Abstract
It has been previously shown that the DHX36 gene product, G4R1/RHAU, tightly binds tetramolecular G4-DNA with high affinity and resolves these structures into single strands. Here, we test the ability of G4R1/RHAU to bind and unwind unimolecular G4-DNA. Gel mobility shift assays were used to measure the binding affinity of G4R1/RHAU for unimolecular G4-DNA-formed sequences from the Zic1 gene and the c-Myc promoter. Extremely tight binding produced apparent Kd’s of 6, 3 and 4 pM for two Zic1 G4-DNAs and a c-Myc G4-DNA, respectively. The low enzyme concentrations required for measuring these Kd’s limit the precision of their determination to upper boundary estimates. Similar tight binding was not observed in control non-G4 forming DNA sequences or in single-stranded DNA having guanine-rich runs capable of forming tetramolecular G4-DNA. Using a peptide nucleic acid (PNA) trap assay, we show that G4R1/RHAU catalyzes unwinding of unimolecular Zic1 G4-DNA into an unstructured state capable of hybridizing to a complementary PNA. Binding was independent of adenosine triphosphate (ATP), but the PNA trap assay showed that unwinding of G4-DNA was ATP dependent. Competition studies indicated that unimolecular Zic1 and c-Myc G4-DNA structures inhibit G4R1/RHAU-catalyzed resolution of tetramolecular G4-DNA. This report provides evidence that G4R1/RHAU tightly binds and unwinds unimolecular G4-DNA structures.
INTRODUCTION
The nucleic acid base guanine has the capacity to base pair with itself, enabling the formation of alternative nucleic acid structures that differ from the canonical Watson–Crick base paired double helix. G4-DNAs (also termed G quadruplex DNAs) have guanines that form Hoogsteen-bonded quartets that coordinately bind a single monovalent cation at the O6 position of each guanine of the quartet (Figure 1A). Guanine quartets form the fundamental building block of G4-nucleic acids (1). Nucleic acid sequences with four appropriately spaced runs of usually three or more guanine bases can form structures of stacked guanine quartet subunits, although in some cases, e.g. the G15D thrombin aptamer (2), as few as four runs of two guanine bases can form this structure. Nucleic acid structures containing stacked guanine quartets demonstrate unusual thermal stability (3). Although all G4-DNAs have the common stacked guanine quartet motif, they are heterogeneous with regard to the number of individual nucleic acid strands involved and the paired direction of the phosphodiester backbones of the four strands of the structure. Stable G4-nucleic acids may contain intra- and/or interstrand hydrogen bonds along with parallel and/or antiparallel strand orientations. Sequence, concentration of strands, and the ionic environment all influence the type of G4-nucleic acid structures formed (4–6). Some sequences (especially fully intramolecular G4-DNA) can form polymorphic structures that are stable under physiological conditions, and, in general, intramolecular G4-DNA must be characterized empirically because ab initio knowledge has not developed sufficiently to predict structures from sequence and solution conditions alone (7).
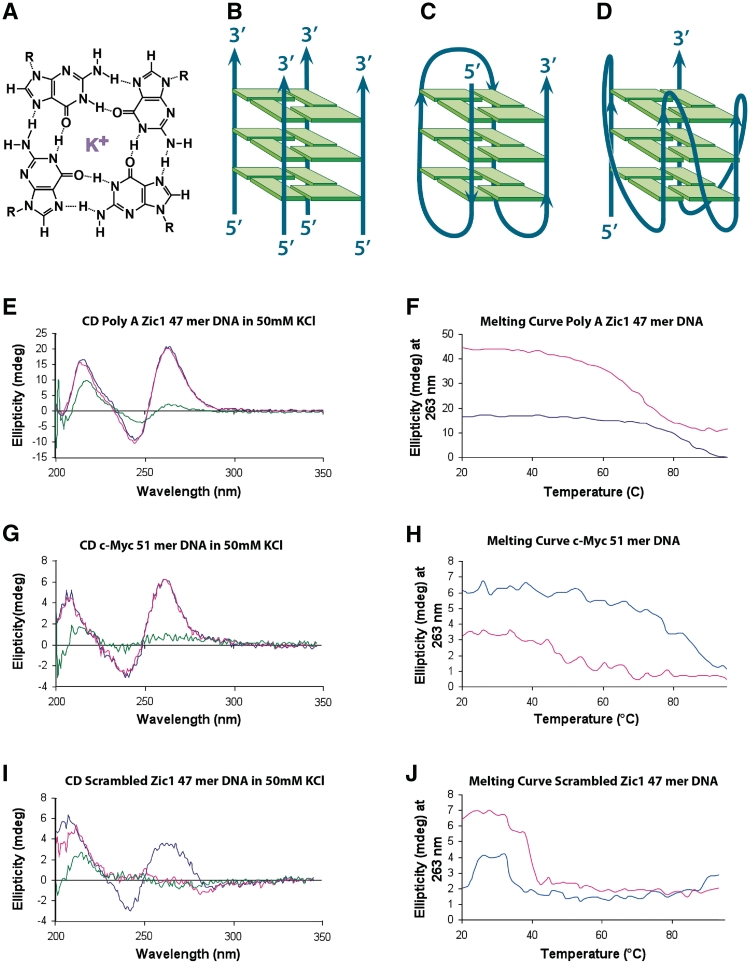
Figure 1.
CD analysis indicates the presence of G4-DNA in two oligonucleotides (Poly A Zic1 DNA 47-mer and c-Myc DNA 51-mer) and the absence of G4-DNA in a third oligonucleotide (Scrambled Zic1 DNA 47-mer). (A) Depiction of guanine quartet structure with each guanine participating in four hydrogen bonds and a potassium cation coordinately bound (purple). (B–D) Schematic depiction of (B) a tetramolecular, parallel G4-DNA structure, (C) a unimolecular antiparallel G4-DNA structure, and (D) a unimolecular parallel G4-DNA structure. (E) CD spectrum of Poly A Zic1 DNA 47-mer in 50 mM KCl (blue trace 25°C, red trace 37°C, green trace 95°C). (F) Melting curve of Poly A Zic1 DNA 47-mer; molar ellipticity at 263 nm versus temperature in 50 mM KCl (blue trace) or 50 mM LiCl (red trace). (G) CD spectrum of c-Myc DNA 51-mer in 50 mM KCl (blue trace 25°C, red trace 37°C, green trace 95°C). (H) Melting curve of c-Myc DNA 51-mer; molar ellipticity at 263 nm versus temperature in 50 mM KCl (blue trace) or 50 mM LiCl (red trace). (I) CD spectrum of Scrambled Zic1 DNA 47-mer in 50 mM KCl (blue trace 25°C, red trace 37°C, green trace 95°C). (J) Melting curve of scrambled Zic1 DNA 47-mer; molar ellipticity at 263 nm versus temperature in 50 mM KCl (blue trace) or 50 mM LiCl (red trace).
The stacked G-quartet structure was first solved in guanylic acid by Gellert et al. (1), and this binding motif was first recognized to form in DNA by Sen and Gilbert (8) working in vitro with DNA oligonucleotide sequences from the immunoglobin switch region. Four separate guanine-rich oligodeoxyribonucleotides formed a parallel, intermolecular, tetramolecular G4-DNA (Figure 1B). Sen and Gilbert (8) proposed such structures could form in the synaptonemal complex of meiotic cells. Although tetramolecular G4-DNA structures are highly thermally stable, they have large kinetic barriers of formation, requiring high concentrations of single-stranded DNA and long periods of time for formation (9). While specialized cellular regions could form tetramolecular G4-DNA, the most common G4-DNA structure predicted to form in cells is the fully intramolecular, unimolecular G4-DNA species. Unimolecular G4-DNA formation was first observed by Williamson et al. (10) in oligodeoxyribonucleotide sequences of the Oxytrichia 3′ telomeric single-stranded overhang. Unimolecular G4-DNA structures have both thermal stability and low first-order kinetic barriers of formation (11). Furthermore, unimolecular G4-DNA is commonly stabilized best by a K+ containing ionic environment (3,12,13), as found in mammalian nuclei. Many of these structures can also be stabilized in an equimolar Na+ environment, albeit to a lesser extent as evidenced by a lowered Tm. Two bioinformatics studies have suggested a canonical sequence predicting unimolecular G4-DNA formation that requires four runs of at least three tandem Gs with three loop regions (Ns) in between the G runs; the canonical putative quadruplex sequence (PQS) is summarized in the formula: d(G3–5N1–6G3–5N1–6G3–5N1–6G3–5) (14,15). These two studies found approximately 375 000 PQS in the human genome using distinctly different sequence search approaches (14,15). These determinations are probably underestimates of the true number of PQS in the human genome, since stable DNA aptamers, e.g. the thrombin aptamer (2), have been found with four guanine runs of as few as two tandem guanines.
The most direct evidence of the presence of G4-nucleic acid structures in vivo comes from studies of unimolecular and bimolecular G4-DNA structures (16,17). Commonly, telomere sequences have a 3′ overhang that remains single stranded, and most telomere sequences meet the canonical sequence requirements to form intramolecular G4-DNA structures. Specific antibodies to G4-DNA were observed to bind to telomeres of the macronuclear chromosomes of the ciliate Stylonychia lemnae. This work included important controls to address the issue of whether the antibody was inducing G4-DNA structures (18,19). A carbazole derivative dye exhibited a G4-DNA binding-dependent fluorescence emission wavelength shift when staining the telomeres of human mitotic chromosomes, suggesting the presence of antiparallel unimolecular G4-DNA (20). An inducible gene containing the immunoglobin Sμ and Sγ3 switch regions cloned into an Escherichia coli plasmid produced G4-DNA structures within the non-template strand of the D-loop created upon transcription induction (21). Furthermore, a number of DNA sequences have been shown in vitro to readily form unimolecular G4-DNA structures under physiological salt conditions. These PQS are located within key control regions of important genes, including the aforementioned immunoglobin switch region (8), the d(pCGG) repeats of the fragile X mental retardation gene (22) and the promoter regions of a number of proliferation-associated genes such as c-Myc (23,24), PDGF-A (25), RET (26), the diabetes susceptibility locus (27) and the human insulin gene (28). Also, within transcribed regions of genes, unimolecular PQS have been found in 5′-UTRs of genes such as Zic1 and NRAS (29,30), and also near intron splicing and poly (A) addition sites of genes (31,32). Although there is interest in the consequences of G4-RNA formation within these gene transcripts (29,30), it is also important to note that the non-template strand of these transcribed genes is capable of forming G4-DNA, which becomes more probable as transcription increases. Such transcription-associated G4-DNA formation has been observed in the non-template strand of transcribed sequences of immunoglobin Sμ and Sγ3 switch regions in a constructed plasmid (21).
Considering that the melting temperature of most G4-DNAs that conform to the PQS sequence are predicted to be above 41°C under physiological salt conditions, it has been hypothesized by a number of researchers that, once G4-DNAs form in vivo, they must be resolved by enzymes that specifically recognize and unwind such structures. In this regard, we identified the first human tetramolecular G4-DNA resolving activity from cell lysates and began to characterize the nucleoside triphosphate (NTP) utilization requirement of this activity (33). Subsequently, a number of genes related to genomic instability syndromes have been demonstrated to code for enzymes possessing G4-DNA unwinding activity in vitro, including the RecQ homologs BLM (34) and WRN (22) and the FANCJ protein (35). However, further purification, mass spectrometric identification and recombinant expression studies revealed that the original G4-DNA resolving activity that we observed in human cell lysates did not reside in one of the above mentioned proteins; rather, it resided in the DHX36 gene product G4R1/RHAU (36). Furthermore, G4R1/RHAU was observed to be responsible for the major tetramolecular G4-DNA resolving activity in HeLa cells (36,37), which conformed to the expectations of isolating a major activity through a biochemical approach rather than a genetic one.
G4R1/RHAU is the protein product of the DHX36 gene, a member of the DEAH box family (36). It is a 115 kDa protein containing a highly conserved helicase core that contains sequence motifs, e.g. the Walker A and Walker B box motifs, in common with RecQ helicases. Full-length recombinant G4R1/RHAU has been shown to bind tightly to tetramolecular G4-DNA and G4-RNA with Kd’s of 77 ± 6 pM and 39 ± 6 pM, respectively, and to efficiently unwind these tetramolecular structures into monomers in the presence of adenosine triphosphate (ATP) (37). G4R1/RHAU requires an unstructured loading region of at least seven bases 3′ of the G4-DNA structure for efficient resolution of tetramolecular molecules; no such extension of nucleotides were required at the 5′-end of the G4-DNA structure for unwinding (S. Creacy, S. Akman and J. Vaughn, unpublished results). These results suggest that the enzyme loads on the 3′-end of tetramolecular G4-DNA structures and translocates in the 3′ to 5′ direction. Recent deletion studies have shown that about a 100 amino acid N-terminal region of the protein is responsible for most of its binding affinity to tetramolecular G4 structures (38). However, our previous work did not address the question of whether G4R1/RHAU can bind and unwind the single-stranded molecules of unimolecular G4-DNA, which are the G4-DNA structures most expected to commonly form in cells. In this study, we assessed the binding and resolution activity of full-length recombinant human G4R1/RHAU upon unimolecular G4-DNA structure, and found that G4R1/RHAU binds tightly to this structure. Moreover, G4R1/RHAU catalyzes unwinding of unimolecular G4-DNA into an unstructured form capable of hybridization with a Watson–Crick complementary peptide nucleic acid (PNA).
