Abstract
PROMoter uPstream Transcripts (PROMPTs) were identified as a new class of human RNAs, which are heterologous in length and produced only upstream of the promoters of active protein-coding genes. Here, we show that PROMPTs carry 3′-adenosine tails and 5′-cap structures. However, unlike mRNAs, PROMPTs are largely nuclear and rapidly turned over by the RNA exosome. PROMPT-transcribing DNA is occupied by RNA polymerase II (RNAPII) complexes with serine 2 phosphorylated C-terminal domains (CTDs), mimicking that of the associated genic region. Thus, the inefficient elongation capacity of PROMPT transcription cannot solely be assigned to poor CTD phosphorylation. Conditions that reduce gene transcription increase RNAPII occupancy of the upstream PROMPT region, suggesting that they reside in a common transcription compartment. Surprisingly, gene promoters that are actively transcribed by RNAPI or RNAPIII also produce PROMPTs that are targeted by the exosome. RNAPIII PROMPTs bear hallmarks of RNAPII promoter-associated RNAs, explaining the physical presence of RNAPII upstream of many RNAPIII-transcribed genes. We propose that RNAPII activity upstream gene promoters are wide-spread and integral to the act of gene transcription.
INTRODUCTION
Human cells harbor approximately 23 000 protein-coding genes, representing ∼2% of the genomic material. These are joined by several thousand RNA genes that produce components involved in translation (tRNAs and rRNAs), maturation of the translation factory (snoRNAs), splicing of protein-coding transcripts (snRNAs), small modulators of protein output (miRNAs) as well as long intergenic non-coding (nc) RNAs (lincRNAs) of mostly unknown function (1). Each of these transcription units is expressed by one of three major RNA polymerases (RNAPs): Protein-coding- and many nc-genes are transcribed by RNAPII. RNAPI produces all rRNAs except 5S rRNA, which is made by RNAPIII that also transcribes tRNAs as well as several other ncRNAs. The remaining parts of the genome were traditionally regarded as inactive rudiments left behind by evolution with little or no function in the present. However, DNA microarray and high-throughput sequencing technologies have allowed large-scale analyses of genome-wide activity unbiased of annotations and challenged this view by demonstrating transcription of intergenic regions at an unexpected level (2,3). The exact fraction of the genome transcribed—let alone its functional entities—remains a subject of debate (4,5).
Adding to the complexity of eukaryotic transcriptomes are RNAs that normally escape detection due to their short-lived nature. In Saccharomyces cerevisiae, cryptic unstable transcripts (CUTs) were discovered as major substrates of the nuclear form of the RNA exosome (6,7), an evolutionary conserved RNA degradation complex with both 3′–5′-exonucleolytic and endonucleolytic activity (8,9). The catalog of CUTs has since been extended to confirm that these ∼200–600 nt long RNAs are found in intergenic regions, where they are principally associated with promoters (10,11). Exosome-regulated RNAs that are co-linear with the 5′-ends of protein-coding transcripts have also been observed in the plant Arabidopsis (12). In humans, a related class of molecules, PROMoter uPstream Transcripts (PROMPTs), has been discovered by tiling ENCODE microarrays covering ∼1% of the genome. Like CUTs, their low abundance only allowed efficient detection after depletion of key components of the human RNA exosome (13). The existence of PROMPTs was also verified for a handful of selected genes outside the ENCODE region, and it was proposed that most, if not all, actively transcribed RNAPII genes have associated PROMPTs, although they may be especially predominant at TATA-less, CpG-rich promoters with broad transcription start site (TSS) regions (13). The average PROMPT is generated in a fairly narrow window between ∼500 and 2500 nt upstream of the TSS and appears in most cases not to extend into the nucleosome-depleted region (and consequently not into the gene). Most PROMPTs are therefore also discontinuous with another class of low-abundant, but substantially shorter, RNAs that arise closer to the TSSs of protein-coding genes. These so-called TSS-associated (TSSa)- (14), or transcription initiation (ti)- (15,16) RNAs were identified in vertebrate and invertebrate cells by high-throughput sequencing of size-selected, uncapped RNAs, and are viewed to be by-products of RNAPII stalling and backtracking (17,18), or to derive from nascent RNA protected by stalled RNAPII against nucleolysis (E. Valen et al., manuscript accepted for publication). Interestingly, TSSa/ti-RNAs are not detectable in S. cerevisiae (16), (our unpublished data), and, as of today, no direct experimental evidence has provided a direct physical link between TSSa/ti-RNAs and PROMPTs [reviewed in (19,20)].
Apart from their labile nature, PROMPTs share other key features with CUTs: (i) they can be transcribed in the sense as well as anti-sense direction with respect to the downstream gene; (ii) they carry 3′-adenosine tails; and (iii) their production is linked to the downstream, protein-coding promoter [reviewed in (19,21)]. A model for the generation of CUTs, aspects of which may also apply to PROMPTs, suggests that general transcription factors (GTFs) recruited to promoter regions by transcriptional activators are responsible for both gene and CUT transcription. In addition to ‘correctly’ positioned pre-initiation complexes (PICs) driving mRNA transcription, cryptic PICs may assemble in the vicinity in either orientation and generate non-genic transcripts (21). The use of such cryptic PICs may be facilitated further by the opening of chromatin by negative supercoiling caused in the wake of RNAPII transcribing into the gene (14). PROMPT regions are generally devoid of transcription initiation factors such as TAFI, TAFII p250 and E2F1 (13), and thus, an alternative possibility would be that transcription initiates without the use of conventional PICs.
While transcription termination of CUTs takes advantage of the Nrd1p/Nab3p/Sen1p termination system of another class of short S. cerevisiae RNA transcripts, the snoRNAs (22–24), these factors are not all well conserved in humans, and it is currently unknown how PROMPT transcription is terminated. It has been speculated that the absence of histone modifications such as H3K79me2 and H3K36me3 prevents productive elongation in the PROMPT region. Because both promoter escape and trimethylation of H3K36 by the Set2 methylase depend, at least in part, on phosphorylation of serine 2 (Ser2P) of the carboxy-terminal domain (CTD) of RPB1, the largest subunit of RNAPII (25,26), it was further suggested that absence of Ser2P could be an important determinant of directionality (14,20).
To get a better understanding of PROMPTs and the relationship to their associated genes, we performed a detailed analysis of the biochemical properties of selected PROMPTs and the regions, from which they are transcribed. We show that RNAPII in PROMPT and gene regions is similar with regard to its CTD phosphorylation status. Indeed, a PROMPT-like region and its downstream gene appear to reside in a shared ‘gene compartment’ with a local concentration of RNAPII and possibly other factors. We also find that all tested genes transcribed by the other two major human RNA polymerases, RNAPI and RNAPIII, have associated PROMPTs, suggesting that cross-regulation between neighboring transcription units, even produced by different RNAPs, is common and might be related to enhancer function.
MATERIALS AND METHODS
Cell culture and handling
HEK293 and HeLa cells were grown in DMEM GlutaMax medium (Invitrogen) supplemented with 10% fetal bovine serum and 1% penicillin/streptomycin where appropriate. Induction of β-globin gene expression in HEK293 cells was with 250 ng/ml tetracycline for 24 h. HeLa cell transfections were done with 20 nM siRNA for 2–3 days and repeated for another 3 days each time using Lipofectamin2000 as transfecting agent according to the manufacturers instructions (Invitrogen). RRP40 and control (eGFP) siRNA sequences were taken from ref. (13). Additional siRNA sequences are listed in Supplementary Table S1. siRNAs were synthesized by RiboTask, Odense, Denmark.
Construction of stable HEK293 cell lines
For the construction of cell lines stably expressing the β-globin gene preceded by a heterologous sequence (TOM1) upstream of the β-globin TSS, a region of the yeast TOM1-coding sequence was first amplified by PCR from S. cerevisiae genomic DNA and digested with MluI. The resulting 2532 bp fragment was ligated into the corresponding site of pcDNA5_β-globin_SV40-LpA (27), a pcDNA5 FRT/TO plasmid containing the β-globin gene followed by the late poly(A) site (LpA) from SV40 virus. In this construct, the β-globin gene is under the control of a tetracycline-inducible human cytomegalovirus (CMV)/TetO2 promoter. The MluI insertion site is situated 573 bp upstream the TATA box of the promoter. A variant, in which the AAUAAA hexamer of the LpA had been mutated to AAGAAA (28) was constructed correspondingly. Both constructs were subsequently stably integrated into HEK293 Flp-In T-rex cells (Invitrogen) as described previously (27).
RT–qPCR assays
Total RNA was extracted using Trizol-reagent (Invitrogen) according to the manufacturers instructions. RNA was subjected to DNaseI treatment at 37°C for 30 min with 1 U DNase I (Ambion) per µg of RNA. The RNA was then re-purified by phenol/chloroform extraction and re-precipitated. Reverse transcription was performed on 3–5 µg total RNA in a volume of 20 µl using 0.5 µg of either random hexamer primer (N6) or of anchored dT primer (dT20VN), and SuperScript II reverse transcriptase (Invitrogen) according to the manufactures instructions. Reactions without reverse transcriptase were included as negative controls. Reactions were diluted 5–10 times, and 1–2 µl (equivalent to 60–100 ng RNA input) were used in each qPCR reaction, which was done with Platinum SYBRGreen (Invitrogen) and 0.2–0.3 μM of specific oligonucleotide primers (see Supplementary Table S2) at an annealing temperature of 59°C. Reactions were run on an Mx3005p machine and analyzed with MxPro software (both from Agilent Technologies, Santa Clara, CA, USA). Samples were normalized to GAPDH as a control where appropriate.
