Abstract
During the development of the vertebrate embryo, genes encoding components of the Notch signaling pathway are required for subdividing the paraxial mesoderm into repeating segmental structures, called somites. These genes are thought to act in the presomitic mesoderm when cells form prospective somites, called somitomeres, but their exact function remains unknown. To address this issue, we have identified two novel genes, called ESR-4 and ESR-5, which are transcriptionally activated in the somitomeres of Xenopus embryos by the Su(H)-dependent Notch signaling pathway. We show that the expression of these genes divides each somitomere into an anterior and posterior half, and that this pattern of expression is generated by a mechanism that actively represses the expression of the Notch pathway genes when paraxial cells enter a critical region and form a somitomere. Repression of Notch signaling during somitomere formation requires a negative feedback loop and inhibiting the activity of genes in this loop has a profound effect on somitomere size. Finally we present evidence that once somitomeres form, ESR-5 mediates a positive feedback loop, which maintains the expression of Notch pathway genes. We propose a model in which Notch signaling plays a key role in both establishing and maintaining segmental identity during somitomere formation in Xenopus embryos.
Keywords: Notch, segmentation, Xenopus, Delta, bHLH, ESR
One of the major events in vertebrate embryonic development is the subdivision of the paraxial mesoderm into repeating, metameric units, called somites (for review, see Gossler and de Angelis 1998). Somite formation begins rostrally and proceeds caudally, passing wave like down the axis of the embryo as successive groups of cells segregate from the paraxial mesoderm to form a somitic unit at regular intervals. Somite formation is preceded by processes that occur within the presomitic mesoderm in which cells are first grouped into prospective somites, called somitomeres. These processes are poorly understood but presumably consist of patterning mechanisms that assign cells to a segmental unit, impose an anterior–posterior polarity on the segment, and specify segmental boundaries. How these patterning mechanisms generate a segmental pattern within the paraxial mesoderm is a fundamental question in vertebrate embryology.
One mechanism that appears to contribute to segmental patterning in vertebrates is cell–cell interactions using the Notch signaling pathway. The Notch receptors are large transmembrane proteins that are bound and activated by a family of related ligands, the Delta–Serrate–Lag2 (DSL) proteins (for review, see Artavanis-Tsakonas et al. 1995). Activation of the receptors leads to a proteolytic liberation of the intracellular domain (ICD), which translocates into the nucleus and forms a complex with an evolutionarily conserved, DNA-binding protein, referred to here as Su(H) but also known as CBF-1, RBP-Jk, and lag-1 in various species (for review, see Chan and Jan 1998). Association with Notch ICD converts Su(H) from a transcriptional repressor to a transcriptional activator of Notch target genes. The Notch signaling pathway has been found to underlie a variety of patterning events that occur during embryogenesis, often by controlling a diverse set of cell decisions made during the development of the same tissue. Notch signaling is used, for example, in the developing Drosophila imaginal wing disc to induce cells to form the wing margin, to control the size of the wing vein primordia, to select out sensory organ precursors (SOP), and to specify the fate of SOP descendants (for review, see Muskavitch 1994). Many, if not all, of these developmental decisions involve transducing a signal via Su(H), indicating that this relatively simple pathway must activate Notch target genes with diverse roles in tissue patterning, although the basis of this diversity remains unclear.
Studies carried out in a variety of vertebrate species have shown that the Notch signaling pathway plays a key role in somitogenesis. For example, somite defects are seen in mouse embryos with mutations in the Notch1 receptor, the Notch ligands, Dll1 or Dll3, or in the mouse homolog of Su(H) (Conlon et al. 1995; Oka 1995; de Angelis et al. 1997; Kusumi et al. 1998). Similar somite defects are generated in Xenopus embryos when the Notch signaling pathway is blocked by ectopic expression of dominant-negative forms of X-Delta-2, or of Xenopus Su(H) [X-Su(H)] (Jen et al. 1997). Finally, somite defects are also generated by inappropriate activation of the Notch signaling pathway, for example, when the Delta homologs are misexpressed in Xenopus and zebrafish embryos (Dornseifer et al. 1997; Jen et al. 1997). Several lines of evidence indicate that Notch signaling is used within the presomitic mesoderm during segmentation, and that this requires segmental activation of Notch signaling along the anterior–posterior (AP) axis of the paraxial mesoderm. For instance, in Xenopus, the putative Notch ligand, X-Delta-2, is expressed in the presomitic mesoderm in a set of stripes that demarcates the anterior half of successive somitomeres (Jen et al. 1997). In the mouse, the expression of the putative Notch ligands within the presomitic mesoderm is more uniform than that in Xenopus. Nonetheless, the segmental activation of the Notch pathway appears to occur in the mouse via segmental expression of lunatic fringe, a vertebrate homolog of a Drosophila protein that restricts the activation of the Notch pathway to cells that lie at compartmental boundaries, perhaps by modifying the ability of Notch ligands to activate the Notch receptor (Irvine and Wieschaus 1994; Evrard et al. 1998; Zhang and Gridley 1998). The segmental expression of mouse lunatic fringe prefigures and overlaps the expression of HES-5, a potential Notch target gene. Embryos with a loss-of-function mutation in lunatic fringe have segmentation defects and fail to express HES-5 in the presomitic mesoderm, suggesting that segmental expression of lunatic fringe promotes segmental activation of the Notch pathway (Evrard et al. 1998; Zhang and Gridley 1998). Thus, the emerging view is that Notch signaling occurs segmentally within the presomitic mesoderm in different vertebrate embryos, and that this is part of an evolutionarily conserved mechanism that underlies segmentation.
If Notch signaling is segmentally activated within the presomitic mesoderm, how does this occur, and does it occur the same way in different vertebrate species? A significant insight into this question comes from the analysis of a gene in the chick, c-hairy1, whose expression oscillates within the presomitic mesoderm with a regular periodicity of 90 min (the formation time of a single somite) (Palmeirim et al. 1997). The oscillating expression of c-hairy1, which occurs even in the absence of protein synthesis, fits the expectations for a molecular clock that determines the number of cells assigned to each segmental unit (for review, see Stern and Vasiliauskas 1998). Subsequent analysis indicated that the expression of lunatic fringe, the putative modulator of Notch signaling, also oscillates with the same periodicity in the presomitic mesoderm of chick and mouse embryos, although this oscillation requires protein synthesis (Forsberg et al. 1998; McGrew et al. 1998). Thus, these observations link the molecular clock in the form of c-hairy1 expression to the periodic activation of the Notch pathway within the presomitic mesoderm via lunatic fringe. The nature of this link, however, is not fully understood, nor is it clear whether this mechanism applies to all vertebrates. How segmental activation of the Notch pathway is achieved in different vertebrate species, and whether this occurs via an evolutionarily conserved mechanism remains an open question.
To determine how the Notch signaling pathway is segmentally activated in the presomitic mesoderm of Xenopus embryos, we have isolated and analyzed two novel WRPW–bHLH genes, called ESR-4 and ESR-5, whose expression in the paraxial mesoderm is activated by the Su(H)-dependent Notch pathway. The expression of ESR-4/ESR-5 indicates that the Notch signaling pathway is activated in a broad domain of cells in the tailbud, but resolves into an ON/OFF state corresponding to A/P half-segments, respectively, when paraxial cells initiate segmentation. Several lines of evidence indicate that this on/off pattern is generated by a dynamic mechanism that represses the expression of the Notch pathway genes in the posterior half-segment. We show that this repression depends in part on a negative feedback loop in which ESR-5 acts as a transcriptional repressor. Finally, we show that whereas ESR-5 restricts the size of anterior half-segments by a negative feedback loop, it then maintains the expression of the Notch pathway genes in the somitomeric region by a positive feedback loop. These results indicate that the dynamic regulation of the Notch pathway genes is required during segmentation in Xenopus embryos for both establishing and maintaining segmental identity.
Results
Isolation of cDNAs encoding novel WRPW–bHLH proteins, ESR-4 and ESR-5
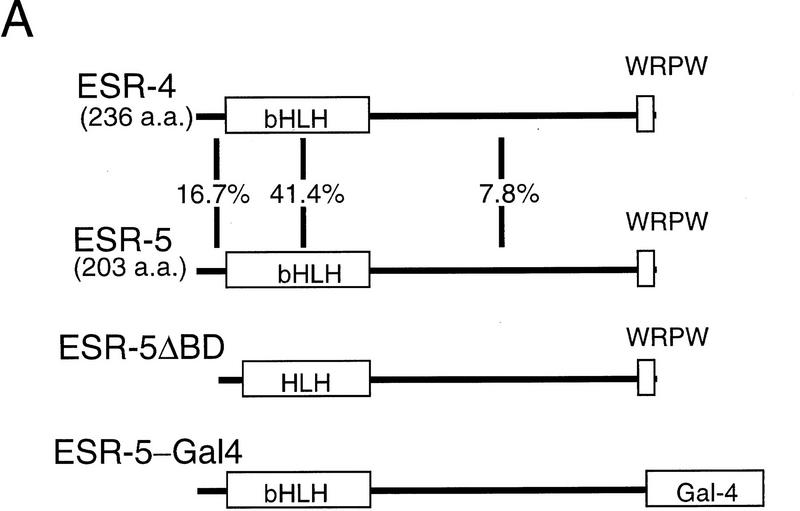
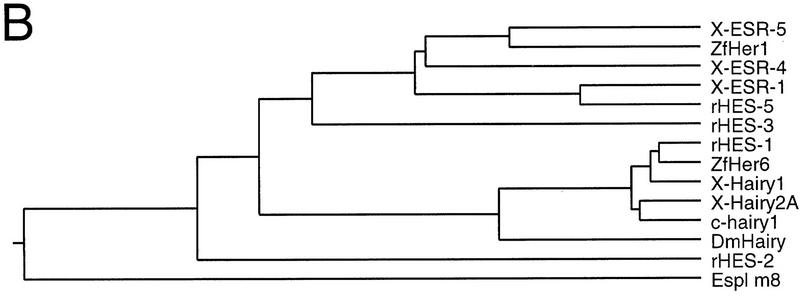
One known class of Notch target genes consists of bHLH proteins that share several characteristic structural features, namely a conserved proline in the basic region and a carboxy-terminal WRPW tetrapeptide necessary for interaction with the corepressor Groucho (Bailey and Posakony 1995). To identify potential targets of Notch signaling during segmentation, sequences corresponding to conserved regions of the WRPW–bHLH proteins were amplified from the presomitic mesoderm of stage 26 neurulae with RT–PCR. RT–PCR clones whose sequence encoded novel WRPW–bHLH proteins were used to isolate full-length cDNAs from a stage 17 library, recovering two novel, related WRPW–bHLH genes, termed ESR-4 and ESR-5. Simultaneously, in a systematic screen of genes expressed in gastrulae stage embryos by in situ hybridization, a cDNA was identified that is expressed in a stripe pattern in the presomitic mesoderm (Gawantka et al. 1998). This partial cDNA, called 32B3.1, encodes the ortholog of ESR-4. The predicted amino acid sequences of ESR-4 and ESR-5 are 41.4% similar in the bHLH domains and only 22.7% overall (Fig. 1A). By comparison to other vertebrate WRPW–bHLH proteins in the database, ESR-4 and ESR-5 are more closely related to proteins encoded by genes that are regulated by Notch signaling, such as ESR-1, than to those that are not, such as Xenopus Hairy1 and Hairy2A (Fig. 1B). Thus, we place ESR-4 and ESR-5 into the Enhancer-of-Split-Related, rather than in the Hairy-related family of WRPW–bHLH proteins. ESR-4 does not appear to be a homolog of any of the published vertebrate WRPW–bHLH proteins. ESR-5 is similar to a zebrafish protein called Her-1 (31.5% identity overall) and shares some features in terms of expression pattern, but enough differences exist to indicate that these genes are not homologs (see below; Müller et al. 1996).
