Abstract
Packaging of the segmented RNA genome of Brome mosaic virus (BMV) into discrete particles is an essential step in the virus life cycle; however, questions remain regarding the mechanism of RNA packaging and the degree to which the viral coat protein controls the process. In this study, we used a plant-derived glycosidase, Pokeweed antiviral protein, to remove 14 specific bases from BMV RNA3 to examine the effect of depurination on virus assembly. Depurination of A771 within ORF3 and A1006 in the intergenic region inhibited coat protein binding and prevented RNA3 incorporation into particles. The disruption of interaction was not based on sequence identity, as mutation of these two purines to pyrimidines did not decrease coat protein-binding affinity. Rather, we suggest that base removal results in decreased thermodynamic stability of local RNA structures required for packaging, and that this instability is detected by coat protein. These results describe a new level of discrimination by coat protein, whereby it recognizes damage to specific viral RNA elements in the form of base removal and selects against incorporating the RNA into particles.
INTRODUCTION
Packaging is an essential step in the life cycle of a virus, whereby assembly of viral genomes with virally encoded coat protein (CP) protects the genome from degradation and serves as the primary means for transfer of the virus from one host to the next. For Brome mosaic virus (BMV), a member of the family Bromoviridae, which infects grasses such as barley, the ability to package its viral genome into particles is also essential for cell-to-cell movement and hence systemic infection in plants (1).
The genome of BMV is composed of three single-stranded, positive-sense messenger RNAs and one subgenomic RNA (2,3). RNA1 (3.2 kb) and RNA2 (2.9 kb) are monocistronic and encode the 1a and 2a replicase proteins, respectively (4,5). RNA3 (2.1 kb) is dicistronic, and encodes the 5′-proximal movement protein, which is translated directly from RNA3, and the 3′-proximal CP, which is translated from a subgenomic RNA4 (0.9 kb), synthesized from the 3′ open reading frame (ORF) of RNA3 (6). RNA1 and RNA2 are packaged independently into separate virus particles, whereas RNA3 and the subgenomic RNA4 are co-packaged into a third virion (7). Each viral particle is comprised of either RNA1, 2 or 3 and sgRNA4 along with 180 molecules of CP, which together form an icosahedral virion (8). Although other viral and cellular components may assist in the formation of particles in vivo, the ability to reconstitute particles in vitro from only viral RNA and CP indicates that particle assembly relies heavily on interactions between these two components.
Particular regions of RNA3 have been shown to be necessary for packaging. For example, the 3′ tRNA-like structure found at the terminus of each BMV RNA is hypothesized to facilitate the pentamer formation of CP dimers, which are the building blocks of the icosahedral particle (9). Because the tRNA-like structure is common to all BMV RNAs and can be functionally replaced by a cellular tRNA, it is not a specific packaging signal for individual RNAs; however, the lack of particle formation in its absence shows the importance of this structure for packaging (10). The fact that viral RNAs are distinguished from cellular RNAs during packaging suggests that sequence- or shape-specific regions of each BMV RNA must exist that are recognized by the CP. A stem–loop region of the movement protein gene of RNA3 has been identified as an RNA element required for efficient packaging, as its deletion substantially diminished particle formation (11,12).
In the current study, we have determined that the CP is selective in packaging of viral RNA, as it is able to distinguish between damaged and intact RNAs. Damage was achieved by depurination, caused by the enzyme activity of Pokeweed antiviral protein (PAP). PAP is a glycosidase that has been shown to remove specific purine bases from RNA3 and to inhibit accumulation of BMV RNAs in barley protoplasts (13,14). Previously, we mapped these depurination sites and determined how missing bases within the intergenic region and at the 3′ end of RNA3 inhibited the initiation and elongation steps of replication, respectively (14). Here, we show that depurination of particular nucleotides within ORF3 and the intergenic region of RNA3 prevents incorporation of RNA3 into particles. Even though CP does not bind to a portion of RNA3 containing only the intergenic region, we demonstrate that when present, the intergenic region enhances CP interaction with RNA3 ORF3. In addition to contributing new information describing how CP recognizes RNA3, we also illustrate how depurination inhibits formation of particles, thereby extending our knowledge of the antiviral activity of PAP to the packaging step of the virus life cycle.
MATERIALS AND METHODS
Isolation and transfection of barley protoplasts
Protoplasts were isolated from 7-day-old barley leaves by enzymatic digestion as described previously (15). Protoplasts (3 × 105) were transfected with capped, in vitro transcripts of BMV RNAs (1.0 µg each) and 25 µg of a PAP cDNA, under the control of a constitutive CaMV promoter, using PEG 1500 as described (13), and incubated at 25°C under constant fluorescent light (165 µmol/m2/s) for 18 h.
Protoplast protein and nucleic acid analyses
Aliquots of protoplasts transfected with BMV RNAs and PAP cDNA were removed at indicated time points. Protoplasts were pelleted at 1000 g for 20 s and cells were lysed by adding 300 µl of extraction buffer (2 × TE, 1% SDS). Total protein was precipitated by adding 1 ml of 100% trichloroacetic acid and the amount of protein was quantified by Bradford assay. Immunoblotting to detect PAP and CP expression was performed as described (16), using PAP and CP polyclonal antibodies at 1:5000. Samples were probed for β-actin (monoclonal antibody, 1:5000) as a control for protein loading. Total nucleic acids were isolated from lysed protoplasts by the addition of 300 µl phenol:chloroform:isoamyl alcohol (PCI). The aqueous layer was re-extracted and nucleic acids were precipitated with 100 µl of 8 M NH4OAc and 1 ml of 95% ethanol and resuspended in ddH2O. Equal amounts of total nucleic acids were separated in a 7 M urea/4.5% acrylamide gel and transferred to nylon membrane. To detect BMV RNAs, the membrane was probed with a 200-nt radiolabeled RNA transcribed from pB3HE1 that was complementary to the 3′ end of all BMV RNAs (13). As a loading control for total cellular RNA, samples from cell lysates were probed with an 80-nt radiolabeled RNA transcript complementary to the 3′ end of 25S rRNA (13). For primer extension analysis, RNA3 was gel purified from total RNA isolated from both PAP-expressing and wild-type cells and depurination sites were identified as described previously (14). To estimate the amount of depurinated wild-type and mutant RNA3 in cells expressing PAP, 200 ng of the gel-purified RNA3 was used as template for reverse transcription. Briefly, a reverse primer that annealed to the last nucleotide of RNA3 was extended using reverse transcriptase in the presence of [α33P]-dATP. cDNA products were separated in a 7 M urea/4.5% acrylamide gel and visualized with a phosphorimager. Depurination of particular nucleotides was evident by truncation of the cDNA products at the expected sizes relative to full-length RNA3. The intensities of truncated bands relative to total RNA3 (truncated + full length) were quantified to estimate the percent of depurinated RNA3 in protoplasts.
Virion particle isolation from barley protoplasts
Following transfection and incubation for 18 h, protoplasts were ground in extraction buffer (0.5 M NaH2PO4, pH 4.5, 80 mM MgCl2, 1% β-mercaptoethanol). The lysate was then transferred to microcentrifuge tubes and centrifuged at 5000 g for 15 min at 4°C. The aqueous layer was centrifuged at 171 500g for 90 min at 4°C through a 10% sucrose cushion. The pellet, containing virion particles, was resuspended and treated with 1 µg/µl of RNase A for 30 min at 30°C, to remove unencapsidated RNAs. Virions were concentrated in a 100-kDa cut-off centrifugal filter and resuspended in virus buffer (50 mM NaH2PO4, pH 4.5, 8 mM MgCl2).
Infectivity of viral particles
Chenopodium quinoa plants were grown to the four-leaf stage before inoculation with BMV virion isolated from barley protoplasts. Isolated virions (15 µg) from either PAP-expressing or wild-type protoplasts were added to 500 µl inoculation buffer (10 mM NaH2PO4, 10 mM MgCl2, 2% silicon carbide). An aliquot of this mixture (20 µl) was rubbed onto a second tier leaf of each plant, and plants were maintained at 25°C/20°C day/night temperature and 16-h day length with 180-µmol/m2/s light intensity. Plants were monitored for the appearance of local lesions. Nine days post infection, lesions were counted and leaves were photographed.