MATERIALS AND METHODS
Circular dichroism spectropolarimetry
Circular dichroism (CD) experiments were performed on an AVIV Model 202 CD spectrometer equipped with a thermoelectrically controlled cell holder. A 1 µM solutions of G4-DNA were prepared in 10 mM Tris–HCl, pH 7.5, 1 mM EDTA ± 0–50 mM KCl or LiCl. Quartz cells with 1 cm path length were used for all experiments. CD spectra were recorded in the UV (200–350 nm) regions with 1 nm increments with an averaging time of 2 s. Thermal stability of G4-DNA was assessed by recording molar ellipticity at 263 nm as a function of temperature. The temperature was raised in 2°C increments; stirred samples were allowed to equilibrate for each temperature setting for 30 s. Molar ellipticity data were collected in the temperature range 25–98°C.
G4-DNA formation and quantification
Oligonucleotides [unlabeled or 5′ 5,6-carboxytetramethylrhodamine (TAMRA)-labeled] were purchased from either Oligos Etc. or Integrated DNA Technologies. Tetramolecular G4-DNA was formed from oligonucleotide Z33 (Table 1) by a previously described procedure (33) that results in over 99% conversion of monomer to tetramolecular G4-DNA. Unimolecular G4-DNA was formed in various oligonucleotides (Table 1) by dissolving them at a concentration of 0.5 mM in 10 mm Tris–HCl, 1 mM EDTA, pH 7.5. Oligonucleotide solutions were aliquoted into polymerase chain reaction (PCR) tubes and incubated in a thermocycler (Eppendorf epGradient S) at 98°C for 10 min, then held at 80°C. Immediately the tubes were opened and KCl was added to a final concentration of 25 mM. The tubes were reclosed and allowed to come slowly to room temperature. Aliquots were combined and stored at 4°C for 2–3 days. Tetramolecular G4-nucleic acids formed from 5′-TAMRA-labeled oligonucleotides were further purified by electrophoresis on a 10% polyacrylamide gel and band excision. Electrophoretic bands containing G4-DNA were recovered by electroelution in a Schleicher and Schuell Elutrap with 1 × TBE (89 mM Tris-Borate, pH 8.3, 2.5 mM EDTA) buffer additionally containing 3 mM KCl. The Elutrap was run with buffer recirculation. 5′-TAMRA-labeled G4 nucleic acids were aliquoted and stored at −20°C or lower.
Table 1.
DNA oligonucleotides used in this work
| Oligo name | Sequence (5′–3′) |
|---|---|
| Poly A Zic1 DNA (47-mer) | 5′-AAA AAA AAA AGG GT GGG GGG GCG GGG GAG GCC GGG GAA AAA AAA AA-3′ |
| c-Myc DNA (51-mer) | 5′-GGC CGC TTA TGG GGA GGG TGG GGA GGG TGG GGA AGG TGG GGA GGA GAC TCA-3′ |
| Scrambled Zic1 DNA (47-mer) | 5′-AGA AGA GAG AGA GTG AGA GAG ACG AGA GAG GCC GAG AAG AGA GAG AG-3′ |
| Poly T Zic1 DNA (47-mer) | 5′-TTT TTT TTT TGG GTG GGG GGG GCG GGG GAG GCC GGG GTT TTT TTT TT-3′ |
| (HHN)11 DNA (33-mer) | 5′-HHN HHN HHN HHN HHN HHN HHN HHN HHN HHN HHN-3′ |
| PolyA DNA (47-mer) | 5′-AAA AAA AAA AAA AAA AAA AAA AAA AAA AAA AAA AAA AAA AAA AAA AA-3′ |
| Poly T DNA (47-mer) | 5′-TTT TTT TTT TTT TTT TTT TTT TTT TTT TTT TTT TTT TTT TTT TTT TT-3′ |
| PNA (11-mer) | H2N-CCC CCC CCA CCC-Lys-COOH |
| Z33 DNA (33-mer) | 5′-AAA GTG ATG GTG GTG GGG GAA GGA TTT TCG AAC-3′ |
G4-DNA concentrations were determined by absorbance of 260 nm light using a Smart Spec 3000 UV spectrophotometer (Bio-Rad). The molar extinction coefficient for each oligonucleotide was determined by the instrumental software after inputting the base composition of each.
5′-[32P]-end labeling of G4 nucleic acids
[32P]-end-labeled G4 oligonucleotides were obtained by incubating unlabeled oligonucleotides previously treated to form G4-DNA as described above, with T4 polynucleotide kinase (Promega Corporation) and [γ-32P] ATP for 0.5 h at 37°C, according to the manufacturer’s instructions. 5′-[32P] end-labeled oligonucleotides were purified with a MicroSpin G25 column (GE Healthcare) equilibrated with TEK (10 mM Tris, 1 mM EDTA and 50 mM KCl) buffer and stored at −20°C.
Gel mobility shift assay and apparent Kd determination for G4R1/RHAU-oligonucleotide complexes
We have previously described the isolation of recombinant G4R1/RHAU from Rosetta 2 cells (36, 37). It should be noted that we have found that the addition of 0.02% β-lactalbumin protein as a stabilizer is important for isolation of enzyme of high-specific activity. However, because β-lactalbumin carrier is the dominant protein in preparations of G4R1, it was necessary to quantify G4R1 with sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS–PAGE) and Coomassie blue staining instead of using solution quantification techniques. Purified recombinant G4R1/RHAU was quantified in the following protocol: 15 µl of purified recombinant G4R1/RHAU was loaded on a standard 5% stacking/12% resolving SDS–polyacrylamide gel and the loading was compared to known amounts of standard; 0.0625, 0.125, 0.250, 0.500 and 0.750 µg of Promega Broad Range Molecular weight markers per band (cat no V849A). The gels were stained with ProtoBlue Safe Colloidal Coomassie G-250 stain (National Diagnostics, EC-722) per manufacturer’s instructions, destained and scanned on an Epson Perfection 2450 Photo scanner with Epson SilverFast TWAIN. Densitometric measurements of bands representing recombinant G4R1/RHAU and three proteins in the marker that were of similar size (150, 100 and 75 kDa) were then made using Fuji Multiguage software. Linear regression analysis of the three standard proteins’ titration series gave similar curves. The concentration of rG4R1/RHAU was then extrapolated from the titration series, yielding a final concentration of 223 ± 18 nM (n = 9). To determine the accuracy of the Promega Broad Range Molecular Weight Markers (V849A) that were used to quantify rG4R1 via Coomassie staining, bovine serum albumin (BSA) (Pierce, # 23209) and β-galactosidase (Sigma, # 5635-1KU) were precisely quantified via spectrophotometry at 280 nm using extinction coefficients. BSA and β-galactosidase were chosen because their histidine, arginine and aromatic amino acid contents are similar to rG4R1. 150 and 300 ng aliquots of each protein (as determined spectrophotometrically) were loaded and run on a typical 5% stacking/12% resolving polyacrylamide gel next to a titration of the Promega Broad Range Molecular Weight markers (31.3, 62.5, 125, 250, 500 ng); these titrations were done in triplicate. Gels were then stained with Coomassie R-250 (50 mg/100 ml of 50:10:40 v/v methanol:acetic acid:H2O) and then destained with 30:10:60 v/v methanol:acetic acid:H2O. The three proteins that were originally used to determine rG4R1 concentration via Coomassie staining (75, 100, 150 kDa bands of Promega # V849A) were used to generate three standard curves (n = 9). Amounts of BSA and β-galactosidase were extrapolated and compared to the amounts that were loaded based upon the spectrophotometrically determined values, respectively. We observed that the two methods (spectrophotometric versus in-gel Coomassie determination of protein concentration) differ by 20–49% (<2-fold, data not shown). The apparent Kd of G4R1/RHAU bound to unimolecular G4-DNA was estimated by GMSA.
Recombinant G4R1/RHAU at concentrations of 2–300 pM was incubated with 1 pM 5′-[32P]end-labeled G4-DNA in K-Res (100 mM KCl, 10 mM NaCl, 3 mM MgCl2, 50 mM Tris acetate, pH 7.8, 70 mM glycine, 0.012% bovine α-lactalbumin, 10% glycerol) buffer with 10 mM EDTA at 37°C for 30 min. Binding mixtures were then loaded and analyzed by 10% non-denaturing PAGE. Electrophoresis was performed at 70 V for 15 h in a cold room (7°C). Gels were imaged on a Typhoon 9210 Imager (GE Healthcare). Band densities were analyzed using Multi Gauge (Fuji) software and statistical analysis was performed with MS Excel. The apparent Kd value was estimated from each model as the concentration of enzyme at which 50% of target DNA substrate was bound for three separate experiments for each DNA. In selected experiments in which data were obtained at closely spaced 0.5–1 pM intervals over a broad range of DNA substrate concentrations, the data were fitted to a saturable single site binding model:
| (1) |
Here Bmax is the fractional degree of saturation of receptor with ligand at large excess, and Kd is the equilibrium affinity constant expressed in molarity. Nonlinear regression analysis (SigmaPlot 11.0, Systat, San Jose, CA, USA) yielded Bmax, Kd and the standard deviation of these fitted parameters. A linear regression analytic fit of 1/[Bound Substrate] as a function of 1/[G4R1/RHAU] was also performed.
koff determination
A 1500 µl solution of 1 pM 5′-[32P]-Poly A Zic1 47-mer G4-DNA and 200 pM G4R1/RHAU was incubated at 37°C for 30 min under standard GMSA conditions (described above). Following incubation, a 200 µl sample of the binding mixture was removed and placed at −80°C to be used as a positive binding control. At this point, 20 µM unlabeled Poly A Zic1 47-mer G4-DNA was added to the binding mixture and maintained at 37°C. At 0, 1, 2, 24, 48 and 72 h following the addition of unlabeled competitor, 200 µl samples of the binding mix were removed and placed at −80°C. As a negative control a reaction in which 20 nM of unlabeled Poly A Zic1 47-mer G4-DNA was added to 1 pM 5′-[32P] end-labeled-Poly A Zic1 47mer G4-DNA prior to incubation with G4R1/RHAU was included to demonstrate that 20 nM of unlabeled Poly A Zic1 47-mer G4-DNA is sufficient to block detection of binding of 5′-[32P] end-labeled-Poly A Zic1 47mer G4-DNA to G4R1/RHAU. Following the completion of the 72 h time point, binding mixtures were simultaneously thawed and analyzed by 10% non-denaturing PAGE. Electrophoresis was performed at 70 V for 15 h in a cold room (7°C). Gels were imaged on a Typhoon 9210 Imager (GE Healthcare). Band densities were analyzed using Multi Gauge (Fuji) software.
Dimethyl sulfate treatment of poly A Zic1 DNA
Poly A Zic1 DNA 47-mer oligonucleotide was treated with dimethyl sulfate using a slight modification of the method of Maxam and Gilbert (39). A Total of 250 pM poly A Zic1 DNA 47-mer was added to a 50 µl reaction volume containing 50 mM sodium cacodylate, pH 8.0, 1 mM EDTA. Samples were chilled on ice for 5 min followed by heating at 95°C for 5 min, after which 2 µl (1:4 ethanol v/v) dimethyl sulfate was added and incubated at 37°C for 30 min.
PNA hybridization and PNA trap assay
Electrophoretic mobility standards for the two isomers of Poly A Zic1 DNA 47-mer G4-DNA (Figure 5 and Supplementary Figure S4, Bands 1 and 2) and the Poly A Zic1 DNA 47-mer:PNA hybrid (Figure 5 and Supplementary Figure S4, Band 3) were prepared by adding 100 pM or 1 nM 5′-[32P]-labeled Poly A Zic1 DNA 47-mer G4-DNA to K-RES buffer, 10 mM ATP and 0, 10, 100,500 nM, or 10 µM PNA (with Watson–Crick complementarity to the internal Poly A Zic1 DNA 47-mer PQS) in a 30 µl final volume. Solutions were held at 98°C for 10 min and then cooled to room temperature. In order to assess G4R1/RHAU-catalyzed unwinding of unimolecular G4-DNA by trapping of the open product with a complementary PNA, 100 pM or 1 nM 5′-[32P]-labeled Poly A Zic1 DNA 47-mer G4-DNA was added to RES buffer, 10 mM ATP [or adenosine 5′-(β, γ-imido)triphosphate (AMP-PNP) in selected experiments], 10 nM complementary PNA and a dilution series of active recombinant G4R1/RHAU enzyme that included 0, 28.5, 463 fM, 1.85, 115 and 7.40 pM, each in a 30 µl final volume. An identical series of reactions were prepared in which recombinant G4R1/RHAU was held at 98°C for 10 min prior to the reaction to destroy the catalytic activity of the enzyme, or wild-type enzyme was replaced with mutant G4R1/RHAU containing an E→A substitution at amino acid position 335 in the Walker B box that abolishes ATPase activity (40), to serve as negative controls. Additionally, a set of 30 µl reactions was made containing 1 nM of 5′-[32P]-labeled Poly A Zic1 DNA 47-mer G4-DNA in K-RES buffer, 10 mM ATP, in the absence of complementary PNA and an identical dilution series of active recombinant G4R1/RHAU as described above. All reactions were incubated at 37°C for 30 min, dropped to 4°C and stopped by adding 5 µl of 0.5 M EDTA and mixing. Reactions were then directly loaded (monitored visually by Schlieren lines) alongside of a set of electrophoretic mobility standards (prepared as described above) and analyzed on a 10% non-denaturing PAGE TBE gel with 10% glycerol. Gels were imaged on a Typhoon 9210 Imager (GE Healthcare).