Figure 1.
Structural features of ESR-4 and ESR-5. (A) ESR-4 and ESR-5 encode closely related WRPW–bHLH proteins. The two bottom diagrams illustrate the mutant forms of ESR-5 that were generated by removing the basic domain, or by replacing the WRPW with the activation domain from Gal4. (B) Phylogenetic tree showing the sequence relationships of ESR-4 and ESR-5 to vertebrate WRPW–bHLH proteins recorded in the GenBank database, including those isolated from Xenopus, ESR-1/X–Hairy1/X-Hairy2A; zebrafish, zfHer1/ZfHer6; rat, rHES-1/rHES-2/rHES-3/rHES-5; chick: c-hairy1; and Drosophila, DmHairy1/Espl (m8).
Expression of ESR-4 and ESR-5 within the presomitic mesoderm
The expression of ESR-4 and ESR-5 in early Xenopus embryos was determined by whole-mount in situ hybridization (Fig. 2A–D). This analysis indicated that both genes are expressed exclusively in the paraxial mesoderm, in contrast to other Xenopus WRPW–bHLH genes that have sites of expression in both mesodermal and ectodermal derivatives. In addition, both genes share a nearly identical pattern of expression that marks the early phases of segmentation within the presomitic mesoderm. The cardinal feature of this pattern is a domain of expression within the posterior paraxial mesoderm, referred to here as a tailbud domain (TBD), and two–three stripes of expression just anterior to the TBD (Fig. 2A–D). This pattern was evident as early as stage 12 when gastrulation is just completed, but somites have not yet formed, and continued throughout development as the paraxial mesoderm undergoes somitogenesis in a rostral–caudal direction. In double-labeling experiments, the staining patterns of ESR-4 and ESR-5 are indistinguishable, and the stripes of expression overlap (data not shown). The only marked difference in their patterns of expression is that the TBD of ESR-5 expression tends to be broader, extending more laterally and anteriorly than the TBD of ESR-4 expression (see Figs. 2C,D and 3C,I).
Figure 2.
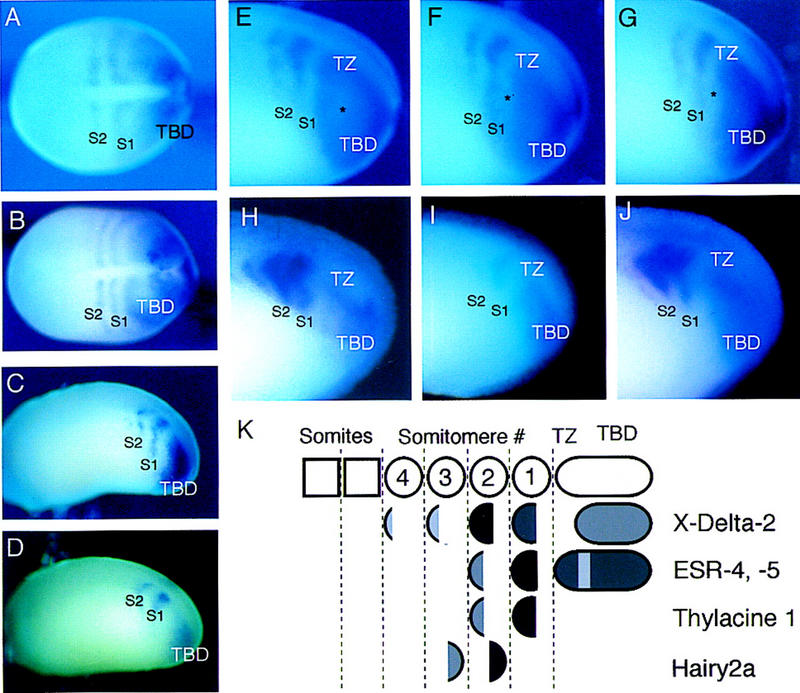
Expression of ESR-4 and ESR-5 in Xenopus embryos. (A–D) Xenopus embryos at different stages were stained for the expression of ESR-4 and ESR-5 with whole-mount, in situ hybridization. All embryos are oriented in this figure as in subsequent figures with anterior to the left. (A) Expression of ESR-5 at late gastrulae stage (st. 12, dorsal view), (B) at early neurulae stage (st. 14/15, dorsal view), and (C) at late neurulae stage (st. 22/24, side view). (D) Expression of ESR-4 at late neurulae stage (st. 22/24, side view). Note that the expression patterns of both ESR-4 and ESR-5 at these different stages consists of a TBD, and two stripes that lie anterior to the TBD in somitomere 1(S1) and 2(S2). The expression patterns of ESR-4 and ESR-5 differ only in one respect: The TBD staining for ESR-5 is more intense and tends to extend more rostrally than that of ESR-4. (C,D; also see Fig. 3C,I). (E–G) Three examples of ESR-5 staining taken from a batch of embryos processed at stage 22. Side views of the posterior third of the embryos are shown. The asterisk marks what appears to be the emergence of a gap in the TBD expression of ESR-5 in a region referred to as the TZ. (H–J) Double labeling of embryos with whole-mount in situ hybridization with probes for ESR-5 and X-Delta-2. (H) Embryo stained for X-Delta-2 alone with Magenta–Phos (MP) as the chromogen, (I) ESR-5 RNA alone with BCIP, or (J) both ESR- 5 with BCIP and X-Delta-2 with MP. Note that the ESR-5 staining in S1 overlaps with the S1 staining in X-Delta-2. (K) Diagram showing the position of ESR-4 and ESR-5 expression relative to that of other genes known to be expressed segmentally in Xenopus embryos (Jen et al. 1997; Sparrow et al. 1998). Because cells are constantly being added posteriorly to the paraxial mesoderm and somites are constantly forming anteriorly, the stripes of gene expression in the somitomeric region represent transient expression in groups of cells as they pass through this domain. For this reason, the S1-4 notation in this and other figures does not denote a fixed group of cells along the AP axis, but rather to different stages of the segmentation process that cells pass through in the paraxial mesoderm.
The stripes of ESR-4/ESR-5 expression within the presomitic mesoderm are reminiscent of several other genes whose expression in Xenopus embryos marks the formation of prospective somites, or somitomeres (Jen et al. 1997; Sparrow et al. 1998). As shown diagrammatically in Figure 2K, the expression of these genes demarcates four somitomeres (S1–S4) in the region of the presomitic mesoderm between the TBD and the most recently formed somite. X-Delta-2 (encoding a putative Notch ligand) and Thylacine1 (encoding a bHLH protein of the MesP family) are primarily expressed in the anterior half of each somitomere, whereas the expression of Hairy2A (encoding a WRPW–bHLH protein) is restricted to the posterior half (Jen et al. 1997; Sparrow et al. 1998). In addition, expression of Thylacine1 and X-Delta-2 in the anterior half of S1 precedes that of Hairy2A, which is first detected in the posterior half of S2.
To determine how the stripes of ESR-4 and ESR-5 expression correspond to the pattern of half-segments within the presomitic mesoderm, embryos were double labeled with probes for ESR-5, and for X-Delta-2, with whole-mount, in situ hybridization (Fig. 2H–J). Staining for ESR-5 and ESR-4 (data not shown) overlaps the staining of X-Delta-2, indicating that ESR-4/ESR-5 are expressed in the anterior half of the prospective somites. This interpretation was confirmed by results obtained by staining embryos for both ESR-5 and Hairy2A expression (data not shown). This analysis also showed that the most posterior stripe of ESR-4/ESR-5 expression overlaps that of X-Delta-2 and Thylacine1 expression in S1 (Fig. 2H–K), thus following the first appearance of segmental gene expression. The expression of ESR-4/ESR-5 in S1 and in the TBD marks the transition in gene expression between these two regions in a manner not evident from Thylacine1 or X-Delta-2 expression. Thylacine1 is not expressed in the TBD. X-Delta-2 is expressed in the TBD but is restricted to the most posterior portion, resulting in a large gap of expression between the TBD and S1 (Fig. 2H,K). In contrast, the gap in the expression of ESR-5 in this region is narrower, and some embryos show a staining pattern in the TBD expected for a forming gap (Fig. 2E–G, asterisk). For simplicity, we will refer to this critical region between the TBD and S1 in which the gap in the expression of ESR-4 and ESR-5 forms as the transition zone (TZ, Fig 2E–G). In summary, these observations indicate that the expression of ESR-4 and ESR-5 evolves as cells emerge from the TBD into the TZ region, closely corresponding to the formation of half-segments in S1. In addition, because ESR-4 and ESR-5 expression overlaps that of X-Delta-2 in the TBD, S1, and to some extent in the TZ, the activation of the Notch pathway may promote their expression during segmentation.
ESR-4 and ESR-5 are regulated by the Su(H)-dependent Notch pathway
To test the role of Notch signaling in promoting the expression of ESR-4 and ESR-5, we first asked whether ectopic expression of X-Delta-2 alters the expression pattern of these genes. RNA encoding X-Delta-2 was injected into the marginal zone of two-cell stage embryos, along with RNA encoding a nuclear-localized form of Β-galactosidase (nlacZ). At stage 20/22, the injected embryos were fixed, reacted with X-gal, which stains the injected side blue, and then double labeled for the expression of ESR-4 and ESR-5 by whole-mount in situ hybridization. The results show that ectopic expression of X-Delta-2 RNA expands the expression of ESR-4 (data not shown) and ESR-5 (Fig. 3A,B), but only by filling in the gaps of expression that would normally appear during segmentation.
