Abstract
Some viral and cellular messages use an alternative mechanism to initiate protein synthesis that involves internal recruitment of the ribosome to an internal ribosome entry site (IRES). The Dicistroviridae intergenic regions (IGR) have been studied as model IRESs to understand the mechanism of IRES-mediated translation. In this study, the in vivo activity of IGR IRESs were compared. Our analysis demonstrates that Class I and II IGR IRESs have comparable translation efficiency in yeast and that Class II is significantly more active in mammalian cells. Furthermore, while Class II IGR IRES activity was enhanced in yeast grown at a higher temperature, temperature did not affect IGR IRES activity in mammalian cells. This suggests that Class II IRESs may not function optimally with yeast ribosomes. Examination of chimeric IGR IRESs, established that the IRES strength and temperature sensitivity are mediated by the ribosome binding domain. In addition, the sequence of the first translated codon is also an important determinant of IRES activity. Our findings provide us with a comprehensive overview of IGR IRES activities and allow us to begin to understand the differences between Classes I and II IGR IRESs.
INTRODUCTION
The vast majority of protein translation initiation in eukaryotes occurs by a cap-dependent mechanism, in which the 40S ribosomal subunit recognizes the m7G cap structure at the 5′-end of the mRNA then scans through the 5′-untranslated region (5′-UTR) until it reaches the start codon. This process requires over 12 eukaryotic initiation factors. During cell stress, e.g. viral infection, global cap-dependent translation is shutdown and translation can occur by a cap-independent mechanism (1). The 5′-UTR of some transcripts contain IRESs, which are highly structured RNA elements that mediate ribosome binding internally to the mRNA, thus bypassing the need for a 5′-cap and reducing the dependency on some or all of the canonical initiation factors. Despite extensive research since the discovery of IRESs over 20 years ago (2,3), the specific mechanism of IRES-mediated initiation remains largely unknown.
Dicistroviridae are a newly described virus family composed of at least 15 RNA viruses that infect invertebrates (Table 1) (4). The genomes of these viruses are characterized by two non-overlapping open reading frames (ORFs, Figure 1A). Translation initiation of the non-structural genes encoded by ORF1 is mediated by a 5′-IRES element similar to the picornaviral-like IRES family (5–8), while structural genes encoded by ORF2 are under the translational control of an IGR IRES. The IGR IRES is capable of binding directly to the 40S ribosomal subunit and recruiting the 60S subunit to generate an 80S complex in the absence of initiation factors, making it the most streamlined form of translation initiation known (9–11).
Table 1.
Dicistroviridae family
| Virus | Abbreviation | Accession No. | Host | IGR–IRESa | References |
|---|---|---|---|---|---|
| Cripavirus | Class I | ||||
| Aphid lethal paralysis virus | ALPV | AF536531 | Aphid | 6648–6822 | (49) |
| Black queen cell virus | BQCV | AF183905 | Honeybee | 5626–5836 | (50) |
| Cricket paralysis virusb | CrPV | AF218039 | Cricket | 6025–6216 | (5) |
| Drosophila C virus | DCV | AF014388 | Fruit fly | 6079–6266 | (51) |
| Himetobi P virusb | HiPV | AB017037 | Planthopper | 6296–6472 | (52) |
| Homalodisca coagulata virus-1b | HoCV-1 | NC008029 | Glass-winged sharpshooter | 5808–5987 | (30) |
| Plautia stali intestine virusb | PSIV | AB006531 | Brown-winged green bug | 6007–6192 | (53) |
| Pteromalus puparum small RNA-containing virusc | PpSRV | EU680971.1 | Wasp | Unknown | (54) |
| Rhopalosiphum padi virus | RhPV | AF022937 | Aphid | 6577–7019 | (55) |
| Triatoma virus | TrV | AF178440 | Kissing bug | 5937–6111 | (56) |
| Aparavirus | Class II | ||||
| Acute bee paralysis virusb | ABPV | JF299264 | Honeybee | 6326–6538 | This study |
| Israeli acute paralysis virusb | IAPV | NC009025 | Honeybee | 6399–6617d | (24) |
| Kashmir bee virusb | KBV | JF299265 | Honeybee | 6411–6629 | This study |
| Solenopsis invicta virus-1b | SINV-1 | AY634314 | Red imported fire ant | 4219–4422d | (57) |
| Taura syndrome virusb | TSV | AF277675 | Shrimp | 6741–6952d | (58) |
aNumbering corresponds to genomic position based on the reference sequence of the full-length genome.
bDenotes viruses used in this study.
cAssignment of PpSRV is based on phylogenetic analysis and secondary structure predictions (data not shown) (54).
dIn-frame AUG in IRES.
Figure 1.
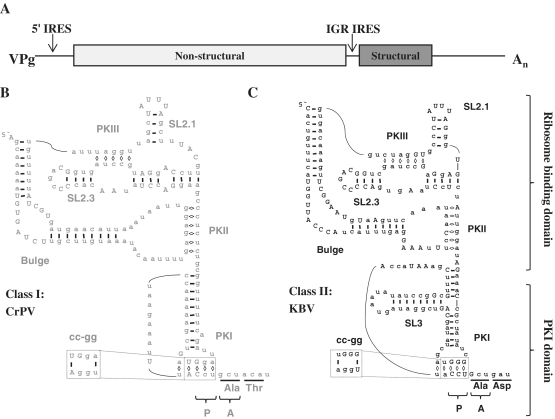
The two classes of IGR IRESs are structurally distinct. (A) Schematic representation of the CrPV genome drawn to scale. (B and C) Diagram of the secondary structure of the Class I IGR IRES from CrPV (B) and Class II IGR IRES from KBV (C) based on secondary structure predictions (22). Conserved bases are in uppercase. The dotted box indicates the location of the cc-gg mutation. The triplet bases positioned in the P- and A-sites of the ribosome are indicated as well as the amino acids encoded by the first two codons of ORF2. The ribosome binding domain and the PKI domain are indicated. SL = stem loop.
Structural studies on IGR IRESs have aided our understanding of how these IRESs recruit the ribosome. The complete crystal structure of the unbound IGR IRES was derived from the ribosome binding domain of the Plautia stali intestine virus (PSIV) IGR IRES and the pseudoknot 1 (PKI) domain of the cricket paralysis virus (CrPV) IGR IRES (12,13). In accordance with previous predictions based on structural probing analysis of multiple IGR IRESs (14–17), the crystal structures verified the presence of three pseudoknots that are necessary for proper folding of the IRES (Figure 1B and C) (14,15). To determine where the IGR IRES interacts with the ribosome, high-resolution cryo-EM reconstructions were determined for the CrPV IGR IRES bound to the 80S ribosome (18,19). These studies demonstrated that the IGR IRES occupies the intersubunit space and interacts primarily with the E-site of the ribosome. Biochemical and structural studies suggest that PKI mimics a tRNA:mRNA interaction in the P-site to set the correct reading frame (13,20). IGR IRESs also contain two stem loops, SL2.1 and SL2.3, which recruit the 40S ribosome (12) and a bulge region that is important for 80S complex formation (16,21) (Figure 1B and C).
Though IGR IRESs are predicted to share a highly conserved secondary structure (22), they are divided into two classes according to differences in two key structures, PKI and the bulge (Figure 1B and C). Class II IGR IRESs have an extra stem loop in the PKI domain (SL3) that may be essential for IRES activity (17,21,23). Additionally, the sequence of the bulge is conserved within each IGR IRES class and the bulge is larger in Class II. Mutations to the bulge region or swapping a Class I bulge region into a Class II IGR IRES background, or vice versa, reduced IRES-mediated initiation, suggesting that the sequence and/or size of the bulge is critical for IRES activity (16,21,24).
The ability of IGR IRESs to initiate translation has been extensively investigated, though most studies have focused on either the CrPV or PSIV IRESs. Additional IGR IRESs were examined either in isolation or in a limited comparative study (24,25). The majority of Dicistroviridae intergenic regions (IGRs) have not been evaluated functionally, and are proposed IRESs only based on bioinformatic analysis or structural probing (17,22). To verify that they are functional in vivo, we performed a broad comparative analysis of the IGR IRESs from 9 of the 15 known Dicistroviridae.