Barley infection and virion preparation
Barley plants were grown to the six-leaf stage before inoculation with BMV virion. Isolated virions (15 µg) were added to 500 µl inoculation buffer (10 mM NaH2PO4, 10 mM MgCl2, 2% silicon carbide). An aliquot of this mixture (20 µl) was rubbed onto the second tier leaf of the plant and plants were grown for 15 days post infection. Plants were maintained under mixed fluorescent and incandescent lighting (230 µmol/m2/s) with 14-h days at 20°C and 10-h nights at 18°C. Following incubation, the systemically infected leaves were ground in 3-ml extraction buffer (0.5 M NaH2PO4, pH 4.5, 80 mM MgCl2, 1% β-mercaptoethanol). The solution was collected by straining through Miracloth and stirred with equal volume of chloroform for 30 min at 4°C. The solution was then transferred to microcentrifuge tubes and centrifuged at 5000g for 15 min at 4°C. The aqueous layer was then centrifuged at 171 500 g for 90 min at 4°C through a 10% sucrose cushion. The pellet was resuspended in virus buffer (50 mM NaH2PO4, pH 4.5, 8 mM MgCl2). The particles contained within this pellet were used as a source of CP for in vitro assembly of particles, described below.
CP preparation, virion assembly assays and electron microscopy
CP was disassembled from isolated BMV particles for in vitro virion assembly assays as described previously (17). Virion assembly reactions were conducted as previously described with minor modifications (18). For each virion assembly reaction, purified BMV CP and wild-type or mutant RNA3 were mixed in a 1:5 (wt/wt) ratio and dialyzed for 24 h at 4°C in assembly buffer (100 µl; 50 mM NaCl, 50 mM Tris–HCl, pH 7.2, 10 mM KCl, 5 mM MgCl2, 1 mM DTT). Following dialysis, assembled particles were isolated by centrifugation at 171 500g for 90 min at 4°C through a 10% sucrose cushion. The pellet, containing virion particles, was resuspended and treated with 1 µg/µl of RNase A for 30 min at 30°C to remove unencapsidated RNA. Assembled virions were concentrated in a 100-kDa cutoff centrifugal filter and resuspended in virus buffer (50 mM NaH2PO4, pH 4.5, 8 mM MgCl2). In some cases, the RNA3 was treated with PAP prior to assembly, as described previously (14). Assembled particles were negatively stained with 1% uranyl acetate and visualized with a transmission electron microscope (Philips EM210). Percentage value indicates the mean number of particles from PAP-treated RNA3 relative to untreated RNA3 ± SE from five fields of view.
Virion RNA analysis
RNA was isolated from virions as described previously (19) and equal volume of RNA was separated in a 7 M urea/4.5% acrylamide gel for northern blot analysis as described above. For primer extension analysis, RNA3 was isolated from gel slices of either total protoplast RNA or total virion RNA separated through 7 M urea/4.5% acrylamide. The extension reaction was performed as described previously (14). For real-time quantitative reverse transcriptase-polymerase chain reaction (RT–PCR and radiolabeled PCR, equal amounts of isolated RNA3 from virions (0.5 µg), along with a positive control depurinated RNA3 (0.5 µg) were reverse-transcribed with SuperScript-II-RT. Quantitative PCR was performed with 200 nM of both sense and antisense primers (5′ GTAAAATACCAACTAATTCTCG 3′; 5′ TGGTCTC TTTTAGAGATTTACAGTG 3′) in a final volume of 25 µl using the SYBR Green PCR core reagent (Invitrogen) in an ABI PRISM 7700 Sequence Detection System Instrument (Applied Biosystems). Known amounts of DNA3 were also amplified to generate a standard curve and the equation Y = 29.56 X−0.0335. Quantitation of the PCR products was determined using the cycle threshold (Ct) values obtained. Radiolabeled PCR was conducted in the presence of [α33P]-dATP (10 mCi/ml; GE Healthcare) using a forward primer that annealed to the first nucleotide of BMV and reverse primer that annealed to the last nucleotide of BMV RNA3, resulting in amplification of a 2113-bp fragment.
Construction of PAP-resistant cDNA3 mutants and A1006 deletion mutant
To create PAP-resistant cDNA3, a series of overlapping PCRs was conducted to mutate all the observed depurinated nucleotides in BMV RNA3 from purines to pyrimidines. The mutations, written as nucleotide change and (amino acid change), were as follows: A116T (S to C), A285C (D to A), A342T (D to V), A371C (M to L), A374T (N to Y), A771T (D to V), G901C (E to A), A1006C (V to V), A1008C (N to T), A1219T (R to S), G1225T (M to I), G1544T (V to F), A1953C (D to A), A1971C(K to T). The resulting resistant cDNA3 was cloned into the BamHI and EcoRI sites of pBluescript vector and capped, in vitro transcripts were synthesized. To produce the cDNA3 mutant for which only A771 could be potentially depurinated (R-RNA3 771), the resistant cDNA3 was used as template for overlapping PCR to re-introduce the wild-type A771 in place of the pyrimidine T771 from R-cDNA3. Using the same strategy, the resistant cDNA3 was used as template for overlapping PCR to re-introduce the wild-type A1006 in place of the pyrimidine C1006 from R-cDNA3 to create R-RNA3 1006, and the two nucleotides together were reverted to wild-type to create the mutant R-RNA3 771 1006, which could be depurinated only at A771 and A1006. To produce the cDNA3 mutant that was resistant to depurination only at A771 (RNA3 771-R) or A1006 (RNA31006-R), wild-type cDNA3 was used as template for overlapping PCR to exchange A771 for T771 or A1006 for C1006, respectively. In the same manner, the double mutant resistant to depurination at A771 and A1006 (RNA3 771 1006-R) was created by overlapping PCR to exchange A771 for T771 and A1006 for C1006, using wild-type cDNA3 as template. The deletion mutant missing A1006 (RNA3 Δ1006) was constructed by overlapping PCR using wild-type cDNA3 as template. These mutant constructs were cloned into the BamHI and EcoRI sites of pBluescript vector and capped, in vitro transcripts were synthesized.
Construction of RNA3 fragments, electrophoretic mobility shift assay and filter binding
The mutant cDNA3 with all observed depurinated nucleotides changed from purines to pyrimidines, except A771 and A1006 (R-RNA3 771 1006), was used as the template DNA for a series of PCRs to create less than full-length RNA3. A forward primer that annealed at nucleotide 92 and reverse primer that annealed at nucleotide 1246 were used to create a DNA3 fragment encoding RNA3 ORF3 and the intergenic region (ORF3 + IGR). A forward primer annealed at nucleotide 92 and reverse primer at nucleotide 1003 were used to produce a DNA3 fragment encoding RNA3 ORF3 only (ORF3 771). A forward primer 1004 and reverse primer 1246 were used to create a DNA3 construct encoding RNA3 intergenic region (IGR). These fragments were cloned into the BamHI and EcoRI sites of pBluescript vector and used as template to transcribe radiolabeled 33P-labeled RNA3 fragments. These in vitro transcripts were treated with PAP or buffer only (14) and used to perform electrophoretic mobility shift assay (EMSA) and filter-binding analysis with isolated BMV CP. Increasing amounts of CP were incubated with PAP-treated or untreated 33P-labeled RNA3 fragments in binding buffer [50 mM Tris–HCl, pH 7.5, 50 mM NaCl, 4 mM MgCl2, 1 mM DTT, 1 mM ethylenediaminetetraacetic acid (EDTA), 5% glycerol] at room temperature for 30 min. For EMSA, the samples were separated through 6% non-denaturing acrylamide gels, and radiolabel was visualized with a phosphorimager. For filter-binding analysis, the samples were passed through a nitrocellulose filter. The amount of retained, labeled RNA was determined by scintillation counting and corrected by subtracting background counts in the absence of BMV CP. The Kd values were extrapolated after the results were fitted with the one site binding equation using Graphpad Prism 4.0.
Construction of RNA3 fragment and UV melting analysis
A 162-nt fragment of RNA3 containing A771 was created by PCR using a forward primer that annealed at nucleotide 679 and reverse primer that annealed at nucleotide 840 on wild-type cDNA3. The only nucleotide that could be depurinated by PAP within the resulting RNA fragment was A771. A PAP-resistant form of this RNA3 fragment was created by PCR using the same primers but R-cDNA3 as template, in which all observed depurinated purines were exchanged for pyrimidines. The resulting cDNA3 fragment contained the mutation A771T, which could not be depurinated in the RNA transcript. The wild-type and mutant cDNA3 fragments were cloned into the BamHI and EcoRI sites of pBluescript vector and capped, in vitro transcripts were synthesized. The in vitro transcripts were PAP-treated or incubated in buffer alone, as described previously (14). Primer extension confirmed that the fragment was as susceptible to depurination at A771 as full-length RNA3 (data not shown). Transcripts were resuspended in buffer (10 mM Tris–HCl, pH 7.0, 5 mM NaCl) and were heated to 85°C and then cooled on ice prior to melting temperature analysis. The absorbance of the samples was monitored at 260 nm from 25 to 85°C. Data were collected with a rate of change in temperature of 1°C/min using a Beckman-Coulter DU 800 UV/VIS spectrophotometer with a Peltier Temperature Programmer. The first temperature derivatives of the absorbance at 260 nM (dA260/dT) were plotted against increasing temperature. The maximum point of each plot represents the melting temperature (Tm) of each fragment.
