. Given that Rad52 is required for the Rad51 filament formation (58), it is surprising to observe a phenotype of rad51 that is more severe than that of rad52. One possible explanation for this result is that in the absence of Rad51 (but in the presence of Rad52), a recombination intermediate is created, which in the absence of both Yen1 and Mms4 is toxic and does not allow recovery from the damage. Another explanation could be that the increased sensitivity of the triple mutant yen1Δ mms4Δ rad51Δ is due to the absence of a Rad52-independent function of the Rad51 filament. One such a function may be the role of Rad51 in the recovery from cell cycle arrest following the repair of DNA damage (40,59). Indeed, Blanco et al. (19) and Ho et al. (20) showed that the yen1 mus81 double mutant exhibits cell cycle defects probably caused by prolonged checkpoint activation. Mutation in the RAD51 gene may reduce the fitness further.
We have shown that Yen1 and Mus81/Mms4, two HJ resolvases, play important roles in meiotic and vegetative cells. Despite the fact that vegetative cells show a much lower association between NCO and CO [10–15%, (32,50)] than meiotic cells, the resolvases still have a central role during vegetative growth. These results suggest that a large fraction of the HJ resolved by these enzymes in vegetative cells end up as NCO events, rather than COs. The current models for the activities of these resolvases (e.g. Figure 1, step B or C) do not account for such a role. In addition, our results show a clear asymmetry between Yen1 and Mms4: strains deleted for the first protein have very mild phenotypes (both in vegetative cells and in meiosis), implying that Mms4/Mus81 can replace it in most events. However, the reciprocal is not true, and Yen1 cannot completely replace the activity of Mms4/Mus81. This suggests the existence of recombination intermediates that are better resolved by the latter than by the former enzyme. Further investigation is needed to identify and characterize these intermediates and to understand their origin.
FUNDING
Israel Cancer Research Foundation (ICRF); Association for International Cancer Research (AICR) (to M.K.); Israeli Ministry of Science, Eshkol Fellowship (to N.A.). Funding for open access charge: ICRF.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank K.J. Myung and Francis Fabre for reagents, and all members of the Kupiec lab for encouragement and ideas.
REFERENCES
- 1.Paques F, Haber JE. Multiple pathways of recombination induced by double-strand breaks in Saccharomyces cerevisiae. Microbiol. Mol. Biol. Rev. 1999;63:349–404. doi: 10.1128/mmbr.63.2.349-404.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Holliday R. A mechanism for gene conversion in fungi. Genet. Res. 1964;5:283–304. doi: 10.1017/S0016672308009476. [DOI] [PubMed] [Google Scholar]
- 3.Liu Y, West SC. Happy Hollidays: 40th anniversary of the Holliday junction. Nat. Rev. Mol. Cell Biol. 2004;5:937–944. doi: 10.1038/nrm1502. [DOI] [PubMed] [Google Scholar]
- 4.Bergerat A, de Massy B, Gadelle D, Varoutas PC, Nicolas A, Forterre P. An atypical topoisomerase II from Archaea with implications for meiotic recombination. Nature. 1997;386:414–417. doi: 10.1038/386414a0. [DOI] [PubMed] [Google Scholar]
- 5.Keeney S, Giroux CN, Kleckner N. Meiosis-specific DNA double-strand breaks are catalyzed by Spo11, a member of a widely conserved protein family. Cell. 1997;88:375–384. doi: 10.1016/s0092-8674(00)81876-0. [DOI] [PubMed] [Google Scholar]
- 6.Szostak JW, Orr-Weaver TL, Rothstein RJ, Stahl FW. The double-strand-break repair model for recombination. Cell. 1983;33:25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- 7.Mizuuchi K, Kemper B, Hays J, Weisberg RA. T4 endonuclease VII cleaves holliday structures. Cell. 1982;29:357–365. doi: 10.1016/0092-8674(82)90152-0. [DOI] [PubMed] [Google Scholar]