Cap-IP assays
Cap-IPs were performed starting with 5 µg of total RNA. As a negative control, the same amount of RNA was treated with 5 U of tobacco acid pyrophosphatase (TAP, Epicentre Biotechnologies) for 90 min at 37°C. Alternatively, RNA was mock treated in parallel by omitting TAP in the reaction. Next, 12 µl of a slurry of agarose-conjugated mouse monoclonal anti-cap antibodies (K121, Calbiochem) was mixed with the RNA samples in 150 µl IPP buffer (150 mM NaCl, 0.1% NP 40, 10 mM Tris, pH 8.0) supplemented with 2.5 mM DTT and 0.2 U/µl RNase inhibitor. After over-night incubation at 4°C on a rotator, the supernatants (unbound fractions) were collected, and the agarose beads were washed four times with 1 ml of ice-cold IPP buffer containing 2.5 mM DTT with a change of tubes after the penultimate wash. Elution of the bound RNA was performed in 200 µl of IPP-150 containing 0.5 mg/ml Proteinase K and incubation at 37°C for 30 min. After phenol/chloroform extraction and ethanol precipitation of the supernatants and the eluates, RNA was subjected to RT–qPCR analyses. The bound fraction of the total RNA (bound and unbound) was determined using the equation 1/1 + 2^(Ct[bound] − Ct[unbound]), where ‘Ct’ is the critical threshold of qPCR amplification.
5′-rapid amplification of cDNA ends
5′-RACE from anchored dT20-primed (dT20VN) cDNA was performed using nested primers and a commercial kit according to the manufacturers instructions (5′-RACE System for Rapid Amplification of cDNA Ends, Version 2.0, Invitrogen). 5′-ends were PCR amplified using the AAP primer from the kit together with a PROMPT-specific primers. The products were re-amplified in a nested PCR using the kit primer AUAP and a nested PROMPT-specific primer. PCR products were gel-purified and ligated into pCR4-TOPO (Invitrogen) followed by subcloning and sequencing of individual clones. Primer sequences are available on request.
Nucleo-cytoplasmic fractionation
Subcellular fractionation was performed from ∼107 HeLa cells grown to near confluency on P10 plates. After two washes with ice-cold phosphate buffered saline (PBS) on plates, cells were collected in 1 ml PBS and pelleted by brief centrifugation at 240g. Buffers were supplemented with a protease inhibitor cocktail (‘Complete Mini’, Roche) without EDTA. Pellets were resuspended in hypotonic buffer RSB-10 (10 mM Tris/HCl, pH 7.4, 10 mM NaCl, 2.5 mM MgCl2) and incubated for 5 min on ice. Next, cells were transferred into a 2 ml glass dounce homogenizer and homogenized by approximately 25 strokes to achieve ∼80% cell breakage as monitored by microscopy. Following centrifugation at 4°C, 240g for 5 min, the cytoplasmic fraction (supernatant) was set aside and the nuclear pellet was resuspended in 900 µl buffer S1 (0.25 M Sucrose, 0.5 mM MgCl2). The nuclear suspension was split and each half was overlaid onto a cushion of 500 µl buffer S3 (0.88 M Sucrose, 0.5 mM MgCl2). After centrifugation at 4°C, 2800g for 10 min, the nuclear RNA was prepared from the pellet by Trizol extraction as described above, while the cytoplasmic fraction was subjected to phenol/chloroform extraction and ethanol precipitation.
Chromatin preparation
Cells, grown to be 90% confluent, were fixed with formaldehyde added directly to the culture medium to a final concentration of 1%. Cross-linking was performed for 10 min at room temperature (HeLa cells) or for 10 min at 37°C followed by 50 min at 4°C (HEK293 cells). Excess formaldehyde was quenched by the addition of glycine to a final concentration of 125 mM and agitation at ambient temperature for 5 min. After washing the cells twice with ice cold PBS, they were scraped off the plate in PBS, pelleted by centrifugation at 2150g for 5 min at 4°C and resuspended in 1 ml cell lysis buffer (20 mM Tris/HCl, pH 8.0, 85 mM KCl, 0.5% NP40) supplemented with a protease inhibitor cocktail (‘Complete Mini’ without EDTA, Roche). After a 10 min incubation on ice, nuclei were collected by centrifugation as above, resuspended in 1 ml nuclei lysis buffer (50 mM Tris/HCl, pH 8.0, 10 mM EDTA, 1% SDS) with protease inhibitors and incubated on ice for another 10 min. Samples were sonicated using a Biorupter sonicator (Diagenode, Liege, Belgium) to generate DNA fragments ranging from ∼150 to 350 bp. Sonicated samples were cleared by centrifugation at 16 900g for 10 min at 4°C, and the supernatant containing the nuclear cell extract (NCE) was collected. Lysis buffers used in chromatin preparation for CTD-Ser2P and CTD-Ser5P ChIP experiments contained phoshatase inhibitors (10 mM b-glycerophosphate and 1 mM Na3VO4).
ChIP assays
ChIP assays were performed after a modification of the protocol described in REF (28). Details can be found in the Supplementary Data for this article. Antibodies against RPB1 (H-224, sc-9001) were purchased from Santa Cruz Biotechnology; anti-histone H3 antibodies (ab1791) were from Abcam. Antibodies against CTD-Ser2P (3E10) and CTD-Ser5P (3E8) were purchased from Dr Elisabeth Kremmer and Dr Dirk Eick (Institute for Molecular Immunology, Helmholtz Zentrum, München, Germany). Per sample, 10 µg of antibody was used, except for ab1791 (2.5 µg) and 3E10 and 3E8 (100 µl of tissue culture supernatant).
Bioinformatic analyses
Locations of RefSeq TSSs were retrieved from the UCSC browser database, assembly hg18. These were collapsed to unique coordinates, and regions spanning from −2000 to +2000 from the TSSs were extracted. RNAPII and CTD-Ser2P data were downloaded from the short read archive (accession numbers GSE12781 and GSE20072, respectively), and all reads mapping within the TSS regions were cumulatively added. Each of the libraries was then normalized to reads per million (rpm) by dividing through the library size. The ratio of the libraries is the normalized CTD-Ser2P divided by RNAPII. The nucleosome data are from (29) that can be retrieved from the Short Read Archive (SRA) under accession number SRA000234 and were processed as in (30) without smoothing.
RESULTS
PROMPTs and mRNAs share basic characteristics
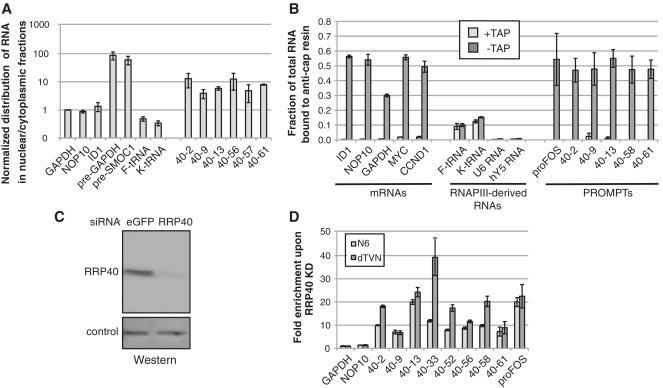
The low steady-state levels of PROMPTs suggest that they are rapidly degraded soon after their production, presumably in the nucleus. In support of this idea, the major cellular pools of two exonucleases (RRP6 and RRP44/DIS3), which interact with the exosome core and affect PROMPT stability synergistically, are nuclear (31,32). A third catalytically active exosome component, DIS3L, is exclusively cytoplasmic and KD of this factor on its own or in combination with KD of RRP6, RRP44/DIS3 or both does not lead to elevated PROMPT levels (31). To corroborate these results, we separated HeLa cells into nuclear- and cytoplasmic-enriched fractions (Supplementary Figure S1A and S1B) and assayed equal amounts of RNA from each fraction by random-primed reverse-transcription and quantitative PCR (RT–qPCR). As shown in Figure 1A, control pre-mRNAs were preferentially enriched in the nuclear fraction, as opposed to mature mRNAs or tRNAs. PROMPTs were enriched 10-fold more in the nuclear fraction than the assayed mRNAs and often became undetectable in the cytoplasmic fraction (Supplementary Figure S1C). Depletion of RRP40 did not significantly affect this nuclear/cytoplasmic distribution (data not shown). Thus, PROMPTs are predominantly nuclear.
Figure 1.
Biochemical characterization of PROMPTs. (A) PROMPTs are nuclear. RNA was prepared from cytoplasmic and nuclear fractions of HeLa cells and quantified by RT–qPCR. The nuclear/cytoplasmic ratios of the indicated PROMPT levels were normalized to that of GAPDH mRNA (the qPCR primers for the GAPDH amplicon span several exons) and are shown on a logarithmic scale. NOP10 and ID1 are additional mRNAs. As positive and negative controls for nuclear enrichment two pre-mRNAs and two tRNAs were used. Error bars show standard deviations of triplicate technical replica of a representative experiment. (B) PROMPTs are capped to the same level as mRNAs. Total RNA from HeLa cells was treated with (‘+TAP’) or without (‘−TAP’), tobacco acid pyrophophatase prior to IP using the K121 anti-cap antibody. Antibody-bound and -unbound (supernatant) fractions were analyzed by RT–qPCR for their relative amounts of various mRNAs, RNAPIII-derived RNAs and PROMPTs (‘40–2’ etc) as indicated. The fraction of the total (bound plus unbound) RNA bound to the antibody was determined (see ‘Materials and Methods’ section) and is plotted with error bars showing standard deviations of triplicate technical replica of a representative experiment. (C) Western blotting analysis confirming KD of RRP40. Equal amounts of protein extracts prepared from HeLa cells, transfected with either a control (eGFP) or a RRP40 siRNA, were separated by SDS–PAGE and transferred to membrane. The blot was probed with a polyclonal anti-RRP40 antibody. The loading control shows a cross-reacting band with different electrophoretic mobility. (D) The 3′-adenylation extent of PROMPTs upon exosome depletion. Total RNA from RRP40 KD and control HeLa cells was reverse-transcribed with either a random hexamer primer (‘N6’) or an anchored oligo-dT primer (‘dTVN’) and the increases of the indicated PROMPT levels upon RRP40 KD were determined by qPCR. GAPDH and NOP10 RNAs were included as controls. Error bars show standard deviations of triplicate technical replica of a representative experiment.
