Abstract
RNA dimerization is an essential step in the retroviral life cycle. Dimerization and encapsidation signals, closely linked in HIV-2, are located in the leader RNA region. The SL1 motif and nucleocapsid protein are considered important for both processes. In this study, we show the structure of the HIV-2 leader RNA (+1–560) captured as a loose dimer. Potential structural rearrangements within the leader RNA were studied. In the loose dimer form, the HIV-2 leader RNA strand exists in vitro as a single global fold. Two kissing loop interfaces within the loose dimer were identified: SL1/SL1 and TAR/TAR. Evidence for these findings is provided by RNA probing using SHAPE, chemical reagents, enzymes, non-denaturing PAGE mobility assays, antisense oligonucleotides hybridization and analysis of an RNA mutant. Both TAR and SL1 as isolated domains are bound by recombinant NCp8 protein with high affinity, contrary to the hairpins downstream of SL1. Foot-printing of the SL1/NCp8 complex indicates that the major binding site maps to the SL1 upper stem. Taken together, these data suggest a model in which TAR hairpin III, the segment of SL1 proximal to the loop and the PAL palindromic sequence play specific roles in the initiation of dimerization.
INTRODUCTION
Retroviruses selectively encapsidate a dimeric RNA genome assembled from two identical positive sense strands interacting near their 5′-ends. It is presumed that in the case of HIV-2, co-translational capture of the full-length RNA by the nucleocapsid (NC) domain of Gag ensures encapsidation specificity (1). The RNA motifs important for interaction with the Gag protein are located in the leader RNA region (5′-UTR) that contains highly structured domains which play various regulatory roles in viral replication (2).
The essential motif for HIV-1 dimerization has been extensively studied and is called the dimerization initiation site (DIS) (3) or stem-loop 1 SL1 (4). In vitro studies using short RNA constructs (5) showed that two DIS RNA molecules interact via a kissing-loop forming a loose dimer that may be further stabilized into an extended duplex (tight dimer). However, it has been shown that HIV-1 can replicate in the absence of the functional DIS (6), and the extended structure of the entire 5′-UTR dimer has not been confirmed (7). Loose dimers could be detected only under native electrophoretic conditions (TBM); they dissociate during semi-native gel electrophoresis (TBE).
Because the mechanism of dimerization and encapsidation of HIV-2 differs somewhat from that of HIV-1 (8–10), HIV-1 RNA cannot be packaged into HIV-2 virions (1). Two dimerization signals within the HIV-2 leader RNA have been proposed: SL1 (similar to HIV-1 SL1) (11,12) and a self-complementary sequence located in the PBS (10). Alternate usage of these sites (13) and involvement of TAR RNA (trans-activation responsive region) (14,15) in the dimerization have also been suggested. Similarly to HIV-1 (16), the LDI/BMH global riboswitch was proposed (17) in which presentation of the SL1 apical loop palindrome and its ability to dimerize are influenced by structural rearrangements of the entire HIV-2 leader RNA. In the case of HIV-1, in vivo RNA structural probing (7,18) did not confirm the riboswitch model. Also similar to HIV-1 (6), HIV-2 can replicate without the SL1 apical loop palindrome. Certain mutants with deletions and/or substitutions in SL1 had no effect on viral replication, while mutations in the Ψ region (encompassing the sequence between the PBS and SL1) were found to be strongly inhibitory (19,20). Within this region, a 10-nt palindromic sequence called PAL (+392–401) is believed to play an important regulatory role in HIV-2 dimerization (12,20). Specifically, it has been proposed that the 3′-end of PAL pairs with a sequence immediately downstream of SL1. This motif, referred to as stem B, effectively stabilizes SL1 and prevents its participation in the extended interstrand base pairing predicted to exist in tight dimers (8,21). A different SL1 secondary-structure model has also been proposed in which only a short hairpin encompassing +409–436 residues is formed and the PAL sequence either remains unconstrained (9,12,22,23) or interacts with the upstream region (+377–386) to form a separate stem-loop motif (19,24). A model of dimerization regulated by alternate presentation of the PAL and SL1 sequences produced by local RNA rearrangements in the monomeric RNA has also been suggested (24).
A regulatory role in HIV dimerization has also been proposed for the long-range U5-AUG interaction, referring to the base-pairing of a C-rich sequence located between the poly(A) and PBS domains with a G-rich sequence around the Gag translation initiation region (9,12,25). The HIV-2 U5-AUG is abbreviated CGI [C-box/G-box interaction; (8)]. In contrast to HIV-1, formation of U5-AUG duplex impairs HIV-2 RNA tight dimerization, probably due to intramolecular entrapment of the SL1 motif (8).
The dimerization and encapsidation signals are closely linked in HIV-2 and the NC protein (abbreviated NCp8), as a domain of Gag, is involved in both processes (19). The NC protein contains two zinc finger motifs. The HIV-1 protein (NCp7), rich in arginine and lysine, has been shown to be a potent chaperone for both nucleic acid folding and unfolding (26), and sites of interaction between NCp7 and HIV-1 RNA have been characterized (7,27,28).
Our knowledge of interactions between NC protein and the HIV-2 leader RNA is based mostly on the results obtained using GST tagged NCp7 together with HIV-2 leader RNA (27). Although NCp7 and NCp8 share 67% similarity, differences in packaging and dimerization have been reported for HIV-1 and HIV-2 (1,8,10). Both zinc finger motifs in HIV-2 NC have been shown to specifically interact with viral RNA during genome encapsidation (29), which is in marked contrast to HIV-1 (30). Moreover, although previous studies have demonstrated site-specific contacts between HIV-1 NC and structural elements within the leader region of HIV-1 RNA (27), similar interactions between NCp8 and HIV-2 leader motifs have yet to be identified.
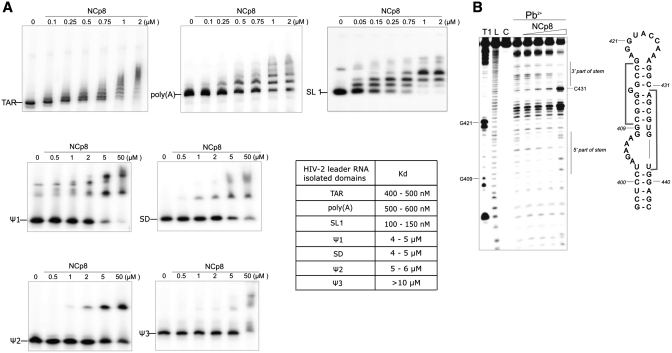
The results of in vitro structural mapping of the HIV-2 leader RNA (+1–560 nt) and shorter transcripts representing motifs considered important for dimerization, encapsidation and interactions with NCp8 protein are presented. The proposed secondary structure of the HIV-2 leader RNA represents a single global fold captured in vitro as a loose dimer. Two contact interfaces in the loose dimer form were identified within the SL1 and TAR motifs. Recombinant HIV-2 NC exhibits tight binding to SL1 (Kd = 125 ± 25 nM) and TAR (Kd = 450 ± 50 nM), but poor binding to hairpin motifs at the leader 3′-end (Ψ1, SD, Ψ2 and Ψ3). Our in vitro results allow us to propose a model for the initiation of HIV-2 RNA dimerization.
MATERIALS AND METHODS
RNA synthesis
The DNA templates for in vitro transcription were generated by PCR amplification of the plasmid HIV-2 Ψ large (31) containing a T7 promoter directly upstream of the +1 position of the wild-type HIV-2 ROD sequence. PCR primers used for template preparation are presented in Supplementary Table S1 (see Supplementary Data). The DNA template for the UTRh3 mutant was generated using the megaprimer PCR method (32). All PCRs were performed under identical conditions using Ambion SuperTaqTM Plus polymerase and conditions described earlier (33). All the RNA transcripts were synthesized with the Ambion T7-MEGAshortscript following the manufacturer’s protocol. Transcripts were purified by denaturing gel electrophoresis (8 M urea), followed by elution and ethanol precipitation. Purified RNAs were dissolved in sterile water and stored at −20°C. Oligoribonucleotides (TAR hairpin III, SL1 upper part, Ψ1, SD, Ψ2 and Ψ3), Cy5-labelled DNA and 2′-O-MeRNA were created by chemical synthesis using solid-support aided phosphoramidite chemistry, and in the case of RNA, 2′-O-TOM protected phosphoramidites (Glen Research) were utilized. Fluorescent labels were introduced at the 3'-end using commercial Cy5-modified solid supports (Glen Research). After deprotection, oligomers were purified by denaturing gel electrophoresis (8 M urea), followed by elution and desalting on NAP columns (Amersham Biosciences).
RNA labelling
RNA molecules were labelled post-transcriptionally with 32P at the 5′-end or intramolecularly during in vitro transcription with [α-32P]-GTP (Ambion T7-MEGAshortscript). 5′-end-labelling of RNA molecules with 32P was performed using [γ-32P]-ATP (ICN) and the Ambion KinaseMax kit according to the protocol. Oligonucleotides obtained by the chemical synthesis were 5′-end-labelled using [γ-32P]-ATP and T4 polynucleotide kinase (Fermentas) according to protocol. Labelled RNAs and DNA primers were purified by denaturing PAGE, followed by elution in water and ethanol precipitation. Samples were dissolved in sterile water and stored at −20°C.
