Abstract
A multifunctional reagent based on a coumarin scaffold was developed for derivatization of naive RNA. The alkylating agent N3BC [7-azido-4-(bromomethyl)coumarin], obtained by Pechmann condensation, is selective for uridine. N3BC and its RNA conjugates are pre-fluorophores which permits controlled modular and stepwise RNA derivatization. The success of RNA alkylation by N3BC can be monitored by photolysis of the azido moiety, which generates a coumarin fluorophore that can be excited with UV light of 320 nm. The azidocoumarin-modified RNA can be flexibly employed in structure-function studies. Versatile applications include direct use in photo-crosslinking studies to cognate proteins, as demonstrated with tRNA and RNA fragments from the MS2 phage and the HIV genome. Alternatively, the azide function can be used for further derivatization by click-chemistry. This allows e.g. the introduction of an additional fluorophore for excitation with visible light.
INTRODUCTION
Functionalization of RNA is of paramount importance in all RNA related research in the life sciences. A plethora of labels and functional groups in RNA are in use e.g. for identification, sequencing and imaging of RNA, as well as for functional and structural studies of RNA and RNA–protein complexes. Co-synthetic approaches to RNA functionalization make use of modified building blocks, carrying either the desired functionality or a functional group that allows selective derivatization in a further step. This concept is widely developed in a huge number of modified phosphoramidites for RNA solid phase synthesis (1–3). For example, the incorporation of fluorescent labels as built-in probes may be achieved via phosphoramidites of fluorescent nucleotide analogs, via building blocks of conjugates of fluorescent dyes and canonical nucleotides or via phosphoramidites carrying a reactive moiety as an attachment site, e.g. a primary aminofunction for later derivatization with NHS-derivatives of fluorophores (1,3,4). Co-synthetic functionalization has also been developed for RNA synthesis via run-off in vitro transcription, e.g. for nucleotide analog interference mapping (NAIM) studies (5). However, the range of nucleotide analog triphosphates (XTPs) successfully incorporated into RNA is severely restricted, due to limitations on size and base pairing properties imposed by the RNA polymerase. Successfully incorporated XTPs include a number of naturally occurring modified nucleotides with unaltered Watson–Crick faces (6), numerous nucleobase analogs carrying additional functionalities attached to the pyrimidine C5 position (7–9), as well as α-phosphorothioates (5) and NTPs modified at the 2′-OH (10,11). Incorporation of XTP building blocks may be quantitative, instead of the respective natural NTP (12), or in a mixture of natural NTP and its analog, where the ratio determines the extent of incorporation. In the latter case, incorporated modifications are randomly distributed over the entire transcript length (5,9). Site-specific incorporation, a typical feature of phosphoramidite chemistry, is much more difficult to achieve for RNA transcripts, because it requires additional information to be processed by the polymerase. In an extension of the genetic code, RNA molecules with enhanced functionality, including fluorescent probes and attachment sites for crosslinking moieties, have been obtained by incorporation of several XTPs, which form unnatural base pairs (13–17). Random incorporation of attachment sites is not restricted to in vitro synthesis of RNA, as is illustrated by the biological incorporation of 5-ethinyluridine in living cells. Such in vivo synthesized RNA was made visible via click conjugation of a fluorescent dye (18).
An area much less developed is the functionalization of naïve RNA, i.e. RNA synthesized in the absence of modified building blocks, be it in vivo, by in vitro transcription or by phosphoramidite chemistry. Post-synthetic functionalization can be site-directed, e.g. by post-transcriptional modification enzymes. Indeed, many early studies on tRNA derivatization make use of pre-existing native modifications (19,20). We have recently used RNA:methyltransferases to introduce alkynylgroups into RNA as an attachment point for further derivatization via click chemistry (21). However, in certain applications, site-specificity is not a desirable attribute, e.g. when not all sequence information is available as is the case when mixtures of various RNA species are to be functionalized. Another application with a particular demand for versatile, random post-synthetic introduction of functional groups into RNA is the identification of RNA protein contacts by UV-irradiation (22). A popular modification for crosslinking studies are thiouridines, which occur as a native modification in tRNA, and can be co-synthetically incorporated into RNA via solid phase synthesis or into transcripts as an XTP. Thiouridines produce UV-dependent crosslinks by themselves (22–24) or can be used as an attachment point for aromatic azides (23). This co-synthetic introduction of modifications as a prerequisite exempts a variety of RNAs from use in crosslinking studies, a drawback that we seek to overcome.
Azides, which are relatively inert under physiological conditions, have lately become very popular functional groups in bioconjugate chemistry. This recent development is mostly due to their efficient orthogonal functionalization by the copper(I)-catalyzed azide–alkyne Huisgen cycloaddition (CuAAC) reaction (25–27). This archetypical ‘click’ reaction (28) is known to proceed between an azide and an alkyne-containing moiety with high selectivity i.e. even in the presence of various functional groups commonly encountered inside a living cell and is therefore also known as ‘bio-orthogonal’ (29). Moreover, the reaction yield is generally very good even at low concentrations of both partners and mild reaction conditions can be applied that are non-destructive with respect to proteins and DNA [e.g. (27)]. Applications to RNA have only very recently become popular (18,21,30–32). On the other hand, aromatic azides are also known as photoaffinity labels in crosslinking experiments (23,33–36). Alkylating agents commonly applied to RNA so far are typically monofunctional, and selective post-synthetic RNA derivatization is hampered by the fact that few reagents show an exploitable degree of selectivity for a singular nucleophilic moiety in the nucleobases (37,38).
Here, we present N3BC [7-azido-4-(bromomethyl)-2H-chromen-2-one] as a versatile multifunctional reagent, which allows the controlled post-synthetic functionalization of RNA. N3BC selectively alkylates uridine residues, thereby introducing an azido-chromophore that allows spectroscopic verification of the reaction efficiency, photochemical crosslinking and versatile post-synthetic derivatization by click chemistry.
MATERIALS AND METHODS
Reaction of N3BC with nucleosides and RNA
The details of the synthesis of N3BC is described in the Supplementary Data. Nucleosides adenosine, guanosine, uridine and cytidine and RNA homopolymers poly-rA, poly-rC, poly-rG and poly-rU were from (Sigma-Aldrich, Munich, Germany) and pseudouridine was from (Berry & Associates, Dexter, MI, USA). RNA binary and ternary oligonucleotides were from Biomers, Ulm, Germany [G/U (3′-GGUUGUGGUGGUUUGUUGGU), G/C (3′-CCGGCCCGGCGGCCGGCGCG), U/C (3′-UUCUUUCUUUCCCUUCCUUU), U/A (3′-UUAUAUUUAUAAAUAUUAAA)], Iba, Göttingen, Germany [CUG: (3′-CUUCGUUCGCUGGUC, GAC: (3′-GACCAGCGAACGAAGCAGG)] and Sigma-Aldrich [G/A (3′-AAAGGAAGGGAAAGAAAGAA), C/A (3′-ACCAACAAACCACCACAACC), G/C (3′-CCGGCCCGGCGGCCGGCGCG)]. In vitro transcription was conducted as previously described for S. cerevisiae tRNAPhe (21), and MS2 and HIV-derived RNAs (39), respectively.
N3BC conjugation to nucleosides and HPLC analysis
Each of the four major ribonucleosides and pseudouridine were separately reacted in the dark at a final concentration of 0.4 mM with N3BC (8 mM) for 1 h at 37°C in a solution containing 50% DMSO and 100 mM sodium phosphate buffer at appropriate pH (e.g. pH 9.0 for reactions analyzed in Figure 2). Reaction mixtures were analyzed on an Agilent HP 1100 series equipped with DAD and FLD (Excitation 350 nm, emission 470 nm). A Synergy Fusion RP column (4 µm particle size, 80 Å pore size, 250 mm length, 2 mm inner diameter) from Phenomenex (Aschaffenburg, Germany) with a guard column was used at 35°C. The solvents consisted of 5 mM ammonium acetate buffer adjusted to pH 5.3 using acetic acid (solvent A) and pure methanol (solvent B). The elution started with isocratic flow of 100% solvent A for 3 min, followed by a linear gradient to 30% solvent B at 10 min, then to 70% solvent B at 20 min and further to 80% solvent B at 25 min. Initial conditions were regenerated by rinsing with 100% solvent A for 10 min. The flow rate was 0.3 ml/min.
Figure 2.
