Abstract
GATA-1 and NF-E2 are erythroid specific activators that bind to the β-globin locus. To explore the roles of these activators in transcription of the human fetal stage specific γ-globin genes, we reduced GATA-1 and p45/NF-E2 using shRNA in erythroid K562 cells. GATA-1 or p45/NF-E2 knockdown inhibited the transcription of the γ-globin genes, hypersensitive site (HS) formation in the LCR and chromatin loop formation of the β-globin locus, but histone acetylation across the locus was decreased only in the case of GATA-1 knockdown. In p45/NF-E2 knockdown cells, GATA-1 binding was maintained at the LCR HSs and γ-globin promoter, but NF-E2 binding at the LCR HSs was reduced by GATA-1 knockdown regardless of the amount of p45/NF-E2 in K562 cells. These results indicate that histone acetylation is dependent on GATA-1 binding, but the binding of GATA-1 is not sufficient for the γ-globin transcription, HS formation and chromatin loop formation and NF-E2 is required. This idea is supported by the distinctive binding pattern of CBP and Brg1 in the β-globin locus. Furthermore GATA-1-dependent loop formation between HS5 and 3′HS1 suggests correlation between histone modifications and chromatin looping.
INTRODUCTION
The human β-globin locus contains the five globin genes, which are transcribed in erythroid cells depending on developmental stage (Figure 1A). The Gγ- and Aγ-globin genes are highly transcribed at the fetal stage, while the transcription of δ- and β-globin genes is great at the adult stage. The mechanisms by which the human adult β-globin gene is transcribed have been studied using the deletion assay of regulatory sequences (1–3) and the knockout or over-expression technique of erythroid specific transcriptional activators (4,5) in transgenic mice and stable cell lines. These studies showed that the locus control region (LCR) and erythroid specific activators binding to the LCR play critical roles in the transcription of the adult β-globin gene by contributing to histone modifications, nucleosome remodeling and chromatin loop formation. However, the transcriptional mechanism of the fetal γ-globin genes is less understood, despite great interest in reactivation of these genes as a means to treat sickle cell anemia (6).
Figure 1.
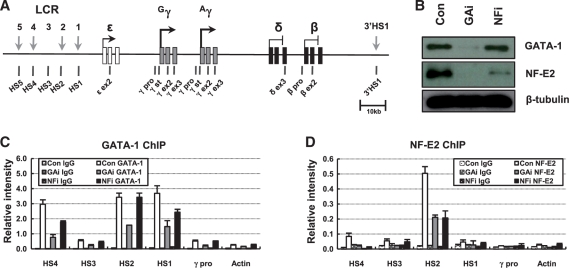
Knockdown of GATA-1 and p45/NF-E2 in erythroid K562 cells. (A) The human β-globin locus is presented. Vertical black arrows indicate DNase I HSs in LCR. The exons of the globin genes are represented by squares. Vertical bars named below the diagram denote the locations of TaqMan amplicons used in real-time PCR. (B) Western blotting was performed using antibodies specific to GATA-1 or p45/NF-E2 in protein extract from K562 cells expressing a control, GATA-1 or p45/NF-E2 shRNA. Blotting with β-tubulin antibody was used an experimental control. ChIP was performed with GATA-1 (C) or p45/NF-E2 (D) antibodies in K562 cells expressing each shRNA. Relative intensity was determined by quantitatively comparing input with immunoprecipitated DNA for the indicated amplicons. Actin served as negative control. Normal rabbit and goat IgG (Con IgG) served as experimental control. The results of two to four independent experiments ± SEM are graphed.
GATA-1 and NF-E2 are erythroid specific transcriptional activators, which bind to LCR DNase I hypersensitive sites (HSs) and gene promoters in the β-globin locus (7,8). The lack of GATA-1 causes failures in generating mature red blood cells in chimaeric mice (9) and in transcribing the mouse βH1 globin gene at normal level in embryo yolk sac cells (10). The stable expression of GATA-1 in non-erythroid HeLa cells induces the transcription of δ- and β-globin genes together with LCR HSs formation in the human β-globin locus (5). In murine erythroleukemia (MEL) cells, the transcription of the β major globin gene is dependent on GATA-1 and NF-E2, which are required for the recruitment of RNA polymerase II (pol II) to the globin promoter (7,11–13). The binding of both activators in the LCR HS2 enhancer is required for the transcription of a linked human ε-globin gene in minichromosomes, but the roles of the activators are distinctive in histone acetylation and DNase I HS formation (14,15).
The human β-globin locus at the fetal stage has a distinct chromatin structure which is different from the structure at the adult stage. The transcriptionally active γ-globin genes are hyperacetylated, but the inactive ε-, δ- and β-globin genes are hypoacetylated in human fetal liver and in mouse embryonic erythroblasts (16,17). In erythroid K562 cells where the γ-globin genes are highly transcribed and the general profile of histone acetylation for the β-globin locus is similar to that in human fetal liver, DNase I HSs are formed in the LCR and the downstream region of the locus (17,18). Erythroid specific activators and histone modifying enzymes associate with the HSs in a HS specific pattern (8). The γ-globin genes are present in close proximity with the LCR and 3′HS1 in transgenic mouse primitive erythrocytes and in K562 cells, while the β-globin gene is looped out from the active chromatin hub containing the LCR, 3′HS1 and the γ-globin genes (19,20).
In previous studies using mutation of GATA-1 and NF-E2 binding sites in the HS2 enhancer, we observed GATA-1-dependent histone modifications and pol II recruitment and NF-E2-dependent HSs formation in minichromosomal loci transcribing a linked embryonic ε-globin gene (14,15). To expand the studies to the expression of fetal γ-globin gene in the endogenous human β-globin locus, here we reduced the expression of GATA-1 and p45/NF-E2 using shRNA in human erythroid K562 cells. The role of erythroid specific activators in transcription of the endogenous human γ-globin gene is not well understood. This study shows that histone modifications, HSs formation, co-activator recruitment and chromatin loop formation are selectively affected by the activator knockdown in the endogenous human β-globin locus transcribing the fetal γ-globin genes. It indicates that GATA-1 and NF-E2 play distinctive roles in the changes of chromatin structure for the γ-globin gene transcription. Furthermore it suggests correlations between gene transcription, HS formation, histone modification and chromatin looping.
MATERIALS AND METHODS
GATA-1 and p45/NF-E2 knockdown using lentiviral short hairpin RNA in K562 cells
GATA-1 and p45/NF-E2 directed TRC lentiviral short hairpin RNA (shRNA) vectors and control shRNA vector (pKLO.1) were purchased from Open Biosystems. Lentiviruses were generated by transfecting shRNA and Virapower packing mix (Invitrogen) into 293FT cells according to the manufacturer’s instructions. After 24 h, complete media were added and viral supernatants were harvested by centrifugation and filtration at 72 h after transfection. Transduction of viral particles into K562 cells was performed in the presence of 6 µg/ml of polybrene and complete media was replaced after 24 h. Two microgram per milliliter of puromycin was added after 72 h. Cells were grown for 2 weeks in RPMI 1640 medium containing 10% FBS. Knockdown of GATA-1 and p45/NF-E2 protein was confirmed by western blot analysis.
