Abstract
Guanine-rich sequences are highly abundant in the human genome, especially in regulatory regions. Because guanine-rich sequences have the unique ability to form G-quadruplexes, these structures may play a role in the regulation of gene transcription. In previous studies, we demonstrated that formation of G-quadruplexes could be induced with peptide nucleic acids (PNAs). PNAs designed to bind the C-rich strand upstream of the human BCL2 gene promoted quadruplex formation in the complementary G-rich strand. However, the question whether G-quadruplex formation was essential for PNA invasion remained unanswered. In this study, we compared PNA invasion in the native and mutant, i.e. not forming G-quadruplex, BCL2 sequences and showed that G-quadruplex is required for effective PNA invasion into duplex DNA. This finding provides strong evidence for not only sequence-specific, but also quadruplex specific, gene targeting with PNA probes. In addition, we examined DNA-duplex invasion potential of PNAs of various charges. Using the gel shift assay, chemical probing and dimethyl sulfate (DMS) protection studies, we determined that uncharged zwitterionic PNA has the highest binding specificity while preserving efficient duplex invasion.
INTRODUCTION
Guanine-rich oligonucleotides have the ability to form various secondary structures, including guanine quadruplexes. Numerous genes possess guanine-rich sequences in promoter and other regulatory segments. Much recent work has shown that these G-rich sequences could play a crucial role in gene transcription regulation. This provides an appealing opportunity for gene regulation techniques by targeting guanine quadruplexes. Quadruplex formation has been studied in PDGF-A (1,2), VEGF (2–4), c-myc (2,5), KRAS (6), C-KIT (7,8), BCL2 (9,10) hTERT (11), Rb (12) and PDGFR-β (13). Most often, G-quadruplexes were observed in models of single-stranded DNA oligonucleotides. Several studies have described evidence of quadruplex formation in supercoiled DNA (14,15); however, their formation is not as robust as that of Z-DNA or H-DNA triplex (16,17).
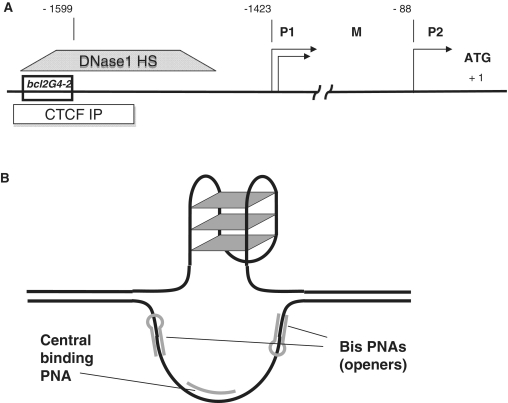
Human BCL2 is an apoptosis inhibitor protein. Due to its crucial role in preserving a balance between cell death and survival, it is considered an important target for anti-cancer treatment strategies. The BCL2 gene has several promoters, named P1, P2 and M (18). The regulatory region upstream of the translation initiation site is highly GC-rich. Several transcription factors were shown to bind to this region suggesting its importance for P1 promoter regulation (19–21). There are several sequences within this region with G-quadruplex-forming potential. One such sequence, which is located from −58 to −19 bp upstream of P1 promoter, was extensively studied and shown to form a G-quadruplex structure (9,10). In our studies, we focused on another sequence (labeled as bcl2G4-2; Figure 1) located in the BCL2 regulatory region 176 bp upstream of P1 promoter. In our previous study, we showed that this sequence could also form a G-quadruplex (22). The bcl2G4-2 sequence flanks the major DNaseI hypersensitive site (23) and coincides with a CTCF binding site (24) upstream of the P1 promoter (25). It was also shown that deletion of the region of the P1 promoter that covers the bcl2G4-2 sequence leads to a reduction in the promoter activity (26). Therefore, we believe that all the above evidence indicates that this sequence is important for regulation of BCL2 expression, and that G-quadruplex formation may play a role in its function.
Figure 1.
(A) Promoter structure of the human BCL2 gene; shown bcl2G4-2 insert is a 51-mer sequence of guanine-rich strand. DNase 1 hypersensitive site (ENCODE: UW Digital Genomic Footprinting K562 cells) and CTCF binding site (ENCODE: UW Histone ChIP Raw signal CTCF in GM12878 cells) are shown as boxes. (B) Proposed design for PNA binding to G-rich DNA region.
Peptide nucleic acids (PNAs) are nucleic acid mimics in which the natural nucleobases are connected to an achiral, uncharged polyamide backbone (27,28). Although PNAs form very stable duplexes with complementary nucleic acids, they are able to invade double-stranded DNA and bind only under certain conditions, such as when the DNA is negatively supercoiled or can form PD-loops (29–31). However, the efficiency of duplex invasion, sequence specificity, and the final stability of such PNA–DNA complexes are still poorly understood. In our previous work, we discovered that certain guanine-rich PNAs can invade naturally supercoiled double-stranded DNA by binding to the complementary cytosine-rich strand. This allows the free guanine-rich strand of DNA to form a quadruplex (22). However, it remained unclear if the ability to form quadruplex in BCL2 region is a strong requirement for PNAs to invade and bind DNA. If so, this phenomenon would highly increase targeting specificity with PNAs and provide new opportunities for efficient gene targeting and regulation. In addition, data from dimethyl sulfate (DMS) protection experiments (22) showed that quadruplexes were formed within 40–60% molecules depending on combination of PNAs used. We propose that PNA properties such as charge at physiological pH could play an important role in efficient DNA targeting.
In this study, we tested the hypothesis that G-quadruplex formation is required for effective PNA invasion into duplex DNA. We incubated PNAs with naturally supercoiled plasmids that had either the native or sequence-modified inserts from the BCL2 gene promoter, and used chemical probing and a DMS protection assay to study PNA invasion efficiency. Comparing invasion into native and mutant (not capable of forming G-quadruplex) BCL2 sequences we showed that G-quadruplex is required for PNA invasion in duplex DNA. This provides strong evidence for not only sequence specific, but also quadruplex specific, gene targeting with PNA probes. We also examined the DNA-duplex invasion abilities of PNAs of various charges. Prior to PNA invasion in plasmid DNA studies, we performed PNA probing using short single- and double-stranded oligonucleotides and plasmid fragments as targets to find the optimal conditions for PNA binding. We show that uncharged PNA possess the highest binding specificity, while simultaneously preserving efficient invading abilities.
MATERIALS AND METHODS
DNA oligonucleotides
The DNA oligonucleotides (Table 2) were synthesized on an ABI394 DNA synthesizer (PE Applied Biosystems, Foster City, CA, USA), and purified by denaturing polyacrylamide gel electrophoresis (PAGE) as described in detail in (32). The concentration of single-stranded oligonucleotides was measured at 260 nm on a HP 8452A Diode Array Spectrophotometer (Agilent Technologies, Hanover, Germany), and was calculated with extinction coefficient calculator software (http://www.basic.northwestern.edu/biotools/oligocalc.html). DNA oligonucleotides were 5′-32P labeled with [γ-32P]-ATP (Perkin Elmer, Waltham, MA, USA) by T4 polynucleotide kinase using standard protocol followed by purification on G-25 microcolumns (GE Healthcare, UK). Duplex DNA oligonucleotides were obtained by annealing of complementary strands by slow cooling from 95°C to room temperature. The DNA oligonucleotides were incubated with PNAs in TE buffer solution (Quality Biological, Gaithersburg, MD, USA) with 20 mM KCl (pH 7.4) at 37°C overnight with varying molar ratios.