We next determined whether PROMPTs posses a methylguanosine cap structure characteristic of RNAPII transcripts. Total RNA was isolated from HeLa cells and subjected to immuno-precipitation (IP) with a monoclonal anti-cap antibody coupled to beads. Unbound and bound fractions were assayed by random-primed RT–qPCR for the presence of selected PROMPTs and other transcripts. As a control, an equal amount of total RNA was treated with tobacco alkaline pyrophosphate (TAP) to remove the cap-structure from RNA prior to the IP (33). Figure 1B shows that five control mRNAs (ID1, NOP10, GAPDH, MYC and CCND1) were efficiently (30–56%) precipitated in a cap-dependent manner. In contrast, hY5 and U6, two uncapped RNAPIII transcripts, were not precipitated above background levels. A minor fraction (∼10%) of tRNAs was recovered in the bound fraction irrespective of TAP-treatment, indicating unspecific binding of this RNA species to the beads. Importantly, all previously identified PROMPTs tested (13) exhibited IP efficiencies (47–54%) comparable to those of mRNAs, and this binding was lost with prior TAP treatment. In addition, we also detected a capped PROMPT upstream the proto-oncogene cFOS (Figure 1B and D). We conclude that PROMPTs resemble mRNAs in having a cap-structure at their 5′-termini.
Depletion of the exosome leads to a robust increase in PROMPT levels as measured by dT-primed RT–qPCR (13). To assess whether part of this increase is attributed to the addition of adenosine residues in the absence of the exosome (as opposed to a change in overall PROMPT levels), we tested whether PROMPT adenylation status was altered upon exosome knockdown (KD). Total RNA from control cells or from cells subjected to siRNA-mediated KD of the exosome core component RRP40 (Figure 1C) were reverse transcribed with either an anchored oligo dT primer (‘dTVN’) that specifically primes from adenosines at their addition site or a random hexamer (‘N6’). Afterwards, PROMPT levels were determined by qPCR and normalized to GAPDH mRNA levels. All PROMPT regions were substantially elevated in the RRP40 KD analyzed with both RT primers (Figure 1D). In contrast, NOP10 mRNA was unaffected. For some PROMPTs dTVN-primed cDNA generated higher levels of stabilization than N6 primed cDNA. However, because of the different mode of dTVN and N6 priming, the results cannot be stringently compared quantitatively, and we can therefore only conclude that increased PROMPT levels are not linked to global changes in the extent, to which these RNAs are adenylated. What feature(s) render PROMPTs efficient exosome substrates remains to be discovered (see ‘Discussion’ section).
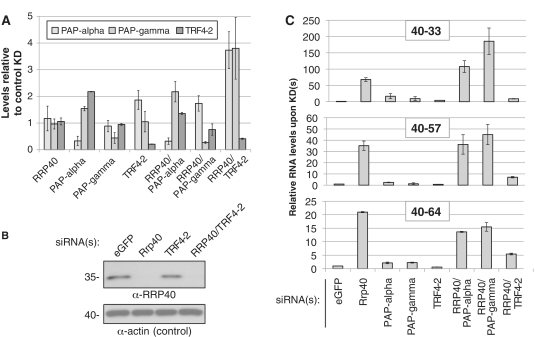
TRF4-2, a putative human homolog of S. cerevisiae Trf4p, adenylates PROMPT 3′-ends
In S. cerevisiae, the RNA exosome is activated by the Trf4p/Air2p/Mtr4p polyadenylation (TRAMP) complex (6,7,34). In fact, the removal of CUTs may be assisted by the Trf4p-catalyzed addition of 3′-adenosines, which appears to provide an entry point for exosomal degradation. We therefore, tested the effect of siRNA-mediated depletion of a putative human homolog of Trf4p, TRF4-2 (also known as PAPD5), on the status of PROMPT 3′-ends. We also carried out co-depletion of TRF4-2 with RRP40, and used as controls the two major nuclear canonical poly(A) polymerases, PAPα and PAPγ (35). Depletion efficiencies were confirmed by RT–qPCR and/or western blotting (Figure 2A and B). None of the individual PAP KD's had a major effect on the steady-state levels of several representative PROMPTs (Figure 2C). Thus, 3′-adenylation by, or the physical presence of, any of these PAPs does not appear to be a requirement for PROMPT degradation. In the double KDs of either PAPα or PAPγ with RRP40, PROMPT RT–PCR levels were similar to those of the single RRP40 KD (Figure 2C). In contrast, co-depletion of RRP40 with TRF4-2 yielded a reduced PROMPT signal compared to the single RRP40 KD. This effect was not the indirect result of changed RRP40 levels upon TRF4-2 KD (Figure 2B). Thus, the result can best be explained by a reduction in the material made available for oligo dT-primed RT–PCR, and we therefore suggest that TRF4-2 adenylates PROMPT 3′-ends, but that this is not a requirement for PROMPT turnover (see ‘Discussion’ section).
Figure 2.
Involvement of TRF4-2 in PROMPT 3′-end adenylation. (A) Decrease in transcript levels upon RNAi-depletion was measured with gene-specific primers by RT–qPCR. Levels were normalized to those of a control KD with siRNAs directed against eGFP and to levels of GAPDH mRNA. Error bars show standard deviations of duplicate technical replica of a representative experiment. (B) Western blotting analysis of RRP40 levels in siRNA-mediated KDs as indicated above each lane. HeLa whole-cell protein extracts were separated on a 10% polyacrylanide gel, blotted to membrane and probed with an anti-RRP40 antibody (top panel). Actin levels served as a loading control (bottom panel). Migration and size of marker proteins is given to the left. (C) Effects on PROMPT levels of KD's of nuclear poly(A) polymerases alone or in combination with an RRP40 KD. RTqPCR was performed as in (A). Fold differences for three representative PROMPTs, as compared to a control KD, are shown on the y-axes.
Characterization of MYC and CCND1 PROMPT regions
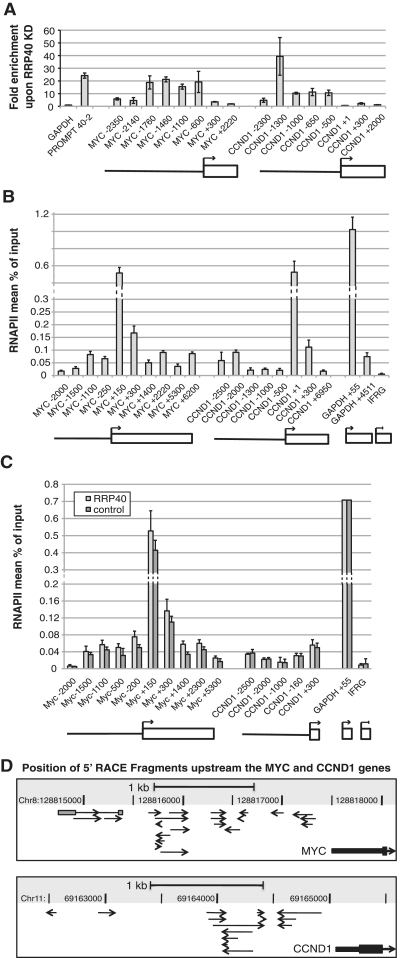
Next we chose the proto-oncogene MYC and CCND1, encoding cyclin D1, for a detailed analysis of their PROMPT regions. To confirm that the MYC and CCND1 loci indeed produce PROMPTs, we initially attempted northern blotting analyses (data not shown). Most likely due to the low abundance of PROMPTs, however, these attempts failed to produce reliable results. As an alternative, we analyzed MYC and CCND1 promoter proximal sequences spanning from their annotated TSSs, as defined by RefSeq, to ∼2.5 kb upstream by RT–qPCR with sets of six and five different amplicons, respectively (Figure 3A). Both regions showed robust (10- to 40-fold) increases in RT–qPCR signals upon RRP40 KD. Notably, farther than ∼2 kb upstream of the respective TSSs less RNA stabilization was observed (amplicons MYC -2350, MYC -2140 and CCND1 -2300). The profiles of these two genes thus follow that of an average PROMPT region with a peak of RNA stabilization ∼1.5 kb upstream the TSS and then tailing off between 2 and 3 kb upstream (13). In addition, two amplicons each in the coding regions of the MYC and CCND1 genes themselves detected no or little RNA stabilization [Figure 3A, (31)].
Figure 3.
