Abstract
Restarting stalled replication forks is vital to avoid fatal replication errors. Previously, it was demonstrated that hydroxyurea-stalled replication forks rescue replication either by an active restart mechanism or by new origin firing. To our surprise, using the DNA fibre assay, we only detect a slightly reduced fork speed on a UV-damaged template during the first hour after UV exposure, and no evidence for persistent replication fork arrest. Interestingly, no evidence for persistent UV-induced fork stalling was observed even in translesion synthesis defective, Polηmut cells. In contrast, using an assay to measure DNA molecule elongation at the fork, we observe that continuous DNA elongation is severely blocked by UV irradiation, particularly in UV-damaged Polηmut cells. In conclusion, our data suggest that UV-blocked replication forks restart effectively through re-priming past the lesion, leaving only a small gap opposite the lesion. This allows continuation of replication on damaged DNA. If left unfilled, the gaps may collapse into DNA double-strand breaks that are repaired by a recombination pathway, similar to the fate of replication forks collapsed after hydroxyurea treatment.
INTRODUCTION
Faithful DNA replication is critical to correctly transmit the genetic information to daughter cells and maintain genomic stability. The DNA replication machinery is continuously challenged by various obstacles, such as loss of replication factors, deprivation of nucleotides, or by physical damage on the DNA template. Stalled replication forks are stabilized through activation of a pathway involving primarily the ATR kinase (1,2), which is activated at RPA coated single-stranded DNA (ssDNA) regions (3) formed after uncoupling of the MCM-helicase at stalled replication forks (4–6). This pathway is activated after replication fork stalling by either hydroxyurea (HU) or physical blockage such as a lesion induced by UV-irradiation. However, other events occurring at stalled replication forks may differ depending on the nature of the fork-stalling agent.
Disrupted replication may be continued through DNA repair or damage tolerance pathways. Repair has been considered as a means of restarting replication, for example through break-induced replication (7,8). The coupling of translesion synthesis (TLS) to the replication machinery is not fully determined. Interestingly, it was recently demonstrated in Saccharomyces cerevisiae that the ubiquitin-dependent TLS of UV-induced lesions is separate from genome replication and may occur post-replicatively (9,10). Consistent with this, there is evidence for separation of DNA repair from replication, for example as demonstrated by the inability of HU-induced collapsed replication forks to restart (11). Instead, replication is resumed by new origin firing (11), which is mediated by firing of dormant origins under replication stress (12,13). The remaining DNA double-strand breaks (DSBs) after replication fork collapse are repaired by a slow homologous recombination (HR) process (11,14,15) and do not provide a substrate for restarting replication (11).
Different to a collapsed fork, a replication fork transiently stalled by HU can effectively restart through several different pathways employing a number of proteins [see (16) for a review]. However, it is not yet known to what extent these pathways are also used for continuation of replication after fork stalling by physical damage. Since HU stalls replication forks by deprivation of nucleotides (17), it is possible that replication forks can simply restart when the nucleotide pool is restored. Here, we studied replication restart after exposure to short-wave ultraviolet radiation (UV), inducing primarily cyclobutane pyrimidine dimers (CPDs), which block replication forks and are bypassed during TLS by Polη (18). We find that UV-induced DNA damage, as opposed to HU, does not result in an increased number of stalled replication forks and causes only a slight reduction in replication fork speed. In contrast, continuous DNA elongation at replication forks is prevented after UV-induced damage, especially in Polη mutated (Polηmut) cells, resulting in gaps. Altogether, our data support a model for restart by re-priming of forks blocked by UV-induced damage, similar to what has been suggested in S. cerevisiae (6). According to this model, replication is quickly resumed on the 5′ side of the lesion, allowing the replication fork to continue but leaving a ssDNA gap in the newly synthesized DNA, opposite the lesion. We show that if left unfilled, the re-priming induced ssDNA gaps collapse into DNA DSBs that are repaired by an HR pathway.
MATERIALS AND METHODS
Cell culture
The XP30RO cells, originally obtained from a patient, have a 13-bp deletion leading to a frameshift in the Polη gene, yielding a 42 amino acid peptide (19). The XP30RO cell line and the restored cell line stably expressing Polη from a vector (20) were a kind gift from Dr Alan Lehmann. Cells were grown in Dulbecco's Eagles minimum essential medium with 10% foetal calf serum and 1% streptomycin–penicillin, restored cells in the presence of 100 μg/ml zeocin. Cells were cultured and allowed to repair in an incubator at 37°C with 5% CO2.
UVC irradiation
Cells were washed with cold Hank's balanced salt solution (HBSS + HEPES without Phenol red) that was removed before irradiation at room temperature under a 254 nm UVC low pressure mercury lamp (Phillips TUV 15 W) at indicated doses. Dose rates used were 0.18 and 0.10 J/m2s. Exposure times were controlled using a fast magnetic shutter mounted within the apparatus.
DNA fibre technique
An amount of 25 μM CldU (Sigma, C6891) and 250 μM IdU (Sigma I7125) in pre-warmed DMEM were incubated at 37°C, 5% CO2 for at least 30 min before labelling. Cells grown for at least 18 h were labelled with CldU for 20 min, washed and UVC irradiated (10 J/m2), and then incubated in IdU media for 30, 60, or 120 min before harvesting by scraping in cold PBS. Harvested cells were diluted to 106 cells/ml. Spreading and staining was performed as described previously (21). Fluorescence images were captured using a Zeiss LSM 510 inverted confocal microscope using planapochromat 63×/NA 1.4 oil immersion objective, excitation wavelengths of 488 and 546 nm, and analysed using the ImageJ software. At least 100 unidirectional forks labelled with both CldU and IdU were measured and at least 500 fork structures were counted for every condition. Conversion factor used is 1 μm = 2.59 kb (22). Three individual experiments were performed and coded samples were analysed.