We found that all of the IGR IRESs tested were functional in yeast and mammalian cells. Notably, we observed that Class II IGR IRESs displayed high activity in vivo, in contrast to the previous assumption that they are less active than Class I (24). We recently determined that the CrPV IGR IRES activity was dependent on the ribosomal protein S25 (Rps25p). Here, we demonstrate that both Classes I and II IGR IRESs require RPS25. The vast majority of IGR IRESs initiate with an alanine in the A-site of the ribosome, here we show that other codons in this position result in lower IGR IRES activity demonstrating that the first translated codon also influences IRES activity. The Class II, but not Class I, IRESs have enhanced activity at elevated temperatures in yeast. Chimeric IRESs that contain the ribosome binding domain of a Class II IRES and the PKI domain of a Class I IRES retain this temperature sensitivity. Taken together, these results demonstrate that the Dicistroviridae family contains functional IGR IRESs and that differences in IRES activity between the two classes of IGR IRESs can be attributed to differences within the ribosome binding domain.
MATERIALS AND METHODS
Yeast strains and general culture techniques
Standard methods were used to grow and transform yeast (26). The Saccharomyces cerevisiae strains used in this study were: wild-type (BY4741: MATa his3Δ1 leu2Δ0 met15Δ0 ura3Δ0) from the Saccharomyces deletion project (27) and rps25aΔbΔ (SRT221: MATα his3Δ1 leu2Δ0 lys2Δ0 ura3Δ0 rps25a ::KanMX rps25b ::KanMX) (28).
Cell culture techniques
HeLa cells (Ambion) were maintained at 37°C, 5% CO2 in DMEM (high-glucose Dulbecco's modified Eagle's medium) supplemented with 10% (v/v) fetal bovine serum, 2 mM l-glutamine, and 0.1 mg/ml penicillin/streptomycin.
RNA isolation from bees
Honeybees from three different sites around Birmingham, Alabama, USA were collected and immediately frozen and stored at −80°C until use. RNA extraction was performed by homogenizing the bees in 1 ml of TRIzol (Invitrogen) per 100 mg tissue and incubating for 5 min at room temperature following the manufacturer's protocol for RNA extraction from tissue (Invitrogen). To generate cDNAs, the RNA was reverse transcribed using Oligo (dT)20 primers (Invitrogen) and the M-MLV reverse transcriptase (Promega) according to the manufacturer's instructions. The IAPV and ABPV IGR IRESs were amplified from the cDNA using specific primers sets (see Supplementary Table S1 for all primer information), P5 and P6, respectively. The primers were flanked with the XhoI and SacII restriction sites to facilitate cloning into the dual luciferase reporter plasmid (see following section). Amplification of the KBV IGR IRES required a two stage nested PCR using primer sets P8 followed by P7. A minimum of five isolates were analyzed by DNA sequencing (Heflin Genomics Core Facility, UAB).
Plasmid manipulations
A high copy dual luciferase reporter, pSRT338, was constructed by cloning the BamHI-SalI fragment from pDualLuc (9) into the BamHI-SalI sites of the pSal6 backbone (29). A SacII restriction site was engineered between the IGR IRES and the firefly luciferase gene by site-directed mutagenesis using the primer set P1 as described previously (9) to facilitate cloning of other IGR IRES sequences. The following IGR IRES sequences were generously provided to us: HoCV-1 from Wayne Hunter (30), PSIV (31) and HiPV (unpublished) from Nobuhiko Nakashima, and IAPV and TSV from Eric Jan (24). Viral sequences from bees collected in the Birmingham, Alabama region (a generous gift from Butch Otwell) were used to obtain the IGR IRESs of KBV and ABPV (GenBank accession numbers JF299265 and JF299264). Long oligo PCR was used to create the SINV-1 and chimeric dicistronic luciferase constructs essentially as described (32). Briefly, long oligonucleotides were designed using the assembly PCR oligomaker. A PCR reaction was used to assemble the long oligomers of DNA [one cycle at 94°C for 4 min, then 8 cycles of (94°C for 60 s, 54°C for 2 min, 72°C for 3 min) followed by a final single cycle at 72°C for 5 min]. A 2 µl aliquot of this reaction was added to the second stage PCR reaction containing 20-mer flanking primers with XhoI and SacII sites on their termini to facilitate cloning into pSRT338 [that was denatured for 94°C for 5 min, then 24 cycles of (94°C for 30 s, 54°C for 2 min, 72°C for 90 s), followed by a final extension cycle at 72°C 5 min]. The CrPV-KBV chimeric IRES is a fusion of the CrPV ribosome binding domain (6027–6171 nt) and KBV PKI domain (6560–6645 nt). The KBV-CrPV chimera consists of KBV RNA binding domain (6408–6559 nt) and CrPV PKI (6172–6232 nt).
Each IGR IRES was amplified with sequence specific primers flanked with XhoI and SacII restriction sites and inserted into the p2.1 TOPO vector (Invitrogen). The XhoI to SacII fragment from the TOPO plasmid was cloned into the XhoI and SacII sites of pSRT338, thus replacing the CrPV IGR IRES with another IGR IRES. Then, the SacII restriction site between the IRES and the firefly luciferase gene was removed by site directed mutagenesis (primer sets P11–P19) because the restriction site decreased the luciferase signal. All clones were confirmed by DNA sequencing using a firefly luciferase antisense primer (P24).
The mammalian expression vector was described previously for the CrPV IGR IRES (28). For the other IGR IRESs, the NheI-BglII fragment of the yeast dual luciferase reporter plasmids, containing the Renilla luciferase, IGR IRES and firefly luciferase was inserted into the NheI-BamHI sites of the mammalian expression vector pΔEMCV_BamHI (pSRT222).
Site-directed mutagenesis was carried out for the PKI mutational analysis (primer sets P25–P46) and to create the IAPVggc, IAPVgcu, KBVggc and IAPV6486aa IRES constructs (primer sets P20–P23).
DNA transfection
DNA transfections were performed using Lipofectamine 2000 (Invitrogen) according to the manufacturer's protocol with slight adjustments. Briefly, 5 × 104 HeLa cells were plated in antibiotic free DMEM one day prior to transfection in a 24-well plate to achieve 80–90% confluency on the day of transfection. Complexes were formed in Opti-MEM (Gibco) with 1 μl of Lipofectamine 2000 and 0.4 μg of plasmid DNA. Complexes were incubated with the cells for 4–6 h at 37°C, then the media was replaced and the cells were incubated at the indicated temperature for 24 h.
Luciferase assays
Luciferase assays from yeast were performed as described previously (9,28). Briefly, yeast strains were transformed with the indicated dual luciferase reporter plasmid and grown in selective SD media at 30°C until mid-log phase (0.5–0.65 OD600). One OD600 unit of cells was pelleted and lysed with 100 μl of 1× passive lysis buffer (PLB; Promega). Luminescence was measured for 2 μl of the lysate using the Dual Luciferase assay kit (Promega) following the manufacturer's protocol using a FB12 Luminometer (Berthold). Each assay was performed in triplicate from independent cultures. IRES activity is presented as a ratio of firefly/Renilla normalized to the wild-type strain or to an IGR IRES (set to 100%) as indicated.
For examination of IRES activity at different incubation temperatures, yeast were transferred from an overnight culture grown at 30°C to a new culture and grown at the indicated temperatures 25, 30 or 37°C for ∼16 h until they reached mid-exponential growth stage (0.5–0.65 OD600) and then luciferase activity was measured as described above.
Luciferase assays from HeLa cell transfections were conducted 24 h post-transfection. Cells from a 24-well plate were washed with phosphate buffered saline (137 mM NaCl, 2.7 mM KCl, 10 mM sodium phosphate dibasic, 2 mM potassium phosphate, pH 7.4) and lysed with 100 μl of 1× PLB for 15 min at room temperature. Lysates were cleared by centrifugation at 16 200g for 2 min. Luminescence was measured for 4 µl of cleared lysate.
RESULTS
The Dicistroviridae family is divided into two clades, the Cripaviruses and Aparaviruses, which correlate with Classes I or II IGR IRESs respectively. Phylogenetic analyses based on the whole genome or the IGR predict the same phylogenetic relationships between the viruses (4,33). In contrast to the majority of the IGR IRES nucleotides, the sequence identity of the single stranded regions in the bulge, SL2.1, and SL2.3 is highly conserved. We chose to study members with sequence variations in these highly conserved regions, since they are likely to affect IRES activity. Additionally, since little is known about the activity of the Class II IGR IRESs, every member of the Class II IGR IRES was included. In total, this study examines 9 of the 15 IGR IRESs (See ‘b’ in Table 1).
