Abstract
Optimal response to environmental stimuli often requires activation of certain genes and repression of others. Dual function regulatory proteins play a key role in the differential regulation of gene expression. While repression can be achieved by any DNA binding protein through steric occlusion of RNA polymerase in the promoter region, activation often requires a surface on the regulatory protein to contact RNAP and thus facilitate transcription initiation. RNAP itself is also a DNA binding protein, therefore it can function as a transcriptional repressor. Searching the Escherichia coli promoter database we found that ∼14% of the identified ‘forward’ promoters overlap with a promoter oriented in the opposite direction. In this article we combine a mathematical model with experimental analysis of synthetic regulatory regions to investigate interference of overlapping promoters. We find that promoter interference depends on the characteristics of overlapping promoters. The model predicts that promoter strength and interference can be regulated separately, which provides unique opportunities for regulation. Our experimental data suggest that in principle any DNA binding protein can be used for both activation and repression of promoter transcription, depending on the context. These findings can be exploited in the construction of synthetic networks.
INTRODUCTION
The logic of regulatory-protein-mediated transcriptional regulation depends on whether the regulatory protein is an activator (positive control) or a repressor (negative control). However, optimization of gene expression often requires that certain promoters are up-regulated while others are turned down. The most compact design for this task is a repression-activation switch (transcriptional switch), where two promoters share a common regulatory region in such a way that one of the two promoters is subjected to positive regulation by the very same protein which represses the activity of the second promoter (1). λ CI is one of the best-known examples of a protein which can simultaneously repress and activate two adjacent promoters (2). Activators are generally more suitable for such differential regulation than repressors because they are typically dual-function regulators (3). Binding of a protein in the core promoter region can interfere with RNAP binding and therefore result in decreased levels of transcription, even if the protein functions as an activator in different contexts. Conversely, some repressors can activate transcription directly in certain contexts by inducing a specific DNA bend (4) or by contacting RNAP (5), or indirectly by repressing another repressor. The two divergent promoters involved in repression-activation switches typically show a certain degree of overlap (1). For example, the major promoter P1 of the aroP gene is repressed by the cofactor-TyrR-RNAP complex formed at the overlapping divergent P3 promoter. Binding of the cofactor-TyrR complex itself is not sufficient for repression of P1, RNAP has to be recruited to the P3 promoter (6). In principle binding of RNAP to one promoter of a repression-activation switch can inhibit RNAP binding to the overlapping other promoter (6,7) or interfere with open complex formation at a nearby promoter (8). Therefore, RNAP itself can act as a transcriptional regulator. In this article we use a mathematical model to investigate interference of overlapping promoters. We explore the unique opportunities of transcriptional regulation provided by overlapping promoters, and provide an experimental example for using a repressor protein as an activator.
MATERIALS AND METHODS
Strain and plasmid construction
Synthetic regulatory regions were created by PCR and inserted between the EcoRI and PstI sites of plasmid pSEM2027 (3). Then the constructed regulatory regions, together with the upstream terminator region, were inserted upstream of the uidA ORF in the Escherichia coli MG1655 chromosome by recombineering as described before (3). Regulation of the uidA (gusA) reporter gene in the resulting cells was evaluated on a population level, by plating cells on two separate LB plates, both containing the chromogenic substrate of β-d-glucuronidase (X-gluc) and one of them also containing 1 mM IPTG. Sequences were verified and are shown in Figure 4. Templates for in vitro transcription were created by inserting the regulatory regions between the EcoRI and PstI sites of plasmid pSEM2008 (9). In this plasmid, transcription initiated at promoter P is terminated by an rpoC terminator, and transcription from PREV is terminated by rrnB T2.
Figure 4.
Synthetic regulatory region used in the in vitro and in vivo studies. Schematic drawings of regulatory regions are shown on the left. Gray boxes represent promoters, in which the −35 elements (yellow) and −10 elements (blue) are highlighted. Transcription start points are indicated by arrows. The red box represents a symmetric LacI operator site (O). The promoter elements are not shown when mutated in the reverse promoter PREV (typed boldface and underlined in the sequence).