In vitro dimerization of HIV-2 RNAs
The homogeneity of all RNA molecules was checked by denaturing PAGE. Aliquots of 32P-labelled RNAs (5–10 pmol) in sterile water were denatured at 90°C for 1 min. A buffer containing 10 mM Tris–HCl (pH 7.5), 130 mM KCl, 0.5 mM EDTA and the appropriate MgCl2 concentration (0–5 mM) was added followed by slow cooling to room temperature. Loose dimer formation was also tested under SHAPE folding and buffers conditions (see below). For tight dimerization, RNA was incubated at 65°C for 10 min and then at 37 or 55°C. For the loose dimer formation, the 65°C incubation was omitted and RNA was incubated at 37°C. The dimerization time was established individually for each transcript and is indicated on the figures. To stabilize the RNA conformers, the samples were kept on ice for 1 min, then 2 µl 30% glycerol or 25% Ficoll were added and samples were loaded directly onto the gel. The process of RNA dimerization was studied using gel electrophoresis under native (TBM, 4°C) or semi-native (TBE, 4°C) conditions. As a control, RNA denatured in formamide (5 min/100°C) was also loaded onto the gel.
HIV-2 RNA structural studies using antisense oligonucleotides
RNA molecules were labelled with 32P, and oligonucleotides with Cy5 or unlabelled. RNAs (5–10 pmol) were folded as described for the dimerization assays and incubated at 37°C for 5 to10 min. The dimerization buffer with 5 mM MgCl2 was used. Subsequently, the appropriate oligonucleotide (DNA or 2′-O-MeRNA) was added in a 10- and 20-fold molar excess and the samples were incubated at 37°C. The loose dimerization was allowed to proceed for an additional 15 min. For analysis of tight dimers the incubation time varied according to transcript length as follows: 2 h for +1–560 RNA and 10 min for shorter transcripts encompassing the leader 3′-end. The samples were kept on ice for 1 min, 2 µl of 30% glycerol or 25% Ficoll were added and the samples were loaded directly onto a TBM or TBE gel (4°C).
In silico two-dimensional RNA structure prediction
Two-dimensional (2D) RNA structures were predicted using RNAstructure PC v. 4.3 or 4.6 with default settings applied (34). We considered all the secondary structures predicted within 20% of free energy of the most stable variant, subsequently changing the window size parameter to generate closely related structures. For the SHAPE-constrained folding, default parameters of RNAstructure PC v. 4.6 were used.
Enzymatic structural probing of RNA
Limited digestions with nucleases S1 (Fermentas), RNase T1 (Boehringer), RNase V1 (Ambion) and RNase A of the 5′-end-labelled target RNAs (30 000 c.p.m. per sample) were performed in a buffer containing 10 mM Tris–HCl (pH 7.2), 40 mM NaCl, 8 mM carrier RNA (Ambion) and 5 mM MgCl2. Prior to the reactions, RNAs were refolded by heating at 90°C for 1 min in reaction buffer and slow cooling to 37°C. The renatured RNAs were incubated for 10 min at 37°C with increasing concentrations of the respective enzyme: T1 (0.2–0.4 U), V1 (0.01–0.0025 U), A (0.1–0.01 ng) or S1 (0.5–1 U). For limited digestion with RNase III (Ambion), the reaction was carried out at 37°C for 5 min with 0.5 or 1 U of enzyme. Reactions were terminated by addition of an equal volume of stop buffer (8 M urea, 20 mM EDTA, 0.02% xylene cyanol and 0.02% bromophenol blue) and freezing on dry ice.
Mapping oligodeoxynucleotide hybridization within the HIV-2 leader RNA
Prior to the reaction, RNAs (30 000 c.p.m.) were subjected to refolding as described above. Subsequently, 2 pmols of appropriate oligodeoxynucleotide were added and the mixture was incubated for 10 min at 37°C. The hybridization sites were detected using 0.25–0.5 U of RNase H at 37°C for 5 min. Reactions were quenched by the addition of an equal volume of stop buffer and freezing on dry ice.
Chemical structural probing of RNA
Probing with DEPC
For chemical probing with diethylpyrocarbonate (DEPC), the target 5′-end-labelled RNAs (50 000 c.p.m) were refolded as described above. Reactions (10 µl) with 0.5 µl DEPC were allowed to proceed for 5, 15 or 30 min at 37°C. Samples were ethanol precipitated twice in the presence of 5 µg carrier RNA (Ambion) and treated with 10 µl of 1 M aniline–acetate buffer (pH 4.5) according to standard protocol (35). Before electrophoresis, the RNAs were dissolved in 5 µl of sterile water and an equal volume of formamide with tracking dyes.
Pb2+ ion-induced cleavage
Prior to Pb2+ ion-induced cleavage, RNA transcripts (30 000 c.p.m.) were subjected to refolding as described above with a reaction buffer containing 10 mM Tris–HCl (pH 8.0), 40 mM NaCl, 8 mM carrier RNA and 5 mM MgCl2. Afterwards, freshly prepared Pb(OAc)2 solution was added to a final concentration of 0.25, 0.5, 1 and 2 mM and the reaction mixtures were incubated for 15 min at room temperature (36,37). Reactions were quenched by the addition of an equal volume of stop buffer and freezing on dry ice.
All products of nuclease digestion and chemical cleavage were analysed by denaturing PAGE in 1× TBE. In order to assign RNA cleavage sites, reaction products were subjected to electrophoresis along with the products of alkaline RNA hydrolysis (formamide with 3 mM MgCl2 at 100°C for 20 min) and limited T1 nuclease digestion [0.2 U, 50 mM sodium citrate (pH 5.3) and 7 M urea at 55°C for 10 min] of the same RNA.
NMIA treatment of RNAs
RNAs (20 pmols) were heated at 90°C for 1 min in 20 μl of renaturation buffer [10 mM Tris–HCl (pH 7.5), 100 mM KCl, 0.1 mM EDTA] and slowly cooled to 4°C. Subsequently, 97 μl of water and 29 μl of 5× folding buffer [200 mM Tris–HCl (pH 7.5), 650 mM KCl, 2.5 mM EDTA, 25 mM MgCl2 and 40U RNase inhibitor] were added and the RNA was incubated at 37°C for 10 min. For NMIA modification reactions taking place in the presence of oligonucleotides, it was at this point that the oligonucleotide was added, followed by the incubation for 10 min at 37°C. The mixture was divided into equal parts, treated with 7.3 μl of 180 mM NMIA in anhydrous DMSO (+) or DMSO alone (−) and allowed to proceed at 37°C for 50 min. The RNA was precipitated and resuspended in 10 μl of water. Modified sites were detected by primer extension, followed by electrophoretic fragment separation.
The primer extension reactions were performed as described earlier (38) except that 5 mM DTT and 1 M betaine were added. Dideoxy sequencing markers were generated using unmodified RNA. Several 5′-end-labelled DNA primers were used to analyse the entire HIV-2 leader RNA or shorter transcripts derived from it. The sequences of these primers are provided in Supplementary Table S2. cDNA band intensities for the (+) and (−) reactions were integrated using SAFA (39) and corrected for stochastic drop-off (40). The control reaction (−) was subtracted from NMIA reaction (+) and SHAPE reactivities were normalized to a scale in which 0 indicates an unreactive residue and the average intensity at highly reactive residues is set to 1.0, as described previously (7).
Cloning of recombinant HIV-2 NC protein
A synthetic gene was designed based on the sequence of the HIV-2ROD NC protein (NCp8) taken from http://ncbi.nml.nih.gov. The gene sequence was codon optimized and engineered to contain the EcoRI restriction site at its 5′-end and the BamHI restriction site at its 3′-end. This gene was synthesized based on a PCR method utilizing single-stranded overlapping oligonucleotides. The PCR product was purified by phenol extraction and ethanol precipitation. After EcoRI and BamHI digestion (37°C/1 h), phenol extraction and ethanol precipitation, this insert was cloned into the pGEX-4 T-3 expression vector as a GST fusion gene. The identity of the clone was confirmed by nucleotide sequencing.
HIV-2 NC protein expression and purification
The GST-NCp8 protein was expressed in Escherichia coli BL21—Codon Plus (DE3)—RIL cells (Stratagene) and purified by affinity chromatography on Glutathione-Sepharose (Amersham Pharmacia Biotech.) using a batch purification method. E. coli cells were grown in the LB medium at 37°C to an A600 of 0.5, after which expression of GST-NCp8 was induced with 1 mM IPTG. The cells were harvested 4 h after induction by centrifugation (4000 rpm, 15 min, 4°C) and resuspended in 50 ml lysis buffer [50 mM Tris–HCl (pH 7.9), 100 mM KCl, 1% Triton X-100, 50 mM DTT, 0,1 mM PMSF, 2 mg/ml benzamidine]. The cells were lysed by three repeated freeze and thaw cycles (−80°C/0°C), followed by sonication. The cell debris was pelleted by centrifugation (15 000 rpm, 40 min, 4°C), the resulting supernatant was mixed with 2 ml Glutathione-Sepharose and incubated with gentle agitation at 4°C for 2 h. The suspension was centrifuged (1500 rpm, 2 min, 4°C) and the beads were washed twice with 10 ml buffer I [PBS (pH 7.4), 1% Triton-X 100, 50 mM DTT, protease inhibitors (Roche)], 10 ml buffer II [50 mM Tris–HCl (pH 7.5), 300 mM NaCl, 50 mM DTT, protease inhibitors], 10 ml buffer III [50 mM Tris–HCl (pH 7.5), 100 mM KCl, 10% glicerol, 50 mM DTT] and 10 ml cleavage buffer [50 mM Tris–HCl (pH 7.5), 20 mM KCl, 10 mM DTT]. Cleavage of the GST-NCp8 fusion protein with thrombin (10 U) occurred overnight at 4°C (mixture volume up to 0.5 ml), and the protein was stored at 4°C in cleavage buffer. The yield was ∼5 mg of purified protein per liter of culture media.
Protein expression and purification were monitored by SDS–PAGE and Coomassie Brilliant Blue staining. Molecular mass determination was performed using an Autoflex MALDI TOF mass spectrometer (Bruker Daltonics).