Figure 3.
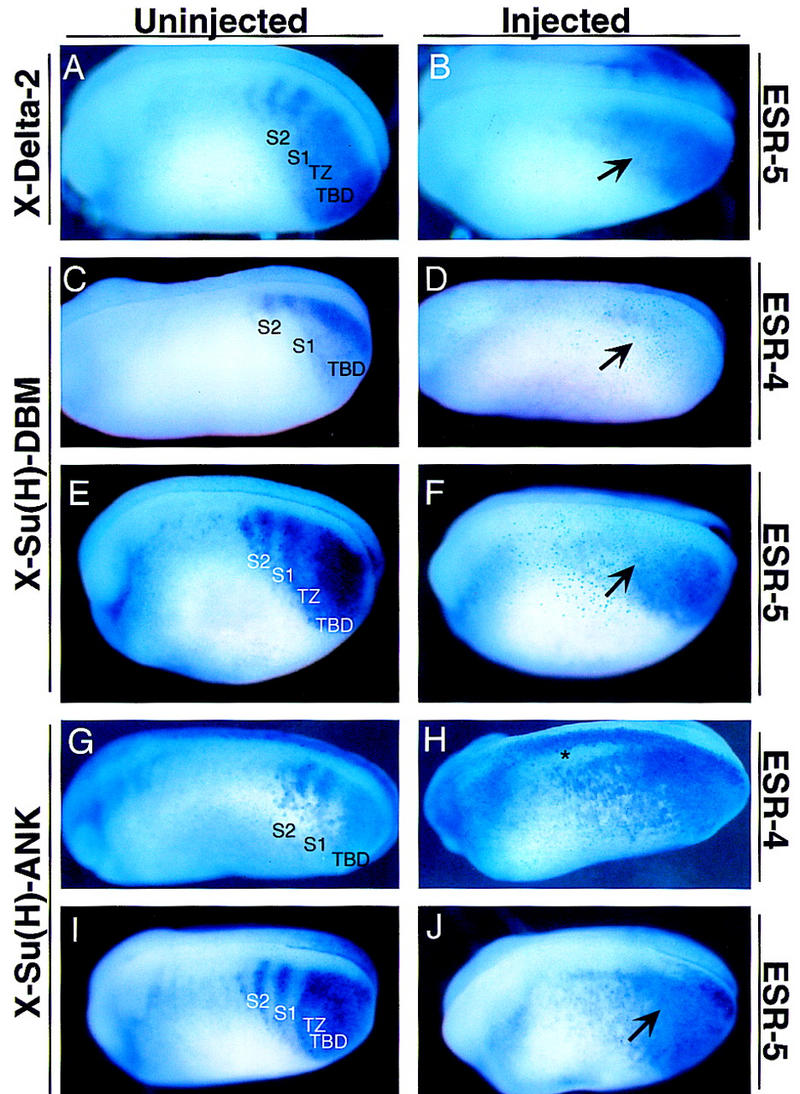
Expression of ESR-4 and ESR-5 during segmentation depends on the Notch signaling pathway. Embryos were injected on one side with RNAs encoding X-Delta-2 (A,B), X-Su(H)–DBM (C– F), or X-Su(H)–Ank (G–J), and processed for expression of ESR-4 (C,D,G,H) or for ESR-5 (A,B,E,F,I,J). A representative embryo from each injection is shown with a view of the uninjected or injected side. (A,B) Note that when embryos ectopically express X-Delta-2, expression of ESR-5 in the TZ increases (B, arrow). X-Su(H)–DBM completely blocks ESR-4 expression (D, arrow) while significantly reducing ESR-5 expression in the somitomeric and TZ region (F, arrow). X-Su(H)–Ank expands the expression of both ESR-4 (H) and ESR-5 (J, arrow). The asterisk in H marks ectopic induction of ESR-4 expression in the neural tube by X-Su(H)–Ank.
We next asked whether X-Delta-2 promotes the expression of ESR-4 and ESR-5 by activating the Su(H)-dependent Notch pathway, because a number of WRPW–bHLH genes have been shown to contain functional Su(H)-binding sites. To address this issue, we used two forms of X-Su(H) that have been shown previously to activate or inhibit Notch target genes. One form is a fusion between X-Su(H) and the ankyrin repeats in the intracellular domain of X-Notch-1 [X-Su(H)–Ank] and behaves as an activated form, whereas the other has mutations in residues required for DNA binding [X-Su(H)–DBM] and behaves as a dominant-negative (Wettstein et al. 1997). For example, these two forms of X-Su(H) have opposite effects on the expression of a Notch target gene, ESR-1, which is activated by the Notch pathway during primary neurogenesis (Wettstein et al. 1997). In addition, as discussed further below, both forms of X-Su(H) cause segmentation defects, and have opposite effects on the expression of X-Delta-2 and Thylacine1 in the presomitic mesoderm (Sparrow et al. 1998).
The results in Figure 3 show that X-Su(H)–DBM and X-Su(H)–Ank also have opposite effects on the expression of ESR-4 and ESR-5 when introduced into embryos by RNA injection. X-Su(H)–DBM completely eliminates the expression of ESR-4 in both the TBD and the somitomeric region (Fig. 3D). Similarly, X-Su(H)–DBM blocks the expression of ESR-5 in the somitomeric region and in the TZ, but does not affect the expression of ESR-5 in the posterior TBD (Fig. 3F). In contrast, X-Su(H)–Ank expands the expression of both ESR-4 and ESR-5 into the somitomeric region, thus eliminating the gaps in the striped expression pattern, as seen in embryos ectopically expressing X-Delta-2 RNA (Fig. 3H,J). Because X-Su(H)–Ank was shown previously to block the expression of X-Delta-2 in the paraxial mesoderm, its effects on ESR-4 and ESR-5 expression seen here cannot be explained by an increase in ligand expression (Sparrow et al. 1998). In addition, the strong effects of X-Su(H)–Ank on ESR-4 and ESR-5 expression also suggest a direct action. For example, X-Su(H)–Ank induces ectopic expression of ESR-4, and to a lesser extent ESR-5, in the neural tube in which neither gene is normally expressed (asterisk in Fig. 3H). Thus, these results indicate that the expression of both ESR-4 and ESR-5 during segmentation is regulated, perhaps directly, by the Su(H)-dependent Notch signaling pathway. ESR-4 expression is promoted by the Notch pathway in the TBD/TZ and somitomeric region. ESR-5 expression is promoted by the Notch signaling only in the TZ and the somitomeric region, whereas other factors appear to promote its expression in the TBD.
Segmental expression of ESR-4 and ESR-5 is regulated by a mechanism intrinsic to the TBD
The results described above show that, as cells emerge from the TBD into the TZ, the expression of ESR-4/ESR-5 takes on a segmental pattern. To analyze the mechanism responsible for generating this pattern, we first asked whether it can arise autonomously by cells within the TBD. This issue was addressed by removing the TBD from stage 24 embryos, and determining whether the cultured TBD regenerates the half-segmental pattern of ESR-5 expression in isolation. The known expression pattern of ESR-5 in similarly staged embryos was used as a guide to estimate the line of dissection to separate away the TBD. Dissected TBDs were either fixed immediately, or fixed after 4 hr in culture, and then processed for ESR-5 staining (Fig. 4). The expression of ESR-5 in the tailbuds fixed immediately after dissection confirmed that in a vast majority of cases, the TBD was isolated away from the rest of the paraxial mesoderm (Fig. 4; Table 1). In the cultured TBDs, a majority also regenerated a segmental pattern of ESR-5 expression (Table 1). Similar results were obtained for ESR-4 (data not shown). Thus, these results indicate that a mechanism acting within the TBD/TZ can autonomously regulate the Notch pathway in a manner that leads to a segmental expression of ESR-4 and ESR-5.
Figure 4.
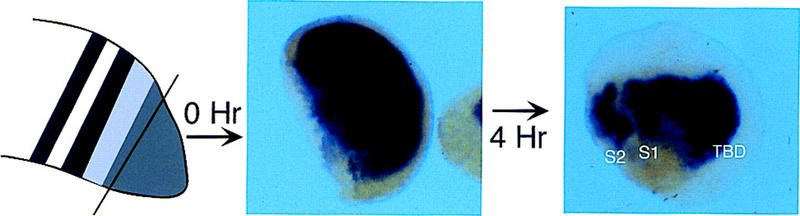
Segmental expression of ESR-5 is generated by mechanisms intrinsic to TBD. The TBD was dissected from a collection of embryos and either fixed immediately (0 hr) or left for 4 hr in culture and then fixed (4 hr). After staining for ESR-5 expression, embryos were scored on the basis of the presence of stripes and gaps (see Table 1). Shown are representative samples after clearing in benzyl benzoate. Identical results were obtained for ESR-4.
Table 1.
Expression of ESR-5 in isolated tailbuds
|
|
Indexa
|
Experiment (%)
|
|||
|---|---|---|---|---|---|
|
1
|
2
|
||||
| TB(Δt = 0) | 0 | 100 | (n = 26) | 95 | (n = 22) |
| 1 | 0 | 5 | |||
| TB(Δt = 4 hr) | 0 | 17 | (n = 30) | 14 | (n = 28) |
| 1 | 23 | 29 | |||
| 2 | 33 | 36 | |||
| 3 | 23 | 7 | |||
| N.S. | 3 | 14 | |||
The TBD was removed form tadpole embryos and either fixed immediately at time 0 or cultured for 4 hr.
After processing for ESR-5 staining, the staining pattern was scored using the following criteria: (0) Tailbud expression only; (1) tailbud expression + gap 1; (2) tailbud expression + gap 1 + stripe 1; (3) tailbud expression + gap 1 + stripe 1 + gap 2 + stripe 2. Examples of 0 and 3 are shown in Fig. 5. Results are shown for two separate experiments. (N.S.) No staining.
Segmental expression of ESR-4, ESR-5, and X-Delta-2 is rapidly altered by inhibitors of protein synthesis
The mechanism that produces the segmental pattern of ESR-4 and ESR-5 could, in principle, act by activating the Notch pathway in cells that form anterior half-segments, or alternatively by inhibiting the pathway in cells that will form posterior half-segments. Moreover, this mechanism could act by regulating the expression of ESR-4/ESR-5 directly, or indirectly by regulating other components of the pathway such as the expression of X-Delta-2. To examine these various possibilities, we asked how the expression of these genes change after a brief exposure to the protein synthesis inhibitor, cycloheximide (CHX). This approach was motivated by experiments in the chick, in which brief CHX treatment does not alter the expression of the Hairy oscillator but instead alters the expression of lunatic fringe, suggesting that the oscillator is upstream of lunatic fringe, which in turn, appears to be linked to the segmental activation of the Notch pathway (McGrew et al. 1998). Accordingly, embryos were treated for 1.0 or 2.0 hr with CHX, and then fixed and stained for the expression of the ESR genes, X-Delta-2 and Thylacine1.
