Abstract
Insertion of a transgene into a defined genomic locus in human embryonic stem cells (hESCs) is crucial in preventing random integration-induced insertional mutagenesis, and can possibly enable persistent transgene expression during hESC expansion and in their differentiated progenies. Here, we employed homologous recombination in hESCs to introduce heterospecific loxP sites into the AAVS1 locus, a site with an open chromatin structure that allows averting transgene silencing phenomena. We then performed Cre recombinase mediated cassette exchange using baculoviral vectors to insert a transgene into the modified AAVS1 locus. Targeting efficiency in the master hESC line with the loxP-docking sites was up to 100%. Expression of the inserted transgene lasted for at least 20 passages during hESC expansion and was retained in differentiated cells derived from the genetically modified hESCs. Thus, this study demonstrates the feasibility of genetic manipulation at the AAVS1 locus with homologous recombination and using viral transduction in hESCs to facilitate recombinase-mediated cassette exchange. The method developed will be useful for repeated gene targeting at a defined locus of the hESC genome.
INTRODUCTION
Human embryonic stem cells (hESCs) and induced pluripotent stem (iPS) cells have the unique ability to generate virtually any differentiated cell type. In the context of regenerative medicine, progenies derived from these human pluripotent cells can be used as transplantable cells for cell-based therapy. Many of such medical applications would benefit from continuous expression of therapeutic transgenes after the genes are integrated into the genome of the transplanted cells.
Random integration is currently commonly used to introduce a transgene into hESCs and their progenies for its stable expression. However, random integration can result in insertional mutagenesis and subsequently tumorigenesis of transplanted cells (1,2). Furthermore, expression of a randomly integrated transgene is unpredictable. Integration within certain DNA sequence contexts can trigger epigenetic silencing causing variegated expression in clonally isolated cells or cessation of transgene expression to occur either over time or upon differentiation (3–6). Hence, there is a compelling need to develop techniques to integrate transgenes into a defined genomic locus that allows stable expression of the genes during expansion and differentiation of hESCs without compromising genomic stability.
Among different genomic sites tested, the adeno-associated virus integration site 1 locus (AAVS1) is one of the few loci that have been successfully targeted. AAVS1 is located within the protein phosphatase 1, regulatory (inhibitor) subunit 12C (PPP1R12C) gene on human chromosome 19 (19q13.3-qter). This site serves as a specific integration locus for AAV serotype 2 (AAV2), a non-pathogenic human parvovirus (7). AAVS1 has been characterized as a transcription-competent environment comprising of an open chromatin structure and contains native insulators that enable resistance of the integrated genes against transgene silencing (8–10). Since there is no known adverse effect on the cell resulting from disruption of the PPP1R12C gene and the transcriptional competence of a transgene cassette inserted into the site remains across diverse cell types, AAVS1 is considered as a ‘safe harbor’ for addition of a transgene into the human genome (11).
To direct gene integration into AAVS1, AAV2 technology using the AAV2 viral inverted terminal repeats (ITRs) and rep 78/68 genes for non-homologous recombination had previously been employed in human somatic cells (12–14). After incorporation of AAV2 ITRs and rep genes into baculovirus, we showed before that transduction of the resulting baculovirus-AAV hybrid system in hESCs led to stable transgene expression, but we were unable to confirm AAVS1 site-specific integration (15). A plasmid transfection-based AAV2 technology was later successfully used in hESCs for persistent transgene expression, although site-specific integration at AAVS1 occurred at a low frequency of 4.16% (6). Recently, zinc-finger nuclease (ZFN)-mediated homologous recombination, a technique using ZFN-induced DNA double-strand breaks to increase the rate of homologous recombination (16), has been employed to direct transgenes into AAVS1, providing targeting efficiencies ranging from 33% to 61% in hESCs and human iPS cells (17). While attractive for gene function studies, the use of ZFNs is still associated with cleavage at off-target sites resulting in genotoxicity and cytotoxicity. This potential issue may complicate the therapeutic use of ZFN technology (18).