Detailed analysis of MYC and CCND1 PROMPT regions. (A) PROMPT stabilization profiles of the MYC and CCND1 genes. Total RNA from HeLa cells treated with RRP40 or control siRNA was analyzed by RT–qPCR using several amplicons spanning the PROMPT- and the coding regions of the two genes as indicated schematically below the graph, as well as by the naming of the amplicons: Negative (upstream) and positive (downstream) numbers designate the approximate midpoint of the amplicon relative to the position of the TSS (+1). As a reference the MYC and CCND1 pA sites are at positions +5364 and +13 370, respectively. After normalization to GAPDH mRNA levels, fold enrichment is shown as the ratio RRP40/control. ‘PROMPT 40–2’ is a PROMPT upstream the STK11IP gene that has been validated previously [(13), Figure 1D]. Error bars show standard deviations of triplicate technical replica of a representative experiment. (B) Similar RNAPII levels in PROMPT and gene regions. The MYC and CCND1 genes were interrogated by RNAPII ChIP using an antibody specific for the N-terminus of the largest subunit of RNAPII (H224) and amplicons covering the PROMPT, promoter and gene regions (shown schematically below the graph). The inactive IFRG gene and GAPDH served as negative and positive controls, respectively. Values show the mean percent of input recovered in the IP as determined by qPCR. Note that for MYC +150, CCND1 +1 and GAPDH +55 values the y-axis is altered. Error bars in this and all following ChIP experiments show standard deviation of triplicate or more technical replica of a representative experiment. (C) RNAPII levels in MYC and CCND1 PROMPT and gene regions are unaffected by RRP40 KD. HeLa cells treated with control (eGFP) or RRP40 siRNA were analyzed by RPB1 ChIP (B). Note that for MYC +150 and GAPDH +55 values the y-axis is altered. (D) 5′-RACE analysis of the MYC and CCND1 PROMPT regions. The position and orientation of individual clones isolated by 5′-RACE is indicated relative to the 5′-end of the MYC and CCND1 genes. Note that one RACE product in the MYC locus was the result of splicing of two exons (gray boxes).
To validate transcription activity in the MYC and CCND1 PROMPT regions, we performed chromatin-immunoprecipitation (ChIP) with a polyclonal antibody against the N-terminus of RPB1, the largest subunit of RNAPII. An amplicon covering the promoter region of the inactive Interferon Responsive Gene (IFRG/RTP4) was used as a negative control. RNAPII levels were highest near the MYC and CCND1 promoters, likely reflecting paused RNAPII at these positions (17) and dropped into the body of the genes (Figure 3B). Notably, in the PROMPT regions RNAPII signals were comparable to that of the genes at the same distance downstream of the TSS. Moreover, RNAPII levels were similar whether or not RRP40 was depleted (Figure 3C), further supporting the conclusion that increased steady-state levels of PROMPTs in exosome-depleted cells are the result of decreased RNA decay, rather than increased transcription. Consequently, RNAPII levels as detected by a particular amplicon do not necessarily correlate strictly with levels of RNA stabilization in the same region because the latter likely also depends on factors other than actual transcription levels, such as substrate recognition by the exosome and/or the position, at which the transcription event was initiated or terminated.
PROMPTs can be transcribed bi-directionally. To map sites of PROMPT transcription initiation more precisely, we performed 5′-rapid amplification of cDNA ends (5′-RACE) analyses on cells depleted of RRP40. We used sets of primers covering the PROMPT regions of MYC and CCND1 (Figure 3D) as well as the DEPDC5 and SPHK1 genes (Supplementary Figure S2) that we had previously shown to exhibit ∼10–15-fold stabilization upon RRP40 KD (13). For each of the four regions analyzed, we identified multiple 5′-ends in both orientations, indicating that initiation of PROMPT transcription is promiscuous and further lending support to the idea that they are not transcribed from their own promoters, but rather rely on the promoter of the associated gene for expression. The 5′-RACE approach proved to be non-saturating, as few 5′-ends were identified more than once. Interestingly, only one cDNA, which was in sense orientation relative to the MYC gene, had undergone removal of a 434-nt intron (Figure 3D), whose splice sites conferred to the mammalian consensus sequences. Although there are no annotated expressed sequence tags in favor, we cannot exclude the possibility that this 5′-end is the result of a rare transcription initiation event upstream of the MYC gene. The RACE products had a median length of 153 (±126 nt; n = 75). Given that the oligonucleotides employed in RACE were randomly positioned, this would indicate that PROMPTs are on the average at least twice that length. However, because RNA features such as secondary structures in the 3′-half of PROMPTs could impede the reverse transcription reaction, this value might be an underestimation of the actual length. Interestingly, yeast CUTs have a median length of 504 nt (11). Taken together, PROMPT transcripts are initiated from multiple sites on both DNA strands, are intron-poor and their size range is in the hundreds of nucleotides, comparable to the size range of CUTs.
PROMPT-transcribing RNAPII is CTD-Ser2 phosphorylated
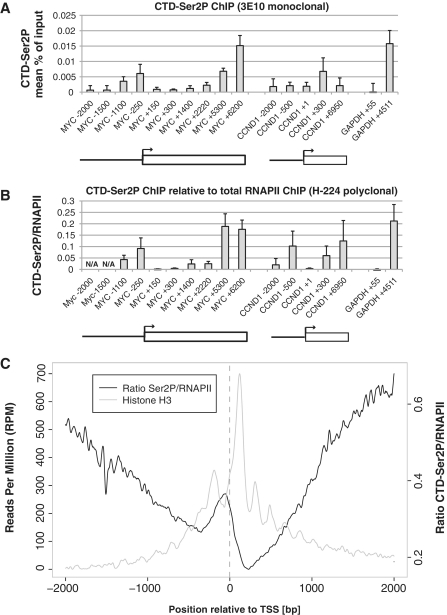
PROMPT transcription units are on the average much shorter than genes, suggesting that PROMPT transcription is more prone to termination or, alternatively, that elongation is impaired. To compare features of PROMPT and gene transcription on a molecular level, we analyzed the phosphorylation status of RPB1 in gene versus PROMPT regions. The CTD of RPB1 undergoes reversible phosphorylation as RNAPII traverses the gene: the CTD first becomes phosphorylated on Ser5 of its heptapeptide repeats immediately after transcription initiation, followed by Ser2P upon entry into the productive elongation phase (36). Consequently, a deficiency in establishing robust Ser2P could contribute to non-productive elongation and/or transcription termination of PROMPTs.
To test the phosphorylation status of the CTD directly, we conducted ChIP using a monoclonal antibody specific for CTD-Ser2P. Under the experimental conditions used, the resolution was <350 bp (see ‘Material and Methods’ section). As previously observed, CTD-Ser2P builds up from the TSS toward the end of the MYC gene [Figure 4A (37)]. A similar pattern is observed when CTD-Ser2P levels are normalized to those of total RPB1 (Figure 4B). In the MYC PROMPT region, the ChIP signal peaks ∼1100 bp upstream the TSS where a ∼16-fold PROMPT RNA stabilization is observed upon RRP40 KD (Figure 3A). At −1500 bp and upstream, the ChIP signal for CTD-Ser2P reaches background levels such that a ratio becomes no longer meaningful. Ser2P/RNAPII signals at the CCND1 PROMPT/gene mimic those of MYC in these regions at comparable distances on either site of the TSS. We also assayed for the presence of Ser5P-RNAPII (Supplementary Figure S3A) and again found that, when normalized to total RNAPII, signals in the PROMPT region are in the same range of those at equally distanced parts of the gene (Supplementary Figure S3B). We note that due to the promiscuous and overlapping nature of PROMPT RNA transcription (Figure 3D and Supplementary Figure S2), Ser2P and Ser5P signals generated independently at many different sites upstream of the gene's TSS will overlap and that this will preclude a linear 5′–3′-order as most often observed at protein-coding genes. Thus, our data appear to exclude the simple hypothesis that the lack of CTD phosphorylation is the major determinant of PROMPT transcription termination.
Figure 4.
PROMPT regions are covered by CTD-Ser2P. (A) CTD-Ser2P levels in the MYC and CCND1 gene and PROMPT regions. ChIP analysis was performed with a monoclonal antibody (3E10) specific for CTD-Ser2P as described in Figure 3B. (B) CTD-Ser2P levels relative to total RNAPII. Ratios were calculated by dividing the results from (A) and Figure 3B. At positions MYC −1500 and −2000 bp, the ChIP signals for CTD-Ser2P are too low to make this calculation meaningful (‘N/A’). (C) Genome wide location of CTD-Ser2P/RNAPII (black) and histone H3 (gray). A composite plot of CTD-Ser2P/RNAPII ratios (right y-axis) over all UCSC TSSs in human T cells was constructed from published data (39). Histone data were taken from Ref. (29).
To assess the generality of this observation, we analyzed published genome-wide ChIP-seq data of CTD-Ser2P from human CD4+ T-cells (38) and compared them to similar data of total RNAPII from HeLa cells (39). Figure 4C shows a composite plot of Ser2P/RNAPII ratio signals (black line graph) over all UCSC TSSs ±2000 bp. This confirms the presence of Ser2P in PROMPT regions of genes and argues that RNAPII machineries in PROMPT regions generally bear characteristics of productive elongation. We also plotted a data set for genome-wide histone H3 occupancy (29) (Figure 4C, gray line graph). As expected the nucleosome patterning is more distinct downstream than upstream the gene TSSs. Interestingly, nucleosome density is generally higher in genic than in PROMPT regions, which may be relevant for the different RNAPII transcription properties of the two regions (see ‘Discussion’ section).