RESULTS
PAP decreases the production of virus from protoplasts
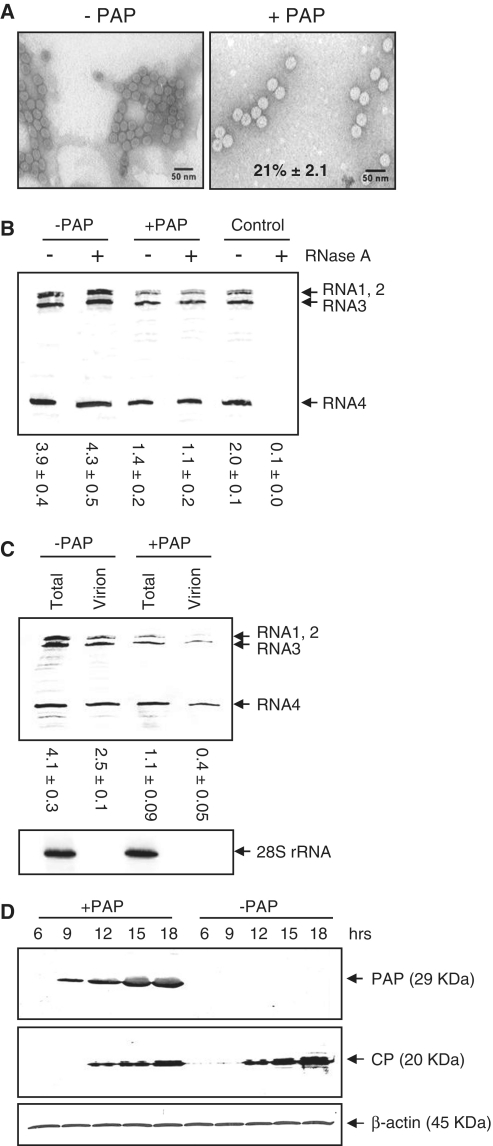
To investigate the effect of PAP on virus packaging, we transfected BMV RNA1, 2 and 3 into barley protoplasts expressing PAP and isolated virus particles from the cells. Transmission electron microscopy indicated no visible difference in morphology of particles isolated from PAP-expressing compared with wild-type protoplasts (Figure 1A). However, fewer particles were isolated from cells expressing PAP (21% relative to −PAP), which was also evident when the same volume of isolated virus was probed by northern blot and less viral RNAs were seen associated with particles from PAP-expressing cells (25% relative to −PAP, compare +PAP with RNase A to −PAP with RNase A; Figure 1B). To ensure that RNA not within particles was excluded from the virion preparation, resuspended particles following centrifugation were treated with RNase A prior to northern blot analysis. The intensities of bands in Figure 1B showed no significant difference between nuclease-treated and -untreated particles, indicating a lack of free unencapsidated RNAs. As a control for RNase A activity, BMV RNAs isolated from virions were also treated with the nuclease and show complete degradation (absence) of the RNA.
Figure 1.
PAP decreases the production of virus from protoplasts. Barley protoplasts were transfected with a plasmid encoding the PAP gene (+PAP) or empty plasmid (−PAP), plus BMV RNA1, 2 and 3 in vitro transcripts and incubated for 18 h. (A) Electron micrographs of virions isolated from PAP-expressing (right) and wild-type protoplasts (left). Viral particles were visualized by staining with uranyl acetate and viewing at 100 000×. The scale bar represents 50 nm. Percentage value indicates the mean number of particles from PAP-expressing cells relative to wild-type cells ± SE from five fields of view. (B) Viral RNAs were purified from equal volume (20 µl) of the isolated viral particles and analyzed by northern blot for the presence of positive-strand BMV RNAs. To ensure that no unincorporated BMV RNAs were included, a portion of the pelleted virion fraction was resuspended and treated with RNase A or left untreated. Control indicates BMV RNAs isolated from virions treated or untreated with RNase A. Values are means of intensities for BMV RNAs ± SE for three separate experiments. (C) Northern blot analysis indicating the levels of total BMV RNAs from protoplast cell lysates and from virus particles isolated from PAP-expressing or wild-type cells, 18 h after transfection. Total RNA from cell lysates was also probed for 28S rRNA as a loading control. Values are means of intensities for BMV RNAs ± SE for three separate experiments. (D) Immunoblot analysis of total cell lysate from protoplasts transfected with a PAP cDNA and in vitro transcripts of BMV RNA1, 2 and 3 over time. Zero hours represent the time at transfection and 18 h represent the time of cell harvest. Equal amounts of total protein (10 µg) were separated by 12% SDS–PAGE, transferred to nitrocellulose and probed with an antibody to PAP (1:5000), BMV CP (1:5000) or β-actin (1:5000).
To assess whether this decrease in particle number correlated with less total BMV RNA in PAP-expressing protoplasts, total RNA and virion RNA were isolated from PAP-expressing protoplasts and their levels compared with wild-type cells (Figure 1C). The amount of total BMV RNA in PAP-expressing cells 18 h after transfection was consistently less than cells not expressing PAP, which agrees with our previous results showing that PAP inhibits the replication, and hence accumulation, of BMV RNAs (13). However, the ratio of signal intensities between virion RNA and total BMV RNA indicates that less RNA was packaged relative to the total amount in cells expressing PAP (0.36) compared with wild-type cells (0.61). This difference in ratio suggests that the lack of virion production was not solely due to less viral RNA in PAP-expressing cells, but that a portion of the total BMV RNA was excluded from particles. To confirm the expression of PAP and CP in barley protoplasts, we tested aliquots of cells over time, from co-transfection of the PAP encoding plasmid with the three infectious transcripts of BMV RNA (0 h) to time of harvest (18 h). The overlap in expression pattern of PAP and CP indicates that the protein components required to test the effect of PAP on packaging were present in protoplasts (Figure 1D). Taken together, these data suggest that the lack of virion production in PAP-expressing cells was not due to insufficient total BMV RNA or CP synthesis in protoplasts.
PAP does not alter the quality of virus from protoplasts
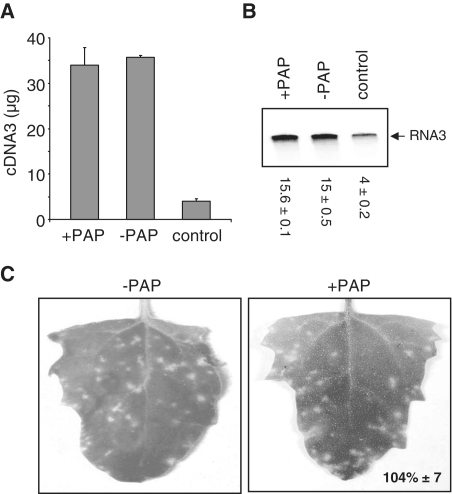
To examine the quality of the viral RNA packaged, quantitative PCR was performed using RNA3 from virions as template. The premise of this test is that if the viral RNA were depurinated by PAP, the presence of a missing base would stall the RT and less than full-length cDNA would be produced which would not be amplified in the real-time PCR reaction. A positive control was generated for this assay by using RNA3 incubated with PAP in vitro as template for the RT (Figure 2A, control). The same amount of RNA3 isolated from virions of PAP-expressing and wild-type protoplasts resulted in similar amounts of PCR product, indicating that the quality of the RNA3 within viral particles was not significantly different. The relative quantity was substantially greater than that of RNA3 treated in vitro with PAP (control), suggesting that depurinated RNA3 was not present in virions of PAP-expressing cells. Results of reverse transcription followed by radiolabeled PCR indicated the same trends; that is, the intensity of PCR products from RNA3 templates isolated from virions of PAP-expressing and wild-type protoplasts was similar and substantially greater than the depurinated control template (Figure 2B). Therefore, the radioactive PCR method was used in future experiments to examine the quality of the packaged RNA3.
Figure 2.
