Abstract
Gene amplification contributes to a variety of biological phenomena, including malignant progression and drug resistance. However, details of the molecular mechanisms remain to be determined. Here, we have developed a gene amplification system in yeast and mammalian cells that is based on double rolling-circle replication (DRCR). Cre-lox system is used to efficiently induce DRCR utilizing a recombinational process coupled with replication. This system shows distinctive features seen in amplification of oncogenes and drug-resistance genes: (i) intra- and extrachromosomal amplification, (ii) intensive chromosome rearrangement and (iii) scattered-type amplification resembling those seen in cancer cells. This system can serve as a model for amplification of oncogenes and drug-resistance genes, and improve amplification systems used for making pharmaceutical proteins in mammalian cells.
INTRODUCTION
Gene amplification contributes to a variety of biological phenomena, including malignant progression (1,2), drug resistance (3,4), certain developmental processes (5), adaptive mutation phenomena (6) and gene evolution (7). However, details of the molecular mechanisms remain to be determined (8–10). In cancer and drug-resistant cells, breakage–fusion–bridge (BFB) cycles (11) form large regular inverted repeats in the early stages of amplification and thereafter these repeats rapidly change into shorter highly amplified units (8,10,12–14). However, it remains largely unknown how complex end products can be rapidly generated after BFB cycles. This rapid process has been difficult to analyze, since previous approaches to understand amplification mechanisms were based on the structural analysis of end products. Therefore, we decided to develop a designed amplification system in vivo to induce a rapid process and analyze the products. Previously, we induced gene amplification in yeast, using recombinational processes induced by chromosomal breaks designed to induce a rapid amplification process, double rolling-circle replication (DRCR) (15). The amplification products resemble two types of product seen in mammalian cells, namely homogeneously staining regions (HSR; intrachromosomal repetitive arrays consisting mainly of inverted repeats) and double minutes (DMs; acentric, autonomous, generally circular extrachromosomal DNA with an inverted structure) (8–10).
In the present study, we used a distinct process, Cre-lox recombination (16), to determine whether DRCR is centrally involved in amplification events in mammalian cells, and to establish an in vivo amplification system that work well in mammalian cells. DRCR is a continuous process in which two replication forks chase each other (Figure 1A) and was confirmed by Volkert and Broach for amplification of yeast 2 µ plasmid (17). We first conceived that DRCR (15,17) can be induced by a recombinational process coupled with replication (Figure 1B and C). The amplification system induced DRCR efficiently in yeast and was shown to be a powerful tool for inducing gene amplification in Chinese hamster ovary (CHO) cells. The features of this amplification closely resembling those seen in mammalian cells strongly suggest the involvement of DRCR in amplification of oncogenes and drug-resistance genes.
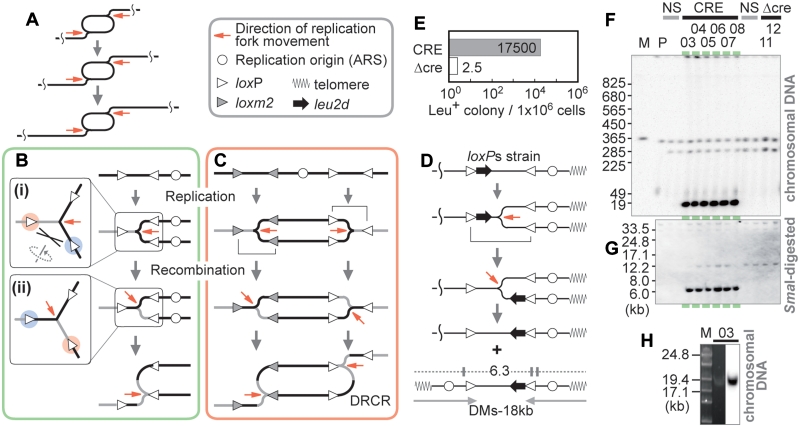
Figure 1.