Table 2.
Sequences of DNA oligonucleotides used in the study
| Name | Sequence |
|---|---|
| BCL2single-C | 5′-AAGGAAACACCCGACCGCCCCCTCCGCCCCGCTCCCTGGCCCGGAAGGAAA-3 |
| BCL2single-G | 5′-TTTCCTTCCGGGCCAGGGAGCGGGGCGGAGGGGGCGGTCGGGTGTTTCCTT-3′ |
| BCL2single-C-m | 5′-AAGGAAACACCCGACCGCCCCATCAGCACCGCTCCCTGGCCCGGAAGGAAA |
| BCL2duplex (original) | 5′-AAGGAAACACCCGACCGCCCCCTCCGCCCCGCTCCCTGGCCCGGAAGGAAA-3′ |
| 3′-TTCCTTTGTGGGCTGGCGGGGGAGGCGGGGCGAGGGACCGGGCCTTCCTTT-5′ | |
| BCL2duplex (mutant) | 5′-AAGGAAACACTTGACCGCTTCCTCCGCCCTGCTCCCTGGCTTGGAAGGAAA-3′ |
| 3′-TTCCTTTGTGAACTGGCGAAGGAGGCGGGACGAGGGACCGAACCTTCCTTT-5′ |
Dashed lines show PNA binding sites, solid lines show runs of guanines, adenines replacing guanines in the mutant sequence are shown in bold.
PNAs
Bis-PNA was purchased from Panagene, South Korea. Central PNA was synthesized via Boc-mediated solid phase synthesis on a 433A Automated Peptide Synthesizer from Applied BioSystems (33). Boc-protected PNA monomers were purchased from PE BioSystems. PNA oligomer quality was tested using an HP 1050 HPLC and spectrophotometrically (HP 8452A Diode Array Spectrophotometer). PNA concentration was measured at 260 nm on a HP 8452A Diode Array Spectrophotometer, and was calculated with extinction coefficient calculator software (http://www.basic.northwestern.edu/biotools/oligocalc.html). Prior to all experiments, PNAs were incubated in a shaker for 15 min at 42°C and their concentration was measured before mixing with DNA samples. PNA sequences are presented in Table 1.
Table 1.
Sequence of PNA oligonucleotides used in the study
| Name | Sequence |
|---|---|
| Bis-PNA (openers) | (Lys)2-TTJ-JTT-T-OOO-TTT-CCT-T—NH2 |
| cPNA1 | Lys-GGGCGGAGG-NH2 |
| cPNA2 | Lys-Glu-GGGCGGAGG-Glu-NH2 |
| cPNA3 | Glu-GGGCGGAGG-Glu-CO-NH2 |
J = pseudoisocytosine.
Plasmids
Plasmids carrying the appropriate inserts were obtained by cloning of the original and mutants BCL2duplex oligonucleotides (Table 2) into pCR-Blunt vector using Zero Blunt PCR Cloning Kit (Invitrogen, Carlsbad, CA, USA). Constructs were incorporated and amplified in One Shot TOP10 Chemically Competent Escherichia coli (Invitrogen) and selected with 50 µg/ml kanamycin. Plasmid DNA was purified using the Plasmid Maxi Kit (Qiagen, Valencia, CA, USA). All procedures were performed according to the manufacturer's recommendations. Plasmids were additionally purified by ultracentrifugation in CsCl gradient (34). All plasmids were sequenced to prove correct insert cloning using Maxam-Gilbert sequencing procedure (35). Incubation of plasmids with PNAs was performed in TE buffer (Quality Biological) with 20 mM potassium chloride (pH 7.4) at 37°C overnight with molar ratio 100:1 PNA to DNA.
Gel shift assay
Precast native polyacrylamide TBE gels (6%, 20%) were purchased from Invitrogen. 5′-32P labeled single stranded oligonucleotides bound with PNAs were run at room temperature in TBE buffer at 120 V. Gels were quantified using BAS-2500 Bioimager (FUJI Medical Systems USA, Stamford, CT, USA). For gel shift studies of plasmid fragments, after incubation with PNAs plasmids were cut by incubation with Fast Digest SpeI and PstI restriction enzymes (Fermentas, Hanover, MD, USA) for 20 min at 37°C followed by 3′-32P labeling with [α-32P]-dCTP (Perkin Elmer) by Klenow fragment of DNA polymerase I (Fermentas). After purification on G-25 microcolumns samples were run with gel electrophoresis.
Chemical Probing of plasmid DNA
Plasmid DNA samples contained 1 μg of plasmid in 40 mM TE buffer (pH 7.4) with 20 mM KCl. Plasmids incubated with PNAs were chemically modified to a total volume of 50 μl with 2.5 mM OsO4 plus 2.5 mM 2.2′-dipyridyl disulfide for 5 min at room temperature, with 2 μl diethylpyrocarbonate (DEPC) for 5 min at room temperature, or with 2 μl 10% DMS in ethanol for 15 min at 15°C. Reactions were stopped with 5× stop solution on ice (1.5 M sodium acetate pH 5.2, 1 M β-mercaptoethanol, 100 μg/ml yeast t-RNA) followed by ethanol precipitation and washing with 70% ethanol. Modified plasmids were cut by incubation with Fast Digest SpeI and Kpn1 restriction enzymes (Fermentas) for 20 min at 37°C followed by 3′-32P labeling with [α-32P]-dCTP (Perkin Elmer) by Klenow fragment of DNA polymerase I (Fermentas). After purification on G-50 microcolumns (GE Healthcare) followed by incubation in 10% piperidine at 95°C for 20 min and repeated lyophilizations, samples were analyzed by electrophoresis on 12% denaturing polyacrylamide gels without fragment purification, allowing the shorter fragment to run off the gel. To measure the intensity of the individual bands, the intensity profile of each lane was generated from the digitized gel image using Image Gauge software (FUJI Medical Systems USA). Intensity of the band corresponding to particular DNA base was normalized on the intensity of whole lane with background subtraction. For DMS protection analysis, profiles of band intensities were acquired and plotted.