Reactivity of N3BC with RNA nucleosides and homopolynucleotides. Reactions of N3BC with free nucleosides and homopolynucleotides were performed as described in ‘Materials and Methods’ section. Reaction products with nucleosides were directly used for HPLC analysis (A), while homopolynucleotides (polyC, polyU etc.) were precipitated to remove unreacted N3BC, and were then digested to free nucleosides for HPLC analysis (B). Only uridine forms a reaction product with N3BC, termed UN3C, which is indicated by an arrow in (A) and (B). Note that N3BC itself is so unpolar that it elutes in the later part of the gradient which is not displayed. (C) Reaction of N3BC with uridine to UN3C. (D) UV-spectra of N3BC, uridine and UN3C. Arrows indicate wavelengths used to monitor aromatic rings (254 nm, upper panels in A and B) and coumarins (320 nm, lower panels in A and B). (E) Fluorescence kinetic of a solution of UN3C upon UV-irradiation at 365 nm. Full emission spectra were recorded at selected time points. (F) Time course of the fluorescence intensity at 450 nm plotted from the data in (E).
N3BC derivatization and workup of RNA
RNA (10–20 µM final concentration) was incubated with 10 mM N3BC (200 mM stock solution in pure DMSO), 100 mM phosphate buffer pH 8.5 and 70% DMSO at 37°C for 180 min under light protection. After the reaction, three volumes of 0.5 M NH4OAc and eight volumes of ethanol were added, the mixture was incubated at −20°C (2 h) and RNA was precipitated by centrifugation at 15000g at +4°C. The RNA was dissolved in 50 µl of pure water, subjected to gel-filtration on an Illustra G-25 column (GE Healthcare, Munich, Germany) and re-precipitated as above.
PAGE analysis of N3BC treated RNA
The concentration of N3BC-conjugated RNA was determined using a Nanodrop-ND-1000 (Peqlab, Erlangen, Germany) and 100–300 pmol (exact amounts are specified in the text) were analyzed on a 15–20% urea gel after exposure to daylight for 1 h. Blue fluorescence of coumarin derivatives was observed upon radiation with UV light (365 nm) and imaged with a GelDoc transilluminator (Peqlab, Erlangen, Germany). Afterwards, the RNA was stained for 10 minutes either with GelRed (according to the instructions of Biotium, Hayward, CA, USA) or with StainsAll solution (0.65× TBE, 0.01% StainsAll, 10% formamide, 25% isopropanol and 65% water), followed by destaining for 2 h in 1× TBE buffer containing 25% isopropanol.
Thin layer chromatography of N3BC treated RNA
N3BC treated RNA (100 pmol) was digested to nucleotides with 0.3 U of nuclease P1 and spotted on a cellulose plate (Merck, Darmstadt, Germany ref#1.05577.0001) and run in NI solvent (isobutyric acid 330 ml, ammonium hydroxide 25% 5 ml, water to 500 ml) for the first dimension and RII solvent (0.1 M Na2HPO4/NaH2PO4 buffer, pH 6.8, 500 ml, ammonium sulfate 300 g, 1-propanol 10 ml) for the second dimension (40). TLC plates were imaged with an AlphaImager (AlphaInnotech, San Leandro, USA) at simultaneous impinging illumination at 254 and 365 nm (Supplementary Figure S4).
Conditions for click reaction with fluorescein-alkyne and AlexaFluor 647–alkyne
AlexaFluor 647-alkyne was from Invitrogen (ref# A10278) and fluorescein-alkyne was synthesized according to (41). The click reaction between N3BC-tRNAPhe and alkyne-containing fluorescent dye was performed in final volume of 10 µl at RT in the dark in PUS buffer (100 mM Tris–HCl, pH 8.0, 100 mM NH4OAc, 1 mM MgCl2). To catalyze the reaction between azide and alkyne moieties, Cu+ was generated in situ by Na-ascorbate reduction of Cu2+. In our tests we used the THPTA ligand (2.5 mM, kindly provided by Dr E. Weinhold, Aachen, Germany), CuSO4 (0.5 mM) and Na-ascorbate (5 mM). Alkyne-coupled fluorescent dye was used at 50 µM final concentration. Reaction time was 2 h, unless indicated otherwise. Reaction products were analyzed by 10% urea–PAGE. Fluorescein-alkyne does not co-migrate with tRNA during electrophoresis, and thus its removal is not necessary, while AlexaFluor 647 co-migrates with the tRNA band and has to be removed by prior gel-filtration through Illustra G-25 columns. Staining with SYBRGold (Invitrogen) was performed according to the manufacturer's instructions. The gels were scanned for fluorescence on a Typhoon (GE Healthcare) using preexisting settings for fluorescein-alkyne (Green laser 532 nm excitation, 526 SP filter, 650 V PM Voltage) and Alexa 647-alkyne (Red laser 634 nm excitation, 670 BP 30 filter, 450 PM Voltage).
N3BC conjugation of α-32P-GTP labeled tRNAPhe and RNase T1 digest
In vitro transcription of tRNAPhe was performed in the presence of [α-32P]-GTP for 3 h at 37°C followed by urea–PAGE for transcript purification. Aliquots (1–3 pmol) of RNA were digested with 500 U of RNase T1 (Fermentas) in 1× T1 buffer (12.5 mM Na-citrate, pH 4.5, 0.5 mM EDTA and 3.5 M urea) for 20 min at 42°C. N3BC treated tRNAPhe (10 µg) was digested under the same conditions. The resulting fragments were then conjugated to AlexaFluor 647-alkyne (as described above). RNase T1 digests were separated on a 20% sequencing gel and signals from radioactive and fluorescent labels were correlated taking into account a relative retardation of fluorescent fragments due to the AlexaFluor647-label (Supplementary Figure S7).
Fluorescence increase measurement of N3BC-polyU and N3BC-tRNAPhe upon UV irradiation
A sample of N3BC-polyU or N3BC-tRNAPhe (∼0.5–1 µg) in PUS buffer was irradiated with an 8 W hand-held UV 365 nm lamp in the spectrofluorimeter cuvette. Fluorescence spectra were recorded at the indicated time points using a JASCO FP-6500 spectrofluorimeter (Excitation 360 nm, 5 nm bandwidth, emission scan 370–500 nm, 10 nm bandwidth).
LC/MS method and analysis
Digestion of RNA for LC/MS (liquid chromatography coupled to mass spectrometry) analysis
Naïve or N3BC-treated RNA (final concentration 1 µg/µl) was dissolved in 20 mM NH4OAc, pH 5.3 and incubated for 5 h at 37°C in the presence of 3 U nuclease P1 (Roche Diagnostics, Mannheim, Germany) per 100 µg RNA. Snake venom phosphodiesterase (Worthington, Lakewood, USA) was then added to a concentration of 0.06 U per 100 µg RNA and the mixture was incubated at 37°C for another 2 h. Finally to convert the resulting mixture of mononucleotides to free nucleosides, 1/10 vol of 10× SAP buffer (Fermentas, St. Leon-Roth, Germany) was added, followed by 3/20 vol of H2O and 1/4 vol of Shrimp Alkaline Phosphatase (SAP stock at 1 U/µl; from Fermentas, St. Leon-Roth, Germany) The mixture was incubated for 1 h at 37°C.
LC/MS/MS instrumental analysis parameters
The LC/MS/MS system (Thermo Electron, Dreieich, Germany) consisted of a Surveyor HPLC (quaternary Surveyor LC pump plus, Surveyor autosampler plus with integrated column heater and cooled sample tray and double-wavelength Surveyor UV-Vis plus) and a triple stage quadrupole mass spectrometer (TSQ 7000 with API-2 ion source and performance kit). Chromatography was performed under the conditions similar to those detailed above (section: N3BC conjugation to nucleosides and HPLC analysis) except that acetonitrile was used as an eluent. The gradient conditions for mass scan and selective reaction monitoring, are given in Supplementary Tables S1 and S2, respectively. UV-detection was performed at 254 and 320 nm. Subsequently, the eluent was introduced without splitting into the electrospray ion source (ESI) of the mass spectrometer. ESI interface parameters were as follows: middle position, spray voltage 4.5 kV, sheath gas (N2) 90 psi, aux gas (N2) 10 scales, capillary heater temperature 350°C. For identification of nucleosides, azides and derivatives, the mass spectrometer scanned the mass range from m/z 160 to m/z 600 (scan time 1 s) in the single MS mode. For selective and sensitive detection as well as quantification, selected reaction monitoring measurements were performed at 1.5 kV multiplier voltage in the MS/MS mode. MS/MS transitions monitored are given in Supplementary Table S3.