Pulsed-field gel electrophoresis
Directly after a 24 h (0.439 mM, 0.925 kBq/ml) or 0.5 h (4.39 mM, 9.25 kBq/ml) labelling with 14C-TdR, cells were UVC irradiated (10 J/m2), and allowed to repair in pre-warmed DMEM for indicated times. Pulsed-field gel electrophoresis (PFGE) was performed essentially as previously described (23). In short, the cells were harvested and 106 cells were melted into 1% InCert agarose (Cambrex) plugs and incubated for 48 h in 0.5 M EDTA, 1% N-laurylsarcosyl and 2 mg/ml proteinase K at 20°C in darkness. After four washings in Tris–EDTA buffer, separation was performed on agarose gel (1% certified megabase agarose, Bio-Rad) on a CHEF DR III apparatus for 20 h (Bio-Rad, 120° angle, 4 V/cm, switch time 60/240 s, 14°C). The gel was stained with ethidium bromide, photographed and then dried to allow exposure onto a phosphoimager plate (FujiFilm) for quantification employing MultiGauge software (FLA- 3000, FujiFilm).
Immunofluorescence
Cells grown on cover slips for 18 h were pulse labelled with 20 µM CldU for 10 min immediately before UVC irradiation (10 J/m2), allowed to repair for 6 h, fixed in 3% paraformaldehyde for 20 min, permeabilized for 10 min with 0.3% Triton X-100 in PBS, blocked with 3% BSA in PBS for at least 30 min, incubated with primary antibodies against RAD51 (Santa Cruz, rabbit, H92, SC-8349) γH2AX (Upstate, mouse, clone JBW301) and RPA (32 kDa subunit, Cell Signalling Technology, rat, 4E4) over night at 4°C, washed, incubated with fluorophore-conjugated secondary antibodies (Alexa 488 goat anti rabbit, Alexa 635 goat anti mouse, Molecular Probes) for 1 h. Incorporated CldU was detected by incubation in 2 N HCl at 37°C for 15 min, followed by neutralization in Borax pH 8.7. After washings, samples were incubated with primary antibody against CldU [Oxford Biotechnology, rat anti BrdU and CldU, clone BU1/75 (ICR1)] for 1 h and secondary antibody (Alexa 555, goat anti rat, Molecular Probes) for 1 h before mounting in ProLong Gold (Invitrogen). All antibodies were diluted 1:1000, except RPA (1:500), in PBS with 3% BSA.
Fluorescence images were captured using a Zeiss LSM 510 inverted confocal microscope using planapochromat 63×/NA 1.4 oil immersion objective, using excitation wavelengths of 488, 546 and 630 nm and processed using the LSM software. Cells were considered as positive for RPA if they contained at least 10 large foci. For determination of co-localization, only cells with distinct CldU foci were analysed for γH2AX and RAD51 foci co-localizing with the CldU foci. Cells were considered as positive for co-localization if at least 80% of γH2AX and RAD51 foci co-localized with CldU foci. Four independent experiments with coded samples were analysed.
Alkaline DNA unwinding technique
Replication fork elongation was measured as described previously (24). Shortly, 3H-thymidine is incorporated into ongoing forks, and the forks are allowed to progress from the labelled area for different times. By addition of alkaline solution, unwinding takes place from the single-stranded ends of the fork or gaps, if present; thus, as the fork moves forward or the gaps are filled, the labelling is removed from the fraction of DNA that becomes single-stranded. Cells plated in 24-well plates (70 000 cells/well) were allowed to grow for 18 h before pulse labelling (0.5 h) with 37 kBq/ml 3H-thymidine (GE Healthcare) in DMEM, directly followed by UVC irradiation with indicated doses and incubated in prewarmed DMEM for indicated times.
RESULTS
Efficient replication fork progression on a UV-damaged template
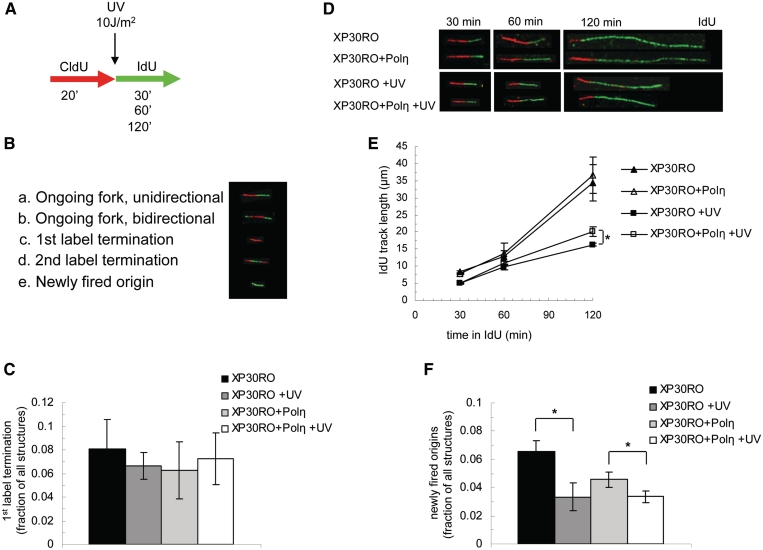
Here, we used the DNA fibre assay to monitor replication after induction of replication fork blocking DNA adducts. We focussed on the first 2 h following exposure to UV, when a large fraction of UV-induced lesions are still present on the DNA. We labelled DNA with CldU for 20 min immediately before exposure to UV, and with IdU for 30, 60 or 120 min after irradiation (Figure 1A) and determined different replication structures (Figure 1B). The UV dose used (10 J/m2) generates about one lesion each 3–4 kb [(25); see Supplementary Data] so that initially, forks will run into damage every five minutes.
Figure 1.