Gene targeting by homologous recombination that uses an extrachromosomal fragment of donor DNA introduced into the cell, without pre-induction of DNA double-strand breaks, has been tested in many different cell types. However, homologous recombination frequency is low in mammalian cells usually occurring in one out of a million treated cells (16). For gene targeting in hESCs, this conventional homologous recombination-based technique has succeeded only in a limited number of genomic loci so far (19–25). There are no reports as yet on conventional homologous recombination-mediated gene targeting at AAVS1, either in differentiated cells or stem cells.
Over the years, the bacteriophage P1-derived Cre/loxP system has proved to be an effective tool for genome modification in human cells owing to the high activity of Cre recombinase in mammalian cells and its high degree of DNA sequence specificity (26). This system has been used for Cre recombinase-mediated cassette exchange (Cre-RMCE) in which Cre recombinases target the loxP sites and exchange a genomic fragment flanked by a pair of heterospecific loxP sequences for a floxed transgene (flanked by the same heterospecific loxP sequences). RMCE has been performed at random genomic sites in hESCs (27,28). When recombinase target sites are inserted into a pre-defined locus in the host cell genome by homologous recombination, RMCE can also be used to introduce a desired transgene into the selected locus. With this strategy, gene targeting has been performed at the ROSA26 and HPRT loci in hESCs (21,22,25). RMCE efficiency is dependent on the relative and absolute quantities of transgene DNA and recombinase present in a cell. As demonstrated in differentiated somatic cells, viral infection is significantly more efficient in mediating RMCE than plasmid transfection (26).
To enable highly efficient and safe gene targeting at AAVS1 in hESCs, we adopted a two-step process in this study. Homologous recombination was first performed to introduce heterospecific loxP sites into AAVS1, followed by Cre-RMCE to introduce a floxed transgene in a site-specific manner. In view of our previous finding that baculoviral vectors are capable of transducing 80% of hESCs with no observable cytotoxicity (15), we tested whether baculoviral transduction-mediated Cre-RMCE (BV-RMCE) could be used to improve integration efficiency.
MATERIALS AND METHODS
Construction of plasmid and baculoviral vectors
To construct pBS-PGK-neo-lox, a AAVS1 targeting vector used for homologous recombination, we first induced a two-base mutation within one of the loxP sites present on PGKneotpAlox2 (Addgene, Cambridge, MA, USA) that converts a loxP site into a lox2722 site containing a new spacer sequence from the original ATGTATGC to AAGTATCC. A 4 kb left homology arm and 3 kb right homology arm pertaining to the PPP1R12C gene were amplified from genomic DNA isolated from H1 hESCs and cloned into pCR BluntII-TOPO (Invitrogen, Carlsbad, CA, USA) independently. The two arms were then excised using ClaI/XhoI and SacI/SacII, respectively, and subcloned into the two ends of PGKneotpAlox2 comprising of the mutated lox2722 site, the phosphoglycerate kinase promoter (PGK), the neomycin resistance gene (neo) and the wild-type loxP site. Primers used in this study are listed in Supplementary Table S1.
To prepare baculoviral vectors for BV-RMCE, 1 kb Cre ORF was amplified from pBS185 (Addgene) and pFB-EF1α-Cre was constructed by inserting the Cre ORF into a pFastBac1 vector previously developed in the lab (15) using HindIII to replace its original 0.7 kb EGFP ORF. To construct pFB-EF1α-EGFP-neo-lox, 2.1 kb fragment containing the elongation factor-1 alpha promoter (EF1-α), the EGFP gene and a SV40 poly(A) tail was amplified from pFB-EF1α-EGFP and cloned into the mutated PGKneotpAlox2 construct using EcoRI. A 4.9 kb fragment comprising of the mutated lox2722 site, the EF1α promoter, the EGFP gene, the neomycin resistance gene and the wild-type loxP site was then excised from this construct and cloned in to pFastBac1 using NotI/SalI. To construct pFB-EF1α-EGFP-hyg-lox, 1.6 kb fragment comprising of the SV40 promoter, the hygromycin resistance gene (hyg) and a SV40 poly(A) tail was amplified from pDsRed-Monomer-Mem Hyg (Clontech, Mountain View, CA, USA) and used to replace the fragment containing the PGK promoter and the neomycin resistance gene in pFB-EF1α-EGFP-neo-lox using BlpI/PflMI. To construct pFB-EF1α-HSVtk-hyg-lox, 1.4 kb fragment containing the HSVtk suicide gene and a SV40 poly(A) tail was amplified from a construct previously developed in the lab (29) and used to replace the fragment comprising of the EGFP gene and the SV40 poly(A) tail in pFB-EF1α-EGFP-hyg-lox using AfeI/SbfI. Recombinant baculoviruses were propagated using the Bac-to-Bac® Baculovirus Expression System (Invitrogen). Recombinant DNA research in this study followed the National Institutes of Health guidelines.