NCp8/HIV-2 RNA gel retardation assay
Prior to the binding reaction, RNA samples labelled with 32P were refolded. RNA–protein complex formation (volume 10 µl) was carried out in a buffer containing 50 mM Tris–HCl (pH 8.0), 20 mM KCl, 100 mM DTT, 0.1% Triton X-100 for 10 min on ice. The binding was tested at different ZnCl2 (0–100 µM) concentrations. In order to test the NCp8 binding specificity reactions were carried out in the presence of 100-fold molar excess of carrier RNA (Ambion). The complexes were mixed with 2 µl of 25% Ficoll with dyes and loaded directly onto a non-denaturing polyacrylamide gel (acrylamide/bisacrylamide, 75:1) containing 0.5× TB with 0.1% Triton X-100. Electrophoresis was carried out at a low, strictly controlled gel temperature (4°C). The equilibrium constants (Kd = [NC][RNA]/[NC–RNA]) were estimated using 1 pmol RNA and dilutions of NCp8 [from 50 nM to 2 µM for TAR, poly(A) and SL1; 500 nM to 50 µM for Ψ1, SD, Ψ2 and Ψ3].
Mapping the RNA binding sites for NCp8
The NCp8 binding sites within the RNAs were detected using Pb2+ ion-induced cleavage. NCp8/HIV-2 RNAs complexes were formed as described above, followed by addition of carrier RNA (0.5 µg, Ambion) and Pb(OAc)2 solution to final concentration 2 mM and by incubation for 10 min at 0°C. The reactions were quenched by the addition of 40 µl of 2 mM EDTA and ethanol precipitation. The reaction products were analysed by denaturing PAGE in 1× TBE.
Results from all PAGE experiments with radiolabelled or fluorescently labelled RNAs and DNAs presented in this study were visualized and quantitatively analysed using a Typhoon 8600 Imager with ImageQuant software (Molecular Dynamics) or an FLA-5100 with MultiGaugeV 3.0 (FujiFilm).
In all cases, at least three independent experiments were performed, and the data presented are representative of the whole.
RESULTS
HIV-2 leader RNA loose dimer structure
The secondary-structure model of the HIV-2 leader RNA is based on the enzymatic and chemical probing data related to the monomer state (17,23). Although several studies attempted to address rearrangements occurring within the leader RNA upon dimerization (17,22,24), the structure of the HIV-2 leader RNA dimer is still unknown. To reveal the HIV-2 leader RNA loose dimer structure, we applied the SHAPE method (41) which had been successfully used to identify kissing-loop interactions (40,42).
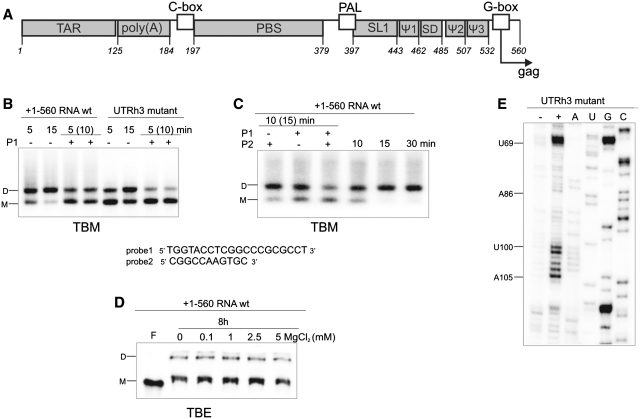
The length of the HIV-2 leader RNA, encompassing +1–546 nt, was extended to 560 nt to include a fragment of the Gag coding sequence proposed to be involved in the U5-AUG duplex formation (Figure 1A) (8,9). Analysis of the 560-mer leader transcript using TBM gels and different folding conditions (Figures 1B, C and 2A) indicates that loose dimers were formed very efficiently at 37°C in the presence of Mg2+. After 10 min, over 50% of the RNA was in the dimeric form (Figure 2A). The comparison of TBM and TBE gel patterns shows that tight dimers, under identical folding conditions, were formed very inefficiently even after prolonged incubation (compare Figure 1C, fourth lane and 1D). Consequently, under SHAPE folding and NMIA modification conditions (38), most of the HIV-2 leader RNA molecules were in the loose dimer form (Figure 2A).
Figure 1.
(A) Organization of the HIV-2 leader RNA. (B) Loose dimer formation by wt HIV-2 leader RNA and its UTRh3 mutant, in the absence or presence of antisense oligonucleotide probe1 (P1) (TBM agarose gel). (C) Influence of antisense oligonucleotides, probe1 (P1; directed to SL1) and probe2 (P2; directed to TAR hairpin III), on the formation of the loose dimers by +1–560 RNA (TBM agarose gel). (D) Tight dimer formation of the +1–560 RNA (TBE PAA gel). For analyses of dimer formation, samples were incubated at 37°C for the times indicated, with some sample incubated for an additional 10 or 15 min (parenthesis) in the presence of P1 and/or P2. Lanes F correspond to the formamide-denatured control sample. RNA monomers are indicated as M, dimers as D. (E) NMIA modification of the leader RNA mutant UTRh3. Lane (−) represents control sample with untreated RNA; lane (+) NMIA modification, A, U, C and G are sequencing lanes.
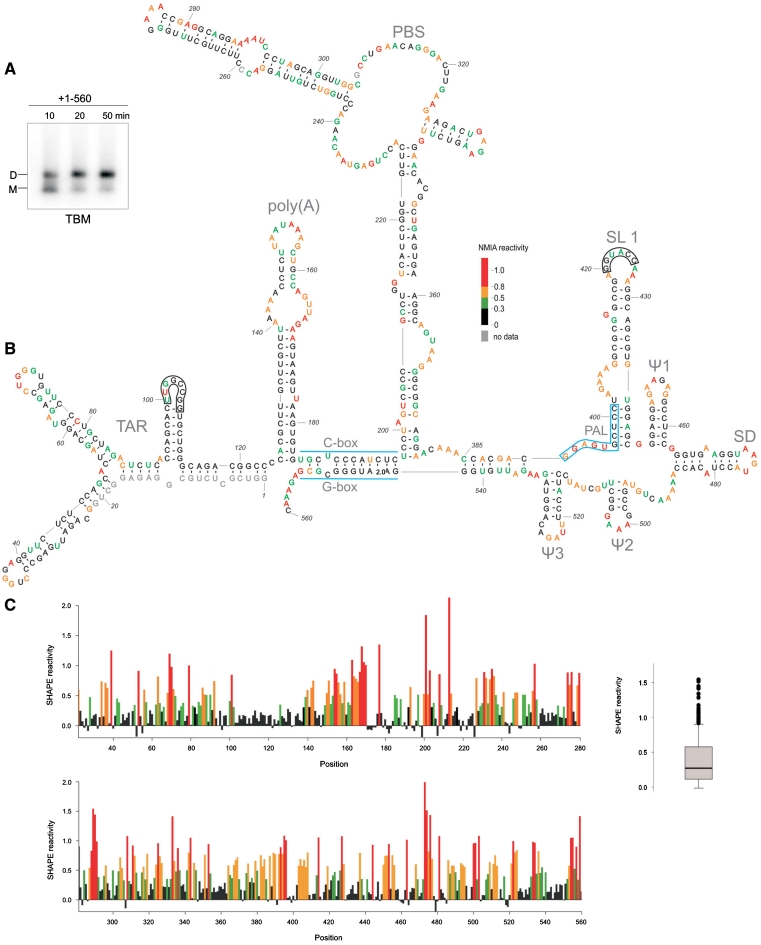
Figure 2.
(A) Analysis of the loose dimer formation by the HIV-2 leader RNA (+1–560) in the SHAPE buffers and folding conditions (TBM agarose gel). (B) Secondary-structure model of the HIV-2 leader RNA captured in the loose dimer form. Two kissing-loop interfaces are indicated as black frames. Nucleotide residues accessibilities to NMIA are given in colours and according to scale. (C) Processed SHAPE reactivities as a function of nucleotide position. Red and orange bars reflect reactive positions in the RNA. Insert on the right represents the box plot analysis of distinct reactivity distributions for the HIV-2 leader RNA. Box outline middle 50% of dataset; the median is shown with heavy line. Circles indicate extreme values (7): 1.5 times the interquartile range (boxed). Horizontal lines above and below the box are the largest or smallest non-outlier values.
Well-resolved SHAPE data, supported by native gel patterns, point to the one global fold of the HIV-2 leader RNA (Figures 1C and 2 and Supplementary Figure S1) captured in vitro in its stable loose dimer state. In this dimeric structure, the TAR domain exists in its branched form (43) (Figure 2B). This domain (+1–123), in its isolated state for HIV-2 (33) and in vivo for SIV (44), undergoes an interconversion between a branched hairpin form and an extended form. The equilibrium state in vitro is highly susceptible to the folding conditions. Analysis of the isolated TAR migration on native gels indicates that under SHAPE folding conditions, the branched hairpin form prevails (Supplementary Figure S2).
SHAPE indicates that the poly(A) domain (+125–184), presented earlier as a hairpin with a large 29-nt apical loop (45), is structured. Residues A144 to U149, C158 and G160 to C162 were not modified by NMIA (Figures 2B, C and 3D), suggesting that they are constrained. Interestingly, three structural models were generated in silico for the poly(A) domain, using RNAstructure software (34), but none of the predicted structures fit the experimental data. To address whether those residues are involved in long-range interactions with other domains of the leader RNA, we performed extensive enzymatic and chemical structure probing of the isolated poly(A) hairpin encompassing +125–184 residues (Figure 3A and C). RNase A, specific for single-stranded pyrimidines, cleaved only 4 of 13 pyrimidines located in the apical loop and none of the residues resistant to NMIA modification were recognized. In addition, residues C145–U149 and C158–C162 were cleaved by RNase V1 and not by nuclease S1. The use of RNase III led to a strong cleavages at U138 and U172 and two weaker hits at C162 and U134, supporting the existence of a structured section within the loop of the poly(A) hairpin. These and other findings are summarized in Figure 3C. Our analyses exclude a possibility that these unexpected structural features of the poly(A) domain result from interactions with other elements of the leader RNA. In addition, our data rule out the formation of a pseudoknot between nucleotide residues within the apical loop of poly(A) and residues located in the coding region, a structural feature which has been recently proposed for HIV-1 (7).