Replicating DNA fibres continues to grow in similar manners in UV irradiated Polηmut and restored cells. (A) Experimental setup. (B) Different structures observed. (C) Fraction of first label terminations of the replication structures, after 30 min IdU labelling. Mean and s.e.m. of three independent experiments. (D) Representative fibres after UV exposure and different times with IdU label. (E) IdU incorporation in UV exposed and unirradiated Polηmut and restored cells. Mean and s.e.m. of three independent experiments. (F) Fraction of newly fired origins among the replication structures, after 30 min IdU labelling. Mean and s.e.m. of three independent experiments. Statistical significance determined in t-test is indicated with one star (P < 0.05).
The Polη TLS polymerase efficiently bypasses CPDs (18) and hence, poor replication arrest may simply be the result of efficient bypass. To test this hypothesis, we used both XP30RO (Polηmut) cells and XP30RO cells with rescue expression of functional Polη (XP30PO + Polη). Despite the tight spacing of the DNA lesions, we observed no increase in replication fork stalling without any incorporation of the second labelling after UV irradiation (first label terminations) even in Polηmut cells (Figure 1C).
Next, we determined replication speed of ongoing forks (those labelled with both CldU and IdU) and found no difference in fork speed in unirradiated XP30RO or XP30PO + Polη cells (Figure 1D and E), confirming earlier results showing that Polη is not required for replication on undamaged DNA (26). When measuring ongoing forks we excluded bidirectional forks and merged replicons (second label terminations) (Figure 1B) as these structures were initiated or terminated during the experiment. Fork speed is slightly reduced on UV-damaged (10 J/m2) templates and surprisingly, in a similar manner in both Polηmut and restored cells (Figure 1D, E and Supplementary Figure S1). However, 120 min after UV exposure a small reduction of IdU fork speed is observed in XP30RO cells compared to restored cells (P < 0.05; Figure 1E), which is in line with previous replication defects observed in Polηmut cells at late time points after UV treatment (27).
Replication forks blocked by prolonged HU treatment restart by an increase in new origin firing (11). In contrast, UV exposed cells show a checkpoint mediated reduction of new origin firing (28,29). In line with this, we observe a reduced level of new origin firing after UV-induced damage in both cell lines (Figure 1F), suggesting that the elongation of replication fibres cannot be explained by new origin firing. Rather, replication fork elongation is able to proceed despite the massive induction of adducts on the template DNA.
Impaired replication-mediated DNA elongation on a UV-damaged template
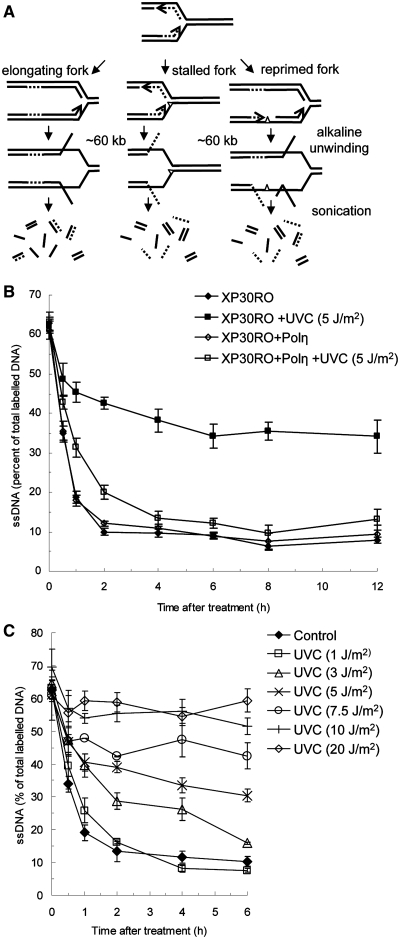
The suggested role of Polη in mediating TLS after UV exposure (18), makes our observation of intact, and Polη independent, replication fork elongation on a template heavily damaged by UV somewhat surprising. However, in S. cerevisiae it has been demonstrated that TLS is uncoupled from genome replication (9,10). Furthermore, ssDNA regions at replication forks have been demonstrated to accumulate after UV-induced damage in S. cerevisiae, which could be formed after a re-priming event where replication continues on the 5′ side of the lesion, leaving a small gap on the newly synthesized strand (6). To investigate if replication fork elongation is continuous after induction of UV-induced damage, we used the DNA alkaline unwinding assay. This assay monitors continuous replication fork elongation from a pulse labelled area, by using ssDNA ends at a replication fork as starting points for DNA unwinding in alkaline solution (24,30). Nascent replication forks are labelled with a 30-min 3H-thymidine pulse and the 3H-labelled DNA is only released into the ssDNA fraction if continuous replication elongation is inhibited (Figure 2A). This method will thus distinguish between continuous or re-primed/gapped replication fork progressions, two scenarios where similar fork movement would be seen with a method that is not sensitive to single-strand gaps, like the fibre assay. In contrast to the DNA fibre assay, we found that continuous elongation of replication forks after UV treatments is delayed in both Polηmut and Polη restored cells (Figure 2B) at early time points. The initial inability of Polη restored cells to grow continuous DNA after UV exposure is close to the levels seen in untreated cells at later time points (Figure 2B). This suggests that replication forks are initially disrupted by UV-induced DNA lesions, leaving behind gaps that are later filled. Interestingly, continuous replication fork elongation is completely impaired in the Polηmut cells even 12 h after UV exposure (Figure 2B). This block in continuous replication fork elongation is in sharp contrast to the results observed in the fibre assay showing similarly intact elongation in both Polηmut and restored cells, altogether suggesting the prolonged existence of gaps in newly synthesized DNA.
Figure 2.