Genetic modification of hESCs
H1 hESCs (WiCell, Madison, WI, USA) were cultured in feeder-free conditions in compliance with the protocol stated in the technical manual: ‘Maintenance of hESCs in mTeSRTM1′ (StemCell Technologies, Vancouver, BC, Canada). For homologous recombination, 2 × 106 hESCs were transfected 4 days following subculture with 2 μg of BsaI linearized pBS-PGK-neo-lox at a 1:3 DNA to reagent ratio (FuGENE® HD Transfection Reagent, Roche, Mannheim, Germany). G418 selection (50 μg/ml, Gibco) in mTeSR1 medium started 48 h after transfection. Medium was changed daily for 3 weeks until resistant colonies propagated. For RMCE, 2 × 106 loxP-hESC2 cells were transduced 4 days following subculture with BV-EF1α-Cre at a MOI of 100. Twenty-four hours later the medium was changed and cells were again transduced at a MOI of 100 with either BV-EF1α-EGFP-hyg-lox or BV-EF1α-HSVtk-hyg-lox. Hygromycin B selection (25 μg/ml, A.G. Scientific, San Diego, CA, USA) in mTeSR1 medium started 48 h after the second viral transduction. Medium was changed daily for 4 weeks. Each colony, propagated through either G418 or hygromycin B selection, was transferred to a single well of a Matrigel-coated 6-well plate and allowed to undergo clonal expansion without drug selection. One week later, each clone was subcultured at 1:6 ratio. DNA of the clones was isolated (DNeasy® Blood and Tissue Kit, Qiagen, Hilden, Germany), and screened for AAVS1 locus modification through PCR and Southern blot analysis.
Stem cell differentiation and characterization of differentiated cells
The protocols for formation and maintenance of neurospheres and differentiation of adherent neural stem cells (NSCs) in to astrocytes and neurons have been previously reported (30,31). Mesenchymal stem cells (MSCs) and dendritic cells were generated as previously reported (32,33).
Primary monoclonal antibodies tested for flow cytometric analysis of EGFP-MSCs include phycoerythrin (PE)-conjugated anti-CD24, anti-CD34, anti-CD45, anti-CD44, anti-CD73, anti-CD90, anti-CD105 and anti-CD166, allophycocyanin (APC)-conjugated anti-HLA ABC, FITC-conjugated anti-HLA DRQP and their respective isotypes (BD Pharmingen, San Diego, CA, USA). After primary antibody incubation, cells were washed and further stained with a TRITC-conjugated secondary antibody (Sigma-Aldrich, St Louis, MO, USA). For EGFP-DC analysis, primary monoclonal antibodies tested were APC-conjugated anti-CD11c, anti-CD40, anti-CD45, anti-CD86, anti-CD209 and their respective isotypes (BD Pharmingen). Before analysis of EGFP-hESCs, dead cells were excluded through PI staining (Sigma-Aldrich). Cells were analyzed using a FACSCaliburTM flow cytometer (BD Biosciences).
For RT–PCR analysis, total RNA was extracted with TRIzol® (Invitrogen) and cDNA was synthesized using the SuperScript® III First-Strand Synthesis System (Invitrogen). Amplified products were analyzed on a 2% agarose gel.