Of the genes examined, the expression of ESR-4 (data not shown) and ESR-5 (Fig. 5) changed the most dramatically in response to a brief CHX treatment. After 1 hr of CHX treatment (the time to form one somite in Xenopus), the expression levels of both ESR-4 and ESR-5 increased significantly in the TBD. Moreover, in the TZ, the nascent gap that normally occurs in the expression of ESR-4 and ESR-5 was filled in (Fig. 5B), whereas the more anterior stripe and gap in S2 remained intact (Fig. 5B, S2). The expression of ESR-4/ESR-5 after 2.0 hr of CHX treatment was similar to that after 1.0 hr, except that the gap of expression in S2 was now reduced as well (Fig. 5C). Together, these results indicate that when cells pass from the TBD through the TZ, de novo protein synthesis is not required for activating, but rather repressing, the expression of ESR-4 and ESR-5. Moreover, de novo protein synthesis appears to be required for generating the gap in ESR-4/ESR-5 expression as cells emerge from the TBD to form S1. Once this gap forms, it apparently persists even in the absence of de novo protein synthesis.
Figure 5.
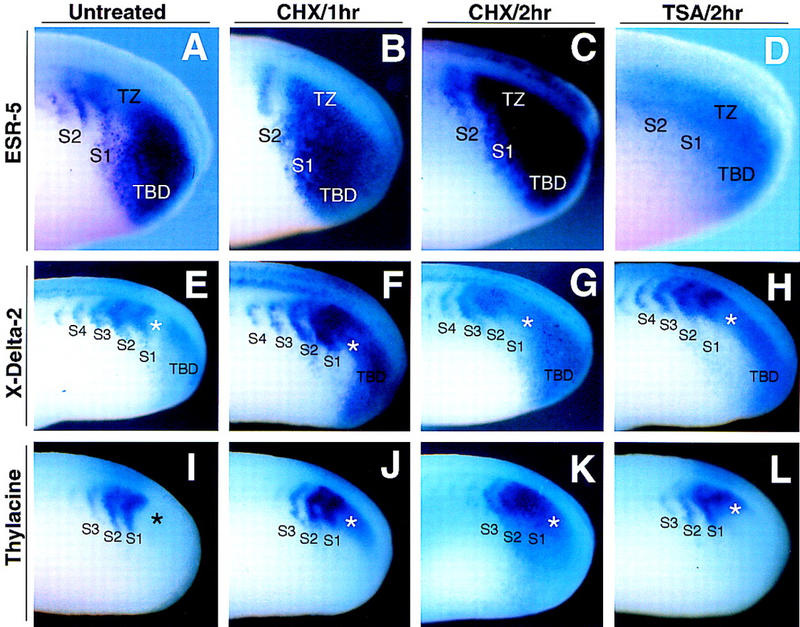
CHX and TSA treatment rapidly alter the segmental expression of ESR-5, X-Delta-2, and Thylacine1. Xenopus embryos at early neurulae stages were left untreated (A,E,I), or treated for 1 hr (B,F,J) or 2 hr (C,G,K) with CHX, or for 2.0 hr with TSA (D,H,L). Embryos were then fixed and stained for ESR-5 expression (A–D), X-Delta-2 expression (E–H) or for Thylacine1 expression (I–L). A side, posterior view of the stained embryos is shown. Note that following 1 hr of CHX treatment, ESR-5, X-Delta-2, and Thylacine1 expression changes by upregulating in the TZ (asterisks), whereas the striped expression of X-Delta-2, Thylacine1, and ESR-5 in the somitomeric region is relatively unaffected. Similar effects on the expression of these genes in the TZ also occur after 2.0 hr of TSA treatment.
Expression of X-Delta-2 also changed rapidly after treatment with CHX, in a manner that mirrored the changes in ESR-4/ESR-5 expression described above. After 1.0 hr of CHX treatment, the expression of X-Delta-2 in the TBD increased significantly, and the gap in expression normally found in the TZ was filled in (Fig. 5F, asterisk), whereas the expression of X-Delta-2 in the somitomeric region was relatively unaffected (Fig. 5F, S2–S4). After 2.0 hr of CHX treatment, the expansion of X-Delta-2 expression occurred in the TBD, S1, and perhaps in S2, but not in S3 and S4 (Fig. 5G). In Figure 5E, the cells in S3 and S4 at 2 hr in Figure 5G correspond to the same cells in S1 and S2 at 0 hr, respectively. Thus, these results indicate that de novo protein synthesis is required to repress X-Delta-2 expression in posterior half-segments as cells pass through the TZ and undergo segmentation. In addition, once a gap in X-Delta-2 expression forms, it apparently can persist in the somitomeric region in the absence of de novo protein synthesis. We note that the effects of CHX treatment on X-Delta-2 expression in the TZ could account for the increased expression of ESR-4 and ESR-5 that occurs following CHX treatment as described above.
Thylacine1 is a member of the MesP family of bHLH transcriptional activators (Saga et al. 1996, 1997; Sparrow et al. 1998) whose expression overlaps that of X-Delta-2 and ESR-4/ESR-5 in the anterior half of somitomere 1 and 2 (Fig. 2). Because Thylacine1 acts as a positive regulator of X-Delta-2 and ESR-5 expression (Sparrow et al. 1998), an increase in Thylacine1 expression might conceivably underlie the changes in X-Delta-2 expression that occur in response to CHX treatment. In agreement with this prediction, the expression of Thylacine1 primarily changed after 1.0 hr of CHX treatment by expanding into the TZ, extending posteriorly from its normal domain of expression in S1 (Fig. 5J, asterisk). This change corresponds directly with the observed changes in X-Delta-2 and ESR-4/ESR-5 expression in the TZ in embryos treated with CHX for 1.0 hr (Fig. 5F, B). As described for X-Delta-2, Thylacine1 expression did not change significantly in S2 or S3, following the brief treatment with CHX. After 2.0 hr of CHX treatment, Thylacine1 expression increased further by expanding into the TZ, extending even further into the TBD, while upregulating in both S1 and S2 (Fig. 5K). Expression as a stripe and gap in S3, however, remains relatively unaffected (Fig. 5K). Thus, Thylacine1, a positive regulator of X-Delta-2 and ESR-4/ESR-5 expression, appears to be upregulated rapidly in response to CHX, particularly in the critical TZ, in which periodic repression of the Notch pathway results in a half-segmental pattern of gene expression. In contrast, once the stripes and gaps of Thylacine1 expression form in the TZ and S1, they appear to persist in the absence of de novo protein synthesis in the somitomeric region.
Segmental expression of ESR-4 and ESR-5 is rapidly altered by inhibitors of histone deacetylase activity
One mechanism that could account for the results above is that de novo protein synthesis is required for producing a transcriptional repressor(s) in the TZ that blocks the expression of X-Delta-2, Thylacine1, and/or ESR-4/ESR-5 when a posterior half-segment forms. Many transcriptional repressors act by recruiting corepressors that contain histone deacetylase activity (for review, see Struhl 1998). Thus, we asked whether the initial pattern of segmental expression depended on the activity of these corepressors, by treating embryos with the drug trichostatin-A (TSA) to inhibit the activity of histone deacetylases. In embryos treated with TSA, the expression of both ESR-4 (data not shown) and ESR-5 (Fig. 5D) does not show the overall increase in intensity, but may even decrease in levels, for example in the TBD, as in the case for embryos treated with CHX (Fig. 5C). Nonetheless, both ESR-4 and ESR-5 expression responds by increasing within the gaps, with a complete loss of the gaps by 2 hr of treatment (Fig. 5D). Similarly, in TSA treated embryos, X-Delta-2 expression also expands into the gap between the TBD and S1, with little changes in expression in the somitomeric region (Fig. 5H). Finally, the expression of Thylacine1 is also upregulated after 2.0 hr of TSA treatment in the region between the TBD and S1 (Fig. 5L). Thus, a brief treatment with TSA does not significantly perturb gene expression in the somitomeric region, but rather in the TZ, in which the first gap appears in the segmental expression pattern of ESR-4/ESR-5, X-Delta-2 and Thylacine1.
Changes in ESR-5 activity alters segmentation
The results of the drug experiments described above suggest that repression of the Notch pathway genes occurs in the TZ, thus establishing their segmental pattern of expression. Results from previous studies also suggested that Notch signaling is repressed in the presomitic mesoderm by a negative feedback loop, because activators of the Notch pathway, such as ICD or X-Su(H)–Ank, repress the expression of X-Delta-2 in the presomitic mesoderm, whereas inhibitors of Notch signaling, such as X-Su(H)–DBM, expand it (Jen et al. 1997; Sparrow et al. 1998). As WRPW–bHLH genes are part of a negative feedback loop during Notch signaling in Drosophila (see, e.g., Heitzler et al. 1996; Huppert et al. 1997), we next asked whether ESR-5 functions as a repressor of Notch pathway genes during segmentation.
To inhibit the activity of ESR-5 in embryos, we generated a mutant form of ESR-5 lacking the basic domain required for DNA binding, called ESR–5ΔBD. Similar mutants of other WRPW–bHLH proteins behave as dominant-negative mutants, presumably because they form nonfunctional homodimers (Fig. 1A) (Jimenez et al. 1996; Giebel and Campos-Ortega 1997). In addition, a second mutant of ESR-5 was generated by replacing the terminal WRPW that interacts with the corepressor Groucho with the activation domain from Gal4. As a first assay, embryos were injected on one side with RNAs encoding ESR-5, ESR-5ΔBD, or ESR-5–Gal4, and examined morphologically at late neurulae stages for tissue defects. Embryos injected with these RNAs gastrulated normally, and formed a normal axis, suggesting that neither the wild-type nor mutant forms of ESR-5 have general nonspecific effects on development. In Xenopus, almost all of the paraxial mesoderm gives rise to myotomal cells that span each somitic unit. In embryos expressing ESR-5, ESR-5ΔBD, or ESR-5–Gal4, the differentiation of paraxial mesoderm into myotomal tissue occurred normally as determined by its morphology and staining for a muscle antigen 12/101 (Fig. 6). However, the somites in these embryos failed to organize into a segmental pattern, showing defects that were indistinguishable from those observed in the previous experiments involving Notch perturbations (Fig. 6A–F; Jen et al. 1997).
Figure 6.