Continuous replication fork elongation is disrupted in Polηmut cells, but not in restored cells, after UV exposure. (A) Schematic illustration of the alkaline DNA unwinding assay used to monitor continuous replication fork progression. Directly before UV exposure, ongoing replication forks are pulse labelled (0.5 h) with 3H-TdR. As replication forks progress the labelled DNA is located further away from the DNA ends at the fork and is not released into the ssDNA fraction by the alkaline unwinding that is initiated from ssDNA ends. However, by this technique, ssDNA will also be released if gaps are formed during replication. (B) Replication progression of replication forks in UV irradiated (5 J/m2) Polηmut XP30RO and restored cells. Replication progression is monitored as loss of 3H activity in the ssDNA fraction (C) Continuous replication fork progression in Polηmut XP30RO cells is slowed down in a dose dependent manner following UV irradiation.
To determine whether the amount of discontinuous replication observed in Polηmut cells is related to the UV dose, we treated XP30RO cells with increasing doses of UV. We find that the apparent rate of replication fork elongation, measured as existence of points for DNA unwinding, in XP30RO cells is dependent on UV dose, such that no apparent elongation is seen at a dose of 10 J/m2 under these conditions (Figure 2C). We also used alkaline sucrose gradients as a separate method to study newly replicated DNA fragments after UV exposure and their elongation into larger DNA molecules over time (26). In Polη proficient cells, pulse labelled DNA fragments synthesized shortly after UV irradiation quickly grow into larger fragments. In contrast, in Polηmut cells, the pulse labelled DNA fragments do not increase considerably in size from 1 to 12 h after labelling (Supplementary Figure S2). This confirms the existence of gaps in DNA replicated shortly after UV exposure, despite the continuation of replication observed with the fibre assay.
UV-induced post-replication ssDNA gaps collapse into DNA DSBs
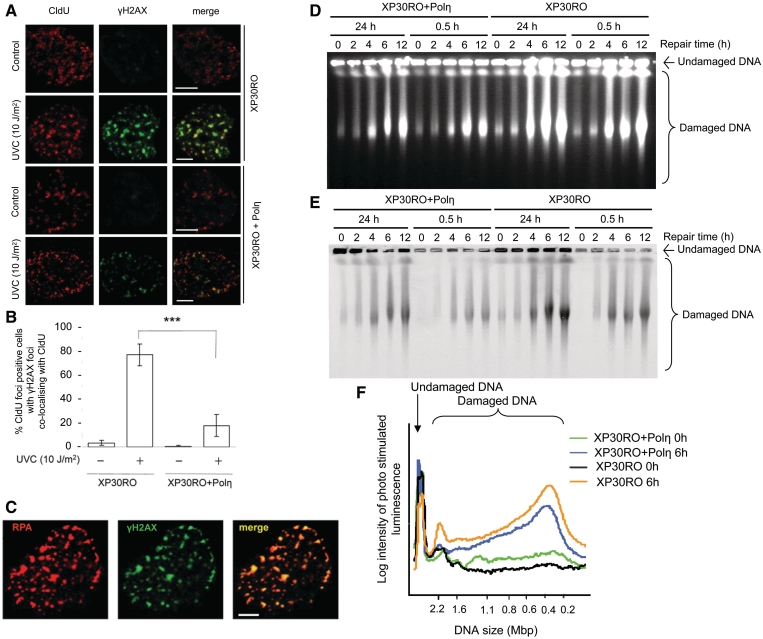
Failure to bypass UV lesions causes initial stalling of one strand, which would generate ssDNA gaps previously visualized in S. cerevisiae (6). Here, we find accumulation of RPA foci after UV treatment, particularly in Polηmut cells (Supplementary Figure S3), in line with previous reports (27) and explaining the increased ATR signalling reported in Polηmut cells (31).
Next, we wanted to determine the fate of these UV associated ssDNA gaps left behind the progressing replication fork. It has previously been demonstrated that γH2AX foci form in response to UV exposure in Polηmut but not to the same extent in restored cells (32). Here, we decided to investigate whether these γH2AX foci form at replication forks. We pulse labelled XP30RO and XP30RO + Polη cells with CldU directly before exposing them to UV and incubated the cells for 6 h before fixation. Staining revealed that γH2AX foci co-localized with CldU-labelled replication sites and RPA in UV exposed Polηmut cells, whereas only a small fraction of restored cells were γH2AX foci positive (Figure 3A–C). Phosphorylation of H2AX to form γH2AX foci can occur either in response to DSB formation, in a nucleotide excision repair (NER) dependent manner after UV exposure independently of DSB formation (33) or at stalled replication forks in the absence of DSBs detectable by PFGE (11).
Figure 3.
Replication forks stalled by UV-induced damage collapse into replication-associated DSBs. (A) Representative images of UV-induced γH2AX foci at replication forks labelled with CldU in Polηmut XP30RO and restored cells, 6 h following UV treatments. One nucleus is shown, bar is 5 μm. (B) Percentage of CldU foci positive cells where γH2AX foci co-localize with CldU. Mean and s.e.m. of four independent experiments is depicted. Statistical significance determined in t-test is indicated with three stars (P < 0.001). (C) UV-induced γH2AX foci in a Polηmut XP30RO cell, co-localizing with RPA 6 h after irradiation (10 J/m2). Bar is 5 μm. (D) UV-induced DSBs produced in Polηmut XP30RO and restored cells after exposure to 10 J/m2, measured by PFGE, and visualized by ethidium bromide or (E) autoradiography. (F) Quantification of the intensity of radioactively labelled DNA fragments released from the 0.5-h pulse labelled regions of XP30RO and XP30RO + Polη cells, 0 h (black and green line, respectively) and 6 h (blue and yellow line, respectively) after UV exposure, depicting remaining damage at previously replicated forks.