RESULTS
G-quadruplex structures in the BCL2 gene and PNA binding design
In our previous work, we demonstrated G-quadruplex formation in a particular guanine-rich sequence in the human BCL2 gene (22). This sequence was chosen from the region of the BCL2 gene 176 bp upstream of the P1 promoter (bcl2G4-2, Figure 1A). Evidence of quadruplex formation was found in single-stranded oligonucleotide, and in naturally supercoiled plasmid DNA. In plasmid DNA, it was achieved by local melting using short invading central PNAs, and triplex-forming bis-PNA (36) binding to areas adjacent to the studied BCL2 insert. Although the binding property of PNAs is usually higher than natural nucleic acids of comparable length, DNA-duplex invasion is dependant on many different factors. It remained unclear if the ability to form a quadruplex in the BCL2 region is a strong requirement for PNAs to invade and bind DNA. In order to answer this question, we slightly mutated the BCL2 insert so that a G-quadruplex cannot form, while the sequence of the PNA binding site is preserved. Also, to address the role of PNA charge at physiological pH on their invading and binding properties we synthesized three different PNA probes with various overall charges (Table 1). cPNA1 is a central invading G-rich probe, with an overall charge +2; cPNA2 is identical to cPNA1 except two glutamic acid moieties are added to make the probe zwitterionic, charge neutral at physiological pH; cPNA3 is similar to cPNA1 and cPNA2, but is negatively charged due to the presence of two glutamic acid moieties. Bis-PNA (‘openers’) are designed to form triplexes at the edges of BCL2 insert, they have an overall charge equal to +3
PNA binding properties
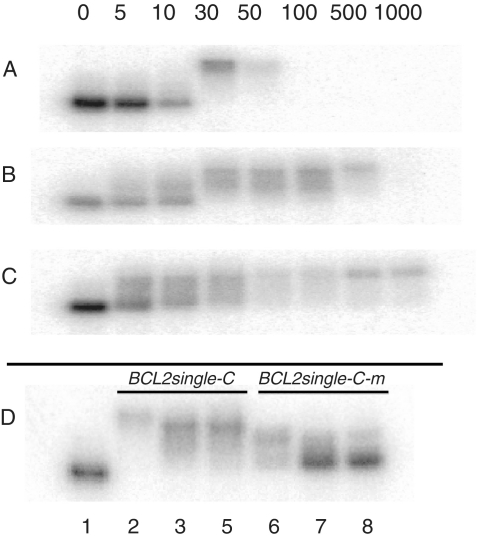
Gel shift studies were performed to evaluate binding of PNAs to complementary DNA oligonucleotides. First, we incubated single stranded oligonucleotide possessing the BCL2 sequence (BCL2single-C, Table 2) with duplex-forming G-rich PNAs with charges varying from +2 to −2 (cPNA1, cPNA2, cPNA3). All three PNA oligomers (cPNA1, cPNA2 and cPNA3) have the same sequence of nucleobases and differ only by charge, modified by introducing lysine and glutamic acid moieties onto the termini. Varying PNA:DNA molar ratios were tested, 5:1, 10:1, 30:1, 50:1, 100:1, 500:1 and 1000:1. Corresponding PNA concentrations were 5, 10, 30, 50, 100, 500 and 1000 nM. The results of gel shift studies are presented on Figure 2. It is evident that starting from 30 nM concentration positively charged cPNA1 binds DNA oligonucleotide (Figure 2A). However, at 100 nM the sample bands were not visualized on gel; instead, samples remained in the wells of the gel. This phenomenon was also observed when cPNA1 was incubated with a non-complementary oligonucleotide and also with an oligonucleotide duplex (data not shown). We attributed this phenomenon to non-specific positively charged cPNA1 binding at higher concentrations to negatively charged DNA. When cPNA2 was incubated with BCL2single-C at similar conditions and concentrations, it bound to the complementary DNA as efficiently as cPNA1 (Figure 2B). Starting from 30 nM PNA concentration DNA is completely bound to PNA. However, in contrast to cPNA1, cPNA2 does not show the non-specific binding at high PNA concentrations/molar ratios (up to 100 nM / 100:1). Incubation of the negatively charged cPNA3 with BCL2single-C showed somewhat lower binding affinity compared cPNA1 or cPNA2 (Figure 2C). Even thought binding of cPNA3 starts at lower 5 nM PNA concentration, at 30 nM the band of free DNA oligonucleotide could still be observed in the case of cPNA3 (Figure 2 C), but not in the case of cPNA1 or cPNA2 (Figure 2A and B). Bands of PNA–DNA complexes for cPNA2 and cPNA3 appear smeared. That may be indicative of their instability during the gel run.
Figure 2.
Gel shift studies of single stranded DNA oligonucleotides bound with PNAs. 5′-32P labeled BCL2single-C was incubated with cPNA1 (A), cPNA2 (B) and cPNA3 (C) in TE buffer with 20 mM KCl (pH 7.4) at 37°C overnight at varying molar ratios; numbers on the top indicate PNA concentration; DNA concentration was 5 nM. Samples were run in 20% native polyacrylamide TBE gels at room temperature. (D) Binding specificity studies. 5′-32P labeled BCL2single-C and BCL2single-C-m were incubated with cPNA1 (2, 6), cPNA2 (3, 7) and cPNA3 (4, 8) in TE buffer with 20 mM KCl (pH 7.4) at 37°C overnight at 30 nM PNA and 5 nM DNA concentration; BCL2single-C without PNAs was used as a control (1).
We also tested specificity of PNAs binding in experiments with a mutated DNA sequence (BCL2single-C-m Table 2). PNAs (30 nM) were incubated with native (lanes 2–4) and mutated (lanes 6–8) DNA oligonucleotides and analyzed in gel-shift assay (Figure 2D). All three PNAs bind to DNA, as expected. cPNA1 showed significant non-specific binding to the mutated sequence (lane 6), while only minor amounts of cPNA2 and cPNA3 bound to the mutated sequence at this concentration. Thus, we assume that positively and neutrally charged G-rich PNAs (cPNA1 and cPNA2) effectively bind complementary single-stranded DNA oligonucleotide at relatively low concentration (30 nM) while the zwitterionic cPNA2 maintains the highest specificity comparing to positively charged cPNA1, which, in addition, forms massive and/or neutrally charged complexes with DNA that could not enter the gel (Figure 2A).
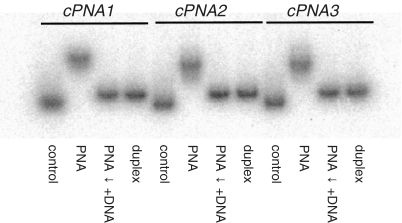
To further evaluate the stability of the aforementioned PNA–DNA complexes with different charge states, we performed concurrent binding studies. We hybridized cPNA1, cPNA2 and cPNA3 to BCL2single-C at 10:1 PNA/DNA molar ratio. After that we added BCL2single-G to formed PNA–DNA complexes at BCL2single-G/BCL2single-C molar ratio 10:1 and incubated at 37°C overnight. BCL2single-C and BCL2duplex oligonucleotides were run on a gel as controls. Results of these experiments are presented on Figure 3. At 10:1 PNA–DNA molar ratio all three PNAs effectively bind BCL2single-C. However, when a longer, complementary, DNA strand is added to the mixture, it completely displaces PNA from PNA–DNA complex and forms a DNA–DNA duplex. Interestingly, PNA displacement was observed for all studied PNAs and was not dependant on PNA charge.
Figure 3.