For immunostaining of differentiated glial cells and neurons, primary monoclonal antibodies tested include anti-GFAP (Sigma-Aldrich) and anti-βIII tubulin (Promega). Detection of Oct-3/4, SOX2, Nanog and SSEA-4 in EGFP-hESC1 was done using the hESC Marker Antibody Panel Plus (R&D Systems, MN, USA). The secondary antibody used was a Texas Red-conjugated secondary antibody (Abcam, Cambridge, MA, USA).
In vitro tumor killing and migration assay
For in vitro tumor killing assays, neurospheres were dissociated and seeded at 1 × 103 cells/well on a Matrigel-coated 96-well plate. U87 cells were seeded into the same plate at 1 × 103 cells/well. NSC medium containing ganciclovir (Invivogen) at 20 µM was added the following day and changed every alternate day for 1 week. A cell viability assay was performed using the CellTiter 96® AQueous One Solution (Promega) and absorbance was read using a Benchmark PlusTM microplate spectrophotometer (Bio-Rad).
The migration ability of TK-NSC was determined using the BD FalconTM HTS FluoroBlokTM 96-Multiwell Insert System (8 μm pore size, BD Biosceinces). U87 cells were seeded in the bottom receiver plate at 6.4 × 104 cells/well in Opti-MEM® I (Invitrogen). NSCs, labeled with Calcein-AM (AnaSpec), were seeded in the inserts at 2.5 × 104 cells/insert. After incubation for 24 h, the fluorescence emitted from the top (corresponding to non-migrating cells) and bottom (corresponding to migrating cells) sides of the plate was measured using a GENios ProTM microplate reader (Tecan, Männedorf, Switzerland).
RESULTS
Generation of loxP-hESC lines
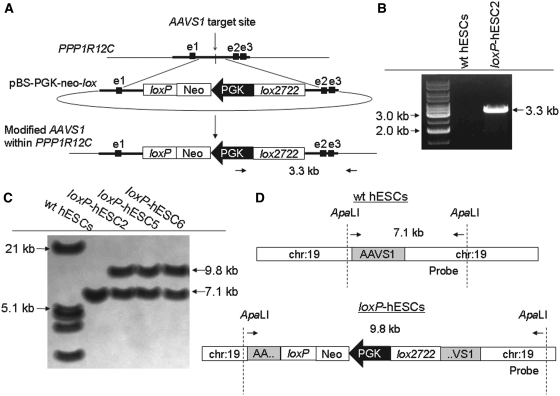
The use of the Cre/loxP system for the integration of transgenes within a specific locus initially requires the insertion of loxP-docking sites within that particular locus through homologous recombination. To target AAVS1 on chromosome 19 (19q13.42), we constructed a targeting vector pBS-PGK-neo-lox containing a floxed neomycin resistance marker flanked by a left and right arm corresponding to ∼7 kb of homology of the PPP1R12C gene. Exon 1 (337 bp) of the gene is present in the left arm while exons 2 (131 bp) and 3 (119 bp) are present on the right. AAVS1 begins 424 bp upstream of the 5′-end of exon 1 and ends 3.35 kb downstream of the 3′-end. The region that we targeted is ∼1.9 kb downstream of the 3′-end of exon 1 (19:60318479–19:60318512) and can also be considered as intron 1.
Following three independent transfection experiments with pBS-PGK-neo-lox in feeder-free culture condition, a total of 39 hESC colonies emerged after G418 selection (Table 1). The DNA of these clones was subjected to PCR genotyping by using a primer specific for the PGK promoter present on the pBS-PGK-neo-lox construct and a primer specific for chromosome 19 downstream of the 3′-end of the right homologous arm. The amplification of 3.3 kb fragment indicated a modified AAVS1 (Figure 1A and B). Through this analysis, 11 clones, out of 39, were identified as containing a modified locus. For further confirmation, Southern blot analysis was performed on three randomly selected loxP-hESC clones (Clones #2, #5 and #6) using a probe specific for chromosome 19. The detection of 9.8 kb fragment indicated a modified AAVS1, as opposed to the detection of 7.1 kb fragment that indicates the intact locus (wild-type). Fragments of both sizes were detected thereby identifying the clones to be heterozygous (Figure 1C and D). loxP-hESC Clone #2 (loxP-hESC2) was used as a master loxP-hESC line subjected to further downstream applications.
