Abstract
DNA polymerase I (pol I) processes RNA primers during lagging-strand synthesis and fills small gaps during DNA repair reactions. However, it is unclear how pol I and pol III work together during replication and repair or how extensive pol I processing of Okazaki fragments is in vivo. Here, we address these questions by analyzing pol I mutations generated through error-prone replication of ColE1 plasmids. The data were obtained by direct sequencing, allowing an accurate determination of the mutation spectrum and distribution. Pol I’s mutational footprint suggests: (i) during leading-strand replication pol I is gradually replaced by pol III over at least 1.3 kb; (ii) pol I processing of Okazaki fragments is limited to ∼20 nt and (iii) the size of Okazaki fragments is short (∼250 nt). While based on ColE1 plasmid replication, our findings are likely relevant to other pol I replicative processes such as chromosomal replication and DNA repair, which differ from ColE1 replication mostly at the recruitment steps. This mutation footprinting approach should help establish the role of other prokaryotic or eukaryotic polymerases in vivo, and provides a tool to investigate how sequence topology, DNA damage, or interactions with protein partners may affect the function of individual DNA polymerases.
INTRODUCTION
ColE1 plasmids constitute a class of plasmids that share regulatory mechanisms of replication [reviewed in refs. (1,2)]. In these plasmids, replication is controlled by a ∼600 bp-long sequence known as plasmid origin of replication or ori (1,3). ColE1 plasmids have been extensively used as a model to study fundamental processes of DNA homeostasis and are present in most Escherichia coli expression and shuttle vectors (4).
ColE1ori sequences encode an RNA primer that is processed by RNAseH and extended by DNA polymerase I (pol I). DNA polymerase I extension unwinds the DNA, exposing a primosome assembly signal (n′ pas) or single-strand initiation A (ssiA) sequence on the leading strand (5). This single-stranded sequence motif allows assembly of the primosome through recruitment and activation of the PriA protein. PriA-dependent initiation represents a distinct form of DNA replication initiation, typically associated with DNA repair. This is in contrast to DnaA-dependent replication initiation at oriC, which mediates replicative DNA synthesis (6). Following PriA-primosome assembly, DnaB helicase and DnaG primase work coordinately to initiate lagging-strand synthesis. Primosome movement facilitates lagging-strand replication, which proceeds discontinuously by extension of short primers laid out by the primase. Leading-strand replication, on the other hand, appears to be continuous, although there are some indications that it may be discontinuous as well (7,8).
Pol I activity is essential for ColE1 plasmid replication (9,10). Three distinct roles have been recognized for this enzyme: (i) extending the processed ori RNA primer; (ii) unwinding the DNA until the primosome is assembled and the DnaB helicase becomes active and (iii) processing Okazaki RNA primers during lagging-strand synthesis. A number of questions regarding the role of pol I during ColE1 plasmid replication in vivo remain unsolved, though. The size of the pol I extension product (corresponding to the 6sL replication intermediate) is unclear, with reports ranging from 100 nt to 1.5 kb (10–13). The extent of pol I processing of short (∼11 nt) RNA primers on the lagging-strand is also unknown due to the nick-translation activity of this polymerase, i.e. to its ability to degrade RNA or DNA in the 5′ to 3′ direction while simultaneously extending it in the 5′ to 3′ direction (14). Likewise, it remains unclear how sharp the transition or ‘switch’ from pol I to pol III replication is in vivo.
Here we use error-prone pol I replication of a ColE1 plasmid to address these questions. The low-fidelity pol I mutant used in this study bears mutations in three key determinants of fidelity, which together increase the mutation frequency of the polymerase in vivo by three orders of magnitude (15). The highly elevated mutation frequency of this error-prone polymerase was essential for the compilation of our database, facilitating the detection of mutations without the use of any selectable reporter. This approach provided an unbiased spectrum and accurate information of the physical distribution of the mutations. We have to assume that the mutations decreasing the fidelity of our error-prone pol I mutant don’t significantly change other aspects of pol I functionality. That having been said, the error-prone replication footprint of pol I suggests that: (i) on the leading-strand, pol I is replaced by pol III very gradually over at least 1.3 kb; (ii) that on the lagging-strand pol I processing of Okazaki fragments is limited to ∼20 nt, which may represent the true size of Okazaki primers in vivo; and (iii) that the size of Okazaki fragments may be shorter for PriA-dependent replicons than that of DnaA-dependent replicons.
Given that ColE1 plasmid, DNA repair associated, and oriC-dependent DNA synthesis differ mainly at the recruitment steps, our findings can likely be generalized to other pol I-dependent replicative processes. In addition, our work shows that mutational footprinting can be utilized to define the template for DNA synthesis by specific DNA polymerases in vivo, and could be used to establish the role of other prokaryotic and eukaryotic polymerases in the cell. Our approach can also be used as a tool to study the impact of sequence topology or of DNA damage on replication by specific polymerases or to study how replication is modulated by interactions between individual DNA polymerases and specific protein partners in vivo.
MATERIALS AND METHODS
Bacterial strains
JS200 (SC-18 recA718 polA12ts uvrA355 trpE65 lon-11 sulA1) cells were used as our host strain. The polA12 allele encodes a point mutation in pol I (G544D) that interferes with the coordination between the polymerase and the 5′ → 3′ exonuclease activities (16). This mutant exhibits reduced temperature stability and activity at 42°C (17). RecA718 is a sensitized allele of RecA, resulting in SOS induction under conditions that are restrictive for polA12 (18).
Plasmid constructs
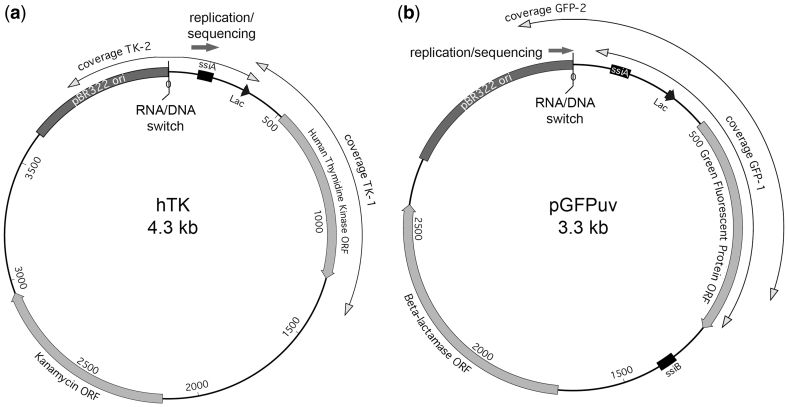
Our mutagenic plasmid expressing low-fidelity pol I (muta-plasmid) was generated by cloning of the mutant pol I sequence into a pHSG576 vector between the HindIII/EcoRI restriction sites, and bears chloramphenicol resistance (19). The human thymidine kinase (hTK) library was generated by cloning in hTK into the pCR 2.1-TOPO vector (Invitrogen, Carlsbad, CA, USA) (20), which carries a carbenicillin gene as a selectable marker. pGFPuv (with carbenicillin resistance) was obtained from Clontech (Mountain View, CA, USA). The annotated sequence of our hTK and GFP plasmids is shown in Supplementary Figure S1.
Media and supplies
Growth media LB Agar and LB broth were purchased from Fisher Scientific and prepared according to vendor specifications. Mutagenesis was carried out in 2XYT rich media containing 0.016 g/ml bacto tryptone, 0.01 g/ml bacto yeast extract and 0.005 g/ml NaCl suspended in deionionized water. The antibiotic concentrations used for marker selection are: 30 µg/ml (chloramphenicol) and 50 µg/ml (hTK) or 100 µg/ml (all other libraries) (carbenicillin). All DNA isolation procedures were performed using Machery Nagel’s Nucleospin Plasmid miniprep. Sequencing was carried out by the sequencing service of the Department of Chemistry of the University of Washington (University of Washington, Seattle) or by Sequetech (Mountain View, CA, USA).