Recombinational process coupled with replication. (A) DRCR. Two replication forks chase each other. One replication fork can replicate a template for the other fork and so amplification proceeds. (B) Recombinational process coupled with replication. The gray and black lines indicate the un-replicated and recently replicated regions at the time of recombination, respectively. If recombination occurs between loxP sites marked red and blue (i), the replication template is switched and thereafter the replicated region is replicated again (ii). (C) DRCR induction. If both bidirectional DNA replications undergo the processes as described in (B), DRCR can be induced. (D) Structure of the loxPs cassette, a model for the production of the minichromosome, and the predicted structure of the 18 kb DM-type products. The SmaI sites (vertical lines), and the sizes (kb) of fragments that hybridize with the leu2d probe are shown. The gray arrows below indicate an inverted structure. (E) Frequency of Leu+ colony formation. The colonies were counted as described in Materials and Methods of Supplementary Data. CRE: Cre-induced condition; Δcre: negative control. (F) Southern analysis of a representative sample of survivors with the leu2d probe. PFGE and Southern analysis were performed as described in Materials and Methods of Supplementary Data. The colony numbers are indicated above. M: Saccharomyces cerevisiae marker; P: the parental strain (LS20); NS: non-selective conditions. (G) Southern analysis of SmaI-digested DNA of the samples from (F) with the leu2d probe. (H) SYBR Green I staining and Southern analysis of the samples from colony #03 by PFGE for lower range.
MATERIALS AND METHODS
Yeast strains, plasmids and cell lines
Yeast strains, plasmids, cell lines and their construction are described in Materials and Methods of the Supplementary Data and Supplementary Figures S5–S7.
Selection of yeast cells with gene amplification
Induction of the Cre recombinase gene in yeast was carried out as follows: cells were grown in SC (synthetic complete) (18) liquid medium with 2% raffinose, lacking Trp, Lys and Ura, to mid-log phase. Then 0.1 volume of 20% galactose was added and culture continued for 90 min. The cells were then harvested by centrifugation, washed twice with sterile distilled water and plated at different dilutions onto SC medium with 2% glucose. SC plates lacking Trp, Lys, Ura and Leu were used for selection of cells with gene amplification and plates lacking Trp, Lys and Ura were used for measurement of the number of viable cells. Cells were grown at 25°C. Colonies on the plates were counted after 4 days of growth. A working protocol is described in Materials and Methods of the Supplementary Data.
Pulsed-field gel electrophoresis and Southern analysis
Cells were embedded in agarose plugs as per the instruction manual of the CHEF plug mold kit (Bio-Rad). The gel-plugs were then treated with 5 mg/ml proteinase K at 37°C for 2 days and washed twice with TE buffer containing 1 mM Pefabloc SC (Roche), twice with TE buffer and once with 1× restriction enzyme buffer. Pulsed-field gel electrophoresis (PFGE) was performed with the CHEF Mapper XA system (Bio-Rad) in 1% agarose gels with 0.5× TBE buffer. The autoalgorithm mode of the system was used with the size range of 150–800 (Figures 1F and 2C; Supplementary Figure S2A and B), 3–25 (Figures 1G and 2D; Supplementary Figures S1B, S1D and S3A), 10–60 (Figures 1H and 2E). Southern blotting was performed onto Hybond-N+ membrane (GE Healthcare) according to Sambrook et al. (19) Hybridization was performed with fluorescein-labeled probes, which were prepared with the Gene Images random-prime labeling module (GE Healthcare), and detected and quantified with a luminescent image analyzer, LAS-1000plus (Fujifilm).
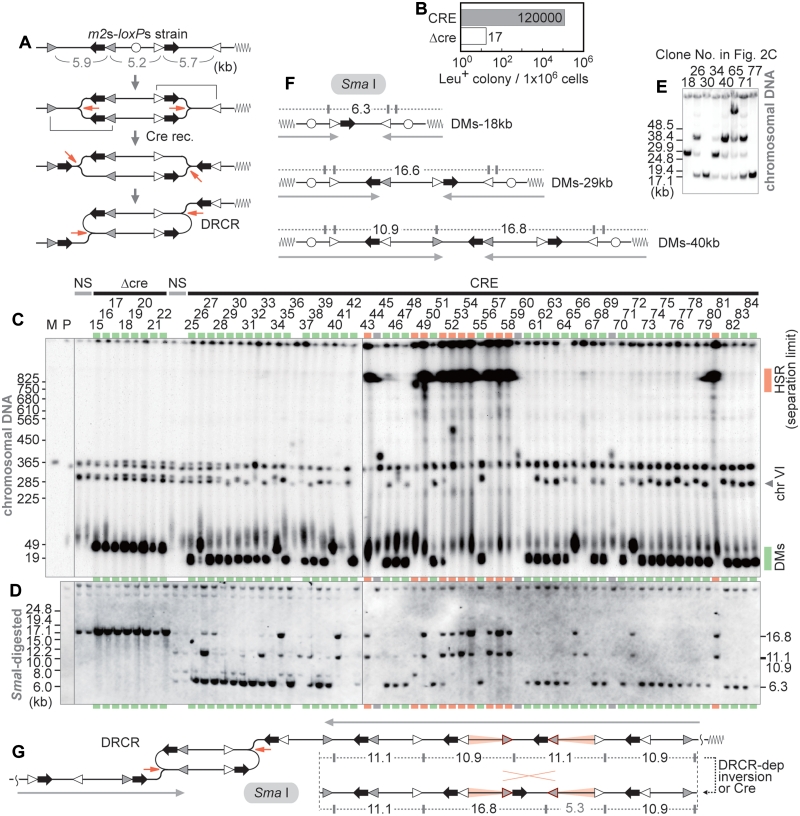
Figure 2.