Table 1.
Efficiencies of gene targeting by homologous recombination and gene insertion by BV-RMCE at AAVS1 in hESCs
| Homologous recombination | Exp1 | Exp2 | Exp3 |
|---|---|---|---|
| No. of transfected cells | 2 000 000 | 2 000 000 | 2 000 000 |
| No. of G418 resistant colonies | 12 | 9 | 18 |
| No. of correctly targeted colonies verified by PCR analysis | 3 | 4 | 4 |
| No. of correctly targeted colonies verified by Southern blot analysis | 3 | nd | nd |
| Homologous recombination frequency as verified by PCR analysis | 1.5 × 10−6 | 2.0 × 10−6 | 2.0 × 10−6 |
| Targeting efficiency as verified by PCR analysis (%) | 25.00 | 44.44 | 22.22 |
| Mean targeting efficiency (%) ± SD | 30.56 ± 12.11 |
| BV-RMCE for EGFP-hESC Colonies | Exp1 | Exp2 | Exp3 |
|---|---|---|---|
| No. of hyg B resistant colonies | 9 | 18 | 6 |
| No. of correctly recombined colonies verified by PCR analysis | 8 | 16 | 6 |
| No. of correctly recombined colonies verified by Southern blot analysis | 8 | 16 | 6 |
| Site-specific integration efficiency (%) | 88.89 | 88.89 | 100 |
| BV-RMCE for TK-hESC Colonies | Exp4 | ||
|---|---|---|---|
| No. of hyg B resistant colonies | 4 | ||
| No. of correctly recombined colonies verified by PCR analysis | 4 | ||
| Site-specific integration efficiency (%) | 100 | ||
| Mean BV-RMCE efficiency (%) ± SD | 94.45 ± 6.41 |
Exp, experiment; PCR, polymerase chain reaction; nd, not determined.
Figure 1.
Homologous recombination to generate loxP-hESCs. (A) Schematic of AAVS1 within the PPP1R12C gene, the targeting vector, and the modified AAVS1 with heterospecific loxP sites following homologous recombination. e1, e2 and e3: the first three exons of the the PPP1R12C gene. (B) PCR analysis for detection of the modified AAVS1. loxP-hESC clones are identified through amplification of 3.3 kb fragment as indicated in (A). (C) Southern blot analysis to detect the modified AAVS1. Following digestion with ApaLI and hybridization, two fragments (7.1 kb and 9.8 kb as indicated in D) are detected in loxP-hESC clones (Clones #2, #5 and #6) opposed to the solitary fragment detected in wild-type (wt) hESCs. This result indicated that the loxP-hESC clones were heterozygous, comprising of one intact AAVS1 and one modified locus. (D) Schematic representation to show the position of the probe used for Southern blot analysis.
