Abstract
Trans-splicing is a powerful approach to reprogram the genome. It can be used to replace 5′, 3′ or internal exons. The latter approach has been characterized by low efficiency, as the requirements to promote internal trans-splicing are largely uncharacterized. The trans-splicing process is induced by engineered ‘RNA trans-splicing molecules’ (RTMs), which target a selected pre-mRNA to be reprogrammed via two complementary binding domains. To facilitate the development of more efficient RTMs for therapeutic applications we constructed a novel fluorescence based screening system. We incorporated exon 52 of the COL17A1 gene into a GFP-based cassette system as the target exon. This exon is mutated in many patients with the devastating skin blistering disease epidermolysis bullosa. In a double transfection assay we were able to rapidly identify optimal binding domains targeted to sequences in the surrounding introns 51 and 52. The ability to replace exon 52 was then evaluated in a more endogenous context using a target containing COL17A1 exon 51–intron 51–exon 52–intron 52–exon 53. Two selected RTMs produced significantly higher levels of GFP expression in up to 61% assayed cells. This novel approach allows for rapid identification of efficient RTMs for internal exon replacement.
INTRODUCTION
Conventional gene therapy strategies are designed to deliver the full-length cDNA copy of a therapeutic gene to produce a functional protein. However, these approaches have disadvantages that may reduce the chance of therapeutic success, including the difficulty to package and deliver large genes and the lack of endogenous regulation of the transgene expression in target cells. Moreover, autosomal dominant disorders may not be amenable to treatment by these conventional therapeutic approaches. These problems can be overcome by using a gene correction strategy, such as spliceosome mediated RNA trans-splicing. This pre-mRNA based method is characterized by the exploitation of naturally occurring splicing events during mRNA maturation (1,2). The endogenous splicing machinery is utilized to recombine two independent RNA molecules by trans-splicing, replacing the disease causing part of a gene by a wild-type version in an exon-wise manner. The trans-splicing process is directed by engineered ‘RNA trans-splicing molecules’ (RTMs), which base-pair with a region of a target pre-mRNA via a complementary binding domain (BD). This interaction between the RTM and the target pre-mRNA confers specificity. Besides the BD, RTMs contain two other elements: a 5′ and/or a 3′ splice site that promote the trans-splicing reaction, and a wild-type coding region that can replace that region with any sequence ranging from a single exon to one or more genes, thus repairing the target gene or reprogramming it to encode any gene of interest.
An RTM can be designed to replace 3′, 5′ or internal sequences of a target pre-mRNA to generate a new gene product (1). After binding to the target pre-mRNA, the trans-splicing process is initiated by the RTM, of which the splicing and coding domains provide recognition sequences for the spliceosome to facilitate what is essentially an alternative splicing reaction in trans. The efficiency of the trans-splicing process depends on the BD and the strength of the splice site sequences of the RTM (3,4). There is a wide range of factors that influence the decision to splice (5). These factors remain largely unexplored in the context of RNA trans-splicing.
Most reports on RNA trans-splicing for gene correction have employed 3′ exon replacement to exchange downstream coding sequences of a gene of interest. 3′ trans-splicing therapy approaches have been successful in restoring potentially therapeutic levels of gene function in patient cells and in animal models of cystic fibrosis, X-linked immunodeficiency and hemophilia A (6–9).
For correction of the upstream regions of a gene, 5′ trans-splicing was first reported using a double transfection model to correct mutations expressed by a plasmid-encoded cystic fibrosis transmembrane receptor (CFTR) pre-mRNA. Those RTMs were designed to replace the 5′ portion of a transcript, inserting the 1.9 kb sequence coding for exon 1–10 of the CFTR gene (3). Similarly, 5′ trans-splicing has been used to replace β-globin exon 1 in cells co-transfected with target and RTM expressing plasmids (10). Endogenous 5′ trans-splicing correction was first reported in 2008 by Wally et al., (11) who demonstrated that an RTM can correct over 1 kb of the upstream coding region of the PLEC gene. Restoration of normal plectin expression patterns were detected by immunofluorescence microscopy after RTM transfection of epidermolysis bullosa (EB) patient fibroblasts. Additionally, Wally et al. developed a 5′ trans-splicing strategy to specifically reprogram exons 1–7 of the keratin 14 gene (KRT14) in an autosomal dominant model of EB-simplex, leading to a phenotypic reversion of RTM treated cells in vitro (12).
RNA trans-splicing can also be used for internal exon replacement (IER) to correct internal coding regions of a gene by using the combination of 5′ and 3′ trans-splicing (1,13,14). Here, an RTM with two BDs base-pairs up and downstream of the region to be replaced (i.e. a mutated exon), inducing the trans-splicing of a wild-type exon from the RTM to the naturally flanking exons of the endogenous target pre-mRNA. The main advantage of this method is the possibility to manufacture and deliver an RTM with a relatively short sequence into patient cells, thereby minimizing the size of the construct and many size-associated limitations.
As a prototypic gene to perform IER, we analyzed the collagen 17A1 (COL17A1) gene, which is mutated in the non-Herlitz junctional form of EB (15). EB is a genetically and clinically heterogeneous group of skin diseases characterized by blister formation and erosions of the skin and mucous membranes after minor trauma. To date, more than 10 different genes that encode for structural proteins within keratinocytes or mucocutaneous basement membranes have been linked to EB. The severity of the disease depends on which gene is mutated and the localization of the mutation within this gene (16,17).
Here, we constructed a novel RTM screening system, in which the reporter molecule acGFP acts as marker for specific double trans-splicing events in the COL17A1 gene in vitro. The acGFP gene was split into three parts. The middle (internal) part was cloned into an RTM capable of performing two targeted trans-splicing reactions. The 5′ and 3′ parts of GFP were cloned into the target molecule flanking the genomic target sequence of COL17A1 intron 51–exon 52–intron 52. We show that this screening system can be used to rapidly identify those RTMs, which produce the highest levels of trans-splicing and function.
MATERIAL AND METHODS
Cell culture and transfection
The human embryonic kidney cell line HEK293AD (Stratagene) was utilized for screening experiments. Cells were grown at 37°C and 5% CO2 in a humidified incubator in DMEM supplemented with 10% FCS and 100 U/ml penicillin/streptomycin (Biochrom). Passaging of the cells was performed every 4–5 days using 1% Trypsin–EDTA (Biochrom), and cells were pelleted by centrifugation at 170g for 5 min. Afterwards cells were seeded to a new tissue culture plate at the desired density. Cells were transiently transfected using jetPEI (Polyplus-transfection SA, Illkirch, France). Various amounts of plasmid DNA (1–10 µg) were applied according to the manufacturer's protocol.