Error-prone pol I mutagenesis
The target plasmid, a ColE1 plasmid bearing the gene of interest, was transformed into JS200 cells carrying muta-plasmid, the pSC101 (pol I-independent) plasmid bearing our low-fidelity pol I. When these transformants are grown under restrictive conditions, low-fidelity pol I is the functional polymerase present in the cell, introducing random errors during replication of the ColE1 target plasmid. Mutagenesis was performed in liquid culture, by switching a culture grown under permissive conditions (LB, 30°C, exponential) to restrictive conditions (2XYT, 37°C, saturation) as described in ref. (15). Briefly, ∼100 ng of the target plasmids (hTK-Topo or pGFPuv) were transformed into electrocompetent JS200 muta-plasmid cells [for preparation of competent cells, see ref. (21)]. The transformants were resuspended in 1 ml LB broth, recovered for 1 h at 30°C, and plated at 30°C on LB agar plates containing 50 µg/ml carb (hTK) or 100 µg/ml carb (pGFPuv). A single colony was picked from each plate, inoculated into 4 ml LB broth and grown at low density at permissive temperature (30°C). For mutagenesis, an aliquot of the overnight culture (dilution factor 1:103 to 1:105) was transferred into 4 ml of 2XYT media (pre-warmed at 37°C), and grown shaking at 37°C for 1 or 3 days to reach complete saturation or hypersaturation (21) (hypersaturated cultures denoted as ‘Day 3’ in Table 1). Following mutagenesis, library plasmid DNA was isolated using Machery Nagel’s Nucleospin Plasmid miniprep kit and put through an additional round of mutagenesis (see below). For sequencing, plasmids were retransformed into a strain that is WT for pol I to separate out individual plasmids and either recovered for sequencing or sequenced by rolling circle amplification (RCA) (22).
Table 1.
Pol I mutation libraries used in this study
| Library | No. of rounds of mutagenesis | No. of clones sequenced | No. of bases sequenced (kb) | Total mutations | Total curated mutations |
|---|---|---|---|---|---|
| Human thymidine kinase | |||||
| TK1 | 5-7 | 88 | 91 | 396 | 280 |
| 5 | 23 | 24 | 81 | 65 | |
| 6 | 22 | 23 | 119 | 88 | |
| 7 | 43 | 44 | 196 | 127 | |
| TK2 | 7 | 65 | 43 | 148 | 113 |
| Green fluorescent protein | |||||
| GFP 1 | 1 | 276 | 305 | 184 | 184 |
| Day 1: 91 | Day 1: 98 | Day 1: 36 | |||
| Day 3: 185 | Day 3: 209 | Day 3: 148 | |||
| GFP 2 | 2 | 63 | 64 | 60 | 60 |
Iteration of mutagenesis and sequencing
The mutagenesis procedure in liquid culture described above was repeated to increase the mutation frequency as described in detail in ref. (21). Briefly, the plasmid library recovered from the initial round of mutagenesis was retransformed into fresh JS200 muta-plasmid cells at 30°C. A plate containing a high density of transformant colonies (>100 000 colonies) was washed with 2 ml LB. For additional mutagenesis, 105 cells from this wash were inoculated into 4 ml of 2XYT media at 37°C and grown to saturation. This procedure was repeated until the desired mutation frequency was reached. Individual plasmids were identified through transformation of a small amount of plasmid DNA (50–100 ng) into a strain that is WT for pol I, DH10beta (hTK) or BL21 (pGFPuv). From this transformation, individual colonies were sequenced. In Supplementary Table S1 for each clone present in our libraries, we list number of mutagenesis cycles, sequence coverage, and mutations found. This information is summarized in Table 1 of the main text.
Library curation
We found evidence for the presence of mutation hotspots in the hTK library. This conclusion was based on distribution of number of mutations/position, which clearly deviated from Poisson (Supplementary Figure S2a) and on the fact that only one type nucleotide substitution was dominant at these sites (Supplementary Figure S2b). Based on this analysis, we conservatively eliminated all mutations present at positions with ≥6 mutations from our database. The positions and nucleotide substitutions involved are listed in Supplementary Figure S2b. Our hTK database also contained clones sharing more than one mutation (Supplementary Table S2), which may have been the result of limited clonal expansion events. To rule out this possibility, we conservatively included only ancestral mutations in our subsequent analyses (Supplementary Table S2). Limited clonal expansions would suggest either a mild positive selection for specific mutations or a stronger selective pressure combined with significant clonal interference (23). While the human thymidine kinase gene was cloned without a promoter, low levels of expression cannot be ruled out in the absence of a repressor. Despite the evidence for limited clonal expansions and the presence of occasional hotspots, the hTK library showed broad genetic diversity and was therefore adequate for our footprinting analysis. Further, the main conclusions drawn from the thymidine kinase library were independently confirmed in a different library (the GFP library). Our final, curated libraries included a total of 393 mutations (TK) and 244 mutations (GFP). These mutations are listed by position, relative to RNA/DNA switch, in Supplementary Table S4.
Statistical methods
All statistical analyses were performed using the R statistical computing package (R-foundation for Statistical Computing: Vienna, Austria, 2009). The significance of biases in the frequency of mutation between complementary pairs was assessed using a two-sided binomial test (Table 2).
Table 2.
hTK library complementary mutation pairs: frequency and significance of bias
| All | P-value | d < 800 | P-value | d = 1100–1300 | d > 3800 | |
|---|---|---|---|---|---|---|
| A to G | 42 | 1.1 × 10−04 | 32 | 4.3 × 10−04 | 2 | 0 |
| T to C | 13 | 9 | 0 | 1 | ||
| C to T | 194 | 2.2 × 10−14 | 140 | 2.2 × 10−16 | 26 | 4 |
| G to A | 71 | 29 | 12 | 7 | ||
| A to T | 24 | 0.20 | 18 | 0.04 | 1 | 1 |
| T to A | 15 | 7 | 2 | 1 | ||
| A to C | 3 | 1 | 2 | 1 | 1 | 0 |
| T to G | 4 | 2 | 1 | 0 | ||
| G to T | 8 | 0.11 | 6 | 0.031 | 0 | 1 |
| C to A | 2 | 0 | 0 | 1 | ||
| C to G | 8 | 0.11 | 6 | 0.29 | 1 | 0 |
| G to C | 2 | 2 | 0 | 0 | ||
| Indels | 7 | 5 | 0 | 0 | ||
| Lagging | 107 | 49 | 15 | 10 | ||
| Other | 279 | 204 | 31 | 6 | ||
| Total | 393 | 258 | 46 | 16 |
RESULTS
Generation of a neutral pol I mutation database
In order to obtain new insights into the function of pol I in the cell, we compiled a database of pol I mutations. Our database contains sequence encoded in a ColE1 plasmid not subject to specific selective pressure, i.e. neutral sequence.
To facilitate our data collection, we decreased the fidelity of pol I replication in vivo by expressing a mutant polymerase in a polA (temperature-sensitive) strain and growing cells under restrictive conditions (37°C and saturation) (15). To raise the mutation density, libraries were submitted to multiple iterations of pol I mutagenesis: 5–7 for hTK, and 1–2 for GFP (Table 1). Our method of pol I mutagenesis in vivo in liquid culture has been described in detail (21), and the generation of our libraries is described in the ‘Materials and Methods’ section. Briefly, following mutagenesis at 37°C, plasmids were recovered and retransformed into wild-type cells to separate out individual plasmids for sequencing analysis. For iteration, the plasmid libraries were retransformed into cells expressing our error-prone pol I at 30°C, and a large number of transformants (>105) was recovered by plate wash, inoculated into a fresh culture, and grown to saturation at 37°C. This process was iterated until the desired mutation frequency was reached. At this point individual plasmids were sequenced.