Cre-lox dependent DRCR amplification in yeast. (A) Structure of the m2s–loxPs cassette and a model for DRCR amplification. The sizes (kb) of the three regions in the structure are indicated below. A possible model for termination of the DRCR process is provided in Supplementary Figure S2D. (B) Frequency of Leu+ colony formation. CRE: Cre-induced condition; Δcre: negative control. (C) Southern analysis of a representative sample of m2s–loxPs survivors with the leu2d probe. The lanes marked in red and green indicate HSR- and DM-type products, respectively. Supplementary information, including the description of the lanes marked in gray, is provided in Supplementary Texts. M: S. cerevisiae marker; P: the parental strain (LS20); NS: non-selective conditions. (D) Southern analysis of SmaI-digested DNA of the samples from (C) with the leu2d probe. (E) Southern analysis of a representative sample with DM-type products by PFGE for lower range. (F) The predicted structure of the 18, 29 and 40 kb DM-type products. The SmaI sites (vertical lines) and the sizes (kb) of fragments that hybridize with the leu2d probe are shown. The gray arrows below indicate an inverted structure. (G) Schematic representation of the expected structure derived through the DRCR process and SmaI-restriction maps of the representative HSR-type structure. Inverted repeats that can engage in DRCR-dependent inversion are marked with red backgrounds. The gray arrows indicate a palindromic structure.
Cell transfection and methotrexate selection
Flp-In CHO cells (Invitrogen) were transfected with the Flp recombinase expression vector pOG44 (Invitrogen) and subsequently transfected with the modified bacterial artificial chromosome (BAC) clone by Targefect-BAC (Targeting systems). After hygromycin B selection, cells with the BAC were cloned. The BAC-CHO clones were transfected with the Cre recombinase expression vector pPGK-Cre-bpA. Methotrexate (MTX) selection was carried out in 96-well plates. MTX-resistant cells were screened for secreted alkaline phosphatase (SEAP), cloned and analyzed by fluorescence in situ hybridization (FISH). Details of these procedures and a working protocol are described in Materials and Methods of the Supplementary Data.
Interphase and metaphase FISH
The samples were fixed, denatured, hybridized and fluorescence detected as described previously (20). For metaphase FISH, cells were treated with colcemid and Chromosome Resolution Additive (Genial Genetic Solutions, Chester, UK) for 2 and 1.5 hours before cell harvest, respectively, and were subjected to hypotonic treatment before fixation. The modified BAC construct and pSV2-dhfr plasmid were biotinylated for use as probes, and visualized by the FITC-conjugated avidin and biotinylated antiavidin antibody combination system (Vector). Details of these techniques are described in Materials and Methods of Supplementary Data.