Target molecule construction
The engineered target molecule consists of a short 5′ region of acGFP sequence (183 bp), a COL17A1 target region consisting of intron 51, exon 52, intron 52 and the 3′-coding region of acGFP (231 bp). The target was constructed by PCR amplification of the acGFP split parts (5′ acGFP forward primer: 5′-gatcggatcccaccatggtgagcaagggcgcc-3′, 5′acGFP reverse primer: 5′-tatgatatcacactcaccagggtgggccagggcac-3′, 3′acGFP forward primer: 5′-gatcgcggccgcttttccctccaggtgaacttcaagatccgc-3′, 3′ acGFP reverse primer: 5′-gatctctagatcacttgtacagctcatcc-3′) and the target region intron 51–exon 52–intron 52 COL17A1 (intron 51 COL17A1 forward primer: 5′-tatgatatcggtgccccgacggtgatgccccacc-3′, intron 52 COL17A1 reverse primer: 5′-gatcgcggccgcgtaaaacaccagagcttgggcacagg-3′) using Pfu Turbo DNA polymerase (Stratagene). The pacGFP vector (Clontech) and genomic DNA from a healthy donor was used as PCR template, respectively. The 5′ acGFP region was cloned into the expression vector pcDNA 3.1D/V5-HIS (Invitrogen), using the restriction enzymes BamHI and EcoRV. A 5′ splice site (ag/gtgagtgt) was included in the reverse amplification primer and cloned at the junction between the 5′ acGFP region and intron 51 of COL17A1. The restriction enzymes EcoRV and NotI were used for cloning of the wild-type sequence of intron 51–exon 52–intron 52 of COL17A1, followed by a 3′ splice site (gcggccgcttttccctccag/g) and the 3′ fragment of acGFP, which was cloned using NotI and XbaI sites. Plasmid preparations were carried out using the Plasmid Mini Prep Kit from Sigma-Aldrich (St. Louis, MO, USA), according to the manufacturer's protocol. All constructs and PCR products were sequenced using a 3130 ABI Prism automated sequencer and ABI PRISM dye terminator cycle sequencing kits (Applied Biosystems, Foster City, CA, USA). PCR products were gel extracted before sequence analysis using the GFX™ PCR DNA and Gel Band Purification Kit (GE Healthcare).
COL17A1 mini-gene construction
The COL17A1 mini-gene target plasmid was constructed by cloning a PCR amplified product containing the COL17A1 target region exon 51–intron 51–exon 52–intron 52–exon 53 into the vector pcDNA 3.1D/V5-HIS (Invitrogen). PCR was performed using an exon 51 COL17A1 specific forward primer (5′-gatcggatccctgccggcttgtcattcatcccaggcc-3′) and an exon 53 COL17A1 specific reverse primer (5′-gatctctagatttggaagaagtccatgaggtccgcagtcacgtt-3′), Pfu Turbo DNA polymerase (Stratagene) and genomic DNA from an EB patient with a homozygous mutation (3898 del TC) in the COL17A1 gene. The resulting PCR product was cloned into the expression vector using the restriction sites for BamHI and XbaI.
Sequential construction to produce a double trans-splicing RTM
Selection of an initial 5′ BD
A 5′ BD targeting the region of COL17A1 exon 52/intron 52 was selected for the first step in construction of a double trans-splicing RTM using a split GFP reporter model system according to the method of Wally et al. (12). In brief, the constructs were produced using the split reporter acGFP instead of dsRED. A 5′ BD library was constructed and ∼40 randomly picked clones were sequenced. Seventeen RTMs with BDs ranging from 78 to 314 nt that were complementary to essentially all regions of the exon 52–intron 52 target were identified and tested for trans-splicing efficiency by quantifying GFP expression using flow cytometry. The most efficient BD was PCR amplified with primers containing flanking NotI sites.
Initial double trans-splicing RTM construction
The double trans-splicing RTM screening cassette vector was created in three steps. First, a pIRES2-AcGFP1 vector (Clontech) was digested with EcoRI and KpnI. Elements of the 3′ trans-splicing arm were introduced as double stranded synthesized oligos into these sites, including an EcoRV site for later cloning in a BD library, a spacer and a branch point (underlined) (5′-gaattcgttaacgatatcgagaacattattatagcgttgctcgagtactaactggtacc-3′). Second, the internal portion of acGFP (306 bp) was generated by PCR using Pfu Turbo DNA polymerase (Stratagene).
The forward primer (5′-tatggtacctcttcttttttttctgcaggtgaccaccctgagctac-3′) contains a KpnI site and the remainders of the 3′ splice site to generate a functional 3′ RTM containing the same elements as previously described (18). The reverse primer (5′-tatgcggccgctgtaataatatcgcaacgagctctcttaccttgatgccattcttggcc-3′) contains a 5′ splice site, a spacer region and a NotI restriction site. These two cloning steps removed the IRES and full-length GFP sequences from the Clontech vector. Finally, the selected 5′ BD was cloned into the NotI restriction site. The resulting clones were sequenced to select one with the correct (complementary) orientation.
Selection of a 3′ binding domain
A 3′ BD library was constructed by fragmenting the PCR amplified target sequence of COL17A1 intron 51/exon 52 using CviJI* (Roboklon; Berlin, Germany).
Partial CviJI* digestion of this 676 bp region was performed for 1–10 s at 37°C and the resulting fragments were pooled. CviJI* cleaves and produces blunt end DNA fragments between the G and C at the sequence 5′-PuGCPy-3′. The resulting fragments were cloned upstream of the 3′ RTM spacer region into the EcoRV site. After cloning, 90 individual bacterial clones were checked for the presence of a BD fragment by colony PCR. DNA sequencing showed that ∼95% of clones contained at least one BD, ∼50% were complementary to the target region. Fifty-four RTMs harboring one or more complementary BDs were tested for trans-splicing efficiency quantifying GFP expression by flow cytometry. Geometric mean calculations were performed using the program FlowJo (Treestar). The most efficient BD targeting intron 51 was chosen for further development.