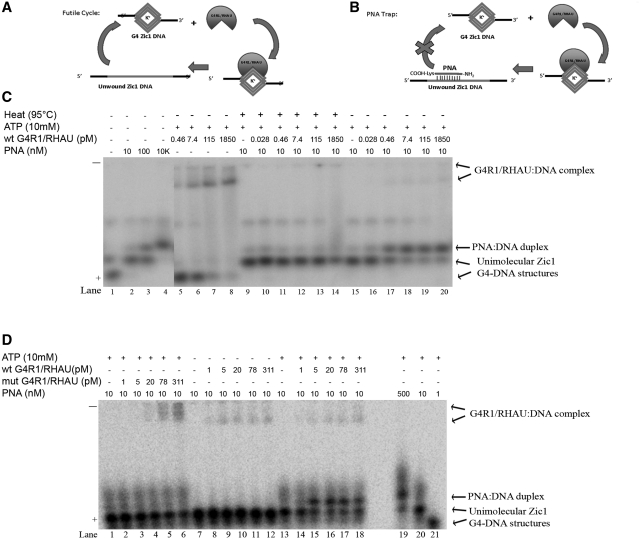
Figure 5.
A G4R1/RHAU resolution assay of Poly A Zic1 DNA 47-mer G4-DNA in the presence of a complementary PNA shows that G4R1/RHAU unwinds the G4-DNA structure. (A) Schematic diagram illustrates the kinetic futile cycle problem of attempting to determine if G4R1/RHAU unwinds the G4-DNA structure in the PQS-containing oligonucleotide. (B) Schematic diagram illustrating how the kinetic futile cycle problem is solved with a PNA trap assay. (C) Phosphorimage of representative (of three repetitions) nondenaturing gel electropherogram of 100 pM 5′-[32P]-labeled Poly A Zic1 DNA 47-mer incubated with increasing concentrations of a PNA with Watson–Crick complementarity to the G4-DNA-forming sequence in the absence of ATP and G4R1/RHAU (lanes 1–4), with 10 mM ATP plus increasing concentrations of G4R1/RHAU, but without PNA (lanes 5–8), with ATP and PNA plus increasing concentrations of G4R1/RHAU that had been pre-heated to 95°C for 5 min (lanes 9–14), or with ATP and PNA plus increasing concentrations of active G4R1/RHAU (lanes 15–20). (D) Phosphorimage of representative (of three repetitions) nondenaturing gel electropherogram of 1 nM 5′-[32P]-labeled Poly A Zic1 DNA 47-mer incubated with PNA and ATP plus increasing concentrations of a G4R1/RHAU mutant that lacks ATPase activity (lanes 1–6), with PNA plus increasing concentrations of wild-type G4R1/RHAU, but without ATP (lanes 7–12), or with PNA, ATP, and increasing concentrations of wild type G4R1/RHAU (lanes 13–18); lanes 19–21 are a titration of 1 nM 5′-[32P]-labeled Poly A Zic1 DNA 47-mer incubated with increasing concentrations of PNA in the absence of G4R1/RHAU and presence of 10 mM ATP.
G4-DNA inhibition assay
Tetramolecular G4-DNA resolving activity was determined as previously described (33) but with K-RES buffer. 2 nM of 5′-TAMRA-labeled tetramolecular G4-DNA and 1 U of recombinant G4R1/RHAU were included per 50-μl reaction. Unlabeled inhibitor unimolecular G4-DNA oligonucleotides, including c-Myc 51-mer and Poly A Zic1 DNA 47-mer (Table 1), were added to some reactions in a 0- to128-fold molar excess of inhibitor to labeled tetramolecular G4-DNA substrate. Reactions were allowed to proceed at 37°C for 30 min, stopped by addition of 5 μl of 200 mM EDTA, and analyzed by electrophoresis through a 10% non-denaturing polyacrylamide TBE gel with 10% glycerol. Gels were scanned on a Typhoon 9210 Imager (GE-Healthcare) and images were analyzed using Multi Gauge version 3.0 imaging software (Fuji). Statistical analysis was done in MS Excel 2003 and standard deviations were calculated by the STDEV function.
RESULTS
Circular dichroism spectropolarimetry was used to determine unimolecular G4-DNA formation in test DNA oligonucleotides
Our first goal in studying the interaction of G4R1/RHAU with unimolecular G4-DNA was to form G4-DNA structures within DNA oligonucleotides of Zic1 and c-Myc and then to confirm and characterize the presence of these G4-DNA structures. CD spectra, combined with melting curve analysis in buffer containing 50 mM K+ or Li+, indicated the formation of G4-DNA.
The differential absorption of circularly polarized light measured by CD is attributable to the presence of chiral structure within the oligonucleotides and has been previously used to characterize G4-DNA in oligonucleotides (41,42). Typically, unimolecular parallel G4-DNA structures (Figure 1D) produce a peak of molar ellipticity at ∼260 nm, while anti-parallel G4-DNA structures (Figure 1C) produce a peak of ellipticity at 290 nm. The Poly A Zic1 DNA 47-mer and the c-Myc DNA 51-mer both contain G-rich sequences that conform to the formulae for formation of unimolecular G4-DNA (14,15). Table 1 shows all sequences used in this study in the order they are reported in the ‘Results’ section. G4-DNA forming regions are bolded. CD spectra indicate that both the Poly A Zic1 DNA 47-mer and the c-Myc 51-mer (Figure 1E and G) show peaks of molar ellipticity at ∼260 nm that are suggestive of parallel G4-DNA structures in 50 mM KCl at both 25°C (blue trace) and 37°C (red trace). The thermal stability of this CD signal is expected from G4-DNA, which is typically stable well above 37°C in KCl solution. However, at 95°C (green trace in Figure 1E and G) the molar ellipticity significantly diminishes at 260 nm, as would be expected for the destabilization of a chiral structure unfolding at the higher temperature and thereby losing its chirality. Note that the CD spectrum of the scrambled Zic1 47-mer sequence (Figure 1I), which is an oligonucleotide not expected to have G4-DNA, shows a similar peak of ellipticity at 260 nm at 25°C (blue trace). However, this structure was destabilized at both 37°C and at 95°C, displaying a significantly diminished ellipticity at 260 nm for both temperatures (Figure 1I, red trace and green trace). The fact that this structure was not stable at 37°C suggests it is unlikely to possess a stable G4-DNA structure. This was further confirmed by melting studies in the presence of K+ and Li+, discussed below.
A hallmark of G4-DNA is stabilization by K+ ions and destabilization by Li+ ions (3,4,7,13,43). Atomic modeling has suggested that K+ coordinately bonds between two stacked G-quartets as an ‘optimal fit’. However, the most accepted explanation of the superior stabilization of G4-DNA by K+ is that it is energetically easier to dehydrate K+ ions (44). Because of enhanced stability, G4 structures are predicted to have higher melting temperatures in K+ solutions than in Li+ solutions. We tested the melting behavior of oligonucleotide structures by monitoring the changes in peak height of molar ellipticity at 263 nm in the presence of K+ and Li+ with increasing temperature (Figure 1F, H and J). Poly A Zic1 DNA 47-mer and the c-Myc 51-mer exhibited the characteristic CD spectral changes for unimolecular G4-DNA structures in the presence of both cations, as expected. The Tm’s, defined as the temperatures of 50% depression of the 263 nm ellipticity peak, were well above 60°C for the G4-DNA structures of both oligonucleotides in K+. However, the Tm in KCl was higher than the Tm in LiCl for each G4-DNA structure. The Poly A Zic1 DNA 47-mer exhibited a Tm in 50 mM KCl of ∼80°C (Figure 1F, blue trace), while its Tm in 50 mM LiCl was ∼70°C (Figure 1F, red trace). The c-Myc 51-mer DNA similarly showed greater stabilization by K+ (Figure 1H), although its Tm in both 50 mM KCl and LiCl was somewhat lower than the Poly A Zic1 DNA oligonucleotide in the presence of the corresponding cation. The CD spectrum of the scrambled Zic1 47-mer control (which has the same nucleotide content but a different sequence order as the Poly A Zic1 DNA 47-mer) exhibited an ellipticity peak at 263 nm; however, this signal was completely lost by raising the temperature to 40°C. Furthermore, the peak of ellipticity at 263 nm observed in the scrambled Zic1 47-mer spectrum diminished by 50% at a lower temperature in KCl than in LiCl (Figure 1J), a behavior which is inconsistent with G4-DNA structure. Therefore, these CD results indicate that G4-DNA was formed in the Poly A Zic1 DNA 47-mer oligonucleotide and the c-Myc 51-mer oligonucleotide, but not in the scrambled Zic1 47-mer control DNA.
The temperature and cation dependence of the CD spectra of a number of other oligonucleotides that were used in G4R1/RHAU binding studies (Figure 3) were similarly assessed (data not shown). In all cases, only single-stranded DNA oligonucleotides with canonical G4-DNA sequences conforming to the unimolecular G4-DNA formulae (14,15) met the criteria of: (i) exhibiting a positive peak of ellipticity at 263 nm; (ii) displaying a CD spectrum that was stable above 37°C but which destabilized at near 95°C; and (iii) exhibiting a 50% decrease in signal of ellipticity at 263 nm (defined as a Tm) at a significantly higher temperature in 50 mM KCl- than in 50 mM LiCl-containing buffer. Oligonucleotides that failed to meet these criteria and also failed to exhibit a positive peak of ellipticity at 290 nm were interpreted as not forming true unimolecular G4-DNA structure. All putative non-G4-DNA-forming control sequences used in these studies exhibited CD spectra that failed to meet these 3-fold criteria for the presence of G4-DNA structure.
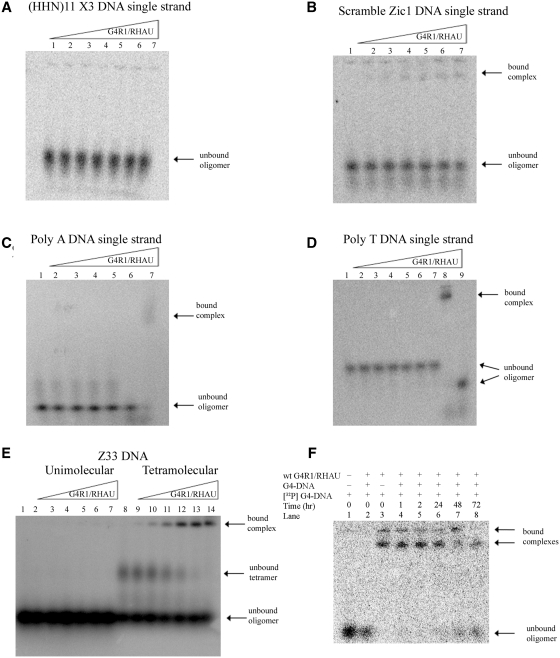
Figure 3.