In vitro transcription
Transcription reactions were performed as described previously (10). The reaction mixture (50 µl) contained 20 mM Tris–acetate pH 7.8, 10 mM magnesium acetate, 200 mM potassium glutamate and 2 nM supercoiled DNA template. LacI was used at 30 nM when present. Twenty nanomolar RNA polymerase was added before incubating the reactions at 37°C for 5 min. Transcription was started by the addition of 1.0 mM ATP, 0.1 mM GTP, 0.1 mM CTP, 0.01 mM UTP and 5 µCi of [α-32P]UTP (3000 Ci/mmol). Reactions were terminated after 10 min by addition of an equal volume of transcription loading buffer (0.025% bromophenol blue, 0.025% xylene cyanol, 0.01 M EDTA and 90% deionized formamide). After heating at 90°C for 3 min, the samples were loaded onto an 8% polyacrylamide–urea DNA sequencing gel. RNA bands were quantified using the ImageQuant™ PhosphorImager (Molecular Dynamics, CA, USA). The RNA1 transcript, which is not affected by LacI binding, was used as an internal control between lanes. Band intensities were background corrected. The uracil content of RNAs was also considered when two different transcripts were compared.
Mathematical modeling
Let θ1 be occupation of P1 and θ2 the occupation of promoter P2. With 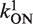 denoting the on rate and
denoting the on rate and 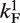 the elongation initiation rate (firing rate) for P1, the total intrinsic activity of the P1 promoter is given by steady-state equation:
the elongation initiation rate (firing rate) for P1, the total intrinsic activity of the P1 promoter is given by steady-state equation:
| (1) |
giving
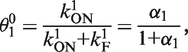 |
(2) |
where the promoter aspect ratio,  . Equivalently the aspect ratio is
. Equivalently the aspect ratio is
 |
(3) |
where the superscript 0 refers to isolated promoters. This aspect ratio approaches ∞ when θ→1. The strength of the promoter P1 is
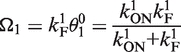 |
(4) |
corresponding to an average time between subsequent elongation initiations:
| (5) |
a time which is the sum of two subsequent times.
In case of two overlapping promoters, P1 and P2, there are three possibilities: (i) P1 is occupied by RNAP (probability = θ1); (ii) P2 is occupied by RNAP (probability = θ2); and (iii) both promoters are free (probability = 1 − θ1 − θ2). The steady state equations for two overlapping promoters are:
| (6) |
| (7) |
which also reads:
| (8) |
| (9) |
with determinant det = (α1 + 1) × (α2 + 1) − α1 × α2 = α1 + α2 + 1 giving
| (10) |
| (11) |
which each should be normalized by the expected occupancy when there is no conflicting promoter [i.e. from Equation (2)] to obtain promoter activities expressed as a fraction of intrinsic promoter activities:
| (12) |
| (13) |
In terms of θ0 the equation expressing the relative P1 activity (Figure 2) reads:
| (14) |
In this model we do not consider the time that the elongating RNAP spends transcribing the region which constitutes the overlapping other promoter. The length of this region depends on the topology of the overlapping promoters. In the case when promoters are oriented tail to tail, this time is negligible, however when promoters are oriented head-to-head, the whole region constituting the overlapping other promoter has to be transcribed, which takes ∼1 s for the elongating RNAP (assuming elongation speed of 50 nt/s). This effect would increase the aggressiveness of strong promoters (which by definition initiate elongation frequently).
Figure 2.
Computed prediction of interference of overlapping promoters. Activity of a promoter (P1) in terms of its own and the opposing promoter's (P2) aspect ratios (α1 and α2) (left panel). Activity of promoter P1 in terms of its own and the opposing promoter's basal occupancies (θ10 and θ20) (right panel). Activities are measured in fraction of the intrinsic promoter activity of P1. Simulations were done with two promoters arranged tail-to-tail, with 1 bp overlap.