We targeted two ColE1 plasmid constructs for mutagenesis: a plasmid bearing human thymidine kinase (hTK) cloned in a TOPO vector (20), and a plasmid bearing ‘cycle 3′ green fluorescent protein (GFP) in pGFPuv (Clontech). The annotated sequence of these plasmids is listed in Supplementary Figure S1 and their graphic maps are shown in Figure 1. Both constructs bear a pBR322 (pMB1) ori, which is a ColE1-like origin of replication (24). Note that ssiA, the primosome assembly site for the lagging strand (25,26), is present in our constructs. SsiB, a second primosome assembly site for the leading strand (which is typically further downstream), is absent in the hTK construct and displaced in pGFPuv (Figure 1). In addition to the ori sequence, the 419 nt downstream of the DNA/RNA switch (including the lac promoter) are identical between the two constructs.
Figure 1.
ColE1 plasmids used for library generation. Diagrams present the ColE1 plasmids used to generate the pol I libraries. Shown are their main features: plasmid ori, ssi primosome assembly site signals, and ORFs (boxes). Sequence coverage (double-headed arrows), and directionality of replication and of sequencing (solid arrow) are also shown. The distance from the RNA/DNA switch is indicated, at 500 bp intervals. (a) hTK libraries: human thymidine kinase gene cloned in the TOPO vector, (b) pGFPuv libraries, bearing the cycle 3 GFP gene.
We sequenced a total of 134 kb from 153 independent clones (hTK libraries) and 369 kb from 339 independent clones (GFP libraries). The sequence represents the leading-strand. Table 1 shows the number of clones (transformant colonies) analyzed, nucleotides sequenced, and mutations identified for each library; the sequence coverage is graphically displayed in Figure 1: it starts 5′ of the DNA/RNA switch and extends to >1 kb 3′ of it (Figure 1).
In the hTK library we found 11 hotpots, i.e. positions with substantially more mutations than would be expected based on a random (Poisson) distribution (n ≥ 6; Supplementary Figure S2a). The mutations present in these hotspots are listed in Supplementary Figure S2b. We also found examples of what appeared to be mild clonal expansions, based on the fact that some clones shared two or more mutations, an event that should be exceedingly rare by chance since it involves a combination of two rare events (Supplementary Table S2). To ensure that only independent TK mutations were included in our analysis, we conservatively removed all mutations present in mutation hotspot positions and also clonal mutations other than putatively ancestral ones. The GFP library, by contrast, produced no significant mutation hotspots or evidence of clonal expansion. Our final, curated libraries included a total of 393 (hTK) and 244 (GFP) mutations. These mutations are listed in Supplementary Table S4, by position relative to RNA/DNA switch.
Pol I mutation frequency decreases with increasing distance from RNA/DNA switch
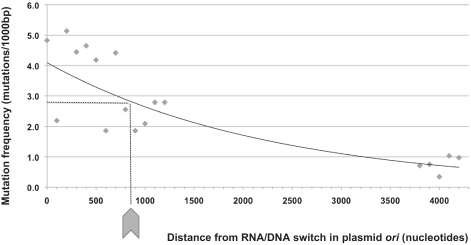
Since pol I is replaced by pol III during plasmid replication, we expect a decreased frequency of pol I mutations with increasing distance to the RNA/DNA switch. To confirm this prediction, we took our largest data set (round 7 hTK library) and plotted the frequency of mutation at increasing distance for 100 bp intervals. The results, presented in Figure 2, show a consistent decrease in pol I mutation frequency as replication proceeds along the plasmid sequence. This decrease is best modeled by an exponential function (r2 = 0.79) (Figure 2, trendline).
Figure 2.
Pol I mutation frequency as a function of distance from RNA/DNA switch. Mutation frequency (expressed as number of mutations per 1000 bp) for the hTK library generated following seven rounds of pol I mutagenesis. The x-axis is the distance (in nucleotides) from the ori RNA/DNA switch. Each point represents a 100-bp interval. The trend line shown represents an optimized exponential fit (r2 = 0.79).
Error rate of pol I in vivo
Table 2 presents the mutation spectrum for our largest library (hTK). Based on our biochemical understanding of ColE1 replication, which involves extension of an ori RNA primer by pol I, we considered these mutations primarily leading-strand replication errors, particularly in proximal areas. Given that the position of mutations relative to RNA/DNA switch is important to determine their origin, we broke down the mutation spectrum by distance intervals in Table 2 and used 800 bp (the distance where the mutation frequency is halfway between that of most proximal and that of most distal sequences) as the threshold distance to classify mutations as either ‘close to ori’ (proximal) or ‘far from ori’ (distal; Figure 2).
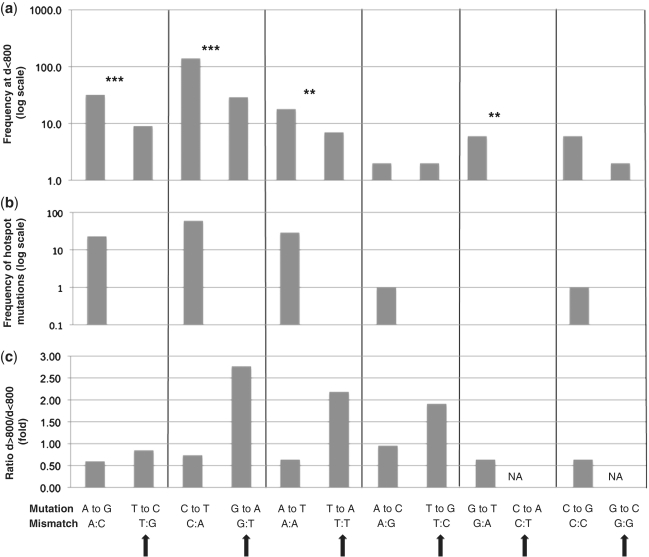
Since mostly one strand is being synthesized in proximal areas (the leading strand), the mutation spectrum approximates the error rate of pol I in vivo (Figure 3a, Scenario 1). This data indicate that complementary pairs of mutations show significant differences in error rate, with one mutation being more frequent than the other within individual pairs. For example, A → G mutations appear 32 times in the proximal hTK library, whereas the complementary T → C mutation appears only nine times (P < 4.3 × 10−4). Likewise, C → T mutations appear a total of 140 times, compared to only 29 times for the complementary G → A mutation (P < 2.2 × 10−16). These differences are statistically significant in four out of six possible complementary pairs (Table 2 and double and triple asterisks in Figure 4a). In all cases, hotspot mutations further increase the observed asymmetry (Figure 4b), consistent with the idea that hotspot mutations are facilitated errors that the polymerase is already prone to making (27).
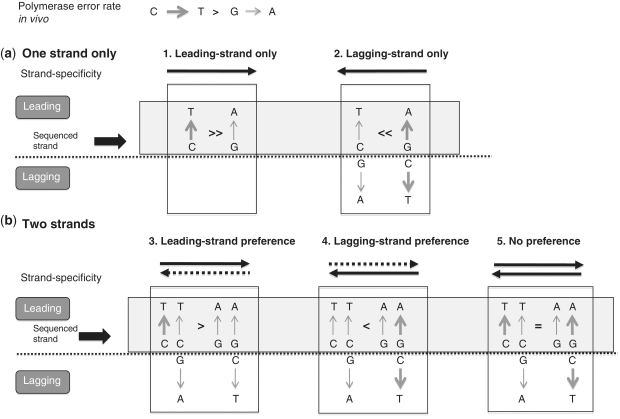
Figure 3.