Levels of RNAPII in a PROMPT region are inversely correlated with those of the downstream gene
Having shown similar transcriptional properties of gene and PROMPT regions, we next sought evidence that these regions might indeed utilize RNAPII stemming from the same ‘pool’. To develop a system amenable to manipulation, we inserted a ∼2.5 kb fragment from the S. cerevisiae TOM1 open reading frame upstream of a tetracycline-inducible CMV promoter (CMV/TetO2) driving expression of the β-globin gene. For better reproducibility, we integrated a single copy of this chimeric construct into a specific site within the genome of HEK293 cells using the Flp-In T-Rex system. RNA levels measured by RT–qPCR showed that the heterologous DNA sequence generated detectable RNA upstream of the CMV promoter that was only modestly (∼2-fold) stabilized in an RRP40 KD (data not shown). We conclude that the TOM1 sequence allows for transcription, but lacks at least part of the information that turns these transcripts into efficient exosome substrates, and thus refer to this sequence as ‘PROMPT like’.
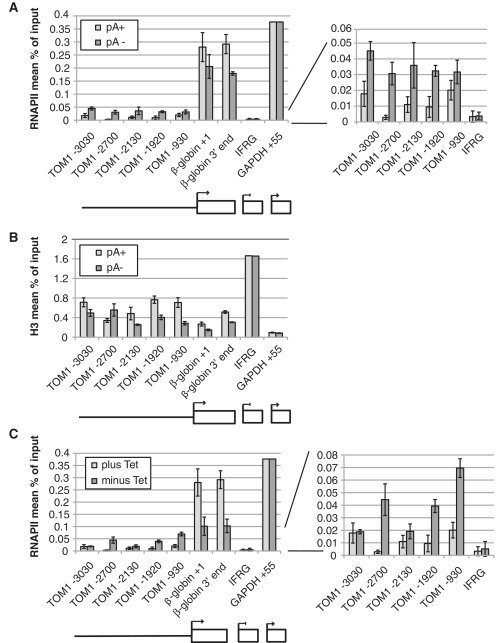
As we have previously shown that a polyadenylation site mutation [AAUAAA (pA+) to AAGAAA (pA−)] leads to decreased transcription initiation in this context, we also integrated an otherwise identical copy containing this mutation (28). To assess the transcriptional activity within gene and upstream regions of the two constructs, RNAPII and H3 occupancies were measured by ChIP under inducing conditions (250 ng/ml tetracycline for 24 h). Consistent with previous results, RNAPII levels decreased 2-fold in the 3′-region of the pA− relative to the pA+ gene [Figure 5A, left panel, (28)]. In contrast, RNAPII levels increased from ∼1.5 to 6-fold in a region from ∼0.9 to 3.0 kb upstream of the β-globin pA− gene TSS (see zoom in of Figure 5A, right panel). This behavior largely anti-correlated with nucleosome density in the same region as measured by H3 ChIP (Figure 5B). Thus, a gene mutation that reduces RNAPII genic density increases RNAPII occupancy upstream in the associated PROMPT-like region.
Figure 5.
Gene and PROMPT compete for the transcription machinery. (A) Lowered gene transcription increases RNAPII occupancy in the adjacent PROMPT region. ChIP analyses of total RNAPII on the TOM1-β-globin pA+ and pA− constructs performed and analyzed as in Figure 3B. The location of amplicons over the PROMPT (solid line) and β-globin gene (box) is shown schematically below the graph. The right panel zooms in on the ChIP results of amplicons located in the TOM1 region upstream of the β-globin TSS and the IFRG control region. (B) ChIP analysis of the same cells as in (A) conducted with an anti-histone H3 antibody. (C) Increased RNAPII occupancy in the adjacent PROMPT region, when transcription of the β-globin gene is decreased in the absence of tetracycline. HEK293 cells containing the TOM1-β-globin pA+ construct were grown in the presence or absence of 250 ng/ml tetracycline for 24 h before harvest and RNAPII ChIP analysis. Data depicted as in (A).
Another way to lower access of RNAPII to a Tet-inducible gene is to reduce its expression in the absence of tetracycline. We therefore measured RNAPII levels on the pA+ gene and its PROMPT-like region in the absence and presence of tetracycline. Again, lowered RNAPII levels inside the gene under repressive conditions were paralleled by increased RNAPII in the PROMPT-like region (Figure 5C). It thus appears that PROMPT- and gene-region belong the same RNAPII-containing ‘gene compartment’ (40) (see ‘Discussion’ section).
RNAPI and RNAPIII genes also harbor PROMPTs
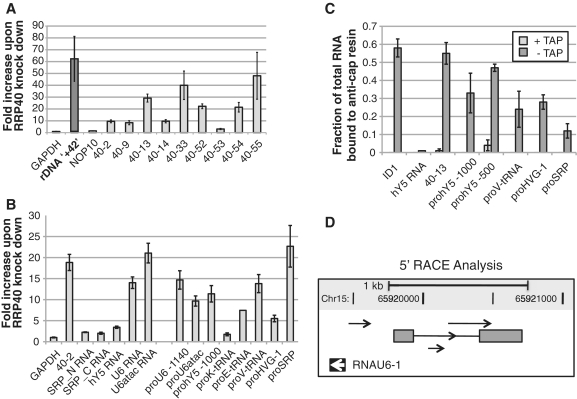
Given the promiscuous nature of PROMPTs, we next asked whether they could arise from promoters other than those serving RNAPII transcription. In line with this possibility, we detected transcript(s) originating from ∼1 kb upstream of ribosomal DNA (rDNA) promoters that were greatly (>60-fold) stabilized in an exosome knock-down (Figure 6A).
Figure 6.
RNAPI and RNAPIII promoters harbor PROMPTs. (A) A PROMPT upstream the RNAPI promoter of rDNAgenes. Transcript levels at ∼1 kb upstream of the TSS (rDNA ‘+42’, second column from the left) were measured in total RNA harvested from control or RRP40-depleted cells by RT–qPCR. The fold increase under exosome-depleted conditions is plotted on the y-axis in comparison to protein-coding genes (GAPDH and NOP10), as well as PROMPTs associated with RNAPII-driven genes (40–2 through 40–55). (B) RNAPIII genes are associated with exosome-targeted PROMPTs. Total RNA from RRP40 KD and control cells was reverse-transcribed with the anchored oligo-dT primer (‘dTVN’) and the changes in steady-state RNA (SRP, hY5, U6 and U6atac) and PROMPT levels upon RRP40 KD were determined by qPCR using the indicated amplicons. ‘pro-’ are amplicons designed for PROMPT regions of the respective RNAPIII genes. ‘40–2’ is a PROMPT upstream the STK11IP gene that has been validated previously [(13), Figures 1D and 3A]. (C) RNAPIII PROMPTs are capped. The fraction of capped RNA of the indicated mRNA, hY5 RNA and PROMPTs was determined as described in the legend to Figure 1B. (D) Adenylated, antisense transcripts present upstream of the U6 gene (RNAU6-1). 5′-RACE products represented by arrows were identified as in Figure 3D. Note that one RACE product was the result of splicing of two exons (gray boxes). Chromosomal co-ordinates and scale bar (1 kb) are shown on top.
Interestingly, it was reported that RNAPII is highly concentrated upstream of all five active, RNAPIII-transcribed U6 genes and that optimal expression of U6 depends on functional RNAPII (41). However, no evidence of RNA produced upstream of U6 was found. More recently, a series of genome-wide studies (38,42–44) found RNAPII upstream of most transcriptionally active RNAPIII genes. Again, none of the studies found evidence of RNAPII activity. We, therefore, conducted control and RRP40 KD experiments and tested by RT–qPCR relative RNA levels arising from regions upstream of one U6 gene (RNAU6-1, chr15) as well as genes encoding three tRNAs, vault RNA (HVG-1, chr5), rho RNA (hY5, chr7), U6atac (chr9) and signal-recognition particle RNA (SRP/7SL, chr14). Amplicons were placed between ∼0.5 and 1.5 kb upstream of the respective TSSs. From all tested regions adenylated transcripts were identified that were from 2- to 20-fold stabilized upon RRP40 depletion (Figure 6B). Thus, production of PROMPTs extends to RNAPIII genes. We also noted the existence of several unspliced ‘orphan ESTs’, i.e. ESTs unlinked to any annotated gene, that overlap the PROMPT region of some RNAPIII genes. Two examples are shown in Supplementary Figure S4A. In the first, 385–576 nt long ESTs overlap the hY5 PROMPT(s) stabilized by RRP40 KD (amplicons ‘hY5 -500’ and ‘hY5 -1000’ in Figure 6B and C). In the second example, ESTs ranging in length from 166 to 555 nt overlap the SRP PROMPT. Such orphan ESTs have also been observed within PROMPT regions of RNAPII genes (13) and their lengths corresponds well to estimates of PROMPT length based on our 5′-RACE analysis (Figure 3D and Supplementary Figure S2). We thus interpret these ESTs as further evidence of PROMPT activity.
Using the anti-cap IP strategy, we could furthermore determine that a sizable fraction of all RNAPIII PROMPTs analyzed are capped (Figure 6C). However, with the exception of the hY5 PROMPT, not to quite the same extent as PROMPTs arising from RNAPII genes (compare Figure 1B). At present, we are unable to distinguish whether this is for technical reasons, such as reduced accessibility of the 5′-end of these RNAs to the anti-cap antibody, or due to genuine differences in their cap status i.e. due to a mixture of capped and un-capped molecules. We conclude, however, that a considerable fraction of these RNAPIII PROMPTs is likely generated by RNAPII. While this is in agreement with recent observations showing RNAPII and its associated epigenetic marks covering regions upstream of RNAPIII promoters (38,42–44), it does not exclude the possibility that some PROMPTs upstream these genes arise from non-RNAPII activity. Like for PROMPTs associated with protein-coding genes, RACE analyses on RNA from RRP40 KD cells revealed the existence of several 5′-ends in the RNAU6-1 PROMPT region (Figure 6D). Like one of the MYC PROMPTs, a single of the cloned products was spliced by the use of canonical splice sites.