PAP does not alter the quality of virus from protoplasts. Barley protoplasts were transfected with a plasmid encoding the PAP gene (+PAP) or empty plasmid (−PAP), plus BMV RNA1, 2 and 3 in vitro transcripts and incubated for 18 h. Virion particles were isolated from cells and BMV RNAs associated with particles were separated through 7 M urea/4.5% acrylamide gel. Gel purified RNA3 (0.5 µg) was reverse transcribed (RT) and used for quantitative PCR (A) and radioactive PCR (B). Quantitative PCR results are plotted as microgram amount of DNA product. The control lane represents in vitro PAP-treated RNA3 used for RT–PCR. Values of radioactive PCR are means of intensities for RNA3 ± SE for three separate experiments. (C) Virion particles were isolated from PAP-expressing and wild-type protoplasts and equal amounts (15 µg) were used to infect Chenopodium quinoa plants. Infectivity of viral particles was assessed by counting the number of lesions, 9 days post infection. Percentage value indicates the mean number of lesions on leaves of plants infected with particles taken from PAP-expressing barley protoplasts (+PAP), relative to particles isolated from wild-type protoplasts (–PAP) ± SE from five leaves.
Equal amounts of virions from PAP-expressing and wild-type protoplasts were used to infect C. quinoa plants, a local lesion host of BMV, to determine whether the two virus preparations were equally infectious. Nine days following infection, the number of lesions was counted and no significant difference existed between particles isolated from PAP-expressing protoplasts compared with wild-type protoplasts (104% ± 7; Figure 2C). Taken together, these data suggest that PAP expression in barley protoplasts does not alter the quality of viral particles synthesized but reduces the number of particles formed.
PAP depurinates BMV RNA3 in vivo
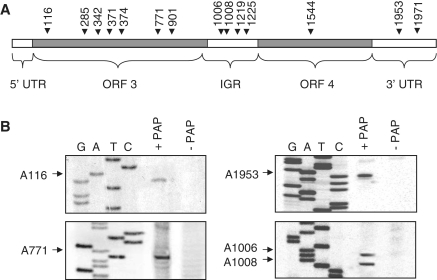
To investigate how PAP expression may be limiting particle formation, we first tested for the presence of depurinated viral RNA in cells. Total RNA from wild-type and PAP-expressing protoplasts transfected with BMV RNA1, 2 and 3 was isolated and RNA3 analyzed by primer extension. Ten primers specific to RNA3 were extended and 14 depurinated nucleotides were identified along its length. Their relative location and nucleotide numbers are outlined in Figure 3A with examples of specific sites (Figure 3B). These same depurinated nucleotides were identified previously by in vitro incubation of RNA3 with PAP (14). Therefore, the depurination pattern of RNA3 seen in vivo from PAP-expressing cells was the same as we observed previously for in vitro treated RNA3.
Figure 3.
PAP depurinates BMV RNA3 in vivo. (A) Schematic of BMV RNA3 illustrating the sites of depurination by their nucleotide number. The 5′ untranslated region (5′-UTR; 1–91 nt), the open reading frame of RNA3 (ORF3; 92–1003 nt), the intergenic region (IGR; 1004–1246 nt), the open reading frame for RNA4 (ORF4; 1247–1813 nt) and the 3′ untranslated region (3′-UTR; 1814–2113 nt) are also indicated. (B) Representative depurination sites as determined by primer extension. Barley protoplasts were transfected with a plasmid encoding the PAP gene (+PAP) or an empty plasmid (−PAP) along with wild-type BMV RNA1, 2 and 3 in vitro transcripts. Following incubation for 18 h, RNA3 was gel purified from total protoplast RNA and 1.0 µg was extended with reverse transcriptase using radiolabeled cDNA primers distributed along the length of RNA3. cDNA products were separated in a 7 M urea/6% acrylamide gel and bands were visualized using a phosphorimager. The bases depurinated by PAP are indicated with arrows and nucleotide numbers. Deoxynucleotide sequencing of BMV DNA3 was conducted using the same primers to identify the depurination sites.
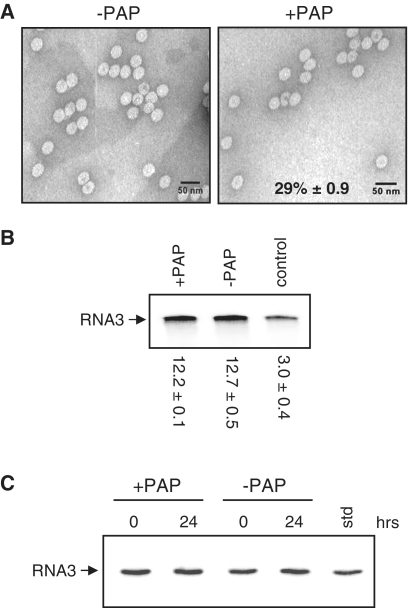
Depurination inhibits packaging of RNA3 in vitro
To investigate whether depurination of BMV RNA directly affected packaging, virus particles were assembled using in vitro transcript of PAP-treated or untreated BMV RNA3 and CP isolated from virions. Assembled viral particles were examined by transmission electron microscopy and fewer particles were observed from samples containing PAP-treated RNA3 (29% relative to −PAP; Figure 4A). No difference in signal intensity of radiolabeled PCR product was detected for RNA3 from virions formed from PAP-treated or untreated RNA3, indicating that the quality of the RNA3 from assembled particles was the same (Figure 4B). The signal intensity of PCR product from PAP-treated RNA3 (control) was substantially less, as would be expected from depurinated RNA as template. The integrity of RNA3, both before its assembly and 24 h following particle formation, was verified by northern blot analysis and no visible degradation was observed (Figure 4C). These data suggest that depurination by PAP inhibits viral RNA packaging and that CP preferentially selects non-depurinated RNA3.
Figure 4.
Depurination inhibits packaging of RNA3 in vitro. (A) Electron micrographs of particles assembled in vitro from isolated CP with either PAP-treated (right) or untreated RNA3 (left). Particles were visualized by staining with uranyl acetate and viewing at 100 000×. The scale bar represents 50 nm. Percentage value indicates the mean number of particles from PAP-treated RNA3 samples relative to untreated samples ± SE from five fields of view. (B) Viral RNAs were isolated from in vitro assembled particles and gel purified RNA3 (0.5 µg) was used for reverse transcription and radioactive PCR. The control lane represents in vitro PAP-treated RNA3 used for RT–PCR. Values are means of intensities for RNA3 ± SE for three separate experiments. (C) Northern blot analysis of RNA3 treated with PAP (+PAP) or buffer alone (−PAP), before its assembly (0 h) and isolated from in vitro particles 24 h following assembly. In vitro transcript of RNA3 was loaded into one lane to serve as a size marker (std).
Depurination inhibits binding of CP to RNA3
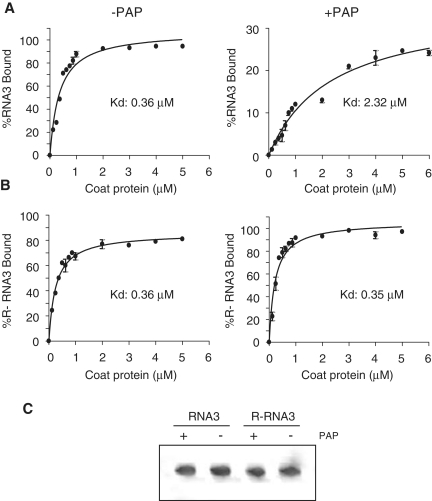
To test whether CP distinguishes depurinated from intact RNA3, PAP-treated or untreated RNA3 was incubated with isolated CP and interaction was assessed by filter binding. The affinity of CP for PAP-treated RNA3 was ∼6-fold less compared with untreated RNA3 (Kd of 2.32 µM versus 0.36 µM; Figure 5A). To verify that inhibition of binding was due to depurination, RNA3 was modified such that all purines susceptible to removal by PAP were substituted with pyrimidines, thereby making an RNA3 that could not be depurinated by PAP (R-RNA3). No significant difference in binding affinity of CP was observed for PAP-treated compared with untreated resistant RNA3, indicating that depurination inhibited interaction of CP with RNA3 (Figure 5B). In addition, the dissociation constants for untreated wild-type compared with untreated resistant RNA3 were the same, showing that modification of these bases from purines to pyrimidines did not decrease CP binding. The radiolabeled RNAs were subjected to denaturing gel electrophoresis and visualized by phosphorimager before incubation with CP, to ensure that differences in binding affinity were not due to degradation of BMV RNA3 upon PAP treatment (Figure 5C).
Figure 5.
Depurination inhibits binding of CP to RNA3. (A) Binding curves of CP with PAP-treated and untreated radiolabeled wild-type BMV RNA3 and (B) PAP-resistant RNA3 (R-RNA3). Transcripts were incubated with increasing concentrations of BMV CP before passing through a nitrocellulose membrane. The amount of retained RNA3 was quantified by scintillation counting and corrected by subtracting background counts in the absence of CP. Points are means ± SE for three separate experiments. The Kd values were obtained after results were fitted to the one site binding equation using Graphpad Prism 4. (C) Radiolabeled wild-type RNA3 (RNA3) and RNA3 resistant to depurination (R-RNA3) were treated with PAP (+PAP) or buffer alone (−PAP). The RNAs were separated through 7 M urea/6% acrylamide gel and visualized with a phosphorimager prior to incubation with CP.