Rationale for using polymerase error bias between complementary mutations to detect strand preference. The CG → TA pair is used as an example, with a relative error rate of C → T >> G → A. Both strands are shown, and the light grey box highlights the leading (sequenced) strand. The thickness of the arrows is proportional to the mutation frequency, factoring in both the error rate and the strand preference of the polymerase. Dashed lines represent decreased strand preference (a) Single-strand synthesis. Since only the leading strand is sequenced, if the lagging strand is used as a template, the most abundant C → T error appears as G → A. This leads to an inversion in frequency bias following a template switch (i.e. G → A >> C → T). (b) Double-strand synthesis. The bias in frequency between the two complementary mutations is decreased depending on the relative frequency of leading versus lagging-strand synthesis (i.e. on the degree of strand preference). In the most extreme case (no strand preference, Scenario 5), the bias between complementary mutations is completely eliminated. In all cases (Scenarios 2–5), lagging-strand synthesis increases the proportion of ‘least frequent’ mutations (in this case, G → A) on the leading-strand compared to the error rate of the polymerase (Scenario 1).
Figure 4.
Identification of marker mutations for lagging-strand replication. Results from the hTK library, showing all six complementary pairs of point mutations, listing the most frequent mutation of the pair first. (a) Mutation frequency of proximal (d < 800) point mutations, in log scale to facilitate comparison across a wide range of frequencies. Significant asymmetry between complementary pairs (listed in Table 2) is denoted with three (P < 0.001) or two (P < 0.05) asterisks. Total number of mutations: 253 (b) Representation of mutations in mutation hotspots. The frequency of each type of point mutation present in mutation hotspots is shown in log scale to facilitate comparison across a wide range of frequencies. Total number of mutation: 114. (c) Enrichment of mutations at distal (d > 800) positions. The ratio of the frequency of a given mutation at d > 800 relative to the frequency at d < 800 is shown. Total number of mutations: proximal (n < 800): 253; distal (n > 800) = 133. Solid arrows indicate marker lagging-strand mutations.
Definition of marker mutations for lagging-strand synthesis
Differences in the error rate of complementary mutations can be used to define strand specificity, because a switch in the template would result in an inversion of the frequency bias (Figure 3a, Scenario 2). Continuing with the examples above, T → C and G → A mutations would be expected to be more frequent than their corresponding complementary A → G and C → T mutations in areas of lagging-strand synthesis (Figure 3a, Scenario 2). Therefore, we reasoned that ‘least frequent’ mutations for each complementary pair can help identify areas of lagging-strand synthesis because they will be more frequent in these areas than in areas undergoing only leading-strand synthesis. The level of enrichment will depend on the strand preference of the polymerase (Figure 3b). Thus, for the remainder of the article we call these ‘least frequent’ mutations, namely T → C, G → A, T → A, C → A and G → C (Figure 4a) markers for lagging-strand synthesis. Please note that they are not signatures in the true sense of the word because we can’t distinguish the strand of origin (Figure 3b), however, we reasoned that since these mutations are enriched in areas of lagging-strand synthesis they can serve as markers to point us to them.
Lagging-strand synthesis by pol I should be associated with processing of RNA primers, which is expected to happen at regular sequence intervals regardless of distance from RNA/DNA switch. The frequency of leading-strand mutations, on the other hand, exhibits an inverse correlation with distance (Figure 2). Therefore, the relative proportion of marker lagging-strand mutations should increase in distal portions of plasmid sequence. Figure 4c shows the relative frequency for each type of point mutation comparing proximal (d < 800) versus distal (d > 800) mutations. Strikingly, marker lagging-strand mutations are consistently overrepresented at distances >800 (compare Figure 4a and c), consistent with our proposed use for these mutations as indicators of strand specificity. This analysis also shows a distal enrichment for T → G mutations, making this mutation an additional marker for lagging-strand synthesis (Figure 4b).
Identification of Okazaki processing sites
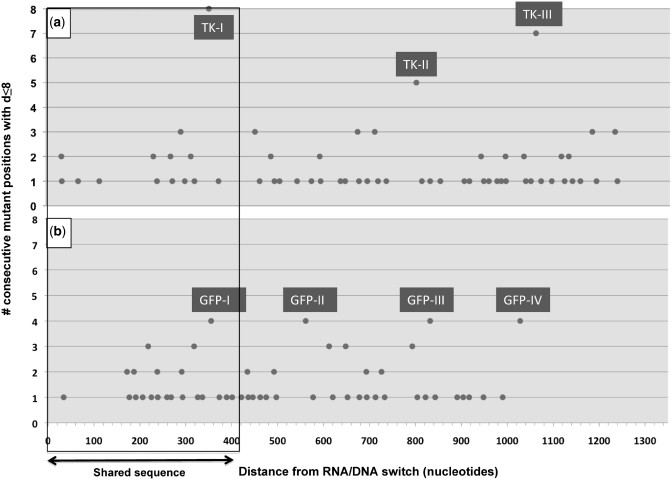
We next looked to see whether the marker mutations for lagging-strand synthesis described could identify a footprint for Okazaki processing (OP) sites. Mutations in these areas can be generated through replication of both strands, particularly in areas close to RNA/DNA switch, where significant leading-strand synthesis is occurring, decreasing the strand specificity of our markers (Figure 3b). Therefore, based on the short length (∼10 nt) of the RNA priming lagging-strand synthesis, we used an additional, more stringent consideration: short distance between marker mutations. Figure 5 shows the number of lagging-strand mutant positions that can be found clustered with a distance between each other of ≤8 nt. This distance is half the average distance between marker lagging-strand mutations within the sequence interval under consideration. Figure 5a shows our clustering analysis of marker lagging-strand mutations for the hTK library (n = 97). Three clusters clearly stand out in this library (TK-I, TK-II and TK-III), with between five and eight mutant positions each. The next observed cluster size is only three (six clusters), suggesting that three mutations are our level of background noise for ∼100 lagging-strand mutations. Figure 5b shows our clustering analysis for the GFP library (n = 80). In this library we found four clusters containing more than three mutations (GFP-I, GFP-II, GFP-III, GFP-IV). Thus, between the two libraries, we tentatively identified seven OP sites (Figures 5 and 6).
Figure 5.
Clustering analysis for marker lagging-strand mutations. Number of consecutive positions with marker lagging-strand mutations that can be found at a distance of ≤8 nt from each other (y-axis) relative to the distance (in nucleotides) of mutant positions from the RNA/DNA switch (x-axis). The portion of sequence shared between the two libraries is boxed. Clusters considered significant (n > 3 mutations) are labeled TKI-III (hTK library) and GFP I–IV (GFP library). (a) hTK library, generated through 5–7 rounds of pol I mutagenesis, and with an average distance between marker lagging-strand mutations (for the interval shown) of 15.9 nt. (b) GFP library, generated through 1–2 rounds of pol I mutagenesis, and with an average distance between marker lagging-strand mutations (for the interval shown) of 15.3 nt.
Figure 6.
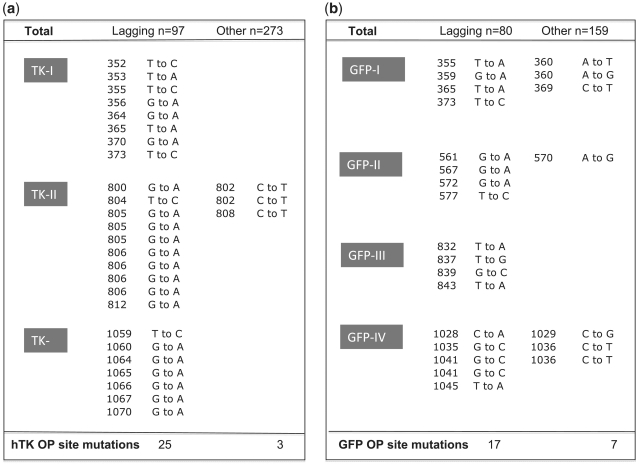
Mutation spectrum within clusters. Listed are the point mutations found in the clusters defined in Figure 5 as putative OP sites. The sequenced strand was the leading strand. Each cluster is labeled, with hTK clusters (TK-I, TK-II and TK-III) shown on the left (a) and GFP clusters (GFP-I, GFP-II, GFP-III and GFP-IV) shown on the right (b). For each library, marker lagging-strand mutations are listed on the left, and their complement on the right. For each mutation, the mutant position relative to the RNA/DNA switch and observed nucleotide substitutions are listed. The reference sequences for the hTK and GFP plasmids can be found in Supplementary Figure S1. Note that the total number of marker lagging-strand mutations listed (n = 42) exceeds the total number of positions shown in Figure 5 within n > 3 clusters (n = 37). This is due to the fact that three positions (804 and 805 of hTK library and 1041 of the GFP library) had more than one mutation.