UV-crosslinking of N3BC-modified RNA
5′-32P-labeling of RNA
About 80 pmol of N3BC- treated RNA or control unmodified transcript were first treated by 8 U of SAP in the appropriate buffer, the phosphatase was inactivated at 65°C for 15 min and transcripts were ethanol precipitated. Labeling was performed with 10 pmol of [γ-32P]-ATP using T4 polynucleotide kinase in labeling buffer A (Fermentas, St. Leon-Roth, Germany). Unincorporated [γ-32P]-ATP was removed by Illustra G-25 spin columns and 5′-32P-labeled tRNA transcripts were purified by 10% urea–PAGE.
Conditions for RNA–RNA crosslinks
All crosslinking experiments were performed in PUS buffer (100 mM Tris–HCl, pH 8.0, 100 mM NH4OAc, 1 mM MgCl2). For internal crosslinking, ∼1 pmol of 5′-32P-labeled transcript was dissolved in PUS buffer. Crosslinking was performed using a hand-held 8 W CAMAG UV Lamp with dual wavelength (254/365 nm) selection, (ref 022.9120, CAMAG, Switzerland) equipped with 254 nm (ref 352.010) and 365 nm (ref 352.011) tubes. Samples for irradiation were placed into a V-shaped polypropylene 96-well plate on ice and the UV-lamp equipped with a UV-transparent glass plate was placed on top at a distance between UV-tube and sample of ∼10 mm. After exposure for the indicated period of time, transcripts were analyzed by 10% urea–PAGE and 32P-imaging was performed using a Typhoon phosphorimaging screen (GE Healthcare).
Conditions for RNA–protein crosslinks
His-tagged recombinant RNA modification enzymes were prepared by Immobilized Metal Ion Affinity Chromatography (IMAC) or GST-affinity chromatography using procedures described previously for scTrm4 (42), tmTruB (43), pfTrm1 (44), paTrmI (45) and paL7ae (46). MS2-MBP protein was purified by affinity chromatography on amylose column followed by Heparine-Sepharose chromatography (47). Recombinant hnRNP A1 protein was prepared as described previously (48). Preparation of HIV-1 derived RNAs SLS2 and SLS123 was described previously (49). Preparation of Pab21 RNA and S 22 mer RNA was done as described in (50). RNA–protein crosslink experiments were performed with 1 pmol of 5′-32P-labeled transcript and ∼1 µg of different RNA binding and control proteins in PUS buffer. Irradiation with hand-held 8 W UV lamp was performed at UV 365 nm or at 254 nm for 30 min. After incubation, RNA–protein crosslinked products were directly loaded onto 10% SDS gel. Protein staining was done by ‘Rapid Stain’ (G-Biosciences, St. Louis, USA) and gels were imaged in a wet state using a Typhoon screen.
RESULTS
The search for a multifunctional RNA derivatization agent was guided by the necessity to accommodate a reactive electrophilic moiety and a photoreactive azide in a small molecule with properties favorable for post-modification analytics. Coumarin appeared as a promising scaffold because it shows absorption ∼320 nm, thus in a UV range where nucleic acids do not absorb. Recent work (51) identified certain dark coumarin azides as pre-fluorophores, which display strong fluorescence after destruction of the quenching azide moiety. These properties conceivably allow identifying coumarin residues in RNA and thus offer a potential to monitor reaction progress.
Synthesis of N3BC and reaction with nucleosides and RNA homopolymers
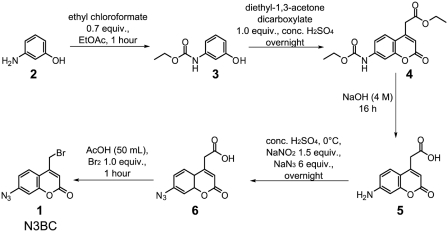
Starting from the known reaction of 4-(bromomethyl)-7-methoxy-2H-chromen-2-one with modified uridines, including in particular thiouridines and pseudouridine (52), we hypothesized that changes in the substitution pattern of the coumarin ring in 4-(bromomethyl)-coumarins might modify the reactivity such that alkylation of otherwise unmodified RNA would become possible. Simultaneously, replacement of the electron donating 7-methoxy group with the electron withdrawing azide would accomplish the accommodation of an azide group for potential later photo-crosslinking and click chemistry. We thus devised a five-step synthesis that yielded N3BC (1) from 3-aminophenol (2) with an overall yield of 44% (Figure 1).
Figure 1.
Synthesis of 7-azido-4-(bromomethyl)-2H-chromen-2-one 1 (N3BC). Experimental details of the Pechmann condensation are described in the supplement.
The light-sensitive compound 1 (termed N3BC) shows low fluorescence, possibly due to quenching by both the bromo- and the azidogroup (51). Its typical chromophore allows the detection of coumarin-containing reaction products by monitoring the absorption at 320 nm in RP18-chromatography runs (Figure 2A). The choice of initial pH and solvent conditions was guided by considerations of reactivity, selectivity and RNA stability. While RNA chains are increasingly prone to phosphodiester hydrolysis as a result of general base catalysis, reactivity is expected to increase with the deprotonation status of nucleobases at higher pH. Initial experiments had indicated a pH range between 8 and 9, a solvent content of 50% (v/v) DMSO and a temperature of 37°C as a reasonable starting point from which to explore reactivity and selectivity. To detect nucleoside alkylation products, each of the four major ribonucleosides was incubated with N3BC and the reaction mixtures were analyzed by LC. As would be expected from a reactive compound, a mock incubation of N3BC without nucleosides showed decomposition into numerous coumarin products, as detected in UV-traces at 320 nm (Figure 2A, lower trace). In comparisons to the other UV-traces in Figure 2A, only uridine shows a covalent adduct containing both nucleoside and coumarin moiety. Pseudouridine, as the most abundant minor nucleoside, was also assayed and found equally reactive (52) (see Supplementary Figure S1).
To the exclusion of light, the reaction product of uridine and N3BC, termed UN3C (Figure 2C), was isolated in microgram quantities from HPLC runs, which allowed its characterization by UV-absorption (Figure 2D), fluorescence (Figure 2E and F) and LC-MS (see below). Intriguingly, LC-traces show very low fluorescence of UN3C, but solutions of the isolated compound become increasingly fluorescent upon irradiation at 365 nm (Figure 2E and F). This strongly suggests that the characteristic coumarin fluorescence is quenched by the azidofunction (51), and that fluorescence is restored by its photochemical destruction. Photochemical conversion to the fluorescent reduction product of a presumed nitrene intermediate (see Supplementary Figure S2) could be monitored in an HPLC/UV/FLD and LC/MS analysis of the irradiated UN3C (See Supplementary Figure S3).
When the investigations were extended to the four RNA homopolymers, only N3BC-treated polyU showed a coumarin chromophore in absorption spectra. Upon UV-irradiation, N3BC-treated polyU, but not the other RNA homopolymers, showed increasing blue fluorescence as well, providing a convenient means to monitor the success of the reaction (See Supplementary Figure S3E). A PAGE analysis of N3BC treated homopolymers failed because of extremely heterogeneous size distribution. After digestion to nucleosides, only the UV-trace of the reaction mixture of N3BC with polyU (Figure 2B) showed the presence of a coumarin-containing product, which corresponded to UN3C in its retention time, UV absorption and fluorescence properties.
Reaction of N3BC with binary RNA oligomers
Because RNA homopolymers do not always behave like typical RNA samples, the analysis was extended to further RNAs, starting with oligonucleotides of binary nucleobase composition. In a re-evaluation of reaction conditions, the DMSO content was increased to 70% in order to denature residual RNA secondary (2D) and tertiary (3D) structure and to promote reactivity. Reacted oligomers were precipitated from the reaction mixture and small molecules were removed by microscale gel filtration.
To exploit fluorescence of the coumarin chromophore by photochemical destruction of the quenching azide moiety (Figure 2E and F), samples were exposed to daylight for at least 1 h prior to electrophoresis and gels were run without light protection. As seen in the PAGE analysis in Figure 3, reactive oligomers show coumarin fluorescence. Figure 3A shows assays of the A/C, A/G and A/U oligomers at pH values between pH 8 and 9. In keeping with the previous analysis, the A/C oligomer does not show any fluorescence, while the A/U oligomer gives rise to a strong signal, originating presumably from UN3C. In contrast to the above results obtained with RNA homopolymers, a weak but measurable signal is apparent for the A/G oligomer, presumably originating from a reaction of guanosines with N3BC. Notably, this signal is much weaker than that from the A/U oligomer, and it is only detectable when at least 200 pmol of the A/G oligomer are loaded onto the gel, a quantity corresponding to ∼1.5 nmol of guanosine residues per band.