Depurination within ORF3 reduces packaging efficiency
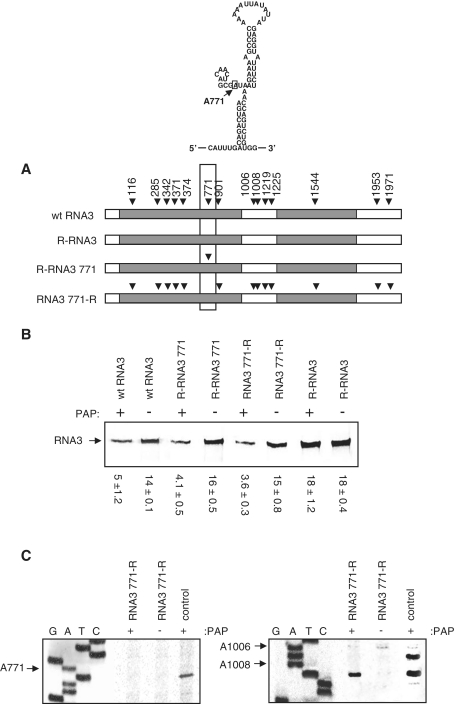
Previous work has shown that an ∼200-nt region within BMV ORF3, nucleotides 5′ 601-817 3′, is important for RNA3 packaging (12; boxed in Figure 6A with structural model of stem–loop C included). This region may fold into a structure containing three loops, called stem–loop A, B and C (12). Deletion of stem–loop B and C completely abolished assembly in vitro. PAP depurinates A771 (14) and because this nucleotide is present within stem–loop C, we hypothesized that depurination of A771 might prevent RNA3 packaging into virions. To test this idea, the resistant RNA3 (R-RNA3) was modified to permit depurination at A771 only (R-RNA3 771), and wild-type RNA3 was modified such that A771 was mutated to a pyrimidine, thereby preventing depurination at this nucleotide but still permitting depurination elsewhere in the RNA3 (RNA3 771-R). These four templates, wild-type RNA3, PAP-resistant RNA3 (R-RNA3), PAP-resistant RNA3 except for A771 (R-RNA3 771) and RNA3 resistant only at A771 (RNA3 771-R), were used to assemble particles in vitro. Line drawings of these RNAs are illustrated in Figure 6A. Based on northern blot analysis of RNA3 isolated from an equal volume of virus particles, fewer particles were formed from PAP-treated wild-type RNA3, whereas no difference was observed for R-RNA3, as expected (Figure 6B). Fewer particles were also assembled from RNA3 that was depurinated only at A771 (R-RNA3 771), indicating that this nucleotide is important for packaging. Surprisingly, fewer particles were seen when RNA3 resistant only at A771 (RNA3 771-R) was used for assembly, suggesting that sequences other than A771 also influence packaging.
Figure 6.
Depurination of A771 inhibits packaging of RNA3. RNA3 constructs were created by site-directed mutagenesis: one that cannot be depurinated by PAP (R-RNA3), one that can be depurinated by PAP only at nucleotide A771 (R-RNA3 771), and RNA3 that cannot be depurinated at A771 only but can be depurinated at other sites (RNA3 771-R). (A) Schematics indicating the sites of depurination for wild-type RNA3, R-RNA3, R-RNA3 771 and RNA3 771-R. An ∼200-nt region of RNA3 is boxed and the modeled secondary structure of a portion containing A771 is shown above. (B) Wt RNA3, R-RNA3 771, RNA3 771-R and R-RNA3 were treated with either PAP or buffer only and used for in vitro particle assembly assays with isolated CP. Viral RNA3 was isolated from equal volume of assembled particles and analyzed by northern blot for the presence of positive-strand BMV RNA3. Values are means of intensities for RNA3 ± SE for three separate experiments. (C) Primer extension analysis of RNA3 771-R isolated from virions. cDNA primers distributed over the length of the viral RNA3 were annealed and extended with reverse transcriptase. Radiolabeled cDNA products were separated in a 7 M urea/6% acrylamide gel and visualized with a phosphorimager. The control lane on the right panel represents PAP-treated RNA3 771-R transcript prior to virion assembly, and on the left panel control represents wild-type RNA3 treated with PAP, to illustrate the location of A771 depurination. The bases depurinated by PAP are indicated with arrows and nucleotide numbers. Deoxynucleotide sequencing of BMV DNA3 was conducted using the same primers to identify the depurination sites.
To identify which nucleotides inhibited packaging when depurinated, apart from A771, primer extension analysis was conducted on PAP-treated RNA3 resistant only at A771 (RNA3 771-R) isolated from assembled particles. All depurinated nucleotides should be detected in particles, except those required for packaging. The nucleotides shown previously to be depurinated by PAP treatment were also depurinated in RNA3 from assembled particles, except A1006 of the intergenic region of RNA3, and A771, even though the source RNA3 was depurinated at A1006 following PAP treatment (right panel control; Figure 6C). The lack of depurination at A771 was expected, given the substitution of this nucleotide to a pyrimidine. However, lack of depurination at A1006 suggests that the intergenic region may be important for packaging, because RNA3 depurinated at this nucleotide was excluded from particles.
Depurination within the intergenic region reduces packaging efficiency
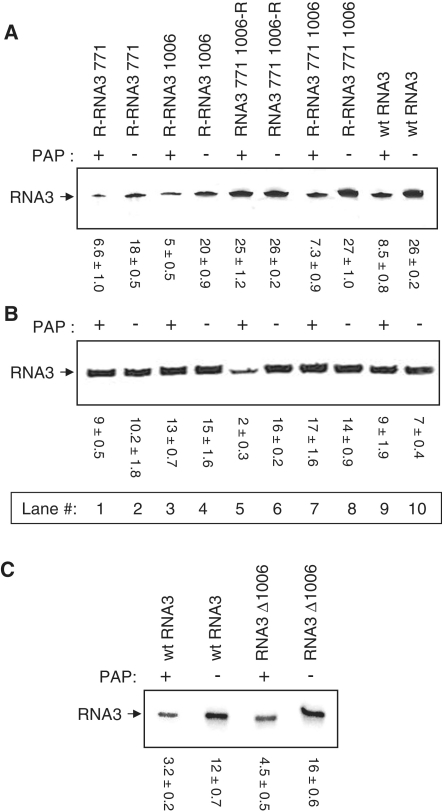
To investigate the possibility that the intergenic region is involved in packaging, RNA3 that permitted depurination only at A1006 (R-RNA3 1006) was incubated with PAP or untreated and used as template for in vitro particle assembly. Less RNA3 from PAP-treated template was isolated from the same volume of particles, suggesting that depurination only within the intergenic region inhibited packaging of RNA3 (Figure 7A, lanes 3 and 4). Therefore, both ORF3 A771 (Figure 7A, lanes 1 and 2) and intergenic region A1006 are independently critical for packaging of BMV RNA3 into virions. We hypothesized that if both these nucleotides are required, then an RNA3 that cannot be depurinated at both A771 and A1006, but retains its ability to be depurinated elsewhere, should allow all of the RNA3 to be packaged, even the depurinated strands. This mutant was synthesized (RNA3 771 1006-R), incubated with PAP and used as template for virion assembly. The amount of RNA3 isolated from virions assembled from PAP-treated compared with untreated RNA3 was similar (Figure 7A, lanes 5 and 6). Therefore, depurination of nucleotides apart from A771 and A1006 did not prevent packaging, i.e. depurinated RNA3 was packaged into particles provided that bases of A771 and A1006 were not removed. Depurination of only A771 and A1006 resulted in less RNA3 assembled, as expected (Figure 7A, lanes 7 and 8) and was similar to PAP-treated or untreated wild-type RNA3 (Figure 7A, lanes 9 and 10), indicating that RNA3 depurinated at A771 and A1006 was excluded from particles.
Figure 7.