Selection of improved 5′ BD
To improve the RTM selected above, an additional round of 5′ BD screening was performed. An EcoRV restriction site was introduced at the 5′ RTM spacer sequence downstream of the internal acGFP fragment by performing a second PCR amplification of the acGFP fragment using an internal acGFP forward primer (5′-tatggtacctcttcttttttttctgcaggtgaccaccctgagctac-3′) and an internal acGFP reverse primer (5′-gatcgcggccgcgatatcttaataatatcgcaacgagctctcttaccttgatgccattcttggcc-3′). The resulting PCR product was subsequently cloned into the RTM vector using the restriction enzymes KpnI and NotI. The PCR amplified COL17A1 target region exon 52/intron 52 was CviJI* digested as previously described and the resulting DNA fragments were cloned into the EcoRV site. Twenty-seven RTMs, harboring one or more complementary BDs for the target region were tested for trans-splicing efficiency by quantifying GFP expression using flow cytometry. Two low efficiency RTMs (RTM 1 and 3) and the two RTMs with the highest trans-splicing efficiency (RTM 2 and 4) were selected for further experiments.
RNA isolation and cDNA synthesis
RNA was isolated from 106 cells using the RNeasy Mini Kit (Qiagen) according to the manufacturer's protocol. Purified RNA (1–2 µg) was digested with DNase I (Sigma) for 30 min at RT, which was subsequently inactivated by incubation for 10 min at 70°C, followed by cDNA synthesis using the iScript™cDNA Synthesis Kit (Biorad) with the provided mixture of oligo(dT) and random hexamer primers for cDNA synthesis.
RT–PCR
Exon replacement was detected by RT–PCR using GoTaq polymerase (Promega) and the following conditions: 95°C for 5 min, 50 cycles of 30 s at 95°C, 30 s at 65°C, 30 s at 72°C, and a final extension of 7 min at 72°C, specific primers for acGFP (5′ acGFP specific forward primer: 5′-gatcggatcccaccatggtgagcaagggcgcc-3′ and 3′acGFP specific reverse primer: 5′-cgccgatgggggtattctgctgg-3′) and cDNA. Expected PCR products were the trans-splicing product acGFP, the cis-splicing product 5′acGFP–exon 52–3′acGFP, and the fusion mRNA 5′acGFP–3′acGFP created by exon skipping on the target molecule. Co-transfection of single RTMs and the COL17A1 mini-gene target plasmid into HEK293AD cells produced two distinct splicing products, a trans-spliced product (exon 51–middle portion of acGFP–exon 53) and a cis-spliced product (exon 51–exon 52–exon 53). These mRNAs were amplified by RT–PCR in one reaction using the COL17A1 mini-gene specific primer pair (exon 51 COL17A1 specific forward primer: 5′-ggagcgagctgatcagctacctcac-3′, and exon 53 COL17A1 specific reverse primer: 5′-gccatcccttgcagtaggccctg-3′) and the same conditions as described above.
Flow cytometry
Fluorescence microscopy and flow cytometric analysis was performed 2–4 days after RTM and target plasmids were co-transfected into HEK293AD cells. For cytometry, the transfected HEK293AD cells were washed with PBS buffer (Dulbecco's PBS), trypsinised to release the cells from the culture plates, and centrifuged for 5 min at 170g. The cell pellet was resuspended in 500–1000 µl PBS and applied to the Beckman Coulter FC500. Approximately 25 000–80 000 HEK293AD cells were analyzed for GFP reporter gene expression. The CXP software was used for data analysis.
FACS sorting and western blot analysis
Two days after HEK293AD cells were co-transfected with RTM and target expression plasmids, acGFP protein expression was quantified by western blot analysis. Western blot analysis was performed using either total cell extracts from transfected HEK293AD cells or FACS selected GFP positive cells (Beckman Coulter Epics Altra). Cells were resuspended in lysis buffer (0.5 M Tris–HCl, pH 6.8, 20% Glycin, 10% SDS, 5% ß-Mercaptoethanol) containing a protease inhibitor cocktail (Roche, Mannheim, Germany). After addition of 4× loading buffer and denaturation at 95°C for 5 min, SDS-gel-electrophoresis was performed at 100–150 V for 1.5 h using a NuPAGE 10% or 4–12% Bis–Tris Gel (1.0 mm × 12 well) and a NuPAGE MOPS SDS running buffer (20×) (Invitrogen). Post-electrophoresis, the SDS gel was equilibrated in standard blotting buffer for up to 30 min at RT, and then the proteins were electro-blotted onto a nitrocellulose membrane (Amersham Hybon-ECL, GE Healthcare) for 75 min at 0.4 Ampere. After the protein transfer, the membrane was soaked in standard blocking buffer containing 1% western blocking reagent (Roche) for 1 h at RT. First antibodies, anti-GFP rabbit IgG (Biomedica) and anti-Annexin I mouse monoclonal IgG (Santa Cruz Biotechnology), were diluted 1:1000 and 1:250 in TBS-T, respectively, containing 0.5% western blocking reagent and incubated with the membrane over night at 4–8°C. The membrane was washed with TBS-T, then incubated with HRP labeled Envision+ anti-rabbit antibody (diluted 1:50–1:100, Dako) and HRP labeled Envision+ anti-mouse antibody (diluted 1:250–1:500, Dako) in TBS-T containing 0.5% Western blocking reagent. After washing with TBS-T the blot was visualized using the Immun-Star WesternC Kit (Biorad). Protein quantification was performed using the program Image Lab 3.0.1. (Biorad).
RESULTS
Rational design, construction and evaluation of RTM-BDs can be an arduous and time consuming process as the requirements for single or double trans-splicing are essentially unknown. To facilitate this process, a rapid method to identify the most efficient trans-splicing RTMs is desirable. We have developed a screening system that can be used to select the most efficient double trans-splicing RTMs from a library containing numerous RTMs with different BDs that are complementary to the target region.
Selection of an initial 5′ BD
To begin, we decided to identify a working 5′ BD targeting COL17A1 exon 52 / intron 52 using the screening system previously published by Wally et al. (12). To minimize the complexity of screening and selecting for two BDs simultaneously, we chose to proceed in a sequential manner, selecting one BD at a time. Seventeen RTMs specific for exon/intron 52 were individually tested for trans-splicing induced restoration of GFP expression by flow cytometry (data not shown). The BD of the 5′ RTM that generated the highest level of GFP expression was incorporated into the 5′ trans-splicing arm of a double trans-splicing RTM vector (version 1).