Equilibrium binding GMSAs of purified recombinant G4R1/RHAU incubated with oligonucleotides not containing unimolecular G4-DNA shows that G4R1/RHAU does not tightly bind these substrates and suggests the enzyme has specificity for G4-DNA structures. (A–D) Phosphorimages of representative (of three repetitions) nondenaturing gel electropherograms of GMSAs; the top triangles indicate the direction of increasing G4R1/RHAU concentration. GMSA of (A) 1 pM 5′-[32P]-labeled d(pHHN)11 randomized DNA oligonucleotide incubated with increasing amounts of G4R1/RHAU (lane 1, 0 pM; lane 2, 30 pM; lane 3, 50 pM; lane 4, 75 pM; lane 5, 100 pM; lane 6, 150 pM; lane 7, 300 pM). (B) 1 pM 5′-[32P]-labeled Scrambled Zic1 single-stranded DNA oligonucleotide incubated with increasing amounts of G4R1/RHAU (lane 1, 0 pM; lane 2, 30 pM; lane 3, 50 pM; lane 4, 75 pM; lane 5, 100 pM; lane 6, 150 pM; lane 7, 300 pM). (C) 1 pM 5′-[32P]-labeled Poly A single-stranded DNA oligonucleotide incubated with increasing amounts of G4R1/RHAU (lanes 1–5, 0–300 pM; lane 6, Poly A DNA Zic1 47-mer G4-DNA incubated in the absence of G4R1/RHAU; lane 7, Poly A Zic1 DNA 47-mer G4-DNA incubated with 50 pM G4R1/RHAU). (D) 1 pM 5′-[32P]-labeled Poly T single-stranded DNA oligonucleotide incubated with increasing amounts of G4R1/RHAU (lanes 1–7, 0–300 pM; lane 8, Poly A Zic1 DNA 47-mer G4-DNA incubated with 30 pM G4R1/RHAU; lane 9, Poly A DNA Zic1 47-mer G4-DNA incubated in the absence of G4R1/RHAU). (E) 10 pM 5′-[32P]-labeled unstructured single-stranded Z33 oligonucleotide (lanes 1–7) or a mixture of unstructured and tetramolecular G4-DNA-structured Z33 (lanes 8–14) incubated with increasing amounts of G4R1/RHAU (lanes 1 and 8, 0 pM; lanes 2 and 9, 30 pM; lanes 3 and 10, 50 pM; lanes 4 and 11, 75 pM; lanes 5 and 12, 100 pM; lanes 6 and 13, 150 pM; lanes 7 and 14, 300 pM). (F) koff determination for a unimolecular G4-DNA bound to G4R1/RHAU. 1 pM 5′-[32P]-labeled Poly A Zic1 47-mer G4-DNA was bound to 200 pM G4R1/RHAU. Lane 1, 1 pM 5′-[32P]-labeled Poly A Zic1 47-mer G4-DNA in the absence of G4R1/RHAU; lane 2, 20 nM unlabeled Poly A Zic1 47-mer G4-DNA was added to G4R1/RHAU prior to the addition of 1 pM 5′-[32P]-labeled Poly A Zic1 47-mer G4-DNA; lane 3, 1 pM 5′-[32P]-labeled Poly A Zic1 47-mer G4-DNA was added to G4R1/RHAU in the absence of unlabeled Poly A Zic1 47-mer G4-DNA; lanes 4–8, 1 pM 5′-[32P]-labeled Poly A Zic1 47-mer G4-DNA was first added to G4R1/RHAU, then at t = 0, 20 nM unlabeled Poly A Zic1 47-mer G4-DNA was added and aliquots removed for GMSA at the times indicated. Half-life of binding was calculated to be 67 ± 9 h.
Although the CD spectra and cation-dependent thermal stability studies indicate that the Poly A Zic1 DNA 47-mer and c-Myc 51-mer oligonucleotides anneal into parallel stranded G4-DNA structures, these studies do not distinguish intramolecular from intermolecular G4-DNA. In this regard, stoichiometry studies were performed to determine whether the G4-DNA structure formed by Poly A Zic1 DNA 47-mer was intra- or intermolecular. A 1 nM Poly A Zic1 DNA 47-mer was mixed with equimolar poly A Zic 1 DNA 37-mer prior to forming G4-DNA by our standard annealing protocol. Poly A Zic1 DNA 37-mer has an identical G4-DNA-forming sequence to Poly A Zic1 DNA 47-mer; the only difference between these two oligonucleotides is that the core sequence in Poly A Zic1 DNA 37-mer is flanked only on the 5′-end by d(pA)10, whereas Poly A Zic1 DNA 47-mer is flanked on both ends. Non-denaturing PAGE after annealing the mixed oligonucleotides revealed only bands whose electrophoretic mobility corresponded to that of Poly A Zic1 37-mer G4-DNA and Poly A Zic1 47-mer G4-DNA (Supplementary Figure S1). No bands of intermediate electrophoretic mobility indicative of intermolecular G4-DNA structure formation between oligonucleotides of different lengths were observed. Thus, under our annealing conditions, poly A Zic DNA forms an intramolecular G4-DNA structure exclusively.
G4R1/RHAU binds unimolecular G4-DNA with an apparent Kd in the picomolar range while non-G4-containing DNA does not display tight binding
The ability of G4R1/RHAU to bind unimolecular G4-DNA was ascertained by the gel mobility shift assay (GMSA). In previous studies, we observed that the addition of EDTA to the G4R1/RHAU resolution buffer inhibits unwinding of, but not binding to, tetramolecular G4-nucleic acid structures, allowing us to determine an apparent Kd for binding tetramolecular G4-DNA (37). This same approach was used to determine apparent Kd’s for unimolecular G4-DNA-containing oligonucleotides and non-G4-DNA-containing control oligonucleotides. Binding reactions were performed by incubating increasing concentrations of recombinant full-length human G4R1/RHAU enzyme with a fixed concentration of 5′-[32P]- end-labeled oligonucleotides. A fine titration of increasing concentrations of the DNA oligonucleotide Poly A Zic1 DNA 47-mer with a fixed concentration of G4R1/RHAU indicated that 70% of G4R1/RHAU molecules possessed DNA binding activity (Supplementary Figure S2; a single site binding model was assumed). Figure 2A, E and I shows representative GMSAs of three DNA oligonucleotides (Poly A Zic1 47-mer DNA, Poly T Zic1 47-mer DNA and c-Myc 51-mer DNA), each of which possess unimolecular G4-DNA by CD criteria; each of these oligonucleotides binds G4R1/RHAU at remarkably low enzyme concentrations.
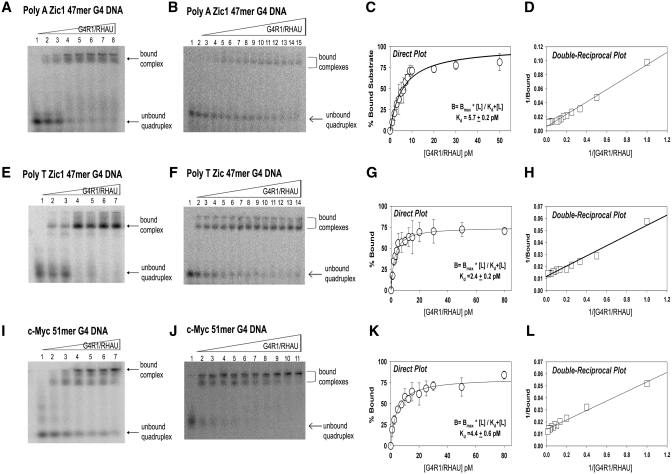
Figure 2.
Equilibrium binding GMSAs of purified recombinant G4R1/RHAU incubated with unimolecular G4-DNA-containing oligonucleotides yield apparent Kd values in the low picomolar range. (A, B, E, F, I and J) are representative (of three repetitions) phosphorimages of non-denaturing gel electropherograms of GMSAs; the top triangles indicate the direction of increasing G4R1/RHAU concentration. GMSA of 1 pM 5′-[32P]-labeled (A) unimolecular Poly A Zic1 47-mer G4-DNA, (E) unimolecular Poly T Zic1 47-mer G4-DNA, (I) c-Myc 51-mer G4-DNA incubated with a broad range of concentrations of G4R1/RHAU (lane 1, 0 pM; lane 2, 1 pM; lane 3, 5 pM; lane 4, 10 pM; lane 5, 20 pM; lane 6, 40 pM; lane 7, 80 pM). GMSA of 1 pM 5′-[32P]-labeled unimolecular (B) Poly A Zic 47-mer G4-DNA, (F) Poly T Zic1 47-mer G4-DNA, (J) c-Myc 51-mer G4-DNA incubated with concentrations of G4R1/RHAU that were increased in small (0.5–1 pM) increments between (B) lanes 1–12, (F) lanes 1–10, or (J) lanes 1–6, followed by larger increments up to 80 pM. Binding data for Poly A Zic1 47-mer G4-DNA, Poly T Zic1 47-mer G4-DNA, and c-Myc 51-mer G4-DNA were directly fit by non-linear regression to a hyperbolic equation (Panels C, G and K, respectively) and to a double reciprocal linear equation by linear regression (Panels D, H and L, respectively). Error bars are Mean ± SD, n = 3 independent experiments.
We initially studied binding of G4R1/RHAU to an oligonucleotide (termed Poly A Zic1 DNA 47-mer) with d(pA)10 tails attached to both the 5′ and 3′of a 27-mer sequence present in the 5′ untranslated region of the coding sequence of the transcription factor Zic1 (Table 1). The d(pA)10 tails were added as unstructured ends that might maximize performance of the enzyme. Nearly 80% of the 1 pM Poly A Zic1 G4-DNA 47-mer was bound at a G4R1/RHAU concentration of 10 pM (Figure 2A, lane 4). The bound complexes formed a broad band of electrophoretic mobility in the higher molecular weight region of the gel, demonstrating retarded migration. In order to determine the apparent Kd for the Poly A Zic1 G4-DNA:G4R1/RHAU interaction, a fine titration was performed and the concentration of G4R1/RHAU was raised in 0.5–1 pM increments in the binding reactions (Figure 2B). The percent bound Poly A Zic1 G4-DNA as a function of G4R1/RHAU concentration was fit directly using Equation (1) (Figure 2C) and via a double reciprocal plot (Figure 2D) to a hyberbolic, saturable, single site binding model. Kd for the Poly A Zic1 DNA:G4R1/RHAU interaction was 5.7 ± 0.2 pM.
To determine if the d(pA)10 sequence influenced the low apparent Kd observed, we replaced the d(pA)10 tails with d(pT)10 tails of the same length and repeated the GMSA. At a concentration of 10 pM of G4R1/RHAU >80% of 1 pM of Poly T Zic1 G4-DNA 47-mer DNA bound the enzyme (Figure 2E, lane 4). The bound Poly T Zic1 G4-DNA-enzyme complex appeared as a main single band with a faster electrophoretic mobility than the Poly A Zic1 G4-DNA 47-mer-enzyme complex (compare Figure 2A and E). In addition to the main band of Poly T Zic1 G4-DNA:G4R1/RHAU complex, slower mobility molecular complexes were also observed in the upper most regions of the gel. Careful titration of the G4R1/RHAU concentration in the binding reactions (Figure 2F) demonstrated a Kd for the Poly T Zic1 G4-DNA:G4R1/RHAU interaction of 2.4 ± 0.2 pM (Figure 2G and H). Binding of the G4-DNA-containing c-Myc 51-mer of complete native sequence (without 3′- or 5′-unstructured sequence tags) to G4R1/RHAU was also evaluated by GMSA. Similar to the Zic1 G4-DNA oligonucleotides, >50% of 1 pM c-Myc G4-DNA 51-mer was shifted in the presence of 10 pM G4R1/RHAU (Figure 2I, lane 3). The bound complexes appeared as two main band regions in the gel, with the most prominent complex observed with the slowest mobility band near the loading origin of the gel, suggesting possible self-association of G4-DNA:G4R1/RHAU bound complexes. Careful titration of the G4R1/RHAU concentration in the binding reactions (Figure 2J) demonstrated a Kd for the c-Myc 51-mer G4-DNA:G4R1/RHAU interaction of 4.4 ± 0.6 pM (Figure 2K and L). Overall, GMSA results indicate the enzyme possesses a remarkably tight and similar dissociation constant for each of the G4-DNA-containing oligonucleotides.
In order to determine whether tight binding of G4R1/RHAU is specific for oligodeoxyribonucleotides that contain G4-DNA structures, we measured apparent binding affinity of the enzyme for a number of 5′-[32P]-end-labeled control oligonucleotides (Figure 3A–D), each of which did not contain G4-DNA structure by the CD criteria discussed above (Figure 1I and J and data not shown). In all cases, G4R1/RHAU concentrations were titrated up to 300 pM. In most cases there was little or no detectable binding. Minimal binding of the scrambled Zic1 oligonucleotide by G4R1/RHAU was observed in Figure 3B (lanes 4–7); however, the percentage of DNA oligonucleotide bound never approached 50% (the concentration for an apparent Kd), even in the presence of 300 pM enzyme. Therefore, it is clear that if G4R1/RHAU binds any of these non-G4-DNA-containing oligonucleotides, the apparent Kd for the bound complex is more than −50-fold higher than the apparent Kd’s observed for G4R1/RHAU-unimolecular G4-DNA complexes.
The question remained, however, whether the tight binding interaction of G4R1/RHAU with G4-DNA sequence-containing oligonucleotides was based solely on recognition and binding of G4-DNA structure, or would a guanine-rich sequence, with or without G4-DNA structure, suffice to interact tightly with G4R1/RHAU? In order to address this question, the 33-mer DNA oligonucleotide Z33, which contains runs of d(pG) (Table 1) that self-anneal into a parallel stranded tetramolecular G4 structure (36), was incubated with increasing concentrations of G4R1/RHAU in either an unstructured single-stranded form, or in tetramolecular G4-DNA form. Oligonucleotide Z33 did not bind G4R1/RHAU when present in unstructured single stranded form (Figure 3E, lanes 1–7). In contrast, G4-DNA-formed Z33 tightly bound G4R1/RHAU, even in the presence of molar excess of unstructured single-stranded Z33 (Figure 3E, lanes 8–14).
An additional test was performed to confirm that the G4R1/RHAU binding interaction with d(pG) run-containing DNA oligonucleotides requires G4-DNA structure. Poly A Zic1 DNA 47-mer was treated with dimethyl sulfate in order to methylate guanine N7, thereby preventing guanine quartet formation by Hoogsteen bonding. Dimethyl sulfate treated Poly A Zic1 DNA 47-mer did not bind G4R1/RHAU (Supplementary Figure S3). Taken together, these data indicate that tight G4R1/RHAU binding of oligonucleotides containing d(pG) runs requires the presence of G4-DNA structure, not simply a guanine-rich sequence.