Co-transduction of p45/NF-E2 expression vector and GATA-1 shRNA using lentivirus in K562 cells
p45/NF-E2 cDNA was cloned into the pLenti6.3/V5-DEST expression vector using the Gateway technology (Invitrogen). p45/NF-E2 expression vector was transfected into 293FT cells with Virapower packing mix (Invitrogen) to produce lentivirus. Lentivirus was harvested as described above and co-transducted with lentivirus having GATA-1 shRNA into K562 cell in the presence of 6 µg/ml of polybrene. Complete media were replaced after 6 h and puromycin (2 µg/ml) and blasticidine (5 µg/ml) were added after 72 h. Cells were grown for 2 weeks in RPMI 1640 medium containing 10% FBS. Knockdown of GATA-1 and expression of p45/NF-E2 were confirmed by western blot analysis. pKLO.1 and pLenti6.3/V5-GW/lacZ were used as the control vectors to make control cells.
Reverse transcription-PCR
RNA was prepared from 2 × 106 cells using the RNeasy Plus Mini Kit (Qiagen). One microgram of RNA was reverse transcribed with random hexamers using the Superscript III first-strand synthesis system as suggested by the manufacturer (Invitrogen). cDNA was diluted to 400 μl, and 2 μl of cDNA was amplified in a 10 μl reaction volume by real-time PCR using TaqMan chemistry. The relative intensity of specific cDNA sequences was compared with a genomic DNA standard using the comparative Ct method and then normalized with the relative intensity for Actin.
Real-time PCR
DNA obtained from all experiments was quantitatively analyzed using 7300 Real-time PCR system (Applied Biosystems). PCR was carried out with 200 nmol of TaqMan probes and 900 nmol of primers in a 10 μl reaction volume for cDNA and DNA obtained from ChIP and DNase I digestion. DNA obtained from 3C and control templates ligated randomly were amplified using SYBR green fluorescence in a 10 μl reaction volume. Data were collected at the threshold where amplification was linear. The locations of amplicon containing TaqMan probe in the β-globin locus are indicated in Figure 1A. Sequences of primers and TaqMan probes are shown in Supplementary Table S1 and sequences of primers for 3C are in Supplementary Table S2.
Chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) was carried out as described (14). Briefly, 2 × 107 K562 cells were incubated in cultured medium containing 1% formaldehyde for 10 min at 25°C, and the cross-linking reaction was quenched by adding glycine. Isolated nuclei were digested with 200 U of MNase at 37°C for 15 min and then sonicated to produce primarily mononucleosome size of chromatin. Fragmented chromatin was reacted with antibodies for 3 h at 4°C after pre-clearing with protein A or G agarose beads. Protein–DNA complexes were recovered with protein A or G agarose beads. Cross-links were reversed at 65°C in 0.25 M NaCl for overnight before purifying DNA.
Antibodies used in this study were purchased from Santa Cruz Biotechnology (Santa Cruz, CA, USA) for GATA-1 (sc-1233), p45/NF-E2 (sc-291), Pol II (sc-899), CBP (sc-369) and Brg1 (sc-10768), from Abcam (Cambridge, UK) for H3K27ac (ab4729) and from Upstate Biotechnology (Lake Placid, NY, USA) for H3ac (06-599), H3K4me2 (07-030), H3K4me3 (07-745), H3K27me2 (07-452), H3K27me3 (07-449) and CTCF (07-729). Normal rabbit IgG (sc-2027) and goat IgG (sc-2028) were purchased from Santa Cruz Biotechnology.
DNase I sensitivity assay
Nuclei were prepared from K562 cells (2 × 107) as described previously (21,22). Aliquots of 3 × 106 nuclei were digested with 150–600 U DNase I in a 100 μl volume for 10 min at 25°C. DNA was purified and run on a 1% agarose gel to visualize the level of digestion. The digested DNA was quantitatively compared with undigested DNA by real-time PCR. Sensitivity was determined by normalizing the difference of amount between digested DNA and undigested DNA in each primer pairs to the difference in Actin gene at each DNase I concentration.
Chromosome conformation capture
Chromosome conformation capture (3C) assays were performed according to the protocol provided with reduced number of cells (23,24). K562 cells were cross-linked with 1% formaldehyde at 25°C for 10 min and the reaction was quenched by adding glycine. Nuclei were prepared from ∼1 to 2 × 106 cells, resuspended in 0.4 ml of restriction enzyme buffer and incubated at 65°C for 1 h with 0.1% SDS and then at 37°C for 1 h with 1% triton X-100. Eight hundred unit of HindIII restriction enzyme was added into the nuclei and digestion was performed overnight at 37°C with shaking at 800 r.p.m. The DNA fragments were treated in 0.16% SDS at 65°C for 25 min and then in 0.1% Triton X-100 at 37°C for 30 min, and ligated with 400 U of T4 ligase for 4 h at 16°C with shaking at 500 r.p.m. in 2.4 ml reaction volume. The ligated DNA was purified by phenol extraction and ethanol precipitation after reverse cross-linking. The 3C products were amplified by real-time PCR using SYBR green as fluorescence dye. To correct the differences of ligation efficiency between fragments and the difference of PCR efficiency between primer sets, control templates were prepared by mixing, digesting and ligating the equimolar amounts of the PCR fragments spanning the restriction enzyme sites and same amount of genomic DNA (24). The ligation between two fragments was analyzed using the reverse direction primer of each fragment except one as shown Figure 5A. The relative cross-linking frequency was determined by comparing DNA ligated between two fragments in 3C samples with DNA ligated randomly in control templates and then by normalizing with the ligation frequency in the Ercc3 gene.
Figure 5.
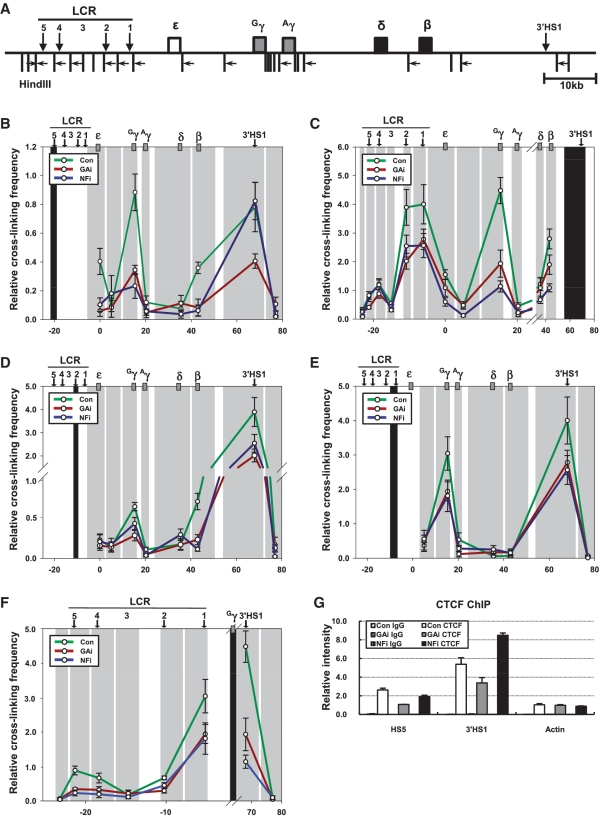
Relative proximity between HSs and the γ-globin gene in the β-globin locus in the GATA-1 or p45/NF-E2 knockdown cells. The 3C assay was performed with HindIII restriction enzyme. (A) HindIII sites and PCR primers in the β-globin locus were represented by vertical bars and horizontal arrows, respectively. The black shading represents the anchor fragment for HS5 (B). 3′HS1 (C). HS2 (D). HS1 (E) and Gγ-globin gene (F) in PCR. The gray shadings are fragments generated by HindIII digestion. Relative cross-linking frequency was determined by quantitatively comparing ligated DNA in cross-linked chromatin with control DNA and then normalizing to the cross-linking frequency at the ERCC gene. The results are averages of four to six independent experiments ± SEM. (G) ChIP was performed using CTCF antibody described in Figure 1.