Concurrent binding studies. cPNA1, cPNA2 and cPNA3 were bound to 5′-32P labeled BCL2single-C at 10:1 PNA/DNA molar ratio (lanes marked ‘PNA’). BCL2single-G was added to PNA–DNA complexes at BCL2single-G/BCL2single-C molar ratio 10:1 and incubated at 37°C overnight (lanes marked ‘PNA ↓ +DNA’). Lanes marked ‘control’ contained BCL2single-C and lanes marked ‘duplex’ contained BCL2duplex oligonucleotides. Samples were run in 20% native polyacrylamide TBE gels at room temperature.
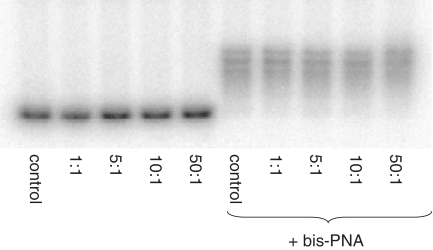
PNA displacement phenomenon would also explain the findings we obtained in the following experiment. We incubated naturally supercoiled plasmid with human BCL2 gene insert in 20 mM KCl with cPNA2, with and without bis-PNA (added with 100:1 PNA–DNA molar ratio). Then, we cut out a plasmid fragment with the target sequence using restriction enzymes, labeled with 32P and performed gel electrophoresis. The following cPNA/DNA molar ratios were used: 1:1, 5:1, 10:1 and 50:1. Results of the gel shift experiments are presented on Figure 4. Interestingly, no band shifts corresponding to cPNA–DNA complexes were observed (left panel). The only complexes present were those between bis-PNA and DNA (right panel). In topologically relaxed DNA (which is plasmid DNA after cutting with restriction enzymes), DNA–DNA duplex appears to be more favorable than PNA–DNA duplex. Thus, due to the observed phenomenon of PNA displacement by complementary strand, the gel shift assay cannot be implemented above as a measurement of effectiveness of PNA invasion within naturally supercoiled plasmid DNA and other assays should used (e.g. chemical probing).
Figure 4.
Gel shift studies of plasmid DNA bound with PNAs. After incubation with PNAs (TE plus 20 mM KCl, pH 7.4, 37°C, overnight) at varying PNA molar ratios, with (right) or without (left) bis-PNA, plasmids were cut with SpeI and Pst1 restriction enzymes followed by 3′-32P labeling with [α-32P]-dCTP (Perkin Elmer) by Klenow fragment of DNA polymerase I. After purification on G-50 microcolumns samples were analyzed in native 6% polyacrylamide gel electrophoresis.
PNA invasion in the plasmid DNA
To study the effectiveness of PNA invasion and binding to plasmid DNA, we performed a chemical probing assay. We used cPNA1, cPNA2 and cPNA3 possessing charge +2, 0 and −2 correspondingly in order to evaluate the effect of PNA charge. Also bis-PNA was used to form triplexes with DNA regions bordering the guanine-rich region to ease central PNA invasion. PNA binding design and promotion of quadruplex formation was similar to previous work (22) and presented on Figure 1B. The main question we addressed in the current study was whether the quadruplex-forming potential in G-strand facilitates PNA invasion. To address this question, we slightly mutated the base sequence in BCL2 insert so that G-quadruplex cannot form, but the sequence of PNA binding site is preserved. Sequences of original and mutant BCL2 inserts are shown in Table 2. Adenines substituted for guanines are shown in bold; runs of guanines are underlined. PNA binding sites are underlined with dashed lines. Figures 5 and 6 represent the results of chemical probing of plasmid DNA with a BCL2 original and mutant inserts incubated with different PNAs. We used several chemical probes to quantify local dsDNA melting at the desired location. First, we probed DNA with OsO4 and DEPC, allowing us to determine if thymines and adenines, respectively, are open within the insert. Second, we used DMS probing to reveal quadruplex formation through guanine protection at the N7 position. Plasmids had natural superhelical density. Only the G-rich strand was analyzed with all probes. Experimental samples were incubated without PNAs, with only central PNAs and with both central PNAs and bis-PNA. Lanes that represent probing with OsO4, DEPC and DMS as well as target plasmids (original and mutant) are labeled. Vertical solid line on the gels indicates central PNA binding site, dashed lines—bis-PNA binding site.
Figure 5.
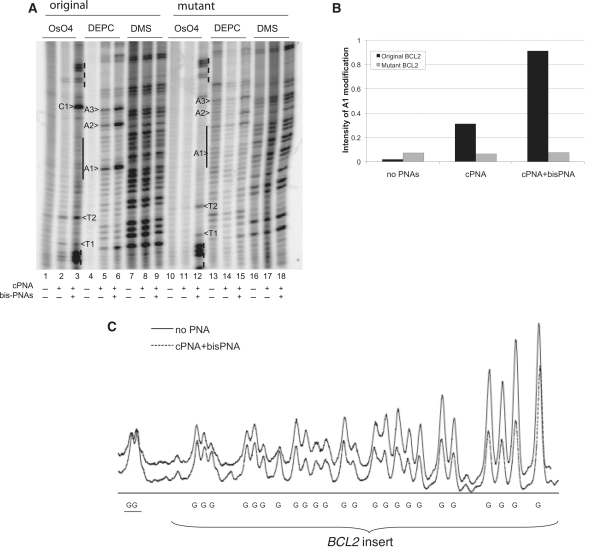
cPNA2 invasion studies. (A) Chemical probing of plasmid with BCL2 insert incubated with cPNA2 only and with both cPNA2 and bis-PNA. Incubation of plasmids with PNAs was performed in 20 mM KCl (pH 7.4) at 37°C overnight with molar ratio 100:1 PNA to DNA. Control sample was incubated only in TE buffer. Samples were chemically modified to a total volume of 50 μl with 2.5 mM OsO4 plus 2.5 mM 2.2′-dipyridyl disulfide for 5 min at room temperature, 2 μl DEPC for 5 min at room temperature, 2 μl 10% DMS in ethanol for 15 min at 15°C. Plasmids with two BCL2 insert were tested: with original quadruplex forming BCL2 sequence and with mutant BCL2 sequence not forming quadruplex. G-rich strand was analyzed. (B) A1 adenine modification. Intensity of the band corresponding to A1 adenine was normalized on the intensity of whole lane with background subtraction and plotted. (C) DMS modification profiles for the plasmid with original BCL2 insert incubated alone or in the presence of cPNA2 and bis-PNA. The two guanines outside of the G-quadruplex-forming region are underlined.
Figure 6.
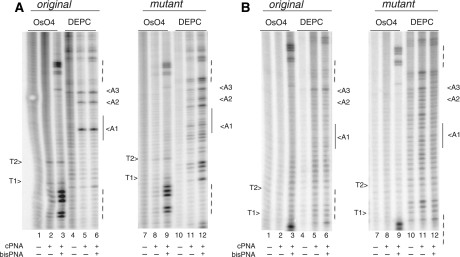
PNA invasion studies. Chemical probing of plasmid with BCL2 insert with PNAs (A—cPNA1; B—cPNA3). Incubation of plasmids with PNAs was performed in TE plus 20 mM KCl (pH 7.4) at 37°C overnight with molar ratio 100:1 PNA to DNA. Experimental samples were incubated with central PNAs only and with both central PNA and bis-PNA. Plasmids with two BCL2 insert were tested: with original quadruplex forming BCL2 sequence and with mutant BCL2 sequence not forming quadruplex. G-rich strand was analyzed.