Finally, it was determined whether the low Kd of the G4-DNA:G4R1/RHAU binding interaction was a result of slow release of bound G4-DNA, as is often the case for tight protein:ligand interactions. Release of bound Poly A Zic1 47-mer G4-DNA from a complex with G4R1/RHAU was determined by incubating a complex containing 1 pM 5′-[32P]-labeled Poly A Zic1 G4-DNA bound to 200 pM G4R1/RHAU at 37°C in the presence of 20 nM unlabeled Poly A Zic1 G4-DNA. This is a 20 000-fold molar excess of unlabeled to labeled G4-DNA and a 100-fold molar excess of unlabeled G4-DNA to enzyme. Figure 3F, lane 2 shows that this added cold blocking material prevents G4R1/RHAU from appreciably binding [32P]-labeled G4-DNA. A nonlinear regression analysis of loss of binding with time produced a t1/2 of binding of 67 ± 9 h. These data indicate a koff of 2.9 × 10−6/s. Given that the GMSA data displayed in Figure 2 indicates a Kd for the Poly A Zic1 G4-DNA:G4R1/RHAU interaction of ∼5 pM, the predicted kon is 5.2 × 105/M/s is reasonable for a diffusion based event. This experiment independently confirms the tight binding of unimolecular G4-DNA by G4R1/RHAU in a manner independent of quantification of the concentration of G4R1/RHAU enzyme.
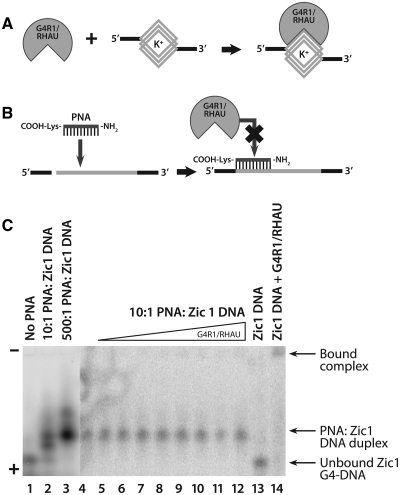
Hybridization of a complementary PNA to the Poly A Zic1 DNA 47-mer forms a complex that does not contain G4-DNA structure and is not bound tightly by G4R1/RHAU
The assertion that G4R1/RHAU binds directly to G4-DNA structure within unimolecular DNA oligonucleotides was further tested by specifically inhibiting the formation of the G4 structure with minimal disruption of the overall base accessibility of the oligonucleotide. Base pair hybridization with a Watson–Crick complementary PNA was used to inhibit G4-DNA formation of the Poly A Zic1 DNA 47-mer. PNAs are synthetic DNA mimics for which N-(2-aminoethyl) glycine moieties replace the sugar phosphate backbone of DNA. In a PNA the bases are arranged on a backbone that typically has no negative charge. Therefore, PNAs bind and base pair to target sequences with significantly greater stability than cognate DNA oligonucleotides, and unlike DNA:DNA duplexes, the stability of PNA:DNA duplexes remains high in low salt conditions. Previously, PNAs have been experimentally utilized for trapping putative G4-forming DNA oligonucleotides in a non-G4 state (45,46).
A 12-mer PNA sequence, 5′-NH2-CCCCCCCCACCC-lysine that is complementary to positions 11–23 d(pGGGTGGGGGGGG) on the Poly A Zic1 DNA 47-mer sequence was tested for its ability to inhibit the formation of G4-DNA by the Poly A Zic1 DNA oligonucleotide. Figure 4C, lane 1 shows the mobility following a boiling and cooling cycle of monomeric Poly A Zic1 DNA 47-mer upon non-denaturing gel electrophoresis. The addition of PNA at a 10:1 PNA:DNA molar ratio prior to a boiling and cooling cycle (Figure 4C, lane 2) caused the appearance of two new bands of slower electrophoretic mobility. CD spectroscopy of these two new bands which were purified electrophoretically (Supplementary Figures S4 and S5), as well as stoichiometric analysis (data not shown) indicated that the lower of the two new bands (denoted as Band 2 in Supplementary Figures S4 and S5) is a unimolecular form of Poly A Zic1 47-mer that may be a structural isomer of unimolecular Poly A Zic1 G4-DNA stabilized by the presence of the PNA, while the upper band (denoted as Band 3 in Supplementary Figures S4 and S5) is a PNA:DNA hybrid that does not possess G4 structure. Band 2 exhibited stabilization by increasing KCl concentration (Supplementary Figure S4B), which is characteristic of G4-DNA. The lack of G4 structure and the presence of the PNA:DNA hybrid was confirmed by its CD spectral behavior under no salt conditions; the molar ellipticity at 263 nm of the PNA:DNA hybrid was not destabilized by increasing temperature in the no salt condition, as expected for a PNA:DNA hybrid that maintains stability under low salts conditions (Supplementary Figure S5, upper right spectra). This behavior is expected from a hybrid containing PNA, but not from a G4-DNA structure. The 263 nm peak of molar ellipticity of the upper band did exhibit temperature dependence in the presence of either LiCl or KCl; this temperature dependence may be due to a destabilization caused by salt shielding of a charged lysine at the carboxyl terminal of the PNA.
Figure 4.
Equilibrium binding GMSA of purified recombinant G4R1/RHAU incubated with Poly A Zic1 DNA 47-mer hybridized to a Watson–Crick complementary PNA shows little binding by the enzyme when G4-DNA structure is inhibited from forming. (A) Schematic drawing of G4R1/RHAU binding an oligonucleotide containing a G4-DNA structure. (B) Schematic drawing showing that hybridization of a complementary PNA to the G4-DNA-forming sequence inhibits high affinity binding by G4R1/RHAU. The GMSA shown in (C) suggests that schematics (A) and (B) are correct. (C) Phosphorimage of a representative (of three repetitions) nondenaturing gel electropherogram of a GMSA; the top triangle indicates the direction of increased G4R1/RHAU concentration. Electrophoretic mobility standards for the two Poly A Zic1 47-mer G4-DNA isomers and the DNA:PNA hybrid were created by incubating 1 nM 5′-[32P]-labeled Poly A Zic1 DNA 47-mer DNA in K-RES buffer (with MgCl2 and ATP) with 0 nM PNA (lane 1, showing fastest mobility G4-DNA isomer), 10 nM PNA (lane 2, showing the slower mobility G4-DNA isomer) and the PNA:DNA duplex, or 500 nM PNA (lane 3, showing PNA:DNA duplex); lanes 4–12, 1 pM 5′-[32P]-labeled PNA:Poly A Zic1 DNA duplex was incubated with increasing concentrations of G4R1/RHAU (lane 4, 0 pM; lane 5, 5 pM; lane 6, 10 pM; lane 7, 15 pM; lane 8, 30 pM; lane 9, 50 pM; lane 10, 100 pM; lane 11, 150 pM; lane 12, 300 pM); lane 13, 1 pM 5′-[32P]-labeled Poly A Zic1 47-mer G4-DNA in the absence of either complementary PNA or G4R1/RHAU; lane 14, 1 pM 5′-[32P]-labeled Poly Zic1 DNA 47-mer G4-DNA incubated with 30 pM G4R1/RHAU in the absence of complementary PNA.
Only the upper, non-G4 PNA:DNA hybrid band was observed after a 500:1 PNA:DNA molar ratio was taken through a boiling and cooling cycle (Figure 4C, lane 3). A slower mobility smear was also observed, which we speculate may represent a second, less stable PNA:DNA hybrid in which the PNA is Watson–Crick paired with the second longest d(pG) run in the Poly A Zic1 DNA 47-mer.
A 10:1 PNA:DNA molar ratio incubated at 55°C for 6 h produced only electrophoretic Band 3 (Figure 4C, lane 4) that was confirmed by CD to represent a PNA:DNA hybrid due to CD signal behavior in the no salt conditions (Supplementary Figure S5). This non-G4 PNA:DNA hybrid was assessed for G4R1/RHAU binding. Figure 4A shows an idealized schematic of G4R1/RHAU recognizing a G4-DNA site and showing the protein binding to that site, thereby forming a protein–nucleic acid complex that retards the electrophoretic mobility of the nucleic acid. We believe this mechanism explains the observed binding with G4R1/RHAU:G4-DNA-containing oligonucleotide complexes (Figure 2). Figure 4B illustrates the expected outcome regarding the interaction of G4R1/RHAU with a PNA:Poly A Zic1 47-mer DNA hybrid. PNA hybridization inhibits G4-DNA formation, which in turn, prevents G4R1/RHAU binding if the enzyme indeed recognizes G4-DNA structure. Consistent with this predicted outcome, titration of G4R1/RHAU concentrations of up to 300 pM with 1 pM PNA:DNA hybrid did not demonstrate any detectable enzyme–hybrid complex formation, as evidenced by GMSA (Figure 4C, lanes 5–12). In contrast, a comparison of G4 structure-containing Poly A Zic1 DNA 47-mer monomer electrophoresed in the absence (Figure 4C, lane 13) or presence (Figure 4C, lane 14) of 30 pM of G4R1/RHAU clearly demonstrates a slow mobility band in the presence of G4R1/RHAU indicative of a bound enzyme:G4-containing monomer complex. These results demonstrate that the Poly A Zic1 DNA 47-mer has a substantially poorer affinity for G4R1/RHAU when hybridized with PNA. The result is consistent with G4R1/RHAU recognizing the G4-DNA structure in the Poly A Zic1 DNA 47-mer. It should be noted that the phosphorimage contrast in Figure 4C, lanes 4–14 was enhanced to better detect additional bands due to gel mobility shifts. Additional controls (Supplementary Figure S6) demonstrated that excess unlabeled PNA does not block G4R1/RHAU binding to G4-DNA and that PNA:DNA hybrids do not bind G4R1/RHAU when produced without excess unlabeled PNA. Therefore, when G4-DNA structure is lost the enzyme does not bind.
A PNA trap assay shows that G4R1/RHAU unwinds G4-DNA structure, enabling it to hybridize to a complementary PNA
In the experiments presented above, a complementary PNA was shown to hybridize to the G4-DNA-forming sequence of the Poly A Zic1 DNA 47-mer upon heat destabilization of the G4-DNA structure. Hybridization occurred within the G4-DNA forming region of the oligonucleotide, producing a PNA:DNA hybrid devoid of G4-DNA structure as determined by CD (Supplementary Figure S5). This finding led us to consider whether active G4R1/RHAU in the presence of ATP could destabilize Poly A Zic1 G4-DNA (i.e. open by unwinding the G4-DNA structure) at 37°C in 30 min and allow significant PNA hybridization, as is accomplished by long term incubation of the Poly A Zic1 DNA 47-mer with PNA at 55° C or by heating (95°C) and cooling cycles in high concentrations of PNA (Figure 4C, lane 3). Observation of a PNA:DNA hybrid in the presence of enzyme would indicate that, not only does G4R1/RHAU bind G4-DNA-containing oligonucleotides, but the enzyme opens the unimolecular G4 structure as well. Our PNA trap assay was largely modeled upon a previous study that assayed the kinetics of the folding and unfolding of telomere DNA oligonucleotide in the presence of a complementary PNA trap (45). Kinetic considerations (7) indicate that if G4R1/RHAU unwound Poly A Zic1 DNA 47-mer G4-DNA structure under physiological conditions and then released the unstructured product, the oligonucleotide would likely snap back within seconds into a G4-DNA structure, producing the futile cycle as diagramed in Figure 5A. However, if a molar excess of base-complementary PNA were present during the resolution reaction, the PNA could hybridize and trap the opened structure oligonucleotide as a PNA:DNA hybrid (Figure 5B).
Non-denaturing gel electrophoretic analysis of the enzymatic PNA:DNA hybrid trap assay demonstrated G4R1/RHAU-catalyzed unwinding of Poly A Zic1 DNA 47-mer G4-DNA (Figure 5C). Total of 100 pM Poly A Zic1 DNA oligonucleotide was incubated at 98°C with 0, 10, 100 nM, or 10 μM of complementary PNA, respectively. After a 10 min incubation the material was cooled to room temperature, then analyzed electrophoretically (Figure 5C, lanes 1–4). In the absence of PNA, two bands that represent two structural isomers of Poly A Zic1 DNA 47-mer were observed (Figure 5C, lane 1). In the presence of 10–100 nM complementary PNA, the slower mobility G4-DNA containing structural isomer of Poly A Zic1 DNA 47-mer is present, but not the faster mobility isomer (Figure 5C, lanes 2 and 3) suggesting that the PNA stabilizes one structural isomer of Poly A Zic1 DNA 47-mer without forming a stable PNA:DNA hybrid. Increasing amounts of a third slower mobility band was observed as the PNA concentration increased from 10 nm to 10 μM (Figure 5C, lanes 2–4). CD of a purified band of similar mobility suggested this band was a PNA:DNA hybrid (Supplementary Figure S5). Nearly all of the Poly A Zic1 DNA 47-mer was found in the PNA:DNA hybrid band in the presence of 10 μM PNA (Figure 5C, lane 4); there was also a smear of even slower mobility species observed in the presence of 10 μM PNA (Figure 5C, lane 4). We speculate that this smear represents other less stable PNA:DNA hybrids. A small amount of a slower migrating band (present at approximately half way down the gel) was also observed in all lanes of the electropherogram except when catalytically active G4R1/RHAU and ATP were present (Figure 5C, lanes 7 and 8, 18–20); it is possible that this band represents a multimolecular Poly a Zic1 G4-DNA that is resolved by active G4R1/RHAU.