Figure 3.
In-gel detection of RNA-coumarin conjugates of binary oligomers. (A) Polyacrylamide gel of binary oligomers A/C, A/G and A/U (200 pmol each) after reaction with N3BC at pH 8.0, 8.5 and 9.0 (indicated on the bottom of the panel). Samples were exposed to daylight for 1 h prior to PAGE. The loading control with StainsAll was imaged with a conventional digital camera, and fluorescence upon excitation at 365 nm was imaged with a GelDoc. Note that the loading control developed with StainsAll does not allow comparison among binary oligomers of different composition (compare Supplementary Figure S4). (B) Polyacrylamide gel of binary oligomers (300 pmol each) of all six permutations of nucleobases (the nucleobase composition is indicated above the gel image) after reaction with N3BC at pH 8.5 (right panels) or mock incubation (left panels). Imaging was done as described in (A) Note that the signal of the GC oligomer smears in the loading control as a consequence of incomplete denaturation.
The results in Figure 3A thus suggest a strong reaction of N3BC with uridine residues in RNA and an additional weak side reaction with guanosines, while cytidines and adenosines remain unreactive. For confirmation, the remaining binary oligomers were investigated. In this and the following reactions, the pH was set to 8.5 and the DMSO content was kept at 70%. Figure 3B shows a PAGE analysis after N3BC modification reactions of 20-mers of all six possible binary permutations of nucleotides. In contrast to homopolymers, single bands of these bipartite oligomers appear in the loading control (Figure 3B, StainsAll stain) except for the species composed of cytidine and guanosine residues (termed G/C-oligomer). The smeared signal observed for the latter species is reproducible for samples of different origin and therefore likely due to incomplete denaturation during electrophoresis. For each sample, the amount of RNA was determined by UV absorption prior to gel loading. Interestingly, some binary RNA compositions led to differential staining using common RNA-staining dyes. A/C and C/U-oligomers could not be efficiently stained with ethidium bromide or GelRed (Supplementary Figure S4). Although StainsAll reveals oligomers of all binary compositions, staining efficiency is disparate and does not allow quantitative comparison among binary oligomers of different composition.
Aliquots of the above reactions were digested to mononucleotides and analyzed by two-dimensional TLC in solvent systems established to detect natural RNA modification. Coumarin fluorescence was observed for all binary oligomers containing uridines (corresponding to photolysis products of UN3C) but not for other binary oligomers (see Supplementary Figure S5). Thus, the in-gel detection is more sensitive, presumably because bands in the gel are more focused than TLC spots.
LC/MS characterization of N3BC reaction products
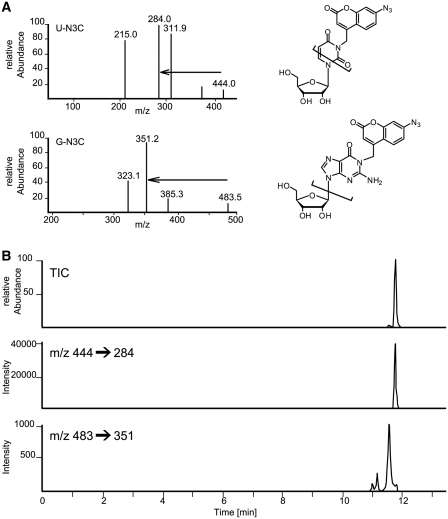
To verify these findings, LC/MS analysis was employed, piloted by characterization of UN3C isolated from the initial nucleoside reactions. In LC/MS runs, the UV-signal of UN3C coincided with a [M + H]+ signal of m/z 444, which is consistent with N3-alkylation (Figure 2C). From the tandem mass spectrum in Figure 4, the m/z 444 → m/z 284 mass transition was identified and used to develop an LC/MS/MS method for the sensitive detection of uridine and its adduct UN3C (see Supplementary Data for details). Using this method we were able to detect UN3C in various N3BC-treated RNA species, including polyU, binary oligomers, ternary oligomers and an in vitro transcript of tRNAPhe from S. cerevisiae (see Supplementary Figures S6 and S7). Figure 4 shows the LC/MS analysis of an N3BC-treated ternary oligonucleotide (CUGGUCGCUUGCUUC), displaying a clearly visible UN3C peak at t = 11.75 min. Screening this RNA for the expected mass of an adduct of N3BC with guanosine, i.e. an [M + H]+ signal of m/z 483, revealed three faint signals at retention times 11.00, 11.12 and 11.55 min. Inspection of the tandem mass spectrum suggested fragmentation behavior similar to the UN3C compound. The origin of three separate peaks may be related to alkylation by N3BC of different nitrogens in guanosine, although this hypothesis cannot be confirmed by LC/MS/MS because fragmentation occurs at or near the glycosidic bond. Assuming a similar response factor for UN3C and the guanosine adducts of N3BC, and taking into account the relative abundance of G and U residues in this oligomer, a conservative estimate of the relative abundance of both compounds (areas under the curves) amounts to a ratio of ∼1:25, which is consistent with fluorescence intensities observed in Figure 3.
Figure 4.
LC/MS/MS analysis of reaction products of N3BC with uridine and guanosine residues in RNA. (A) Mass spectrum, structure and main fragmentation of positively charged [M + H]+ of conjugates of N3BC to uridine (upper panel) and guanosine (lower panel). Mass transitions used in (B) are indicated by arrows. The structure of the guanosine product is one out of three possible reaction products of nitrogen alkylation. (B) LC/MS/MS transition trace for guanosine adducts (lower trace) and the uridine adduct (middle trace). The total ion count (TIC) in the upper trace is the weighed sum of the lower two traces. Note the disparate relative intensities in the TIC trace: the smaller peak corresponds to guanosine adducts and resolves into three species seen in the lower trace.
Influence of RNA structure on reactivity towards N3BC
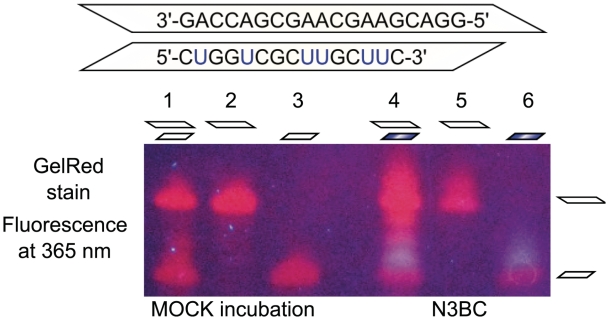
To evaluate the influence on UN3C formation of potential blocking of uridine-N3 by base pairing in the helical RNA structure, 100 pmol of each of two N3BC-treated tripartite CUG and GAC oligomers were imaged after PAGE. These conditions were deliberately chosen such that coumarin fluorescence resulting from guanosine adducts is not detectable (400 and 700 pmol of G residues per band, respectively). Accordingly, only the blue fluorescence of the oligomer containing uridine residues is visible (Figure 5, lanes 4 and 6). These oligomers, which are complementary with only a small 5′-overhang of the GAC oligomer, were treated with N3BC separately and as an equimolar mixture. Under both conditions, the CUG oligomer in the corresponding lanes 4 and 6 in Figure 5 shows identical blue coumarin fluorescence, which can be distinguished by the naked eye. Since the presence of the complementary GAC oligomer does not affect the reaction of N3BC with the CUG oligomer, it is concluded that the denaturing effect of 70% DMSO content is sufficient to render all uridines accessible to alkylation. Interestingly, lowering the DMSO content to 40% protects uridines against alkylation by N3BC in the same experimental setting (Supplementary Figure S8), suggesting that elements of secondary structure are forming in 40% DMSO.
Figure 5.
Influence of RNA secondary structure on UN3C formation investigated by in-gel detection of RNA-coumarin conjugates. The two ternary GAC and CUG oligomers shown in the upper part are complementary save for a 5′-overhang on the GAC oligomer. The oligomers were reacted with N3BC separately (lanes 5 and 6), and as an equimolar mixture (lane 4). Lanes 1–3 show the corresponding mock incubation controls. The samples were run on a denaturing PAGE and stained with GelRed. Upon excitation at 365 nm, the fluorescence was imaged with a conventional digital camera. The blue coumarin fluorescence can be clearly distinguished from the red fluorescence of the GelRed stain. Note that coumarin conjugates migrate more slowly than unreacted RNA.