RESULTS
NF-E2 binding at the β-globin LCR HS2 is affected by GATA-1 knockdown in erythroid K562 cells, but GATA-1 binding is not affected by p45/NF-E2 knockdown
Erythroid specific activator GATA-1 and NF-E2 bind to the β-globin LCR HSs. To explore the roles of these activators in the transcription of the human fetal stage specific γ-globin genes, we performed knockdown of GATA-1 and the p45 subunit of the NF-E2 (p45/NF-E2) heterodimer using shRNA in erythroid K562 cells. The severe reduction of GATA-1 or p45/NF-E2 was confirmed by western blot in the knockdown cells (Figure 1B). We observed that p45/NF-E2 was also strongly reduced in the GATA-1 knockdown cells, which might be due to failure to transcribe the p45 gene in the absence of GATA-1 (25).
To analyze the change of GATA-1 and NF-E2 binding in the human β-globin locus depending on the knockdown of the proteins, ChIP assay was carried out using antibodies specific to GATA-1 or p45/NF-E2. In GATA-1 knockdown K562 cells, its binding was reduced at LCR HSs and the γ-globin promoter as expected (Figure 1C). NF-E2 binding was also reduced at LCR HSs (Figure 1D) consistent with the reduction of p45/NF-E2 protein in these cells. In p45/NF-E2 knockdown cells, NF-E2 binding was reduced to less than half at LCR HS2 (Figure 1D), but GATA-1 binding was unaffected at the HS2 and the γ-globin promoter and was only modestly reduced at the HS4 and HS1 (Figure 1C), which is consistent with the unaffected expression of GATA-1 protein. In addition, sMaf proteins that can form an NF-E2 heterodimer with p45 or form a homodimer were less detected at the HS2 in both knockdown cells (Supplementary Figure S1). These complex results can be summarized that GATA-1 knockdown inhibits the expression of both GATA-1 and p45/NF-E2 and their binding in the β-globin locus, but p45/NF-E2 knockdown decreases only the expression and binding of p45/NF-E2 itself. Thus effects by GATA-1 knockdown may be both direct and indirect. NF-E2-independent GATA-1 binding to the β-globin locus in p45/NF-E2 knockdown cells provides a model situation to study whether GATA-1 binding alone is enough for the transcription of the human fetal γ-globin genes or whether NF-E2 is also required. The direct and indirect roles of GATA-1 and p45/NF-E2 in the various chromatin changes can be distinguished using the knockdown cells.
GATA-1 binding is not enough for the transcription of the human γ-globin genes and NF-E2 binding is required
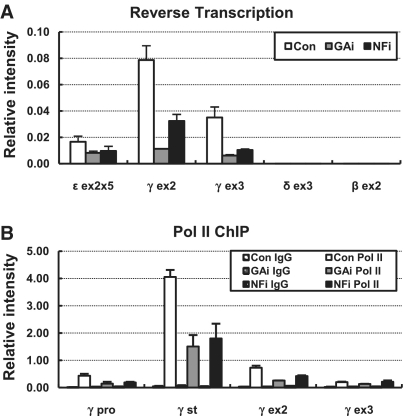
The fetal γ-globin genes are highly transcribed in human erythroid cell line K562 where the embryonic ε-globin gene are transcribed at low level but the transcripts of the adult δ- and β-globin genes are not detected. To analyze the effect of GATA-1 and NF-E2 knockdown on the transcription of the γ-globin genes, RNA transcripts were measured by real time reverse transcription (RT)–PCR in the Exons 2 and 3. In the GATA-1 knockdown K562 cells where p45/NF-E2 level is also reduced, the γ-globin transcripts were reduced to <20% compared to control cells (Figure 2A). The level of transcripts was also reduced by p45/NF-E2 knockdown, although less strongly compared to the reduction by GATA-1 knockdown. The transcription of the embryonic ε-globin gene was also decreased in the knockdown cells and the δ- and β-globin genes were still silent. The transcriptional reduction of the γ-globin genes was paralleled by reduced pol II detection across the gene (Figure 2B). Thus these results show that GATA-1 binding is not sufficient for robust transcription of the human fetal γ-globin genes and both erythroid specific transcriptional activators, GATA-1 and NF-E2 are required for optimal transcription.
Figure 2.
Transcription of the globin genes in GATA-1 or p45/NF-E2 knockdown K562 cells. (A) cDNA was prepared from RNA isolated from K562 expressing each shRNA and then amplified by real-time PCR using primers and probes for exons of the ε-, γ-, δ- and β-globin genes. Transcript levels of the globin genes were compared to transcript levels of the Actin control gene. The level of the ε-globin gene was multiplied by five to show changes in the knockdown cells. The results of two to four independent experiments ± SEM are graphed. (B) ChIP was performed with antibody specific to RNA polymerase II in K562 cells expressing each shRNA. Relative intensity was determined by quantitatively comparing immunoprecipitated DNA with input for the indicated γ-globin gene amplicons and then normalizing to the intensity at the Actin. Normal rabbit IgG (Con IgG) served as experimental control. The results of two to four independent experiments ± SEM are graphed.