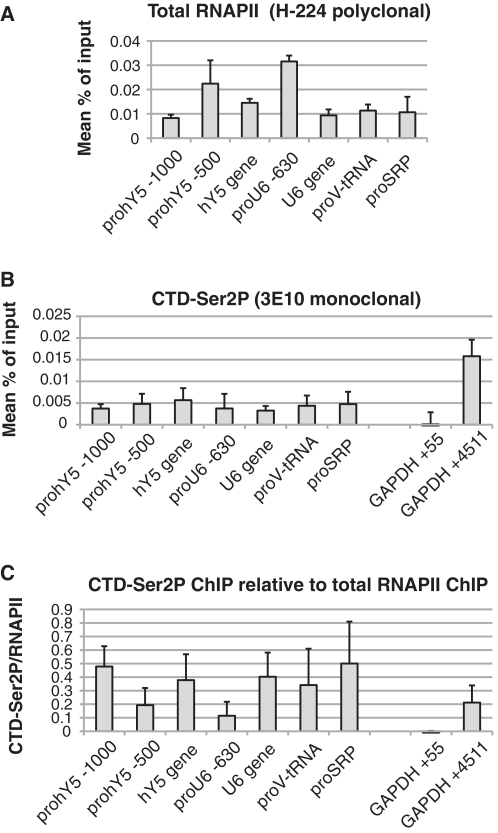
ChIP analyses of four RNAPIII PROMPT regions confirmed the presence of RNAPII, and moreover revealed the presence of Ser2P (Figure 7), underscoring the similarity between PROMPTs upstream of RNAPII- and RNAPIII-driven genes. Surprisingly, RNAPII also ChIP'd to the body of both the U6 and hY5 genes (Figure 7A). Perhaps, RNAPII is engaged in the production of transcripts from at least one of the U6 and Y-RNA gene copies. Another interesting finding was that both U6 and U6atac RNAs were greatly stabilized upon RRP40 KD (Figure 6B). Notably, U6 transcription switches from RNAPIII to –II driven when its TATA box is mutated (45). Moreover, insertion of a TATA box upstream the U2 gene converts it into an RNAPIII promoter. Likewise, perturbations in the rDNA processing machinery allow ribosomal RNAs to be transcribed by RNAPII (46). Thus, there appears to be considerable plasticity in RNA transcription, which is also reflected by the fact that the different RNA polymerases share common co-factors as well as similar factor binding sites [reviewed in (43,47)], and most RNAPIII promoters utilize the transcription factor MYC (48).
Figure 7.
RNAPIII PROMPT regions are covered by Ser2P-RNAPII. (A) RNAPIII PROMPT regions are covered by RNAPII. The indicated gene and PROMPT regions were subjected to RNAPII ChIP analysis as described in the legend to Figure 3B. (B) Ser2P ChIP analysis of the indicated gene and PROMPT regions as described in the legend to Figure 4A. (C) Ser2P levels relative to total RNAPII. Ser2p/RNAPII ratios were calculated by dividing the results from (B) and (A).
Thus both, RNAPIII type 2 genes (tRNAs, HVG-1 and SRP), containing internal promoter elements (A- and B-box), as well as type 3 genes (hY5, U6 and U6atac), which rely on upstream regulatory elements rather than gene-internal elements (47) have PROMPTs, at least a fraction, of which is produced by RNAPII. We conclude that PROMPTs, or PROMPT-like molecules, are produced upstream of promoters utilized by all three human RNA polymerases.
DISCUSSION
We describe here a biochemical characterization of human PROMPTs and their relation to the neighboring transcription unit. PROMPTs carry a 5′-cap and a 3′-adenosine tail. Although PROMPT 3′-end adenylation still needs further characterization, it is unlikely to follow conventional mRNA 3′-processing, but instead utilizes TRF4-2, a putative human homolog (35) of the yeast TRAMP-component Trf4p. However, in contrast to yeast where both the exosome and TRAMP are required for efficient CUT-degradation, and mutations in the genes encoding Trf4p and the exosome component Rrp6p are synthetic lethal (49), it appears that PROMPTs do not require TRF4-2 for their degradation. Instead, we suggest that the reduction in PROMPT signals, observed by dT-primed RT–qPCR in the RRP40/TRF4-2 double KD, is due to a partial loss of PROMPT poly(A) tail upon TRF4-2-depletion.
PROMPTs are mostly restricted to the nucleus (Figure 1A). Together with the estimated relatively small sizes of PROMPTs (∼200–600 nt), this raises the question why transcription productively elongates in the direction of the known gene to yield stable transcripts, while PROMPT transcription, regardless of its orientation with respect to the gene, is unproductive and rapidly terminates. It has been suggested that this choice would be dictated by the phosphorylation status of the RNAPII CTD (20). Indeed in yeast, termination of CUTs is promoted by the Nrd1p/Nab3p/Sen1p complex and coupled to rapid degradation by the exosome. The Nrd1p subunit directly binds the nascent CUT as well as the CTD-Ser5P, whereas CTD-Ser2P is antagonistic to Nrd1p binding (22,24). However, we show here that CTD-Ser2P is present in comparable amounts in gene and PROMPT regions of the MYC and CCND1 loci, a trend which is also apparent from genome-wide CTD-Ser2P profiles (Figure 4C). In addition, it is currently not known whether an Nrd1/Nab3p/Sen1p complex-like termination system functions in higher eukaryotes. We therefore disfavor the simple hypothesis that CTD phosphorylation status is directly responsible for PROMPT termination and degradation.
Nucleosome-depleted regions (NDRs) are a common feature of promoters and enhancers. The exposed DNA allows access to relevant DNA binding factors, but as a trade-off, these same regions may invite promiscuous transcriptional initiation events that not only compete for available resources, but whose products also need to be effectively recognized and discarded. This appears most obvious in S. cerevisiae, where CUTs almost exclusively originate from NDRs (10,11). In humans, PROMPT transcription is preferentially initiated upstream of the NDR, however, in a region where the nucleosome pattern appears less well phased than in the body of the gene (Figure 4C). Conceivably, it could, therefore, be this lack of structure that not only allows for PROMPT initiation (see below), but also leads to their default termination. In addition, another possible determinant for PROMPT transcription termination is the status of the underlying chromatin, e.g. the nature of their post-translational modifications. While many histone modifications associated with active transcription show, like RNAPII itself, symmetrical distribution over the TSS, some are clearly biased toward the gene. These include H3K79me2 (20) H3K36me3, H4K20me1 and H2BK5me1 (50). Whether any of these modifications (or lack thereof) are causal to PROMPT metabolism or rather a consequence of PROMPT activity remains to be tested. Finally, unlike genes, cloned PROMPTs and ‘orphan’ ESTs that overlap PROMPT regions are largely devoid of introns (Figure 3D, Supplementary Figure S4A and data not shown). Intron removal from pre-mRNAs occurs mostly co-transcriptionally and stimulates gene expression by promoting export of the mature mRNA (51). A simple hypothesis therefore would be that PROMPTs are retained in association with chromatin partly due to a failure to undergo splicing, and that this might contribute to activation of the exosome (52,53). Moreover, splicing can feed back to stimulate transcription (54,55), which may positively impact transcription elongation properties of genes over PROMPTs. Further experiments are required to test these hypotheses.
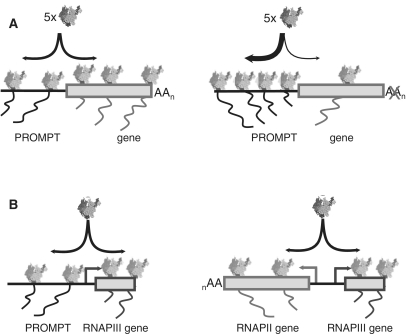
By comparing RNAPII profiles over a single-copy stably integrated β-globin gene and its associated PROMPT-like region, we found evidence for transcriptional anti-correlation. Taken together with our observation that RNAPII CTD phosphorylation is not grossly different between the two regions, this implies that PROMPT and gene regions share the same type of RNAPII machinery. This notion is akin to the recently proposed ‘gene compartments’ with a local concentration of RNAPII and other transcription factors (40). While used to explain the relationship between initiation and termination of transcription (28,40), such an environment might also encompass the PROMPT region, which then could experience increased RNAPII density under conditions that impede gene transcription (Figure 8A). Regardless the mechanistic details, gene/PROMPT ‘competition’ might even take the form of a co-operation. Rather than ‘selfish’ PROMPT transcription merely taking advantage of available resources, conceivably any event that would shift usage of the RNAPII transcription machinery between gene and PROMPT could serve a regulatory role by redirecting such resources. Although the general function of PROMPTs remains obscure, it might incur fine-tuning by the amount of available transcription factors. By extension, a related mechanism might also be in place between genes with closely divergent TSSs, each serving as a PROMPT for the other. In this respect, it is interesting to note that bioinformatic studies indicate that in vertebrates, divergent genes statistically tend to perform related functions, which subjects functionally related genes to correlated transcriptional regulation (56).
Figure 8.
Schematic representation of PROMPT/RNAPII gene, PROMPT/RNAPIII gene and RNAPII gene/RNAPIII gene relationships. (A) Gene and PROMPT transcription reside in the same gene compartment. This can lead to competition for available RNAPII and other transcription factors as conditions that reduce transcription of the protein-coding gene—here shown to be the result of a pA site mutation—allow the PROMPT to attract more RNAPII. (B) RNAPII activity upstream of RNAPIII genes can either take the form of PROMPT transcription (left) or expression of a neighboring, divergently transcribed protein-coding gene (right). See ‘Discussion’ section for more details.