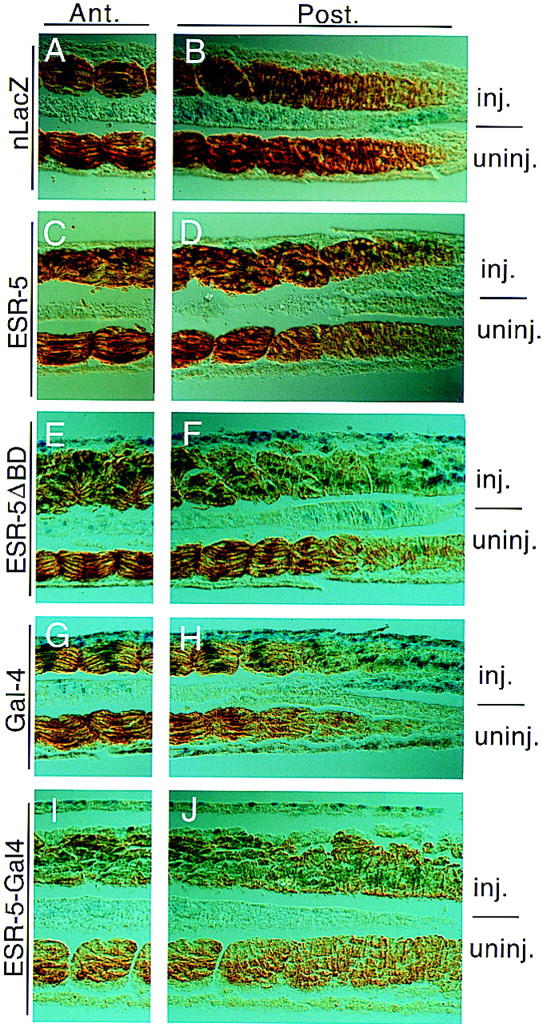
Altered activity of ESR-5 produces somite defects in Xenopus embryos. Embryos were injected with lacZ RNA alone (A,B) or along with RNAs encoding ESR-5 (C,D), ESR-5ΔBD (E,F), Gal-4 (G,H), or ESR-5–Gal4 (I,J). Embryos were processed at tadpoles stages by fixation, then staining with X-gal and 12/101. Shown are longitudinal sections of the processed embryos, in which the right panels are taken from the region of the paraxial mesoderm where somites are just forming, whereas the left panels are taken from a region more anterior. Note that in embryos injected with lacZ alone (A,B) or with Gal4 as a control (G,H), expression of 12/101 is initiated prior to somite formation and, after somites form, the myotomal cells line up along the AP axis with their nuclei aligned along the middle of each somitic unit. In contrast, embryos injected with ESR-5 (C,D), ESR-5ΔBD (E,F), or ESR-5–Gal4 RNA (I,J), 12/101 expression is initiated on schedule, but the myotomal cells fail to organize into a segmental pattern. In the more extreme cases, such as ESR-5–Gal4, the myotomal cells do not show signs of initiating somite formation (J).
As a specificity control, we compared phenotypes produced by ESR-5ΔBD with those produced by a related Xenopus ESR gene, called ESR-6e. ESR-6e is not expressed in the presomitic mesoderm but is expressed in the skin in which it acts downstream of Notch to control the differentiation of ectodermal cells into ciliated cells (data not shown). The DNA-binding mutant of ESR-6e leads to an overproduction of ciliated cells, whose numbers are controlled by Notch signaling, but does not affect the morphology of the somites, including their segmentation (data not shown). Conversely, ESR-5ΔBD which alters segmentation (Fig. 6) does not alter the number of ciliated cells (data not shown). Thus, these data indicate that DNA-binding mutants of these two closely related WPRW-bHLH act specifically to perturb the function of these two Notch target genes in these separate contexts.
Negative feedback represses gene expression in the TZ
To test the role of negative feedback in establishing the segmental expression of Notch pathway genes, we first examined X-Delta-2 expression in embryos injected with RNA encoding ESR-5ΔBD, ESR-5–Gal4, or X-Su(H)–DBM. As shown previously, when Notch signaling is inhibited by X-Su(H)–DBM, the expression of X-Delta-2 expands in the TBD, filling in and eliminating the gap of X-Delta-2 expression in the TZ (Fig. 7F). Both ESR-5ΔBD and ESR-5–Gal4 produce a similar phenotype (Fig. 7B,D). Although similar, the ESR-5 mutants tend to fill in the gap of X-Delta-2 expression in the TZ less effectively than X-Su(H)–DBM (Fig. 7, cf., F with B and D). This difference may indicate that the ESR-5 mutants are not as effective as X-Su(H)–DBM in eliminating ESR-5 function, or that X-Su(H)–DBM blocks the expression of more genes than ESR-5, for example, ESR-4, whose function is not inhibited by the ESR-5 mutants. Nonetheless, these results indicate that ESR-5 is part of a negative feedback loop that represses the expression of X-Delta-2 in the region in which the somitomeres first form. In addition, when Notch signaling is blocked with X-Su(H)–DBM, expression of X-Delta-2 in the somitomeric region occurs in one large stripe, rather than in the normal pattern of stripes and gaps (Fig. 7F, som). Again, both ESR-5ΔBD and ESR-5–Gal4 produce the exact same phenotype (Fig. 7B,D). Thus, Notch signaling is required for repressing the expression of X-Delta-2 in posterior half-segments, and this repression involves ESR-5 acting in a negative feedback loop. To extend these results, we next examined the expression of Thylacine1 in the somitomeric region, because the results from the drug experiments indicated that this pattern also arises by repression in the TZ. As predicted, Thylacine1 expression changes in a similar way in embryos expressing RNA encoding ESR-5ΔBD, ESR-5–Gal4, or X-Su(H)–DBM, by expanding into the gaps in S1 and S2 in which expression would normally be excluded (Fig. 7H,J,L). Finally, in embryos expressing RNAs encoding ESR-5ΔBD (data not shown), or ESR-5–Gal4 (Fig. 8B), the expression of ESR-4 is derepressed in the TZ. Thus, together these results indicate that ESR-5 is part of a negative feedback loop that represses Notch pathway genes during segmentation.
Figure 7.
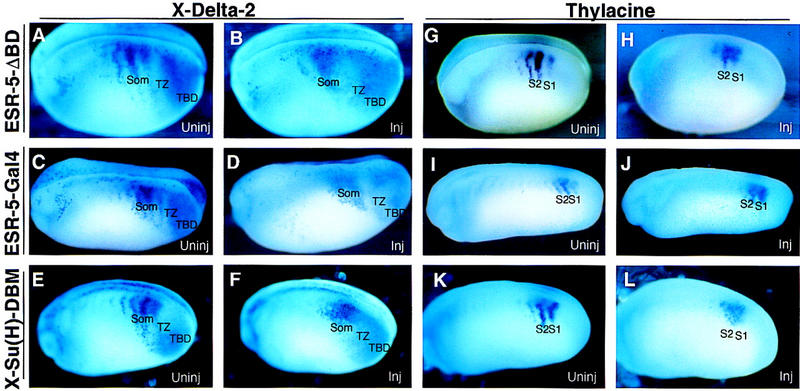
Negative feedback represses Notch pathway genes in the TZ. Embryos at the two-cell stage were injected on one side with RNAs encoding ESR-5ΔBD (A,B, G,H), ESR-5–Gal4 (C,D,I,J), or X-Su(H)–DBM (E,F,K,L), and then processed at stage 22/26 for the expression of X-Delta-2 (A–F) or Thylacine1 (G–L). A representative embryo from each injection is shown with a view of the uninjected (uninj) or injected side (inj). (Som) somitomeric.
Figure 8.
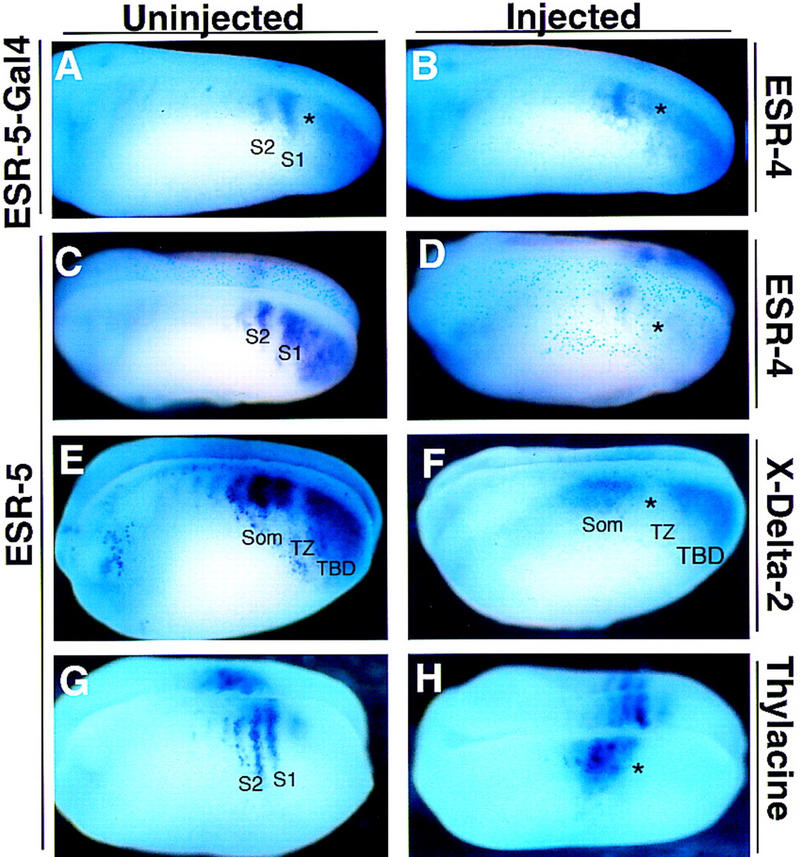
ESR-5 activity alters segmental gene expression. Embryos at the two-cell stage were injected on one side with RNAs encoding ESR- ESR-4 (A–D), X-Delta-2 (E,F), or Thylacine1 (G,H). A representative embryo from each injection is shown with a view of the uninjected or injected side.