Mutation profile is consistent with OP sites
Figure 6 lists the marker lagging-strand mutations in the clusters shown in Figure 5 to have more than three mutations. Note that all types of marker mutations for lagging-strand synthesis are represented in these clusters, including the three most infrequent ones: T → G, C → A and G → C, suggesting that OP sequences are enriched for all types of lagging-strand mutations rather than for specific ones. Table 3 shows the representation of lagging-strand marker mutations within clusters relative to total sequence coverage. Merging the hTK and GFP library data, we demonstrate a substantial enrichment for all six types of lagging-strand mutations. Even after correcting for biases in nucleotide representation within the OP sites (bias calculations are shown in Supplementary Table S3), we see an overrepresentation of all six types of marker lagging strand mutations, with levels ranging between 2.3- and 10.6-fold. Their complementary mutations, on the other hand, show decreased representation, being 8.3- (hTK) and 2.4-fold (GFP) less frequent, even though they are overall 2- to 3-fold more abundant (Figure 6). This inversion in the frequency pattern of mutations, with all ‘least frequent’ ones being enriched and ‘most frequent’ ones being underrepresented strongly suggests a switch in polymerase template.
Table 3.
Overrepresentation of marker lagging-strand mutations within putative OP sites
| Lagging-strand mutations | T → C | G → A | T → A | T → G | C → A | G → C | Total |
|---|---|---|---|---|---|---|---|
| hTK | |||||||
| Total | 12 | 64 | 14 | 4 | 1 | 2 | 97 |
| OP sites | 5 | 18 | 2 | 0 | 0 | 0 | 25 |
| Overrepresentation (fold) | 10.4 | 7.0 | 3.6 | 0.0 | 0.0 | 0.0 | |
| Normalized for nucleotide composition (fold) | 7.8 | 4.1 | 2.7 | N/A | N/A | N/A | |
| GFP | |||||||
| Total | 12 | 38 | 16 | 2 | 7 | 5 | 80 |
| OP sites | 2 | 5 | 5 | 1 | 1 | 4 | 18 |
| Overrepresentation (fold) | 2.9 | 2.3 | 5.4 | 8.6 | 2.5 | 13.8 | |
| Normalized for nucleotide composition (fold) | 2.7 | 1.3 | 5.1 | 8.1 | 4.3 | 8.1 | |
| hTK and GFP | |||||||
| Total | 24 | 102 | 30 | 6 | 8 | 7 | 177 |
| OP sites | 7 | 23 | 7 | 1 | 1 | 4 | 43 |
| Overrepresentation (fold) | 5.4 | 4.2 | 4.3 | 3.1 | 2.3 | 10.6 | |
| Normalized for nucleotide composition (fold) | 4.5 | 2.5 | 3.6 | 2.6 | 4.9 | 6.4 |
Overrepresentation is defined as the percent of a given marker lagging-strand mutation within OP sites relative to the percent of the total, considering the fraction of sequence represented by these sites; in the case of hTK OP sites represent 4% of the total coverage, and in the case of GFP OP sites represent 5.8%. These values for overrepresentation were normalized by dividing the overrepresentation values by a ‘nucleotide composition bias factor’, a fudge factor expressing the relative abundance of individual nucleotides within OP sites (Supplementary Table S3). Values we were unable to calculate due to gaps in our data are indicated as N/A.
Lagging-strand marker mutations are very evenly distributed along OP sequence: 37 positions are mutant (30% of the total OP sequence), and only three of them have more than one mutation. This strongly argues against local sequence context (hotspot) effects on the fidelity of pol I synthesis; instead it is more consistent with a change in template altering the spectrum of mutations without changing its overall frequency. Thus, overall the mutation profile of the lagging-strand mutation clusters identified in Figure 5 suggests that these clusters most likely represent OP sites.
Sequence context of putative OP sites
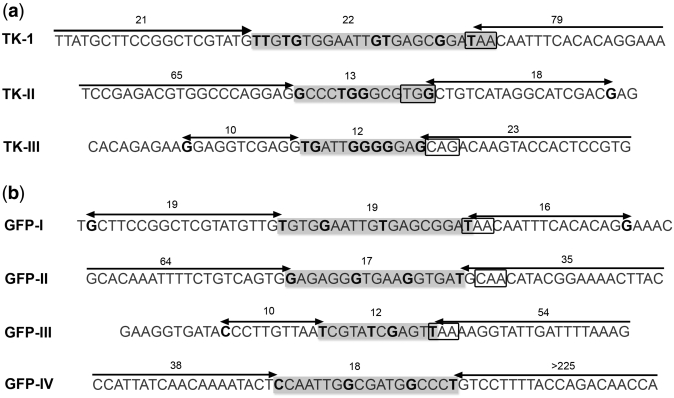
Figure 7 shows the sequences identified as putative OP sites, with ∼20 nt of additional flanking sequence on each side. The sequence is grayed out and mutant positions are highlighted in black. We also indicate the estimated length (in nucleotides) of the site, as well as the distance to the next marker lagging-strand mutation. To facilitate visualizing these sequences in a wider sequence context, we also highlighted putative OP sites in Supplementary Figure S1. OP sites are, on average, 16 nt long (varying in size between 22 and 12 nt) and often far (>30 nt) from the next lagging-strand mutation.
Figure 7.
Okazaki processing site sequence context. Mutant positions are highlighted in bold black on a grayed out sequence. Sequence intervals defined by marker lagging-strand mutation clusters (Figure 5) are highlighted (light gray box) and the total number of nucleotides within each interval is listed. Arrows represent the distance (in nucleotides, listed on top) from the Okazaki processing site to the next lagging-strand mutation marker. 3′-PuPyPy-5′ primase recognition sequences at the 5′ end of the putative RNA primer (3′-end of the complementary sequence shown) are boxed. To help placing OP sites in a wider sequence context, these sites are also highlighted in Supplementary Figure S1, which lists the complete sequence of the hTK and pGFPuv plasmids.
We also looked for signature 3′-PuPyPy-5′ primase recognition sequences (28) at the 5′-end of the putative RNA primer, which corresponds to the 3′-end of our leading-strand sequence. We were able to identify a 3′-PuPyPy-5′ motif at the expected position in all but one OP site (Figure 7, boxed). Thus, the sequence context of lagging-strand mutation clusters is also consistent with their interpretation as OP sites.
DISCUSSION
DNA polymerase I is one of five known polymerases expressed by E. coli (29). Even though pol I was the first polymerase to be discovered, some questions regarding its function in vivo remain. Here, we address some of these questions by using the footprint of pol I–generated mutations in neutral sequence to define pol I replication templates.
We increased the mutation frequency of pol I by genetically altering the fidelity of this polymerase. Low fidelity pol I mutations may admittedly have pleiotropic effects such as altering the processivity of the polymerase, its efficiency to exchange with other polymerases, or its nick-translation activity. However, the dramatic increase in mutation frequency produced by our low-fidelity polymerase was critical for our experimental approach for two reasons:
Minimal background from other mutation sources: the mutation rate of the muta-plasmid system is so far above that of spontaneous mutagenesis that it virtually guarantees that all mutations sequenced are produced by pol I. There is no question about the source of mutations in our system because the frequency of ColE1 plasmid mutation in vivo correlates directly with the fidelity of individual error-prone pol I alleles expressed (19).
Direct sequencing: our elevated mutation frequency allowed efficient data collection by direct sequencing, bypassing the need for reporters. The absence of any significant functional selection allowed the generation of an accurate spectrum of mutations in vivo, and (even more importantly) of accurate data on the physical distribution of mutations along the plasmid sequence. This level of resolution was essential for the identification of a footprint for lagging-strand processing by pol I, which as it turned out is restricted to only ∼5% of the sequence.