The Classes I and II IRESs are functional in yeast
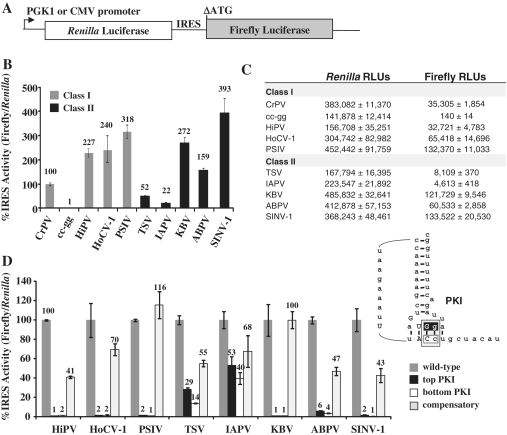
To examine IGR IRES activity in vivo, each IRES was cloned into a dual luciferase reporter and assayed for IRES activity in yeast (Figure 2A). The IRES activity was quantified as a function of firefly luciferase expression normalized to cap-dependent translation of Renilla luciferase and was expressed as a percentage of CrPV IGR IRES activity (Figure 2B, raw luciferase values Figure 2C). The Class I Himetobi P virus (HiPV), Homalodisca coagulata virus-1 (HoCV-1) and PSIV IGR IRESs demonstrated higher IRES activity than the CrPV IGR IRES (Figure 2B, gray bars).
Figure 2.
The family of IGR IRESs is functional in yeast. (A) A diagram of the DNA dual luciferase reporter. Transcription of the bicisictronic reporter is controlled by the PGK1 promoter in yeast and by the CMV promoter in HeLa cells. The Renilla luciferase activity measures the level of cap-dependent translation, while the firefly luciferase is under the control of the IGR IRES located in the intercistronic region and therefore measures the IRES activity. The first start codon in the firefly luciferase gene has been deleted (ΔATG) to decrease background translation (9,28). (B) Activity of Class I (gray) and Class II (black) IGR IRESs. IRES activity was normalized to cap-dependent translation and presented relative to the CrPV IGR IRES activity, which was set to 100%. (C) A table of the raw luciferase values from (B). (D) PKI mutational analysis for IGR IRESs. Mutations that disrupt PKI change the sequence of either the top (black) or bottom (white) of the pseudoknot. The compensatory mutant restores base pairing in the pseudoknot (light gray). See inset for a diagram of the mutations in PKI. The IRES activity of each mutant was normalized to the cap-dependent translation and presented as a percentage of the wild-type IGR IRES activity (dark gray). Standard error (SE) for n = 3 is shown.
The Class II Taura syndrome virus (TSV) IGR IRES had approximately half the activity of the CrPV IGR IRES and the IAPV IGR IRES had ∼20% of CrPV IGR IRES activity (Figure 2B, black bars). However, all other Class II IGR IRESs tested, specifically, the acute bee paralysis virus (ABPV), Kashmir bee virus (KBV) and Solenopsis invicta virus-1 (SINV-1), were more active than the CrPV IGR IRES and demonstrated a similar level of activity to the other Class I IGR IRESs (Figure 2B, black bars). This contradicts previous assumptions based only on the activity of TSV and IAPV, that Class I IRESs are inherently more active than Class II (24,34).
Integrity of PKI is necessary for IGR IRES activity
If a cryptic promoter element is present in the IRES or Renilla luciferase gene, this can produce a monocistronic firefly luciferase message that will register as a false positive for IRES activity. Therefore, it was important to verify for each IGR IRES construct that the firefly luciferase expression was derived only from a functional IRES. A 2-nt mutation (CC to gg) that disrupts PKI in the CrPV IGR IRES (cc-gg, dashed box in Figure 1B), results in a complete loss in IRES activity, and demonstrates that the firefly luciferase activity is solely dependent upon an active IGR IRES for translation (Figure 2B) (9,20,35). A similar PKI mutational analysis was performed on each IGR IRES (schematic for the mutations is presented in the inset of Figure 2D) and the activity of the mutant IRESs were assayed in yeast. The activity in the PKI mutations for TSV ranged from 14 to 29% and for IAPV ranged from 40 to 53% suggesting that cryptic promoter or splicing activity is responsible for about one third to one half of the reporter activity, respectively. For all of the other IGR IRESs tested, disruption of PKI resulted in a loss of firefly activity (ranging from 1 to 6%) indicating that the firefly luciferase activity is dependent upon a functional IGR IRES (Figure 2D, white and black bars). Additionally, when the compensatory mutations were made to restore base pairing in PKI, a partial to complete rescue of IRES activity was observed (Figure 2D, light gray bars). Incomplete restoration is consistent with previous analysis of the CrPV IGR IRES and suggests that although formation of the PKI structure is sufficient to partially restore IRES activity, the specific sequences in PKI are necessary for full IRES activity (20).
Class I and II IGR IRES activity is dependent on Rps25
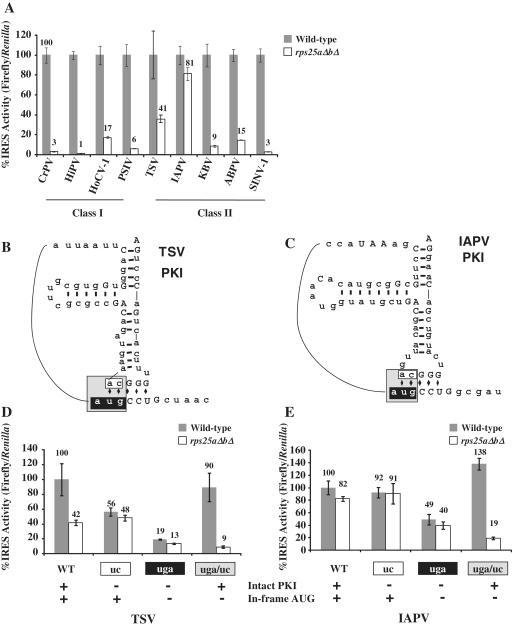
We have shown previously that Rps25p is required for CrPV IGR IRES-mediated but not cap-dependent translation (28). In order to determine if other IGR IRES members shared this requirement, IRES activity was measured in the yeast strain containing genomic deletions of RPS25 (rps25aΔbΔ). Consistent with the CrPV IGR IRES, all of the IGR IRESs required Rps25p for efficient translation initiation, with the exception of IAPV (Figure 3A). The high activity in the rps25aΔbΔ deletion yeast for IAPV is consistent with the PKI disruption studies that suggest that the reporter activity is not solely due to IRES-mediated translation.
Figure 3.
Both Classes I and II IGR IRESs are dependent on RPS25 in yeast. (A) IRES activity was assayed in the wild-type (gray bars) and rps25aΔbΔ (white bars) yeast strains and reported as a percentage of IRES activity in the isogenic wild-type strain. (B and C) Diagrams of PKI from TSV and IAPV. The mutations are color coded as follows, black = AUG to uga; white = CA to uc; gray = AUG to uga and CA to uc. (D and E) Rps25p dependent activity is driven by PKI structure, not by in-frame AUG. Analysis of the PKI mutants of TSV and IAPV in the wild-type (gray bars) and rps25aΔbΔ yeast (white bars) strains recorded as a percentage of the IRES activity of the wild-type IRES. The color of the axis titles correspond to the sequences of the same color in (B and C). Error bars shown are SE for n = 3.
Since IGR IRESs do not initiate at the canonical AUG start codon, the start codon of the firefly reporter used in these studies was deleted (Figure 2A, see ΔATG) thereby eliminating background firefly activity if monocistronic cap-dependent firefly transcripts are generated. These transcripts must initiate at downstream AUGs in the firefly coding region and would not generate an active firefly luciferase since the N-terminal region is required for firefly luciferase activity (9,36). In fact, a northern blot for firefly luciferase demonstrates that smaller firefly luciferase transcripts are produced in addition the full-length dicistronic transcript for the CrPV, IAPV and TSV reporters; however cryptic activity was only detectable for IAPV and TSV (Supplementary Figure S1). This may be because TSV and IAPV have in-frame start codons within their IGR IRESs, which could allow for translation of a full-length firefly luciferase protein from a cap-dependent monocistronic message. Since we have shown previously that cap-dependent translation is unaffected by deletion of RPS25 (28), these results suggest that cap-dependent translation initiation at the start codon within the TSV or IAPV IGR IRES is responsible for their elevated firefly luciferase activity in the rps25aΔbΔ yeast strain (Figures 2D and 3A).