Cre recombinase-mediated cassette exchange using baculoviral vectors
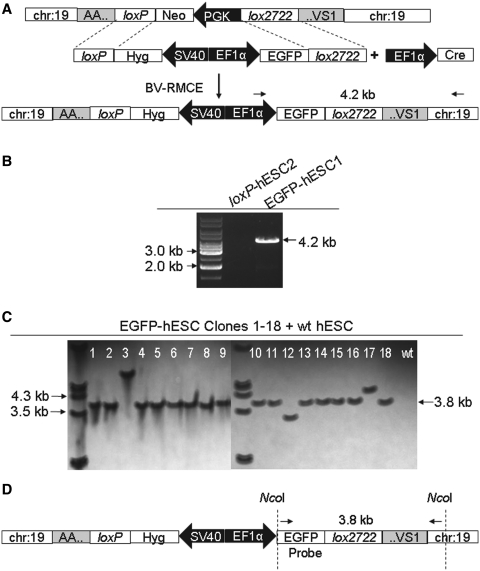
Once a loxP-hESC line was generated, it was subjected to Cre-RMCE. We generated two baculoviral vectors, BV-EF1α-Cre to express Cre recombinase and BV-EF1α-EGFP-hyg-lox as the transgene donor. The constitutive EF1α promoter was used to facilitate transgene expression in both un-differentiated and differentiated states. BV-RMCE was then performed in loxP-hESC2 using the two viral vectors. Two days after transduction, cells were subjected to hygromycin B selection for 4 weeks. Most of the cells died off in the first week. Small colonies remained in the second week, which expanded in the third and fourth week. Following three independent transduction experiments, a total of 33 EGFP-hESC colonies stably propagated in feeder-free culture condition through hygromycin B selection (Table 1). The DNA of these clones was subjected to PCR genotyping by using one primer specific for chromosome 19 and another specific for the EF1α promoter present in BV-EF1α-EGFP-hyg-lox. The amplification of 4.2 kb fragment indicated the integration of the EGFP gene within AAVS1 by BV-RMCE. Through this analysis, 30 EGFP-hESC clones were identified as having undergone site-specific integration at AAVS1 (Figure 2A and B; Supplementary Figure S1).
Figure 2.
Baculoviral transduction to introduce the EGFP gene into AAVS1 in loxP-hESCs. (A) Schematic representation of the generation of an EGFP-hESC line through BV-RMCE. loxP-hESC2 with a modified AAVS1 was transduced with two baculoviral vectors for site-specific transgene integration. (B) PCR analysis for detection of site-specific transgene integration. EGFP-hESC clones that underwent site-specific integration are identified through amplification of 4.2 kb fragment indicated in A. (C) Southern blot analysis to detect site-specific transgene integration and EGFP gene copy number. Following digestion with NcoI and hybridization, a solitary fragment (3.8 kb) is detected in most of EGFP-hESC clones with site-specific integration. A fragment of dissimilar size is detected in EGFP-hESC Clones #3, #12 and #17, which underwent random integration. (D) Schematic representation to show the position of the probe used for Southern blot analysis.
We also confirmed that the transduction of wild-type hESCs (wt hESCs) with BV-EF1α-Cre and BV-EF1α-EGFP-hyg-lox and also the transduction of loxP-hESC2 with BV-EF1α-EGFP-hyg-lox alone yielded no colonies through hygromycin B selection (unpublished observation). These results indicate that loxP sites within the genome and the expression of Cre recombinase are both necessary for BV-RMCE.
To dismiss the possible existence of multiple EGFP gene copies within the clones, Southern blot analysis was performed on all the EGFP-hESC clones using a probe specific for the EGFP gene. The detection of 3.8 kb fragment indicated the integration of the EGFP gene within AAVS1 and the inability to detect fragments of other sizes indicated the absence of random integration. This analysis confirmed that all 30 EGFP-hESC clones that were identified as having undergone site-specific integration through the PCR analysis contained only one EGFP gene copy (Figure 2C and D; Supplementary Figure S2). The EGFP-hESC clones that lack 4.2 kb fragment in PCR analysis (Clones #3, #12 and #17 in Supplementary Figure S1) also contained one EGFP gene copy as revealed by Southern blotting, but showed distinct band positions that clearly differed from that of 3.8 kb in length (Figure 2C), suggesting random chromosome integration of the cassette in the three clones. EGFP-hESC Clone #1 (EGFP-hESC1) with a single copy of the EGFP gene at AAVS1 was used for downstream characterizations.