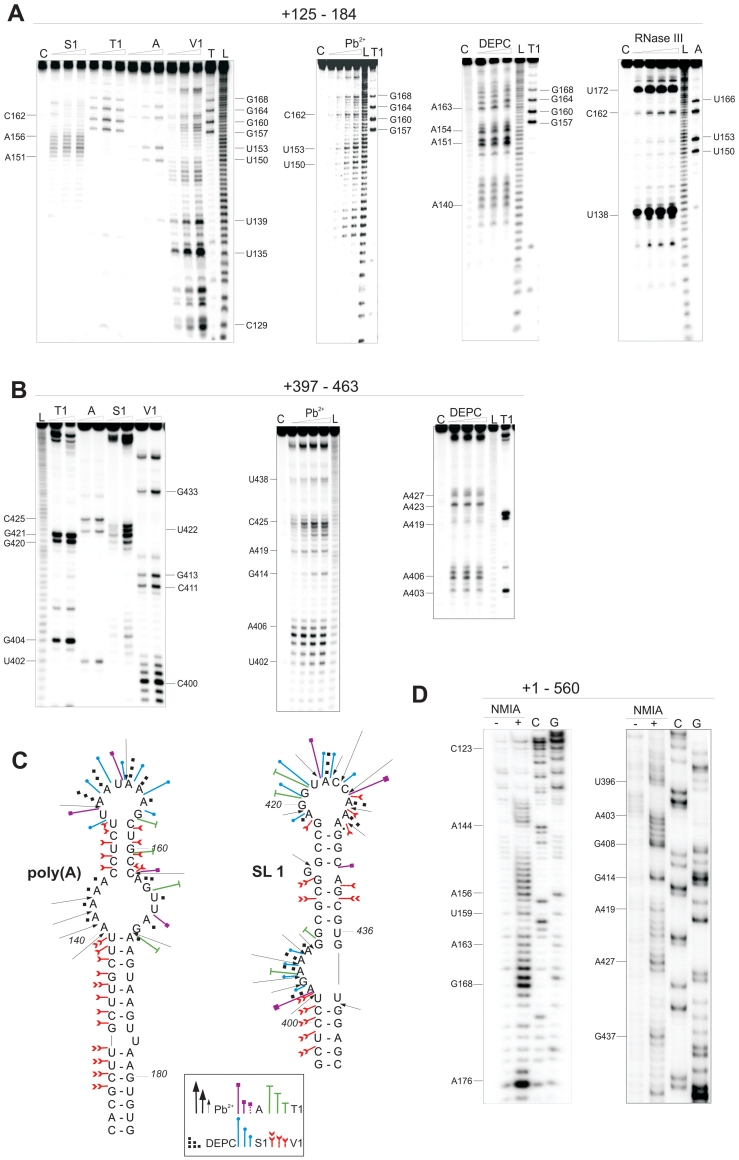
Figure 3.
Structural probing of HIV-2 transcripts encompassing regions: (A) +125–184 and (B) +397–463. The RNAs were treated with single-strand specific reagents (S1, T1, A, Pb2+, DEPC) and double-strand specific RNase V1 and RNase III. Lanes C represent control samples with untreated RNA; lanes L, formamide ladder. (C) Summary of enzymatic and chemical cleavages and modifications viewed on the secondary-structure models of poly(A) and the SL1 domains. (D) NMIA modification of poly(A) and SL1 regions in the HIV-2 leader RNA (+1–560). Lanes (−) represent control sample with untreated RNA; lanes (+) NMIA modification, C and G are the sequencing lanes.
The data obtained point to the extended SL1 forming stem B (8,11,21), rather than the short hairpin (+409–436) (9,12,22,23). Residues of the 3′-end of PAL (G397CUCC401) were not modified by NMIA and they form the stem B of SL1 (Figures 2B, C and 3D). The in vitro data obtained for the captured loose dimer exclude the presence of in silico predicted stem-loop structures encompassing the Ψ region containing the PAL sequence (19,24). Based on enzymatic and chemical probing data obtained for the shorter transcripts containing SL1, we propose the existence of several non-canonical base pairs closing bulges and loops within the SL1 structure (Figure 3B, C and Supplementary Figure S3).
Analysis of the data obtained for the HIV-2 leader RNA 3′-end (+380–560) (Figure 2B and C and Supplementary Figure S1) confirms the presence of characteristic short hairpins Ψ1, SD, Ψ2 and Ψ3 presented in a previous model (23). In addition, the U5-AUG (CGI) interaction (8,9) is confirmed. The lack of NMIA modification indicates that residues G543 to C553 (G-box) are paired with G186 to C196 (C-box). As presented in Figure 2B and C and Supplementary Figure S1, the analysis of the SHAPE data also supports base-pairing between segments C385CACGAC391 and G535AUUGUGG542.
The secondary-structure model of the HIV-2 leader RNA proposed here is based on the modification pattern where single-stranded regions are reactive towards NMIA (Figure 2B and C). However, two exceptions were found that are consistent with the presence of tertiary interactions involved in the loose dimer formation. Both of them represent a kissing-loop interaction. Specifically, the SL1 palindromic sequence G420GUACC425 and a self-complementary sequence within the TAR (U100UGGCCGG107) were largely unreactive towards NMIA. The latter of those sites is located within the short 3′ hairpin of the TAR domain, and is characteristic of both TAR forms (abbreviated herein as hairpin III). Within the aforementioned sequence, only U101 which is involved in an intermolecular G–U pair formation, was found to be susceptible to NMIA modification (Figure 2C and Supplementary Figure S1). Similar to the data obtained with SL1 (Figure 3B and C), the hairpin III loop in the monomeric state was fully accessible to several enzymes and chemical reagents used to probe the isolated motif (+93–113) and a longer RNA encompassing TAR (+1–123) (33). In addition, both short constructs representing the upper part of the SL1 and the isolated hairpin III of TAR were found to undergo the transition from loose to tight dimers very efficiently (Supplementary Figure S4). To further confirm the participation of the TAR hairpin III apical loop in formation of the kissing-loop complex, an RNA mutant was constructed. The U101GGCC105 sequence of the TAR hairpin III apical loop was changed to A101AAAA105 in the UTRh3 mutant of the HIV-2 leader RNA. The NMIA modification pattern obtained for the UTRh3 mutant transcript (Figure 1E) shows that, contrary to the wild-type leader RNA, residues of the hairpin III apical loop were reactive towards NMIA. SHAPE data also showed that the secondary structure of the TAR domain in the mutant RNA remained unaffected. The analysis of the dimerization properties of UTRh3 RNA using TBM gels, indicates that loose dimers were formed less efficiently relative to the wild type HIV-2 leader RNA (Figure 1B). Furthermore, the loose dimer formation was assayed in the presence of an oligodeoxynucleotide complementary to the apical loop and upper stem of SL1 (probe1) (Figure 1B). The oligodeoxynucleotide was added to the pre-folded RNA. Although the strong inhibitory effect of probe1 was observed for both wild type and mutant RNA, it should be emphasized that the presence of probe1 almost completely abolished UTRh3 loose dimerization. In this case, one contact site is disrupted by the mutations, and the second by hybridization with probe1. To extend our findings on the interaction interfaces in the loose dimer, we also tested the ability of the oligodeoxynucleotide complementary to TAR hairpin III (probe2) and SL1 (probe1) to suppress loose dimer formation of the wild-type leader RNA (Figure 1C). While some reduction of loose dimerization was observed in the presence of probe2, the effect was not as pronounced as for probe1. However, when both probes were included in the hybridization mixture, the inhibitory effects on dimer formation were additive (Figure 1C).
PAL/SL1 rearrangement
It has been proposed that the PAL sequence plays a regulatory role in SL1-mediated HIV-2 RNA dimerization and monomeric RNA can adopt two alternative conformations (24). The first conformation, with an extended SL1 and PAL partially base-paired in the stem B, impairs dimerization. The second, with a short SL1 (+409–436) and PAL involved in formation of an additional stem-loop motif (19), favours dimerization (24). Because our studies of the leader RNA (+1–560) in the conditions applied do not show indications of PAL/SL1 rearrangement, we decided to study this region within different structural contexts. A structural interconversion that involves only a limited number of base pairs within a relatively small region might be energetically more favourable in a shorter transcript. Transcripts +397–445, +397–463, +377–463, +377–560 (Figure 3B and C and Supplementary Figure S3) were subjected to structural probing using selected single-strand specific (S1, T1, A) and double-strand specific (RNase V1) enzymes and chemical reagents (DEPC and Pb2+ ions). The data obtained show that in our in vitro conditions, the secondary structure of the extended SL1 remains unchanged regardless of the presence of adjacent elements at the 5′ and/or 3′-ends.
To allow direct comparison to the SHAPE data obtained for the full-length leader RNA, two transcripts, +377–560 and +397–560 were probed with NMIA (Supplementary Figure S5). Only the +377–560 RNA contains the stretch of nucleotide residues that would allow the proposed PAL/SL1 structural rearrangements (24). NMIA reactivity profiles indicate that the structures of transcripts +377–560 and +397–560 are the same, and match that of the 3′-end of the entire HIV-2 leader RNA in vitro.
The structure of the HIV-2 leader RNA 3′-end (+380–560) was also analysed in the context of the +1–891 transcript. Although this transcript encompasses a much larger fragment of the Gag coding sequence, no differences in the organization of leader RNA were found (Supplementary Figure S1).