To determine if this upstream start codon is responsible for the residual activity in the rps25aΔbΔ deletion yeast, IRESs with the start codon mutated were analyzed (Figure 3B–E). The two TSV reporters that retain the upstream AUG, wild-type TSV and TSVuc, had over 40% residual firefly luciferase activity (Figure 3D). However, mutants without the upstream start codon, TSVuga and TSVuga/uc, have only background levels (<19% IRES activity, 860 RLUs) of firefly luciferase activity (Figure 3D, compare TSVuga/uc in wild-type and rps25ΔaΔb strains). These results suggest that the removal of in-frame AUGs within the IGR IRES abolishes all firefly reporter activity that is not IRES-dependent. Similarly, when PKI was disrupted in the IAPV IGR IRES high levels of firefly activity are observed consistent with the level of activity of the wild-type IRES in the rps25ΔaΔb strain. This is suggestive of cryptic promoter activity. Indeed, mutation of an upstream inframe start codon significantly reduced firefly luciferase activity. Similar to the TSV mutations, when the AUG mutations are present in IAPV in combination with compensatory mutations that restore the PKI basepairing (IAPVuga/uc) IRES activity is restored and it is RPS25-dependent (Figure 3E) like the other IGR IRESs. Taken, together these results suggest that there is cryptic activity originating from an in-frame start codon, and that the TSV and IAPV IRESs are still active after removal of this codon, along with appropriate compensatory mutations to maintain PKI basepair interactions.
Class II IGR IRESs are more active in HeLa cells
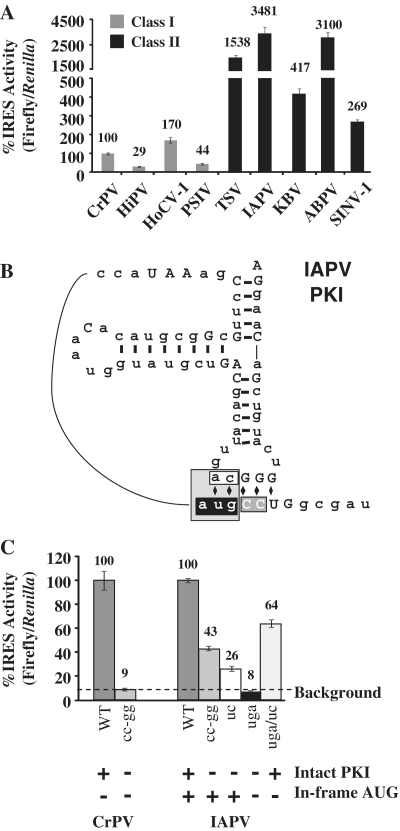
To determine if the IGR IRES activities observed in yeast are conserved in mammalian cells, the IGR IRESs were cloned into a mammalian dual luciferase reporter with a CMV promoter (Figure 2A) and the IRES activities were measured following transient transfection into HeLa cells. Among the Class I IGR IRESs, HoCV-1 was 170% as active as the CrPV IGR IRES, while HiPV and PSIV both exhibited <50% of CrPV activity (Figure 4A, gray bars). In contrast, all of the Class II IGR IRESs were highly active in HeLa cells ranging from 270 to 3500% of the CrPV IGR IRES activity (Figure 4A, black bars). The most striking change in activity was for IAPV, which is the least active in yeast (Figure 2B), but the most active in HeLa cells (Figure 4A).
Figure 4.
The Class II IGR IRESs are more active in HeLa cells. (A) IRES activities in HeLa cells are shown for Class I (gray) and Class II (black) IGR IRESs. IGR IRES activity was normalized to cap-dependent translation and presented as a percentage of the CrPV IGR IRES activity. Error bars are SE for n = 5. (B) A diagram of the IAPV PKI mutations tested. The shading on the diagram indicates the four mutants and corresponds to the bar graph in (C): gray = cc-gg, black = AUG to uga, white = CA to uc, and light gray = AUG to uga and CA to uc. (C) The IAPV IGR IRES is still highly active in HeLa cells after removing the in-frame AUG. IRES activity was normalized to cap-dependent translation and presented as a percentage of wild-type activity for each IRES. The dotted line on the bar graph indicates the background level of firefly luciferase activity (1300 RLUs) determined by the CrPV cc-gg IGR IRES mutant. Under each construct, plus and minus signs specify whether the PKI is intact and whether there is an in-frame start codon (AUG). Error bars shown are SE for n = 3.
Since the IAPV IGR IRES reporter exhibited a high level of cryptic activity in yeast (Figures 2D and 3E), the contribution of cryptic activity for the IAPV IGR IRES reporter was examined in HeLa cells by measuring the activity of ΔAUG and compensatory PKI mutations (Figure 4B). The IAPVuga mutant (black boxed nucleotides, Figure 4B) replaced the start codon with a stop codon and disrupted PKI. It has 8% activity in HeLa cells (black bar, Figure 4C), which is comparable to the 9% background level seen with the CrPVcc-gg mutant. The IAPV mutants with a disrupted PKI and an intact AUG, IAPVcc-gg and IAPVuc, had significant background activity, likely due to the in-frame AUG (Figure 4B and C, medium gray and white bars).
In order to determine if the IAPV IGR IRES can mediate PKI-dependent translation in HeLa cells without the in-frame start codon, a compensatory mutant that restores base-pairing in PKI was tested. The IAPVuga/uc compensatory mutant has 64% of firefly luciferase activity compared to the wild-type IAPV IRES (Figure 4C, light gray bar). This activity is due to the restoration of the PKI structure and constitutes true IRES activity since the IRES lacks the in-frame start codon. Taken together, these results demonstrate that the IAPV IGR IRES is highly active in mammalian cells.
IGR IRES activity is dependent on the first codon of ORF2
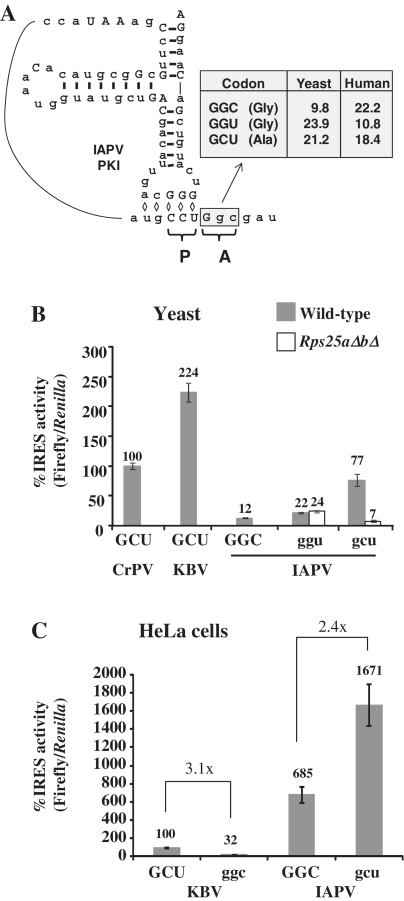
In the Dicistroviridae family, the bee viruses IAPV, KBV and ABPV have the most similar IGR IRES sequences; therefore, it was surprising that IAPV was the least active IGR IRES in yeast while ABPV and KBV exhibited high activity (Figure 2B). A closer examination of the sequences from these three IGR IRESs revealed two regions that may factor into the reduced activity of the IAPV IGR IRES in yeast: the first translated codon in the A-site of the ribosome and the PKIII sequence (Figure 5A, Supplementary Figure S2A). Since small changes in or adjacent to the IRES can lead to large effects on activity, we examined the collection of IAPV sequences in the NCBI database to find candidate IRESs that may exhibit more activity than the reference sequence. As of January 2011, NCBI had 166 entries for IAPV, 66 of which contained the IGR IRES sequence. Of the 66 IRES sequences, exactly half were the same as the reference sequence, which suggests that this sequence is replication competent in bees. The other 33 ranged from one to 19 deviations from the reference sequence. Thirty percent of these divergent IGR IRESs (10/33) encoded GGU (glycine) instead of GGC (glycine) as the first codon of ORF2. There were no other deviations within the first five codons of ORF2 or in the region surrounding PKIII in the database.
Figure 5.