Screening for double trans-splicing efficiency
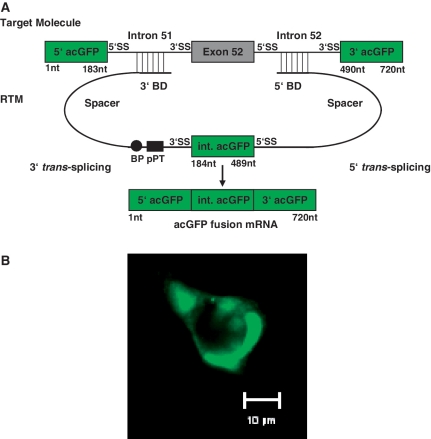
Since 3′ and 5′ trans-splicing have to work in synchrony, we feel that screening for the most efficient BDs for double trans-splicing should be done in the context of a double trans-splicing RTM. Our double trans-splicing fluorescence based screening system utilizes two pre-mRNA components, a target molecule containing a disease associated region of the COL17A1 gene and a library of RTMs that can potentially trans-splice and restore the coding sequence of a reporter gene flanking the target sequence (Figure 1). Trans-splicing efficiency can be evaluated by restoration of reporter gene expression. For this screen, the reporter acGFP was divided into three portions and strategically cloned into the target molecule and the RTM. The 5′ and 3′ parts of acGFP were cloned into the target vector flanking the COL17A1 intron 51–exon 52–intron 52 target region. The double trans-splicing RTMs carry the middle portion of acGFP flanked by a 3′ and a 5′ trans-splicing arm, each consisting of a BD and a 5′ or 3′ splice site. A diversity of RTMs, differing in their target specific BD sequences was created by fragmenting the target region of COL17A1 and blunt end cloning of the resulting fragments into the RTM vector.
Figure 1.
Schematic description of the fluorescence based RTM screening system. (A) The fluorescence based RTM screen utilizes cells expressing both an exon specific target molecule and an RTM. Trans-splicing induced RNA recombination between the target and the RTM pre-mRNAs creates the fusion of all three acGFP fragments, thus restoring the full coding sequence and expression of functional acGFP. The target molecule contains exon 52 and the flanking introns of COL17A1, along with 5′ and 3′ fragments of acGFP. The RTM contains the internal coding region of acGFP (the missing coding sequence) flanked by a 3′ and 5′ splice site and other features required for the double-trans-splicing reaction. Each RTM has two arms, containing an acceptor (3′) or donor (5′) splice site, a short spacer region and a ‘random’ BD created by fragmenting the COL17A1 target region. The randomized fragments were blunt ended, and cloned into the RTM backbone using the restriction site for EcoRV. (B) AcGFP expression was visualized by fluorescence microscopy in HEK293AD cells co-transfected with RTM and target molecule plasmids (40× magnification). BP: Branch Point, pPT: Polypyrimidine Tract, SS: Splice Site, nt: nucleotides.
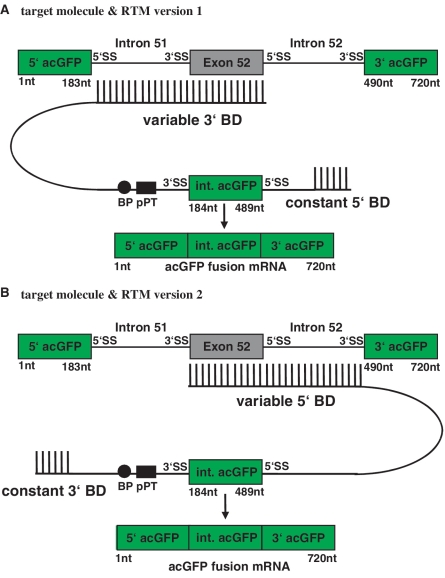
Screening for an efficient 3′ BD
A random 3′ BD library targeting COL17A1 intron 51–exon 52 was constructed as described in the methods section and cloned into the double trans-splicing RTM vector (version 1) harboring the initial working 5′ BD (Figure 2A). From a library of thousands of independent RTMs, 90 unique RTMs were sequenced and 54 with complementary BDs were identified. Each of these RTMs was individually tested by co-transfection with target plasmid into HEK293AD cells and analyzed for trans-splicing efficiency by GFP expression using flow cytometry. After calculating the average GFP expression of RTM treated cells, the 3′ BD of the most efficient RTM was chosen for further RTM improvement.
Figure 2.
Sequential construction of a double trans-splicing RTM. Since 3′ and 5′ trans-splicing have to work in synchrony, we believe that screening for the most efficient BDs for double trans-splicing should be done in the context of a double trans-splicing RTM. A random 3′ BD library targeting COL17A1 intron 51–exon 52 was constructed and cloned into the double trans-splicing RTM vector (version 1) harboring the initial working 5′ BD identified using the method of Wally et al. (A). To further improve the efficiency of the double trans-splicing RTM (version 2), a new 5′ BD screen was performed using the efficient 3′ BD identified in the 3′ BD screen (A) and a variable 5′ BD specific for the COL17A1 exon/intron 52 target region (B).
Screening for an efficient 5′ BD
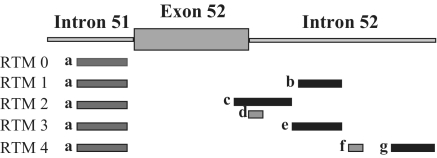
The main aim of this additional screen was the further improvement of the existing RTM by screening for a new 5′ BD that works well together with the selected 3′ BD specific for intron 51 and better than the initial 5′ BD specific for exon/intron 52 (Figure 2B). Twenty-seven different RTMs, with the selected 3′ BD (Figure 3) and a variable BD for 5′ trans-splicing, were evaluated for efficient trans-splicing efficiency by flow cytometry. The geometric mean of GFP expression in RTM treated cells ranged from 2.6 to 3.5. Geometric mean calculation was performed only for RTMs that induced GFP expression in >5% of all analyzed cells. Five RTMs (RTM 0, 1, 2, 3, 4) from the 5′ double trans-splicing RTM library were selected as candidates for further experiments shown in this study (Figure 3). RTM 0 lacks a BD for the 5′ trans-splicing reaction and was therefore included as negative control in the experiment. RTMs 1 and 3 were representatives for less functional RTMs, as the level of acGFP expression was decreased in comparison to the most efficient RTMs 2 and 4.
Figure 3.