Inhibiting Notch signaling or ESR-5 activity causes a derepression of Notch pathway genes during somitomere formation (Figs. 7 and 8). Next, we asked whether the converse is true; whether increased levels of Notch signaling or ESR-5 activity repress the expression of Notch pathway genes. Previous studies showed that agents that activated the Notch pathway, such as Notch ICD or X-Su(H)–Ank, repress the expression of X-Delta-2 and Thylacine1 in the presomitic mesoderm (Jen et al. 1997; Sparrow et al. 1998). To determine whether increased ESR-5 activity also inhibits the expression of Notch pathway genes, the expression of X-Delta-2, Thylacine1 and ESR-4 was assayed in embryos injected with the synthetic ESR-5 RNA described above. In accordance with a negative feedback model, the expression of ESR-4 was markedly reduced in embryos injected with ESR-5 RNA (Fig. 8D; asterisk). Moreover, in embryos injected with ESR-5 RNA, the expression of X-Delta-2 in the TBD diminishes, and the gap of expression in the TZ between the TBD and S1 increases (Fig. 8F, TZ). Thus, ectopic expression of ESR-5 represses the expression of the Notch pathway genes in the region of paraxial mesoderm in which the first somitomere forms.
Positive feedback promotes gene expression in the somitomeres
Activating the Notch pathway with X-Su(H)–Ank or ICD also eliminates the expression of X-Delta-2 and Thylacine1 in the somitomeric region (Jen et al. 1997; Sparrow et al. 1998). However, in embryos injected with ESR-5 RNA, the expression of X-Delta-2 in the somitomeric region is reduced, but not eliminated, and more surprisingly, shifts anteriorly and shows a loss of segmentation (Fig. 8F). Thus, ESR-5 has a repressive effect on the levels of X-Delta-2 expression in the somitomeric region, but also leads to ectopic expression anterior to its normal expression domain (Fig. 8F, som). A reason for this became apparent when we examined embryos injected with ESR-5 RNA for Thylacine1 expression (Fig. 8H). Strikingly, in embryos injected with ESR-5 RNA, the level of Thylacine1 expression is increased, mostly medially, and more significantly, the expression domain shifts anteriorly within the presomitic mesoderm (Fig. 8H, asterisk). Together, these results indicate that ESR-5 activity contributes to the repression of the Notch pathway genes that occurs when segments form in TZ. However, anterior to the TZ, ESR-5 activity appears to have an opposite role. By promoting the expression of Thylacine1, ESR-5 appears to promote the expression of X-Delta-2. Thus, these results suggest that in the TZ, ESR-5 acts in a negative feedback loop to limit the size of segments, as marked by the expression of Notch pathway genes, but then acts as part of a positive feedback loop to maintain segmental domains in the somitomeric region.
Discussion
The expression pattern of X-Delta-2 in the presomitic mesoderm was one of the first indications that segmental activation of the Notch pathway was required for somitogenesis in Xenopus embryos (Jen et al. 1997). On the basis of this pattern, the presomitic mesoderm could be divided into two regions; a somitomeric region in which X-Delta-2 is expressed in anterior half-segments, and the TBD region in which expression is unsegmented. However, on the basis of this expression pattern alone, or even the phenotypes that resulted from blocking the Notch pathway, it was still unclear when, during the process of segmentation, Notch signaling is required and what function it might serve. In other words, is Notch signaling required in the somitomeric region to maintain or elaborate further on a segmental pattern generated earlier, or is Notch signaling required even earlier, in the TBD, when an initial segmental pattern is established? To address this question, we isolated and analyzed two novel Notch target genes called ESR-4 and ESR-5. On the basis of the overlap between the expression of ESR-4/ESR-5 and X-Delta-2, as well as how the expression of these genes change in response to ectopic expression of X-Delta-2 or to the two forms of X-Su(H), we conclude that ESR-4 and ESR-5 are transcriptionally activated by the Su(H)-dependent Notch signaling pathway. As Notch target genes, the expression pattern of ESR-4 and ESR-5 confirmed that Notch signaling occurs in the TBD, and more importantly, is modulated in the TZ, a critical region between the TBD and S1 in which cells appear to initiate segmentation. These observations suggest that Notch signaling acts much earlier in the process of segmentation than appreciated previously, and led us to analyze the mechanisms that regulate Notch signaling within the TBD/TZ.
Periodic repression of ESR-4/ESR-5
On the basis of the expression of ESR-4 and ESR-5 in the presomitic mesoderm, Notch signaling is active in a broad domain in the TBD but is regulated in the TZ to produce an ON/OFF pattern corresponding to the anterior/posterior half-segments, respectively. This regulation is achieved by mechanisms that are intrinsic to the cells in the TBD, which can regenerate this ON/OFF pattern in isolation. In addition, the response of the Notch pathway genes to treatment with CHX or TSA leads to several conclusions about how regulation of Notch signaling occurs during segmentation. After CHX treatment over a time period when one segment forms (1.0 hr), the expression of ESR-4 and ESR-5 in the TBD and the TZ is dramatically derepressed. We conclude from this result that de novo protein synthesis is required to repress Notch signaling in the TZ, thus generating the OFF state corresponding to posterior half-segments. Because CHX treatment also derepresses the expression of X-Delta-2 and Thylacine1 in this same region, this OFF state could be produced by repression on all genes involved, or it could be a cascade effect as Thylacine1 is an activator of X-Delta-2 expression (Sparrow et al. 1998), which in turn is an essential activator of ESR-4/ESR-5 via the Notch pathway (Fig. 3). In either case, the Notch pathway genes clearly have a marked susceptibility to CHX treatment in terms of forming an ON/OFF pattern of expression in the TBD/TZ. In contrast, the ON/OFF pattern of X-Delta-2 and Thylacine1 is still intact in S2–S4 after 1.0 hr of CHX treatment, and in S3 and S4 after 2 hr of treatment, indicating that the regulation of these genes in the somitomeric region is relatively insensitive to CHX treatment. Our interpretation of these results is that de novo protein synthesis is required to set the OFF state for the Notch pathway genes in the TZ, but that once this state is set, it is maintained in the absence of de novo protein synthesis. Finally, treatment with TSA, an inhibitor of histone deacetylase activities (HDACs) associated with corepressors, mirrors the results obtained with CHX, in that the most sensitive region to this treatment is the TZ, in which once again, the expression of X-Delta-2, Thylacine1, and ESR-4/ESR-5 is derepressed. Thus, generation of the OFF state in the TZ may require remodeling of chromatin or other regulators of transcription by HDACs, but once this regulation occurs it becomes dispensable in the somitomeric region. Together these results indicate that whereas cells are in the TBD/TZ, the regulation of the Notch pathway genes is extremely plastic. Within this region, an OFF state is generated by factors that require de novo protein synthesis and HDAC activity. Once this ON/OFF state is generated, it propagates into the somitomere region in which it is relatively more stable.
Repression of the Notch pathway genes in the TBD/TZ
Repression of the Notch pathway genes by a negative feedback loop commonly occurs in many developmental processes in which Notch signaling acts (Chitnis et al. 1995; Heitzler et al. 1996; Huppert et al. 1997). For example, during lateral, or mutual inhibition in Drosophila, the Notch signaling pathway activates the expression of WRPW–bHLH proteins encoded in the E-SPL-C, which negatively feeds back on the Notch pathway by inhibiting the expression of the ligand, Delta, the expression of bHLH genes that promotes differentiation, or even their own expression. In this example, negative feedback is thought to dampen the levels of Notch signaling to a point at which some cells can differentiate. A similar form of negative feedback was also proposed to repress the expression of X-Delta-2 in the presomitic mesoderm, on the basis of the changes in X-Delta-2 expression that occur when Notch signaling is inhibited or activated with X-Su(H)–DBM and X-Su(H)–Ank, respectively (Jen et al. 1997; Sparrow et al. 1998). The results reported here extend the evidence for a negative feedback loop in several ways. First, as Notch target genes, the expression of ESR-4/ESR-5 is a strong indication that the Notch pathway is active in the TBD/TZ, and thus is positioned to repress the Notch pathway genes when cells begin to segment in the TZ. Second, X-Su(H)–DBM blocks the expression of ESR-4/ESR-5 in the TZ but expands the expression of X-Delta-2, a hallmark of negative feedback. Third, ESR-5, whose expression is promoted by Notch signaling, regulates the expression of X- Delta-2 and ESR-4 in the TZ as predicted for a negative feedback loop: Expression of X-Delta-2 and Thylacine1 increases when ESR-5 activity is inhibited and decreases when ESR-5 activity is increased. Thus, negative feedback via Notch signaling represses the expression of Notch pathway genes in the TZ, and this repression appears to be mediated by the activation of ESR-5. We note, however, that the results obtained in both types of experiments with ESR-5 may be due to mimicking of the activity of other WPRW-bHLH proteins, including ESR-4. ESR-4 and ESR-5 differ in terms of how their expression is regulated in the TBD, but share similar patterns of expression when a somitomere forms, and thus could have redundant functions. Whether or not ESR-4 has the same activity as ESR-5, or whether there are other WRPW–bHLH proteins that mediate negative feedback, are questions that need to be resolved in the future.
Combining the evidence for negative feedback with the results from the CHX/TSA experiments described above, we propose that negative feedback is one of the CHX/TSA-sensitive factors required in the TZ to produce an OFF state in the Notch pathway genes during segmentation (Fig. 9). One attractive feature of this model is that it also accounts for the expression pattern of the Notch pathway genes in the somitomeric region of embryos when Notch signaling is altered. For example, when the Notch pathway is blocked with either X-Su(H)–DBM, or the mutant forms of ESR-5, the expression of both X-Delta-2 and Thylacine1 expands in the somitomeric region, creating one large stripe. Conversely, activation of Notch signaling in the presomitic mesoderm with ICD or X-Su(H)–Ank represses the expression of X-Delta-2 or Thylacine1 in the somitomeric region. At first glance, these results are paradoxical because in the somitomeric region, the expression of X-Delta-2 and Thylacine1 is associated with regions in which Notch signaling takes place. However, these are the results predicted by a model in which Notch signaling is acting earlier within the TZ, primarily to repress the Notch pathway genes in the posterior half-segments. If this repression fails in the TZ, in which the regulation of the Notch pathway genes is plastic, then a gap never forms, and the consequences are propagated forward as a large stripe of gene expression, lacking the proper gaps. Conceptually, this model is similar to the role of Notch in the establishment of wing vein size in Drosophila, in which Notch activity is promoted within the protein, but acts by inhibiting vein formation (Huppert et al. 1997).
Figure 9.

Model for Xenopus vertebrate segmentation. Segmental regulation of Notch pathway genes in the paraxial mesoderm of Xenopus embryos. The presomitic mesoderm of Xenopus embryos can be divided into three regions, TBD (red shading), TZ (blue shading), and somitomeric region (green shading) in which the Notch pathway is regulated by different factors. The Notch pathway is active when paraxial cells are in the TBD but is repressed in posterior half- segments when cells enter the TZ. This repression is likely to be due to several factors including negative feedback via ESR-5, as well as segmental repressors such as the Hairy-oscillator. At this point in the process, Notch signaling acts to establish segment size by restricting the domain in which the Notch pathway genes are active. In the somitomeric region, Notch signaling appears to take on another role in which ESR-5 appears to promote the expression of Thylacine1 and X-Delta-2 in a positive feedback loop. Positive feedback would act to maintain segmental identity by preserving the domains in which Notch signaling occurs.