- 8.Symington LS, Kolodner R. Partial purification of an enzyme from Saccharomyces cerevisiae that cleaves Holliday junctions. Proc. Natl Acad. Sci. USA. 1985;82:7247–7251. doi: 10.1073/pnas.82.21.7247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.West SC, Korner A. Cleavage of cruciform DNA structures by an activity from Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA. 1985;82:6445–6449. doi: 10.1073/pnas.82.19.6445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kleff S, Kemper B, Sternglanz R. Identification and characterization of yeast mutants and the gene for a cruciform cutting endonuclease. EMBO J. 1992;11:699–704. doi: 10.1002/j.1460-2075.1992.tb05102.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boddy MN, Gaillard PH, McDonald WH, Shanahan P, Yates JR, 3rd, Russell P. Mus81-Eme1 are essential components of a Holliday junction resolvase. Cell. 2001;107:537–548. doi: 10.1016/s0092-8674(01)00536-0. [DOI] [PubMed] [Google Scholar]
- 12.Chen XB, Melchionna R, Denis CM, Gaillard PH, Blasina A, Van de Weyer I, Boddy MN, Russell P, Vialard J, McGowan CH. Human Mus81-associated endonuclease cleaves Holliday junctions in vitro. Mol. Cell. 2001;8:1117–1127. doi: 10.1016/s1097-2765(01)00375-6. [DOI] [PubMed] [Google Scholar]
- 13.Ciccia A, Constantinou A, West SC. Identification and characterization of the human mus81-eme1 endonuclease. J. Biol. Chem. 2003;278:25172–25178. doi: 10.1074/jbc.M302882200. [DOI] [PubMed] [Google Scholar]
- 14.Kaliraman V, Mullen JR, Fricke WM, Bastin-Shanower SA, Brill SJ. Functional overlap between Sgs1-Top3 and the Mms4-Mus81 endonuclease. Genes Dev. 2001;15:2730–2740. doi: 10.1101/gad.932201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.de los Santos T, Hunter N, Lee C, Larkin B, Loidl J, Hollingsworth NM. The Mus81/Mms4 endonuclease acts independently of double-Holliday junction resolution to promote a distinct subset of crossovers during meiosis in budding yeast. Genetics. 2003;164:81–94. doi: 10.1093/genetics/164.1.81. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Osman F, Dixon J, Doe CL, Whitby MC. Generating crossovers by resolution of nicked Holliday junctions: a role for Mus81-Eme1 in meiosis. Mol. Cell. 2003;12:761–774. doi: 10.1016/s1097-2765(03)00343-5. [DOI] [PubMed] [Google Scholar]
- 17.Ip SC, Rass U, Blanco MG, Flynn HR, Skehel JM, West SC. Identification of Holliday junction resolvases from humans and yeast. Nature. 2008;456:357–361. doi: 10.1038/nature07470. [DOI] [PubMed] [Google Scholar]
- 18.Lieber MR. The FEN-1 family of structure-specific nucleases in eukaryotic DNA replication, recombination and repair. Bioessays. 1997;19:233–240. doi: 10.1002/bies.950190309. [DOI] [PubMed] [Google Scholar]
- 19.Blanco MG, Matos J, Rass U, Ip SC, West SC. Functional overlap between the structure-specific nucleases Yen1 and Mus81-Mms4 for DNA-damage repair in S. cerevisiae. DNA Repair. 2010;9:394–402. doi: 10.1016/j.dnarep.2009.12.017. [DOI] [PubMed] [Google Scholar]
- 20.Ho CK, Mazon G, Lam AF, Symington LS. Mus81 and Yen1 promote reciprocal exchange during mitotic recombination to maintain genome integrity in budding yeast. Mol. Cell. 2010;40:988–1000. doi: 10.1016/j.molcel.2010.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tay YD, Wu L. Overlapping roles for Yen1 and Mus81 in cellular Holliday junction processing. J. Biol. Chem. 2010;285:11427–11432. doi: 10.1074/jbc.M110.108399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nassif N, Penney J, Pal S, Engels WR, Gloor GB. Efficient copying of nonhomologous sequences from ectopic sites via P-element-induced gap repair. Mol. Cell. Biol. 1994;14:1613–1625. doi: 10.1128/mcb.14.3.1613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Orr-Weaver TL, Nicolas A, Szostak JW. Gene conversion adjacent to regions of double-strand break repair. Mol. Cell. Biol. 1988;8:5292–5298. doi: 10.1128/mcb.8.12.5292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jessop L, Lichten M. Mus81/Mms4 endonuclease and Sgs1 helicase collaborate to ensure proper recombination intermediate metabolism during meiosis. Mol. Cell. 2008;31:313–323. doi: 10.1016/j.molcel.2008.05.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ira G, Malkova A, Liberi G, Foiani M, Haber JE. Srs2 and Sgs1-Top3 suppress crossovers during double-strand break repair in yeast. Cell. 2003;115:401–411. doi: 10.1016/s0092-8674(03)00886-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wu L, Hickson ID. The Bloom's syndrome helicase suppresses crossing over during homologous recombination. Nature. 2003;426:870–874. doi: 10.1038/nature02253. [DOI] [PubMed] [Google Scholar]