Because cPNA2 showed the most promising results in our binding studies with DNA oligonucleotides, we first used cPNA2 for the invasion experiments. We performed probing of plasmids incubated with neutrally charged cPNA2 with and without bis-PNA (Figure 5A). There are no significant modifications of any Ts and As by OsO4 and DEPC correspondingly within the insert in the control plasmids without PNAs (lanes 1, 4, 10 and 13). There are two thymines in the central part of the insert marked as T1 and T2 and five Ts on each flank of the insert corresponding to bis-PNA binding regions. We observed effective modification of Ts corresponding to bis-PNA binding sites indicating their effective invasion into plasmids (lanes 3 and 12). Modification of T1 and T2 is weak in plasmid incubated with cPNA2 only (lane 2) but became significantly stronger after incubation with both cPNA2 and bis-PNA (lane 3). Surprisingly, the strongest modification with OsO4 after incubation with cPNA2 and bis-PNA was observed at the cytosine (C1). Even though thymines are the primary targets, modification of cytosines by OsO4 was also described (37). These findings indicate on effective invasion of cPNA2 even though T1 and T2 are not located within cPNA2 binding site, only adjacent to it. Further evidence of successful cPNA2 invasion is modification of adenines. DEPC probing is more useful as one of adenines is located within the cPNA1 binding site (labeled as A1) and can be used to estimate the efficiency of local melting, i.e. the efficiency of PNA invasion. There are also two other adenines within the native insert, marked as A2 and A3. We can see that cPNA2 successfully invades plasmid DNA with the original quadruplex forming sequence (lane 5); invasion is more pronounced when bis-PNA is added (lane 6).
In the plasmid with mutant BCL2 sequence, OsO4 modification of the thymines in the bis-PNA binding sites indicates the effective invasion of bis-PNA (lane 12). However, internal T1, T2 and C1 were modified in a smaller extent, and only in the presence of both PNAs (lane 12). These slight modifications are most likely due to the effect of bis-PNA invasion nearby. In addition, DEPC modification of adenines in the area corresponding to cPNA2 binding site is almost absent (lanes 14 and 15). The graph, representing intensity of A1 modification is shown in Figure 5B. It demonstrates that no local melting takes place in the plasmid with the mutant BCL2 sequence while it is present in original quadruplex-forming sequence and it is the most prominent when both, cPNA2 and bis-PNA, are used.
We also performed DMS protection studies for the plasmid incubated with cPNA2 with and without bis-PNA. It can be seen that combination of cPNA2 and bis-PNA leads to protection of several guanines within the BCL2 insert (lane 9). No protection was detected in the plasmid with mutated insert (lanes 16–18). Quantification of the DMS probing results in the original plasmid incubated with both PNAs (lane 9) and control plasmid (lane 7) is shown in Figure 5C. It shows that while modification of guanines outside of the insert was almost equal (2 G's on the left), within the insert modification of guanines in the plasmid incubated with cPNA2 and bis-PNA was reduced by ∼50–60%.
Results of plasmid probing using positively charged cPNA1 (Figure 6A) also show evidence of its invasion in plasmid with original BCL2 sequence. There is only background modification of thymines by OsO4 in the sample with cPNA only (lane 2). This indicates an inefficient opening of the insert outside the cPNA1 binding site. After incubation with both cPNA1 and bis-PNA, there is slightly increased modification of T1, T2 and C1; and pronounced modification of thymines in the bis-PNA binding site indicative of invasion of both PNAs (lane 3) with invasion of bis-PNA being more effective. This conclusion is further supported by DEPC probing. All three adenines, A1, A2 and A3 were unpaired in plasmids incubated with cPNA1, and with both cPNA1 and bis-PNA (lanes 5 and 6). The strongest signal is observed for A1. Chemical probing of the plasmid with the mutant BCL2 sequence revealed important differences. While probing of thymines revealed their modification only in the region of bis-PNA binding (lane 9), probing with DEPC shows non-specific adenine modifications within the whole BCL2 insert (lanes 11 and 12). Local melting takes place relatively far from central PNA binding site, and is also present outside of insert region. Modification of adenines is more prominent when bis-PNA is added (lane 12). Thus, there are several distinctions from the results of experiments with cPNA1 and cPNA2, namely: (i) cPNA2 invades plasmid DNA with the original BCL2 sequence more efficiently when bis-PNA is added; (ii) local melting in plasmid with the mutant BCL2 sequence is absent completely in case of cPNA2 alone. This indicates that invasion of cPNA2 depends to a larger extent on quadruplex formation in the G-rich strand than that for invasion of cPNA1.
The results of experiments with negatively charged central PNA (cPNA3) are presented on Figure 6B. In both plasmids, with original and mutant BCL2 sequence, local DNA melting was not observed within the PNA binding site, which indicates the inability of negatively charged cPNA3 to invade plasmid DNA and form a stable duplex. We also performed DMS probing of plasmids incubated with cPNA1 and cPNA3. However, we did not see conclusive DMS protection within the BCL2 insert sequence (data not shown).
Based on these data, we conclude that effective PNA invasion requires the G-rich strand to possess quadruplex-forming potential, which provides evidence of not only sequence-specific PNA targeting, but also quadruplex-specific. Positively charged (cPNA1) and zwitterionic (cPNA2) are able to sequence specifically invade naturally supercoiled plasmid with the original quadruplex-forming BCL2 sequence, and cPNA2 appeared to provide better target specificity while preserving DNA duplex invasion properties.
DISCUSSION
The human BCL2 gene is one of the most studied oncogenes due to its high importance in apoptosis regulation. Named after B-cell lymphoma 2, in which it was first discovered, the BCL2 gene encodes an apoptosis inhibitor protein. BCL2 protein is localized on the mitochondrial membrane and maintains a delicate balance between programmed cell death and survival. Due to its crucial role in cell fate regulation the BCL2 gene is an important target in anti-cancer treatment at present. Targeting BCL2 gene expression has been approached in various ways, such as interfering with transcription or post-translational regulation. Triplex-forming oligonucleotides (38), antisense oligonucleotides (39), miRNAs (miR-15 and miR-16 induce apoptosis by targeting BCL2) inhibiting protein–protein interactions (40) have been proposed. Some methods showed effectiveness in in vitro experiments, but issues concerning delivery of oligonucleotides to the target, stability within an intracellular environment, and target specificity are still major obstacles. One of the promising techniques which may overcome classical issues of DNA–RNA oligonucleotide delivery is implementation of PNAs as targeting probes. Their advantages, including charge neutrality (or weak charge due to possible chemical modifications), backbone flexibility, resistance to cellular nucleases and their ability to form base pairs with DNA and RNA, make them potentially promising agents for gene targeting. It has been demonstrated that PNAs can be successfully delivered into cells via conjugation with nuclear localization and cell penetrating peptides, and inhibit expression of target genes (41–44). Targeting based on structure recognition proposed in this study could significantly increase specificity of PNA probes and reduce off-target effects.