When Poly A Zic1 G4-DNA 47-mer was incubated in the presence of G4R1/RHAU in resolution buffer containing 100 mM KCl and 10 mM ATP, but without complementary PNA, only substrate Poly A Zic1 47-mer G4-DNA (principally faster migrating G4-DNA isomer, with a very small amount of slower mobility G4-DNA isomer) and increasing amounts of slow mobility G4R1/RHAU-oligonucleotide complexes were observed as the enzyme concentration increased (Figure 5C. lanes 5–8). This result was similar to the GMSAs of G4R1/RHAU incubated with PolyA Zic1 G4-DNA in the presence of EDTA discussed earlier (compare Figure 5C, lanes 5–8 with Figure 2A). The addition of 10 nM complementary PNA under identical conditions changed the products observed after G4R1/RHAU-Poly A Zic1 G4-DNA 47-mer incubation (Figure 5C, lanes 15–20). In the presence of complementary PNA, but without G4R1/RHAU, Poly A Zic1 DNA 47-mer only adopted a conformation with the electrophoretic mobility of the upper G4-DNA band (compare Figure 5C, lanes 9 and 15). However, as increasing amounts of G4R1/RHAU were titrated into the reaction (Figure 5C, lanes 16–20), the amount of the G4-DNA band decreased, while the PNA:DNA duplex band increased in intensity. This observation suggests that the complementary PNA hybridizes and traps the DNA oligonucleotide in a PNA:DNA hybrid once the G4-DNA structure has been resolved by G4R1/RHAU. In order to assess whether formation of the PNA:DNA hybrid was dependent upon the catalytic activity of intact G4R1/RHAU, the Poly A Zic1 47-mer containing G4-DNA was incubated with complementary PNA in the presence of increasing concentrations of boiled (denatured) G4R1/RHAU. In these reactions there was no increase in the appearance of the PNA:DNA duplex (Figure 5C, lanes 10–14), although a slow mobility smear was observed at the highest G4R1/RHAU concentration (Figure 5C, lane 14), suggesting the boiled enzyme was still capable of some binding of DNA, albeit not as efficiently as the native enzyme (Figure 5C, lanes 5–8). These results indicate that G4R1/RHAU catalytically opens the G4 structure of the Poly A Zic1 DNA 47-mer, allowing the PNA to hybridize to the complementary sequence in the DNA oligonucleotide; denatured G4R1/RHAU cannot open the G4-DNA structure to allow PNA/DNA duplex formation.
In order to further demonstrate that unwinding of Poly A Zic1 DNA 47-mer with formation of a PNA:DNA duplex in the presence of G4R1/RHAU required catalytic activity of G4R1/RHAU, resolution reactions were carried out in which wild-type G4R1/RHAU was replaced by G4R1/RHAU containing a E→A substitution mutation at amino acid position 335 in the Walker B box. The ATPase activity of G4R1/RHAU is abolished by this mutation (40). No PNA:DNA duplex band was observed after incubation with mutant G4R1/RHAU with 1 nM G4-DNA (Figure 5D, lanes 2–6), although slow mobility bound mutant G4R1/RHAU:G4-DNA complexes were observed. In contrast, PNA:DNA duplex formation was observed after incubation with wild type G4R1/RHAU at concentrations identical to those of mutant G4R1/RHAU (Figure 5D, lanes 14–18). PNA:DNA duplex formation in the presence of wild-type G4R1/RHAU was not observed if ATP was absent from the reaction (Figure 5D, lanes 8–12), or if 10 mM AMP-PNP, the non-hydrolyable ATP analog, was substituted for ATP (Supplementary Figure S7). These data confirm that formation of PNA:DNA duplex in the presence of G4R1/RHAU, indicative of G4-DNA unwinding, requires G4R1/RHAU ATPase activity.
Competition studies indicate that unimolecular G4-DNA containing-oligonucleotides inhibit G4R1/RHAU-catalyzed resolution of tetramolecular G4-DNA; the c-Myc oligonucleotide is an effective competitor at competitor:substrate molar ratios <<1
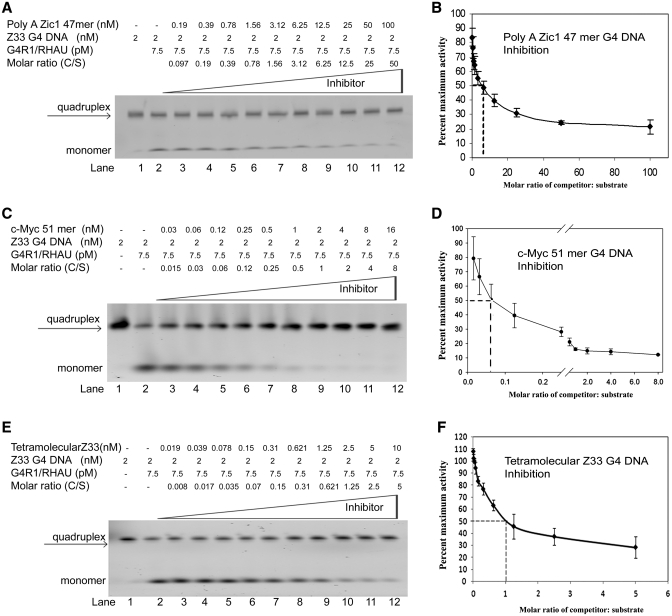
Inhibition of the resolution of 5′-TAMRA-labeled tetramolecular G4-DNA Z33 oligonucleotide into single-stranded DNA was assessed by titrating increasing molar ratios of unlabeled unimolecular G4-DNA-containing oligonucleotides into resolution reactions consisting of a fixed amount of G4R1/RHAU and 2 nM 5′-TAMRA-labeled tetramolecular G4-DNA substrate (Figure 6). Upon completion of the resolution reaction, the extent of unwinding of the tetramolecular structure was analyzed by non-denaturing gel electrophoresis (Figure 6A, C and E). The substrate target was preformed tetramolecular G4-DNA of >95% purity (Figure 6A, lane 1; Figure 6C, lane 1; Figure 6E, lane 1). Sufficient G4R1/RHAU was present in each reaction to produce ∼50% resolution of tetramer into monomer in the absence of competitor (Figure 6A, lane 2; Figure 6C, lane 2; Figure 6E, lane 2), corresponding to a G4R1/RHAU concentration of 7.5 pM. We have previously defined 1 U of enzyme to be the amount of enzyme that can unfold 0.2 pmol of tetramolecular Z33 quadruplex in 30 min at 37°C (36). Addition of increasing amounts of unlabeled unimolecular G4-DNA-containing Poly A Zic1 DNA 47-mer (Figure 6A, lanes 3–12), c-Myc 51-mer (Figure 6C, lanes 3–12), or tetramolecular pZ33 G4-DNA (Figure 6E, lanes 3–12) caused a competitor concentration-dependent decrease in the rate of G4R1/RHAU-catalyzed tetramolecular G4-DNA resolution. The c-Myc 51-mer unimolecular G4-DNA quadruplex inhibited the enzymatic resolution of tetramolecular G4-DNA substrate at lower inhibitor:substrate molar ratios than did the Poly A Zic DNA 47-mer (compare Figure 6C to A).
Figure 6.
G4R1/RHAU-catalyzed resolution of 5′-TAMRA-labeled tetramolecular G4-DNA substrate is inhibited most efficiently by unlabeled unimolecular c-Myc 51-mer G4-DNA. (A, C and E) Phosphorimage of representative (of three repetitions) nondenaturing gel electropherograms; the top triangles indicate increasing concentrations of unlabeled (A) Poly A Zic1 47-mer G4-DNA, (C) c-Myc 51-mer G4-DNA, or (E) tetramolecular Z33 G4-DNA added to G4R1/RHAU-catalyzed resolving reactions of 5′-TAMRA-labeled tetramolecular Z33 G4-DNA substrate. Lane 1 of panels A, C and E: 2 nM 5′-TAMRA-labeled tetramolecular Z33 G4-DNA substrate in the absence of enzyme or unlabeled unimolecular G4-DNA. Lanes 2–12 of panels A, C and E: 2 nM 5′-TAMRA-labeled tetramolecular Z33 G4-DNA substrate after incubation with 1 unit of G4R1/RHAU plus increasing concentrations of unlabeled inhibitor G4-DNA. (B, D and F) Graphic representation of concentration-dependent inhibition of G4R1/RHAU-catalyzed resolution of 5′-TAMRA-labeled tetramolecular Z33 G4-DNA substrate by unlabeled (B) Poly A Zic1 47-mer G4-DNA, (D) c-Myc 51-mer G4-DNA, or (F) tetramolecular Z33 G4-DNA. Inhibition is represented by the percentage of maximal resolution (error bars are Mean ± SD, n = 3 independent experiments). Inhibition of 50% is observed at a 6.25:1 unimolecular Poly A Zic1 DNA 47-mer G4-DNA:Z33 tetramer G4-DNA molar ratio, at a 0.06:1 unimolecular c-Myc DNA 51-mer:Z33 tetramer G4-DNA molar ratio, and at a 1:1 unlabeled Z33 tetramer G4-DNA:labeled Z33 tetramer G4-DNA molar ratio.
The data were analyzed graphically as the percent of maximal activity (([activity measured with competitor at given molar ratio]/[activity measured without competitor]) × 100) as a function of the unlabeled competitor:labeled substrate molar ratio (Figure 6B, D and F). We have previously used this assay to demonstrate that 300-fold molar excesses of Y form DNA, 5′ overhang duplex DNA and 3′ overhang duplex DNA were required to inhibit resolution of 5′-TAMRA-labeled tetramolecular Z33 G4-DNA as effectively as unlabeled tetramolecular G4-DNA (36). As expected, addition of unlabeled tetramolecular pZ33 G4-DNA inhibited resolution of 5′-TAMRA-labeled tetramolecular Z33 G4-DNA by 50% at a 1:1 molar ratio of unlabeled:labeled DNA (Figure 6F). Poly A Zic1 47-mer DNA caused 50% inhibition of G4R1/RHAU-catalyzed resolution of tetramolecular G4-DNA at a 6:1 competitor:substrate molar ratio (see dotted line at abscissa intercept in Figure 6B), indicating that it is a much better inhibitor than Y form DNA and duplex DNA from previous studies (36). However, the c-Myc 51-mer caused 50% inhibition at a 0.06:1 unlabeled competitor:labeled substrate molar ratio, indicating that it inhibits resolution of tetramolecular G4-DNA ∼100-fold better than the Poly A Zic1 DNA 47-mer G4-DNA.
DISCUSSION
Previously our laboratory identified G4R1/RHAU as the protein responsible for the majority of HeLa cell tetramolecular G4-DNA resolving activity by using a highly stringent affinity chromatography approach selective for tetramolecular G4-DNA binding proteins (36). Selection for specificity was achieved by utilizing over a 100-fold ratio of blocking single-stranded d(pHHN)11 DNA to target tetramolecular G4-DNA (attached to paramagnetic beads) (36). d(pHHN)11 DNA was chosen as a non-specific blocking DNA because the sequence could not form intramolecular or intermolecular G4-DNA structures. We expected the isolated protein would bind tightly to tetramolecular G4-DNA. Indeed, recombinant G4R1/RHAU was shown to bind tightly to tetramolecular G4-DNA with a Kd of 77 ± 6 pM (37). However, it remained unknown whether G4R1/RHAU could bind and unwind unimolecular G4-DNA. Although tetramolecular G4-DNA and unimolecular G4-DNA share many structural commonalities, there are differences between the structures, such as DNA looping found in unimolecular G4-DNA structures, which could present steric interference to enzymatic recognition. In addition, the issue of whether G4R1/RHAU recognizes the unimolecular G4-DNA structure addresses the potential physiologic role(s) of this enzyme because the formation of unimolecular G4-DNA has a low kinetic barrier of formation under intracellular conditions, and unimolecular G4-DNA is the most common G4-DNA structure expected to form in vivo. Therefore, the current study was essential to determine whether G4R1/RHAU could specifically bind to unimolecular G4-DNA and open that structure.