RESULTS AND DISCUSSION
Analyzing the recently published transcription unit architecture database of the E. coli genome (11), we found that ∼314 of 2281 forward promoters overlap with a reverse promoter (Supplementary Table S1). RNAP occupies ∼75 bp (from −55 to +20) when sitting on the promoter (12), however, our list is more conservative, considering overlaps only in the regions from −40 to +10. Promoters can be arranged head-to-head or tail-to tail (Figure 1). We found three fundamentally different arrangements: (i) each promoter transcribes a gene and one or more regulatory proteins are identified which affect transcription of at least one of the promoters directly (marked red in Supplementary Table S1); (ii) each promoter transcribes a gene but no regulatory proteins are known to bind the promoter region (marked green in Supplementary Table S1); and (iii) only one of the promoters transcribe an annotated gene (marked yellow in Supplementary Table S1). In the latter case the other promoter could transcribe a regulatory antisense RNA (13) or act as a regulatory RNAP binding site which interferes with the promoter transcribing the annotated gene. A significant proportion of promoters in E. coli are bound by RNAP but lack transcriptional activity (14), which also supports the hypothesis that RNAP plays an active role in regulation. Because overlapping promoters are common in the E. coli genome, we have built a mathematical model to investigate how the level of interference depends on the characteristics of the two promoters involved. A related phenomenon, interference of an elongating RNAP with a convergent but non-overlapping promoter, has been described and modeled recently (15,16). Our model shows that the mutual interference of overlapping promoters is independent of the relative intrinsic strength of promoters, it only depends on the aspect ratios (16) [tendency of promoter to be occupied by RNAP, see Equation (3) in ‘Materials and Methods’ section]. Promoters which successfully inhibit the other overlapping promoter have a high aspect ratio (Figure 2). Therefore, the capacity of a promoter to inhibit an overlapping promoter, termed the aggressiveness of the promoter, is determined by the fraction of time it is occupied by RNAP. For large aspect ratios (>>1), the mutual influence scales with the ratio of the two aspect ratios.
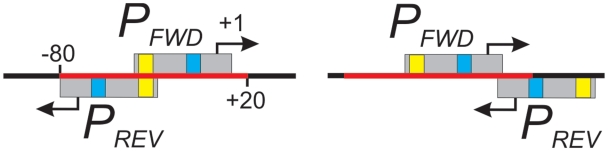
Figure 1.
Arrangements of overlapping promoters. Promoters can be arranged head-to-head (right) or tail-to tail (left). Transcription start points (tsp) are indicated by arrows. The tsp of the forward promoter (PFWD) serves as +1 in the numbering system. Gray boxes show the DNA regions occupied by RNAP (from −40 to +10). The −10 (blue) and −35 (yellow) promoter elements are indicated. All the reverse promoters (PREV) with a tsp falling into the red region (from −80 to +20) are considered to be overlapping promoters with PFWD.
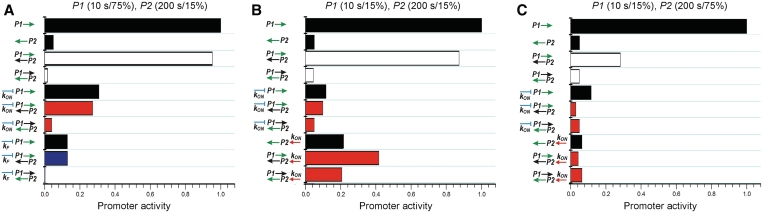
Activities of overlapping promoters depend on (i) the intrinsic activity of the individual promoters, (ii) promoter interference and (iii) the effect of regulatory proteins. Therefore, the activity of overlapping promoters can be regulated in different ways. The simplest case is when no regulatory proteins are involved. In this case RNAP partitioning between the two promoters is determined by the concentration of the free RNAP holoenzyme and its affinity to the promoters. For example, the lower affinity promoter will inhibit the higher affinity promoter to a lesser extent as RNAP availability decreases. When a regulatory protein affects only one of the promoters directly, the indirect effect of regulation on the two promoters depends on whether the regulated promoter becomes more or less aggressive and not whether it becomes stronger (activated) or weaker (repressed). Therefore depending on the characteristics of the two promoters, the interference can enhance or suppress the effect of the regulator. The change in the aggressiveness of the regulated promoter depends on how the regulator changes the time RNAP spends at the promoter DNA. For example, activators can act by enhancing promoter binding and/or increasing the rates of the downstream steps of transcription initiation. Increasing the affinity of the promoter means that RNAP spends more time on DNA, however, faster transcription initiation results in less time spent