We also assayed for PROMPT activity associated with RNAPI and RNAPIII-driven genes. RNAPII had previously been found upstream of the U6 gene family member, which is transcribed by RNAPIII. RNAPII's presence is important to facilitate U6 transcription, however, no evidence of actual RNAPII activity was reported (41). Upon exosome KD, we detected capped RNA species that were greatly stabilized not only upstream of the U6 gene, but also of all other RNAPIII genes tested (Figure 6). Very recently, a series of studies found RNAPII upstream of many transcriptonally active RNAPIII genes (38,42–44). Again, none of the studies found evidence of RNAPII activity. We suggest that this is because the produced transcripts are rapidly turned over and thus are bona fide PROMPTs. Inspection of individual RNAPIII transcripts throughout the genome, revealed that these are frequently located immediately upstream of canonical RNAPII TSSs, and that their transcription is convergent with the RNAPII genes (examples in Supplementary Figure S4B and data not shown). Moreover, a genome-wide study recently came to the conclusion that 20% of tRNA genes reside within 1 kb of the TSS of a RNAPII-transcribed transcription unit (43). We speculate that efficient expression of RNAPIII genes requires active RNAPII upstream, and that this activity can either lead to the production of a stable mRNA or unstable PROMPTs (Figure 8B). In this vein, PROMPT regions could possibly be viewed as enhancers/modulators of both RNAPII and RNAPIII-driven genes. In support of this idea, two recent studies reported the surprising finding that large fractions of mouse transcriptional enhancer domains in both macrophages (57) and cortical neurons (58) are covered by RNAPII and generate non-coding RNAs [dubbed enhancer RNAs (eRNAs)]. While the two studies disagreed on the directionality and the 3′-adenylation status of these transcripts, they both found them to be of extremely low abundance. Interestingly, eRNAs posses heterogeneous 5′-ends (58), depend on the presence of the gene promoter, and their levels are correlated with expression levels of the adjacent genes (57); features reminiscent of PROMPTs. It thus appears that enhancers, even though they can be located tens of thousands of base pairs upstream of their cognate promoter, PROMPT regions and promoters themselves share at least some common characteristics. Notably, it has been suggested that all of these transcription events might create a chromatin landscape favorable for transcription of the associated gene, possibly by attracting chromatin modifying enzymes and/or providing a pool of readily available RNAPII and/or GTFs (19,20,57,58). Whether functionality in general is conveyed by the transcript, the act of transcription, or both remains an open question.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The Danish National Research Foundation and The Danish Cancer Society. P. P. is the recipient of a grant from the Lundbeck Foundation. Funding for open access charge: Budget of the corresponding author.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Dorthe C. Riishøj for excellent technical assistance. Drs Elisabeth Kremmer, Dirk Eick, Ger Pruijn and Douglas Black are thanked for antibodies. We are grateful to Drs Søren Lykke-Andersen and Manfred Schmid for critical reading of the manuscript.
REFERENCES
- 1.Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, Thurman RE, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermuller J, Hofacker IL, et al. RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science. 2007;316:1484–1488. doi: 10.1126/science.1138341. [DOI] [PubMed] [Google Scholar]
- 4.Hayashizaki Y, Carninci P. Genome Network and FANTOM3: assessing the complexity of the transcriptome. PLoS Genet. 2006;2:e63. doi: 10.1371/journal.pgen.0020063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.van Bakel H, Nislow C, Blencowe BJ, Hughes TR. Most ‘dark matter’ transcripts are associated with known genes. PLoS Biol. 2010;8:e1000371. doi: 10.1371/journal.pbio.1000371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wyers F, Rougemaille M, Badis G, Rousselle JC, Dufour ME, Boulay J, Regnault B, Devaux F, Namane A, Seraphin B, et al. Cryptic pol II transcripts are degraded by a nuclear quality control pathway involving a new poly(A) polymerase. Cell. 2005;121:725–737. doi: 10.1016/j.cell.2005.04.030. [DOI] [PubMed] [Google Scholar]
- 7.LaCava J, Houseley J, Saveanu C, Petfalski E, Thompson E, Jacquier A, Tollervey D. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell. 2005;121:713–724. doi: 10.1016/j.cell.2005.04.029. [DOI] [PubMed] [Google Scholar]
- 8.Houseley J, LaCava J, Tollervey D. RNA-quality control by the exosome. Nat. Rev. Mol. Cell. Biol. 2006;7:529–539. doi: 10.1038/nrm1964. [DOI] [PubMed] [Google Scholar]
- 9.Schmid M, Jensen TH. The exosome: a multipurpose RNA-decay machine. Trends Biochem. Sci. 2008;33:501–510. doi: 10.1016/j.tibs.2008.07.003. [DOI] [PubMed] [Google Scholar]
- 10.Neil H, Malabat C, d'Aubenton-Carafa Y, Xu Z, Steinmetz LM, Jacquier A. Widespread bidirectional promoters are the major source of cryptic transcripts in yeast. Nature. 2009;457:1038–1042. doi: 10.1038/nature07747. [DOI] [PubMed] [Google Scholar]
- 11.Xu Z, Wei W, Gagneur J, Perocchi F, Clauder-Munster S, Camblong J, Guffanti E, Stutz F, Huber W, Steinmetz LM. Bidirectional promoters generate pervasive transcription in yeast. Nature. 2009;457:1033–1037. doi: 10.1038/nature07728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Chekanova JA, Gregory BD, Reverdatto SV, Chen H, Kumar R, Hooker T, Yazaki J, Li P, Skiba N, Peng Q, et al. Genome-wide high-resolution mapping of exosome substrates reveals hidden features in the Arabidopsis transcriptome. Cell. 2007;131:1340–1353. doi: 10.1016/j.cell.2007.10.056. [DOI] [PubMed] [Google Scholar]
- 13.Preker P, Nielsen J, Kammler S, Lykke-Andersen S, Christensen MS, Mapendano CK, Schierup MH, Jensen TH. RNA exosome depletion reveals transcription upstream of active human promoters. Science. 2008;322:1851–1854. doi: 10.1126/science.1164096. [DOI] [PubMed] [Google Scholar]
- 14.Seila AC, Calabrese JM, Levine SS, Yeo GW, Rahl PB, Flynn RA, Young RA, Sharp PA. Divergent transcription from active promoters. Science. 2008;322:1849–1851. doi: 10.1126/science.1162253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Taft RJ, Glazov EA, Cloonan N, Simons C, Stephen S, Faulkner GJ, Lassmann T, Forrest AR, Grimmond SM, Schroder K, et al. Tiny RNAs associated with transcription start sites in animals. Nat. Genet. 2009;41:572–578. doi: 10.1038/ng.312. [DOI] [PubMed] [Google Scholar]
- 16.Taft RJ, Simons C, Nahkuri S, Oey H, Korbie DJ, Mercer TR, Holst J, Ritchie W, Wong JJ, Rasko JE, et al. Nuclear-localized tiny RNAs are associated with transcription initiation and splice sites in metazoans. Nat. Struct. Mol. Biol. 2010;17:1030–1034. doi: 10.1038/nsmb.1841. [DOI] [PubMed] [Google Scholar]
- 17.Nechaev S, Adelman K. Promoter-proximal Pol II: when stalling speeds things up. Cell Cycle. 2008;7:1539–1544. doi: 10.4161/cc.7.11.6006. [DOI] [PubMed] [Google Scholar]
- 18.Taft RJ, Kaplan CD, Simons C, Mattick JS. Evolution, biogenesis and function of promoter-associated RNAs. Cell Cycle. 2009;8:2332–2338. doi: 10.4161/cc.8.15.9154. [DOI] [PubMed] [Google Scholar]
- 19.Preker P, Nielsen J, Schierup MH, Jensen TH. RNA polymerase plays both sides: vivid and bidirectional transcription around and upstream of active promoters. Cell Cycle. 2009;8:1106–1107. [PubMed] [Google Scholar]
- 20.Seila AC, Core LJ, Lis JT, Sharp PA. Divergent transcription: a new feature of active promoters. Cell Cycle. 2009;8:2557–2564. doi: 10.4161/cc.8.16.9305. [DOI] [PubMed] [Google Scholar]
- 21.Jacquier A. The complex eukaryotic transcriptome: unexpected pervasive transcription and novel small RNAs. Nat. Rev. Genet. 2009;10:833–844. doi: 10.1038/nrg2683. [DOI] [PubMed] [Google Scholar]
- 22.Gudipati RK, Villa T, Boulay J, Libri D. Phosphorylation of the RNA polymerase II C-terminal domain dictates transcription termination choice. Nat. Struct. Mol. Biol. 2008;15:786–794. doi: 10.1038/nsmb.1460. [DOI] [PubMed] [Google Scholar]
- 23.Thiebaut M, Kisseleva-Romanova E, Rougemaille M, Boulay J, Libri D. Transcription termination and nuclear degradation of cryptic unstable transcripts: a role for the nrd1-nab3 pathway in genome surveillance. Mol. Cell. 2006;23:853–864. doi: 10.1016/j.molcel.2006.07.029. [DOI] [PubMed] [Google Scholar]