GATA-1 plays a role in establishing histone acetylation, but NF-E2 may not be required
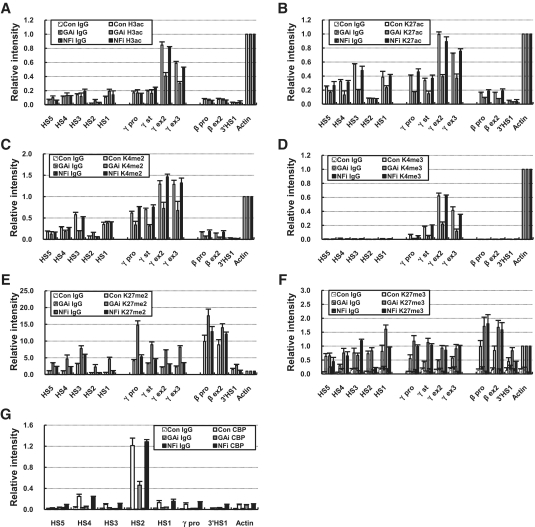
GATA-1 plays a role in histone acetylation of the mouse β-globin locus and minichromosomal β-globin locus by recruiting CBP (14,26). To ask the role of GATA-1 and NF-E2 in establishing histone modifications in the human β-globin locus transcribing the fetal γ-globin genes, various histone modifications were analyzed by ChIP assay in the knockdown K562 cells. Active histone marks including H3K9/K14ac, H3K27ac, H3K4me2 and H3K4me3, were markedly decreased at the LCR and γ-globin genes by GATA-1 knockdown also leading to p45/NF-E2 reduction, while inactive marks, H3K27me2 and H3K27me3, were increased in the regions (Figure 3A–F). However, these changes were not observed in p45/NF-E2 knockdown cells; most histone marks were maintained in the γ-globin genes and LCR HSs. Only H3K27me3 was similarly increased in the γ-globin genes by p45/NF-E2 knockdown with GATA-1 knockdown (Figure 3F). In the transcriptionally inactive β-globin gene, active marks, H3K27ac and H3K4me2, were further decreased but inactive marks, H3K27me2 and H3K27me3, were increased by GATA-1 knockdown. Histone acetyltransferase CBP was detected at LCR HSs and the γ-globin promoter as much in p45/NF-E2 knockdown cells as in control cells (Figure 3G), which parallels the histone acetylation pattern of the globin genes. Thus, CBP appears to be recruited to the human β-globin locus in a NF-E2-independent manner but in a GATA-1-dependent manner. This binding pattern of CBP supports the idea that GATA-1 is responsible for establishing histone acetylation in the β-globin locus transcribing the fetal γ-globin genes.
Figure 3.
Histone modification of the β-globin locus in the GATA-1 or p45/NF-E2 knockdown cells. ChIP was performed with antibodies specific to H3K9/K14ac (A), H3K27ac (B), H3K4me2 (C), H3K4me3 (D), H3K27me2 (E) and H3K27me3 (F) in K562 cells expressing each shRNA. Relative intensity was determined by normalizing to the intensity at the Actin as described in Figure 2. (G) The relative intensity of CBP ChIP was determined as described in Figure 1. Normal rabbit IgG (Con IgG) served as experimental control. The results of two to four independent experiments ± SEM are graphed.
NF-E2 is required for HSs formation in the β-globin LCR and GATA-1 binding alone is not sufficient
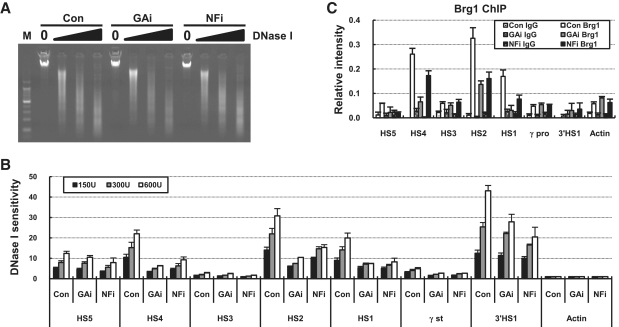
Four DNase I HSs, HS1, HS2, HS4 and HS5, are formed on the human β-globin LCR in K562 cells, and one more HS, 3′HS1 is found in the downstream region of the β-globin gene. We asked whether the formation of HSs is affected by GATA-1 or p45/NF-E2 knockdown. Sensitivity to DNase I attack was quantitatively determined by amplifying genomic DNA digested with DNase I using real-time PCR and then comparing with undigested genomic DNA (Figure 4A). DNase I sensitivity was strongly reduced at the HS4, HS2, HS1 and the γ-globin transcription start site in both GATA-1 and p45/NF-E2 knockdown cells (Figure 4B). The LCR HS5 and 3′HS1 showed relatively modest reduction of sensitivity. In agreement with the reduction of DNase I sensitivity, the association of Brg1, the catalytic subunit of a nucleosome remodeling complex, was reduced at HS2 and HS1 in both GATA-1 and p45/NF-E2 knockdown cells (Figure 4C). Thus these results indicate that GATA-1 binding alone is not sufficient for HSs formation in the fetal human β-globin locus and NF-E2 is required.
Figure 4.
DNase I sensitivity at the β-globin LCR HSs, γ-globin promoter and 3′HS1 in the GATA-1 or p45/NF-E2 knockdown cells. (A) Nuclei of K562 cells expressing each shRNA were digested with 150–600 U DNase I. DNA extracted from the digest was run on 1% agarose gel. (B) DNase I sensitivity was determined by quantitatively comparing digested DNA with undigested DNA and normalizing to the sensitivity at the Actin gene at each concentration. The results are averages of two to four nuclei preparations ± SEM. (C) ChIP was performed using Brg1 antibody described in Figure 1.
Chromatin looping between the Gγ-globin gene and LCR HSs requires NF-E2
Next to illuminate the chromatin looping state of the fetal stage β-globin locus and to analyze the role of these activators in the loop formation, the 3C assay was performed using HindIII restriction enzyme which cuts such that each HS and each globin gene are resolved on a separate fragment (Figure 5A). The fragments containing HS5, HS4, HS2, HS1 and 3′HS1 were frequently ligated with a fragment containing the Gγ-globin gene in the K562 control cells transcribing the γ-globin genes compared to non-erythroid 293T cells, indicating close proximity between these fragments (Supplementary Figure S2). In particular, the ligation of the Gγ-globin gene fragment with 3′HS1 fragment was remarkable. However a high level of ligation was not observed with a fragment containing the Aγ-globin gene. This result is not due to the transcriptional inactivation of the Aγ-globin gene, because the second intron regions of the Gγ and Aγ-globin gene, where nucleotide sequences are distinctive between the γ-globin genes, were amplified at a similar level by RT–PCR (Supplementary Figure S3). Interestingly, a fragment containing the transcriptionally inactive β-globin gene showed some ligation with most HSs fragments (Supplementary Figure S2A–C). A fragment containing HS3 was ligated at a very low level with the Gγ-globin gene or 3′HS1 fragments compared to other LCR HSs fragments (Supplementary Figure S2B and E). These results from 3C assay indicate that the transcriptionally active Gγ-globin gene is frequently in close proximity with LCR HSs and 3′HS1 in K562 cells. In addition, the β-globin gene which is not transcribed appears to achieve some proximity with LCR HSs albeit at a lower frequency.