on the DNA. Similarly, repressors can decrease the rate of promoter loading by inhibiting RNAP binding (steric hindrance) (17) or decrease the firing rate by trapping a promoter-bound RNAP (contact inhibition) (5). Steric hindrance can efficiently reduce the time RNAP spends on DNA, thus allowing the overlapping promoter to be transcribed. However, a trapped RNAP would sit on the promoter for a longer time and inhibit transcription of the overlapping promoter. We explored these scenarios by simulating how repression or activation changes transcription of overlapping promoters of different nature. Repression of strong promoters was simulated by 10-fold reduction of the RNAP binding rate (kON) or by a 10-fold decrease in the firing rate (kF). Activation of weak promoters was simulated by a 10-fold increase in the RNAP binding rate (kON) or in the firing rate (kF) (Figure 3, Supplementary Figure S1). In general, changing the RNAP binding rate has a differential effect on the activity of two promoters (Figure 3, red bars), while both promoter activities are changed in the same direction when the firing rate is altered (Figure 3, blue bars). Differential regulation obtained by the direct and indirect regulatory effects can result in a transcriptional switch. However, a successful transcriptional switch requires different activities of the overlapped promoters in the absence of the regulatory protein combined with a strong effect of differential regulation on both promoters. Promoter activities in the absence of the regulator are determined by the intrinsic promoter activities and by the aggressiveness of the promoters. Therefore high activity differences can be obtained by e.g. (i) overlapping a strong and aggressive promoter with a non-aggressive promoter (Figure 3A), or (ii) overlapping a strong and a weak promoter which do not interfere with each other (both being non-aggressive) (Figure 3B). In the first case repression directly decreases the activity and aggressiveness of the strong promoter, allowing transcription of the overlapping non-aggressive promoter (Figure 3A). In the second case increasing the loading rate of the weak promoter increases its activity and makes the promoter more aggressive, thus repressing the overlapping strong and non-aggressive promoter (Figure 3B). However, in this setup, repression of the strong promoter by decreasing the loading rate does not affect the activity of the overlapping weak promoter (Figure 3B). This example shows that direct and indirect regulation can affect promoters to different extents, and allow practically independent regulation of one of the promoters. In certain cases the regulated promoter remains unchanged while the indirect effect on the overlapping promoter is significant. For instance, increasing the RNAP binding rate for a weak promoter, which is limited by the firing rate, has little effect on the promoter's activity. However, it increases the time RNAP spends at the promoter, therefore it can strongly repress an overlapping promoter (Figure 3C). The possibility of independent regulation of overlapping promoters can contribute to the compact genome organization of prokaryotes.
Figure 3.
Examples of simulated responses of overlapping promoters to regulation of one of the promoters. Regulation of the promoters (arrows) is shown on the left. Bars show activities of promoters labeled with green arrows. Black bars show intrinsic promoter activities. Unregulated promoter strengths (firing frequencies in seconds) and aggressiveness of promoters (the fraction of time RNAP spends at the promoter, %) are shown on the top of each panel. White bars show promoter activities when the two promoters, P1 and P2 overlap. Red bars indicate differential effect of regulation while blue bars indicate that both promoter activities change in the same direction.
The third possibility is that a regulatory protein affects both promoters directly. In this case the indirect effect of altered promoter interference adds to the direct effect of the regulatory protein. Therefore, it provides a suitable mechanism to enhance the contrast of gene expression levels in transcriptional switches. Such a phenomenon has been observed at the λ PR/PRM promoters, where the CI repressor inhibits PR and activates PRM directly (2). PRM is also activated indirectly through removing the PR-bound polymerase which inhibits PRM activity (8).