Another issue in gene targeting is the availability of the target sequence for a probe. This especially applies to the targets in chromosomal DNA as it is packed within the chromatin. Secondary DNA structures which ‘protrude’ from chromatin might be more vulnerable to nucleic acid probes. Furthermore, these secondary structures might even indicate or be crucial in active gene expression (45–48). Of these types of DNA secondary structures, guanine quadruplexes are believed to play an important role in gene regulation; as mentioned earlier, guanine-rich sequences with the potential to form G-quadruplexes are frequently found in promoter regions of many genes, especially in regulatory genes (1–6,9,10,49).
In our study, we addressed the question of the possibility of quadruplex targeting and stabilization by short PNAs. Our results showed that when added in equimolar ratio with the PNA probe, the DNA strand effectively displaces PNA from a PNA–DNA complex. This concerns all three central PNAs: cPNA1, cPNA2 and cPNA3 (Figure 3). This observation might explain the previously described (50,51) inability of PNAs to invade relaxed plasmid DNA, and requires further studies on PNA structure to achieve more stable PNA–DNA complexes. Our further studies also supported the observation of PNA displacement by DNA strand. We incubated cPNA1 with the original plasmid with BCL2 insert and no band shifts corresponding to PNA–DNA complexes were observed (Figure 4). However, in our previous studies, chemical probing showed efficient PNA invasion (22). The possible explanation may be the displacement of PNAs by complementary DNA strand once superhelical stress is relieved after the plasmid is cut by restriction enzymes. Based on these results, we concluded that gel shift assay cannot serve as a good method to study PNA invasion efficiency in plasmid DNA, so we proceeded with chemical probing.
PNA invasion studies enabled us to answer the main question for this study—what is the role of quadruplex formation on PNA invasion? In addition to plasmid with the original BCL2 sequence, we used also a plasmid with mutant sequence so that quadruplex cannot form while the sequence of PNA binding site is preserved (Table 2). We used chemical probing to test the opened state of G-strand. It revealed that only plasmid DNA with the original BCL2 sequence allowed the PNA oligomers to invade and bind. This also provides a means of assessing the stability of the G-quadruplex. Among the tested PNAs, positively charged (cPNA1) and neutral (cPNA2) have the highest potential to sequence specifically invade a naturally supercoiled plasmid with the BCL2 sequence. cPNA2 showed higher specificity to the BCL2 sequence compared to cPNA1. cPNA2 invasion was absent completely, as judged by local melting in the plasmid containing the mutant BCL2 insert. However, in the plasmid containing the original BCL2 sequence, we observed strong evidence of local melting within the BCL2 sequence, especially within the PNA binding site. Moreover, according to DMS protection studies, G-runs within the BCL2 sequence tend to form a quadruplex in the case of a combination of cPNA2 and bis-PNA.
There are several studies performed where formation of secondary structures in DNA is related with the increased ability to bind complementary oligonucleotide probes. Similar results were shown by Zhang et al. (30) where another secondary structure, cruciform, facilitated PNA invasion. Nielsen et al. (52) showed the role of multiple t-loops to increase the ability of single-stranded DNA to invade plasmid DNA; however, telomeric protein TRF2 was required for effective invasion. Duquette et al. (53) showed quadruplex formation in the G-strand of plasmid DNA during transcription while the C-strand was bound to de novo synthesized RNA along a relatively high length of transcribed DNA (up to 500 bp). Belotserkovskii et al. (54) show that Bis-PNAs hybridized to the (GAA/CTT) repeats of the frataxin gene results in varying degrees of transcription blockage, although no secondary DNA structures were necessary for effective DNA duplex invasion or PNA/DNA triplex stability. We have shown in the BCL2 sequence that G-quadruplex forming potential facilitates PNA invasion, while PNAs, once binding to the cytosine-rich complementary strand, promotes quadruplex stabilization. This provides strong evidence for PNA probes that are not only sequence specific, but also quadruplex specific. We also show that zwitterionic cPNA2 maintained binding affinity while improving the sequence and quadruplex specificity by avoiding large, non-specific aggregates. In future studies, we plan to implement these type of PNAs for in vivo studies on the regulation of the BCL2 promoter through G-quadruplex stabilization.
In conclusion, we show G-quadruplex formation in the guanine-rich promoter region of the human BCL2 gene is a prerequisite for a stable invasion of C-strand binding PNAs. Our results demonstrate a new mode of sequence-specific targeting with short, duplex-forming PNAs as means of stabilizing G-quadruplexes through targeting the complementary C-rich strand. This approach could provide a basis for future applications of gene expression regulation through G-quadruplex stabilization.
FUNDING
The study was partially sponsored by the Imaging Sciences Training Program supported in part by the Radiology and Imaging Sciences Department, Clinical Center and Intramural Research Program at the National Institute of Biomedical Imaging and Bioengineering, National Institute of Health. Funding for open access charge: Intramural Research Program of the National Institutes of Health, USA.
Conflict of interest statement. None declared.