Effect of depurination and deletion of A1006 on packaging efficiency. RNA3 constructs, R-RNA3 771, R-RNA3 1006, RNA3 771 1006-R and R-RNA3 771 1006 were created by site directed mutagenesis. Transcripts of these four RNA3 mutants and wild-type RNA3 were treated with PAP or buffer only. PAP-treated and untreated RNA3 were used to assemble particles in vitro with isolated CP. (A) RNA was isolated from equal volume of in vitro assembled particles and analyzed by northern blot for the presence of positive-strand BMV RNA3. Values are means of intensities for RNA3 ± SE for three separate experiments. (B) Equal amounts of gel purified RNA3 (0.5 µg) from in vitro assembled particles were used for reverse transcription and radioactive PCR. Values are means of intensities for RNA3 ± SE for three separate experiments. (C) The deletion construct RNA3 Δ1006, missing A1006 from RNA3, was created by site-directed mutagenesis and its in vitro transcript was treated with PAP or buffer alone. PAP-treated and untreated RNA3 were used to assemble particles in vitro with isolated CP. RNA was isolated from equal volume of assembled particles and analyzed by northern blot for the presence of positive-strand BMV RNA3. Values are means of intensities for RNA3 ± SE for three separate experiments.
To test if depurinated RNA3 was present within virion particles assembled from the mutant that did not permit depurination of A771 and A1006 (RNA3 771 1006-R), reverse transcription followed by radioactive PCR was conducted on equal amounts of RNA3 isolated from particles. Substantially less PCR product was visualized from PAP-treated RNA3 compared with untreated RNA3, indicating that damaged RNA3 was packaged into virions assembled from the transcript that could not be depurinated at nucleotides A771 and A1006, but could be depurinated elsewhere (Figure 7B, lanes 5 and 6). As expected, an equal amount of PCR product was observed from RNA isolated from virions made from wild-type RNA3 or RNA3 susceptible to depurination at A771 or A1006 (R-RNA3 771, R-RNA3 1006 or R-RNA3 771 1006) treated with PAP, as RNA3 depurinated at either nucleotide would not be packaged into particles. These data confirm that nucleotides A771 and A1006 are essential for RNA3 packaging and damage to either or both inhibits virion assembly.
We were interested to determine whether deletion of one of these important nucleotides altered the efficiency of RNA3 packaging. We deleted A1006 from the cDNA of RNA3 and the resulting mutant transcript was used for in vitro particle assembly. Northern blot analysis of RNA isolated from the same volume of particles indicated that RNA3 Δ1006 packaged as well as wild-type RNA3 (Figure 7C). Moreover, PAP treatment of the mutant prior to assembly reduced the total amount of RNA3 packaged, indicating that depurination of A771 still prevented packaging (confirmed by primer extension, data not shown). Therefore, the effect on packaging of deleting one of these nucleotides did not mimic depurination of the same nucleotide.
CP interaction with RNA3 depurinated at A771 and A1006
Filter-binding analysis was used to test whether the exclusion from particles of RNA3 depurinated at A771 or A1006 was due to decrease in CP affinity for these RNA molecules. CP bound significantly less to RNA3 depurinated only at A771 (R-RNA3 771) compared with non-depurinated RNA3 (Kd of 2.50 µM versus 0.23 µM; Table 1). PAP treatment of RNA3 resistant to depurination except at A1006 (R-RNA3 1006) also significantly reduced the affinity of CP; the dissociation constant for untreated RNA3 was 0.30 µM compared to 2.63 µM for PAP-treated RNA3. Finally, CP bound to RNA3 depurinated only at A771 and A1006 (R-RNA3 771 1006) with similar affinity as depurinated wild-type RNA3 (Kd of 2.89 µM versus 2.32 µM; Table 1 and Figure 5A). Therefore, A771 and A1006 are important for efficient CP binding to RNA3.
Table 1.
Coat protein affinity for RNA3 depurinated at specific nucleotides, as measured by filter-binding assay
| RNA3 Mutant |
Kd µM |
|
|---|---|---|
| +PAP | −PAP | |
| R-RNA3 771 | 2.50 | 0.23 |
| R-RNA3 1006 | 2.63 | 0.30 |
| R-RNA3 771 1006 | 2.89 | 0.20 |
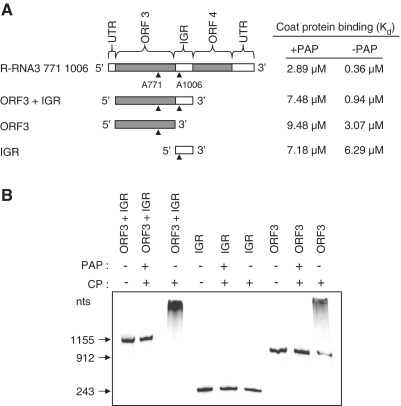
BMV CP binds to the 3′-UTR of all BMV RNAs (17,20). To determine whether CP could bind to RNA3 containing only ORF3 and the intergenic region, and if depurination at nucleotides A771 and A1006 affected binding, RNA3 fragments were constructed from nucleotides 92–1246 (ORF3 + IGR), containing both A771 and A1006; RNA3 from nucleotides 92–1003 (ORF3), containing A771; and RNA3 from nucleotides 1004–1246 (IGR), containing A1006 (Figure 8A). These three constructs were designed to be resistant to depurination except for A771 and A1006 and the affinity of CP for these transcripts was analyzed by filter binding. CP bound less to the ORF3 + IGR fragment relative to full-length RNA3; the Kd for ORF3 + IGR was 0.94 µM compared with Kd of 0.36 µM for full-length RNA3 (Figure 8A and 5A), which is consistent with the results of Yi et al., (17) indicating the importance of the 3′ end in binding. Our results do not preclude the possibility that CP binds to ORF4, we did not test this. However, depurination at A771 and A1006 substantially reduced CP binding to ORF3 + IGR (Kd of 7.48 µM), suggesting that this fragment does contain a binding site. CP binding to the ORF3 fragment, in the absence of PAP treatment, was substantially less than ORF3 + IGR, indicating that the intergenic region contributed to binding of CP to ORF3 (Kd of 3.07 µM for ORF3 compared to 0.94 µM for ORF3 + IGR). PAP treatment of ORF3 inhibited binding, indicating that this region has a CP binding site that is disrupted by depurination at A771. The CP bound with low affinity to the intergenic region, suggesting that this fragment does not contain a CP-binding site. Moreover, PAP treatment did not alter binding (7.18 µM compared with 6.29 µM); therefore, depurination at A1006 does not affect CP binding to the intergenic region alone. Taken together, these data show that fragments of RNA3 in addition to the 3′ end are important for CP binding. Moreover, the intergenic region contributes to the binding of CP to ORF3, even though CP does not appear to bind to the intergenic region alone.
Figure 8.
CP interaction with RNA3 fragments depurinated at A771 and A1006. (A) Schematics showing regions of RNA3 (R-RNA3 771 1006, depurinated only at A771 and A1006) tested for their binding affinity to isolated CP by filter binding assay and EMSA. RNA3 fragments were treated with PAP or buffer only and incubated with increasing concentration of BMV CP before passing through a nitrocellulose membrane. The amount of retained RNA was quantified by scintillation counting and corrected by subtracting background counts in the absence of CP. The Kd values were obtained after results were fitted to the one site binding equation using Graphpad Prism 4, and appear in the table to the right. (B) Radiolabeled RNA3 fragments were incubated with PAP or buffer only, followed by incubation with BMV CP. Samples were separated through a 6% acrylamide non-denaturing gel and the bands were visualized using a phosphorimager.
To further illustrate that ORF3 has a CP-binding site and that the intergenic region does not, EMSA was conducted with isolated CP and the same fragments, ORF3 + IGR, ORF3 and IGR from R-RNA3 771 1006. CP caused a shift in ORF3 + IGR and a partial shift for ORF3 and no shift for IGR, indicating that the binding of CP to movement protein ORF is enhanced by the presence of IGR, even though CP does not bind to IGR alone (Figure 8B). No shift was observed following PAP treatment of ORF3 + IGR and ORF3, indicating that depurination of A771 and A1006 abolished binding of CP to these fragments.
Thermal stability of RNA3 fragment depurinated at A771
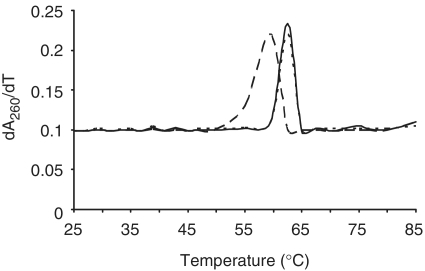
We hypothesized that the difference in binding affinity of CP for depurinated compared with non-depurinated RNA was due to decrease in stability of the local RNA structure. To test this possibility, a 162-nt fragment of RNA3 (nt 679–840) containing A771 was treated with PAP or buffer alone, and its thermal stability measured by UV spectrophotometry. Depurination of the RNA fragment at A771 was confirmed by primer extension (data not shown). The average melting temperature of the depurinated RNA3 fragment was 57.6°C (long broken line) compared with the non-depurinated fragment at 62.5°C (solid line; Figure 9). Interestingly, the melting temperature of the RNA fragment in which A771 was mutated to T771 was the same as wild-type, at 62.5°C (short broken line). Therefore, substitution of the purine for a pyrimidine did not alter the overall thermal stability of the RNA fragment. The lower melting temperature of depurinated RNA indicates a higher thermal instability and flexibility relative to both non-depurinated and mutated RNA.