Although our results indicate that negative feedback is one factor in regulating the Notch pathway in the TZ, negative feedback alone is unlikely to be sufficient to produce the periodic repression of Notch signaling that is required for segmentation. We suggest, therefore, that the repression of the Notch pathway genes in the TZ is also dependent on the Xenopus homolog of the Hairy oscillator that has been described in the chick, called c-hairy1 (Palmeirim et al. 1997). Unfortunately, the Xenopus WRPW–bHLH gene that is most closely related to c-hairy1 at present is Hairy2A (See Fig. 1), which is expressed in posterior half-segments but only detectable in S2 and S3 (Fig. 2K) (Jen et al. 1997). Because Hairy2A expression is not detectable in the TBD, at least in our hands, we cannot address whether or not it contributes to the repression of Notch target genes in the TZ. Nonetheless, an oscillating transcriptional repressor has obvious appeal in terms of explaining how the Notch pathway genes are segmentally repressed in a CHX-sensitive manner. In this model, negative feedback lowers the levels of Notch signaling in the TZ, but the expression levels of the Notch pathway genes are only pushed, periodically, below a certain threshold by the activity of the oscillating Hairy repressor. If the negative feedback loop is blocked, the levels of Notch signaling in the TZ are initially too high, and the ability of the oscillator to repress the pathway is compromised. One prediction of this model is that the Notch pathway genes are targets of the Hairy repressor. In preliminary experiments we have found that ectopic Hairy2A expression is very potent at repressing X-Delta-2, and ESR-4 expression in the TBD and TZ.
Finally, another potential mechanism for repressing Notch target genes in the TZ is by changing the state of X-Su(H) from an activator to a repressor. Recent evidence indicates that activation of Su(H) by ICD involves displacing a corepressor complex containing SMRT/HDAC activity (Kao et al. 1998). Although the in vivo significance of this corepressor complex in the regulation of Notch target genes remains uncertain, one site of action could be the transcriptional regulation of ESR-4 and ESR-5 expression in the TBD/TZ.
Evidence for positive feedback in the somitomeric region
Ectopic expression of ESR-5 inhibits the expression of X-Delta-2 in the TZ and TBD, and reduces the levels of X-Delta-2 expression in the somitomeric region as predicted by a negative feedback model. However, ESR-5 also expands the expression of X-Delta-2 in the somitomeric region, shifting its domain of expression anteriorly along the axis of the paraxial mesoderm. The expression of Thylacine1 in the somitomeric region responds even more dramatically to ESR-5 expression, by increasing in levels of expression and also shifting anteriorly. These results are a strong indication that when paraxial cells enter the more anterior portion of the somitomeric region, ESR-5 takes on an element of positive feedback and promotes the expression of genes in the Notch pathway, (Fig. 9). This interpretation also explains the complex phenotypes produced in the somitomeric region when Notch signaling or ESR-5 activity is inhibited. These phenotypes are best explained by a model in which loss of negative feedback in the TZ increases the expression of Notch pathway genes in the gaps of S1 and S2, whereas a loss of positive feedback in the somitomeric region decreases levels of expression in the stripes and gaps in S3 and S4 (e.g., See Fig. 7L and F).
The role of Notch signaling during segmentation
Current evidence from the study of segmentation in different vertebrate embryos indicates that Notch signaling is segmentally activated during somitogenesis and that this pattern is likely to be linked to the activity of the molecular clock that gates the number of cells that comprise each segmental unit (Jiang et al. 1998). Our results indicate that in Xenopus, establishment of a segmental pattern occurs by the regulation of Notch signaling periodically, thus producing an on/off pattern corresponding to anterior and posterior half-segments, respectively. A key element in generating this on/off pattern is the repression of Notch pathway genes in a critical region in which somitomeres arise, so that too much or too little repression at this point appears to alter the gating of cells into segments. One function of the molecular clock, therefore, could be the repression of Notch pathway genes, perhaps directly, as cells are gated into segments. Finally, the molecular clock is thought to act in combination with a wavefront, which determines the point along the anterior–posterior axis of the paraxial mesoderm in which segmentation occurs (Stern and Vasiliauskas 1998). Although the molecular nature of the wavefront remains unclear, we note that our results indicate that the paraxial cells regulate Notch pathway genes differently depending on where they lie along the anterior–posterior axis. A major goal is to determine how the activity of this pathway is modulated as cells leave the tailbud, enter the paraxial mesoderm, segment, and form somites.
Materials and methods
Isolation of ESR-4 and ESR-5
Novel sequences encoding WRPW–bHLH proteins were isolated by RT–PCR with two sets of forward degenerate oligonucleotide primers, 5′-TCACTCGAG(A/C)GNG(A/C)N(A/C)GNATNAA and 5′-TCACTCGAGAAA(A/G)CCNNTNNTNGA, and one set of reverse degenerate oligonucleotide primer, 5′-TCGGATCCNATNTCNGC(C/T)TTNTC.
Sequences were amplified from RNA prepared from presomitic mesoderm isolated from stage 26 Xenopus embryos after gentle treatment with Dispase. Products of the RT–PCR reaction (125 bp) were subcloned in pBluescript and sequenced. Clones with novel sequences were used to screen a stage 17 (neurula) library (Kintner and Melton 1987), yielding cDNAs encoding ESR-4 and ESR-5. The nucleotide sequences of ESR-4 and ESR-5 are submitted to GenBank under the accession numbers AF137072 and AF137073, respectively.
RNA synthesis and injection
For generating synthetic RNA in vitro, the coding region of the ESR-4 and ESR-5 cDNAs were individually inserted into the CS2+ vector (Turner and Weintraub 1994). Care was taken to remove almost all of the 5′ and 3′ untranslated regions of the ESR-4 and ESR-5 cDNAs, because in preliminary experiments these sequences led to rapid RNA breakdown in embryos. ESR-5ΔBD is a version of ESR-5 with the basic domain (RKILKPVVEKQRRDR) deleted, whereas ESR-5–Gal4 is a version of ESR-5 with its carboxyl terminus WRPWP residues removed and replaced with Gal4, with a CS2/Gal4 vector provided by Dave Turner (University of Michigan, Ann Arbor). Both the basic domain deletion, and the substitution of Gal4 were carried out by a PCR-based approach, and the resulting constructs were sequenced to confirm that unintended nucleotide changes were not introduced.
RNA was transcribed in vitro with SP6 polymerase, in the presence of GpppG, from templates linearized with NotI. The templates for the synthesis of nlacZ RNA (Turner and Weintraub 1994), of X-Delta-2 (Jen et al. 1997), of X-Su(H)1–DBM and of X-Su(H)1–Ank were described previously (Wettstein et al. 1997). Capped RNAs were phenol/chloroform extracted, ethanol precipitated, and resuspended in a final concentration of at least 40 ng/μl. Integrity of the mRNAs was assayed by formaldehyde-agarose gel electrophoresis. RNA was injected into the equatorial region of a single blastomere of a two-cell embryo. For each set of experiments, nlacZ RNA alone was injected as a negative control.
To assay the effects of RNA injection on gene expression, embryos injected with RNAs encoding genes of interest, along with nLacZ, were harvested at tailbud stage (between stage 22 and 24) and fixed in MEMFA for 1 hr. After staining with X-Gal, embryos were then assayed for gene expression by whole-mount in situ hybridization with the appropriate probe. For each RNA injection, at least 20 embryos were analyzed for each probe and the phenotypes reported occurred in a majority of cases. Each RNA injection was also repeated at least once independently.
In situ hybridization
Whole-mount in situ hybridization was performed on staged embryos as described by Harland (1991). The probes for ESR-4 and ESR-5 encompassed the entire cDNA, whereas those for X-Delta-2, Thylacine1, and Hairy2A have been described previously (Jen et al. 1997; Sparrow et al. 1998). To determine the relative spatial expression patterns of ESR-4 and ESR-5, of ESR-5 and X-Delta-2, and of ESR-5 and Hairy2A, double-labeled in situ hybridization was performed with digoxigenin-labeled X-Delta-2, Hairy2A, and ESR-5 (in the case of ESR-4 and ESR-5 double labeling) riboprobes and fluorescein-labeled ESR-4 and ESR-5 riboprobes. Chromogenic reactions were carried out on X-Delta-2, Hairy2A, and ESR-5 (in the case of ESR-4 and ESR-5 double labeling) with Magenta-Phos (Biosynth) and tetrazolium red (Sigma), followed by reaction for ESR-4 and ESR-5 with BCIP (Biosynth). Stained embryos were postfixed in MEMFA (0.1 m MOPS at pH 7.4, 2 mm EGTA, 1 mm MgSO4, 3.7% formaldehyde) and photographed with a Wild M33 microscope.
Immunohistochemistry and histology
To assay the effects of changing ESR-5 activity on somite formation, embryos injected with RNAs encoding ESR-5 or ESR-5ΔBD or ESR-5–Gal4, along with nΒ-gal, were harvested at tadpole stage (approximately stage 26) and fixed in MEMFA for 1 hr. Β-galactosidase expression was detected by staining in X-Gal. Embryos were then stained for the formation of somitic tissue with the monoclonal antibody 12/101 (Kintner and Brockes 1985). For best antibody penetration into the paraxial mesoderm, embryos were incubated with a 1:10 dilution of 12/101 culture supernatant, in the presence of blocking solution (20% goat serum, 0.5% Triton X-100, and 1× PBS) for 48 hr or longer and with a 1:250 dilution of HRP-conjugated goat anti-mouse secondary antibody for 48 hr or longer. Following staining in DAB, embryos were postfixed in MEMFA, embedded in paraffin, and sectioned at 10 μm.
Tailbud isolation
Tailbud tissues were dissected from stage 24 embryos with a micrometer to remove a piece of tissue ∼0.5 mm in length. These tissues were fixed either immediately or after culturing for 4 hr in 0.1× MMR medium, and then were assayed for ESR-4 or ESR-5 expression, by whole-mount in situ hybridization.
CHX and TSA treatments
Embryos were treated with 10 μg/ml of CHX (Sigma) or 400 nm TSA (Sigma) for either 1 or 2 hr at 25°C, followed by MEMFA fixation. In control experiments, protein synthesis in isolated Xenopus animal cap tissue, as measured by the uptake of 35S into acid insoluble radioactivity was reduced to 20% of control levels (Wettstein et al. 1997).