To determine whether Polη-dependent gap-filling opposite DNA adducts protects from the formation of UV-induced DSBs, we determined the formation of replication-induced DSBs by PFGE. Prior to UV exposure, we labelled cells with 14C-thymidine either homogeneously for 24 h or for 0.5 h to only label nascent replication forks. Cells were then harvested at different time points (0, 2, 4, 6, 12 h) after UV exposure, and the DNA was separated using PFGE. We stained the gel with ethidium bromide and found an increased amount of DSBs formed in XP30RO cells as compared with the Polη restored cells (Figure 3D). We next analysed the DSBs that are associated with replication, by measuring the 14C-labelled DNA. In the 0.5 h-labelled cells most of the 14C-labelled DNA is within replication bubbles, which do not migrate into the gel unless opened by a DSB produced adjacent to a replication fork [see (15) for details]. We found an increase in replication associated DSBs produced by UV treatments in Polηmut cells (Figure 3E and F). The total fractions of radioactivity released were 93% in Polηmut cells and 61% in complemented cells (Figure 3F), showing that many forks present at the time of UV exposure, were collapsed into DSBs 6 h after UV treatment. Thus, despite the initial lengthening of replication fibres in both Polηmut and restored cells, there is a considerable induction of replication-associated DSBs after UV exposure (10 J/m2).
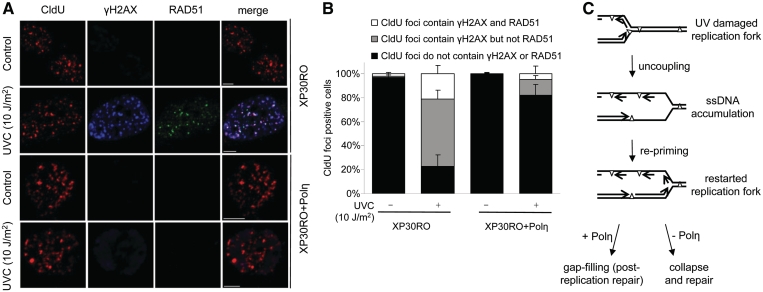
It is well established that RAD51-dependent HR is triggered in order to repair collapsed replication forks after HU treatment (11,15). This is in contrast to yeast, where recombination is suggested to be required for bypassing replication blocks (6). The Cleaver lab has shown that MRE11 foci are formed in Polηmut cells after UV irradiation and that these co-localize with PCNA (32). Furthermore, Polη co-localizes with RAD51 after UV irradiation (18). These data support both models: that HR is involved in mediating bypass of blocked replication forks, which is done instantaneously and/or that recombination is involved in repairing collapsed post-replication ssDNA gaps in Polηmut cells. To test whether RAD51 foci form at collapsed replicative gaps, we co-stained CldU, γH2AX foci and RAD51 in cells 6 h after UV treatment, when the active replication fork has extended past the lesion. We find that the post-replication damaged regions co-localize with RAD51 (Figure 4A), suggesting RAD51-dependent repair of collapsed replicative gaps following UV exposure. In order to only study cells that were replicating at the time of UV exposure, we selectively looked at CldU foci positive cells, and quantified the number of cells in which RAD51 co-localized with CldU and γH2AX foci (Figure 4B). In both the Polηmut and the restored cells, 25% of the cells showing colocalization of γH2AX and CldU, also show co-localization of RAD51 (Figure 4B). However, the total proportion of cells with co-localization of γH2AX, RAD51 and CldU foci is four times larger in Polηmut cells (Figure 4B). This clearly shows the importance of recombination-mediated repair of collapsed gaps after replication of DNA with UV-induced damage in mammalian cells.
Figure 4.
RAD51 co-localizes with replication sites of after UV exposure of Polηmut cells. (A) Representative images of UV-induced γH2AX and RAD51 foci at replication forks in Polηmut XP30RO and restored cells pulse labelled (10 min) with CldU directly before UV exposure (10 J/m2), and allowed to repair for 6 h before fixation. γH2AX was used to visualize DSBs and RAD51 as an indicator of homologous recombination. Bar is 5 μm. (B) Quantification of co-localization of γH2AX and RAD51 with CldU foci, in cells fixed 6 h after UV exposure (10 J/m2). Only cells with distinct CldU foci were checked for co-localization, to only study cells replicating when irradiated. Mean and s.e.m. of four independent experiments are shown. (C) Model for the continuation of DNA replication on damaged DNA. When the replication fork runs into a UV-induced DNA lesion, the PCNA molecule is left and can be ubiquitinated to allow bypass with Polη. Replication is resumed on the 5′ side of the lesion, leaving a gap. In the absence of Polη, the gaps are not bypassed, but collapsed into DSBs that are repaired with a pathway involving RAD51.
DISCUSSION
Although UV-induced DNA damage can block replication elongation, we observe no increase in the immediate stalling of replication forks after a high UV exposure, using the DNA fibre assay. Rather, we show that the elongation speed on a template with UV-induced damage is only marginally reduced and independent on the TLS polymerase Polη, which is responsible for bypass of UV-induced adducts. In contrast, using two different replication assays we demonstrate that the continuous elongation of DNA molecules on a template damaged by UV is severely impaired, particularly in Polηmut cells. Taken together, these results demonstrate that after exposure to UV, the replication fork is able to proceed, but without properly elongating the DNA molecule. We propose a model to explain this phenomenon, where replication continues efficiently by re-priming at UV-induced DNA damage, leaving behind gaps that are left unsealed in Polηmut cells (Figure 4C). Although postreplicative gaps have been previously demonstrated in Polηmut cells (26) and in Escherichia coli defective in NER (34), replication fork uncoupling and efficient replication fork elongation on UV-damaged DNA has not previously been demonstrated in mammalian cells. Data from S. cerevisiae show the presence of ssDNA gaps after UV exposure visualized by electron microscopy (6) and that gap filling after UV exposure can occur separately from genome replication (9,10). This supports a re-priming model for restart of UV stalled replication forks, which is conserved from lower eukaryotes. In addition, the data presented here argue that the re-priming kinetics are very fast and only marginally slow down the fork speed. Re-priming may be important to continue replication on damaged DNA, differing from replication restart of forks stalled after nucleotide deprivation or loss of replication factors.