Our first observation was a decline in pol I mutation frequency with increasing distance from RNA/DNA switch (Figure 2). We have recently reported a very similar decrease in mutation frequency with increasing distance from ColE1 ori using a streamlined muta-plasmid mutagenesis protocol (21). This mutation frequency profile is consistent with a switch to pol III replication, as pol III is a high-fidelity polymerase and therefore not expected to leave a detectable mutation footprint in our system. However, this ‘switch’ is not as sharp as its name suggests; instead it occurs gradually over at least 1.3 kb. This gradual transition to pol III replication may be a default mechanism in the absence of the ssiB primosome assembly site for the leading strand (25,26); alternatively, in ColE1 plasmids the switch between pol I and pol III polymerases may be far more gradual than previously thought.
We observed an asymmetric distribution of complementary point mutations that is more pronounced in areas that are proximal to the DNA/RNA switch (d < 800) (Table 2). This pronounced asymmetry agrees with pol I’s known role of mediating initiation of leading-strand synthesis because if there was no strand preference, we would expect the frequency of every pair of complementary mutations to be symmetrical (Figure 3b, Scenario 5) (30).
Given that we sequenced the leader strand, we can assume that the mutation spectrum in proximal areas of the plasmid (shown in Figure 4a) approximates the error rate of the polymerase in vivo, after proofreading and mismatch repair (Figure 3a, Scenario 1). We have been unable to determine the mutation spectrum of our error-prone polymerase in vitro, but the in vivo data, we present here is consistent with what would be expected for an 3′ → 5′ exonuclease-deficient pol I, with a predominance of transitions (83%) (31), a low frequency of frameshift errors (1.3% if we include hotspots) (31), and the lowest frequency of mutation corresponding to mismatches involving pyrimidine opposite pyrimidine (5.8% between all four mutations) (32,33).
The contribution of mismatch repair, which preferentially resolves frameshifts and transitions (34) to the error rates of pol I in vivo, is unclear. The polA12 strain we used as a host is mismatch repair-proficient. Widespread mutagenesis can saturate mismatch repair (35). In our case, mutagenesis is largely targeted to ColE1 plasmid sequences (15) and is therefore less likely to saturate the mismatch repair capacity of the cell than non-targeted in vivo mutagenesis. On the other hand, our muta-plasmid mutagenesis protocol involves prolonged culture under saturation conditions, which are known to deplete mismatch repair function through induction of the stress/starvation response (36). The high representation of transitions, particularly of AT → GC (the preferred substrate for mismatch repair) (34), and the striking predominance of transitions in mutation hotspots (86%; 16% A → G) (Figure 4b) suggests that our system may exhibit decreased mismatch repair capacity (34).
The asymmetry between complementary mutations mentioned above can be interpreted as a difference in the error rate of the polymerase for the two complementary mutations in vivo (Figure 3a, Scenario 1). We exploited these differences to establish strand preference, as a switch in template strand should produce an inversion in the frequency bias between ‘least frequent’ and ‘most frequent’ mutations of the pair (Figure 3a, Scenario 2).
At d = 1100–1350 we detected the same bias as in proximal mutations, with 26 C → T mutations, compared to 11 G → A, (P < 0.033) (Table 2). This indicates that pol I leading-strand synthesis continues for long distances (up to 1350 bp) in a significant fraction of the plasmids. This observation contrasts with in vitro studies, where a size of only between 100 and 300 bp was reported for the pol I extension product (11,12). It agrees, however, with another study of ColE1 plasmid replication in vivo showing that inhibition of primosome assembly through a dnaT mutation or by treatment with anti-dnaT antibodies results in 0.5–1 kb-long early replication intermediates (13). This suggests that ori RNA primer extension by pol I may be longer in vivo than in vitro, possibly through recruitment of processivity factors such as the β-clamp, which stimulates pol I processivity in vitro (37).
By contrast, at far distal (d > 3.5 kb) positions we see an inversion of the asymmetry, with more G → A mutations (n = 7) than C → T mutations (n = 4) (Table 2; nine and five mutations, respectively, if GFP library data is included). This inversion suggests that leading-strand synthesis in this area is negligible (Figure 3a, Scenario 2). This observation agrees with reports showing that pol III is essential for completion of ColE1 plasmid replication (38) and supports our use of marker mutations to identify strand preferences in replication.
Next, we looked for a mutation footprint that may correspond to Okazaki primer processing. We reasoned that mutations in proximal areas that show a negative bias in frequency compared to their complementary ones (Figure 4a) should be enriched in areas of lagging-strand synthesis (Figure 3). We confirmed this approach showing that these marker mutations are enriched at distal positions (Figure 4c). Combining frequency bias (‘least frequent’) and distal enrichment information we designated the following mutations as markers for lagging-strand synthesis: T → C, G → A, T → A, T → G, C → A and G → C, although the numbers were low for most transversions. In addition to an enrichment for marker lagging-strand mutations, we hypothesized that these mutations should be close to each other, as Okazaki primers are small (11 ± 1 nt) (39). Combining spectrum and distance criteria we identified seven putative OP sites, three in the hTK library, and four in the GFP library (Figures 5 and 6).
The distribution and spectrum of mutations at these sites argues strongly against clustering due to a local increase in polymerase error associated with sequence context (hotspot effects). We can distinguish four lines of evidence supporting our proposition that instead lagging-strand mutation clusters represent OP sites.
Inverted bias: marker lagging-strand mutations are overrepresented and their complementary mutations underrepresented at these sites (Figure 6). Strikingly, the enrichment for marker lagging-strand mutations was not limited to one or two types; instead we found that all six types of marker point mutations were enriched, between 2.3- and 10.6-fold (Table 3). The most parsimonious explanation in this comprehensive shift in mutation pattern is a template switch.
No evidence for local increase in error rate: a local increase in polymerase error rate typically results in multiple hits in one or a few (two to three) adjacent positions, such as we saw in the hTK hotspots (Supplementary Figure S2) or as previously reported for the lacI reporter gene (34). In contrast, at our putative OP sites mutations are remarkably evenly distributed: 37 positions (30% of the total number of positions) show marker lagging-strand mutations, and only three of these show more than one mutation.
Right sequence context: the DnaG primase is known to recognize the 3′-PuPyPy-5′ motif for initiation of primer synthesis (28). Therefore the complementary sequence for this motif would be expected to be at the 3′ end of the OP (leading-strand) sequence (the 5′ end of the RNA primer). We found this motif at the expected location in all OP sites but one (highlighted in Figure 7).
Positional enrichment: whereas at OP sites the frequency of leading-strand replication by pol I will be <100% (as it is partially replaced by pol III) and will vary depending on distance from ori, the frequency of pol I lagging-strand replication is 100%, because all Okazaki primers need to be processed for successful replication. Therefore each iterative cycle of mutagenesis should increase the differential between frequencies of leading- versus lagging-strand replication. This prediction agrees with our experimental data: for comparable numbers of lagging-strand mutations (97 versus 80) we see a much higher number mutations per site in the hTK library relative to the pGFPuv library (8.3 mutations/site versus 4.2 mutations/site, respectively), correlating with the number of iteration cycles (5–7 versus 1–2, respectively).
The size of our clusters [12–22 nt (Figure 7)] is consistent with our interpretation of representing footprints for Okazaki primer processing. While we used proximity between lagging-strand mutations to identify these sites, we did not assume any particular size. The value we obtained is probably moderately under-estimated because the limits are defined by stochastic events (mutations), so our best guess is that OP sites are ∼20 nt in length. This size is significantly larger than the 11 ± 1 nt reported by Kitani et al. for Okazaki RNA primers (39). That previously reported work was done in a rnhA strain of E. coli and the primer may have been partially processed by pol I, which has 5′ → 3′ exonuclease activity. Other known prokaryote primases synthesize primers of between 17 and 30 bp in length (5), more in line with our ∼20 nt Figure 5. Alternatively, mutations located at positions >12 nt from the primer 5′ end may represent very limited nick-translation into DNA. Regardless of the origin of these additional nucleotides, our data indicate that Okazaki fragment processing by pol I is very limited, in agreement with a recent report estimating the contribution of pol I to chromosomal replication at 2% (40).