3′ and 5′ BDs of selected RTMs. The BD sequences have a great influence on the efficiency of trans-splicing. Five RTMs selected from an RTM library with the same 3′ BD for intron 51 (a; binding position from nt 106 of intron 51 to nt 257 of intron 51) of COL17A1 but with different BDs for the exon 52—intron 52 target region were selected for further testing. RTM 0 had no 5′-BD, whereas RTM 2 and 4 harbored two separate BDs specific for exon 52—intron 52. One region of complementarity of RTM 2 masks the exon/intron boundary of exon 52—intron 52 (c; binding position from nt 360 of exon 52 to nt 190 of intron 52). The second domain repeats some of same region of the 5′-end of intron 52 (d; binding position from nt 20 to nt 63 of intron 52). RTM 4 binds to the central (f; binding position from nt 318 to nt 338 of intron 52) and 3′ (g; binding position from nt 457 to nt 555 of intron 52) portion of intron 52. RTMs 1 and 3 each have one 5′ BD complementary to the central portion of intron 52 (b; nt 217 to nt 297 of intron 52, e; nt 191 to nt 297 of intron 52).
Exon replacement on mRNA level
A COL17A1 specific double trans-splicing RTM is able to recombine with a target pre-mRNA by two synchronous trans-splicing reactions (5′ and 3′). Thus the exon on the target molecule should be replaced by the middle portion of acGFP, restoring the open reading frame of acGFP leading to the expression of the reporter molecule in RTM treated cells.
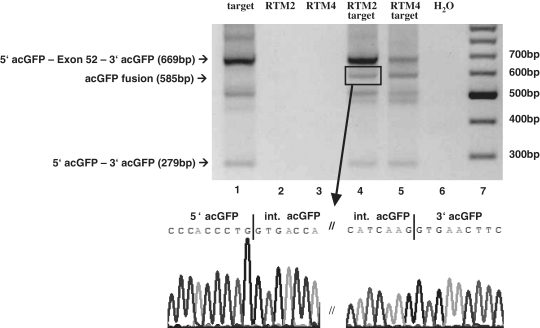
One day after transfecting double trans-splicing RTMs into HEK293AD cells, different splicing patterns were observed by RT–PCR. Cis-splicing within the target molecule generated an mRNA consisting of the 5′ and 3′ acGFP with exon 52 of COL17A1 in the middle. This demonstrates that the acceptor (3′) and donor (5′) splice sites at the exon / intron junctions were recognized by the endogenous splicing machinery and that the target introns were excised. The cis-spliced 5′ acGFP–COL17A1 exon 52–3′ acGFP fusion product was detected by RT–PCR amplification and agarose gel electrophoresis. This band was visible in all HEK293AD cells transfected with target plasmid (Figure 4, lanes 1, 4 and 5). Double trans-splicing between the RTM and target molecule pre-mRNAs lead to the replacement of exon 52 of the target molecule by the internal acGFP sequence carried by the RTM. The resulting full-length GFP mRNA was exclusively detected in HEK293AD cells co-transfected with both target and RTM expression plasmids (Figure 4, lanes 4 and 5). Since the products created by cis- or trans-splicing differ in size, discrimination by RT–PCR is possible, as shown in Figure 4, lanes 4 and 5, indicated bands. Skipping of exon 52 was also detected in cells transfected with the target plasmid (Figure 4, lanes 1, 4 and 5). PCR products were excised from the gel and splicing products were confirmed by DNA sequence analysis. The PCR primers used were designed to hybridize to the 5′ and 3′ terminal regions of the target molecule. No trans-spliced products were detected in HEK293AD cells transfected exclusively with RTM (Figure 4, lanes 2 and 3).
Figure 4.
Detection of acGFP restoration by double RNA trans-splicing on mRNA level. Trans-splicing on mRNA level can be detected by RT–PCR in HEK293AD cells co-transfected with RTM 4 or RTM 2 and target molecule expressing plasmids. COL17A1 exon 52 replacement by the internal acGFP fragment contributed by the RTM was detected using a 5′ acGFP and 3′ acGFP specific primer pair to discriminate between cis- and trans-splicing events. The cDNA templates included: lane 1: target molecule alone, lane 2: RTM 2 plasmid only, lane 3: RTM 4 plasmid only, lane 4: target molecule + RTM 2 plasmids, lane 5: target molecule + RTM 4 plasmids, lane 6: water control. The cis-splicing product 5′acGFP - exon 52 - 3′acGFP (lanes 1, 4, 5), the trans-splicing product acGFP (lanes 4 and 5; The marked band in lane 4, indicated by an arrow, was sequenced after recovery and gel extraction) or the exon skipping product 5′ acGFP - 3′ acGFP (lanes 1, 4, 5) was visualized on a 2% agarose gel after gel electrophoresis. Lane 7: DNA ladder mix (Fermentas).
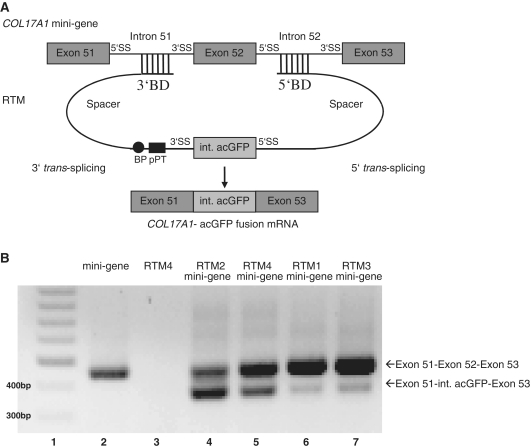
Double trans-splicing into the COL17A1 gene was modeled using a mini-gene target plasmid that was constructed to express the endogenous pre-mRNA region from exon 51 through exon 53. Successful double trans-splicing should cause the replacement of COL17A1 exon 52 with the internal portion of acGFP (Figure 5A). HEK293AD cells were co-transfected with plasmids expressing the COL17A1 mini-gene target and a single RTM (1–4). Trans-splicing efficiency was evaluated by RT–PCR using RNA extracted from these cells one day post transfection. Trans-splicing between the target exons and the middle portion of acGFP demonstrates the efficiency of the RTMs tested in the context of the endogenous COL17A1 sequence (Figure 5B). Both splice products (cis and trans) were amplified by RT–PCR and visualized by agarose gel electrophoresis. These PCR products were gel purified and confirmed by DNA sequence analysis.
Figure 5.