The sequence of the first codon of ORF2 is important for IGR IRES activity. (A) Secondary structure of the IAPV IGR IRES based on the predicted structure (22). The first codon of ORF2 that is positioned in the A-site of the ribosome is indicated by a gray box. A table with the coding frequencies of glycine and alanine codons in yeast and humans is shown (39). (B) Mutant IAPV IGR IRESs with various A-site codons were assayed for activity in wild-type (gray) and rps25aΔbΔ (white) yeast strains and normalized to the CrPV IGR IRES activity. (C) IRES activity of mutant IRESs in HeLa cells normalized to the KBV IGR IRES activity. Error bars shown are SE for n = 3.
Previous reports have shown that the sequence encoding the first 5 amino acids can enhance IGR IRES-mediated translation (37,38). If the IAPV GGC codon is negatively affecting firefly luciferase production, it could do so by directly decreasing IGR IRES activity, or indirectly, by reducing the translation efficiency due to codon bias. This was a particular concern because codon usage of GGC in yeast is only 9.8/1000 making it a rare codon [Figure 5A, codon usage database (39)]. GGU is a preferred codon in yeast for glycine (Figure 5A) likely due to an increased number of hydrogen bonds formed during the codon:anticodon interaction (40). To address this, the naturally occurring GGU sequence, present in 10 out of 66 IAPV isolates, was assayed for translation activity in yeast. IAPVggu only slightly increased activity in wild-type yeast, but a concomitant increase was also seen in the rps25aΔbΔ strain indicating that this increase in activity is not IRES-dependent (Figure 5B). This finding suggests that codon usage bias is not the reason for low IAPV IGR IRES activity in yeast, because simply changing the rare GGC codon to the common GGU did not increase IRES activity. Consistent with this the GGC and GCU codons are used in a similar frequency in bees (39) thereby providing no selective advantage for one codon over the other.
Twelve of the 15 known IGR IRESs encode alanine as the first amino acid of ORF2, nine of which use a GCU triplet. The preference for GCU suggests it may be important for efficient IGR IRES activity; thus, the first codon after the IAPV IGR IRES was mutated to GCU to create IAPVgcu. The activity of this mutant in yeast was increased 10-fold over wild-type IAPV IGR IRES activity (Figure 5B). When IAPVgcu activity is measured in the rps25aΔbΔ yeast a dramatic reduction in firefly luciferase activity is observed suggesting that the activity was IRES-mediated (Figure 5B, see white bars). Since changing the GGC to a GCU codon dramatically increased IRES-mediated translation there is now an observable and significant difference in IRES activity in the wild-type yeast relative to the rps25ΔaΔb yeast strain. These data suggest that the IAPV IGR IRES functions better with a GCU codon as the first translated codon and may explain the apparent bias towards GCU as the first ORF2 codon in Dicistroviridae.
The effect of switching the first codon to GCU was tested in HeLa cells to determine whether it would influence IAPV IGR IRES activity in cells that already display high IRES activity. The IAPVgcu IGR IRES has ∼2.5-fold enhanced activity in HeLa cells (Figure 5C). GGC and GCU have comparable coding frequency in humans (Figure 5A), which suggests that the enhanced IRES activity mediated by GCU is not simply due to differential codon:anticodon interactions, but rather a direct influence on the activity of the IGR IRES. To further examine the importance of the sequence of the first codon of ORF2, the reverse mutational strategy was used to test if the replacement of the first codon with GGC would decrease the activity of an IRES that naturally contains GCU. KBVggc exhibited a 3-fold decrease in IRES activity compared to the wild-type (Figure 5C), which is similar to the fold change in IRES activity between the IAPVGGC and IAPVgcu IGR IRESs in HeLa cells.
An alternative explanation for differences in IAPV and KBV activities could be due to structural differences, which potentially extend PKIII and reduce the base-pairing in SL2.3 in the IAPV IGR IRES (Supplementary Figure S2A and Supplementary Data). However, mutations predicted to prevent extension of PKIII had no effect on IAPV IGR IRES activity (Supplementary Figure S2B). Taken together, these results suggest that the IGR IRESs function better with a GCU (alanine) codon in the A-site.
Class II IGR IRES activity is dependent on temperature
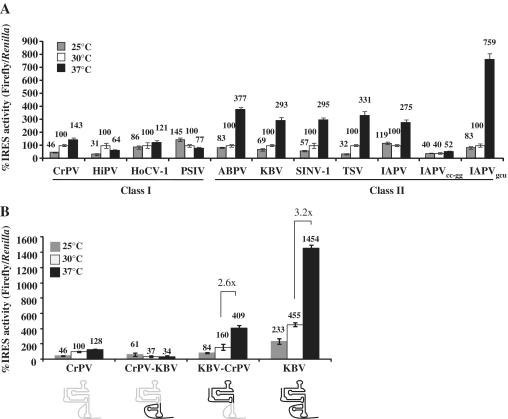
Since the IGR IRESs exhibited different IRES activities in yeast and human cells, this suggests that either the ribosomes differ in some significant manner or that the temperature that the cells are grown affects the IRES activity. Temperature changes are known to influence RNA folding and ribosome conformation (41), therefore the IGR IRES activity was examined in yeast grown at 25, 30 or 37°C. The Class I IGR IRESs activities do not correlate with changes in temperature (Figure 6A). In contrast, there is a 3-fold increase in activity for all of the Class II IGR IRESs at 37°C and the IAPVgcu IGR IRES mutant is eight times more active (Figure 6A). The increase in activity is not due to a change in global protein synthesis at higher temperatures because cap-dependent translation, as measured by Renilla luciferase values, remained constant for all of the reporters at each temperature (Supplementary Figure S3). Additionally, the increase in firefly luciferase activity is dependent on the presence of an active IRES upstream of the firefly luciferase gene. When the inactive IRES, IAPVcc-gg, reporter was assayed there was no change in the firefly luciferase values across the temperature gradient (Figure 6A). Interestingly, in HeLa cells there was no correlation between IGR IRES activity and temperature at 25, 30, 37 or 42°C (data not shown). Suggesting that in the context of the HeLa ribosomes, the IGR IRES functions efficiently.
Figure 6.
The ribosome binding domain imparts temperature sensitivity onto the Class II IGR IRESs in yeast. (A) Luciferase activity was measured for each of the IGR IRES reporters in yeast grown at 25, 30 and 37°C. The IRES activity was normalized to cap-dependent translation and shown as a percentage of the IGR IRES activity at 30°C for individual IRESs. Error bars represent SE for n = 3. (B) IRES activity of chimeric IGR IRESs was measured and normalized to cap-dependent translation and the activity was compared to the CrPV IGR IRES at 30°C. A diagram of the IRESs is shown below the graph. Domains in gray correspond to CrPV sequences, domains in black are from KBV. The fold enhancement of IRES activity between 30 and 37°C is indicated for the KBV and KBV-CrPV IGR IRESs. Error bars represent SE for n = 3.
IGR IRES activity is determined by the ribosome binding domain
Chimeric IGR IRESs were engineered to examine if the ribosome binding domain or the PKI domain has a more dominant role in determining IRES activity. The CrPV-KBV chimera is a fusion of the Class I ribosome binding domain from the CrPV IGR IRES to the Class II PKI domain from KBV. The KBV-CrPV chimera has the ribosome binding domain of KBV and PKI domain of CrPV. Each chimeric IRES is functional in yeast at 30°C (Figure 6B, white bars). The CrPV-KBV chimera had less activity than CrPV, while the KBV-CrPV chimera demonstrated intermediate activity between the CrPV and KBV. Consistent with previous findings, the ribosome binding domain plays the major role in determining the IGR IRES activity (21).
To determine which domain mediates temperature sensitivity, the activity of each chimeric IRES was tested in yeast at 25 and 37°C (Figure 6B). The CrPV-KBV chimera was unaffected by the temperature changes similar to the Class I IGR IRESs, while the KBV-CrPV chimeric IGR IRES activity increased 2.6-fold at 37°C similar to KBV. Taken, together these findings suggest that the temperature dependence of the IGR IRES in yeast resides in the ribosome binding domain.
DISCUSSION
Our data demonstrate that the Dicistroviridae IGR IRESs are active in both yeast and mammalian cells. Furthermore, the Class II IGR IRESs are as active, or more active, than the Class I. This contradicts the previous generalization based on studies of the IAPV and TSV IGR IRESs in vitro (21,24,34), which turn out to be atypical Class II IGR IRESs. Mutational analysis demonstrated that IGR IRES activity was influenced by the sequence of the first codon of ORF2 and may explain why the majority of IGR IRESs encode GCU (alanine) as the first translated codon. The Class II ribosome binding domain mediated enhanced IGR IRES activity in yeast grown at a higher temperature, however, temperature did not affect IGR IRES activity in mammalian cells. This suggests that Class II IRESs may not function optimally with yeast ribosomes and require a higher temperature to perform optimally.