The results presented here indicate that G4R1/RHAU binds unimolecular G4-DNA with remarkable tightness (Kd of 5.7 ± 0.2 pM for Poly A Zic1 47-mer and 4.4 ± 0.6 pM for c-Myc). These estimations of Kd have at least one fundamental caveat: the accuracy of the estimates of Kd is dependent upon accurate protein quantification through estimating Coomassie staining of G4R1/RHAU protein against standards of known protein quantities on an SDS gel. Comparison of mass estimation of purified proteins, e.g. bovine serum albumin or E. coli β-galactosidase, by ultraviolet light absorption spectroscopy with estimates obtained by Coomasie staining of these proteins in SDS gels indicate that the two methods differ by 20%–49% (<2-fold, data not shown). Therefore, the Kd estimate reported here could be off by as much as 2-fold due to the method of estimation of protein content. Additionally, the modest free enzyme concentrations required for measuring these low Kd’s can be diminished due to significant amounts of protein being bound to DNA during binding reactions, thus limiting the precision of the Kd determinations to upper boundary estimates in these cases. However, despite these qualifications, the conclusion stands that G4R1/RHAU binds G4-DNA with remarkably tight binding affinity. In addition, a determination of the t1/2 of binding at 67 ± 7 h made in this study is protein concentration independent and further affirms the low Kd and tight binding results found for G4R1/RHAU interaction with unimolecular G4-DNA.
To our knowledge, these are the lowest Kd’s reported for a protein binding to G4-DNA. The next lowest Kd’s in the literature for protein binding to G4-DNA is 77 pM for G4R1/RHAU binding to tetramolecular DNA (37) and 79 pM for the mouse otholog mXRN1p of the yeast Xrn1 or Kem1 protein binding with tetramolecular DNA (47). The affinity found between G4R1/RHAU and unimolecular G4-DNA is tighter than that found in most antigen-antibody binding interactions (48). The G4R1/RHAU unimolecular G4-DNA binding interaction has substantial ligand specificity, considering that control DNA sequences not possessing G4-DNA structure bind G4R1/RHAU with at least a 50-fold poorer magnitude of affinity. Even DNA oligonucleotides with G-rich regions capable of forming quadruplex DNA but not participating in a G4-DNA structure, were not bound tightly by G4R1/RHAU. Furthermore, inhibition of G4-DNA formation within the PQS of Poly A Zic1 DNA 47-mer by a PNA 12-mer whose base sequence was complementary to the PQS of the oligonucleotide abolished the tight affinity for G4R1/RHAU observed when the oligonucleotide possesses G4-DNA structure. These observations indicate that the presence of G4-DNA structure is required by G4R1/RHAU binding and that the G4R1/RHAU binding site within Poly A Zic1 DNA 47-mer is localized to within its unimolecular G4-DNA forming sequence.
G4R1/RHAU specifically unwinds G4-DNA, allowing a complementary PNA to hybridize within the G4-forming sequence. This ‘PNA trap’ approach has been used previously to study the kinetics of unimolecular G4-DNA opening and folding (46). This study represents the first use of a PNA trap to demonstrate enzymatic unwinding of unimolecular G4-DNA. However, at least three other methods have been used to study unimolecular G4-DNA unwinding by proteins; these approaches include CD spectroscopy, fluorescence quenching and surface plasmon resonance (SPR) (49–54). All of these approaches are relatively insensitive in comparison to the PNA trap method that can demonstrate resolution of 1 pmol of unimolecular G4-DNA by 0.5 pmol of enzyme in a 30 μl reaction volume. In comparison to these previously reported techniques for assessing resolution of unimolecular G4-DNA, the PNA trap assay retains the advantages of: (i) direct observation of the resolved DNA; (ii) enhanced sensitivity of detection of enzyme-catalyzed G4-DNA resolution; and (iii) the ability to demonstrate complete unwinding events. One disadvantage that we have observed with the PNA trap is that, in the presence of Mg2+, PNA can impede binding of G4-DNA to G4R1/RHAU (data not shown), possibly slowing the kinetics of the unwinding reaction. Blocking of the G4-DNA:G4R1/RHAU binding interaction does not occur in the absence of free Mg2+ ions (Supplementary Figure S6, lanes 14 and 15).
Comparison with other proteins such as RPA, BLM and T-Ag (52–54) that have been characterized for binding unimolecular G4-DNA highlights the unique G4-DNA binding properties of G4R1/RHAU. First, G4R1/RHAU shows significantly more specificity in binding G4-DNA than unstructured single-stranded DNA (Figure 3). In contrast, RPA (52) and T-Ag (54) do not preferentially bind G4 versus unstructured single-stranded DNA. With regard to BLM (52), there is current debate whether this enzyme has a greater specificity for G4-DNA structures versus Watson–Crick duplex DNA. G4R1/RHAU also displays a much tighter binding affinity for unimolecular G4-DNA (Kd in the low pM range) than does RPA (Kd in the low µM range) (52), or BLM (Kd of ∼4 nM for binding of tetramolecular G4 DNA (55). The SPR technique could not accurately report a Kd for G4-DNA binding by T antigen; however, the working concentration range for T antigen interaction was in the low nanomolar range (54). It is also of interest that G4R1/RHAU tightly binds unimolecular G4-DNA structures without a requirement for divalent cations or ATP. In contrast, RPA, BLM and T-Ag require divalent cations for G4-DNA binding (52–54); T-Ag requires both Mg2+ and ATP (54). Although ATP is not required for G4 DNA binding by G4R1/RHAU, the PNA trap demonstrates that ATP is required for G4R1/RHAU-catalyzed G4-DNA resolution.
We tested two unimolecular-G4-DNA-containing oligonucleotides to determine how well each oligonucleotide could inhibit the ability of G4R1/RHAU to unwind a tetramolecular G4-DNA substrate. The stoichiometric ratio of c-Myc oligonucleotidecompetitor:G4R1/RHAU concentration (50% inhibition at an inhibitor:substrate molar ratio of 0.06:1) is consistent with a simple competitive binding model of inhibition. However, the poor inhibition of resolution of tetramolecular G4-DNA substrate by Poly A Zic1 G4-DNA, relative to the ∼10-fold lower Kd of G4R1/RHAU for unimolecular versus tetramolecular G4-DNA is not consistent with this model. The explanation for why inhibition of G4R1/RHAU catalyzed unwinding of tetramolecular G4-DNA by Poly A Zic1 G4-DNA is weak despite its tighter binding to G4R1/RHAU is currently unknown; however, a possibility that may be considered (among many) is that the efficiency of inhibition is influenced by the catalytic unwinding rate of the G4-DNA. The unlabeled competing unimolecular G4-DNAs are substrates competing for the enzyme active sites with labeled tetramolecular G4-DNA substrate. It is possible that Poly A Zic1 G4-DNA is being catalytically unwound by and released as single-stranded DNA from G4R1/RHAU at a faster rate than is the tetramolecular G4-DNA, thereby making Poly A Zic1 DNA a less efficient inhibitor. In contrast, G4R1/RHAU may unwind c-Myc more slowly, in which case the enzyme spends a greater residence time engaged catalytically to the c-Myc 51-mer, making it a better competitor.
G4R1/RHAU is the protein product of the DHX36 gene, a member of the DEAH-box family of helicases (36). Some members of the DEAH-box family of helicases are known to have helicase activity on duplex RNA or DNA substrates and in some cases both RNA and DNA. The closest characterized enzyme homolog to G4R1/RHAU is the nuclear DNA helicase II (NDH II), which is also known as DHX9 or RNA helicase A. NDH II appears to have roles in both DNA and RNA metabolism (56). A number of lines of evidence have already suggested that G4R1/RHAU is involved with RNA processing. G4R1/RHAU was originally identified by affinity chromatography associated with mRNA of the AU-rich element derived from the urokinase plasminogen activator gene (40). G4R1/RHAU was shown to interact with PARN, the exosome, and target RNAs to enhance decay of ARE(uPA)-mRNAs (40). More recently is has been shown that G4R1/RHAU also accumulates in stress granules after arsenite stress, suggesting it is involved in modulating changes in RNA metabolism in response to stress (57). However, the majority of G4R1/RHAU-GFP fusion protein is observed in the nucleus (58) suggesting the potential of DNA interactions and the potential of early transcript RNA processing interactions as roles for the enzyme. It should be noted that RNA readily forms unimolecular G4 structures; G4R1/RHAU may also have a role in processing unimolecular G4-RNA in cells as well, and we have previously shown that it can recognize and unwind tetramolecular G4-RNA (37). However, preliminary experiments with RNA in our laboratory have suggested that, although G4R1/RHAU does not tightly bind RNA non-specifically, it does recognize other sequences or structural motifs in RNA in addition to tightly binding to G4-RNA. These preliminary observations suggest a significantly more complicated binding interaction with RNA than DNA. These G4R1/RHAU-RNA binding interactions are currently under further investigation in our laboratory.
Our data suggest that it is most likely that G4R1/RHAU interacts with both DNA and RNA in cells. Although DNA interaction may be statistically less common per protein binding event, it may still potentially be of great biological importance. The amplifying nature of control of G4-DNA promoters or the control of telomere structures could have profound biological effects. If the enzyme does not act upon DNA, one would have to speculate the existence of special cellular mechanisms to exclude interaction with DNA. This study of G4R1/RHAU interactions with unimolecular G4-DNA highlights the ability of this enzyme to recognize and process the unimolecular G4-DNA structure. In the case of binding to G4-DNA, G4R1/RHAU exhibits extraordinarily tight binding affinity for G4-DNA, and the capability of directly destabilizing G4 structures, thereby enabling the G4 forming nucleotide sequence to be available for Watson–Crick type duplex hybridization. The data in this report open up a number of unimolecular G4-DNA sequence-containing targets as potential substrates for G4R1/RHAU. One can speculate that G4R1/RHAU might have direct activity at substrates such as the human telomere, G-rich promoters and G-rich nontranscribed strands of active genes. These potential targets of action suggest interesting future experiments to test the biological actions of this enzyme.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The National Institutes of Health (Grant P30-CA12197) and a gift from the Ware Foundation (to J.V.). National Institutes of Health Institutional Training (Grant T32-CA079448) (to B.G.). Funding for open access charge: A gift from the Sara Self Foundation.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Terrence Oas of the Duke University Medical Center for kindly allowing us to use their AVIV 202 CD spectrometer and Yu-Chu Chang for technical assistance with it. We thank Mark Lively of Wake Forest Health Sciences for helpful comments on the manuscript. We thank Nancy Maizels of University of Washington Medical Center for allowing us to use her G4-DNA schematics.