Proximity between the globin genes and LCR HSs was generally reduced by GATA-1 and p45/NF-E2 knockdown in K562 cells. When a fragment containing the Gγ-globin gene was used as anchor in 3C assay, relative cross-linking frequency with HSs was reduced by the knockdowns (Figure 5F). The cross-linking frequency of the β-globin gene with HSs was also decreased (Figure 5B–D). In addition, cross-linking between 3′HS1 and LCR HS2 or HS1 was affected in the knockdown cells (Figure 5C–E). Interestingly, cross-linking between HS5 and 3′HS1 was reduced by GATA-1 knockdown, but not by p45/NF-E2 knockdown that does not affect GATA-1 expression and its binding at LCR HSs (Figure 5B). These results are accompanied by reduced CTCF binding in HS5 and 3′HS1 insulator sequences (Figure 5G). The requirement of CTCF binding for loop formation was already demonstrated in the β-globin locus (27,28). Thus these results indicate that GATA-1 appears to influence chromatin looping between HS5 and 3′HS1, even though the relationship between GATA-1 and CTCF is not clear. NF-E2 appears to be required for active chromatin hub formation by juxtaposing LCR HSs and the Gγ-globin gene with HS5 and 3′HS1 in the human β-globin locus.
p45/NF-E2 restoration did not restore its binding at the LCR HSs in the absence of GATA-1
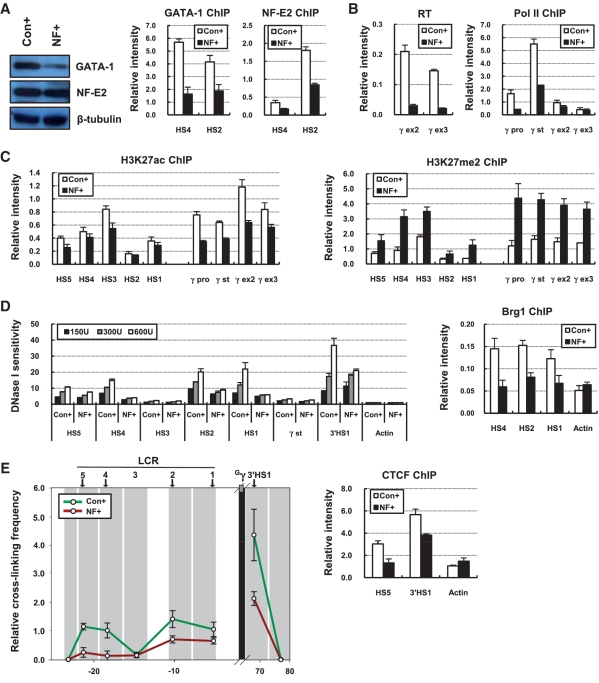
GATA-1 knockdown reduced the expression of p45/NF-E2 and its binding at the LCR HSs, generating a phenotype lacking both proteins (Figure 1). This condition did not allow us to explain the roles of GATA-1 independent from NF-E2. We generated a new condition where GATA-1 is reduced but p45/NF-E2 is present at normal level by ectopic expression using lentiviruses. The amount of p45/NF-E2 was restored in the GATA-1 knockdown cells as shown by western blot analysis (Figure 6A). However the binding of NF-E2 was not restored at the LCR HSs in the absence of GATA-1, which is a similar condition with the GATA-1 knockdown cells. Here transcription of the γ-globin genes was reduced and RNA polymerase was less detected in the γ-globin genes, as expected (Figure 6B). Active histone modifications including H3K27ac were decreased in the LCR HSs and the γ-globin genes and H3K27me2 was increased in these regions (Figure 6C) (data not shown). DNase I sensitivity was not restored in the LCR HSs by the restoration of p45/NF-E2 expression, which was in agreement with reduced Brg1 binding (Figure 6D). In addition, 3C and CTCF ChIP experiments showed decreased ligation frequency between the Gγ-globin gene and HSs fragments and the reduction of CTCF binding in HS5 and 3′HS1, respectively (Figure 6E). Therefore NF-E2 itself does not appear to be able to bind to the LCR HSs without GATA-1. The roles of GATA-1 in the γ-globin genes transcription may include the mediation of NF-E2 binding at LCR HSs.
Figure 6.
Chromatin structure in the GATA-1 knockdown cells with restoration of p45/NF-E2 protein. (A) Western blotting was performed using antibodies specific to GATA-1 or p45/NF-E2 in protein extract from K562 cells where co-transduction of control shRNA and control expression vector (Con+) or co-transduction of GATA-1 shRNA and p45/NF-E2 cDNA expression vector (NF+) was performed. ChIP was performed with GATA-1 or p45/NF-E2 antibodies in Con+ and NF+ cells and analyzed as described in Figure 1. (B) RT–PCR and ChIP using antibody specific RNA polymerase II were performed in Con+ and NF+ cells and analyzed as described in Figure 2. (C) ChIP was performed with antibodies specific to H3K27ac and H3K27me2 in Con+ and NF+ cells and analyzed as described in Figure 2. (D) DNase I sensitivity was measured in Con+ and NF+ cells as described in Figure 4 and ChIP was performed with Brg1 antibody and analyzed as described in Figure 1. (E) The 3C assay was performed with HindIII restriction enzyme in Con+ and NF+ cells and fragment containing the Gγ-globin gene was used as anchor. Relative cross-linking frequency was determined as described in Figure 5. ChIP was performed with CTCF antibody and analyzed as described in Figure 1.
DISCUSSION
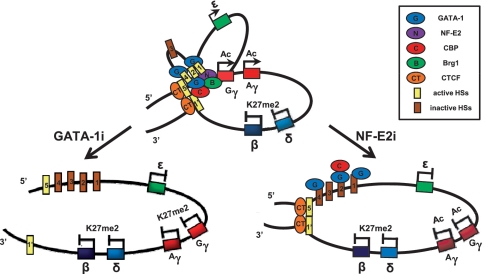
Our study shows that erythroid specific activators GATA-1 and NF-E2 are required for the expression of the human fetal γ-globin gene using human K562 erythroid cell line (Figure 7). GATA-1 plays a role in establishing a high level of histone acetylation by probably recruiting CBP, while NF-E2 is required for HS formation by probably contributing to Brg1 recruitment. Chromatin looping between LCR HS5 and 3′HS1 requires GATA-1, but the gathering of Gγ-globin gene and other LCR HSs to the HS5 and 3′HS1 is dependent on NF-E2. These results indicate that GATA-1 and NF-E2 have distinctive roles in the fetal γ-globin gene transcription.
Figure 7.
Summary of the changes by GATA-1 or p45/NF-E2 knockdown in the human fetal β-globin locus in K562 cells. The results presented in Figures 1–6 were combined and represented in model of the human β-globin locus.
Distinctive roles of GATA-1 and NF-E2 in histone modifications and HSs formation in the human fetal β-globin locus
GATA-1 and p45/NF-E2 knockdown in K562 cells generate complex situations in their phenotypes. By GATA-1 knockdown, the amount of p45/NF-E2 is also decreased because this gene is a target of GATA-1 (25,29) and NF-E2 binding is reduced at the β-globin LCR HSs, even though the NF-E2 binding is not restored with normal level of p45/NF-E2 by ectopic expression. However, GATA-1 is not affected by p45/NF-E2 knockdown. Thus, results by GATA-1 knockdown include direct and indirect effects, while results by p45/NF-E2 knockdown show its more direct roles. The changes caused by p45/NF-E2 knockdown are more directly dependent on NF-E2, because GATA-1 binding is maintained at LCR HS2 and the γ-globin promoter. The results from these situations complement our previous study which explains the role of GATA-1 independent from NF-E2 binding at the HS2 enhancer (14).