The Dicistroviridae IGR IRESs are functional in vivo
In this study we used two mechanisms, disruption of PKI (Figure 2D) and deletion of RPS25 (Figure 3) to demonstrate that the HiPV, HoCV-1, ABPV, KBV and SINV-1 IGR IRESs have IRES activity. Loss of RPS25 or disruption of PKI results in a reduction in reporter expression to background levels (<2000 RLUs), demonstrating that all of the IGR IRESs are functional. Our analysis of PKI mutants, IRES activity in the rps25ΔaΔb deletion strain, mutation of upstream in frame AUGs with and without compensatory mutations and the northern analysis of transcripts generated from the dicistronic reporter in vivo are all consistent with the non-specific firefly luciferase activity observed for TSV and IAPV being due to translation of firefly luciferase from a monocistronic message using a cap-dependent mechanism. However, the IAPV and TSV IGRs were still shown to have functional IRESs, because they retain a high level of activity dependent on both an intact PKI domain and RPS25 when the start codons are mutated to stop codons (Figures 3D and E and 4C). Therefore, all 9 of the assayed IGR IRES, including IAPV and TSV, are functional in yeast and mammalian cells.
The role of the first codon of ORF2 in IGR IRES-mediated translation
Previous studies have shown that the first 6–11 nucleotides of the Dicistroviridae ORF2 enhance IGR IRES-mediated translation and that the single-stranded nature of the sequence following PKI functions as a barrier to allow the IGR IRES to fold properly (37,38). However our data takes this one step further, showing that IRES activity is affected by the specific sequence of the first codon that is translated in the A-site of the ribosome. The IAPV IGR IRES demonstrated a dramatic 10-fold increase in expression in yeast when the first codon was mutated from a GGC (glycine) to a GCU (alanine). The possibility that this difference in activity was due to codon bias was ruled out because exchanging the rare glycine codon, GGC, to the more abundant GGU glycine codon did not enhance IAPV IGR IRES activity since the marginal increase in expression of the reporter was not RPS25 dependent. Consistent with this finding, it has been shown that introduction of rare codons into the firefly luciferase protein in yeast even at the N-terminus, does not have a large effect on overall activity (42). Furthermore, GGU (glycine) and GCU (alanine) have very similar codon usage in yeast, but only IAPVgcu exhibits a dramatic increase in RPS25 dependent activity, demonstrating that the IAPV IGR IRES functions better with a GCU (alanine) codon in the A-site (Figure 5B).
In HeLa cells, the GGC glycine codon and the GCU alanine codon have similar codon usage but GCU increases IGR IRES activity. The reciprocal two-nucleotide change from GCU to GGC in the KBV IGR IRES decreased IRES activity to ∼30% of the wild-type level. Taken together these results demonstrate that IGR IRESs functions better with a GCU (alanine) codon as the first translated codon. However, it cannot be ruled out that the presence of an alanine tRNA in the A-site is responsible for the enhancement. Nonetheless, the majority of IGR IRESs are enriched for a GCU (alanine) codon as the first codon of ORF2 perhaps as a way to maximize expression of the viral structural proteins.
Differences between the Classes I and II IGR IRESs
In general, the Classes I and II IGR IRESs exhibit similar activity in yeast (Figure 2B). However, in mammalian cells, Class II are significantly more active (Figure 4A). This finding contradicts the previous assumption that the Class II IGR IRESs were less active than Class I based on accurate, though limited, studies on the atypical TSV and IAPV IGR IRESs (21,24,34) and underscores the importance of testing multiple IGR IRESs from each class in order to establish trends. Carter et al. (25) has performed a similar analysis of various IGR IRESs in insect and mammalian cells. However, they did not observe significant differences between Classes I and II IGR IRES activities. One explanation for this discrepancy could be that the large differences in activities that we observe are observable due to the fact that we have reduced the background expression (no IRES activity) to two orders of magnitude below the CrPV IGR IRES activity. Thus, allowing for a more sensitive measurement of differences in IRES activities.
The Class II IGR IRESs function more robustly at higher temperatures in yeast, which suggested that the temperature the cells are grown at could explain the enhanced activity observed in HeLa cells. However, the Class II IGR IRES activity did not correlate with temperature in mammalian cells (data not shown). Alternatively, the Class II IGR IRESs may function better with mammalian ribosomes and the temperature dependence in yeast may reflect differences in the IRES, or yeast ribosome, which can be overcome at higher temperatures where the ribosome has been shown to be more flexible (41).
The chimeric IGR IRES data demonstrated that the ribosome binding domain of the IGR IRES is the primary functional distinction that is responsible for the differences in IRES activity between the two classes. As SL2.1 and SL2.3 are absolutely conserved between KBV and CrPV, the most likely region responsible for differences in activity is the bulge region because it is the major class-specific difference in the ribosome binding domain. The bulge is larger in the Class II IGR IRESs (11 bases in the bottom loop and 7 in the top) compared to the Class I IGR IRESs (7 bases in the bottom loop and 5 in the top) (see Figure 1B and C). In addition, the bulge region is composed of different bases, which are conserved within, but not between, the two classes.
In order for a virus to be successful it must be able to replicate at the temperature of the insect host. Therefore, it is interesting to note that the majority of Class II IRESs are found in viruses that infect social insects (see Table 1) which live in hives maintained at high temperatures, between 32–36°C (43). The environment of the hive may have added a selective pressure to the virus to replicate at higher temperatures. In contrast, the majority of Class I IGR IRESs are found in viruses that infect non-social insects, which would maintain the temperature of their environment and consequently, they would have no selective pressure for viral replication at higher temperatures.
Revisiting the role of SL3
SL3 represents a striking structural difference between Classes I and II IGR IRESs, however little is known about how it contributes to differences in IGR IRES activity. It was proposed that SL3 is required for activity because mutations or deletions of SL3 led to a reduction in IRES activity (17,23,24). Efforts to establish the importance of SL3 are complicated by the fact that mutations may disrupt the global folding of PKI (14,15,20). The study of chimeric IGR IRESs has brought new insight into the role of SL3 in the Class II IGR IRESs. The TSV-CrPV chimeras (21), and the KBV-CrPV chimeras (Figure 6B), demonstrate that SL3 is not essential for IRES activity because when a Class I PKI is fused to the ribosome binding domain of a Class II IRES, it is active, thus demonstrating that the Class II ribosome binding domain does not require a PKI domain with SL3 in order to function. However, chimeric IRESs appear to be less fit than the wild-type IRESs i.e. KBV is more active than the KBV-CrPV chimera, suggesting that the two domains have functional interactions.
Ribosome recruitment by IGR IRESs
The current model for ribosome recruitment by IGR IRESs suggests that SL2.1 and SL2.3 are responsible for binding the 40S ribosome (15,18,44). Recent crystal structures and cryo-EM reconstructions of the eukaryotic ribosome have modeled Rps25p in the E-site of the ribosome in the region where the CrPV IGR IRES binds (45–47). This agrees with biochemical data that demonstrates that the PSIV IGR IRES cross-links to Rps25p (44). Our previous findings demonstrated that ribosomes lacking Rps25p were unable to bind to the CrPV IGR IRES, suggesting this is an essential interaction for ribosome binding (28). In chemical protection assays, both SL2.1 and 2.3 are protected after the addition of free Rps25p (48). Based on these data it appears that stem loops SL2.1 and/or SL2.3 interact with Rps25p (28). This model is further supported by the findings that Rps25p is required for both Classes I and II IGR IRESs (Figure 3A) because SL2.1 and SL2.3 are conserved between the two IGR IRES classes. While ribosome recruitment and Rps25p are essential for IRES activity, ribosome recruitment may not be the defining difference between Classes I and II IGR IRESs. Rather, differences in IGR IRES activity between the Classes I and II IRESs may be explained by a downstream step in initiation such as 80S complex formation (16,24) or pseudotranslocation (the movement of the ribosome by one codon in the absence of GTP hydrolysis following tRNA delivery to the A-site) (20).
ACCESSION NUMBERS
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (R01GM084547 and 3R01GM084547-01A1S1 to S.R.T.) Funding for open access charge: National Institutes of Health (R01GM084547).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Wayne Hunter, Nobuhiku Nakashima, Jeffrey Kieft and Eric Jan for plasmids containing IGR IRESs and thank Mr Butch Otwell for his generous gift of honeybees. We thank R. Curtis Hendrickson for critical reading of this article.