The features of the extended SL1 structure characteristic of the isolated hairpin are retained within all RNAs analysed, regardless of the additional 5′ and/or 3′ fragments of leader RNA, indicating that adjacent structural motifs fold independently. There is no evidence in these data to suggest that multiple thermodynamically stable forms of PAL/SL1 region exist in the monomeric RNA.
The SL1 palindrome is dispensable for the stability of the tight dimer
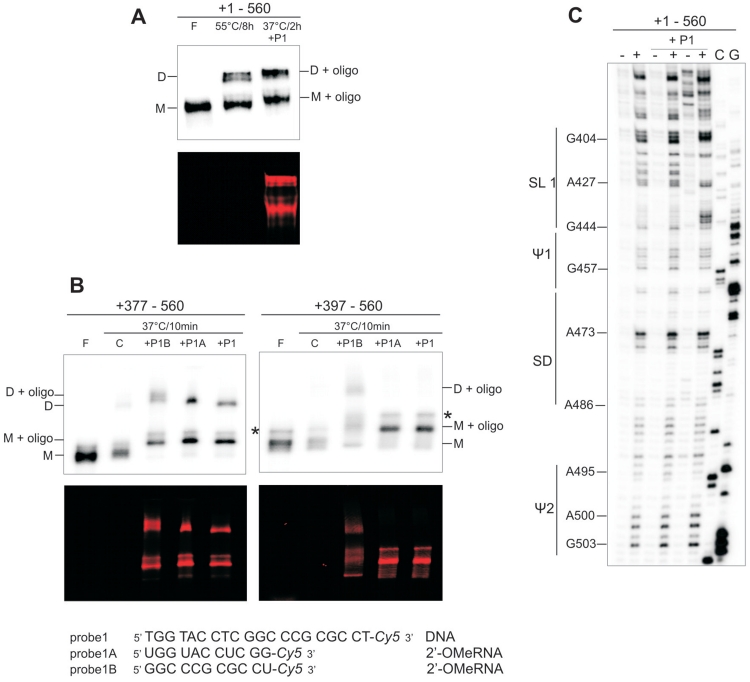
The HIV-2 leader RNA forms tight dimers in vitro very inefficiently (11,12). However, as reported earlier, formation of tight dimers was induced by folding the target RNA in the presence of antisense oligonucleotides directed against such regions as U5-AUG duplex, PAL or SL1 (8,12). To investigate the structural properties of the SL1 within the pre-folded leader RNA and to resolve the influence of oligonucleotide probes on tight dimer formation, a set of hybridization experiments was performed. Initially we used an oligodeoxynucleotide (probe1) complementary to the 5′ upper stem and apical loop of SL1 (+407–426) of the same sequence as reported earlier for the probe asSL1 (8,12). Probe1, labelled at the 3′-end with the fluorescent marker Cy5, readily associated with the pre-folded RNA present in a 1:1 ratio of monomer to loose dimer. Inclusion of the probe1 in the mixture led to an increase in leader RNA tight dimerization (Figure 4A). The amount of dimer formed in the presence of probe1 after 2 h at 37°C is comparable to that obtained without probe1 after 8 h at 55°C. TBE gel patterns show that the fluorescent probe1 co-migrates with both the monomer and the tight dimer. An RNase H assay confirmed that probe1 was bound at the expected location within the SL1 domain. Residues G411 to C425 were cleaved within the 377–560 RNA, indicating strand invasion (Supplementary Figure S6).
Figure 4.
Influence of antisense oligonucleotide probes directed to SL1 domain on HIV-2 leader RNA tight dimerization. (A) Probe1-mediated activation of +1–560 RNA tight dimerization (TBE). Time and temperature of incubation are indicated to describe differences in the conditions necessary to achieve similar dimerization efficiency in the dimerization buffer with 5 mM MgCl2. (B) The tight dimerization (TBE) of the +377–560 and +397–560 transcripts mediated by probes 1, 1A or 1B. Transcripts were 32P-labelled, oligonucleotide probes 3′-end-labelled with Cy5 fluorophore, the upper gel panels show radioactivity scans, the bottom, fluorescence scans. Lanes F correspond to formamide-denatured control samples; lanes C, control RNA dimerization in the absence of oligonucleotides. Monomers are indicated as M, dimers as D. Asterisks indicate RNA produced by non-templated addition of nucleotides by T7 RNA polymerase. (C) NMIA modification of the leader RNA after hybridization with probe1 (5- and 10-fold molar excess). Lanes (−) represent control sample with untreated RNA; lanes (+) NMIA modification, C and G are sequencing lanes.
To reveal the mode of action for probe1, two shorter 2′-O-Me oligoribonucleotides 3′-end-labelled with Cy5 were prepared: probe1A complementary to the apical loop (+416–426) and probe1B complementary to the 5′ upper stem of SL1 (+407–417). Each of them was hybridized to transcripts representing the 3′-end of the leader RNA but differing with respect to the presence of the Ψ region encompassing the PAL palindrome (RNAs +377–560 and +397–560). Transcript +377–560 contains the entire PAL sequence, whereas +397–560 includes only the 3′ part of PAL (i.e. that involved in SL1 stem B formation). Probe1 and probe1A significantly increased the level of tight dimers for the +377–560 transcript, but not for the +397–560 RNA (Figure 4B), indicating that the Ψ region plays a role in tight dimerization, as previously proposed (20). In contrast, probe1B triggered an increase in tight dimer formation for both transcripts, indicating that tight dimerization induced by this probe is Ψ independent. However, in the case of the full-length leader RNA (+1–560), the influence of probes 1A and 1B on tight dimerization was not as pronounced as in the case of probe1 (data not shown).
SHAPE was applied to reveal probe1-induced changes within the full-length leader RNA structure. Probe1 was added to the pre-folded leader RNA (monomer to loose dimer ratio 1:1) and the resultant complexes were treated with NMIA. Although the SHAPE readout represents the superimposed reactivities of both free and bound leader forms, some structural features are apparent (Figure 4C). Interesting structural rearrangements are observed in the SL1 stem B. Nucleotide residues G437–C443 showed an increase of their reactivity towards NMIA in relation to the leader alone. Residues G397–C401 remained unreactive. These two stretches of residues in the monomeric leader or its loose dimer fold into the SL1 lower stem (stem B). In addition, the 5′-end of the PAL was protected from NMIA modification and residues immediately upstream gained reactivity. No additional changes in the reactivity pattern of the leader RNA in the presence of probe1 were observed (Figure 4C, Supplementary Figure S7). This indicates that the rearrangements of the secondary structure induced by probe1 are restricted to the PAL/SL1 region. In addition, it shows that the SL1 apical loop is not involved in base pairing with other regions of HIV-2 leader RNA.
The SL1 motif is a high-affinity binding site for NCp8
An expression vector was constructed that generates the HIV-2 ROD NCp8. The gene was cloned into the pGEX-4T-3 vector as a GST fusion. The clone over-expresses in E. coli efficiently and yields ∼5 mg of pure NCp8 protein after GST removal with thrombin (Supplementary Figure S8A). The resultant protein contains two additional residues at the N-terminus due to the protease specificity. MALDI-TOF showed the expected NCp8 molecular weight (Supplementary Figure S8B).
The specificity of NCp8 interaction with 32P-end-labelled RNA transcripts corresponding to isolated domains of the 5′-UTR has been analysed using an electrophoretic mobility shift assay (EMSA). Titration of the transcripts encompassing isolated domains: TAR, poly(A), SL1, Ψ1, SD, Ψ2 and Ψ3 with NCp8 resulted in the formation of a new, distinct bands, albeit with differing intensities (Figure 5A and Supplementary Figure S9). NCp8 has the highest affinity for SL1 with a dissociation constant of 125 ± 25 nM. At least three discrete bands were formed, in accord with multiple proteins interacting with RNA. Complexes remained unaffected in the presence of 100-fold molar excess of an unrelated competitor RNA, confirming the specificity of the interaction (Supplementary Figure S9). The NCp8/SL1 dissociation constant is similar to that reported for HIV-1 NCp7 and SL3 RNA (28).
Figure 5.
NCp8 interactions with the isolated domains of the HIV-2 leader RNA. (A) Electrophoretic mobility shift assays for leader domains and NCp8. The protein concentrations are indicated above the lanes. (B) Mapping the NCp8 binding sites within the +397–463 transcript using Pb2+-ions. Residues protected from cleavage in the presence of NCp8 are indicated by grey brackets.
Footprinting analysis of the NCp8/SL1 complex using Pb2+ induced RNA cleavage was performed under stringent conditions that allow most of the base-paired nucleotides to be cleaved. Titration of the +397–463 transcript with NCp8 resulted in a decrease in RNA cleavages at residues C410–C417 and A432–U438 at low-protein concentration (Figure 5B). This indicates that the SL1 upper stem is the NCp8 binding site. Our structure probing data obtained for isolated SL1 in the absence of protein show that this binding site contains a purine–purine base pair (G413–A432) next to the unpaired G414. The G414 residue was susceptible to Pb2+ induced hydrolysis. The lack of reactivity of the A432 residue towards DEPC suggests that its Hoogsteen site is involved in base-pairing with G413.
Footprinting analysis also demonstrates that cleavages at C425 and C431 were enhanced at low protein concentration, while in the presence of high concentrations of NCp8 most of the residues previously protected became reactive. The data are consistent with the destabilizing activity of NC proteins on the RNA structure (46,47).
The affinity of NCp8 for the other isolated hairpin motifs present at the 3′-end of HIV-2 leader RNA is much lower than that for SL1. Kds for Ψ1 and SD were ∼4–5 µM, for Ψ2 5–6 µM and >10 µM for Ψ3, suggesting that the observed associations are likely the product of nonspecific binding. Interestingly, a much higher affinity was observed for TAR and poly(A) isolated domains (Kd = 450 ± 50 and 550 ± 50 nM, respectively).