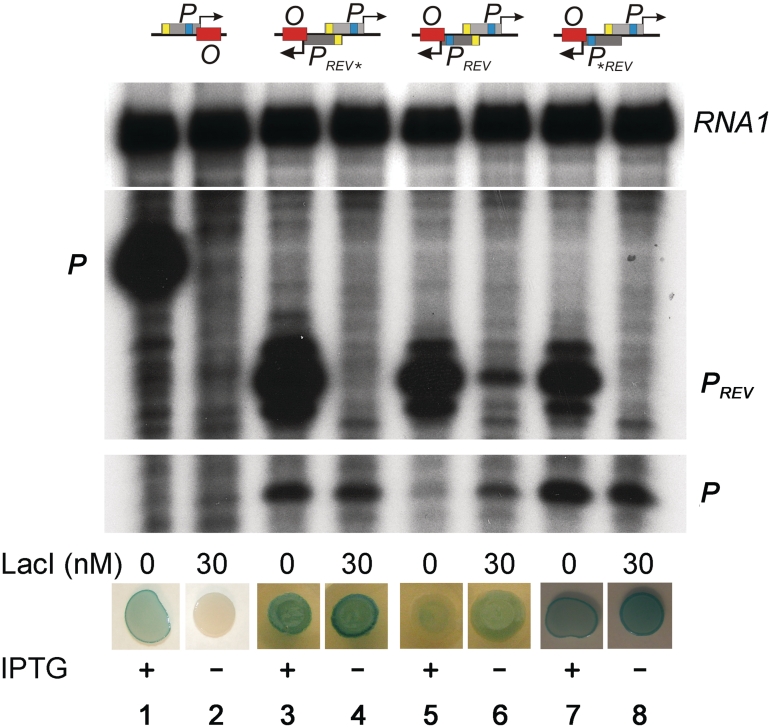
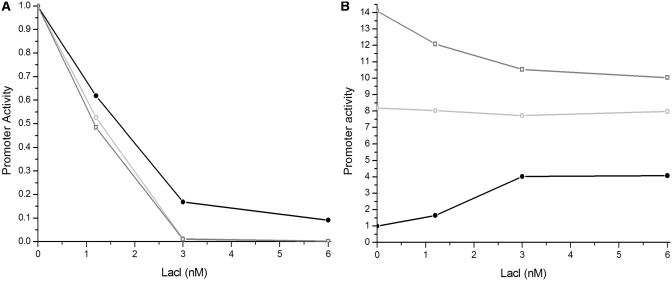
Our simulations suggest that overlapping promoters can provide unique opportunities of regulation, which can be exploited in the construction of synthetic genetic circuits. For example, a non-aggressive promoter is inhibited when overlapped with a strong and aggressive promoter. In this case the non-aggressive promoter can be activated (derepressed) by a repressor which inhibits RNAP binding to the strong promoter. The potential advantage of this setup is that practically any DNA binding protein can be used for repression of the aggressive promoter, and no direct effects are required for the activation of the non-aggressive promoter. To test the feasibility of such regulation we constructed the synthetic system depicted in Figure 4. We overlapped a promoter (P) which has a weak −35 and a consensus −10 element with a reverse promoter (PREV) which has consensus −10 and −35 elements and also contains the extended −10 element. The transcription start site of PREV overlaps with a symmetric LacI operator site (O). The position of the operator site was chosen to be neutral for transcription of P (18). As a first step, activity of P was assayed qualitatively in vivo using a P promoter-uidA (encoding β-glucuronidase) transcriptional fusion in the presence and absence of IPTG (Figure 5). We observed a decrease in P activity in the presence of IPTG, suggesting that as opposed to the natural logic of the lac system (lanes 1 and 2), here P is inactivated in the presence of the inducer (lanes 5 and 6). To understand the system more quantitatively, we measured activities of both P and PREV in the presence and absence of LacI in vitro (Figures 5 and 6). Figure 5 shows that PREV inhibits P activity, and this inhibition depends on having an accessible transcription start site in PREV because it is alleviated when LacI inhibits RNAP binding to PREV by binding to the overlapping lac operator site (compare lanes 5 and 6). We observed ∼4-fold higher P activity in the presence of LacI (Figure 6), demonstrating that LacI can indeed function as an activator of P through the repression of PREV. We found that a single base pair mutation in the consensus −10 element (PREV*) or a two base-pair mutation in the −35 element (P*REV) of PREV results in loss of repression of P (compare lanes 3 and 4, and 7 and 8, respectively), confirming the model prediction that overlapping promoters do not necessarily interfere with each other. While this observation demonstrates that the genome size could be reduced by using overlapping promoters, analysis of natural overlapping promoters is needed to estimate the potential extent of genome compaction that can be achieved by this mechanism. In addition, these control experiments demonstrate that LacI binding does not control P directly, since the addition of LacI does not have a substantial effect on P when PREV is mutated (Figures 5 and 6B). Interestingly, these mutations affect the aggressiveness of PREV but not its overall strength. The mutant promoters are easier to repress by LacI than the wild-type PREV promoter (Figure 6A), likely because the affinity of RNAP to the mutant DNA sequences is decreased. The decreased on rate itself would result in reduced promoter activity if the elongation initiation rate remains unchanged. However, we did not observe such a decrease in the activity of the mutant promoters (Figure 6A), suggesting that mutations in the consensus promoter elements speed up elongation initiation. Our results are in agreement with the previous observation that strong RNA polymerase-promoter interactions stall the polymerase at the promoter, thereby reducing the rate of promoter escape (19). We suggest that the mutant promoters are less successful in inhibition of the P promoter because both the decreased on rate and the faster elongation initiation reduce the time RNAP spends on the promoter DNA. Unlike these mutations, LacI binding reduces only the promoter availability (on rate), resulting in lower promoter activity because of the unchanged elongation initiation rate. Our results show that when PREV has only 65% of its activity, P becomes 1.65 times stronger, and when PREV activity is reduced to 16% than promoter interference is practically eliminated, further repression of PREV by LacI does not increase P activity substantially (Figure 6).