Acknowledgments
We thank Drs. Noako Koyano-Nakagawa, Anne Bang, Ajay Chitnis, Elise Lamar, and Martyn Goulding for comments on the manuscript. We are grateful to Dr. David Turner for providing the primer sequences used to amplify WRPW-bHLH genes. This work was supported by National Institutes of Health grant to C.K.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked ‘advertisement’ in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL kintner@salk.edu; FAX (619) 552-8285.
References
- Artavanis-Tsakonas S, Matsuno K, Fortini ME. Notch signaling. Science. 1995;268:225–232. doi: 10.1126/science.7716513. [DOI] [PubMed] [Google Scholar]
- Bailey AM, Posakony JW. Suppressor of hairless directly activates transcription of enhancer of split complex genes in response to Notch receptor activity. Genes & Dev. 1995;9:2609–2622. doi: 10.1101/gad.9.21.2609. [DOI] [PubMed] [Google Scholar]
- Chan YM, Jan YN. Roles for proteolysis and trafficking in notch maturation and signal transduction. Cell. 1998;94:423–426. doi: 10.1016/s0092-8674(00)81583-4. [DOI] [PubMed] [Google Scholar]
- Chitnis A, Henrique D, Lewis J, Ish-Horowicz D, Kintner C. Primary neurogenesis in Xenopus embryos regulated by a homologue of the Drosophila neurogenic gene Delta. Nature. 1995;375:761–766. doi: 10.1038/375761a0. [DOI] [PubMed] [Google Scholar]
- Conlon RA, Reaume AG, Rossant J. Notch 1 is required for the coordinate segmentation of somites. Development. 1995;121:1533–1545. doi: 10.1242/dev.121.5.1533. [DOI] [PubMed] [Google Scholar]
- de Angelis MH, McIntyre J, II, Gossler A. Maintenance of somite borders in mice requires the Delta homologue Dll1. Nature. 1997;386:717–721. doi: 10.1038/386717a0. [DOI] [PubMed] [Google Scholar]
- Dornseifer P, Takke C, Campos-Ortega JA. Overexpression of a zebrafish homologue of the Drosophila neurogenic gene Delta perturbs differentiation of primary neurons and somite development. Mech Dev. 1997;63:159–171. doi: 10.1016/s0925-4773(97)00037-3. [DOI] [PubMed] [Google Scholar]
- Evrard YA, Lun Y, Aulehla A, Gan L, Johnson RL. lunatic fringe is an essential mediator of somite segmentation and patterning. Nature. 1998;394:377–381. doi: 10.1038/28632. [DOI] [PubMed] [Google Scholar]
- Forsberg H, Crozet F, Brown NA. Waves of mouse Lunatic fringe expression, in four-hour cycles at two-hour intervals, precede somite boundary formation. Curr Biol. 1998;8:1027–1030. doi: 10.1016/s0960-9822(07)00424-1. [DOI] [PubMed] [Google Scholar]
- Gawantka V, Pollet N, Delius H, Vingron M, Pfister R, Nitsch R, Blumenstock D, Niehrs C. Gene expression screening in Xenopus identifies molecular pathways, predicts gene function, and provides a global view of embryonic patterning. Mech Dev. 1998;77:95–142. doi: 10.1016/s0925-4773(98)00115-4. [DOI] [PubMed] [Google Scholar]
- Giebel B, Campos-Ortega JA. Functional dissection of the Drosophila enhancer of split protein, a suppressor of neurogenesis. Proc Natl Acad Sci. 1997;94:6250–6254. doi: 10.1073/pnas.94.12.6250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gossler A, de Angelis MH. Somitogenesis. San Diego, CA: Academic Press; 1998. [Google Scholar]
- Harland RM. In situ hybridization: An improved whole-mount method for Xenopus embryos. Methods Cell Biol. 1991;36:685–695. doi: 10.1016/s0091-679x(08)60307-6. [DOI] [PubMed] [Google Scholar]
- Heitzler P, Bourouis M, Ruel L, Carteret C, Simpson P. Genes of the Enhancer of split and achaete-scute complexes are required for a regulatory loop between Notch and Delta during lateral signalling in Drosophila. Development. 1996;122:161–171. doi: 10.1242/dev.122.1.161. [DOI] [PubMed] [Google Scholar]
- Huppert SS, Jacobsen TL, Muskavitch MA. Feedback regulation is central to Delta-Notch signalling required for Drosophila wing vein morphogenesis. Development. 1997;124:3283–3291. doi: 10.1242/dev.124.17.3283. [DOI] [PubMed] [Google Scholar]
- Irvine KD, Wieschaus E. fringe, a boundary-specific signaling molecule, mediates interactions between dorsal and ventral cells during Drosophila wing development. Cell. 1994;79:595–606. doi: 10.1016/0092-8674(94)90545-2. [DOI] [PubMed] [Google Scholar]
- Jen WC, Wettstein D, Turner D, Chitnis A, Kintner C. The Notch ligand, X-Delta-2, mediates segmentation of the paraxial mesoderm in Xenopus embryos. Development. 1997;124:1169–1178. doi: 10.1242/dev.124.6.1169. [DOI] [PubMed] [Google Scholar]
- Jiang Y-J, Smithers L, Lewis J. The clock is linked to Notch signaling. Curr Biol. 1998;8:868–871. doi: 10.1016/s0960-9822(07)00547-7. [DOI] [PubMed] [Google Scholar]
- Jimenez G, Pinchin SM, Ish-Horowicz D. In vivo interactions of the Drosophila Hairy and Runt transcriptional repressors with target promoters. EMBO J. 1996;15:7088–7098. [PMC free article] [PubMed] [Google Scholar]
- Kao HY, Ordentlich P, Koyano-Nakagawa N, Tang Z, Downes M, Kintner CR, Evans RM, Kadesch T. A histone deacetylase corepressor complex regulates the Notch signal transduction pathway. Genes & Dev. 1998;12:2269–2277. doi: 10.1101/gad.12.15.2269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kintner CR, Brockes JP. Monoclonal antibodies to the cells of a regenerating limb. J Embryol Exp Morphol. 1985;89:37–55. [PubMed] [Google Scholar]
- Kintner CR, Melton DA. Expression of Xenopus N-CAM RNA in ectoderm is an early response to neural induction. Development. 1987;99:311–325. doi: 10.1242/dev.99.3.311. [DOI] [PubMed] [Google Scholar]
- Kusumi K, Sun ES, Kerrebrock AW, Bronson RT, Chi D-C, Bulotsky MS, Spencer JB, Birren BW, Frankel WN, Lander ES. The mouse pudgy mutation disrupts Delta homologue Dll3 and initiation of early somite boundaries. Nat Genet. 1998;19:274–278. doi: 10.1038/961. [DOI] [PubMed] [Google Scholar]
- McGrew MJ, Dale JK, Fraboulet S, Pourquié O. The lunatic Fringe gene is a target of the molecular clock linked to somite segmentation in avian embryos. Curr Biol. 1998;8:979–982. doi: 10.1016/s0960-9822(98)70401-4. [DOI] [PubMed] [Google Scholar]
- Müller M, v. Weizsäcker E, Campos-Ortega JA. Expression domains of a zebrafish homologue of the Drosophila pair-rule gene hairy correspond to primordia of alternating somites. Development. 1996;122:2071–2078. doi: 10.1242/dev.122.7.2071. [DOI] [PubMed] [Google Scholar]
- Muskavitch MA. Delta-Notch signaling and Drosophila cell fate choice. Dev Biol. 1994;166:415–430. doi: 10.1006/dbio.1994.1326. [DOI] [PubMed] [Google Scholar]
- Oka C, Nakano T, Wakeham A, de la Pompa JL, Mori C, Sakai T, Okazaki S, Kawaichi M, Shiota K, Mak TW, Honjo T. Disruption of the mouse RBP-JK gene results in early embryonic death. Development. 1995;121:3291–3301. doi: 10.1242/dev.121.10.3291. [DOI] [PubMed] [Google Scholar]
- Palmeirim I, Henrique D, Ish-Horowicz D, Pourquié O. Avian hairy gene expression identifies a molecular clock linked to vertebrate segmentation and somitogenesis. Cell. 1997;91:639–648. doi: 10.1016/s0092-8674(00)80451-1. [DOI] [PubMed] [Google Scholar]
- Saga Y, Hata N, Kobayashi S, Magnuson T, Seldin MF, Taketo MM. MesP1: A novel basic helix-loop-helix protein expressed in the nascent mesodermal cells during mouse gastrulation. Development. 1996;122:2769–2778. doi: 10.1242/dev.122.9.2769. [DOI] [PubMed] [Google Scholar]
- Saga Y, Hata N, Koseki H, Taketo MM. Mesp2: A novel mouse gene expressed in the presegmented mesoderm and essential for segmentation initiation. Genes & Dev. 1997;11:1827–1839. doi: 10.1101/gad.11.14.1827. [DOI] [PubMed] [Google Scholar]
- Sparrow DB, Jen WC, Kotecha S, Towers N, Kintner C, Mohun TJ. Thylacine 1 is expressed segmentally within the paraxial mesoderm of the Xenopus embryo and interacts with the Notch pathway. Development. 1998;125:2041–2051. doi: 10.1242/dev.125.11.2041. [DOI] [PubMed] [Google Scholar]
- Stern CD, Vasiliauskas D. Clocked gene expression in somite formation. BioEssays. 1998;20:528–531. doi: 10.1002/(SICI)1521-1878(199807)20:7<528::AID-BIES2>3.0.CO;2-G. [DOI] [PubMed] [Google Scholar]
- Struhl K. Histone acetylation and transcriptional regulatory mechanisms. Genes & Dev. 1998;12:599–606. doi: 10.1101/gad.12.5.599. [DOI] [PubMed] [Google Scholar]
- Turner DL, Weintraub H. Expression of achaete-scute homolog 3 in Xenopus embryos converts ectodermal cells to a neural fate. Genes & Dev. 1994;8:1434–1447. doi: 10.1101/gad.8.12.1434. [DOI] [PubMed] [Google Scholar]
- Wettstein D, Turner D, Kintner C. The Xenopus homolog of Drosophila Suppressor of Hairless mediates Notch signaling during primary neurogenesis. Development. 1997;124:693–702. doi: 10.1242/dev.124.3.693. [DOI] [PubMed] [Google Scholar]
- Zhang N, Gridley T. Defects in somite formation in lunatic fringe-deficient mice. Nature. 1998;394:374–377. doi: 10.1038/28625. [DOI] [PubMed] [Google Scholar]