RESULTS
Recombinational process coupled with replication
We first predicted that, if recombination occurs between un-replicated and recently replicated regions during replication (Figure 1B), the replication fork will make an additional copy of the replicated region. To demonstrate this process, we constructed the strain loxPs in which the right terminus region of chromosome VI was modified (Figure 1D, top). An autonomously replicating sequence (ARS) is naturally located in the subtelomeric region (21). The amplification marker, leu2d (black arrows in Figure 1D), has slight transcriptional activity and can complement leucine auxotrophy only if amplified. If the recombinational process occurs, a minichromosome should be produced (Figure 1D, bottom) whose copy number will increase under selection. To induce Cre-lox recombination, a galactose-inducible Cre expression vector (CRE) or a control vector (Δcre) was used. We induced Cre expression in galactose medium and then plated these strains on glucose plates lacking leucine to select Leu+ survivors (those with amplification). The Cre induction caused a 7000-fold increase in the frequency of survivors (Figure 1E). Chromosomal and SmaI-digested DNA from Leu+ survivors was separated by PFGE and hybridized with the leu2d gene as the probe. As expected, a linear minichromosome of ∼18 kb in length (Figure 1F and H), yielding a 6.3 kb leu2d SmaI fragment (Figure 1G), was produced in a Cre-dependent manner. The signal intensity indicated approximately 27 copies of minichromosome (clone #03, Supplementary Figure S2A). The frequent appearance of this minichromosome demonstrates that Cre-lox recombination can efficiently cause the recombinational process coupled with replication.
Cre-lox-dependent gene amplification system in yeast
The above results imply that two recombinational processes could induce DRCR (Figure 1C). To explore this further, we used two different types of lox sequence, the wild-type loxP (lox for short) and a mutant-type loxm2 (m2 for short). Cre recombination occurs between identical sites (lox–lox or m2–m2) but not between different sites (lox–m2). Using these cis-elements (lox and m2) and the leu2d gene, we constructed an amplification cassette (Figure 2A). A natural ARS consensus sequence that exhibits ARS activity (21) is located in the region between the lox pair and m2 pair. Using the strain designated m2s–loxPs, we repeated the experiment as described above.
The Cre induction caused a >7000-fold increase in the frequency of survivors (Figure 2B) and 12% of the Cre-induced cells survived. Structural analysis of the chromosomes revealed the generation of two distinct leu2d amplification products resembling HSR and DMs in Cre-dependent Leu+ clones (Figure 2C and D). Detailed analysis of DM-type products indicated linear minichromosomes of at least three sizes (18, 29 and 40 kb; Figure 2E and F), whose formation can be explained by three types of the recombinational processes (Supplementary Figure S1). Interestingly, even under Δcre conditions (clones #15–22 in Figure 2C), homology-based recombination can produce 29 kb minichromosomes. The signal intensity of these DMs products indicated ∼15–30 mini chromosome copies (Supplementary Figure S2A). The HSR-type products generated a quite different pattern; the original chromosome VI band containing the m2s-loxs constructs (∼290 kb) disappeared and very dense DNA spots appeared above the separation limit and at the well position, indicating that extensive amplification of leu2d results in elongation of chromosome VI. The amplification occurs within the chromosome, since a chromosome VI-specific probe, RET2, hybridized to the same chromosomal fragments as the leu2d probe (Supplementary Figure S2B). The signal intensity of leu2d from the HSR-type products indicated the presence of ∼90–140 copies of the leu2d gene (Supplementary Figure S2A), corresponding to a 3.6–5.6-fold increase in the length of the original (275 kb) chromosome VI (Supplementary Figure S2C). Thus, our yeast system generated HSR- or DM-type products at high frequency (>10%).
We previously found that sequences flanked by inverted repeats, which are formed by DRCR amplification, were subject to frequent inversion (15). We call this phenomenon DRCR-dependent inversion, which can explain the production of ∼17 kb SmaI fragments in addition to the 10.9 and 11.1 kb SmaI fragments that DRCR originally amplifies (Figure 2G). This type of inversion could also explain the results using other restriction enzymes (EcoRI, HindIII, PstI and XbaI) (Supplementary Figure S3).
Gene amplification system in CHO cells induced by Cre-lox recombination
Next, we attempted to induce gene amplification in CHO cells in this way. We constructed an amplification cassette on a rat genomic BAC (Figure 3A) and integrated it into a specific site on a CHO cell chromosome using the Flp-FRT (Flp recombination target site) system (16). The resulting structure is equivalent to that in the yeast system. An amplification marker, a mouse dihydrofolate reductase (DHFR) gene, provides MTX resistance when amplified.