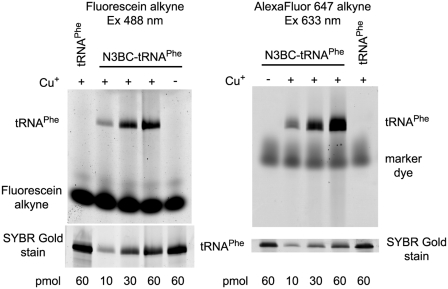
To further elucidate, if strongly structured elements in RNA prevents its reaction with N3BC in 70% DMSO, we have investigated yeast tRNAPhe, generated by in vitro transcription. This tRNA is one of the working horses of RNA structural biology, and a paradigm of a strongly structured RNA, whose intricate 3D structure is well understood from crystallography (53). In order to increase the sensitivity of coumarin azide detection in N3BC-treated RNA, it was decided to exploit the inherent potential of the aromatic azide for click reactions. Recently, we demonstrated the application of click-reaction to alkyne-derivatized tRNAs and optimized reaction conditions in order to avoid RNA degradation (21). Thus, N3BC-treated in vitro transcript of tRNAPhe, estimated by LC/MS analysis to contain ∼10% of N3BC-modified uridines, was incubated with a fluorescein terminal alkyne derivative under Huisgen-type cycloaddition conditions. These include, in particular, the presence of copper-(I) ions, which are typically generated in situ by reduction of Cu-(II) with ascorbate. Copper-(I) ions were stabilized by addition of the chelating ligand THPTA. To analyze derivatization by Huisgen-type cycloaddition, the RNA was subjected to urea–PAGE and the gel was scanned for fluorescence. The left panel in Figure 6 shows a fluorescence emission scan upon fluorescein excitation at 532 nm. The scan reveals a distinct fluorescent band co-migrating with the tRNA. The appearance of this click-adduct is dependent on the presence of Cu(I) in the reaction mixture, product formation is proportional to the amount of N3BC-modified RNA in the reaction, and the reaction does not occur without prior treatment of the RNA with N3BC. Unreacted dye can be detected migrating in the gel. The SYBR Gold stain in the lower panel clearly shows that RNA is stable under click conditions. Identical results were obtained with a terminal alkyne derivative of the AlexaFluor 647 dye, except that the unreacted alkyne-dye co-migrates with tRNA and was therefore removed by spin column gel filtration prior to PAGE analysis (Figure 6, right panel). We conclude that serial treatment of RNA with N3BC and a fluorescent terminal alkyne under click conditions constitutes a new way for post-synthetic fluorescent labeling of RNA that takes place selectively on the N3 of uridines.
Figure 6.
Click-modification of N3BC-treated tRNAPhe with alkyne-coupled fluorescent dyes. N3BC-treated tRNA and untreated control tRNA were incubated with fluorescein-alkyne (left panel) or with AlexaFluor 647-alkyne (right panel) in the presence and absence of copper ions, as indicated above the gel images. For incubation, 10, 30 and 60 pmol of N3BC-treated tRNA and 60 pmol of untreated tRNA transcript (as indicated below the gel image) were used. Analysis was done by urea–PAGE and subsequent fluorescence imaging. Gels were stained by SYBR Gold to provide an RNA loading control.
To trace the distribution of coumarin azide residues throughout the tRNA sequence, material obtained from in vitro transcription in the presence of [α-32P]-GTP was submitted to complete digestion with RNase T1. This combination results in the generation of short RNA fragments containing one 32P-labeled guanosine residue on their respective 3′-ends. Separation of such a ‘G-catalog’ on a denaturing 20% PAGE resolves most of the fragments of 1–8 nt in size (See Supplementary Figure S9). When a G-catalog of N3BC-treated tRNA was click-labeled with Alexa 647 azide, fluorescence signals were detected for at least seven fragments. Mapping of the G-catalog onto the fluorescence signals (See Supplementary Figure S6 for details of the mapping) reveals that, although the technique does not allow complete coverage of all fragments, coumarin residues are clearly not confined to any particular region of the tRNA. Clearly, a content of 70% DMSO is sufficient to denature even highly structured RNAs such as this tRNA, and to render all uridines equally accessible to alkylation by N3BC. Application of the ‘click-detection’ developed above also allowed us to trace low amounts of N3BC-conjugates obtained with RNA oligomers at DMSO concentrations as low as 20% (see Supplementary Figure S10). Although a decrease in DMSO content dramatically decreases alkylation yield, these data suggest, that for a given RNA, an optimization of the DMSO content should allow to choose between alkylation under denaturing or under near-native conditions. Consequently, the resulting RNAs would bear UN3C equally distributed over the RNA sequence, or preferentially at those positions, whose uridine-N3 is not involved in features of the native structure.
Application of N3BC-treated RNA in crosslinking studies
During initial investigations of the light sensitivity of N3BC derivatives isolated from nucleoside reactions, a variety of fluorescent peaks were observed in LC analysis of irradiated material (Supplementary Figure S2), presumably arising from various reaction pathways undergone by a coumarin-nitrene intermediate, which was generated by photochemical elimination of N2 from UN3C (35). A peak at 18.2 min displays high coumarin fluorescence and a molecular mass of m/z 418 (See Supplementary Figure S2), which is consistent with an aromatic amine, a known reduction product of nitrenes (35). Nitrenes are an electron deficient species known to undergo a variety of insertion reactions, which leads to an important application in crosslinking studies (23,33,35,36). Because the observed photoreactivity suggested the potential application of UN3C-containing RNA in crosslinking reactions to cognate proteins, this possibility was exploited in detail. 5′-32P-labeled aliquots of the above N3BC-treated tRNAPhe were irradiated with UV-light (365 nm) in the presence of proteins known to bind this particular tRNA. The SDS–PAGE in the Figure 7A clearly shows efficient formation of intermolecular crosslink bands with a variety of cognate tRNA modification enzymes, including methyltransferases and pseudouridine synthases known to specifically recognize features of tRNA architecture (42–45). No crosslink was observed with untreated tRNAPhe (Figure 7A, right panel) and with control proteins known to not bind RNA (see Supplementary Figure S7), confirming the specificity of the reaction. Analysis by urea–PAGE shows that the N3BC-treated tRNA also forms inter- or intramolecular RNA–RNA crosslinks in the absence of protein (see Supplementary Figure S12).
Figure 7.
Use of N3BC-treated RNA in RNA–protein crosslinking studies. (A) N3BC-treated tRNAPhe crosslinks with RNA binding proteins upon irradiation. The secondary tRNA structure is shown on the left. 5′-32P-labeled N3BC-treated tRNAPhe or similar amounts of control tRNAPhe transcript were irradiated (as indicated above the gel image) for 0 min or 30 min at 365 nm. RNA–protein crosslinked products were analyzed by SDS–PAGE shown in the middle panel. The following RNA binding proteins were used: P. furiosus Trm1 (pfTrm1), P. abyssi TrmI (paTrmI), S. cerevisiae Trm4 (scTrm4) and T. maritima TruB (tmTruB). Gels were Coomassie Blue stained to verify similar loading of different proteins (shown at the bottom). The lane labeled ‘C’ shows a tRNA sample irradiated in the absence of protein. The negative control in the right panel shows an SDS–PAGE of samples which were not treated with N3BC prior to crosslinking. A control experiment with proteins known not to bind to RNA is shown in Supplementary Figure S11. (B) Secondary structure of MS2 RNA (left) and crosslinking of N3BC-treated MS2 RNA with the recombinant MS2-MBP fusion protein (right). Incubations, crosslinking and analysis were performed as described above for tRNAPhe. (C) Secondary structure of HIV-1 derived SLS2 and SLS123 RNAs (left). The region corresponding to SLS2 RNA is shaded. Numbering corresponds to nucleotide numbering in the HIV-1 genome. Crosslinking of SLS2 and SLS123 RNAs to the recombinant hnRNP A1 protein is shown in the right panel. Incubations, crosslinking and analysis were performed as described above for tRNAPhe.