Double RNA trans-splicing between RTM and COL17A1 mini-gene. (A) A COL17A1 mini-gene expression plasmid was constructed by cloning the endogenous gene region exon 51–intron 51–exon 52–intron 52–exon 53 of COL17A1 into a pcDNA 3.1D/V5-HIS expression vector (Invitrogen). Co-transfection of the COL17A1 mini-gene expression plasmid and a functional RTM generated RNA trans-splicing between the middle portion of acGFP carried by the RTM to exon 51 and 53 of the target molecule. (B) One day after HEK293AD cells were co-transfected with either RTM 1 (lane 6), 2 (lane 4), 3 (lane 7), or 4 (lane 5) and COL17A1 target mini-gene expression plasmids, double trans-splicing was detected by RT–PCR. Using a COL17A1 mini-gene specific primer pair (exon 51 COL17A1 specific forward primer, exon 53 COL17A1 specific reverse primer) it is possible to discriminate between the product of cis-splicing (exon 51–exon 52–exon 53 = 442 bp) or trans-splicing (exon 51—middle acGFP fragment—exon 53 = 358 bp). No trans-splicing products were detected by RT–PCR of RNA from HEK293AD cells transfected with COL17A1 mini-gene expression plasmid (lane 2) or RTM 4 (lane 3) alone. Lane 1: DNA ladder mix (Fermentas).
Flow cytometric analysis of treated HEK293AD cells
To detect acGFP repair and expression at the protein level, fluorescence microscopy and flow cytometry were performed on HEK293AD cells co-transfected with RTM and target plasmids.
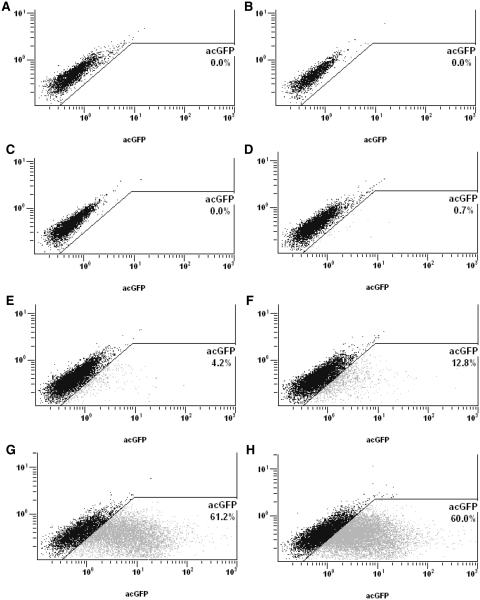
Two or more days post co-transfection with RTM and target molecule the cells were evaluated for acGFP expression by fluorescence microscopy. The fraction of acGFP expressing cells and the expression level of GFP reporter was also quantified by flow cytometry. The sequences included as BDs have a profound effect on the efficiency of double trans-splicing RTMs as quantified by the number of GFP positive cells and the level of GFP produced. Compared with the less efficient RTMs 1 (4.2% GFP positive cells) and 3 (12.8%), RTMs 2 and 4 produced significantly higher levels of acGFP expression in up to 61% of transfected HEK293AD cells (Figure 6G). Background expression levels were less than 1% as detected in cells co-transfected with target and RTM expression plasmid 0, which had no 5′ BD (Figure 6D). No acGFP expression was detected after single transfection with target molecule, RTM 2, or RTM 4 (Figure 6A–C). The geometric mean of RTM 1 and 3 was 2.17 and 2.53. The most efficient RTMs 2 and 4 had significantly higher values (geometric mean: 6.77 and 5.28) for the average GFP expression.
Figure 6.
Detection of acGFP expression by flow cytometry. Flow cytometric analysis of HEK293AD cells co-transfected with one RTM and target expression plasmids demonstrates the ability to discriminate between weak and highly efficient RTMs. Single transfection of HEK293AD cells with target molecule plasmid (A), RTM 2 (B) or RTM 4 (C) did not generate any expression of acGFP. Transfection of target molecule in combination with RTM 0, which does not have a specific BD for intron 52, generates weak acGFP expression in ∼0.7% of analyzed cells (D). The RTMs 1 (E) and 3 (F) generated acGFP expression in 4.2 and 12.8% of co-transfected cells. RTM 2 (G) and 4 (H) demonstrated the greatest trans-splicing efficiency, up to 61% of co-transfected HEK293AD cells were green, and the geometric mean level of acGFP expression was significant greater. All RTMs harbor the same 3′ BD specific for intron 51, but differ in the binding sequences complementary to intron 52 of COL17A1 (Figure 3).
Detection of acGFP by western blot analysis
Western Blot analysis was performed to demonstrate that the portion of acGFP carried by the RTM trans-spliced twice into target pre-mRNA in treated cells and correctly assembled all three of the split parts of acGFP. The repaired acGFP should be detected at the same position as a full-length acGFP after immunostaining.
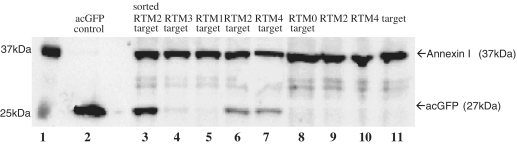
Western blot analysis was performed at least two days after transfection using total cell extracts from transfected HEK293AD cells to quantify the restoration of full-length acGFP (27 kDa) by trans-splicing (Figure 7). A full-length acGFP-expressing plasmid was used as positive control in the experiment (Figure 7, lane 2). AcGFP was visualized after blotting and immuno-staining of HEK293AD total protein extracts from cells co-transfected with target molecule along with either RTM 2, 3, or 4 (Figure 7, lanes 4, 6 and 7). RTMs 0 and 1 were not able to generate a detectable acGFP band (Figure 7, lanes 5, 8). These RTMs also produced less than ∼5% GFP positive cells as quantified by flow cytometric analysis. No acGFP expression was detected after single transfection of target molecule, RTM 4 or RTM 2 alone into HEK293AD cells (Figure 7, lanes 9–11). HEK293AD cells co-transfected with RTM 2 and target expression plasmids were also selected by FACS sorting with a purity of ∼90% (data not shown). Removal of the untransfected cells increased the intensity of the acGFP band significantly (Figure 7, lane 3).
Figure 7.
Detection of full-length acGFP restoration by western blot analysis. After co-transfection of target molecule and either RTM 2 (lane 6), 4 (lane 7), or 3 (lane 4), acGFP protein (27 kDa) was detected by western blot analysis. As positive control, total-cell extract from acGFP control vector (acGFP cloned in pcDNA 3.1D/V5-HIS vector, Invitrogen) transfected HEK293AD cells was also included (lane 2). Lane 3 demonstrates a significant increase (∼3-fold) in the amount of acGFP protein in acGFP positive cells collected by FACS sorting of HEK293AD cells co-transfected with RTM 2 and target expression plasmids (lane 3) compared to total cell extract from co-transfected HEK293AD cells without sorting (lane 6). No detectable acGFP expression was visible after single transfection of target molecule (lane 11), RTM 4 (lane 10) or RTM 2 (lane 9) into HEK293AD cells. No expression of acGFP was detected by western blot analysis of HEK293AD cells co-transfected with target molecule and either RTM 1 (lane 5) or RTM 0 (which is missing a specific BD for intron 52, lane 8). Annexin I (37 kDa) was included as the loading control for this experiment. Lane 1: Precision Plus Protein WesternC Standards (Biorad).