Replication restart is not likely to be achieved by new origin firing, since origin firing is decreased after UV exposure (28,29), which we also see in our cells (Figure 1F). Furthermore, due to the resolution of the fibre technique, a newly fired origin would have to be within <2 kb of the fork-stalling lesion in order to be interpreted as continuous, while the number of licensed origins are believed to be spaced 30–150 kb apart (35,36).
Efficient restart of a HU-stalled fork requires several proteins including RAD51, XRCC3, MUS81, BLM, PARP1, MRE11 and SMARCAL1 (11,37–41). Are there any specific factors required for restart of UV stalled forks by re-priming? This may not be the case since long stretches of ssDNA is essentially the natural substrate for DNA Polα-primase mediated priming and start of another Okazaki fragment on the lagging strand. Thus, re-priming may simply be triggered by the formation of ssDNA regions, requiring no other factors than those present in a normal replication factory.
Interestingly, continued primer synthesis was recently demonstrated at replication forks stalled by aphidicolin in Xenopus egg extracts (42). Although several factors have been identified to assist restart of HU-stalled replication forks, a majority of the forks (80%) restart without need of an active restarting process (11,41). With the presence of primers, there is a possibility that restart of most replication forks are carried out by re-priming also after HU treatment. However, the ssDNA gaps left behind a HU-stalled fork would be likely to close rapidly, as there is no physical block and hence such short persisting gaps would be difficult to demonstrate experimentally.
We find a large portion of replication-associated DNA present in DNA fragments 6 h after UV exposure, in particular in Polηmut cells. For fragment separation using PFGE, two DSBs are needed for fragment release. The sizes of the fragments range from 0.2 to 5 Mb in length, which is considerably larger than the average replicon. Since replication origin firing occurs in clusters (43), it is possible that two DSBs in distant replicons within one cluster would release the whole cluster into the PFGE. Thus, although a large amount of replication-associated DNA fragments are released into the PFGE, the number of collapsed replication forks is likely to be fewer.
We find RAD51 foci associated with replication-associated DSBs. rad51 mutants in S. cerevisiae accumulate ssDNA gaps, visualized by electron microscopy, to a higher degree than wild-type cells suggesting that Rad51 is involved in immediate bypassing of the lesion (44). In contrast, we earlier demonstrated that DNA molecule elongation after UV-induced damage is not impaired in the RAD51 paralogue XRCC3 mutant mammalian cells (24), suggesting that gap closure in mammalian cells is not dependent on HR. This may be explained by slow kinetics of recombination repair in mammals. Here, we observed markedly higher levels of RAD51 foci in Polηmut cells as compared to the Polη rescued cells (Figure 4). If RAD51 is involved in Polη-independent re-priming, one would not expect more RAD51 foci in Polηmut cells. Rather, the increased amount of RAD51 foci correlate with the increase in ssDNA gaps in Polηmut cells. Furthermore, the timing and location of the foci suggest that RAD51 foci form at the ssDNA gaps and not for re-priming the blocked fork, as suggested for S. cerevisiae. The absence of RAD51 at many replication site associated γH2AX foci may be because they are not sites of DSBs, but RPA coated ssDNA regions, or alternatively that such DSBs are processed by an MRE11-dependent repair pathway, as suggested earlier (32). Thus, we suggest that HR in mammalian cells is activated for repair of UV-collapsed ssDNA gaps but not restart of UV-blocked forks.
In conclusion, we demonstrate that immediate continuation of DNA replication fork progression on a UV-damaged template is efficient and independent of TLS. In contrast, we observe discontinuous elongation of replication forks, which is very pronounced in Polηmut cells, demonstrating distinct mechanisms for fork progression and DNA molecule extension on a UV-damaged template. Furthermore, we demonstrate that replication-induced gaps on DNA damaged by UV readily collapse into DSBs, which are substrates for RAD51-mediated recombination repair.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Swedish Cancer Society, the Swedish Children's Cancer Foundation; the Swedish Research Council, the Swedish Pain Relief Foundation’ and the Medical Research Council. Funding for open access charge: Swedish Research Council.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Drs Alan Lehmann and Simone Sabbioneda, University of Sussex, for the XP30RO and XP30RO + Polη cell lines, for welcoming I.E. for a visit in their laboratory, and for assistance with the sucrose gradient experiments.