In order to extend the N3BC application to RNAs differing from the canonical tRNA scaffold, we included several further RNA–protein interaction pairs in our study. One pair consisted in phage MS2 RNA (Figure 7B) and its partner MS2 protein (47,54) used as a fusion protein with Maltose Binding Protein (MS2-MBP). As a second model, we used two fragments of HIV-1 virus RNA containing the A7 splice site and its specific regulator, hnRNP A1 protein (49,55) (Figure 7C). The first fragment (SLS2 RNA) of ∼80 nt contained an HIV-1 RNA region spanning nt 7965–8046 and exhibits an extended stem-loop structure. The second HIV-1 derived transcript (SLS123 RNA) contained the SLS2 region along with two additional stem-loop structures at 5′- and 3′-extremities (nt 7903–8092 in HIV-1 RNA). Both fragments had previously been shown to bind hnRNP A1 protein with high affinity and the binding sites for hnRNP A1 had been determined by footprinting studies (39,49).
5′-32P-labeled in vitro transcripts of these model RNAs were N3BC-treated, and, after UV-crosslinking at 365 nm, the resulting RNA–protein covalent adducts were analyzed by SDS–PAGE. As shown in Figure 7 (Panels B and C), all three studied RNAs were successfully crosslinked with their corresponding protein partners, demonstrating that N3BC-treated RNA transcripts may be used for such studies. Similar data were also obtained for one other binding partner of SLS2 and SLS123 RNAs, namely hnRNP K protein (see Supplementary Figure S13).
In order to assess the specificity of X-link using N3BC-treated RNA, we also performed experiments with the paL7ae protein, which specifically binds K-turn containing RNAs (like sRNA guides). This protein shows a faint but specific crosslink signal with Pab21 RNA, which contains such a structural motif. On the contrary, RNAs lacking a K-turn, such as tRNAPhe or the 22mer fragment of P. abyssi rRNA, did not crosslink to L7ae upon irradiation at UV365 nm (see Supplementary Figure S14). The above findings clearly identify crosslinking reactions as a useful feature of the coumarin azide in various types of RNA.
DISCUSSION
Alkylation and acylation reactions of nucleobases have a long standing scientific history that includes the development of RNA sequencing techniques (56) and of structural probing [(57); for reviews see (37,58)]. Classical electrophiles such as dimethylsulfate are monofunctional and rather unspecific with respect to the targeted nucleophile. While modern electrophilic reagents are more selective, e.g. for ribose 2′-OH (59) or the N2 of guanosine (60), the specificity of N3BC for uridine is a unique feature among electrophiles; CMCT, the only commonly employed structural probe for uridines, also reacts with guanosine to an extend that allows its application in structural probing of guanosine residues, while N3BC shows only trace reactivity with guanosine (37).
The bidentate chloroacetaldehyde (61) is still one of the few electrophilic reagents, which contain or form chromophores of analytical benefit. More recently developed non-alkylating structural probes make use of light induced scission (62,63). Certain alkylating derivatives of aromatic azides have been used for crosslinking purposes (64). On the other hand, modern bioconjugate chemistry features the development of reagents with multiple functionalities in application such as activity-based protein profiling which may include functionalities for target binding, crosslinking, spectroscopic detection, affinity purification and others [e.g. (65), for reviews see (66,67)].
The N3BC reagent developed in this study combines alkylating reactivity with analytically useful spectroscopic properties and multifunctionality; its coumarin chromophore can be detected in UV absorption, and, after photolysis of the azido function, by fluorescence. We have found the properties of N3BC and its RNA conjugates very convenient in investigations of RNA–protein interactions. RNA can be treated in large quantities, and the resulting RNA conjugates are stable over several months at −20°C, when thoroughly protected from light. Aliquots may be removed to verify the success of the modification reaction, for labeling or for crosslinking purposes.
Previous azide-based RNA crosslinking methods required the presence of sulfur as reactive moiety for the introduction of the photoactive compound (23). Here, N3BC offers a much wider range of applications, because it can be used on native RNA, synthetic RNA, and in vitro transcripts alike. The crosslinking is conducted at 365 nm, a wavelength that does not damage the nucleobases. The RNA used here carried UN3C residues randomly distributed throughout the entire sequence, because the RNA structure was denatured in 70% DMSO during the reaction with N3BC. However, we have also shown, that elements of secondary structure remain intact at 40% DMSO content, and that UN3C can be detected even in reactions conducted in 20% DMSO. Hence, future applications of N3BC may possibly be developed, which contain crosslinking UNC3 moieties only at positions that are accessible in the native RNA structure.
In addition to being a crosslinking moiety, the aromatic azide doubles as a conjugation module, onto which almost any desired functionality can be attached by click chemistry. Although here exemplified with a fluorescent dye, click conjugation to alkyne derivatives conceivably allows attachment of other functional groups such as biotin. Advantages over other types of chemistry offered by azide–alkyne click derivatization include the low concentration of azide compounds and mild reaction conditions. While numerous examples of DNA derivatization by click chemistry have recently been published (27,68,69), its application in the RNA field is only about to become popular (18,21,30–32). We find that RNA is stable under in vitro click conditions and therefore we anticipate a widespread future use of the N3BC reagent.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
This work was supported by the Deutsche Forschungsgemeinschaft (HE 3397/4) and the Volkswagen Foundation. Funding for open access charge: Institute of Pharmacy and Biochemistry, Mainz University.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Chandrabali Bhattacharya for technical assistance, Henri Grosjean (Paris, France), Louis Droogmans (Brussels, Belgium) and Robert Stroud (San Francisco, CA, USA) for plasmids and Andres Jäschke for constant support. Georges Khoury and Anne-Sophie Tillault (Laboratoire AREMS, Nancy) are acknowledged for providing recombinant hnRNP proteins and paL7ae as well as appropriate RNA substrates for control crosslinking experiments.
REFERENCES
- 1.Gait MJ. DNA/RNA synthesis and labelling. Curr. Opin. Biotechnol. 1991;2:61–68. doi: 10.1016/0958-1669(91)90062-a. [DOI] [PubMed] [Google Scholar]
- 2.Sproat BS. Chemistry and applications of oligonucleotide analogues. J. Biotechnol. 1995;41:221–238. doi: 10.1016/0168-1656(95)00012-f. [DOI] [PubMed] [Google Scholar]
- 3.Verma S, Eckstein F. Modified oligonucleotides: synthesis and strategy for users. Annu. Rev. Biochem. 1998;67:99–134. doi: 10.1146/annurev.biochem.67.1.99. [DOI] [PubMed] [Google Scholar]
- 4.Sinkeldam RW, Greco NJ, Tor Y. Fluorescent analogs of biomolecular building blocks: design, properties, and applications. Chem. Rev. 2010;110:2579–2619. doi: 10.1021/cr900301e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cochrane JC, Strobel SA. Probing RNA structure and function by nucleotide analog interference mapping. Curr. Protoc. Nucleic Acid. Chem. 2004 doi: 10.1002/0471142700.nc0609s17. Chapter 6, Unit 6.9. [DOI] [PubMed] [Google Scholar]