REFERENCES
- 1.Gebauer F, Hentze MW. Molecular mechanisms of translational control. Nat. Rev. Mol. Cell Biol. 2004;5:827–835. doi: 10.1038/nrm1488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jang SK, Krausslich HG, Nicklin MJ, Duke GM, Palmenberg AC, Wimmer E. A segment of the 5′ nontranslated region of encephalomyocarditis virus RNA directs internal entry of ribosomes during in vitro translation. J. Virol. 1988;62:2636–2643. doi: 10.1128/jvi.62.8.2636-2643.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pelletier J, Sonenberg N. Internal initiation of translation of eukaryotic mRNA directed by a sequence derived from poliovirus RNA. Nature. 1988;334:320–325. doi: 10.1038/334320a0. [DOI] [PubMed] [Google Scholar]
- 4.Bonning BC, Miller WA. Dicistroviruses. Ann. Rev. Entomol. 2010;55:129–150. doi: 10.1146/annurev-ento-112408-085457. [DOI] [PubMed] [Google Scholar]
- 5.Wilson JE, Powell MJ, Hoover SE, Sarnow P. Naturally occurring dicistronic cricket paralysis virus RNA is regulated by two internal ribosome entry sites. Mol. Cell Biol. 2000;20:4990–4999. doi: 10.1128/mcb.20.14.4990-4999.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Domier LL, McCoppin NK. In vivo activity of Rhopalosiphum padi virus internal ribosome entry sites. J. Gen. Virol. 2003;84:415–419. doi: 10.1099/vir.0.18781-0. [DOI] [PubMed] [Google Scholar]
- 7.Groppelli E, Belsham GJ, Roberts LO. Identification of minimal sequences of the Rhopalosiphum padi virus 5′ untranslated region required for internal initiation of protein synthesis in mammalian, plant and insect translation systems. J. Gen. Virol. 2007;88:1583–1588. doi: 10.1099/vir.0.82682-0. [DOI] [PubMed] [Google Scholar]
- 8.Shibuya N, Nakashima N. Characterization of the 5′ internal ribosome entry site of Plautia stali intestine virus. J. Gen. Virol. 2006;87:3679–3686. doi: 10.1099/vir.0.82193-0. [DOI] [PubMed] [Google Scholar]
- 9.Deniz N, Lenarcic EM, Landry DM, Thompson SR. Translation initiation factors are not required for Dicistroviridae IRES function in vivo. RNA. 2009;15:932–946. doi: 10.1261/rna.1315109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hertz MI, Thompson SR. Mechanism of translation initiation by Dicistroviridae IGR IRESs. Virology. 2011;411:355–361. doi: 10.1016/j.virol.2011.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pestova TV, Hellen CU. Translation elongation after assembly of ribosomes on the cricket paralysis virus internal ribosomal entry site without initiation factors or initiator tRNA. Genes Dev. 2003;17:181–186. doi: 10.1101/gad.1040803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Costantino D, Kieft JS. A preformed compact ribosome-binding domain in the cricket paralysis-like virus IRES RNAs. RNA. 2005;11:332–343. doi: 10.1261/rna.7184705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Costantino DA, Pfingsten JS, Rambo RP, Kieft JS. tRNA-mRNA mimicry drives translation initiation from a viral IRES. Nat. Struct. Mol. Biol. 2008;15:57–64. doi: 10.1038/nsmb1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jan E, Sarnow P. Factorless ribosome assembly on the internal ribosome entry site of cricket paralysis virus. J. Mol. Biol. 2002;324:889–902. doi: 10.1016/s0022-2836(02)01099-9. [DOI] [PubMed] [Google Scholar]
- 15.Nishiyama T, Yamamoto H, Shibuya N, Hatakeyama Y, Hachimori A, Uchiumi T, Nakashima N. Structural elements in the internal ribosome entry site of Plautia stali intestine virus responsible for binding with ribosomes. Nucleic Acids Res. 2003;31:2434–2442. doi: 10.1093/nar/gkg336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pfingsten JS, Castile AE, Kieft JS. Mechanistic role of structurally dynamic regions in Dicistroviridae IGR IRESs. J. Mol. Biol. 2010;395:205–217. doi: 10.1016/j.jmb.2009.10.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pfingsten JS, Costantino DA, Kieft JS. Conservation and diversity among the three-dimensional folds of the Dicistroviridae intergenic region IRESes. J. Mol. Biol. 2007;370:856–869. doi: 10.1016/j.jmb.2007.04.076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schuler M, Connell SR, Lescoute A, Giesebrecht J, Dabrowski M, Schroeer B, Mielke T, Penczek PA, Westhof E, Spahn CM. Structure of the ribosome-bound cricket paralysis virus IRES RNA. Nat. Struct. Mol. Biol. 2006;13:1092–1096. doi: 10.1038/nsmb1177. [DOI] [PubMed] [Google Scholar]
- 19.Spahn CM, Jan E, Mulder A, Grassucci RA, Sarnow P, Frank J. Cryo-EM visualization of a viral internal ribosome entry site bound to human ribosomes: the IRES functions as an RNA-based translation factor. Cell. 2004;118:465–475. doi: 10.1016/j.cell.2004.08.001. [DOI] [PubMed] [Google Scholar]
- 20.Wilson JE, Pestova TV, Hellen CU, Sarnow P. Initiation of protein synthesis from the A site of the ribosome. Cell. 2000;102:511–520. doi: 10.1016/s0092-8674(00)00055-6. [DOI] [PubMed] [Google Scholar]
- 21.Jang CJ, Jan E. Modular domains of the Dicistroviridae intergenic internal ribosome entry site. RNA. 2010;16:1182–1195. doi: 10.1261/rna.2044610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakashima N, Uchiumi T. Functional analysis of structural motifs in dicistroviruses. Virus Res. 2009;139:137–147. doi: 10.1016/j.virusres.2008.06.006. [DOI] [PubMed] [Google Scholar]
- 23.Hatakeyama Y, Shibuya N, Nishiyama T, Nakashima N. Structural variant of the intergenic internal ribosome entry site elements in dicistroviruses and computational search for their counterparts. RNA. 2004;10:779–786. doi: 10.1261/rna.5208104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Jang CJ, Lo MC, Jan E. Conserved element of the dicistrovirus IGR IRES that mimics an E-site tRNA/ribosome interaction mediates multiple functions. J. Mol. Biol. 2009;387:42–58. doi: 10.1016/j.jmb.2009.01.042. [DOI] [PubMed] [Google Scholar]
- 25.Carter JR, Fraser TS, Fraser MJ., Jr Examining the relative activity of several dicistrovirus intergenic internal ribosome entry site elements in uninfected insect and mammalian cell lines. J. Gen. Virol. 2008;89:3150–3155. doi: 10.1099/vir.0.2008/003921-0. [DOI] [PubMed] [Google Scholar]
- 26.Becker DM, Lundblad V. Current Protocols in Molecular Biology. Somerset, NJ: John Wiley & Sons, Inc.; 2000. pp. 13.17.11–13.17.10. [Google Scholar]
- 27.Winzeler EA, Shoemaker DD, Astromoff A, Liang H, Anderson K, Andre B, Bangham R, Benito R, Boeke JD, Bussey H, et al. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- 28.Landry DM, Hertz MI, Thompson SR. RPS25 is essential for translation initiation by the Dicistroviridae and hepatitis C viral IRESs. Genes Dev. 2009;23:2753–2764. doi: 10.1101/gad.1832209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Mascorro-Gallardo JO, Covarrubias AA, Gaxiola R. Construction of a CUP1 promoter-based vector to modulate gene expression in Saccharomyces cerevisiae. Gene. 1996;172:169–170. doi: 10.1016/0378-1119(96)00059-5. [DOI] [PubMed] [Google Scholar]
- 30.Hunnicutt LE, Hunter WB, Cave RD, Powell CA, Mozoruk JJ. Genome sequence and molecular characterization of Homalodisca coagulata virus-1, a novel virus discovered in the glassy-winged sharpshooter (Hemiptera: Cicadellidae) Virology. 2006;350:67–78. doi: 10.1016/j.virol.2006.02.034. [DOI] [PubMed] [Google Scholar]