- 27.Aylon Y, Liefshitz B, Bitan-Banin G, Kupiec M. Molecular dissection of mitotic recombination in the yeast Saccharomyces cerevisiae. Mol. Cell. Biol. 2003;23:1403–1417. doi: 10.1128/MCB.23.4.1403-1417.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smith S, Hwang JY, Banerjee S, Majeed A, Gupta A, Myung K. Mutator genes for suppression of gross chromosomal rearrangements identified by a genome-wide screening in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA. 2004;101:9039–9044. doi: 10.1073/pnas.0403093101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schild D. Suppression of a new allele of the yeast RAD52 gene by overexpression of RAD51, mutations in srs2 and ccr4, or mating-type heterozygosity. Genetics. 1995;140:115–127. doi: 10.1093/genetics/140.1.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Basile G, Aker M, Mortimer RK. Nucleotide sequence and transcriptional regulation of the yeast recombinational repair gene RAD51. Mol. Cell. Biol. 1992;12:3235–3246. doi: 10.1128/mcb.12.7.3235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reynolds P, Prakash L, Prakash S. Nucleotide sequence and functional analysis of the RAD1 gene of Saccharomyces cerevisiae. Mol. Cell. Biol. 1987;7:1012–1020. doi: 10.1128/mcb.7.3.1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Inbar O, Liefshitz B, Bitan G, Kupiec M. The relationship between homology length and crossing over during the repair of a broken chromosome. J. Biol. Chem. 2000;275:30833–30838. doi: 10.1074/jbc.C000133200. [DOI] [PubMed] [Google Scholar]
- 33.St Onge RP, Mani R, Oh J, Proctor M, Fung E, Davis RW, Nislow C, Roth FP, Giaever G. Systematic pathway analysis using high-resolution fitness profiling of combinatorial gene deletions. Nat. Genet. 2007;39:199–206. doi: 10.1038/ng1948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Prakash L, Prakash S. Isolation and characterization of MMS-sensitive mutants of Saccharomyces cerevisiae. Genetics. 1977;86:33–55. doi: 10.1093/genetics/86.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Interthal H, Heyer WD. MUS81 encodes a novel helix-hairpin-helix protein involved in the response to UV- and methylation-induced DNA damage in Saccharomyces cerevisiae. Mol. Gen. Genet. 2000;263:812–827. doi: 10.1007/s004380000241. [DOI] [PubMed] [Google Scholar]
- 36.Lee KY, Myung K. PCNA modifications for regulation of post-replication repair pathways. Mol. Cells. 2008;26:5–11. [PMC free article] [PubMed] [Google Scholar]
- 37.Hoege C, Pfander B, Moldovan GL, Pyrowolakis G, Jentsch S. RAD6-dependent DNA repair is linked to modification of PCNA by ubiquitin and SUMO. Nature. 2002;419:135–141. doi: 10.1038/nature00991. [DOI] [PubMed] [Google Scholar]
- 38.Stelter P, Ulrich HD. Control of spontaneous and damage-induced mutagenesis by SUMO and ubiquitin conjugation. Nature. 2003;425:188–191. doi: 10.1038/nature01965. [DOI] [PubMed] [Google Scholar]
- 39.Sung P, Reynolds P, Prakash L, Prakash S. Purification and characterization of the Saccharomyces cerevisiae RAD1/RAD10 endonuclease. J. Biol. Chem. 1993;268:26391–26399. [PubMed] [Google Scholar]
- 40.Agmon N, Pur S, Liefshitz B, Kupiec M. Analysis of repair mechanism choice during homologous recombination. Nucleic Acids Res. 2009;37:5081–5092. doi: 10.1093/nar/gkp495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ivanov EL, Haber JE. RAD1 and RAD10, but not other excision repair genes, are required for double-strand break-induced recombination in Saccharomyces cerevisiae. Mol. Cell. Biol. 1995;15:2245–2251. doi: 10.1128/mcb.15.4.2245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liefshitz B, Parket A, Maya R, Kupiec M. The role of DNA repair genes in recombination between repeated sequences in yeast. Genetics. 1995;140:1199–1211. doi: 10.1093/genetics/140.4.1199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Liefshitz B, Steinlauf R, Friedl A, Eckardt-Schupp F, Kupiec M. Genetic interactions between mutants of the 'error-prone' repair group of Saccharomyces cerevisiae and their effect on recombination and mutagenesis. Mutat. Res. 1998;407:135–145. doi: 10.1016/s0921-8777(97)00070-0. [DOI] [PubMed] [Google Scholar]