- 6.Kariko K, Buckstein M, Ni H, Weissman D. Suppression of RNA recognition by Toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity. 2005;23:165–175. doi: 10.1016/j.immuni.2005.06.008. [DOI] [PubMed] [Google Scholar]
- 7.Tarasow TM, Tarasow SL, Eaton BE. RNA-catalysed carbon-carbon bond formation. Nature. 1997;389:54–57. doi: 10.1038/37950. [DOI] [PubMed] [Google Scholar]
- 8.Vaught JD, Dewey T, Eaton BE. T7 RNA polymerase transcription with 5-position modified UTP derivatives. J. Am. Chem. Soc. 2004;126:11231–11237. doi: 10.1021/ja049009h. [DOI] [PubMed] [Google Scholar]
- 9.Srivatsan SG, Tor Y. Synthesis and enzymatic incorporation of a fluorescent pyrimidine ribonucleotide. Nat. Protoc. 2007;2:1547–1555. doi: 10.1038/nprot.2007.222. [DOI] [PubMed] [Google Scholar]
- 10.Padilla R, Sousa R. Efficient synthesis of nucleic acids heavily modified with non-canonical ribose 2′-groups using a mutant T7 RNA polymerase (RNAP) Nucleic Acids Res. 1999;27:1561–1563. doi: 10.1093/nar/27.6.1561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sousa R, Padilla R. A mutant T7 RNA polymerase as a DNA polymerase. EMBO J. 1995;14:4609–4621. doi: 10.1002/j.1460-2075.1995.tb00140.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gugliotti LA, Feldheim DL, Eaton BE. RNA-mediated metal-metal bond formation in the synthesis of hexagonal palladium nanoparticles. Science. 2004;304:850–852. doi: 10.1126/science.1095678. [DOI] [PubMed] [Google Scholar]
- 13.Hirao I. Unnatural base pair systems for DNA/RNA-based biotechnology. Curr. Opin. Chem. Biol. 2006;10:622–627. doi: 10.1016/j.cbpa.2006.09.021. [DOI] [PubMed] [Google Scholar]
- 14.Kawai R, Kimoto M, Ikeda S, Mitsui T, Endo M, Yokoyama S, Hirao I. Site-specific fluorescent labeling of RNA molecules by specific transcription using unnatural base pairs. J. Am. Chem. Soc. 2005;127:17286–17295. doi: 10.1021/ja0542946. [DOI] [PubMed] [Google Scholar]
- 15.Kimoto M, Mitsui T, Yamashige R, Sato A, Yokoyama S, Hirao I. A new unnatural base pair system between fluorophore and quencher base analogues for nucleic acid-based imaging technology. J. Am. Chem. Soc. 2010;132:15418–15426. doi: 10.1021/ja1072383. [DOI] [PubMed] [Google Scholar]
- 16.Kimoto M, Hirao I. Site-specific incorporation of extra components into RNA by transcription using unnatural base pair systems. Methods Mol. Biol. 2010;634:355–369. doi: 10.1007/978-1-60761-652-8_25. [DOI] [PubMed] [Google Scholar]
- 17.Kimoto M, Mitsui T, Yokoyama S, Hirao I. A unique fluorescent base analogue for the expansion of the genetic alphabet. J. Am. Chem. Soc. 2010;132:4988–4989. doi: 10.1021/ja100806c. [DOI] [PubMed] [Google Scholar]
- 18.Jao CY, Salic A. Exploring RNA transcription and turnover in vivo by using click chemistry. Proc. Natl Acad. Sci. USA. 2008;105:15779–15784. doi: 10.1073/pnas.0808480105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kasai H, Shindo-Okada N, Noguchi S, Nishimura S. Specific fluorescent labeling of 7-(aminomethyl)-7-deazaguanosine located in the anticodon of tRNATyr isolated from E. coli mutant. Nucleic Acids Res. 1979;7:231–238. doi: 10.1093/nar/7.1.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wintermeyer W, Schleich HG, Zachau HG. Incorporation of amines or hydrazines into tRNA replacing wybutine or dihydrouracil. Methods Enzymol. 1979;59:110–121. doi: 10.1016/0076-6879(79)59073-9. [DOI] [PubMed] [Google Scholar]
- 21.Motorin Y, Burhenne J, Teimer R, Koynov K, Willnow S, Weinhold E, Helm M. Expanding the chemical scope of RNA: methyltransferases to site-specific alkynylation of RNA for click labeling. Nucleic Acids Res. 2011;39:1943–1952. doi: 10.1093/nar/gkq825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hafner M, Landthaler M, Burger L, Khorshid M, Hausser J, Berninger P, Rothballer A, Ascano M, Jungkamp AC, Munschauer M, et al. PAR-CliP–a method to identify transcriptome-wide the binding sites of RNA binding proteins. J. Vis. Exp. 2010;41:pii: 2034. doi: 10.3791/2034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Juzumiene DI, Shapkina TG, Wollenzien P. Distribution of cross-links between mRNA analogues and 16 S rRNA in Escherichia coli 70 S ribosomes made under equilibrium conditions and their response to tRNA binding. J. Biol. Chem. 1995;270:12794–12800. doi: 10.1074/jbc.270.21.12794. [DOI] [PubMed] [Google Scholar]
- 24.Juzumiene D, Shapkina T, Kirillov S, Wollenzien P. Short-range RNA-RNA crosslinking methods to determine rRNA structure and interactions. Methods. 2001;25:333–343. doi: 10.1006/meth.2001.1245. [DOI] [PubMed] [Google Scholar]
- 25.Rostovtsev VV, Green LG, Fokin VV, Sharpless KB. A stepwise huisgen cycloaddition process: copper(I)-catalyzed regioselective “ligation” of azides and terminal alkynes. Angew. Chem. Int. Ed. 2002;41:2596–2599. doi: 10.1002/1521-3773(20020715)41:14<2596::AID-ANIE2596>3.0.CO;2-4. [DOI] [PubMed] [Google Scholar]
- 26.Meldal M, Tornoe CW. Cu-catalyzed azide-alkyne cycloaddition. Chem. Rev. 2008;108:2952–3015. doi: 10.1021/cr0783479. [DOI] [PubMed] [Google Scholar]
- 27.Gramlich PM, Wirges CT, Manetto A, Carell T. Postsynthetic DNA modification through the copper-catalyzed azide-alkyne cycloaddition reaction. Angew. Chem. Int. Ed. 2008;47:8350–8358. doi: 10.1002/anie.200802077. [DOI] [PubMed] [Google Scholar]
- 28.Becer CR, Hoogenboom R, Schubert US. Click chemistry beyond metal-catalyzed cycloaddition. Angew. Chem. Int. Ed. 2009;48:4900–4908. doi: 10.1002/anie.200900755. [DOI] [PubMed] [Google Scholar]
- 29.Sletten EM, Bertozzi CR. Bioorthogonal chemistry: fishing for selectivity in a sea of functionality. Angew. Chem. Int. Ed. 2009;48:6974–6998. doi: 10.1002/anie.200900942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Paredes E, Das SR. Click chemistry for rapid labeling and ligation of RNA. Chembiochem. 2011;12:125–131. doi: 10.1002/cbic.201000466. [DOI] [PubMed] [Google Scholar]
- 31.Jayaprakash KN, Peng CG, Butler D, Varghese JP, Maier MA, Rajeev KG, Manoharan M. Non-nucleoside building blocks for copper-assisted and copper-free click chemistry for the efficient synthesis of RNA conjugates. Org. Lett. 2010;12:5410–5413. doi: 10.1021/ol102205j. [DOI] [PubMed] [Google Scholar]
- 32.Aigner M, Hartl M, Fauster K, Steger J, Bister K, Micura R. Chemical synthesis of site-specifically 2′-azido-modified RNA and potential applications for bioconjugation and RNA interference. Chembiochem. 2011;12:47–51. doi: 10.1002/cbic.201000646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hixson SH, Hixson SS. P-Azidophenacyl bromide, a versatile photolabile bifunctional reagent. Reaction with glyceraldehyde-3-phosphate dehydrogenase. Biochemistry. 1975;14:4251–4254. doi: 10.1021/bi00690a016. [DOI] [PubMed] [Google Scholar]
- 34.Millon R, Olomucki M, Le Gall JY, Golinska B, Ebel JP, Ehresmann B. Synthesis of a new reagent, ethyl 4-azidobenzoylaminoacetimidate, and its use for RNA-protein cross-linking within Escherichia coli ribosomal 30-S subunits. Eur. J. Biochem. 1980;110:485–492. doi: 10.1111/j.1432-1033.1980.tb04890.x. [DOI] [PubMed] [Google Scholar]
- 35.Buchmueller KL, Hill BT, Platz MS, Weeks KM. RNA-tethered phenyl azide photocrosslinking via a short-lived indiscriminant electrophile. J. Am. Chem. Soc. 2003;125:10850–10861. doi: 10.1021/ja035743+. [DOI] [PubMed] [Google Scholar]
- 36.Wombacher R, Jäschke A. Probing the active site of a diels-alderase ribozyme by photoaffinity cross-linking. J. Am. Chem. Soc. 2008;130:8594–8595. doi: 10.1021/ja802931q. [DOI] [PubMed] [Google Scholar]
- 37.Ehresmann C, Baudin F, Mougel M, Romby P, Ebel J-P, Ehresmann B. Probing the structure of RNAs in solution. Nucleic Acids Res. 1987;15:9109–9128. doi: 10.1093/nar/15.22.9109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Giegé R, Helm M, Florentz C. Chemical and Enzymatic Probing of RNA Structure. In: Söll D, Nishimura S, Moore P, editors. Prebiotic Chemistry, Molecular Fossils, Nucleosides, and RNA. Vol. 6. Oxford: Pergamon; 1999. pp. 63–80. [Google Scholar]