REFERENCES
- 1.Qin Y, Rezler EM, Gokhale V, Sun D, Hurley LH. Characterization of the G-quadruplexes in the duplex nuclease hypersensitive element of the PDGF-A promoter and modulation of PDGF-A promoter activity by TMPyP4. Nucleic Acids Res. 2007;35:7698–7713. doi: 10.1093/nar/gkm538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Qin Y, Hurley LH. Structures, folding patterns, and functions of intramolecular DNA G-quadruplexes found in eukaryotic promoter regions. Biochimie. 2008;90:1149–1171. doi: 10.1016/j.biochi.2008.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sun D, Guo K, Rusche JJ, Hurley LH. Facilitation of a structural transition in the polypurine/polypyrimidine tract within the proximal promoter region of the human VEGF gene by the presence of potassium and G-quadruplex-interactive agents. Nucleic Acids Res. 2005;33:6070–6080. doi: 10.1093/nar/gki917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Guo K, Gokhale V, Hurley LH, Sun D. Intramolecularly folded G-quadruplex and i-motif structures in the proximal promoter of the vascular endothelial growth factor gene. Nucleic Acids Res. 2008;36:4598–4608. doi: 10.1093/nar/gkn380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Siddiqui-Jain A, Grand CL, Bearss DJ, Hurley LH. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl Acad. Sci. USA. 2002;99:11593–11598. doi: 10.1073/pnas.182256799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Cogoi S, Paramasivam M, Spolaore B, Xodo LE. Structural polymorphism within a regulatory element of the human KRAS promoter: formation of G4-DNA recognized by nuclear proteins. Nucleic Acids Res. 2008;36:3765–3780. doi: 10.1093/nar/gkn120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rankin S, Reszka AP, Huppert J, Zloh M, Parkinson GN, Todd AK, Ladame S, Balasubramanian S, Neidle S. Putative DNA quadruplex formation within the human c-kit oncogene. J. Am. Chem. Soc. 2005;127:10584–10589. doi: 10.1021/ja050823u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hsu ST, Varnai P, Bugaut A, Reszka AP, Neidle S, Balasubramanian S. A G-rich sequence within the c-kit oncogene promoter forms a parallel G-quadruplex having asymmetric G-tetrad dynamics. J. Am. Chem. Soc. 2009;131:13399–13409. doi: 10.1021/ja904007p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dexheimer TS, Sun D, Hurley LH. Deconvoluting the structural and drug-recognition complexity of the G-quadruplex-forming region upstream of the bcl-2 P1 promoter. J. Am. Chem. Soc. 2006;128:5404–5415. doi: 10.1021/ja0563861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dai J, Chen D, Jones RA, Hurley LH, Yang D. NMR solution structure of the major G-quadruplex structure formed in the human BCL2 promoter region. Nucleic Acids Res. 2006;34:5133–5144. doi: 10.1093/nar/gkl610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Palumbo SL, Ebbinghaus SW, Hurley LH. Formation of a unique end-to-end stacked pair of G-quadruplexes in the hTERT core promoter with implications for inhibition of telomerase by G-quadruplex-interactive ligands. J. Am. Chem. Soc. 2009;131:10878–10891. doi: 10.1021/ja902281d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu Y, Sugiyama H. Formation of the G-quadruplex and i-motif structures in retinoblastoma susceptibility genes (Rb) Nucleic Acids Res. 2006;34:949–954. doi: 10.1093/nar/gkj485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Qin Y, Fortin JS, Tye D, Gleason-Guzman M, Brooks TA, Hurley LH. Molecular cloning of the human platelet-derived growth factor receptor beta (PDGFR-beta) promoter and drug targeting of the G-quadruplex-forming region to repress PDGFR-beta expression. Biochemistry. 49:4208–4219. doi: 10.1021/bi100330w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sun D, Hurley LH. The importance of negative superhelicity in inducing the formation of G-quadruplex and i-motif structures in the c-Myc promoter: implications for drug targeting and control of gene expression. J. Med. Chem. 2009;52:2863–2874. doi: 10.1021/jm900055s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sun S, Chen J, Li W, Altintas I, Lin A, Peltier S, Stocks K, Allen EE, Ellisman M, Grethe J, et al. Community cyberinfrastructure for advanced microbial ecology research and analysis: the CAMERA resource. Nucleic Acids Res. 39:D546–551. doi: 10.1093/nar/gkq1102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hanvey JC, Klysik J, Wells RD. Influence of DNA sequence on the formation of non-B right-handed helices in oligopurine.oligopyrimidine inserts in plasmids. J. Biol. Chem. 1988;263:7386–7396. [PubMed] [Google Scholar]
- 17.Panyutin IG, Wells RD. Nodule DNA in the (GA)37.(CT)37 insert in superhelical plasmids. J. Biol. Chem. 1992;267:5495–5501. [PubMed] [Google Scholar]
- 18.Young RL, Korsmeyer SJ. A negative regulatory element in the bcl-2 5′-untranslated region inhibits expression from an upstream promoter. Mol. Cell. Biol. 1993;13:3686–3697. doi: 10.1128/mcb.13.6.3686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Seto M, Jaeger U, Hockett RD, Graninger W, Bennett S, Goldman P, Korsmeyer SJ. Alternative promoters and exons, somatic mutation and deregulation of the Bcl-2-Ig fusion gene in lymphoma. EMBO J. 1988;7:123–131. doi: 10.1002/j.1460-2075.1988.tb02791.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Heckman C, Mochon E, Arcinas M, Boxer LM. The WT1 protein is a negative regulator of the normal bcl-2 allele in t(14;18) lymphomas. J. Biol. Chem. 1997;272:19609–19614. doi: 10.1074/jbc.272.31.19609. [DOI] [PubMed] [Google Scholar]
- 21.Gomez-Manzano C, Mitlianga P, Fueyo J, Lee HY, Hu M, Spurgers KB, Glass TL, Koul D, Liu TJ, McDonnell TJ, et al. Transfer of E2F-1 to human glioma cells results in transcriptional up-regulation of Bcl-2. Cancer Res. 2001;61:6693–6697. [PubMed] [Google Scholar]
- 22.Onyshchenko MI, Gaynutdinov TI, Englund EA, Appella DH, Neumann RD, Panyutin IG. Stabilization of G-quadruplex in the BCL2 promoter region in double-stranded DNA by invading short PNAs. Nucleic Acids Res. 2009;37:7570–7580. doi: 10.1093/nar/gkp840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sabo PJ, Kuehn MS, Thurman R, Johnson BE, Johnson EM, Cao H, Yu M, Rosenzweig E, Goldy J, Haydock A, et al. Genome-scale mapping of DNase I sensitivity in vivo using tiling DNA microarrays. Nat. Methods. 2006;3:511–518. doi: 10.1038/nmeth890. [DOI] [PubMed] [Google Scholar]
- 24.Sabo PJ, Hawrylycz M, Wallace JC, Humbert R, Yu M, Shafer A, Kawamoto J, Hall R, Mack J, Dorschner MO, et al. Discovery of functional noncoding elements by digital analysis of chromatin structure. Proc. Natl Acad. Sci. USA. 2004;101:16837–16842. doi: 10.1073/pnas.0407387101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Celniker SE, Dillon LA, Gerstein MB, Gunsalus KC, Henikoff S, Karpen GH, Kellis M, Lai EC, Lieb JD, MacAlpine DM, et al. Unlocking the secrets of the genome. Nature. 2009;459:927–930. doi: 10.1038/459927a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Liu YZ, Boxer LM, Latchman DS. Activation of the Bcl-2 promoter by nerve growth factor is mediated by the p42/p44 MAPK cascade. Nucleic Acids Res. 1999;27:2086–2090. doi: 10.1093/nar/27.10.2086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nielsen PE, Egholm M, Berg RH, Buchardt O. Sequence-selective recognition of DNA by strand displacement with a thymine-substituted polyamide. Science. 1991;254:1497–1500. doi: 10.1126/science.1962210. [DOI] [PubMed] [Google Scholar]