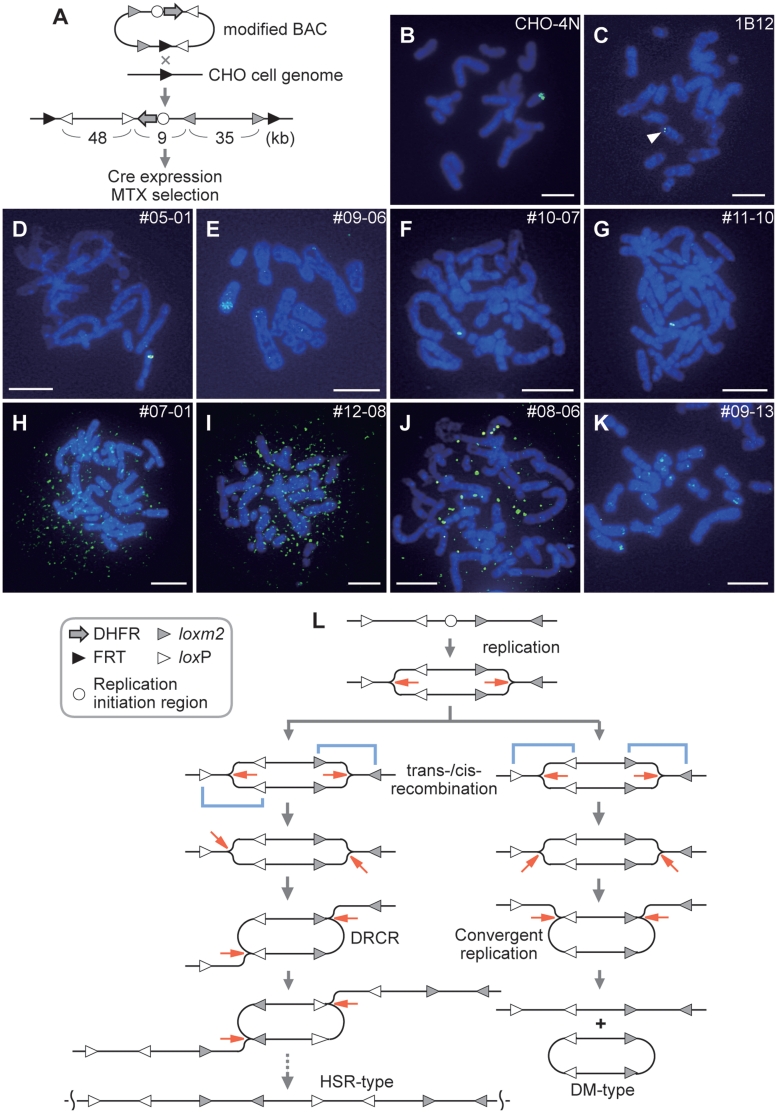
Figure 3.
Gene amplification in CHO cells induced by Cre recombination. See details in the text and Supplementary Materials and Methods. (A) Structure of the modified BAC and construction of the CHO strain for gene amplification. The sizes (kb) of the three regions in the structure are indicated below. (B–K) Metaphase FISH analysis with FITC-labeled probes (green). The CHO DR1000L-4N (CHO-4N) strain (B) that contains approximately 170 copies of DHFR was probed with a pSV2-dhfr plasmid. The BAC-CHO strain (C; 1B12) without Cre induction and MTX selection and MTX-resistant clones (D–K) were probed with the BAC in (A). DNA is counterstained with DAPI (blue). The scale bars represent 10 µm. These amplified products would be derived from the integrated BAC construct because the BAC probe could not detect the original DHFR locus (Supplementary Texts). (L) A model for the HSR amplification and DMs production by Cre recombination coupled with replication. See details in the text.