DISCUSSION
In this study we demonstrate that IER is a technology that has potential applications in treating genetic diseases. However, at present there are no rules or guidelines for designing highly efficient double trans-splicing RTMs. Building RTMs one at a time and testing them for efficiency is an arbitrary and arduous process. Thus we have developed a rapid screening system to screen for efficient BDs to two introns flanking a given exon. As a model system we chose to replace exon 52 of the COL17A1 gene, a gene mutated in the devastating disease of the skin and mucous membranes EB.
The methodology of RNA trans-splicing provides many important advantages over conventional gene therapy approaches. Mutated RNA molecules can be reprogrammed to wild-type, thus reducing the amount of cis-spliced disease associated gene expression. RNA trans-splicing can be used to replace 5′, 3′ or internal exon(s), referred to as 5′, 3′ or IER. To date, several groups have shown functional RNA repair using either 3′ or 5′ RNA trans-splicing in genetic diseases (6–12).
Still, there are many distinct genetic conditions in which combining 3′ and 5′ trans-splicing might be advantageous or even the only feasible therapeutic strategy. Such situations occur for example when the gene segment to be replaced by 3′ or 5′ RNA trans-splicing is too large to be delivered by existing vectors. IER permits the use of RTMs, which are only ∼200 nt larger than the exon(s) to be replaced. Double trans-splicing RTMs do not need to deliver the full-length coding sequence of a gene, untranslated regions (UTRs) and start or termination codons. Alternatively spliced exons and regulatory sequences within the 5′- and 3′-UTRs, which are essential for mRNA stability and translation regulation, are left intact in the target pre-mRNA. One such gene is PLEC, which has multiple mRNA isoforms some that exceed 14 kb. Mutations in this gene lead to a subgroup of EB that is associated with progressive form of late onset muscular dystrophy (EBS-MD). In this case, 3′ or 5′ RNA trans-splicing repair can only be applied to a fraction of EB patients that carry mutations in exons located at the 5′- and 3′-end of the gene. The terminal exon of PLEC, exon 32, is >6.5 kb by itself. Therefore, double trans-splicing to replace the 3.38 kb coding sequence of exon 31, is required to effect delivery and repair of mutations in this major EBS-MD disease associated region. Additionally, the expression of PLEC isoforms, should not be altered, except for the correction of mutations within the targeted exon.
Other promising target genes for IER are the EB associated genes COL17A1 and COL7A1. Both of these genes have been corrected using 3′ trans-splicing (6,18). In vitro studies by Murauer et al. demonstrated a phenotypical correction of COL7A1 in dystrophic EB (DEB) patient cells. Primary keratinocytes from a DEB patient were transduced with a retrovirus encoding a 3′ RTM designed to repair COL7A1 exons 65–118. Significant levels of normal type VII collagen expression was restored. The treated keratinocytes secreted type VII collagen at the basement membrane zone in skin equivalents. A double RNA trans-splicing strategy may be needed to repair many of the internal exons (exons upstream of exon 65) of COL7A1 as the 3′ trans-splicing strategy described above would require an RTM that exceeds the packaging limit of many delivery systems.
A double trans-splicing RTM is a tool that can replace one or more internal exons of a targeted pre-mRNA. The exons to be reprogrammed by the RTM are chosen on the basis of their clinical relevance. The design of the other RTM elements is much more empiric. Alternative sequences for the spacers, 3′ and 5′ splice sites can modulate trans-splicing efficiency or specificity; however, the sequence of the BDs and their interactions with the pre-mRNA target clearly has a profound effect (1–3). At present the rules for building optimal double trans-splicing RTMs are unknown. Therefore we created a fluorescence based screening system to rapidly identify the most efficient double trans-splicing RTMs from a library of RTMs with target specific, but random position and length of BDs.
In the fluorescence based screening system, the repair of the reporter molecule acGFP acts as marker for specific double trans-splicing events in co-transfected cells. Using flow cytometry, we were able to rapidly evaluate and compare the trans-splicing efficiency of RTMs differing in their target binding sequences. Up to 60% of analyzed HEK293AD cells expressed high levels of acGFP after co-transfection with target and either double trans-splicing RTM 2 or RTM 4. Under the same co-transfection conditions, other RTMs with an identical 3′ BD but no or a different 5′ BD generated 0.7–13% green cells. There are several possible explanations for the observed difference in efficiency: (i) It is likely that binding to the target molecule requires that the target region is accessible, that it does not strongly interact with itself or other molecules in the nucleus; (ii) two discontinuous BDs may be more stable or more difficult to completely disrupt than a single BD; (iii) blockage of the exon/intron boundary of the target molecule, accomplished by the longer BD of RTM 2, may interfere with recognition of the exon 52 5′ splice site and facilitate trans-splicing; and (iv) BDs with higher GC content may be more stable in binding to the target region. The BDs of RTM 2 (BD c = 61% G+C, BD d = 66%) and RTM 4 (BD f = 57%, BD g = 56%) have a higher GC content in comparison to those of RTM 1 (47% G+C) and RTM 3 (52%). The essential point is that the BDs for both trans-splicing reactions (3′ and 5′) must work together well for efficient trans-splicing replacement of the target exon. Immunostaining was performed to confirm the production of full-length 27-kDa acGFP protein. Very low-level expression of acGFP was detected by flow cytometric analysis of HEK293AD cells co-transfected with target molecule and RTM 0, which has no specific 5′ BD. This may have been caused by the RTM performing a specific 3′ trans-splicing reaction into the target and a relatively non-specific 5′ trans-splicing reaction into the target, facilitated by the interaction of the single BD of the RTM with the target. We also created a trans-splicing model system that more closely resembles the endogenous gene. A COL17A1 target mini-gene was created with fully endogenous sequence from exon 51 through exon 53, with the intervening exon/intron boundaries. This model allows for further characterization and selection of double trans-splicing RTMs in a standard cell line (e.g. HEK293AD cells) prior to final selection in EB patient cells. Since there are no general rules for predicting optimal binding regions of a pre-mRNA of interest, the described RTM screen represents a promising tool to optimize gene correction by spliceosome mediated RNA trans-splicing. Different RTM-BDs and spacer region sequences can be rapidly evaluated by this screen. This should reduce time and effort required to select optimal RTMs prior to testing them in the more complex, but highly relevant endogenous gene expression model systems. In this report we have demonstrated that 5′ and 3′ RNA trans-splicing can be combined as a promising tool to repair large genes or those that occur in multiple isoforms. Our fluorescence based RTM screening system should help to more rapidly optimize RNA correction by trans-splicing to higher levels, which should have a greater probability to correct genetic diseases.