- 28.Larsen HJ, Bentin T, Nielsen PE. Antisense properties of peptide nucleic acid. Biochim. Biophys. Acta. 1999;1489:159–166. doi: 10.1016/s0167-4781(99)00145-1. [DOI] [PubMed] [Google Scholar]
- 29.Nielsen PE, Christensen L. Strand displacement binding of a duplex-forming homopurine PNA to a homopyrimidine duplex DNA target. J. Am. Chem. Soc. 1996;118:2287–2288. [Google Scholar]
- 30.Zhang X, Ishihara T, Corey DR. Strand invasion by mixed base PNAs and a PNA-peptide chimera. Nucleic Acids Res. 2000;28:3332–3338. doi: 10.1093/nar/28.17.3332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bukanov NO, Demidov VV, Nielsen PE, Frank-Kamenetskii MD. PD-loop: a complex of duplex DNA with an oligonucleotide. Proc. Natl Acad. Sci. USA. 1998;95:5516–5520. doi: 10.1073/pnas.95.10.5516. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.He Y, Neumann RD, Panyutin IG. Intramolecular quadruplex conformation of human telomeric DNA assessed with 125I-radioprobing. Nucleic Acids Res. 2004;32:5359–5367. doi: 10.1093/nar/gkh875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Englund EA, Appella DH. Gamma-substituted peptide nucleic acids constructed from L-lysine are a versatile scaffold for multifunctional display. Angew. Chem. Int. Edn Engl. 2007;46:1414–1418. doi: 10.1002/anie.200603483. [DOI] [PubMed] [Google Scholar]
- 34.Sambrook J, Fritsch EF, Maniatis T. Molecular cloning: a laboratory manual. 2nd edn. NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 35.Maxam AM, Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Methods Enzymol. 1980;65:499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- 36.Panyutin IG, Panyutin IV, Demidov VV. Targeting linear duplex DNA with mixed-base peptide nucleic acid oligomers facilitated by bisPNA openers. Anal. Biochem. 2007;362:145–147. doi: 10.1016/j.ab.2006.11.014. [DOI] [PubMed] [Google Scholar]
- 37.Dizdaroglu M, Holwitt E, Hagan MP, Blakely WF. Formation of cytosine glycol and 5,6-dihydroxycytosine in deoxyribonucleic acid on treatment with osmium tetroxide. Biochem. J. 1986;235:531–536. doi: 10.1042/bj2350531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shen C, Rattat D, Buck A, Mehrke G, Polat B, Ribbert H, Schirrmeister H, Mahren B, Matuschek C, Reske SN. Targeting bcl-2 by triplex-forming oligonucleotide–a promising carrier for gene-radiotherapy. Cancer Biother. Radiopharm. 2003;18:17–26. doi: 10.1089/108497803321269296. [DOI] [PubMed] [Google Scholar]
- 39.Klasa RJ, Gillum AM, Klem RE, Frankel SR. Oblimersen Bcl-2 antisense: facilitating apoptosis in anticancer treatment. Antisense Nucleic Acid Drug Dev. 2002;12:193–213. doi: 10.1089/108729002760220798. [DOI] [PubMed] [Google Scholar]
- 40.Oltersdorf T, Elmore SW, Shoemaker AR, Armstrong RC, Augeri DJ, Belli BA, Bruncko M, Deckwerth TL, Dinges J, Hajduk PJ, et al. An inhibitor of Bcl-2 family proteins induces regression of solid tumours. Nature. 2005;435:677–681. doi: 10.1038/nature03579. [DOI] [PubMed] [Google Scholar]
- 41.Cutrona G, Carpaneto EM, Ulivi M, Roncella S, Landt O, Ferrarini M, Boffa LC. Effects in live cells of a c-myc anti-gene PNA linked to a nuclear localization signal. Nature Biotechnol. 2000;18:300–303. doi: 10.1038/73745. [DOI] [PubMed] [Google Scholar]
- 42.Boffa LC, Scarfi S, Mariani MR, Damonte G, Allfrey VG, Benatti U, Morris PL. Dihydrotestosterone as a selective cellular/nuclear localization vector for anti-gene peptide nucleic acid in prostatic carcinoma cells. Cancer Res. 2000;60:2258–2262. [PubMed] [Google Scholar]
- 43.Cutrona G, Carpaneto EM, Ponzanelli A, Ulivi M, Millo E, Scarfi S, Roncella S, Benatti U, Boffa LC, Ferrarini M. Inhibition of the translocated c-myc in Burkitt's lymphoma by a PNA complementary to the E mu enhancer. Cancer Res. 2003;63:6144–6148. [PubMed] [Google Scholar]
- 44.Cogoi S, Codognotto A, Rapozzi V, Meeuwenoord N, van der Marel G, Xodo LE. Transcription inhibition of oncogenic KRAS by a mutation-selective peptide nucleic acid conjugated to the PKKKRKV nuclear localization signal peptide. Biochemistry. 2005;44:10510–10519. doi: 10.1021/bi0505215. [DOI] [PubMed] [Google Scholar]
- 45.Halder R, Halder K, Sharma P, Garg G, Sengupta S, Chowdhury S. Guanine quadruplex DNA structure restricts methylation of CpG dinucleotides genome-wide. Mol. BioSyst. 2010;6:2439–2447. doi: 10.1039/c0mb00009d. [DOI] [PubMed] [Google Scholar]
- 46.Gonzalez V, Hurley LH. The c-MYC NHE III(1): function and regulation. Ann. Rev. Pharmacol. Toxicol. 50:111–129. doi: 10.1146/annurev.pharmtox.48.113006.094649. [DOI] [PubMed] [Google Scholar]
- 47.Brooks TA, Kendrick S, Hurley L. Making sense of G-quadruplex and i-motif functions in oncogene promoters. FEBS J. 277:3459–3469. doi: 10.1111/j.1742-4658.2010.07759.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Strauss J, Reyes-Dominguez Y. Regulation of secondary metabolism by chromatin structure and epigenetic codes. Fungal Genet. Biol. 2011;48:62–69. doi: 10.1016/j.fgb.2010.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Todd AK, Johnston M, Neidle S. Highly prevalent putative quadruplex sequence motifs in human DNA. Nucleic Acids Res. 2005;33:2901–2907. doi: 10.1093/nar/gki553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lundin KE, Ge R, Svahn MG, Tornquist E, Leijon M, Branden LJ, Smith CI. Cooperative strand invasion of supercoiled plasmid DNA by mixed linear PNA and PNA-peptide chimeras. Biomol. Eng. 2004;21:51–59. doi: 10.1016/j.bioeng.2003.10.003. [DOI] [PubMed] [Google Scholar]
- 51.Nielsen PE. Peptide nucleic acid targeting of double-stranded DNA. Methods Enzymol. 2001;340:329–340. doi: 10.1016/s0076-6879(01)40429-0. [DOI] [PubMed] [Google Scholar]
- 52.Amiard S, Doudeau M, Pinte S, Poulet A, Lenain C, Faivre-Moskalenko C, Angelov D, Hug N, Vindigni A, Bouvet P, et al. A topological mechanism for TRF2-enhanced strand invasion. Nature Struct. Mol. Biol. 2007;14:147–154. doi: 10.1038/nsmb1192. [DOI] [PubMed] [Google Scholar]
- 53.Duquette ML, Handa P, Vincent JA, Taylor AF, Maizels N. Intracellular transcription of G-rich DNAs induces formation of G-loops, novel structures containing G4 DNA. Genes Dev. 2004;18:1618–1629. doi: 10.1101/gad.1200804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Belotserkovskii BP, Liu R, Hanawalt PC. Peptide nucleic acid (PNA) binding and its effect on in vitro transcription in friedreich's ataxia triplet repeats. Mol. Carcinogen. 2009;48:299–308. doi: 10.1002/mc.20486. [DOI] [PMC free article] [PubMed] [Google Scholar]