The specific location of the OP sites varies between the two libraries, with the exception of the site closest to the ssiA site at positions 352–373 (Figure 5), which is in an area of sequence shared between the two libraries. This suggests that primase recognition is sequence-context dependent. We found the 3′-PuPyPy-5′ motif for initiation of primer synthesis (28) where we expected it in six out of seven sites. The preferred primase recognition motif is (on the lagging strand) GTC (28,39,41). We detected this motif in one of our sites (TK-III). The most frequent motif we saw as 3′-PuPyPy-5′ was ATT (three times). We ignore whether ATT (TAA on the leading-strand sequence) represents a preferred primase motif in ColE1 plasmids or whether this is a serendipitous finding due to the small number of OP sites represented in our study.
In sum, based on at least five different criteria regarding mutation distribution and spectrum, positional enrichment, size and flanking sequence context we have identified a mutational footprint very likely corresponding to lagging-strand processing by pol I.
Given that the different modalities of pol I replication in the cell differ mostly at the initiation steps, our observations regarding the transition between pol I and pol III replication and Okazaki fragment processing likely apply to pol I genomic replication and DNA repair as well.
The size of Okazaki fragments that we inferred for ColE1 replication based on the spacing between RNA processing sites is, on average, 260 nt. This size is remarkably short compared to the 1000–2000 bp range described for oriC (chromosomal) replicons (42). The primosome assembled for initiation of ColE1 plasmid replication is essentially identical to the PriA-dependent replisome recruited to R- or D-loops during DNA repair (6,16). Gaps that form during lagging-strand synthesis are known to play a major role in processing replication blocks by facilitating strand-switch and replication fork reversal (43–45). If confirmed, the presence of short Okazaki fragments during PriA-dependent replication could assist in the processing of replication blocks.
Mutational footprinting has allowed us to follow the switch from pol I to pol III replication during leading-strand synthesis and even RNA processing by pol I on the lagging strand, which is restricted to very short sequences. Deep-sequencing and other improvements in sequencing technologies should enable the use of this genetic strategy to map templates for other prokaryotic or eukaryotic polymerases in vivo so long as a mutation signature can be defined (46) and/or the mutation frequency of the polymerase can be sufficiently elevated [(46) and this study]. Finally, our mutational footprinting approach can also be used more broadly to study processing of specific lesions by individual polymerases in vivo and to investigate how polymerase activity may be affected by sequence topology or by interactions with protein partners such as DNA repair or processivity factors.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The National Institute of Health K08 award (CA116429-04 to M.C.); National Cancer Institute R01 awards (CA102029 and CA115802 to L.L.); Ellison Medical Foundation (AG-NS-0577-09 to J.B.); National Science Foundation (DBI-0845275 to R.K.). Funding for open access charge: NIH-K08 Award.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENT
The authors would like to thank Dr Abel Rodríguez (UCSC) for help during the initial stages of this manuscript.
REFERENCES
- 1.Camps M. Modulation of ColE1-like plasmid replication for recombinant gene expression. Rec. Pat. DNA & Gene Seq. 2010;4:58–73. doi: 10.2174/187221510790410822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.del Solar G, Giraldo R, Ruiz-Echevarria MJ, Espinosa M, Diaz-Orejas R. Replication and control of circular bacterial plasmids. Microbiol. Mol. Biol. Rev. 1998;62:434–464. doi: 10.1128/mmbr.62.2.434-464.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cesareni G, Helmer-Citterich M, Castagnoli L. Control of ColE1 plasmid replication by antisense RNA. Trends Genet. 1991;7:230–235. doi: 10.1016/0168-9525(91)90370-6. [DOI] [PubMed] [Google Scholar]
- 4.Balbas P, Bolivar F. Back to basics: pBR322 and protein expression systems in E. coli. Methods Mol. Biol. 2004;267:77–90. doi: 10.1385/1-59259-774-2:077. [DOI] [PubMed] [Google Scholar]
- 5.Masai H, Nomura N, Kubota Y, Arai K. Roles of phi X174 type primosome- and G4 type primase-dependent primings in initiation of lagging and leading strand syntheses of DNA replication. J. Biol. Chem. 1990;265:15124–15133. [PubMed] [Google Scholar]
- 6.Masai H, Arai K. DnaA- and PriA-dependent primosomes: two distinct replication complexes for replication of Escherichia coli chromosome. Front Biosci. 1996;1:d48–d58. doi: 10.2741/a115. [DOI] [PubMed] [Google Scholar]
- 7.Sakakibara Y. Discontinuous replication of colicin E1 plasmid DNA in a cell extract containing thermolabile DNA ligase. J. Mol. Biol. 1978;124:373–389. doi: 10.1016/0022-2836(78)90305-4. [DOI] [PubMed] [Google Scholar]
- 8.Wang TC. Discontinuous or semi-discontinuous DNA replication in Escherichia coli? Bioessays. 2005;27:633–636. doi: 10.1002/bies.20233. [DOI] [PubMed] [Google Scholar]
- 9.Kingsbury DT, Helinski DR. Temperature-sensitive mutants for the replication of plasmids in Escherichia coli: requirement for deoxyribonucleic acid polymerase I in the replication of the plasmid ColE 1. J. Bacteriol. 1973;114:1116–1124. doi: 10.1128/jb.114.3.1116-1124.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tacon W, Sherratt D. ColE plasmid replication in DNA polymerase I-deficient strains of Escherichia coli. Mol. Gen. Genet. 1976;147:331–335. doi: 10.1007/BF00582885. [DOI] [PubMed] [Google Scholar]
- 11.Itoh T, Tomizawa J. Initiation of replication of plasmid ColE1 DNA by RNA polymerase, ribonuclease H, and DNA polymerase I. Cold Spring Harb. Symp. Quant. Biol. 1979;43(Pt 1):409–417. doi: 10.1101/sqb.1979.043.01.047. [DOI] [PubMed] [Google Scholar]
- 12.Itoh T, Tomizawa J. Formation of an RNA primer for initiation of replication of ColE1 DNA by ribonuclease H. Proc. Natl Acad. Sci. USA. 1980;77:2450–2454. doi: 10.1073/pnas.77.5.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Masai H, Arai K. Initiation of lagging-strand synthesis for pBR322 plasmid DNA replication in vitro is dependent on primosomal protein i encoded by dnaT. J. Biol. Chem. 1988;263:15016–15023. [PubMed] [Google Scholar]
- 14.Hartman CP, Rabussay D. E. coli DNA polymerase I: enzymatic functions and their application in polymer formation, nick translation and DNA sequencing. Gene Amplif. Anal. 1981;2:17–39. [PubMed] [Google Scholar]
- 15.Camps M, Naukkarinen J, Johnson BP, Loeb LA. Targeted gene evolution in Escherichia coli using a highly error-prone DNA polymerase I. Proc. Natl Acad. Sci. USA. 2003;100:9727–9732. doi: 10.1073/pnas.1333928100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Camps M, Loeb LA. Critical role of R-loops in processing replication blocks. Front Biosci. 2005;10:689–698. doi: 10.2741/1564. [DOI] [PubMed] [Google Scholar]