Our study indicates that GATA-1 has a role in histone acetylation by recruiting CBP into the LCR HSs and γ-globin promoter in the fetal stage β-globin locus. To play this role GATA-1 does not appear to require NF-E2. This result is comparable to reduced histone acetylation and CBP binding in minichromosomal loci in K562 cells where GATA-1 association at the HS2 is inhibited by its binding site mutation but NF-E2 associates at normal level (14). Physical interaction between GATA-1 and CBP supports this GATA-1-dependent histone acetylation (30). However the acetylation levels and association levels of GATA-1 and CBP across the LCR HSs are not perfectly matched, suggesting also indirect control by GATA-1 on histone acetylation. GATA-1-dependent histone acetylation and CBP recruitment were reported in mouse erythroleukemia MEL cells expressing the adult β major globin gene (26,31). In addition, other modifications such as H3K4me2, H3K4me3 and H3K27me2 are also dependent on GATA-1 rather than NF-E2, even though we do not have results showing the association of the enzyme responsible for each modification. NF-E2 does not appear to be essential for the various histone modifications on the fetal stage β-globin locus in K562 cells, although several studies on the mouse β-globin locus report histone acetylation and the recruitment of histone methyltransferase MLL and G9a in a NF-E2-dependent manner (11,13,32,33). The GATA-1-dependent histone modification patterns in the human β-globin locus may be related with the GATA-1-dependent loop formation between HS5 and 3′HS1 (see below).
NF-E2 appears to play a more direct role in HSs formation on the fetal stage β-globin locus, even thought GATA-1 is required for the NF-E2 binding (Figure 6A). GATA-1 binding itself is not enough for the HS formation. The binding pattern of Brg1 also supports the NF-E2 dependence of HSs formation in the LCR HSs. This is consistent with our previous findings obtained from the manipulable minichromosomal locus; failure of HS2 formation by the mutation of NF-E2 binding sites, but normal formation of HS2 after mutation of GATA-1 biding site (14,15). GATA-1-independent Brg1 association and hypersensitivity to restriction enzymes are observed at the mouse LCR HS4 and HS2 in MEL cells (31). Thus NF-E2 is believed to contribute more directly to HS formation in the human fetal β-globin locus, which might mediate the recruitment of Brg1 to LCR HSs, even though there is no evidence that NF-E2 interacts with Brg1 directly.
Distinctive roles of GATA-1 and NF-E2 in chromatin loop formation in the fetal β-globin locus
The transcriptionally active Gγ-globin is in close proximity with LCR and 3′HS1 insulators in K562 cells as revealed by our 3C analysis (Figure 5). However, HS3 among the LCR HSs is not substantially juxtaposed with the Gγ-globin gene or 3′HS1 as revealed by comparison with 3C results in non-erythroid 293T cells (Supplementary Figure S2). It suggests that HS3 is not necessary for the transcription of the γ-globin genes, even if K562 cells may not always reflect mechanism in primary erythroid cells. The deletion of HS3 core sequences does not affect the formation of other HSs in the LCR, the histone acetylation pattern of the β-globin locus or the expression of γ-globin gene in transgenic mice embryonic yolk sac (16,34). HS3 region is not a strong HS in K562 cells as shown Figure 4. In contrast, HS3 play a critical role in the transcription of the adult β-globin gene (35,36).
Chromatin loop formation of the human fetal β-globin locus is changed by GATA-1 and p45/NF-E2 knockdown (Figure 7). The close proximity between the β major globin gene and HS2 in the mouse locus requires GATA-1 and FOG-1 (37), and NLI/Ldb1, which forms complex with GATA-1, is also involved in the long-range loop formation (38). In the human fetal β-globin locus, GATA-1 is believed to plays a role in loop formation between two insulator regions, HS5 and 3′HS1. However GATA-1 binding at HS2 is not enough to form loop between the Gγ-globin gene and other LCR HSs and NF-E2 is required. NF-E2 binding appears to correlate with loop formation in the mouse β-globin locus, because its binding level increases to 1.5 times at HS2 after GATA-1 restoration in GATA-1 deficient mouse G1E cells (7) and to 2.5 times after induction in I/11 mouse erythroid cells (39). Even though p45/NF-E2 is dispensable for the interaction between the β major globin gene and LCR HSs in knockout mice, this can be a result of compensation by other proteins such as Nrf-2 (39).
Correlation between gene transcription, HSs formation, histone modification and chromatin looping
Gene transcription appears to relate with the looping. In the human and mouse β-globin loci, transcriptional inhibition by insertion of insulator sequences or absence of transcriptional activators/co-activators accompanies the loss of loop between LCR HSs (HS1–HS4) and the gene (27,38–41). Our study in here shows similar results. However it is not clear whether a high frequency of contact between the LCR and globin gene is necessary for the gene transcription, because the Aγ-globin gene is transcribed at a similar level to the Gγ-globin gene without frequent LCR interaction and one of the completely repressed genes in this locus, β-globin gene is in relatively frequent contact with most HSs (HS5, HS4, HS2, 3′HS1). A study on the mouse globin locus shows that proximity between HS2 and β major globin gene is coincidently increased with an increase of pol II in the gene promoter and intron, implying correlation of loop formation with the onset of the gene transcription (41). However, proximity between LCR HSs and the β-globin gene is almost maintained when the gene is not transcribed by the promoter deletion and when ongoing transcription is inhibited by DRB or α-amanitin, suggesting that loop formation is not an indicator of transcription and not a result of it (2,42).
Chromatin loop formation might relate with HS formation and histone modification. In our study, the decrease of hypersensitivity at LCR HSs and gene promoter accompanies reduced proximity between the LCR HSs and the γ-globin gene in the human β-globin locus. The HS formation might be required for the association of various proteins mediating chromatin loop formation. Histone modification also appears to relate with the chromatin loop. Locus wide histone modification patterns observed in this study except for H3K27me3 correlate with looping between two insulator regions, HS5 and 3′HS1, not with interaction between the LCR HSs and Gγ-globin gene. Loss of looping between HS5 and 3′HS1 accompany a decrease of active histone marks and an increase of inactive histone marks in the LCR and globin genes. Possibly, this is due to the loss of active chromatin status by the failure of insulation. On the other hand, H3K27me3 appears to be involved with the active chromatin hub formation of the Gγ-globin gene with LCR HSs and 3′HS1, because its level is increased when the active hub is disrupted by GATA-1 or p45/NF-E2 knockdown. The locus wide depletion of H3K27me3 and EZH2 leads the change of chromatin loop in the GATA-4 locus (43). However, it is still not clear whether histone modification affects chromatin loop formation or the reverse. The knockdown of histone modifying enzymes such as HDAC4 and LSD1 has been reported to disrupt chromatin looping between distal regulator regions and promoter/transcription start site (44,45). Thus it might be worthwhile to clarify the correlation between various histone modifications and chromatin loop formation.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Grant of the Korea Healthcare Technology R&D Project, Ministry for Health, Welfare & Family Affairs, Republic of Korea (A090172). Funding of open access charge: Pusan National University, Republic of Korea.
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors are grateful to Ann Dean for critical reading and comments. The authors thank Sang-Hyun Song for helpful suggestion for shRNA experiment and Kyung-Sun Kang for a kind gift of pLenti6.3/V5-DEST vector. Y.W.K., S.K., C.G.K. and A.K designed research. Y.W.K. and S.K. performed research. Y.W.K. and A.K. analyzed data and wrote the article.