REFERENCES
- 1.Gellert M, Lipsett MN, Davies DR. Helix formation by guanylic acid. Proc. Natl Acad. Sci. USA. 1962;48:2013–2018. doi: 10.1073/pnas.48.12.2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Macaya RF, Schultze P, Smith FW, Roe JA, Feigon J. Thrombin-binding DNA aptamer forms a unimolecular quadruplex structure in solution. Proc. Natl Acad. Sci. USA. 1993;90:3745–3749. doi: 10.1073/pnas.90.8.3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Mergny JL, Phan A-T, Lacroix L. Following G-quartet formation by UV-spectroscopy. FEBS Lett. 1998;435:74–78. doi: 10.1016/s0014-5793(98)01043-6. [DOI] [PubMed] [Google Scholar]
- 4.Sen D, Gilbert W. A sodium-potassium switch in the formation of four-stranded G4-DNA. Nature. 1990;344:410–414. doi: 10.1038/344410a0. [DOI] [PubMed] [Google Scholar]
- 5.Prislan I, Lah J, Vesnaver G. Diverse polymorphism of G-quadruplexes as a kinetic phenomenon. J. Am. Chem. Soc. 2008;130:14161–14169. doi: 10.1021/ja8026604. [DOI] [PubMed] [Google Scholar]
- 6.Arthanari H, Bolton PH. Functional and dysfunctional roles of quadruplex DNA in cells. Chem. Biol. 2001;8:221–230. doi: 10.1016/s1074-5521(01)00007-2. [DOI] [PubMed] [Google Scholar]
- 7.Lane AN, Chaires JB, Gray RD, Trent JO. Stability and kinetics of G-quadruplex structures. Nucleic Acids Res. 2008;36:5482–5515. doi: 10.1093/nar/gkn517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sen D, Gilbert W. Formation of parallel four-stranded complexes by guanine-rich motifs in DNA and its implications for meiosis. Nature. 1988;334:364–366. doi: 10.1038/334364a0. [DOI] [PubMed] [Google Scholar]
- 9.Mergny JL, De Cian A, Ghelab A, Saccà B, Lacroix L. Kinetics of tetramolecular quadruplexes. Nucleic Acids Res. 2005;33:81–94. doi: 10.1093/nar/gki148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Williamson JR, Raghuraman MK, Cech TR. Monovalent cation-induced structure of telomeric DNA: the G-quartet model. Cell. 1989;59:871–880. doi: 10.1016/0092-8674(89)90610-7. [DOI] [PubMed] [Google Scholar]
- 11.Green JJ, Ying L, Klenerman D, Balasubramanian S. Kinetics of unfolding the human telomeric DNA quadruplex using a PNA trap. J. Am. Chem. Soc. 2003;125:3763–3767. doi: 10.1021/ja029149w. [DOI] [PubMed] [Google Scholar]
- 12.Marathias VM, Bolton PH. Determinants of DNA quadruplex structural type: sequence and potassium binding. Biochemistry. 1999;38:4355–4364. doi: 10.1021/bi982604+. [DOI] [PubMed] [Google Scholar]
- 13.Guedin A, Alberta P, Mergny JL. Stability of intramolecular quadruplexes: sequence effects in the central loop. Nucleic Acids Res. 2009;37:5559–5567. doi: 10.1093/nar/gkp563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Todd AK, Johnston M, Neidle S. Highly prevalent putative quadruplex sequence motifs in human DNA. Nucleic Acids Res. 2005;33:2901–2907. doi: 10.1093/nar/gki553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Huppert JL, Balasubramanian S. Prevalence of quadruplexes in the human genome. Nucleic Acids Res. 2005;33:2908–2916. doi: 10.1093/nar/gki609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maizels N. Dynamic roles for G4 DNA in the biology of eukaryotic cells. Nat. Struct. Mol. Biol. 2006;13:1055–1059. doi: 10.1038/nsmb1171. [DOI] [PubMed] [Google Scholar]
- 17.Lipps HJ, Rhodes D. G-quadruplex structures: in vivo evidence and function. Trends Cell Biol. 2009;19:414–422. doi: 10.1016/j.tcb.2009.05.002. [DOI] [PubMed] [Google Scholar]
- 18.Schaffitzel C, Berger I, Postberg J, Hanes J, Lipps HJ, Pluckthun A. In vitro generated antibodies specific for telomeric guanine-quadruplex DNA react with Stylonychia lemnae macronuclei. Proc. Natl Acad. Sci. USA. 2001;98:8572–8577. doi: 10.1073/pnas.141229498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Paeschke K, Juranek S, Simonsson T, Hempel A, Rhodes D, Lipps HJ. Telomerase recruitment by the telomere end binding protein-beta facilitates G-quadruplex DNA unfolding in ciliates. Nat. Struct. Mol. Biol. 2008;15:598–604. doi: 10.1038/nsmb.1422. [DOI] [PubMed] [Google Scholar]
- 20.Chang CC, Chu JF, Kao FJ, Chiu YC, Lou PJ, Chen HC, Chang TC. Verification of antiparallel G-quadruplex structure in human telomeres by using two-photon excitation fluorescence lifetime imaging microscopy of the 3,6-Bis(1-methyl-4-vinylpyridinium)carbazole diiodide molecule. Anal. Chem. 2006;78:2810–2815. doi: 10.1021/ac052218f. [DOI] [PubMed] [Google Scholar]
- 21.Duquette ML, Handa P, Vincent JA, Taylor AF, Maizels N. Intracellular transcription of G-rich DNAs induces formation of G-loops, novel structures containing G4 DNA. Genes Dev. 2004;18:1618–1629. doi: 10.1101/gad.1200804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Fry M, Loeb LA. Human Werner Syndrome DNA helicase unwinds tetrahelical structures of the Fragile X Syndrome repeat sequence d(CGG)n. J. Biol. Chem. 1999;274:12797–12802. doi: 10.1074/jbc.274.18.12797. [DOI] [PubMed] [Google Scholar]
- 23.Simonsson T, Pecinka P, Kubista M. DNA tetraplex formation in the control region of c-myc. Nucleic Acids Res. 1998;26:1167–1172. doi: 10.1093/nar/26.5.1167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Siddiqui-Jain A, Grand CL, Bearss DJ, Hurley LH. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl Acad. Sci. USA. 2002;99:11593–11598. doi: 10.1073/pnas.182256799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Qin Y, Rezler EM, Gokhale V, Sun D, Hurley LH. Characterization of the G-quadruplexes in the duplex nuclease hypersensitive element of the PDGF-A promoter and modulation of PDGF-A promoter activity by TMPyP4. Nucleic Acids Res. 2007;35:7698–7713. doi: 10.1093/nar/gkm538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Guo K, Pourpak A, Beetz-Rogers K, Gokhale V, Sun D, Hurley LH. Formation of pseudosymmetrical G-quadruplex and i-motif structures in the proximal promoter region of the RET oncogene. J. Am. Chem. Soc. 2007;129:10220–10228. doi: 10.1021/ja072185g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Connor AC, Frederick KA, Morgan EJ, McGown LB. Insulin capture by an insulin-linked polymorphic region G-quadruplex DNA oligonucleotide. J. Am. Chem. Soc. 2006;128:4986–4991. doi: 10.1021/ja056097c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lew A, Rutter WJ, Kennedy GC. Unusual DNA structure of the diabetes susceptibility locus IDDM2 and its effect on transcription by the insulin promoter factor Pur-1/MAZ. Proc. Natl Acad. Sci. USA. 2000;97:12508–12512. doi: 10.1073/pnas.97.23.12508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Arora A, Dutkiewicz M, Scaria V, Hariharan M, Maiti S, Kurreck J. Inhibition of translation in living eukaryotic cells by an RNA G-quadruplex motif. RNA. 2008;14:1290–1296. doi: 10.1261/rna.1001708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kumari S, Bugaut A, Huppert JL, Balasubramanian S. An RNA G-quadruplex in the 5′ UTR of the NRAS proto-oncogene modulates translation. Nat. Chem. Biol. 2007;3:218–221. doi: 10.1038/nchembio864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Eddy J, Maizels N. Conserved elements with potential to form polymorphic G-quadruplex structures in the first intron of human genes. Nucleic Acids Res. 2008;36:1321–1333. doi: 10.1093/nar/gkm1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kikin O, D’Antonio L, Bagga PS. QGRS Mapper: a web-based server for predicting G-quadruplexes in nucleotide sequences. Nucleic Acids Res. 2006;34:W676–W682. doi: 10.1093/nar/gkl253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harrington C, Lan Y, Akman SA. The identification and characterization of a G4-DNA resolvase activity. J. Biol. Chem. 1997;272:24631–24636. doi: 10.1074/jbc.272.39.24631. [DOI] [PubMed] [Google Scholar]
- 34.Sun H, Karow JK, Hickson ID, Maizels N. The Bloom’s syndrome helicase unwinds G4 DNA. J. Biol. Chem. 1998;273:27587–27592. doi: 10.1074/jbc.273.42.27587. [DOI] [PubMed] [Google Scholar]
- 35.Wu Y, Shin-Ya K, Brosh RM., Jr FANCJ helicase defective in Fanconia anemia and breast canceruUnwinds G-quadruplex DNA to defend genomic atability. Mol. Cell. Biol. 2008;28:4116–4128. doi: 10.1128/MCB.02210-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Vaughn JP, Creacy SD, Routh ED, Joyner-Butt C, Jenkins GS, Pauli S, Nagamine Y, Akman SA. The DEXH protein product of the DHX36 gene is the major source of tetramolecular quadruplex G4-DNA resolving activity in HeLa cell lysates. J. Biol. Chem. 2005;280:38117–38120. doi: 10.1074/jbc.C500348200. [DOI] [PubMed] [Google Scholar]
- 37.Creacy SD, Routh ED, Iwamoto F, Nagamine Y, Akman SA, Vaughn JP. G4 Resolvase 1 binds both DNA and RNA tetramolecular quadruplex with high affinity and is the major source of tetramolecular quadruplex G4-DNA and G4-RNA resolving activity in HeLa cells. J. Biol. Chem. 2008;283:34626–34634. doi: 10.1074/jbc.M806277200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lattmann S, Giri B, Vaughn JP, Akman SA, Nagamine Y. Role of the amino terminal RSM domain in the recognition and resolution of guanine quadruplex RNA by the DEAH-box RNA helicase RHAU. Nucleic Acids Res. 2010;38:6219–6233. doi: 10.1093/nar/gkq372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maxam A, Gilbert W. Sequencing end-labeled DNA with base-specific chemical cleavages. Meth. Enzymol. 1980;65:499–560. doi: 10.1016/s0076-6879(80)65059-9. [DOI] [PubMed] [Google Scholar]
- 40.Tran H, Schilling M, Wirbelauer C, Hess D, Nagamine Y. Facilitation of mRNA deadenylation and decay by the exosome-bound, DExH protein RHAU. Mol. Cell. 2004;13:101–111. doi: 10.1016/s1097-2765(03)00481-7. [DOI] [PubMed] [Google Scholar]
- 41.Giraldo R, Suzuki M, Chapman L, Rhodes D. Promotion of parallel DNA quadruplexes by a yeast telomere binding protein: a circular dichroism study. Proc. Natl Acad. Sci. USA. 1994;91:7658–7662. doi: 10.1073/pnas.91.16.7658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Paramasivan S, Rujan I, Bolton PH. Circular dichroism of quadruplex DNAs: applications to structure, cation effects and ligand binding. Methods. 2007;43:324–331. doi: 10.1016/j.ymeth.2007.02.009. [DOI] [PubMed] [Google Scholar]
- 43.Bardin C, Leroy JL. The formation pathway of tetramolecular G-quadruplexes. Nucleic Acids Res. 2008;36:477–488. doi: 10.1093/nar/gkm1050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hud NV, Smith FW, Anet FA, Feigon J. The selectivity for K+ versus Na+ in DNA quadruplexes is dominated by relative free energies of hydration: a thermodynamic analysis by 1 H NMR. Biochemistry. 1996;35:15383–15390. doi: 10.1021/bi9620565. [DOI] [PubMed] [Google Scholar]
- 45.Datta B, Armitage BA. Hybridization of PNA to structured DNA targets: quadruplex invasion and the overhang effect. J. Am. Chem. Soc. 2001;123:9612–9619. doi: 10.1021/ja016204c. [DOI] [PubMed] [Google Scholar]
- 46.Green JJ, Ying L, Klenerman D, Balasubramanian S. Kinetics of unfolding the human telomeric DNA quadruplex using a PNA trap. J. Am. Chem. Soc. 2003;125:3763–3767. doi: 10.1021/ja029149w. [DOI] [PubMed] [Google Scholar]
- 47.Bashkirov VI, Scherthan H, Solinger JA, Buerstedde JM, Heyer WD. A mouse cytoplasmic exoribonuclease (mXRN1p) with preference for G4 tetraplex substrates. J. Cell Biol. 1997;136:761–773. doi: 10.1083/jcb.136.4.761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Boder ET, Midelfort KS, Wittrup KD. Directed evolution of antibody fragments with monovalent femtomolar antigen-binding affinity. Proc. Natl Acad. Sci. USA. 2000;97:10701–10705. doi: 10.1073/pnas.170297297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kankia BI, Barany G, Musier-Forsyth K. Unfolding of DNA quadruplexes induced by HIV-1 nucleocapsid protein. Nucleic Acids Res. 2005;33:4395–4403. doi: 10.1093/nar/gki741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Torigoe H, Furukawa A. Tetraplex structure of fission yeast telomeric DNA and unfolding of the tetraplex on the interaction with telomeric DNA binding protein Pot1. J. Biochem. 2007;141:57–68. doi: 10.1093/jb/mvm011. [DOI] [PubMed] [Google Scholar]
- 51.Paramasivam M, Membrino A, Cogoi S, Fukuda H, Nakagama H, Xodo LE. Protein hnRNP A1 and its derivative Up1 unfold quadruplex DNA in the human KRAS promoter: implications for transcription. Nucleic Acids Res. 2009;37:2841–2853. doi: 10.1093/nar/gkp138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Fan J-H, Bochkareva E, Bochkarev A, Gray DM. Circular dichroism spectra and electrophoretic mobility shift assays show that human replication protein A binds and melts intramolecular G-quadruplex structures. Biochemistry. 2009;48:1099–1111. doi: 10.1021/bi801538h. [DOI] [PubMed] [Google Scholar]
- 53.Liu J-Q, Chen C-Y, Xue Y, Hao Y-H, Zheng T. G-quadruplex hinders translocation of BLM helicase on DNA: a real-time fluorescence spectroscopic unwinding study and comparison with duplex substrates. J. Am. Chem. Soc. 2010;132:10521–10527. doi: 10.1021/ja1038165. [DOI] [PubMed] [Google Scholar]
- 54.Plyler J, Jasheway K, Tuesuwan B, Karr J, Brennan JS, Kerwin SM, David WM. Real-time investigation of SV40 large T-antigen helicase activity using surface plasmon resonance. Cell Biochem. Biophys. 2009;53:43–52. doi: 10.1007/s12013-008-9038-z. [DOI] [PubMed] [Google Scholar]
- 55.Huber MD, Lee DC, Maizels N. G4 DNA unwinding by BLM and Sgs1p: substrate specificity and substrate-specific inhibition. Nucleic Acids Res. 2002;30:3954–3961. doi: 10.1093/nar/gkf530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhang S, Grosse F. Multiple functions of nuclear DNA helicase II (RNA helicase A) in nucleic acid metabolism. Acta Biochim. Biophys. 2004;36:177–183. doi: 10.1093/abbs/36.3.177. [DOI] [PubMed] [Google Scholar]
- 57.Chalupníková K, Lattmann S, Selak N, Iwamoto F, Fujiki Y, Nagamine Y. Recruitment of the RNA Helicase RHAU to Stress Granules via a Unique RNA-binding Domain. J. Biol. Chem. 2008;283:35186–35198. doi: 10.1074/jbc.M804857200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Iwamoto F, Stadler M, Chalupnikova K, Oakeley E, Nagamine Y. Transcription-dependent nucleolar cap localization and possible nuclear function of DExH RNA helicase RHAU. Exp. Cell Res. 2008;314:1378–1391. doi: 10.1016/j.yexcr.2008.01.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.