REFERENCES
- 1.Guo Z, Kumagai A, Wang SX, Dunphy WG. Requirement for Atr in phosphorylation of Chk1 and cell cycle regulation in response to DNA replication blocks and UV-damaged DNA in Xenopus egg extracts. Genes Dev. 2000;14:2745–2756. doi: 10.1101/gad.842500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Hekmat-Nejad M, You Z, Yee MC, Newport JW, Cimprich KA. Xenopus ATR is a replication-dependent chromatin-binding protein required for the DNA replication checkpoint. Curr. Biol. 2000;10:1565–1573. doi: 10.1016/s0960-9822(00)00855-1. [DOI] [PubMed] [Google Scholar]
- 3.Zou L, Elledge SJ. Sensing DNA damage through ATRIP recognition of RPA-ssDNA complexes. Science. 2003;300:1542–1548. doi: 10.1126/science.1083430. [DOI] [PubMed] [Google Scholar]
- 4.Pacek M, Walter JC. A requirement for MCM7 and Cdc45 in chromosome unwinding during eukaryotic DNA replication. EMBO J. 2004;23:3667–3676. doi: 10.1038/sj.emboj.7600369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Byun TS, Pacek M, Yee MC, Walter JC, Cimprich KA. Functional uncoupling of MCM helicase and DNA polymerase activities activates the ATR-dependent checkpoint. Genes Dev. 2005;19:1040–1052. doi: 10.1101/gad.1301205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lopes M, Foiani M, Sogo JM. Multiple mechanisms control chromosome integrity after replication fork uncoupling and restart at irreparable UV lesions. Mol. Cell. 2006;21:15–27. doi: 10.1016/j.molcel.2005.11.015. [DOI] [PubMed] [Google Scholar]
- 7.Kraus E, Leung WY, Haber JE. Break-induced replication: a review and an example in budding yeast. Proc. Natl Acad. Sci. USA. 2001;98:8255–8262. doi: 10.1073/pnas.151008198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arnaudeau C, Lundin C, Helleday T. DNA double-strand breaks associated with replication forks are predominantly repaired by homologous recombination involving an exchange mechanism in mammalian cells. J. Mol. Biol. 2001;307:1235–1245. doi: 10.1006/jmbi.2001.4564. [DOI] [PubMed] [Google Scholar]
- 9.Daigaku Y, Davies AA, Ulrich HD. Ubiquitin-dependent DNA damage bypass is separable from genome replication. Nature. 2010;465:951–955. doi: 10.1038/nature09097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Karras GI, Jentsch S. The RAD6 DNA damage tolerance pathway operates uncoupled from the replication fork and is functional beyond S phase. Cell. 2010;141:255–267. doi: 10.1016/j.cell.2010.02.028. [DOI] [PubMed] [Google Scholar]
- 11.Petermann E, Orta ML, Issaeva N, Schultz N, Helleday T. Hydroxyurea-stalled replication forks become progressively inactivated and require two different RAD51-mediated pathways for restart and repair. Mol. Cell. 2010;37:492–502. doi: 10.1016/j.molcel.2010.01.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ge XQ, Jackson DA, Blow JJ. Dormant origins licensed by excess Mcm2-7 are required for human cells to survive replicative stress. Genes Dev. 2007;21:3331–3341. doi: 10.1101/gad.457807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Woodward AM, Gohler T, Luciani MG, Oehlmann M, Ge X, Gartner A, Jackson DA, Blow JJ. Excess Mcm2-7 license dormant origins of replication that can be used under conditions of replicative stress. J. Cell. Biol. 2006;173:673–683. doi: 10.1083/jcb.200602108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Saintigny Y, Delacote F, Vares G, Petitot F, Lambert S, Averbeck D, Lopez BS. Characterization of homologous recombination induced by replication inhibition in mammalian cells. EMBO J. 2001;20:3861–3870. doi: 10.1093/emboj/20.14.3861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lundin C, Erixon K, Arnaudeau C, Schultz N, Jenssen D, Meuth M, Helleday T. Different roles for nonhomologous end joining and homologous recombination following replication arrest in mammalian cells. Mol. Cell. Biol. 2002;22:5869–5878. doi: 10.1128/MCB.22.16.5869-5878.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Petermann E, Helleday T. Pathways of mammalian replication fork restart. Nat. Rev. Mol. Cell. Biol. 2010;11:683–687. doi: 10.1038/nrm2974. [DOI] [PubMed] [Google Scholar]
- 17.de Feraudy S, Limoli CL, Giedzinski E, Karentz D, Marti TM, Feeney L, Cleaver JE. Pol eta is required for DNA replication during nucleotide deprivation by hydroxyurea. Oncogene. 2007;26:5713–5721. doi: 10.1038/sj.onc.1210385. [DOI] [PubMed] [Google Scholar]
- 18.Kannouche P, Broughton BC, Volker M, Hanaoka F, Mullenders LH, Lehmann AR. Domain structure, localization, and function of DNA polymerase eta, defective in xeroderma pigmentosum variant cells. Genes Dev. 2001;15:158–172. doi: 10.1101/gad.187501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Masutani C, Kusumoto R, Yamada A, Dohmae N, Yokoi M, Yuasa M, Araki M, Iwai S, Takio K, Hanaoka F. The XPV (xeroderma pigmentosum variant) gene encodes human DNA polymerase eta. Nature. 1999;399:700–704. doi: 10.1038/21447. [DOI] [PubMed] [Google Scholar]
- 20.Stary A, Kannouche P, Lehmann AR, Sarasin A. Role of DNA polymerase eta in the UV mutation spectrum in human cells. J. Biol. Chem. 2003;278:18767–18775. doi: 10.1074/jbc.M211838200. [DOI] [PubMed] [Google Scholar]
- 21.Groth P, Auslander S, Majumder MM, Schultz N, Johansson F, Petermann E, Helleday T. Methylated DNA causes a physical block to replication forks independently of damage signalling, O(6)-methylguanine or DNA single-strand breaks and results in DNA damage. J. Mol. Biol. 2010;402:70–82. doi: 10.1016/j.jmb.2010.07.010. [DOI] [PubMed] [Google Scholar]
- 22.Henry-Mowatt J, Jackson D, Masson JY, Johnson PA, Clements PM, Benson FE, Thompson LH, Takeda S, West SC, Caldecott KW. XRCC3 and Rad51 modulate replication fork progression on damaged vertebrate chromosomes. Mol. Cell. 2003;11:1109–1117. doi: 10.1016/s1097-2765(03)00132-1. [DOI] [PubMed] [Google Scholar]