Figure 5.
Regulation of overlapping synthetic promoters in vitro and in vivo. Schematic drawings of regulatory regions are shown on top (see also Figure 4). Results of in vitro transcription from the promoter P and PREV in the presence and absence of LacI are shown below each drawing. The RNA1 transcript, which is not affected by LacI binding, was used as an internal control between lanes (see also Supplementary Figure S2). The regulatory regions were inserted into the E. coli chromosome in such orientation that the promoter P transcribes the uidA reporter gene, encoding β-glucuronidase. Expression of the reporter gene in the presence and absence of IPTG is indicated by the blue color of colonies, resulted from degradation of X-gluc by the β-glucuronidase enzyme.
Figure 6.
The effect of LacI on promoter activities. In vitro transcription was performed at different LacI concentrations and the amount of P and PREV transcripts were quantified as described in the ‘Materials and Methods’ section. (A) PREV (black dots), PREV* (gray circles), P*REV (gray squares) activities as a function of LacI concentration. The background corrected and RNA1 normalized values were normalized for the corresponding promoter activities in the absence of LacI (=1). The relative activities of PREV, PREV* and P*REV in the absence of LacI are 1.00, 2.38 and 0.99, respectively. (B) P promoter activities as a function of LacI concentration when the P promoter is overlapped with PREV (black dots), PREV* (gray circles) and P*REV (gray squares). The background corrected and RNA1 normalized values were normalized for the promoter activity of the P promoter when overlapped with PREV, in the absence of LacI (=1). For comparison of activities in panels (A) and (B), PREV shows ∼80 times higher activity than P when LacI is absent.
In summary our results show that given the proper sequence context, RNAP binding to one promoter can repress an other overlapping promoter (P). Inhibition of RNAP binding to (PREV) by a DNA binding protein (LacI) occupying a site overlapping with the PREV promoter can activate the transcription of (P), resulting in a compact and simple activation-repression switch.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The Danish Council for Independent Research/Natural Sciences, the Hungarian Scientific Research Fund Grant PD75496 (to S.S.), the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research, and the Danish National Research Foundation. Funding for open access charge: Danish National Research Foundation.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank their colleagues in the laboratory for various inputs, in particular Sankar Adhya and László Orosz for useful discussions, and Takácsné Botond Judit for excellent technical assistance.