DISCUSSION
The HIV-2 leader RNA structure in the loose dimer form
It has been recently demonstrated that in vitro transcribed HIV-1 leader RNA folds into the same structure as RNA present in virus particles, and that this structure represents single fold (7). It was suggested that the HIV-2 leader RNA monomer may adopt an alternative conformations with different abilities to dimerize (17,24). Our results of enzymatic and chemical structural probing, supported by native gel electrophoresis, show that in the presence of magnesium, the HIV-2 leader RNA strand shows preference to exist in vitro as a single conformer. The data collected from the SHAPE probing correspond to the leader RNA loose dimer structure. This is due to the preferential formation of the loose dimer by the HIV-2 leader RNA and to the mild action of the NMIA reagent, which is non-disruptive to the loose dimer (38,41,42).
To our knowledge, this is the first study to structurally characterize the HIV-2 leader RNA captured in one of the potential kissing-loop complex forms. Here, we document the existence of two contact interfaces involving palindromes: the SL1 apical loop and the TAR hairpin III apical loop. Although the involvement of the SL1 in dimerization has been studied extensively, the role of the SL1 apical loop has not been fully established yet. Some reports underlined the importance of the SL1 apical loop (11,12,17,24) while others disputed its involvement in dimerization (10,20). Our data indicate that in vitro the SL1 apical loop palindrome contributes to the loose dimer formation. Participation of the TAR domain in HIV-2 RNA dimerization has also been suggested (14,15), but has not been proven, as is the case for HIV-1 (15,48). However, it was shown that deletion of the TAR caused a significant drop in the efficiency of HIV-2 leader RNA loose and tight dimer formation in vitro (17). Based on our results, we are able to support the involvement of TAR in dimerization and to propose that TAR hairpin III serves as a kissing-loop site. As we presented previously, a phylogenetic comparison of TAR sequences of HIV-2 and SIV isolates revealed that this palindromic sequence is highly conserved (33). The only notable change (present in three different isolates) is the G106A substitution that would not interfere with the palindromic character of this sequence. Mutations in the TAR hairpin III palindrome markedly reduced the amount of the loose dimer formed in vitro (Figure 1B). Furthermore, the residues of hairpin III apical loop were reactive towards NMIA in the UTRh3 mutant (Figure 1E), contrary to the wild type HIV-2 leader RNA. The oligodeoxynucleotides complementary to the SL1 (probe1) or TAR hairpin III (probe2) suppressed the formation of loose dimers by the wild type HIV-2 leader RNA (Figure 1C) supporting the involvement of both motifs (TAR and SL1) in in vitro dimerization. Less pronounced effect of probe2 may reflect a supplementary role of the TAR hairpin III in the dimerization or less efficient hybridization of this probe. In addition, the abolished formation of loose dimers by the UTRh3 mutant in the presence of probe1 is an expected result in the light of our findings concerning two main contact interfaces within the HIV-2 loose dimer. Although our data indicate two contact sites in the HIV-2 RNA loose dimer, our focus was not on the Nar I sequence and its participation in dimerization (10). It is known that local RNA rearrangements occur upon tRNALys3 binding (49). Therefore, structural analysis of that region is not emphasized here. However, we noted that three of six residues (G304, G305 and C308) from the Nar I palindromic sequence were highly reactive towards NMIA (Figure 2C).
Within the loose dimer, the global RNA fold merges structural features of the previously proposed models of the HIV-2 leader monomers (9,12,17,23). We found that the structures of the individual domains within the full-length HIV-2 leader RNA are similar of those of the isolated ones. Results of detailed enzymatic and chemical probing of the isolated domains are fully consistent with the SHAPE data for the +1–560 and +1–891 transcripts (Figures 2B, C and 3; Supplementary Figures S1 and S3). Formation of the extended structure of SL1 is independent of the structural context in which it is present, indicating the stability of the SL1 stem B. The extended SL1 had been previously shown to be necessary for HIV-2 replication and RNA encapsidation (21). We showed that the extended SL1 and the intramolecular U5-AUG are characteristic of the HIV-2 leader RNA loose dimer structure (Figure 2B). A previous report, based on the use of antisense oligonucleotide probes concluded that the presence of U5-AUG duplex favoured the extended SL1 motif formation (8,12). Although we do not dispute the U5-AUG regulatory role in HIV-2 RNA dimerization, our structural probing data using transcripts of different lengths clearly show that the extended SL1 is formed irrespective of U5-AUG duplex in vitro.
The SL1 RNA domain is a high-affinity binding site for NCp8
The NC proteins cover the entire retroviral leader RNA with high occupancy. To address their binding specificity and the structure of RNA/protein complexes, it is useful to perform studies on models representing isolated RNA domains (28,50). All individual HIV-2 leader RNA domains except the PBS were incubated with recombinant NCp8 protein (Figure 5A). Although we cannot exclude the possibility that the binding affinities of full length Gag protein for the various constructs may differ from those presented here for NCp8, the fact that SL1 RNA is bound with an affinity more than an order of magnitude higher than for Ψ1, SD, Ψ2 or Ψ3 is strongly suggestive of specific recognition of this structural element by HIV-2 NC.
Footprinting experiments reveal that the upper stem of SL1 is the binding site for NCp8 (Figure 5B). An interesting feature of this binding site is the presence of a purine–purine base pair (G413–A432) next to the unpaired G414. It has been demonstrated that the HIV-1 NC protein prefers binding to unpaired G residues (28,51). Moreover, our footprinting data demonstrate that NCp8 binding destabilizes SL1, in accord with the accepted opinion that NC proteins destabilize continuous double-stranded RNA structures (46,47,52).
The HIV-1 NC protein NCp7 binds the HIV-1 DIS (SL1) and Ψ (SL3) domains with similar affinity, with Kd’s ranging from 100 to 200 nM (28). Interestingly, the NCp8 dissociation constant for SL1 is much lower than for other hairpins of the leader 3′-end (Figure 5A). It has been shown (27) that the heterologous NCp7 protein preferentially binds the Ψ3 domain within HIV-2 leader RNA which structurally resembles the HIV-1 hairpin Ψ (SL3). The complex between the Ψ3 hairpin and NCp8 was studied by NMR (50). We observed that Ψ3 cannot be considered a specific binding site for NCp8 in vitro due to the high Kd measured (above 10 µM). Furthermore, the Ψ3 motif is not essential for HIV-2 dimerization or packaging (19). One can speculate that tight and preferential binding of SL1 by NCp8 supports the hypothesis that the failure of HIV-2 to package the HIV-1 RNA may in part be due to the differing specificities of the two NC proteins.
Interestingly, NCp8 showed a much higher affinity for the HIV-2 TAR and poly(A) isolated domains than for the hairpin motifs located downstream of SL1. The relatively tight binding of NCp8 to the TAR domain is especially interesting in view of the data presented here indicating the involvement of the TAR hairpin III in the formation of loose dimers. In addition, in clinical isolate from HIV-2 infected patients, a gag mRNA species exists that differs from the genomic RNA due to the absence of the +61–202 intron in the leader region (53). An important question in this case is how unspliced viral RNAs are differentiated by Gag protein from those of the spliced species. The intron contains half of the TAR structure and the poly(A) hairpin. Our results with respect to NCp8 binding to the TAR and poly(A) domains may indicate that these two important binding sites are missing in the spliced RNA resulting in differentiation between both RNA species.
Structural changes during the initiation of HIV-2 leader RNA dimerization
Our results indicate that the HIV-2 leader RNA prefers to form loose dimers in vitro, and in accordance with previous reports (11,12), tight dimers were shown to form very inefficiently (Figure 1D). However, tight dimerization had been induced by antisense oligonucleotides added prior to denaturation and subsequent folding of the HIV-2 leader RNA (8,12). As this approach might lead to substantial changes in the RNA fold, we added the oligonucleotide probes to the pre-folded HIV-2 leader RNA at an equilibrium of 1:1 monomer to loose dimer. The fluorescent oligodeoxynucleotide probe1 promoted tight dimer formation, and stayed bound to both the monomer and tight dimer species of the leader RNA (Figure 4A). This allows us to conclude that the SL1 structures in both the monomer and loose dimer are susceptible to hybridization. However, the mechanism by which probe1 participates in tight dimer formation was difficult to unravel in detail. Our data obtained for two shorter 2′-O-MeRNA probes 1A and 1B (Figure 4B) show not only that both parts of probe1 are necessary for efficient induction of tight dimer formation, but also that both the upper stem and apical loop of SL1 are involved. Furthermore, the SHAPE probing revealed that upon probe1-induced HIV-2 leader RNA tight dimerization, SL1 stem B undergoes opening (Figure 4C), experimentally confirming the previous hypothesis (8). However, it has been suggested that the promotion of dimerization is due to the formation of dimerization-competent short SL1 (+409–436) (8). Based on our data, we conclude that this is a consequence of PAL release. In other words, PAL becomes free to participate in alternate interstrand base pairing interactions that stabilize the tight dimer (Figure 6). Data obtained from experiments with shorter transcripts representing the HIV-2 leader RNA 3′-end confirmed the importance of PAL in SL1 mediated tight dimerization. The presence of the entire PAL palindrome is necessary for the induction of RNA tight dimerization by oligonucleotides complementary to the SL1 apical loop (probes 1 and 1A). Interestingly, tight dimerization of the transcript lacking the 5′ part of PAL sequence was promoted exclusively by probe1B, complementary to the SL1 upper stem. In this case, the tight dimer could be formed only by the SL1 apical loop palindrome. These results suggest a possible functional exchange between the PAL and SL1 palindromic sequences in the formation of intermolecular interactions in vitro. It is also possible that for this transcript, destabilization of the SL1 upper stem by probe1B induces a conformation of the apical loop that favours tight dimer formation.