- 31.Shibuya N, Nishiyama T, Nakashima N. Cell-free synthesis of polypeptides lacking an amino-terminal methionine by using a dicistroviral intergenic internal ribosome entry site. J. Biochem. 2004;136:601–606. doi: 10.1093/jb/mvh167. [DOI] [PubMed] [Google Scholar]
- 32.Rydzanicz R, Zhao XS, Johnson PE. Assembly PCR oligo maker: a tool for designing oligodeoxynucleotides for constructing long DNA molecules for RNA production. Nucleic Acids Res. 2005;33:W521–W525. doi: 10.1093/nar/gki380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hertz MI, Thompson SR. Mechanism of translation initiation by Dicistroviridae IGR IRESs. Virology. 2011;411:355–361. doi: 10.1016/j.virol.2011.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cevallos RC, Sarnow P. Factor-independent assembly of elongation-competent ribosomes by an internal ribosome entry site located in an RNA virus that infects penaeid shrimp. J. Virol. 2005;79:677–683. doi: 10.1128/JVI.79.2.677-683.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Thompson SR, Gulyas KD, Sarnow P. Internal initiation in Saccharomyces cerevisiae mediated by an initiator tRNA/eIF2-independent internal ribosome entry site element. Proc. Natl Acad. Sci. USA. 2001;98:12972–12977. doi: 10.1073/pnas.241286698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sung D, Kang H. The N-terminal amino acid sequences of the firefly luciferase are important for the stability of the enzyme. Photochem. Photobiol. 1998;68:749–753. doi: 10.1562/0031-8655(1998)068<0749:tntaas>2.3.co;2. [DOI] [PubMed] [Google Scholar]
- 37.Sasaki J, Nakashima N. Translation initiation at the CUU codon is mediated by the internal ribosome entry site of an insect picorna-like virus in vitro. J. Virol. 1999;73:1219–1226. doi: 10.1128/jvi.73.2.1219-1226.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shibuya N, Nishiyama T, Kanamori Y, Saito H, Nakashima N. Conditional rather than absolute requirements of the capsid coding sequence for initiation of methionine-independent translation in Plautia stali intestine virus. J. Virol. 2003;77:12002–12010. doi: 10.1128/JVI.77.22.12002-12010.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Nakamura Y, Gojobori T, Ikemura T. Codon usage tabulated from international DNA sequence databases: status for the year 2000. Nucleic Acids Res. 2000;28:292. doi: 10.1093/nar/28.1.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Percudani R, Ottonello S. Selection at the wobble position of codons read by the same tRNA in Saccharomyces cerevisiae. Mol. Biol. Evol. 1999;16:1752–1762. doi: 10.1093/oxfordjournals.molbev.a026087. [DOI] [PubMed] [Google Scholar]
- 41.Fischer N, Konevega AL, Wintermeyer W, Rodnina MV, Stark H. Ribosome dynamics and tRNA movement by time-resolved electron cryomicroscopy. Nature. 2010;466:329–333. doi: 10.1038/nature09206. [DOI] [PubMed] [Google Scholar]
- 42.Letzring DP, Dean KM, Grayhack EJ. Control of translation efficiency in yeast by codon-anticodon interactions. Rna. 2010;16:2516–2528. doi: 10.1261/rna.2411710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jones JC, Myerscough MR, Graham S, Oldroyd BP. Honey bee nest thermoregulation: diversity promotes stability. Science. 2004;305:402–404. doi: 10.1126/science.1096340. [DOI] [PubMed] [Google Scholar]
- 44.Nishiyama T, Yamamoto H, Uchiumi T, Nakashima N. Eukaryotic ribosomal protein RPS25 interacts with the conserved loop region in a dicistroviral intergenic internal ribosome entry site. Nucleic Acids Res. 2007;35:1514–1521. doi: 10.1093/nar/gkl1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Armache JP, Jarasch A, Anger AM, Villa E, Becker T, Bhushan S, Jossinet F, Habeck M, Dindar G, Franckenberg S, et al. Localization of eukaryote-specific ribosomal proteins in a 5.5-A cryo-EM map of the 80S eukaryotic ribosome. Proc. Natl Acad. Sci. USA. 2010;107:19754–19759. doi: 10.1073/pnas.1010005107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Ben-Shem A, Jenner L, Yusupova G, Yusupov M. Crystal structure of the eukaryotic ribosome. Science. 2010;330:1203–1209. doi: 10.1126/science.1194294. [DOI] [PubMed] [Google Scholar]
- 47.Rabl J, Leibundgut M, Ataide SF, Haag A, Ban N. Crystal Structure of the Eukaryotic 40S Ribosomal Subunit in Complex with Initiation Factor 1. Science. 2010;330:730–736. doi: 10.1126/science.1198308. [DOI] [PubMed] [Google Scholar]
- 48.Muhs M, Yamamoto H, Ismer J, Takaku H, Nashimoto M, Uchiumi T, Nakashima N, Mielke T, Hildebrand PW, Nierhaus KH, et al. Structural basis for the binding of IRES RNAs to the head of the ribosomal 40S subunit. Nucleic Acids Res. 2011 doi: 10.1093/nar/gkr114. 4 March (Epub ahead of print; doi:10.1093/nar/gkr114) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Van Munster M, Dullemans AM, Verbeek M, Van Den Heuvel JF, Clerivet A, Van Der Wilk F. Sequence analysis and genomic organization of Aphid lethal paralysis virus: a new member of the family Dicistroviridae. J. Gen. Virol. 2002;83:3131–3138. doi: 10.1099/0022-1317-83-12-3131. [DOI] [PubMed] [Google Scholar]
- 50.Leat N, Ball B, Govan V, Davison S. Analysis of the complete genome sequence of black queen-cell virus, a picorna-like virus of honey bees. J. Gen. Virol. 2000;81:2111–2119. doi: 10.1099/0022-1317-81-8-2111. [DOI] [PubMed] [Google Scholar]
- 51.Johnson KN, Christian PD. The novel genome organization of the insect picorna-like virus Drosophila C virus suggests this virus belongs to a previously undescribed virus family. J. Gen. Virol. 1998;79(Pt 1):191–203. doi: 10.1099/0022-1317-79-1-191. [DOI] [PubMed] [Google Scholar]
- 52.Nakashima N, Sasaki J, Toriyama S. Determining the nucleotide sequence and capsid-coding region of himetobi P virus: a member of a novel group of RNA viruses that infect insects. Arch. Virol. 1999;144:2051–2058. doi: 10.1007/s007050050726. [DOI] [PubMed] [Google Scholar]
- 53.Sasaki J, Nakashima N, Saito H, Noda H. An insect picorna-like virus, Plautia stali intestine virus, has genes of capsid proteins in the 3′ part of the genome. Virology. 1998;244:50–58. doi: 10.1006/viro.1998.9094. [DOI] [PubMed] [Google Scholar]
- 54.Zhu JY, Ye GY, Fang Q, Wu ML, Hu C. A pathogenic picorna-like virus from the endoparasitoid wasp, Pteromalus puparum: initial discovery and partial genomic characterization. Virus Res. 2008;138:144–149. doi: 10.1016/j.virusres.2008.09.001. [DOI] [PubMed] [Google Scholar]
- 55.Moon JS, Domier LL, McCoppin NK, D'Arcy CJ, Jin H. Nucleotide sequence analysis shows that Rhopalosiphum padi virus is a member of a novel group of insect-infecting RNA viruses. Virology. 1998;243:54–65. doi: 10.1006/viro.1998.9043. [DOI] [PubMed] [Google Scholar]
- 56.Czibener C, Alvarez D, Scodeller E, Gamarnik AV. Characterization of internal ribosomal entry sites of Triatoma virus. J. Gen. Virol. 2005;86:2275–2280. doi: 10.1099/vir.0.80842-0. [DOI] [PubMed] [Google Scholar]
- 57.Valles SM, Strong CA, Dang PM, Hunter WB, Pereira RM, Oi DH, Shapiro AM, Williams DF. A picorna-like virus from the red imported fire ant, Solenopsis invicta: initial discovery, genome sequence, and characterization. Virology. 2004;328:151–157. doi: 10.1016/j.virol.2004.07.016. [DOI] [PubMed] [Google Scholar]
- 58.Mari J, Poulos BT, Lightner DV, Bonami JR. Shrimp Taura syndrome virus: genomic characterization and similarity with members of the genus Cricket paralysis-like viruses. J. Gen. Virol. 2002;83:915–926. doi: 10.1099/0022-1317-83-4-915. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.