This CHO-derived cell line, designated BAC-CHO, was transiently transfected with a Cre expression vector, pPGK-Cre and grown under selection for resistance to MTX. Cre-induced cells showed an increased expression of a reporter gene located in the BAC construct (Materials and Methods in Supplementary Data). The MTX-resistant cells were then screened and cloned. Two foci derived from the BAC-CHO strain without Cre-induction showed very slow growth in the concentration of MTX, consequently resulting in growth arrest. To determine whether the MTX-resistant clones had undergone gene amplification, we used FISH on metaphase spreads with the BAC construct probe. Controls for the FISH detection of chromosomal amplification were CHO DR1000L-4N (CHO-4N) cells that contain approximately 170 copies of DHFR (Figure 3B); the BAC-CHO line (1B12) tested before Cre induction acted as a control for cells with a single BAC copy (Figure 3C). In the samples of MTX-resistant clones analyzed (Supplementary Table S1); three characteristic types of product were observed. The first type of products (Figure 3D–G) exhibited localization of fluorescent signals similar to the CHO-4N strain (Figure 3C). These cells appear to have amplified a region within the chromosome, indicating HSR amplification. Some of the HSR-type cells showed approximately a 20–50-fold increase in fluorescence intensity (Supplementary Figure S4). Detailed analysis of some of the HSR-type clones showed that the amplified regions contained the BAC sequence but that this was associated with chromosome rearrangements (Watanabe T., Horiuchi T., unpublished data). The second type of products (Figure 3H–J) were characterized by fluorescent signals dispersed over the nuclei, and are presumed to represent extrachromosomal products, DMs. The third type of products (Figure 3K) yielded scattered signals on chromosomes similar to those seen in cancer cells. The formation of HSR/DM-type products can be explained by Cre recombination coupled with replication in two alternative ways, by trans- or cis-recombination, which can induce either DRCR or convergent replication, respectively (Figure 3L). The scattered-type amplification may be generated by reintegration of DM-type products into ectopic chromosomes through interspersed repetitive elements.
DISCUSSION
The data described above indicate that the Cre-lox system can amplify any selectable genes via DRCR in both yeast and mammalian cells and further show scattered-type products resembling those seen in caner cells. Previously, a distinct recombinational process induced by chromosomal breaks was designed to induce DRCR and consequently produced HSR/DM-type products in yeast (15). These findings strongly suggest that DRCR is directly involved in the amplification events. In gene amplification in mammalian cells, BFB cycles would form megabase-sized inverted repeats, which may induce DRCR if homology-based recombination is coupled with DNA replication. Recently, a similar process, replication template exchange, was reported to lead to acentric or dicentric chromosome formation in yeast, indicating an important contribution to genome instability (22,23). We propose that such processes can occur in cultured cells and tumor cells through genome instability associated with deregulated replication (24,25). We have recently found that two pairs of inverted repeats (∼3 kb) induced HSR/DM-type products in yeast (Watanabe T., Horiuchi T., unpublished data), similarly to the Cre-lox system, strongly suggesting DRCR can be spontaneously induced by homology-based recombination. This involvement of DRCR can explain a recent data that HSR was lengthened more rapidly than expected from BFB cycle model (26).
Our Cre-lox system can induce tissue-specific amplification of gene of interest, and therefore may allow a direct approach to examine which genetic elements contribute to oncogenesis or malignant potential in each tissue when amplified. In addition, our CHO system showed scattered-type amplification products resembling those seen in cancer cells, although in non-cancerous cell line. Thus, the system in this study should serve as a useful model for amplification of oncogenes and drug-resistance genes, and contribute to a better understanding of oncogene amplification and development of anticancer strategies in future.
DRCR-dependent inversion is an interesting phenomenon, but the mechanism remains unknown. DRCR is expected to form an unstable structure, a palindromic structure. We propose that DRCR-dependent inversion may disrupt the palindromic structure and substantially stabilize the highly repetitive array. In our yeast system, the inversions were frequently found, and the final amplification level of leu2d (approximately 100 copies) far exceeded that required for complementation (10–20 copies). Our CHO system showed an adequate magnitude of amplification, given that approximately a 20- to 50-fold increase in fluorescence intensity and chromosome rearrangements was found in the HSR-type products. Furthermore, DRCR may be associated with deletions, as well as inversions, since intensive rearrangement was found in the amplified region of our CHO system. We have recently shown that DRCR process activates inversion or deletion/duplication using yeast 2-micron plasmid and an amplification system on a yeast chromosome (27). This DRCR-dependent rearrangement may occur via interspersed repetitive elements abundant in higher eukaryotic genomes, and serve as a good model for the secondary rearrangements in mammalian gene amplification.