Figure 9.
Thermal stability of RNA3 fragment depurinated at A771. The 162-nt cDNA constructs of RNA3 containing either A771 (wt cDNA3 fragment) or this nucleotide mutated to T771 (R-cDNA3 fragment) were produced by PCR. In vitro transcripts of these cDNAs were treated with PAP or buffer alone. The melting temperatures of the RNA fragments were measured by UV spectrophotometry. The first derivative of the absorbance at 260 nm was plotted as a function of temperature. Wild-type RNA3 fragment is represented by a continuous line, PAP-treated wild-type RNA3 fragment by long broken lines and R-RNA3 fragment by short broken lines. The absorbance of each fragment was measured independently and values are means of each ± SE for three separate experiments.
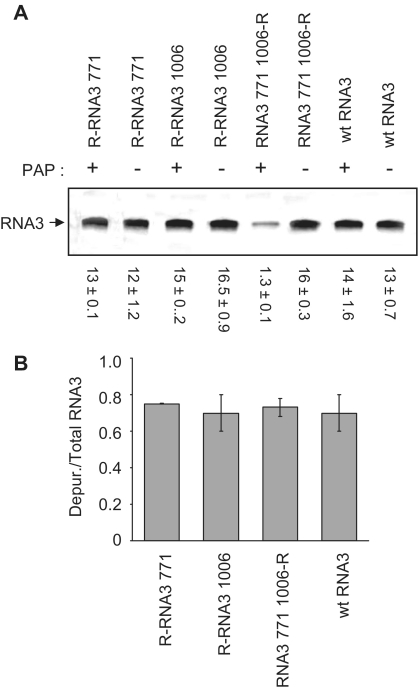
RNA3 depurinated at A771 and A1006 is not packaged in vivo
To determine whether depurination at nucleotides A771 and A1006 was also critical for RNA3 packaging in vivo, the RNA3 mutants tested for in vitro assembly were transfected, along with wild-type RNA1 and 2, into barley protoplasts expressing PAP. Assessment by radiolabeled PCR of the quality of the RNA packaged indicated that RNA3 depurinated at only A771 or A1006 was excluded from particles, given that a similar amount of PCR product was observed from equal amount of virion RNA3 template from PAP-expressing and wild-type protoplasts (Figure 10A). In contrast, RNA3 that was resistant to depurination at A771 and A1006 but could be depurinated elsewhere (RNA3 771 1006-R) was incorporated into particles, as seen by the decreased amount of PCR product, indicative of incomplete reverse transcription due to damaged RNA3 in viral particles from PAP-expressing protoplasts. To test whether the lack of incorporation of RNA3 depurinated at A771 or A1006 was due to insufficient depurinated RNA3 in cells, total RNA3 from protoplasts was extracted and the level of depurination estimated by reverse transcription. Radiolabeled cDNA products were extended using a primer that annealed to the 3′-end of RNA3, and depurination resulted in fragments of expected sizes that were less than full-length RNA3. The intensities of bands representing depurination at A771 (for R-RNA3 771), depurination at A1006 (R-RNA3 1006) and depurination at A1971 (the most 3′ depurinated nucleotide for RNA3 771 1006-R and wt RNA3) were plotted relative to total RNA3 (Figure 10B). Approximately 70% of each mutant RNA3 was depurinated, suggesting that lack of depurinated RNA3 in particles containing R-RNA3 771 or R-RNA3 1006 was not due to lack of depurinated template inside cells.
Figure 10.
Effect of depurination at A771 and A1006 on the quality of RNA3 packaged in vivo. In vitro transcripts of RNA1, 2 and either wild-type or mutant RNA3 were transfected into barley protoplasts along with a plasmid encoding the PAP gene (+PAP) or empty plasmid (−PAP) and allowed to replicate for 18 h before viral particles were isolated. (A) RNAs were isolated from viral particles and equal amounts of gel purified RNA3 (0.5 µg) were used for reverse transcription and radioactive PCR. Values are means of intensities for RNA3 ± SE for three separate experiments. (B) Total RNA was isolated from protoplasts, RNA3 was gel purified and reverse transcribed in the presence of [α33P]-dATP. The intensities of truncated bands, due to depurination, were plotted relative to total RNA3. Bars represent mean percent depurination ± SE for three separate experiments of wtRNA3 and each mutant.
DISCUSSION
We observed that PAP expression inhibits the production of virion particles in barley protoplasts transfected with BMV RNAs. Fewer virus particles were formed in PAP-expressing cells; however, their quality and infectivity were not reduced. A decrease in number was also observed when particles were assembled in vitro from isolated CP and PAP-treated RNA3 transcript, suggesting that PAP decreased the fitness of the RNA for packaging. Examination of RNA3 from particles revealed that of the 14 possible depurinated nucleotides, only depurination of A771 in ORF3 and A1006 in the intergenic region prevented incorporation of the RNA. Some specific interactions between CP and RNA3 appear to regulate packaging. Initial competition experiments between BMV RNA, yeast tRNA and Alfalfa mosaic virus RNA resulted in the preferential packaging of BMV RNAs by BMV CP (21). It has been shown since that a 187-nt fragment of the ORF3 of RNA3 is a specific recognition element for the CP (12). Deletion of this fragment from RNA3 resulted in a mutant that was not packaged. We have identified a single nucleotide within this region, A771, which when depurinated, inhibits binding of CP to RNA3. CP detects damage to this nucleotide and does not efficiently package the depurinated RNA3. We suggest that A771, along with A1006 of the intergenic region, are located within RNA elements that impart specificity during initial recognition by the CP.
Deletion mutations within RNA3 have shown that complete removal of the intergenic region did not negatively affect packaging (12). We have observed the same result (data not shown); however, depurination of A1006 within this region inhibited CP binding and excluded the RNA from packaging. Even though packaging occurs in vitro in the absence of the intergenic region, perhaps when present, this intergenic structure regulates the specific interaction of CP with the region of ORF3 containing A771. We have noted previously an unexpected role for the intergenic region in viral replication; namely, that replicase binding to the intergenic region is required for initiation of negative strand synthesis, even though this region is distant from the 3′ terminal core promoter for the replicase. Depurination of A1006 prevented replicase binding to RNA3 and initiation of RNA synthesis (14). CP also co-purifies with active replicase complex (22) and the replicase has been shown to play a role in assembly as a specificity filter, blocking cellular mRNA packaging into particles (23). Taken together, we speculate that in vivo, the CP associates with the replicase at the intergenic region, and that this complex distinguishes viral RNAs. In vitro, in the absence of replicase, CP can still recognize damage to the intergenic region and will not bind. However, if the IGR is removed, CP looses this form of regulation and binding is achieved by interaction with the specific site within ORF3 containing A771. The existence of two distant regions that bind CP suggests that the tertiary structure of the viral RNA contributes to specificity.
We have shown here that CP can distinguish between damaged RNA3, in the form of depurination at particular sites, and intact RNA3. It was of interest to us that this distinction does not rely on the identity of individual nucleotides because mutation of A771 and A1006 to pyrimidines did not decrease CP binding. Secondary structure predictions based on M-fold analysis indicated that in wild-type RNA3, A771 and A1006 are within loop regions and their mutation to pyrimidines would likely create stem regions that incorporate these nucleotides. The potential establishment of base-pairing and the resulting change in shape of the RNA were also not sufficient to affect CP binding. We hypothesize that the fundamental difference between an intact nucleotide, regardless of identity, and an abasic nucleotide is stability. Overlap between pi orbitals of bases in single-stranded RNA provides substantial stability to the molecule (24,25). Depurination of a nucleotide will disorder the local structure and increase its flexibility due to disruption of base-stacking from the missing purine. We tested this assertion by measuring the melting temperature of a fragment of RNA3 that was depurinated at A771. The melting temperature of the depurinated fragment was consistently lower than the intact fragment. The increased flexibility of a depurinated RNA may mean that recognition structures are not always accessible for CP binding. Therefore, we suggest that this instability is detected by CP, given that its affinity for depurinated nucleotides within these RNA packaging signals was significantly reduced. Interestingly, no difference in melting temperature was detected between the intact wild-type fragment and the same fragment for which the purine was mutated to a pyrimidine. CP also bound with the same affinity to full-length resistant RNA3 as wild-type RNA3, supporting our view that the stability of particular RNA regions, rather than their nucleotide sequences, influences whether the RNA will be packaged.