- 23.Lundin C, North M, Erixon K, Walters K, Jenssen D, Goldman AS, Helleday T. Methyl methanesulfonate (MMS) produces heat-labile DNA damage but no detectable in vivo DNA double-strand breaks. Nucleic Acids Res. 2005;33:3799–3811. doi: 10.1093/nar/gki681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Johansson F, Lagerqvist A, Erixon K, Jenssen D. A method to monitor replication fork progression in mammalian cells: nucleotide excision repair enhances and homologous recombination delays elongation along damaged DNA. Nucleic Acids Res. 2004;32:e157. doi: 10.1093/nar/gnh154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Williams JI, Cleaver JE. Excision repair of ultraviolet damage in mammalian cells. Evidence for two steps in the excision of pyrimidine dimers. Biophys. J. 1978;22:265–279. doi: 10.1016/S0006-3495(78)85488-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lehmann AR, Kirk-Bell S, Arlett CF, Paterson MC, Lohman PH, de Weerd-Kastelein EA, Bootsma D. Xeroderma pigmentosum cells with normal levels of excision repair have a defect in DNA synthesis after UV-irradiation. Proc. Natl Acad. Sci. USA. 1975;72:219–223. doi: 10.1073/pnas.72.1.219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Despras E, Daboussi F, Hyrien O, Marheineke K, Kannouche PL. ATR/Chk1 pathway is essential for resumption of DNA synthesis and cell survival in UV-irradiated XP variant cells. Hum. Mol. Genet. 2010;19:1690–1701. doi: 10.1093/hmg/ddq046. [DOI] [PubMed] [Google Scholar]
- 28.Heffernan TP, Simpson DA, Frank AR, Heinloth AN, Paules RS, Cordeiro-Stone M, Kaufmann WK. An ATR- and Chk1-dependent S checkpoint inhibits replicon initiation following UVC-induced DNA damage. Mol. Cell. Biol. 2002;22:8552–8561. doi: 10.1128/MCB.22.24.8552-8561.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chastain PD, 2nd, Heffernan TP, Nevis KR, Lin L, Kaufmann WK, Kaufman DG, Cordeiro-Stone M. Checkpoint regulation of replication dynamics in UV-irradiated human cells. Cell Cycle. 2006;5:2160–2167. doi: 10.4161/cc.5.18.3236. [DOI] [PubMed] [Google Scholar]
- 30.Jansen JG, Tsaalbi-Shtylik A, Hendriks G, Gali H, Hendel A, Johansson F, Erixon K, Livneh Z, Mullenders LH, Haracska L, et al. Separate domains of Rev1 mediate two modes of DNA damage bypass in mammalian cells. Mol. Cell. Biol. 2009;29:3113–3123. doi: 10.1128/MCB.00071-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bomgarden RD, Lupardus PJ, Soni DV, Yee MC, Ford JM, Cimprich KA. Opposing effects of the UV lesion repair protein XPA and UV bypass polymerase eta on ATR checkpoint signaling. EMBO J. 2006;25:2605–2614. doi: 10.1038/sj.emboj.7601123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Limoli CL, Giedzinski E, Bonner WM, Cleaver JE. UV-induced replication arrest in the xeroderma pigmentosum variant leads to DNA double-strand breaks, gamma -H2AX formation, and Mre11 relocalization. Proc. Natl Acad. Sci. USA. 2002;99:233–238. doi: 10.1073/pnas.231611798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Marti TM, Hefner E, Feeney L, Natale V, Cleaver JE. H2AX phosphorylation within the G1 phase after UV irradiation depends on nucleotide excision repair and not DNA double-strand breaks. Proc. Natl Acad. Sci. USA. 2006;103:9891–9896. doi: 10.1073/pnas.0603779103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rupp WD, Howard-Flanders P. Discontinuities in the DNA synthesized in an excision-defective strain of Escherichia coli following ultraviolet irradiation. J. Mol. Biol. 1968;31:291–304. doi: 10.1016/0022-2836(68)90445-2. [DOI] [PubMed] [Google Scholar]
- 35.Blow JJ, Dutta A. Preventing re-replication of chromosomal DNA. Nat. Rev. Mol. Cell. Biol. 2005;6:476–486. doi: 10.1038/nrm1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Blow JJ, Gillespie PJ. Replication licensing and cancer–a fatal entanglement? Nat. Rev. Cancer. 2008;8:799–806. doi: 10.1038/nrc2500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yang YG, Cortes U, Patnaik S, Jasin M, Wang ZQ. Ablation of PARP-1 does not interfere with the repair of DNA double-strand breaks, but compromises the reactivation of stalled replication forks. Oncogene. 2004;23:3872–3882. doi: 10.1038/sj.onc.1207491. [DOI] [PubMed] [Google Scholar]
- 38.Hanada K, Budzowska M, Davies SL, van Drunen E, Onizawa H, Beverloo HB, Maas A, Essers J, Hickson ID, Kanaar R. The structure-specific endonuclease Mus81 contributes to replication restart by generating double-strand DNA breaks. Nat. Struct. Mol. Biol. 2007;14:1096–1104. doi: 10.1038/nsmb1313. [DOI] [PubMed] [Google Scholar]
- 39.Davies SL, North PS, Hickson ID. Role for BLM in replication-fork restart and suppression of origin firing after replicative stress. Nat. Struct. Mol. Biol. 2007;14:677–679. doi: 10.1038/nsmb1267. [DOI] [PubMed] [Google Scholar]
- 40.Ciccia A, Bredemeyer AL, Sowa ME, Terret ME, Jallepalli PV, Harper JW, Elledge SJ. The SIOD disorder protein SMARCAL1 is an RPA-interacting protein involved in replication fork restart. Genes Dev. 2009;23:2415–2425. doi: 10.1101/gad.1832309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bryant HE, Petermann E, Schultz N, Jemth AS, Loseva O, Issaeva N, Johansson F, Fernandez S, McGlynn P, Helleday T. PARP is activated at stalled forks to mediate Mre11-dependent replication restart and recombination. EMBO J. 2009;28:2601–2615. doi: 10.1038/emboj.2009.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Van C, Yan S, Michael WM, Waga S, Cimprich KA. Continued primer synthesis at stalled replication forks contributes to checkpoint activation. J. Cell. Biol. 2010;189:233–246. doi: 10.1083/jcb.200909105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Blow JJ, Gillespie PJ, Francis D, Jackson DA. Replication origins in Xenopus egg extract Are 5-15 kilobases apart and are activated in clusters that fire at different times. J. Cell. Biol. 2001;152:15–25. doi: 10.1083/jcb.152.1.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hashimoto Y, Chaudhuri AR, Lopes M, Costanzo V. Rad51 protects nascent DNA from Mre11-dependent degradation and promotes continuous DNA synthesis. Nat. Struct. Mol. Biol. 2010;17:1305–1311. doi: 10.1038/nsmb.1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.