Gene amplification is a widely used strategy for making pharmaceutical proteins in mammalian cells (28). Since our DRCR system shares features with gene amplification seen in CHO cells, we speculate that our system could produce as much recombinant protein as a conventional CHO cell expression system. Our DRCR system can shorten the process of developing high-producing cell lines with gene amplification, and be applied to a systematic production of large variety of proteins or tissue-/time-specific amplification. Improvement of the CHO system to form longer inverted repeats that facilitate DRCR-dependent inversion may allow a much higher level of amplification. Our system may be applied to other systems, such as an artificial chromosome system (28) and transgenic animals and plants for protein production (29). Finally, a unique feature of this study is the explosive nature of the HSR-type amplification. If the amplification yields multiple mutations, this type of process may contribute importantly to gene evolution.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The Ministry of Education, Culture, Sports, Science and Technology of Japan, Grant-in-Aid for Scientific Research (18058023 and 18207013 to T.H.); Japan Society for the Promotion of Science (JSPS), Grant-in-Aid for Young Scientists (18770161 and 21770194 to T.W.); Grant for Research Projects from Hayama Center for Advanced Studies (to T.H., H.T. and T.W.); Support in part by the Center for the Promotion of Integrated Sciences (CPIS) of Sokendai (to T.H., H.T. and T.W.). Funding for open access charge: JSPS, Grant-in-Aid for Young Scientists (21770194) and Support in part by CPIS of Sokendai.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank J. Roth (University of California, Davis) for critical reading of the article, D. Butler (Montana State University-Billings) for providing a yeast strain, T. Omasa (Osaka University) for providing the CHO DR1000L-4N strain and N. Shimizu (Hirosima University) for providing a plasmid. We thank Cell Biology Research Facility, NIBB Bioresource Center and Spectrography and Bioimaging Facility, NIBB Core Research Facilities for technical support.
REFERENCES
- 1.Lengauer C, Kinzler KW, Vogelstein B. Genetic instabilities in human cancers. Nature. 1998;396:643–649. doi: 10.1038/25292. [DOI] [PubMed] [Google Scholar]
- 2.Albertson DG. Gene amplification in cancer. Trends Genet. 2006;22:447–455. doi: 10.1016/j.tig.2006.06.007. [DOI] [PubMed] [Google Scholar]
- 3.Schimke RT. Gene amplification in cultured animal cells. Cell. 1984;37:705–713. doi: 10.1016/0092-8674(84)90406-9. [DOI] [PubMed] [Google Scholar]
- 4.Devonshire AL, Field LM. Gene amplification and insecticide resistance. Annu. Rev. Entomol. 1991;36:1–23. doi: 10.1146/annurev.en.36.010191.000245. [DOI] [PubMed] [Google Scholar]
- 5.Tower J. Developmental gene amplification and origin regulation. Annu. Rev. Genet. 2004;38:273–304. doi: 10.1146/annurev.genet.37.110801.143851. [DOI] [PubMed] [Google Scholar]
- 6.Roth JR, Kugelberg E, Reams AB, Kofoid E, Andersson DI. Origin of mutations under selection: the adaptive mutation controversy. Annu. Rev. Microbiol. 2006;60:477–501. doi: 10.1146/annurev.micro.60.080805.142045. [DOI] [PubMed] [Google Scholar]
- 7.Kimura M, Ota T. On some principles governing molecular evolution. Proc. Natl Acad. Sci. USA. 1974;71:2848–2852. doi: 10.1073/pnas.71.7.2848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Debatisse M, Malfor B. In: Genome Instability in Cancer Development. Nigg EA, editor. Netherlands: Springer; 2005. pp. 343–361. [Google Scholar]
- 9.Tanaka H, Yao MC. Palindromic gene amplification–an evolutionarily conserved role for DNA inverted repeats in the genome. Nat. Rev. Cancer. 2009;9:216–224. doi: 10.1038/nrc2591. [DOI] [PubMed] [Google Scholar]
- 10.Mondello C, Smirnova A, Giulotto E. Gene amplification, radiation sensitivity and DNA double-strand breaks. Mutat. Res. 2010;704:29–37. doi: 10.1016/j.mrrev.2010.01.008. [DOI] [PubMed] [Google Scholar]