- 24.Vasiljeva L, Kim M, Mutschler H, Buratowski S, Meinhart A. The Nrd1-Nab3-Sen1 termination complex interacts with the Ser5-phosphorylated RNA polymerase II C-terminal domain. Nat. Struct. Mol. Biol. 2008;15:795–804. doi: 10.1038/nsmb.1468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Buratowski S. Progression through the RNA polymerase II CTD cycle. Mol. Cell. 2009;36:541–546. doi: 10.1016/j.molcel.2009.10.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li J, Moazed D, Gygi SP. Association of the histone methyltransferase Set2 with RNA polymerase II plays a role in transcription elongation. J. Biol. Chem. 2002;277:49383–49388. doi: 10.1074/jbc.M209294200. [DOI] [PubMed] [Google Scholar]
- 27.Eberle AB, Lykke-Andersen S, Muhlemann O, Jensen TH. SMG6 promotes endonucleolytic cleavage of nonsense mRNA in human cells. Nat. Struct. Mol. Biol. 2009;16:49–55. doi: 10.1038/nsmb.1530. [DOI] [PubMed] [Google Scholar]
- 28.Mapendano CK, Lykke-Andersen S, Kjems J, Bertrand E, Jensen TH. Crosstalk between mRNA 3′-end processing and transcription initiation. Mol. Cell. 2010;40:410–422. doi: 10.1016/j.molcel.2010.10.012. [DOI] [PubMed] [Google Scholar]
- 29.Schones DE, Cui K, Cuddapah S, Roh TY, Barski A, Wang Z, Wei G, Zhao K. Dynamic regulation of nucleosome positioning in the human genome. Cell. 2008;132:887–898. doi: 10.1016/j.cell.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Spies N, Nielsen CB, Padgett RA, Burge CB. Biased chromatin signatures around polyadenylation sites and exons. Mol. Cell. 2009;36:245–254. doi: 10.1016/j.molcel.2009.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tomecki R, Kristiansen MS, Lykke-Andersen S, Chlebowski A, Larsen KM, Szczesny RJ, Drazkowska K, Pastula A, Andersen JS, Stepien PP, et al. The human core exosome interacts with differentially localized processive RNases: hDIS3 and hDIS3L. EMBO J. 2010;29:2342–2357. doi: 10.1038/emboj.2010.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Staals RH, Bronkhorst AW, Schilders G, Slomovic S, Schuster G, Heck AJ, Raijmakers R, Pruijn GJ. Dis3-like 1: a novel exoribonuclease associated with the human exosome. EMBO J. 2010;29:2358–2367. doi: 10.1038/emboj.2010.122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Maruyama K, Sugano S. Oligo-capping: a simple method to replace the cap structure of eukaryotic mRNAs with oligoribonucleotides. Gene. 1994;138:171–174. doi: 10.1016/0378-1119(94)90802-8. [DOI] [PubMed] [Google Scholar]
- 34.Vanacova S, Wolf J, Martin G, Blank D, Dettwiler S, Friedlein A, Langen H, Keith G, Keller W. A new yeast poly(A) polymerase complex involved in RNA quality control. PLoS Biol. 2005;3:e189. doi: 10.1371/journal.pbio.0030189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Martin G, Keller W. RNA-specific ribonucleotidyl transferases. RNA. 2007;13:1834–1849. doi: 10.1261/rna.652807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Buratowski S. Transcription. Gene expression–where to start? Science. 2008;322:1804–1805. doi: 10.1126/science.1168805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Glover-Cutter K, Kim S, Espinosa J, Bentley DL. RNA polymerase II pauses and associates with pre-mRNA processing factors at both ends of genes. Nat. Struct. Mol. Biol. 2008;15:71–78. doi: 10.1038/nsmb1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Barski A, Chepelev I, Liko D, Cuddapah S, Fleming AB, Birch J, Cui K, White RJ, Zhao K. Pol II and its associated epigenetic marks are present at Pol III-transcribed noncoding RNA genes. Nat. Struct. Mol. Biol. 2010;17:629–634. doi: 10.1038/nsmb.1806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Rozowsky J, Euskirchen G, Auerbach RK, Zhang ZD, Gibson T, Bjornson R, Carriero N, Snyder M, Gerstein MB. PeakSeq enables systematic scoring of ChIP-seq experiments relative to controls. Nat. Biotechnol. 2009;27:66–75. doi: 10.1038/nbt.1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yao J, Ardehali MB, Fecko CJ, Webb WW, Lis JT. Intranuclear distribution and local dynamics of RNA polymerase II during transcription activation. Mol. Cell. 2007;28:978–990. doi: 10.1016/j.molcel.2007.10.017. [DOI] [PubMed] [Google Scholar]
- 41.Listerman I, Bledau AS, Grishina I, Neugebauer KM. Extragenic accumulation of RNA polymerase II enhances transcription by RNA polymerase III. PLoS Genet. 2007;3:e212. doi: 10.1371/journal.pgen.0030212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Moqtaderi Z, Wang J, Raha D, White RJ, Snyder M, Weng Z, Struhl K. Genomic binding profiles of functionally distinct RNA polymerase III transcription complexes in human cells. Nat. Struct. Mol. Biol. 2010;17:635–640. doi: 10.1038/nsmb.1794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Oler AJ, Alla RK, Roberts DN, Wong A, Hollenhorst PC, Chandler KJ, Cassiday PA, Nelson CA, Hagedorn CH, Graves BJ, et al. Human RNA polymerase III transcriptomes and relationships to Pol II promoter chromatin and enhancer-binding factors. Nat. Struct. Mol. Biol. 2010;17:620–628. doi: 10.1038/nsmb.1801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Raha D, Wang Z, Moqtaderi Z, Wu L, Zhong G, Gerstein M, Struhl K, Snyder M. Close association of RNA polymerase II and many transcription factors with Pol III genes. Proc. Natl Acad. Sci. USA. 2010;107:3639–3644. doi: 10.1073/pnas.0911315106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lobo SM, Hernandez N. A 7 bp mutation converts a human RNA polymerase II snRNA promoter into an RNA polymerase III promoter. Cell. 1989;58:55–67. doi: 10.1016/0092-8674(89)90402-9. [DOI] [PubMed] [Google Scholar]
- 46.Conrad-Webb H, Butow RA. A polymerase switch in the synthesis of rRNA in Saccharomyces cerevisiae. Mol. Cell. Biol. 1995;15:2420–2428. doi: 10.1128/mcb.15.5.2420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dieci G, Fiorino G, Castelnuovo M, Teichmann M, Pagano A. The expanding RNA polymerase III transcriptome. Trends Genet. 2007;23:614–622. doi: 10.1016/j.tig.2007.09.001. [DOI] [PubMed] [Google Scholar]
- 48.Gomez-Roman N, Felton-Edkins ZA, Kenneth NS, Goodfellow SJ, Athineos D, Zhang J, Ramsbottom BA, Innes F, Kantidakis T, Kerr ER, et al. Activation by c-Myc of transcription by RNA polymerases I, II and III. Biochem. Soc. Symp. 2006;73:141–154. doi: 10.1042/bss0730141. [DOI] [PubMed] [Google Scholar]
- 49.Reis CC, Campbell JL. Contribution of Trf4/5 and the nuclear exosome to genome stability through regulation of histone mRNA levels in Saccharomyces cerevisiae. Genetics. 2007;175:993–1010. doi: 10.1534/genetics.106.065987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Barski A, Cuddapah S, Cui K, Roh TY, Schones DE, Wang Z, Wei G, Chepelev I, Zhao K. High-resolution profiling of histone methylations in the human genome. Cell. 2007;129:823–837. doi: 10.1016/j.cell.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 51.Valencia P, Dias AP, Reed R. Splicing promotes rapid and efficient mRNA export in mammalian cells. Proc. Natl Acad. Sci. USA. 2008;105:3386–3391. doi: 10.1073/pnas.0800250105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Schmid M, Jensen TH. Quality control of mRNP in the nucleus. Chromosoma. 2008;117:419–429. doi: 10.1007/s00412-008-0166-4. [DOI] [PubMed] [Google Scholar]
- 53.West S, Proudfoot NJ. Transcriptional termination enhances protein expression in human cells. Mol. Cell. 2009;33:354–364. doi: 10.1016/j.molcel.2009.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Furger A, O'Sullivan JM, Binnie A, Lee BA, Proudfoot NJ. Promoter proximal splice sites enhance transcription. Genes Dev. 2002;16:2792–2799. doi: 10.1101/gad.983602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Damgaard CK, Kahns S, Lykke-Andersen S, Nielsen AL, Jensen TH, Kjems J. A 5′-splice site enhances the recruitment of basal transcription initiation factors in vivo. Mol. Cell. 2008;29:271–278. doi: 10.1016/j.molcel.2007.11.035. [DOI] [PubMed] [Google Scholar]
- 56.Li YY, Yu H, Guo ZM, Guo TQ, Tu K, Li YX. Systematic analysis of head-to-head gene organization: evolutionary conservation and potential biological relevance. PLoS Comput. Biol. 2006;2:e74. doi: 10.1371/journal.pcbi.0020074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.De Santa F, Barozzi I, Mietton F, Ghisletti S, Polletti S, Tusi BK, Muller H, Ragoussis J, Wei CL, Natoli G. A large fraction of extragenic RNA pol II transcription sites overlap enhancers. PLoS Biol. 2010;8:e1000384. doi: 10.1371/journal.pbio.1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Kim TK, Hemberg M, Gray JM, Costa AM, Bear DM, Wu J, Harmin DA, Laptewicz M, Barbara-Haley K, Kuersten S, et al. Widespread transcription at neuronal activity-regulated enhancers. Nature. 2010;465:182–187. doi: 10.1038/nature09033. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.