- 17.Uyemura D, Lehman IR. Biochemical characterization of mutant forms of DNA polymerase I from Escherichia coli. I. The polA12 mutation. J. Biol. Chem. 1976;251:4078–4084. [PubMed] [Google Scholar]
- 18.Fijalkowska I, Jonczyk P, Ciesla Z. Conditional lethality of the recA441 and recA730 mutants of Escherichia coli deficient in DNA polymerase I. Mutat. Res. 1989;217:117–122. doi: 10.1016/0921-8777(89)90063-3. [DOI] [PubMed] [Google Scholar]
- 19.Shinkai A, Loeb LA. In vivo mutagenesis by Escherichia coli DNA polymerase I. Ile(709) in motif A functions in base selection. J. Biol. Chem. 2001;276:46759–46764. doi: 10.1074/jbc.M104780200. [DOI] [PubMed] [Google Scholar]
- 20.Bielas JH, Schmitt MW, Icreverzi A, Ericson NG, Loeb LA. Molecularly evolved thymidylate synthase inhibits 5-fluorodeoxyuridine toxicity in human hematopoietic cells. Hum. Gene Ther. 2009;20:1703–1707. doi: 10.1089/hum.2009.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Troll CJ, Alexander DL, Allen JM, Marquette JT, Camps M. Mutagenesis and functional selection protocols for directed evolution of proteins in. E. coli. J. Visual. Exp. 2011;49 doi: 10.3791/2505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Predki PF, Elkin C, Kapur H, Jett J, Lucas S, Glavina T, Hawkins T. Rolling circle amplification for sequencing templates. Methods Mol. Biol. 2004;255:189–196. doi: 10.1385/1-59259-752-1:189. [DOI] [PubMed] [Google Scholar]
- 23.Campos PR, Wahl LM. The effects of population bottlenecks on clonal interference, and the adaptation effective population size. Evolution. 2009;63:950–958. doi: 10.1111/j.1558-5646.2008.00595.x. [DOI] [PubMed] [Google Scholar]
- 24.Selzer G, Som T, Itoh T, Tomizawa J. The origin of replication of plasmid p15A and comparative studies on the nucleotide sequences around the origin of related plasmids. Cell. 1983;32:119–129. doi: 10.1016/0092-8674(83)90502-0. [DOI] [PubMed] [Google Scholar]
- 25.Nomura N, Low RL, Ray DS. Identification of ColE1 DNA sequences that direct single strand-to-double strand conversion by a phi X174 type mechanism. Proc. Natl Acad. Sci. USA. 1982;79:3153–3157. doi: 10.1073/pnas.79.10.3153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zipursky SL, Marians KJ. Identification of two Escherichia coli factor Y effector sites near the origins of replication of the plasmids (ColE1 and pBR322. Proc. Natl Acad. Sci. USA. 1980;77:6521–6525. doi: 10.1073/pnas.77.11.6521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Maki H. Origins of spontaneous mutations: specificity and directionality of base-substitution, frameshift, and sequence-substitution mutageneses. Annu. Rev. Genet. 2002;36:279–303. doi: 10.1146/annurev.genet.36.042602.094806. [DOI] [PubMed] [Google Scholar]
- 28.Yoda K, Okazaki T. Specificity of recognition sequence for Escherichia coli primase. Mol. Gen. Genet. 1991;227:1–8. doi: 10.1007/BF00260698. [DOI] [PubMed] [Google Scholar]
- 29.Jarosz DF, Beuning PJ, Cohen SE, Walker GC. Y-family DNA polymerases in Escherichia coli. Trends Microbiol. 2007;15:70–77. doi: 10.1016/j.tim.2006.12.004. [DOI] [PubMed] [Google Scholar]
- 30.Albrecht-Buehler G. The spectra of point mutations in vertebrate genomes. Bioessays. 2009;31:98–106. doi: 10.1002/bies.080081. [DOI] [PubMed] [Google Scholar]
- 31.Bebenek K, Joyce CM, Fitzgerald MP, Kunkel TA. The fidelity of DNA synthesis catalyzed by derivatives of Escherichia coli DNA polymerase I. J. Biol. Chem. 1990;265:13878–13887. [PubMed] [Google Scholar]
- 32.Carroll SS, Cowart M, Benkovic SJ. A mutant of DNA polymerase I (Klenow fragment) with reduced fidelity. Biochemistry. 1991;30:804–813. doi: 10.1021/bi00217a034. [DOI] [PubMed] [Google Scholar]
- 33.Loh E, Choe J, Loeb LA. Highly tolerated amino acid substitutions increase the fidelity of Escherichia coli DNA polymerase I. J. Biol. Chem. 2007;282:12201–12209. doi: 10.1074/jbc.M611294200. [DOI] [PubMed] [Google Scholar]
- 34.Schaaper RM, Dunn RL. Spontaneous mutation in the Escherichia coli lacI gene. Genetics. 1991;129:317–326. doi: 10.1093/genetics/129.2.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schaaper RM, Radman M. The extreme mutator effect of Escherichia coli mutD5 results from saturation of mismatch repair by excessive DNA replication errors. EMBO J. 1989;8:3511–3516. doi: 10.1002/j.1460-2075.1989.tb08516.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Galhardo RS, Hastings PJ, Rosenberg SM. Mutation as a stress response and the regulation of evolvability. Crit. Rev. Biochem. Mol. Biol. 2007;42:399–435. doi: 10.1080/10409230701648502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Maul RW, Sanders LH, Lim JB, Benitez R, Sutton MD. Role of Escherichia coli DNA polymerase I in conferring viability upon the dnaN159 mutant strain. J. Bacteriol. 2007;189:4688–4695. doi: 10.1128/JB.00476-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Staudenbauer WL. Replication of small plasmids in extracts of Escherichia coli: requirement for both DNA polymerases I and II. Mol. Gen. Genet. 1976;149:151–158. doi: 10.1007/BF00332883. [DOI] [PubMed] [Google Scholar]
- 39.Kitani T, Yoda K, Ogawa T, Okazaki T. Evidence that discontinuous DNA replication in Escherichia coli is primed by approximately 10 to 12 residues of RNA starting with a purine. J. Mol. Biol. 1985;184:45–52. doi: 10.1016/0022-2836(85)90042-7. [DOI] [PubMed] [Google Scholar]
- 40.Makiela-Dzbenska K, Jaszczur M, Banach-Orlowska M, Jonczyk P, Schaaper RM, Fijalkowska IJ. Role of Escherichia coli DNA polymerase I in chromosomal DNA replication fidelity. Mol. Microbiol. 2009;74:1114–1127. doi: 10.1111/j.1365-2958.2009.06921.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Blattner FR, Plunkett G, 3rd, Bloch CA, Perna NT, Burland V, Riley M, Collado-Vides J, Glasner JD, Rode CK, Mayhew GF, et al. The complete genome sequence of Escherichia coli K-12. Science. 1997;277:1453–1462. doi: 10.1126/science.277.5331.1453. [DOI] [PubMed] [Google Scholar]
- 42.Okazaki R, Okazaki T, Sakabe K, Sugimoto K, Sugino A. Mechanism of DNA chain growth. I. Possible discontinuity and unusual secondary structure of newly synthesized chains. Proc. Natl Acad. Sci. USA. 1968;59:598–605. doi: 10.1073/pnas.59.2.598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.McGlynn P, Lloyd RG. Recombinational repair and restart of damaged replication forks. Nat. Rev. Mol. Cell Biol. 2002;3:859–870. doi: 10.1038/nrm951. [DOI] [PubMed] [Google Scholar]
- 44.Michel B, Grompone G, Flores MJ, Bidnenko V. Multiple pathways process stalled replication forks. Proc. Natl Acad. Sci. USA. 2004;101:12783–12788. doi: 10.1073/pnas.0401586101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kogoma T. Recombination by replication. Cell. 1996;85:625–627. doi: 10.1016/s0092-8674(00)81229-5. [DOI] [PubMed] [Google Scholar]
- 46.Pursell ZF, Isoz I, Lundstrom EB, Johansson E, Kunkel TA. Yeast DNA polymerase epsilon participates in leading-strand DNA replication. Science. 2007;317:127–130. doi: 10.1126/science.1144067. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.