REFERENCES
- 1.Fang X, Xiang P, Yin W, Stamatoyannopoulos G, Li Q. Cooperativeness of the higher chromatin structure of the beta-globin locus revealed by the deletion mutations of DNase I hypersensitive site 3 of the LCR. J. Mol. Biol. 2007;365:31–37. doi: 10.1016/j.jmb.2006.09.072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Patrinos GP, de Krom M, de Boer E, Langeveld A, Imam AM, Strouboulis J, de Laat W, Grosveld FG. Multiple interactions between regulatory regions are required to stabilize an active chromatin hub. Genes Dev. 2004;18:1495–1509. doi: 10.1101/gad.289704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schubeler D, Francastel C, Cimbora DM, Reik A, Martin DI, Groudine M. Nuclear localization and histone acetylation: a pathway for chromatin opening and transcriptional activation of the human beta-globin locus. Genes Dev. 2000;14:940–950. [PMC free article] [PubMed] [Google Scholar]
- 4.Bottardi S, Ross J, Bourgoin V, Fotouhi-Ardakani N, Affar el B, Trudel M, Milot E. Ikaros and GATA-1 combinatorial effect is required for silencing of human gamma-globin genes. Mol. Cell. Biol. 2009;29:1526–1537. doi: 10.1128/MCB.01523-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Layon ME, Ackley CJ, West RJ, Lowrey CH. Expression of GATA-1 in a non-hematopoietic cell line induces beta-globin locus control region chromatin structure remodeling and an erythroid pattern of gene expression. J. Mol. Biol. 2007;366:737–744. doi: 10.1016/j.jmb.2006.11.094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pace BS, Zein S. Understanding mechanisms of gamma-globin gene regulation to develop strategies for pharmacological fetal hemoglobin induction. Dev. Dyn. 2006;235:1727–1737. doi: 10.1002/dvdy.20802. [DOI] [PubMed] [Google Scholar]
- 7.Johnson KD, Grass JA, Boyer ME, Kiekhaefer CM, Blobel GA, Weiss MJ, Bresnick EH. Cooperative activities of hematopoietic regulators recruit RNA polymerase II to a tissue-specific chromatin domain. Proc. Natl Acad. Sci. USA. 2002;99:11760–11765. doi: 10.1073/pnas.192285999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim A, Song SH, Brand M, Dean A. Nucleosome and transcription activator antagonism at human beta-globin locus control region DNase I hypersensitive sites. Nucleic Acids Res. 2007;35:5831–5838. doi: 10.1093/nar/gkm620. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pevny L, Simon MC, Robertson E, Klein WH, Tsai SF, D’Agati V, Orkin SH, Costantini F. Erythroid differentiation in chimaeric mice blocked by a targeted mutation in the gene for transcription factor GATA-1. Nature. 1991;349:257–260. doi: 10.1038/349257a0. [DOI] [PubMed] [Google Scholar]
- 10.Fujiwara Y, Browne CP, Cunniff K, Goff SC, Orkin SH. Arrested development of embryonic red cell precursors in mouse embryos lacking transcription factor GATA-1. Proc. Natl Acad. Sci. USA. 1996;93:12355–12358. doi: 10.1073/pnas.93.22.12355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Johnson KD, Christensen HM, Zhao B, Bresnick EH. Distinct mechanisms control RNA polymerase II recruitment to a tissue-specific locus control region and a downstream promoter. Mol. Cell. 2001;8:465–471. doi: 10.1016/s1097-2765(01)00309-4. [DOI] [PubMed] [Google Scholar]
- 12.Kotkow KJ, Orkin SH. Dependence of globin gene expression in mouse erythroleukemia cells on the NF-E2 heterodimer. Mol. Cell. Biol. 1995;15:4640–4647. doi: 10.1128/mcb.15.8.4640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhou Z, Li X, Deng C, Ney PA, Huang S, Bungert J. USF and NF-E2 cooperate to regulate the recruitment and activity of RNA polymerase II in the {beta}-globin gene locus. J. Biol. Chem. 2010;285:15894–15905. doi: 10.1074/jbc.M109.098376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cho Y, Song SH, Lee JJ, Choi N, Kim CG, Dean A, Kim A. The role of transcriptional activator GATA-1 at human beta-globin HS2. Nucleic Acids Res. 2008;36:4521–4528. doi: 10.1093/nar/gkn368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kim A, Dean A. A human globin enhancer causes both discrete and widespread alterations in chromatin structure. Mol. Cell. Biol. 2003;23:8099–8109. doi: 10.1128/MCB.23.22.8099-8109.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fang X, Yin W, Xiang P, Han H, Stamatoyannopoulos G, Li Q. The higher structure of chromatin in the LCR of the beta-globin locus changes during development. J. Mol. Biol. 2009;394:197–208. doi: 10.1016/j.jmb.2009.09.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yin W, Barkess G, Fang X, Xiang P, Cao H, Stamatoyannopoulos G, Li Q. Histone acetylation at the human beta-globin locus changes with developmental age. Blood. 2007;110:4101–4107. doi: 10.1182/blood-2007-05-091256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kim A, Kiefer CM, Dean A. Distinctive signatures of histone methylation in transcribed coding and noncoding human beta-globin sequences. Mol. Cell. Biol. 2007;27:1271–1279. doi: 10.1128/MCB.01684-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Dostie J, Richmond TA, Arnaout RA, Selzer RR, Lee WL, Honan TA, Rubio ED, Krumm A, Lamb J, Nusbaum C, et al. Chromosome Conformation Capture Carbon Copy (5C): a massively parallel solution for mapping interactions between genomic elements. Genome Res. 2006;16:1299–1309. doi: 10.1101/gr.5571506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Palstra RJ, Tolhuis B, Splinter E, Nijmeijer R, Grosveld F, de Laat W. The beta-globin nuclear compartment in development and erythroid differentiation. Nat. Genet. 2003;35:190–194. doi: 10.1038/ng1244. [DOI] [PubMed] [Google Scholar]
- 21.Gong QH, McDowell JC, Dean A. Essential role of NF-E2 in remodeling of chromatin structure and transcriptional activation of the epsilon-globin gene in vivo by 5' hypersensitive site 2 of the beta-globin locus control region. Mol. Cell. Biol. 1996;16:6055–6064. doi: 10.1128/mcb.16.11.6055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.McArthur M, Gerum S, Stamatoyannopoulos G. Quantification of DNaseI-sensitivity by real-time PCR: quantitative analysis of DNaseI-hypersensitivity of the mouse beta-globin LCR. J. Mol. Biol. 2001;313:27–34. doi: 10.1006/jmbi.2001.4969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tolhuis B, Palstra RJ, Splinter E, Grosveld F, de Laat W. Looping and interaction between hypersensitive sites in the active b-globin locus. Mol. Cell. 2002;10:1453–1465. doi: 10.1016/s1097-2765(02)00781-5. [DOI] [PubMed] [Google Scholar]