- 44.Habraken Y, Sung P, Prakash L, Prakash S. Holliday junction cleavage by yeast Rad1 protein. Nature. 1994;371:531–534. doi: 10.1038/371531a0. [DOI] [PubMed] [Google Scholar]
- 45.West SC. Holliday junctions cleaved by Rad1? Nature. 1995;373:27–28. doi: 10.1038/373027a0. [DOI] [PubMed] [Google Scholar]
- 46.Fekairi S, Scaglione S, Chahwan C, Taylor ER, Tissier A, Coulon S, Dong MQ, Ruse C, Yates JR, 3rd, Russell P, et al. Human SLX4 is a Holliday junction resolvase subunit that binds multiple DNA repair/recombination endonucleases. Cell. 2009;138:78–89. doi: 10.1016/j.cell.2009.06.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Toh GW, Sugawara N, Dong J, Toth R, Lee SE, Haber JE, Rouse J. Mec1/Tel1-dependent phosphorylation of Slx4 stimulates Rad1-Rad10-dependent cleavage of non-homologous DNA tails. DNA Repair. 2010;9:718–726. doi: 10.1016/j.dnarep.2010.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Borts RH, Haber JE. Meiotic recombination in yeast: alteration by multiple heterozygosities. Science. 1987;237:1459–1465. doi: 10.1126/science.2820060. [DOI] [PubMed] [Google Scholar]
- 49.Lee PS, Greenwell PW, Dominska M, Gawel M, Hamilton M, Petes TD. A fine-structure map of spontaneous mitotic crossovers in the yeast Saccharomyces cerevisiae. PLoS Genet. 2009;5:e1000410. doi: 10.1371/journal.pgen.1000410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bzymek M, Thayer NH, Oh SD, Kleckner N, Hunter N. Double Holliday junctions are intermediates of DNA break repair. Nature. 2010;464:937–941. doi: 10.1038/nature08868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Branzei D, Vanoli F, Foiani M. SUMOylation regulates Rad18-mediated template switch. Nature. 2008;456:915–920. doi: 10.1038/nature07587. [DOI] [PubMed] [Google Scholar]
- 52.Karras GI, Jentsch S. The RAD6 DNA damage tolerance pathway operates uncoupled from the replication fork and is functional beyond S phase. Cell. 2010;141:255–267. doi: 10.1016/j.cell.2010.02.028. [DOI] [PubMed] [Google Scholar]
- 53.Michel B, Flores MJ, Viguera E, Grompone G, Seigneur M, Bidnenko V. Rescue of arrested replication forks by homologous recombination. Proc. Natl Acad. Sci. USA. 2001;98:8181–8188. doi: 10.1073/pnas.111008798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Postow L, Ullsperger C, Keller RW, Bustamante C, Vologodskii AV, Cozzarelli NR. Positive torsional strain causes the formation of a four-way junction at replication forks. J. Biol. Chem. 2001;276:2790–2796. doi: 10.1074/jbc.M006736200. [DOI] [PubMed] [Google Scholar]
- 55.Branzei D, Foiani M. The checkpoint response to replication stress. DNA Repair. 2009;8:1038–1046. doi: 10.1016/j.dnarep.2009.04.014. [DOI] [PubMed] [Google Scholar]
- 56.Prakash L. Characterization of postreplication repair in Saccharomyces cerevisiae and effects of rad6, rad18, rev3 and rad52 mutations. Mol. Gen. Genet. 1981;184:471–478. doi: 10.1007/BF00352525. [DOI] [PubMed] [Google Scholar]
- 57.Mortensen UH, Lisby M, Rothstein R. Rad52. Curr. Biol. 2009;19:R676–677. doi: 10.1016/j.cub.2009.06.001. [DOI] [PubMed] [Google Scholar]
- 58.Seong C, Sehorn MG, Plate I, Shi I, Song B, Chi P, Mortensen U, Sung P, Krejci L. Molecular anatomy of the recombination mediator function of Saccharomyces cerevisiae Rad52. J. Biol. Chem. 2008;283:12166–12174. doi: 10.1074/jbc.M800763200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Dubrana K, van Attikum H, Hediger F, Gasser SM. The processing of double-strand breaks and binding of single-strand-binding proteins RPA and Rad51 modulate the formation of ATR-kinase foci in yeast. J. Cell Sci. 2007;120:4209–4220. doi: 10.1242/jcs.018366. [DOI] [PubMed] [Google Scholar]