- 11.McClintock B. The stability of broken ends of chromosomes in Zea Mays. Genetics. 1941;26:234–282. doi: 10.1093/genetics/26.2.234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smith KA, Stark MB, Gorman PA, Stark GR. Fusions near telomeres occur very early in the amplification of CAD genes in Syrian hamster cells. Proc. Natl Acad. Sci. USA. 1992;89:5427–5431. doi: 10.1073/pnas.89.12.5427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Toledo F, Le Roscouet D, Buttin G, Debatisse M. Co-amplified markers alternate in megabase long chromosomal inverted repeats and cluster independently in interphase nuclei at early steps of mammalian gene amplification. EMBO J. 1992;11:2665–2673. doi: 10.1002/j.1460-2075.1992.tb05332.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ma C, Martin S, Trask B, Hamlin JL. Sister chromatid fusion initiates amplification of the dihydrofolate reductase gene in Chinese hamster cells. Genes Dev. 1993;7:605–620. doi: 10.1101/gad.7.4.605. [DOI] [PubMed] [Google Scholar]
- 15.Watanabe T, Horiuchi T. A novel gene amplification system in yeast based on double rolling-circle replication. EMBO J. 2005;24:190–198. doi: 10.1038/sj.emboj.7600503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Stark WM, Boocock MR, Sherratt DJ. Catalysis by site-specific recombinases. Trends Genet. 1992;8:432–439. [PubMed] [Google Scholar]
- 17.Volkert FC, Broach JR. Site-specific recombination promotes plasmid amplification in yeast. Cell. 1986;46:541–550. doi: 10.1016/0092-8674(86)90879-2. [DOI] [PubMed] [Google Scholar]
- 18.Burke D, Dawson D, Stearns T. Methods in Yeast Genetics. New York: Cold Spring Harbor Laboratory Press; 2000. [Google Scholar]
- 19.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. 2nd edn. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 20.Tanabe H, Nakagawa Y, Minegishi D, Hashimoto K, Tanaka N, Oshimura M, Sofuni T, Mizusawa H. Human monochromosome hybrid cell panel characterized by FISH in the JCRB/HSRRB. Chromosome Res. 2000;8:319–334. doi: 10.1023/a:1009283529392. [DOI] [PubMed] [Google Scholar]
- 21.Katou Y, Kanoh Y, Bando M, Noguchi H, Tanaka H, Ashikari T, Sugimoto K, Shirahige K. S-phase checkpoint proteins Tof1 and Mrc1 form a stable replication-pausing complex. Nature. 2003;424:1078–1083. doi: 10.1038/nature01900. [DOI] [PubMed] [Google Scholar]
- 22.Mizuno K, Lambert S, Baldacci G, Murray JM, Carr AM. Nearby inverted repeats fuse to generate acentric and dicentric palindromic chromosomes by a replication template exchange mechanism. Genes Dev. 2009;23:2876–2886. doi: 10.1101/gad.1863009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Paek AL, Kaochar S, Jones H, Elezaby A, Shanks L, Weinert T. Fusion of nearby inverted repeats by a replication-based mechanism leads to formation of dicentric and acentric chromosomes that cause genome instability in budding yeast. Genes Dev. 2009;23:2861–2875. doi: 10.1101/gad.1862709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Branzei D, Foiani M. Maintaining genome stability at the replication fork. Nat. Rev. Mol. Cell. Biol. 2010;11:208–219. doi: 10.1038/nrm2852. [DOI] [PubMed] [Google Scholar]
- 25.Aguilera A, Gomez-Gonzalez B. Genome instability: a mechanistic view of its causes and consequences. Nat. Rev. Genet. 2008;9:204–217. doi: 10.1038/nrg2268. [DOI] [PubMed] [Google Scholar]
- 26.Harada S, Sekiguchi N, Shimizu N. Amplification of a plasmid bearing a mammalian replication initiation region in chromosomal and extrachromosomal contexts. Nucleic Acids Res. 2011;39:958–969. doi: 10.1093/nar/gkq882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Okamoto H, Watanabe T, Horiuchi T. Double rolling circle replication (DRCR) is recombinogenic. Genes Cells. 2011;16:503–513. doi: 10.1111/j.1365-2443.2011.01507.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cacciatore JJ, Chasin LA, Leonard EF. Gene amplification and vector engineering to achieve rapid and high-level therapeutic protein production using the Dhfr-based CHO cell selection system. Biotechnol. Adv. 2010;28:673–681. doi: 10.1016/j.biotechadv.2010.04.003. [DOI] [PubMed] [Google Scholar]
- 29.Dove A. Uncorking the biomanufacturing bottleneck. Nat. Biotechnol. 2002;20:777–779. doi: 10.1038/nbt0802-777. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.