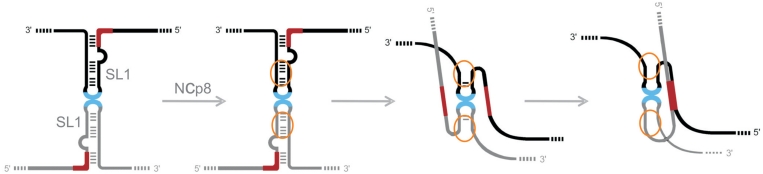
Figure 6.
Model of the HIV-2 dimerization initiation in vitro showing rearrangement in the PAL/SL1 region. Palindromic sequences within the SL1 apical loop and PAL are indicated in blue and in red, respectively. The NCp8 interaction region is presented as ellipse in orange.
The local conformational switch in the PAL/SL1 region in the leader RNA monomer had been previously proposed to regulate HIV-2 RNA dimerization (8,12,24), but a compatible and comprehensive model showing the structural rearrangements in this region was not established. The importance of PAL in HIV-2 dimerization is generally accepted, but its role is less understood (12,20,24), and the participation of the SL1 apical loop in this process is still a matter of dispute (10,11,13,17,20). It should be emphasized that overall, our data indicate that in monomeric leader RNA, PAL forms stem B rather than the intramolecular structures with nucleotides located upstream (19,24), while PAL in the tight dimer is involved in the intermolecular PAL/PAL interaction, which does not support the regulatory mechanism formerly proposed (24). We have also demonstrated that the palindromic sequences in the SL1 apical loop and TAR hairpin III, participate in loose dimer formation in vitro. On the other hand, our observation that fluorescent probe1 is bound to the species in the tight dimer form (Figure 4A), supports the statement that the SL1 loop palindrome is not necessary in vivo for the formation of the dimers (20). We hypothesize that if the SL1 apical loop palindrome is not present, the TAR hairpin III can substitute for its role in the loose dimer formation. These dimers could be held together by base pairing between TAR hairpins, while SL1 hairpins remain in close proximity and conducive to maturation by interaction with Gag/NC. It is likely that at the initiation stage of the dimerization process, several regions are recognized by the NC domain of Gag, with SL1 and TAR being the most likely candidates. The fact that the main binding sites for NCp8 map to the upper stem, but not to the apical loop of SL1, suggests that recognition of this motif by NC is possible regardless of the presence of a SL1/SL1 kissing-loop interaction in the loose dimer. It had been demonstrated that NC shows duplex destabilizing activity upon interaction with distorted RNA structures (46,47,52,54). Indeed, destabilization caused by the binding of oligonucleotides and/or NCp8 to the upper part of the SL1 could be translated downstream to stem B leading to PAL release. The PAL/PAL interaction can now enhance the stability of the maturating HIV-2 RNA dimer. Our data most probably reveal only the most stable interfaces within the immature dimer and we believe that many more RNA/RNA and RNA/protein interactions are involved in the dimer maturation process as previously suggested (6).
In conclusion, we propose a model in which the upper part of SL1 serves as a binding site for the NC domain of Gag thus stimulating the initiation of HIV-2 dimerization, without necessarily being a part of the tight dimer structure (Figure 6). Moreover, the kissing-loop formation between TAR hairpins III in addition to the interaction between SL1 apical loop palindromic sequences plays a role in stabilizing HIV-2 loose dimers. However, the SL1, once destabilized by NCp8, frees PAL for participation in a new type of intermolecular interaction which appears to be involved in the formation of tight dimers. It remains to be seen whether the TAR kissing-loop interaction is maintained throughout dimerization or is further transformed to a more stable dimer element.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The Ministry of Education and Science (grants PBZ-MNiSW-07/I/2007 and N30101432/0925) (K.J.P. dissertation). Funding for open access charge: Institute of Bioorganic Chemistry, Polish Academy of Sciences.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Kaye JF, Lever AM. Nonreciprocal packaging of human immunodeficiency virus type 1 and type 2 RNA: a possible role for the p2 domain of Gag in RNA encapsidation. J. Virol. 1998;72:5877–5885. doi: 10.1128/jvi.72.7.5877-5885.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rabson AB, Graves BJ. Synthesis and processing of viral RNA. In: Coffin JM, Hughes SH, Vink M, editors. Retroviruses. New York: Cold Spring Harbor Laboratory Press; 1997. pp. 205–262. [PubMed] [Google Scholar]
- 3.Skripkin E, Paillart JC, Marquet R, Ehresmann B, Ehresmann C. Identification of the primary site of the human immunodeficiency virus type 1 RNA dimerization in vitro. Proc. Natl Acad. Sci. USA. 1994;91:4945–4949. doi: 10.1073/pnas.91.11.4945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.McBride MS, Panganiban AT. The human immunodeficiency virus type 1 encapsidation site is a multipartite RNA element composed of functional hairpin structures. J. Virol. 1996;70:2963–2973. doi: 10.1128/jvi.70.5.2963-2973.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Paillart JC, Marquet R, Skripkin E, Ehresmann C, Ehresmann B. Dimerization of retroviral genomic RNAs: structural and functional implications. Biochimie. 1996;78:639–653. doi: 10.1016/s0300-9084(96)80010-1. [DOI] [PubMed] [Google Scholar]
- 6.Berkhout B, van Wamel JL. Role of the DIS hairpin in replication of human immunodeficiency virus type 1. J. Virol. 1996;70:6723–6732. doi: 10.1128/jvi.70.10.6723-6732.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wilkinson KA, Gorelick RJ, Vasa SM, Guex N, Rein A, Mathews DH, Giddings MC, Weeks KM. High-throughput SHAPE analysis reveals structures in HIV-1 genomic RNA strongly conserved across distinct biological states. PLoS Biol. 2008;6:e96. doi: 10.1371/journal.pbio.0060096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Baig TT, Strong CL, Lodmell JS, Lanchy JM. Regulation of primate lentiviral RNA dimerization by structural entrapment. Retrovirology. 2008;5:65. doi: 10.1186/1742-4690-5-65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lanchy JM, Rentz CA, Ivanovitch JD, Lodmell JS. Elements located upstream and downstream of the major splice donor site influence the ability of HIV-2 leader RNA to dimerize in vitro. Biochemistry. 2003;42:2634–2642. doi: 10.1021/bi0271190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Jossinet F, Lodmell JS, Ehresmann C, Ehresmann B, Marquet R. Identification of the in vitro HIV-2/SIV RNA dimerization site reveals striking differences with HIV-1. J. Biol. Chem. 2001;276:5598–5604. doi: 10.1074/jbc.M008642200. [DOI] [PubMed] [Google Scholar]
- 11.Dirac AM, Huthoff H, Kjems J, Berkhout B. The dimer initiation site hairpin mediates dimerization of the human immunodeficiency virus, type 2 RNA genome. J. Biol. Chem. 2001;276:32345–32352. doi: 10.1074/jbc.M103462200. [DOI] [PubMed] [Google Scholar]
- 12.Lanchy JM, Ivanovitch JD, Lodmell JS. A structural linkage between the dimerization and encapsidation signals in HIV-2 leader RNA. RNA. 2003;9:1007–1018. doi: 10.1261/rna.5590603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lanchy JM, Lodmell JS. Alternate usage of two dimerization initiation sites in HIV-2 viral RNA in vitro. J. Mol. Biol. 2002;319:637–648. doi: 10.1016/S0022-2836(02)00369-8. [DOI] [PubMed] [Google Scholar]
- 14.Berkhout B, Essink BB, Schoneveld I. In vitro dimerization of HIV-2 leader RNA in the absence of PuGGAPuA motifs. FASEB J. 1993;7:181–187. doi: 10.1096/fasebj.7.1.8422965. [DOI] [PubMed] [Google Scholar]
- 15.Andersen ES, Contera SA, Knudsen B, Damgaard CK, Besenbacher F, Kjems J. Role of the trans-activation response element in dimerization of HIV-1 RNA. J. Biol. Chem. 2004;279:22243–22249. doi: 10.1074/jbc.M314326200. [DOI] [PubMed] [Google Scholar]
- 16.Huthoff H, Berkhout B. Two alternating structures of the HIV-1 leader RNA. RNA. 2001;7:143–157. doi: 10.1017/s1355838201001881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dirac AM, Huthoff H, Kjems J, Berkhout B. Regulated HIV-2 RNA dimerization by means of alternative RNA conformations. Nucleic Acids Res. 2002;30:2647–2655. doi: 10.1093/nar/gkf381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paillart JC, Dettenhofer M, Yu XF, Ehresmann C, Ehresmann B, Marquet R. First snapshots of the HIV-1 RNA structure in infected cells and in virions. J. Biol. Chem. 2004;279:48397–48403. doi: 10.1074/jbc.M408294200. [DOI] [PubMed] [Google Scholar]