- 39.Marchand V, Santerre M, Aigueperse C, Fouillen L, Saliou JM, Van Dorsselaer A, Sanglier-Cianférani S, Branlant C, Motorin Y. Identification of protein partners of the human immunodeficiency virus 1 tat/rev exon 3 leads to the discovery of a new HIV-1 splicing regulator, protein hnRNP K. RNA Biol. 2011;8:325–342. doi: 10.4161/rna.8.2.13984. [DOI] [PubMed] [Google Scholar]
- 40.Grosjean H, Droogmans L, Roovers M, Keith G. Detection of enzymatic activity of transfer RNA modification enzymes using radiolabeled tRNA substrates. Methods Enzymol. 2007;425:55–101. doi: 10.1016/S0076-6879(07)25003-7. [DOI] [PubMed] [Google Scholar]
- 41.Hurd C, Schmerling L. Alkenyl derivatives of fluorescein. J. Am. Chem. Soc. 1937;59:112–117. [Google Scholar]
- 42.Motorin Y, Grosjean H. Multisite-specific tRNA:m5C-methyltransferase (Trm4) in yeast Saccharomyces cerevisiae: identification of the gene and substrate specificity of the enzyme. RNA. 1999;5:1105–1118. doi: 10.1017/s1355838299982201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pan H, Agarwalla S, Moustakas DT, Finer-Moore J, Stroud RM. Structure of tRNA pseudouridine synthase TruB and its RNA complex: RNA recognition through a combination of rigid docking and induced fit. Proc. Natl Acad. Sci. USA. 2003;100:12648–12653. doi: 10.1073/pnas.2135585100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Constantinesco F, Benachenhou N, Motorin Y, Grosjean H. The tRNA(guanine-26,N2-N2) methyltransferase (Trm1) from the hyperthermophilic archaeon Pyrococcus furiosus: cloning, sequencing of the gene and its expression in Escherichia coli. Nucleic Acids Res. 1998;26:3753–3761. doi: 10.1093/nar/26.16.3753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Roovers M, Wouters J, Bujnicki JM, Tricot C, Stalon V, Grosjean H, Droogmans L. A primordial RNA modification enzyme: the case of tRNA (m1A) methyltransferase. Nucleic Acids Res. 2004;32:465–476. doi: 10.1093/nar/gkh191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Charpentier B, Muller S, Branlant C. Reconstitution of archaeal H/ACA small ribonucleoprotein complexes active in pseudouridylation. Nucleic Acids Res. 2005;33:3133–3144. doi: 10.1093/nar/gki630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zhou Z, Reed R. Purification of Functional RNA-Protein Complexes using MS2-MBP. Curr. Protoc. Mol. Biol. 2003;Unit 27.3.127.7 doi: 10.1002/0471142727.mb2703s63. [DOI] [PubMed] [Google Scholar]
- 48.Portman DS, Dreyfuss G. RNA annealing activities in HeLa nuclei. EMBO J. 1994;13:213–221. doi: 10.1002/j.1460-2075.1994.tb06251.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Marchand V, Mereau A, Jacquenet S, Thomas D, Mougin A, Gattoni R, Stevenin J, Branlant C. A Janus splicing regulatory element modulates HIV-1 tat and rev mRNA production by coordination of hnRNP A1 cooperative binding. J. Mol. Biol. 2002;323:629–652. doi: 10.1016/s0022-2836(02)00967-1. [DOI] [PubMed] [Google Scholar]
- 50.Charpentier B, Fourmann JB, Branlant C. Reconstitution of archaeal H/ACA sRNPs and test of their activity. Methods Enzymol. 2007;425:389–405. doi: 10.1016/S0076-6879(07)25016-5. [DOI] [PubMed] [Google Scholar]
- 51.Sivakumar K, Xie F, Cash BM, Long S, Barnhill HN, Wang Q. A fluorogenic 1,3-dipolar cycloaddition reaction of 3-azidocoumarins and acetylenes. Org. Lett. 2004;6:4603–4606. doi: 10.1021/ol047955x. [DOI] [PubMed] [Google Scholar]
- 52.Yang C, Söll D. Covalent attachment of fluorescent groups to transfer ribonucleic acid. Reactions with 4-bromomethyl-7-methoxy-2-oxo-2H-benzopyran. Biochemistry. 1974;13:3615–3621. doi: 10.1021/bi00714a033. [DOI] [PubMed] [Google Scholar]
- 53.Shi H, Moore PB. The crystal structure of yeast phenylalanine tRNA at 1.93 A resolution: a classic structure revisited. RNA. 2000;6:1091–1105. doi: 10.1017/s1355838200000364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Witherell GW, Gott JM, Uhlenbeck OC. Specific interaction between RNA phage coat proteins and RNA. Prog. Nucleic Acid Res. Mol. Biol. 1991;40:185–220. doi: 10.1016/s0079-6603(08)60842-9. [DOI] [PubMed] [Google Scholar]
- 55.Tazi J, Bakkour N, Marchand V, Ayadi L, Aboufirassi A, Branlant C. Alternative splicing: regulation of HIV-1 multiplication as a target for therapeutic action. FEBS J. 2010;277:867–876. doi: 10.1111/j.1742-4658.2009.07522.x. [DOI] [PubMed] [Google Scholar]
- 56.Peattie DA. Direct chemical method for sequencing RNA. Proc. Natl Acad. Sci. USA. 1979;76:1760–1764. doi: 10.1073/pnas.76.4.1760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Peattie DA, Gilbert W. Chemical probes for higher-order structure in RNA. Proc. Natl Acad. Sci. USA. 1980;77:4679–4682. doi: 10.1073/pnas.77.8.4679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Giegé R, Helm M, Florentz C. Classical and Novel Chemical Tools for RNA Structure Probing. In: Söll D, Nishimura S, Moore P, editors. RNA. Oxford: Pergamon Press; 2001. pp. 71–89. [Google Scholar]
- 59.Merino EJ, Wilkinson KA, Coughlan JL, Weeks KM. RNA structure analysis at single nucleotide resolution by selective 2′-hydroxyl acylation and primer extension (SHAPE) J. Am. Chem. Soc. 2005;127:4223–4231. doi: 10.1021/ja043822v. [DOI] [PubMed] [Google Scholar]
- 60.Mortimer SA, Johnson JS, Weeks KM. Quantitative analysis of RNA solvent accessibility by N-silylation of guanosine. Biochemistry. 2009;48:2109–2114. doi: 10.1021/bi801939g. [DOI] [PubMed] [Google Scholar]
- 61.Kimura K, Nakanishi M, Yamamoto T, Tsuboi M. A correlation between the secondary structure of DNA and the reactivity of adenine residues with chloroacetaldehyde. J. Biochem. 1977;81:1699–1703. doi: 10.1093/oxfordjournals.jbchem.a131629. [DOI] [PubMed] [Google Scholar]
- 62.Burgstaller P, Famulok M. Flavin-Dependent Photocleavage of RNA at G.U Base Pairs. J. Am. Chem. Soc. 1997;119:1137–1138. [Google Scholar]
- 63.Burgstaller P, Hermann T, Huber C, Westhof E, Famulok M. Isoalloxazine derivatives promote photocleavage of natural RNAs at G.U base pairs embedded within helices. Nucleic Acids Res. 1997;25:4018–4027. doi: 10.1093/nar/25.20.4018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Demeshkina N, Repkova M, Ven'yaminova A, Graifer D, Karpova G. Nucleotides of 18S rRNA surrounding mRNA codons at the human ribosomal A, P, and E sites: a crosslinking study with mRNA analogs carrying an aryl azide group at either the uracil or the guanine residue. RNA. 2000;6:1727–1736. doi: 10.1017/s1355838200000996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Bi X, Schmitz A, Hayallah AM, Song JN, Famulok M. Affinity-based labeling of cytohesins with a bifunctional SecinH3 photoaffinity probe. Angew. Chem. Int. Ed. 2008;47:9565–9568. doi: 10.1002/anie.200803962. [DOI] [PubMed] [Google Scholar]
- 66.Nomura DK, Dix MM, Cravatt BF. Activity-based protein profiling for biochemical pathway discovery in cancer. Nat. Rev. Cancer. 2010;10:630–638. doi: 10.1038/nrc2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Puri AW, Bogyo M. Using small molecules to dissect mechanisms of microbial pathogenesis. ACS Chem. Biol. 2009;4:603–616. doi: 10.1021/cb9001409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Amblard F, Cho JH, Schinazi RF. Cu(I)-catalyzed Huisgen azide-alkyne 1,3-dipolar cycloaddition reaction in nucleoside, nucleotide, and oligonucleotide chemistry. Chem. Rev. 2009;109:4207–4220. doi: 10.1021/cr9001462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gramlich PM, Wirges CT, Gierlich J, Carell T. Synthesis of modified DNA by PCR with alkyne-bearing purines followed by a click reaction. Org. Lett. 2008;10:249–251. doi: 10.1021/ol7026015. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.