REFERENCES
- 1.Perez-Martin J, Rojo F, de Lorenzo V. Promoters responsive to DNA bending: a common theme in prokaryotic gene expression. Microbiol. Rev. 1994;58:268–290. doi: 10.1128/mr.58.2.268-290.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ptashne M, Gann A. Genes and Signals. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2002. [Google Scholar]
- 3.Hunziker A, Tuboly C, Horvath P, Krishna S, Semsey S. Genetic flexibility of regulatory networks. Proc. Natl Acad. Sci. USA. 2010;107:12998–13003. doi: 10.1073/pnas.0915003107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Perez-Martin J, Espinosa M. Protein-induced bending as a transcriptional switch. Science. 1993;260:805–807. doi: 10.1126/science.8387228. [DOI] [PubMed] [Google Scholar]
- 5.Choy HE, Park SW, Aki T, Parrack P, Fujita N, Ishihama A, Adhya S. Repression and activation of transcription by Gal and Lac repressors: involvement of alpha subunit of RNA polymerase. EMBO J. 1995;14:4523–4529. doi: 10.1002/j.1460-2075.1995.tb00131.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang P, Yang J, Ishihama A, Pittard AJ. Demonstration that the TyrR protein and RNA polymerase complex formed at the divergent P3 promoter inhibits binding of RNA polymerase to the major promoter, P1, of the aroP gene of Escherichia coli. J. Bacteriol. 1998;180:5466–5472. doi: 10.1128/jb.180.20.5466-5472.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Shearwin KE, Callen BP, Egan JB. Transcriptional interference–a crash course. Trends Genet. 2005;21:339–345. doi: 10.1016/j.tig.2005.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hershberger PA, Mita BC, Tripatara A, deHaseth PL. Interference by PR-bound RNA polymerase with PRM function in vitro. Modulation by the bacteriophage lambda cI protein. J. Biol. Chem. 1993;268:8943–8948. [PubMed] [Google Scholar]
- 9.Mitarai N, Benjamin JA, Krishna S, Semsey S, Csiszovszki Z, Masse E, Sneppen K. Dynamic features of gene expression control by small regulatory RNAs. Proc. Natl Acad. Sci. USA. 2009;106:10655–10659. doi: 10.1073/pnas.0901466106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Semsey S, Geanacopoulos M, Lewis DE, Adhya S. Operator-bound GalR dimers close DNA loops by direct interaction: tetramerization and inducer binding. EMBO J. 2002;21:4349–4356. doi: 10.1093/emboj/cdf431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cho BK, Zengler K, Qiu Y, Park YS, Knight EM, Barrett CL, Gao Y, Palsson BO. The transcription unit architecture of the Escherichia coli genome. Nat. Biotechnol. 2009;27:1043–1049. doi: 10.1038/nbt.1582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schickor P, Metzger W, Werel W, Lederer H, Heumann H. Topography of intermediates in transcription initiation of E.coli. EMBO J. 1990;9:2215–2220. doi: 10.1002/j.1460-2075.1990.tb07391.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Dornenburg JE, Devita AM, Palumbo MJ, Wade JT. Widespread antisense transcription in Escherichia coli. MBio. 2010;1:pii: e00024–10. doi: 10.1128/mBio.00024-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Reppas NB, Wade JT, Church GM, Struhl K. The transition between transcriptional initiation and elongation in E. coli is highly variable and often rate limiting. Mol. Cell. 2006;24:747–757. doi: 10.1016/j.molcel.2006.10.030. [DOI] [PubMed] [Google Scholar]
- 15.Palmer AC, Ahlgren-Berg A, Egan JB, Dodd IB, Shearwin KE. Potent transcriptional interference by pausing of RNA polymerases over a downstream promoter. Mol. Cell. 2009;34:545–555. doi: 10.1016/j.molcel.2009.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sneppen K, Dodd IB, Shearwin KE, Palmer AC, Schubert RA, Callen BP, Egan JB. A mathematical model for transcriptional interference by RNA polymerase traffic in Escherichia coli. J. Mol. Biol. 2005;346:399–409. doi: 10.1016/j.jmb.2004.11.075. [DOI] [PubMed] [Google Scholar]
- 17.Jacob F, Monod J. Genetic regulatory mechanisms in the synthesis of proteins. J. Mol. Biol. 1961;3:318–356. doi: 10.1016/s0022-2836(61)80072-7. [DOI] [PubMed] [Google Scholar]
- 18.Ryu S, Fujita N, Ishihama A, Adhya S. GalR-mediated repression and activation of hybrid lacUV5 promoter: differential contacts with RNA polymerase. Gene. 1998;223:235–245. doi: 10.1016/s0378-1119(98)00237-6. [DOI] [PubMed] [Google Scholar]
- 19.Vo NV, Hsu LM, Kane CM, Chamberlin MJ. In vitro studies of transcript initiation by Escherichia coli RNA polymerase. 3. Influences of individual DNA elements within the promoter recognition region on abortive initiation and promoter escape. Biochemistry. 2003;42:3798–3811. doi: 10.1021/bi026962v. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.