- 19.Griffin SD, Allen JF, Lever AM. The major human immunodeficiency virus type 2 (HIV-2) packaging signal is present on all HIV-2 RNA species: cotranslational RNA encapsidation and limitation of Gag protein confer specificity. J. Virol. 2001;75:12058–12069. doi: 10.1128/JVI.75.24.12058-12069.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.L'Hernault A, Greatorex JS, Crowther RA, Lever AM. Dimerisation of HIV-2 genomic RNA is linked to efficient RNA packaging, normal particle maturation and viral infectivity. Retrovirology. 2007;4:90. doi: 10.1186/1742-4690-4-90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lanchy JM, Lodmell JS. An extended stem-loop 1 is necessary for human immunodeficiency virus type 2 replication and affects genomic RNA encapsidation. J. Virol. 2007;81:3285–3292. doi: 10.1128/JVI.02025-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lanchy JM, Szafran QN, Lodmell JS. Splicing affects presentation of RNA dimerization signals in HIV-2 in vitro. Nucleic Acids Res. 2004;32:4585–4595. doi: 10.1093/nar/gkh800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Berkhout B. Structure and function of the human immunodeficiency virus leader RNA. Prog. Nucleic Acid Res. Mol. Biol. 1996;54:1–34. doi: 10.1016/s0079-6603(08)60359-1. [DOI] [PubMed] [Google Scholar]
- 24.Baig TT, Lanchy JM, Lodmell JS. HIV-2 RNA dimerization is regulated by intramolecular interactions in vitro. RNA. 2007;13:1341–1354. doi: 10.1261/rna.483807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Abbink TE, Berkhout B. A novel long distance base-pairing interaction in human immunodeficiency virus type 1 RNA occludes the Gag start codon. J. Biol. Chem. 2003;278:11601–11611. doi: 10.1074/jbc.M210291200. [DOI] [PubMed] [Google Scholar]
- 26.Rein A, Henderson LE, Levin JG. Nucleic-acid-chaperone activity of retroviral nucleocapsid proteins: significance for viral replication. Trends Biochem. Sci. 1998;23:297–301. doi: 10.1016/s0968-0004(98)01256-0. [DOI] [PubMed] [Google Scholar]
- 27.Damgaard CK, Dyhr-Mikkelsen H, Kjems J. Mapping the RNA binding sites for human immunodeficiency virus type-1 gag and NC proteins within the complete HIV-1 and -2 untranslated leader regions. Nucleic Acids Res. 1998;26:3667–3676. doi: 10.1093/nar/26.16.3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.De Guzman RN, Wu ZR, Stalling CC, Pappalardo L, Borer PN, Summers MF. Structure of the HIV-1 nucleocapsid protein bound to the SL3 psi-RNA recognition element. Science. 1998;279:384–388. doi: 10.1126/science.279.5349.384. [DOI] [PubMed] [Google Scholar]
- 29.Komatsu H, Tsukahara T, Tozawa H. Viral RNA binding properties of human immunodeficiency virus type-2 (HIV-2) nucleocapsid protein-derived synthetic peptides. Biochem. Mol. Biol. Int. 1996;38:1143–1154. [PubMed] [Google Scholar]
- 30.De Rocquigny H, Gabus C, Vincent A, Fournie-Zaluski MC, Roques B, Darlix JL. Viral RNA annealing activities of human immunodeficiency virus type 1 nucleocapsid protein require only peptide domains outside the zinc fingers. Proc. Natl Acad. Sci. USA. 1992;89:6472–6476. doi: 10.1073/pnas.89.14.6472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Oude Essink BB, Das AT, Berkhout B. HIV-1 reverse transcriptase discriminates against non-self tRNA primers. J. Mol. Biol. 1996;264:243–254. doi: 10.1006/jmbi.1996.0638. [DOI] [PubMed] [Google Scholar]
- 32.Barik S. Mutagenesis and gene fusion by megaprimer PCR. In: White BA, editor. PCR Cloning Protocols. Totawa, NJ: Humana Press; 1997. pp. 173–182. [DOI] [PubMed] [Google Scholar]
- 33.Pachulska-Wieczorek K, Purzycka KJ, Adamiak RW. New, extended hairpin form of the TAR-2 RNA domain points to the structural polymorphism at the 5' end of the HIV-2 leader RNA. Nucleic Acids Res. 2006;34:2984–2997. doi: 10.1093/nar/gkl373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mathews DH, Disney MD, Childs JL, Schroeder SJ, Zuker M, Turner DH. Incorporating chemical modification constraints into a dynamic programming algorithm for prediction of RNA secondary structure. Proc. Natl Acad. Sci. USA. 2004;101:7287–7292. doi: 10.1073/pnas.0401799101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Peattie DA, Gilbert W. Chemical probes for higher-order structure in RNA. Proc. Natl Acad. Sci. USA. 1980;77:4679–4682. doi: 10.1073/pnas.77.8.4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ciesiolka J, Michalowski D, Wrzesinski J, Krajewski J, Krzyzosiak WJ. Patterns of cleavages induced by lead ions in defined RNA secondary structure motifs. J. Mol. Biol. 1998;275:211–220. doi: 10.1006/jmbi.1997.1462. [DOI] [PubMed] [Google Scholar]
- 37.Krzyzosiak WJ, Marciniec T, Wiewiorowski M, Romby P, Ebel JP, Giege R. Characterization of the lead(II)-induced cleavages in tRNAs in solution and effect of the Y-base removal in yeast tRNAPhe. Biochemistry. 1988;27:5771–5777. doi: 10.1021/bi00415a056. [DOI] [PubMed] [Google Scholar]
- 38.Wilkinson KA, Merino EJ, Weeks KM. Selective 2'-hydroxyl acylation analyzed by primer extension (SHAPE): quantitative RNA structure analysis at single nucleotide resolution. Nat. Protoc. 2006;1:1610–1616. doi: 10.1038/nprot.2006.249. [DOI] [PubMed] [Google Scholar]
- 39.Das R, Laederach A, Pearlman SM, Herschlag D, Altman RB. SAFA: semi-automated footprinting analysis software for high-throughput quantification of nucleic acid footprinting experiments. RNA. 2005;11:344–354. doi: 10.1261/rna.7214405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Badorrek CS, Weeks KM. Architecture of a gamma retroviral genomic RNA dimer. Biochemistry. 2006;45:12664–12672. doi: 10.1021/bi060521k. [DOI] [PubMed] [Google Scholar]
- 41.Merino EJ, Wilkinson KA, Coughlan JL, Weeks KM. RNA structure analysis at single nucleotide resolution by selective 2'-hydroxyl acylation and primer extension (SHAPE) J. Am. Chem. Soc. 2005;127:4223–4231. doi: 10.1021/ja043822v. [DOI] [PubMed] [Google Scholar]
- 42.Gherghe C, Leonard CW, Gorelick RJ, Weeks KM. Secondary structure of the mature ex virio Moloney murine leukemia virus genomic RNA dimerization domain. J. Virol. 2010;84:898–906. doi: 10.1128/JVI.01602-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Berkhout B. Structural features in TAR RNA of human and simian immunodeficiency viruses: a phylogenetic analysis. Nucleic Acids Res. 1992;20:27–31. doi: 10.1093/nar/20.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Das AT, Klaver B, Centlivre M, Harwig A, Ooms M, Page M, Almond N, Yuan F, Piatak M, Jr, Lifson JD, et al. Optimization of the doxycycline-dependent simian immunodeficiency virus through in vitro evolution. Retrovirology. 2008;5:44. doi: 10.1186/1742-4690-5-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Berkhout B, Schoneveld I. Secondary structure of the HIV-2 leader RNA comprising the tRNA-primer binding site. Nucleic Acids Res. 1993;21:1171–1178. doi: 10.1093/nar/21.5.1171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Beltz H, Azoulay J, Bernacchi S, Clamme JP, Ficheux D, Roques B, Darlix JL, Mely Y. Impact of the terminal bulges of HIV-1 cTAR DNA on its stability and the destabilizing activity of the nucleocapsid protein NCp7. J. Mol. Biol. 2003;328:95–108. doi: 10.1016/s0022-2836(03)00244-4. [DOI] [PubMed] [Google Scholar]
- 47.Williams MC, Rouzina I, Wenner JR, Gorelick RJ, Musier-Forsyth K, Bloomfield VA. Mechanism for nucleic acid chaperone activity of HIV-1 nucleocapsid protein revealed by single molecule stretching. Proc. Natl Acad. Sci. USA. 2001;98:6121–6126. doi: 10.1073/pnas.101033198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Huthoff H, Berkhout B. Mutations in the TAR hairpin affect the equilibrium between alternative conformations of the HIV-1 leader RNA. Nucleic Acids Res. 2001;29:2594–2600. doi: 10.1093/nar/29.12.2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Freund F, Boulme F, Litvak S, Tarrago-Litvak L. Initiation of HIV-2 reverse transcription: a secondary structure model of the RNA-tRNA(Lys3) duplex. Nucleic Acids Res. 2001;29:2757–2765. doi: 10.1093/nar/29.13.2757. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Matsui T, Tanaka T, Endoh H, Sato K, Tanaka H, Miyauchi E, Kawashima Y, Nagai-Makabe M, Komatsu H, Kohno T, et al. The RNA recognition mechanism of human immunodeficiency virus (HIV) type 2 NCp8 is different from that of HIV-1 NCp7. Biochemistry. 2009;48:4314–4323. doi: 10.1021/bi802364b. [DOI] [PubMed] [Google Scholar]
- 51.Fisher RJ, Rein A, Fivash M, Urbaneja MA, Casas-Finet JR, Medaglia M, Henderson LE. Sequence-specific binding of human immunodeficiency virus type 1 nucleocapsid protein to short oligonucleotides. J. Virol. 1998;72:1902–1909. doi: 10.1128/jvi.72.3.1902-1909.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Turner KB, Hagan NA, Fabris D. Understanding the isomerization of the HIV-1 dimerization initiation domain by the nucleocapsid protein. J. Mol. Biol. 2007;369:812–828. doi: 10.1016/j.jmb.2007.03.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Strong CL, Lanchy JM, Dieng-Sarr A, Kanki PJ, Lodmell JS. A 5'UTR-spliced mRNA isoform is specialized for enhanced HIV-2 gag translation. J. Mol. Biol. 2009;391:426–437. doi: 10.1016/j.jmb.2009.06.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Berglund JA, Charpentier B, Rosbash M. A high affinity binding site for the HIV-1 nucleocapsid protein. Nucleic Acids Res. 1997;25:1042–1049. doi: 10.1093/nar/25.5.1042. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.