FUNDING
DEBRA Austria. Funding for open access charge: Division of Molecular Dermatology and EB House Austria, Department of Dermatology, Paracelsus Medical University Salzburg.
Conflict of interest statement. Dr Mitchell has an ownership interest in a biotechnology company.
REFERENCES
- 1.Mitchell LG, McGarrity GJ. Gene therapy progress and prospects: reprograming gene expression by trans-splicing. Gene Ther. 2005;12:1477–1485. doi: 10.1038/sj.gt.3302596. [DOI] [PubMed] [Google Scholar]
- 2.Puttaraju M, Jamison SF, Mansfield SG, Garcia-Blanco MA, Mitchell LG. Spliceosome-mediated RNA trans-splicing as a tool for gene therapy. Nat. Biotechnol. 1999;17:246–252. doi: 10.1038/6986. [DOI] [PubMed] [Google Scholar]
- 3.Mansfield SG, Clark RH, Puttaraju M, Kole J, Cohn JA, Mitchell LG, Garcia-Blanco MA. 5′ exon replacement and repair by spliceosome-mediated RNA trans-splicing. RNA. 2003;9:1290–1297. doi: 10.1261/rna.5101903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Puttaraju M, DiPasquale J, Baker CC, Mitchell LG, Garcia-Blanco MA. Messenger RNA repair and restoration of protein function by spliceosome-mediated RNA trans-splicing. Mol. Ther. 2001;4:105–114. doi: 10.1006/mthe.2001.0426. [DOI] [PubMed] [Google Scholar]
- 5.Shepard PJ, Hertel KJ. Conserved RNA secondary structures promote alternative splicing. RNA. 2008;14:1463–1469. doi: 10.1261/rna.1069408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Murauer EM, Gache Y, Gratz IK, Klausegger A, Muss W, Gruber C, Meneguzzi G, Hintner H, Bauer JW. Functional correction of type VII collagen expression in dystrophic Epidermolysis bullosa. J. Invest. Dermatol. 2011;131:74–83. doi: 10.1038/jid.2010.249. [DOI] [PubMed] [Google Scholar]
- 7.Chao H, Mansfield SG, Bartel RC, Hiriyanna S, Mitchell LG, Garcia-Blanco MA, Walsh CE. Phenotype correction of hemophilia A mice by spliceosome-mediated RNA trans-splicing. Nat. Med. 2003;9:1015–1019. doi: 10.1038/nm900. [DOI] [PubMed] [Google Scholar]
- 8.Liu X, Jiang Q, Mansfield SG, Puttaraju M, Zhang Y, Zhou W, Cohn JA, Garcia-Blanco MA, Mitchell LG, Engelhardt JF. Partial correction of endogenous Delta F508 CFTR in human cystic fibrosis airway epithelia by spliceosome-mediated RNA trans-splicing. Nat. Biotechnol. 2002;20:47–52. doi: 10.1038/nbt0102-47. [DOI] [PubMed] [Google Scholar]
- 9.Tahara M, Pergolizzi RG, Kobayashi H, Krause A, Luettich K, Lesser ML, Crystal RG. Trans-splicing repair of CD40 ligand deficiency results in naturally regulated correction of a mouse model of hyper-IgM X-linked immunodeficiency. Nat. Med. 2004;10:835–841. doi: 10.1038/nm1086. [DOI] [PubMed] [Google Scholar]
- 10.Kierlin-Duncan MN, Sullenger BA. Using 5′-PTMs to repair mutant beta-globin transcripts. RNA. 2007;13:1317–1327. doi: 10.1261/rna.525607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wally V, Klausegger A, Koller U, Lochmüller H, Krause S, Wiche G, Mitchell LG, Hintner H, Bauer JW. 5′ trans-splicing repair of the PLEC1 gene. J. Invest. Dermatol. 2008;128:568–574. doi: 10.1038/sj.jid.5701152. [DOI] [PubMed] [Google Scholar]
- 12.Wally V, Brunner M, Lettner T, Wagner M, Koller U, Trost A, Murauer EM, Hainzl S, Hintner H, Bauer JW. K14 mRNA reprogramming for dominant epidermolysis bullosa simplex. Hum. Mol. Genet. 2010;19:4715–4725. doi: 10.1093/hmg/ddq405. [DOI] [PubMed] [Google Scholar]
- 13.Garcia-Blanco MA, Puttaraju M, Mansfield SG, Mitchell LG. Spliceosome-mediated RNA trans-splicing in gene therapy and genomics. Gene Ther. Reg. 2000;1:141–163. doi: 10.1038/sj.gt.3301307. [DOI] [PubMed] [Google Scholar]
- 14.Lorain S, Peccate C, Le Hir M, Garcia L. Exon exchange approach to repair duchenne dystrophin transcripts. PloS One. 2010;5:e10894. doi: 10.1371/journal.pone.0010894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bauer JW, Lanschuetzer C. Type XVII collagen gene mutations in junctional epidermolysis bullosa and prospects for gene therapy. Clin. Exp. Dermatol. 2003;28:53–60. doi: 10.1046/j.1365-2230.2003.01192.x. [DOI] [PubMed] [Google Scholar]
- 16.Fine JD, Hintner H. Life with Epidermolysis bullosa (EB): Etiology, Diagnosis, Multidisciplinary Care and Therapy. NewYork: SpringerWien; 2009. [Google Scholar]
- 17.Laimer M, Lanschützer CM, Nischler E, Klausegger A, Diem A, Pohla-Gubo G, Bauer JW, Hintner H. Hereditary blistering diseases. Symptoms, diagnosis and treatment of epidermolysis bullosa. Hautarzt. 2009;60:378–388. doi: 10.1007/s00105-008-1686-9. [DOI] [PubMed] [Google Scholar]
- 18.Dallinger G, Puttaraju M, Mitchell LG, Yancey KB, Yee C, Klausegger A, Hintner H, Bauer JW. Development of spliceosome-mediated RNA trans-splicing (SMaRT) for the correction of inherited skin diseases. Exp. Dermatol. 2003;12:37–46. doi: 10.1034/j.1600-0625.2003.120105.x. [DOI] [PubMed] [Google Scholar]