- 24.Hagège H, Klous P, Braem C, Splinter E, Dekker J, Cathala G, de Laat W, Forné T. Quantitative analysis of chromosome conformation capture assays (3C-qPCR) Nat. Protoc. 2007;2:1722–1733. doi: 10.1038/nprot.2007.243. [DOI] [PubMed] [Google Scholar]
- 25.Toki T, Arai K, Terui K, Komatsu N, Yokoyama M, Katsuoka F, Yamamoto M, Ito E. Functional characterization of the two alternative promoters of human p45 NF-E2 gene. Exp. Hematol. 2000;28:1113–1119. doi: 10.1016/s0301-472x(00)00523-3. [DOI] [PubMed] [Google Scholar]
- 26.Letting DL, Rakowski C, Weiss MJ, Blobel GA. Formation of a tissue-specific histone acetylation pattern by the hematopoietic transcription factor GATA-1. Mol. Cell. Biol. 2003;23:1334–1340. doi: 10.1128/MCB.23.4.1334-1340.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hou C, Zhao H, Tanimoto K, Dean A. CTCF-dependent enhancer-blocking by alternative chromatin loop formation. Proc. Natl Acad. Sci. USA. 2008;105:20398–20403. doi: 10.1073/pnas.0808506106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Splinter E, Heath H, Kooren J, Palstra RJ, Klous P, Grosveld F, Galjart N, de Laat W. CTCF mediates long-range chromatin looping and local histone modification in the beta-globin locus. Genes Dev. 2006;20:2349–2354. doi: 10.1101/gad.399506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fujiwara T, O’Geen H, Keles S, Blahnik K, Linnemann AK, Kang YA, Choi K, Farnham PJ, Bresnick EH. Discovering hematopoietic mechanisms through genome-wide analysis of GATA factor chromatin occupancy. Mol. Cell. 2009;36:667–681. doi: 10.1016/j.molcel.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Blobel GA, Nakajima T, Eckner R, Montminy M, Orkin SH. CREB-binding protein cooperates with transcription factor GATA-1 and is required for erythroid differentiation. Proc. Natl Acad. Sci. USA. 1998;95:2061–2066. doi: 10.1073/pnas.95.5.2061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Im H, Grass JA, Johnson KD, Kim S, Boyer ME, Imbalzano AN, Bieker JJ, Bresnick EH. Chromatin domain activation via GATA-1 utilization of a small subset of dispersed GATA motifs within a broad chromosomal region. Proc. Natl Acad. Sci. USA. 2005;102:17065–17070. doi: 10.1073/pnas.0506164102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chaturvedi C, Hosey AM, Palii C, Perez-Iratxeta C, Nakatani Y, Ranish JA, Dilworth FJ, Brand M. Dual role for the methyltransferase G9a in the maintenance of beta-globin gene transcription in adult erythroid cells. Proc. Natl Acad. Sci. USA. 2009;106:18303–18308. doi: 10.1073/pnas.0906769106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Demers C, Chaturvedi CP, Ranish JA, Juban G, Lai P, Morle F, Aebersold R, Dilworth FJ, Groudine M, Brand M. Activator-mediated recruitment of the MLL2 methyltransferase complex to the beta-globin locus. Mol. Cell. 2007;27:573–584. doi: 10.1016/j.molcel.2007.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Navas PA, Peterson KR, Li Q, Skarpidi E, Rohde A, Shaw SE, Clegg CH, Asano H, Stamatoyannopoulos G. Developmental specificity of the interaction between the locus control region and embryonic or fetal globin genes in transgenic mice with an HS3 core deletion. Mol. Cell. Biol. 1998;18:4188–4196. doi: 10.1128/mcb.18.7.4188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fang X, Sun J, Xiang P, Yu M, Navas PA, Peterson KR, Stamatoyannopoulos G, Li Q. Synergistic and additive properties of the beta-globin locus control region (LCR) revealed by 5'HS3 deletion mutations: implication for LCR chromatin architecture. Mol. Cell. Biol. 2005;25:7033–7041. doi: 10.1128/MCB.25.16.7033-7041.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Jia CP, Huang SZ, Yan JB, Xiao YP, Ren ZR, Zeng YT. Effects of human locus control region elements HS2 and HS3 on human beta-globin gene expression in transgenic mouse. Blood Cells Mol. Dis. 2003;31:360–369. doi: 10.1016/j.bcmd.2003.07.001. [DOI] [PubMed] [Google Scholar]
- 37.Vakoc CR, Letting DL, Gheldof N, Sawado T, Bender MA, Groudine M, Weiss MJ, Dekker J, Blobel GA. Proximity among distant regulatory elements at the beta-globin locus requires GATA-1 and FOG-1. Mol. Cell. 2005;17:453–462. doi: 10.1016/j.molcel.2004.12.028. [DOI] [PubMed] [Google Scholar]
- 38.Song SH, Hou C, Dean A. A positive role for NLI/Ldb1 in long-range beta-globin locus control region function. Mol. Cell. 2007;28:810–822. doi: 10.1016/j.molcel.2007.09.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Kooren J, Palstra RJ, Klous P, Splinter E, von Lindern M, Grosveld F, de Laat W. Beta-globin active chromatin Hub formation in differentiating erythroid cells and in p45 NF-E2 knock-out mice. J. Biol. Chem. 2007;282:16544–16552. doi: 10.1074/jbc.M701159200. [DOI] [PubMed] [Google Scholar]
- 40.Kim SI, Bresnick EH, Bultman SJ. BRG1 directly regulates nucleosome structure and chromatin looping of the alpha globin locus to activate transcription. Nucleic Acids Res. 2009;37:6019–6027. doi: 10.1093/nar/gkp677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vakoc CR, Mandat SA, Olenchock BA, Blobel GA. Histone H3 lysine 9 methylation and HP1gamma are associated with transcription elongation through mammalian chromatin. Mol. Cell. 2005;19:381–391. doi: 10.1016/j.molcel.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 42.Palstra RJ, Simonis M, Klous P, Brasset E, Eijkelkamp B, de Laat W. Maintenance of long-range DNA interactions after inhibition of ongoing RNA polymerase II transcription. PLoS One. 2008;3:e1661. doi: 10.1371/journal.pone.0001661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tiwari VK, McGarvey KM, Licchesi JD, Ohm JE, Herman JG, Schübeler D, Baylin SB. PcG proteins, DNA methylation, and gene repression by chromatin looping. PLoS Biol. 2008;6:2911–2927. doi: 10.1371/journal.pbio.0060306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Perillo B, Ombra MN, Bertoni A, Cuozzo C, Sacchetti S, Sasso A, Chiariotti L, Malorni A, Abbondanza C, Avvedimento EV. DNA oxidation as triggered by H3K9me2 demethylation drives estrogen-induced gene expression. Science. 2008;319:202–206. doi: 10.1126/science.1147674. [DOI] [PubMed] [Google Scholar]
- 45.Saramäki A, Diermeier S, Kellner R, Laitinen H, Vaïsänen S, Carlberg C. Cyclical chromatin looping and transcription factor association on the regulatory regions of the p21 (CDKN1A) gene in response to 1alpha,25-dihydroxyvitamin D3. J. Biol. Chem. 2009;284:8073–8082. doi: 10.1074/jbc.M808090200. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.