Examining the effects of damage to RNA3 through depurination has provided us with new understanding of the regulatory role of CP during packaging of virus particles. Namely, we show that depurination of two nucleotides, one within the ORF3 and the other within the intergenic region of RNA3, prevents CP binding to the RNA. Depurination of 12 other nucleotides along the RNA3 did not inhibit packaging; therefore, the CP recognizes specific regions of RNA3 that may serve as initial recognition elements. Not all nucleotides within these regions are sequence dependent however, as substitution of the abasic purines to pyrimidines did not diminish CP interaction with the RNA. Rather, we suggest that the damage to specific packaging signals of RNA3 containing these two nucleotides results in localized instability of the RNA molecule, which is detected by CP and rejected for incorporation into particles. Future work will focus on the role of the replicase, together with CP for packaging in vivo and will identify whether a structural connection between A771 of ORF3 and A1006 of the intergenic region exists to facilitate RNA3 packaging.
ACKNOWLEDGEMENTS
We thank Dr. Cheng Kao for the vectors pB3TP8 bearing BMV DNA3 and pB3HE1 containing a fragment of the tRNA-like structure from the 3′-end of BMV RNA3. We are grateful to Ms. Karen Rethoret for assistance with electron microscopy of virus particles.
FUNDING
A Discovery Grant from the Natural Sciences and Engineering Research Council of Canada; a Premier’s Research Excellence Award; and infrastructural support by the Canada Foundation for Innovation (CFI) and the Ontario Innovation Trust (OIT) (to K.A.H.). Funding for open access charges: Natural Sciences and Engineering Research Council of Canada.
Conflict of interest statement. None declared.
REFERENCES
- 1.Schmitz I, Rao AL. Molecular studies on bromovirus capsid protein. I. Characterization of cell-to-cell movement-defective RNA3 variants of brome mosaic virus. Virology. 1996;226:281–293. doi: 10.1006/viro.1996.0656. [DOI] [PubMed] [Google Scholar]
- 2.Goldbach R, LeGall O, Wellink J. Alpha-like viruses in plants. Semin. Virol. 1991;2:19–25. [Google Scholar]
- 3.Ahlquist P. Bromovirus RNA replication and transcription. Curr. Opin. Genet. Dev. 1992;2:71–76. doi: 10.1016/s0959-437x(05)80325-9. [DOI] [PubMed] [Google Scholar]
- 4.Ahola T, den Boon JA, Ahlquist P. Helicase and capping enzyme active site mutations in brome mosaic virus protein 1a cause defects in template recruitment, negative-strand RNA synthesis, and viral RNA capping. J. Virol. 2000;74:8803–8811. doi: 10.1128/jvi.74.19.8803-8811.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kao CC, Quadt R, Hershberger RP, Ahlquist P. Brome mosaic virus RNA replication proteins 1a and 2a from a complex in vitro. J. Virol. 1992;66:6322–6329. doi: 10.1128/jvi.66.11.6322-6329.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Miller WA, Dreher TW, Hall TC. Synthesis of brome mosaic virus subgenomic RNA in vitro by internal initiation on (-)-sense genomic RNA. Nature. 1985;313:68–70. doi: 10.1038/313068a0. [DOI] [PubMed] [Google Scholar]
- 7.Rao AL. Bromoviruses. In: Maloy OC, Murray TD, editors. Encyclopedia of Plant Pathology. New York, NY: Wiley; 2000. pp. 155–158. [Google Scholar]
- 8.Kao CC, Sivakumaran K. Brome mosaic virus, good for an RNA virologist's basic needs. Mol. Plant Pathol. 2000;1:91–97. doi: 10.1046/j.1364-3703.2000.00017.x. [DOI] [PubMed] [Google Scholar]
- 9.Zlotnick A, Aldrich R, Johnson JM, Ceres P, Young MJ. Mechanism of capsid assembly for an icosahedral plant virus. Virology. 2000;277:450–456. doi: 10.1006/viro.2000.0619. [DOI] [PubMed] [Google Scholar]
- 10.Choi YG, Dreher TW, Rao AL. tRNA elements mediate the assembly of an icosahedral RNA virus. Proc. Natl Acad. Sci. USA. 2002;99:655–660. doi: 10.1073/pnas.022618199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Damayanti TA, Tsukaguchi S, Mise K, Okuno T. cis-acting elements required for efficient packaging of brome mosaic virus RNA3 in barley protoplasts. J. Virol. 2003;77:9979–9986. doi: 10.1128/JVI.77.18.9979-9986.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Choi YG, Rao AL. Packaging of brome mosaic virus RNA3 is mediated through a bipartite signal. J. Virol. 2003;77:9750–9757. doi: 10.1128/JVI.77.18.9750-9757.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Picard D, Kao CC, Hudak KA. Pokeweed antiviral protein inhibits brome mosaic virus replication in plant cells. J. Biol. Chem. 2005;280:20069–20075. doi: 10.1074/jbc.M413452200. [DOI] [PubMed] [Google Scholar]
- 14.Karran RA, Hudak KA. Depurination within the intergenic region of Brome mosaic virus RNA3 inhibits viral replication in vitro and in vivo. Nucleic Acids Res. 2008;36:7230–7239. doi: 10.1093/nar/gkn896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kroner P, Richards D, Traynor P, Ahlquist P. Defined mutations in a small region of the brome mosaic virus 2 gene cause diverse temperature-sensitive RNA replication phenotypes. J. Virol. 1989;63:5302–5309. doi: 10.1128/jvi.63.12.5302-5309.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tourlakis ME, Karran R, DeSouza L, Siu KWM, Hudak KA. Homodimerization of pokeweed antiviral protein as a mechanism to limit depurination of pokeweed ribosomes. Mol. Plant Pathol. 2010;11:757–767. doi: 10.1111/j.1364-3703.2010.00640.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yi G, Vaughan RC, Yarbrough I, Dharmaiah S, Kao CC. RNA binding by the brome mosaic virus capsid protein and the regulation of viral RNA accumulation. J. Mol. Biol. 2009;391:314–326. doi: 10.1016/j.jmb.2009.05.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Choi YG, Rao ALN. Molecular studies on bromovirus capsid protein VII. Selective packaging of BMV RNA4 by specific N-terminal arginine residues. Virology. 2000;275:207–217. doi: 10.1006/viro.2000.0513. [DOI] [PubMed] [Google Scholar]
- 19.Rao ALN, Duggal R, Lahser FC, Hall TC. Analysis of RNA replication in plant viruses. In: Adolph KW, editor. Methods in Molecular Genetics: Molecular Virology Techniques, Part A. Vol. 4. San Diego, CA: Academic Press; 1994. pp. 216–236. [Google Scholar]
- 20.Zhu J, Gopinath K, Murali A, Yi G, Hayward SD, Zhu H, Kao C. RNA-binding proteins that inhibit RNA virus infection. Proc. Natl Acad. Sci. USA. 2007;104:3129–3134. doi: 10.1073/pnas.0611617104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cuillel M, Herzog M, Hirth L. Specificity of in vitro reconstitution of bromegrass mosaic virus. Virology. 1979;95:146–153. doi: 10.1016/0042-6822(79)90409-4. [DOI] [PubMed] [Google Scholar]
- 22.Bujarski JJ, Hardy SF, Miller WA, Hall TC. Use of dodecyl-beta-D-maltoside in the purification and stabilization of RNA polymerase from brome mosaic virus-infected barley. Virology. 1982;119:465–473. doi: 10.1016/0042-6822(82)90105-2. [DOI] [PubMed] [Google Scholar]
- 23.Annamalai P, Rao AL. Replication-independent expression of genome components and capsid protein of brome mosaic virus in planta: a functional role for viral replicase in RNA packaging. Virology. 2005;338:96–111. doi: 10.1016/j.virol.2005.05.013. [DOI] [PubMed] [Google Scholar]
- 24.Sponer J, Riley KE, Hobza P. Nature and magnitude of aromatic stacking of nucleic acid bases. Phys. Chem. Chem. Phys. 2008;10:2595–2610. doi: 10.1039/b719370j. [DOI] [PubMed] [Google Scholar]
- 25.Freier SM, Petersheim M, Hickey DR, Turner DH. Thermodynamic studies of RNA stability. J. Biomol Struct. Dyn. 1984;1:1229–1242. doi: 10.1080/07391102.1984.10507514. [DOI] [PubMed] [Google Scholar]