AAVS1 locus-directed transgene integration results in stable expression in hESCs
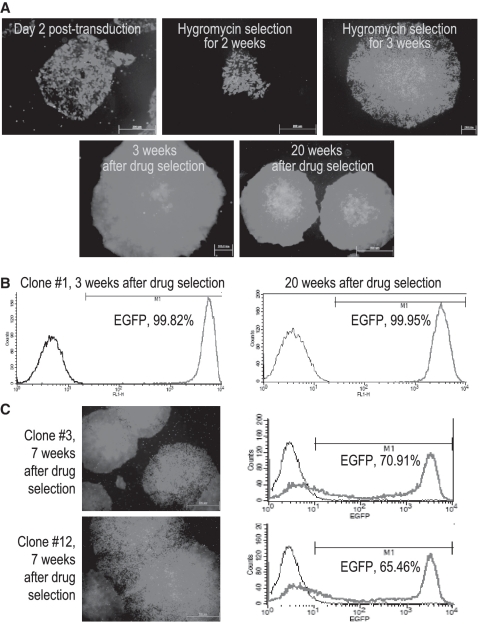
Six hours after baculoviral transduction, we started to observe EGFP-positive hESCs. But most of the EGFP-positive cells underwent cell death together with EGFP-negative cells in the first 2 weeks of drug selection. Drug-resistant EGFP-positive colonies emerged after 2 weeks of selection and EGFP-positive cells increased over time (Figure 3A). By 4 weeks, the hESC colonies appeared almost entirely green. Flow cytometric analysis revealed that 99.82% of EGFP-hESC1 collected 3 weeks after stopping drug selection was EGFP-positive (Figure 3A and B). The EGFP transgene in the selected hESCs maintained stable expression at the same intensity for at least 20 weeks over 20 passages (Figure 3B).
Figure 3.
Stable transgene expression after AAVS1 integration. (A) EGFP expression as demonstrated with fluorescence imaging in EGFP-hESC1, a hESC clone, in which AAVS1 integration of the EGFP transgene was confirmed. Cell samples photographed before, during or after drug selection are shown. (B) EGFP expression quantified using flow cytometric analysis of EGFP-hESC1. The cells were collected 3 or 20 weeks after stopping drug selection for analysis. (C) EGFP expression in EGFP-hESCs of Clones #3 and #12 with random integration of the EGFP transgene. The cells were collected and analyzed 7 weeks after stopping drug selection.
Two EGFP-hESC clones that underwent random integration (Clones #3 and #12) were also subjected to hygromycin B selection. Similar to hESC colonies with the EGFP gene integrated into AAVS1, pure EGFP-expressing hESC populations were obtained from the two clones after 4 weeks of drug selection. However, after stopping drug selection, we observed a decrease in EGFP expression as early as 7 weeks after drug selection. Flow cytometric analysis indicated that transgene silencing occurred in 30–35% of the selected hESCs within 7 weeks following the withdrawal of drug selection and a significant portion of the EGFP-expressing cells displayed low fluorescence intensity (Figure 3C). Our results are consistent with previous reports that random integration of transgenes is subject to silencing that leads to variegated transgene expression (3–5) and confirm that integration within AAVS1 does in fact result in persistent gene expression (6,11,17).
AAVS1 locus-directed transgene integration had no effects on hESC pluripotency
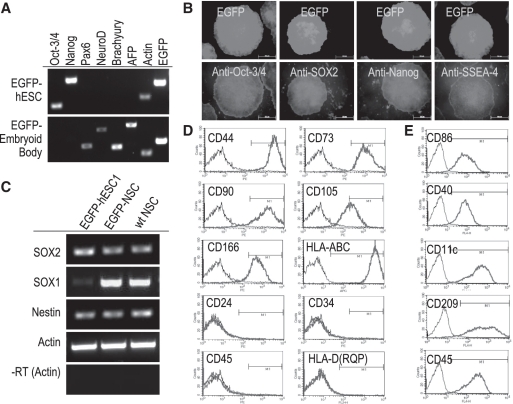
As revealed by RT–PCR analysis, EGFP-hESC1 continued to express pluripotent markers, but not ectoderm, mesoderm and endoderm markers (Figure 4A). Immunostaining also demonstrated the expression of several pluripotent marker proteins in EGFP-hESC1 (Figure 4B and Supplementary Figure S3), further confirming the undifferentiated state of these genetically modified hESCs.
Figure 4.
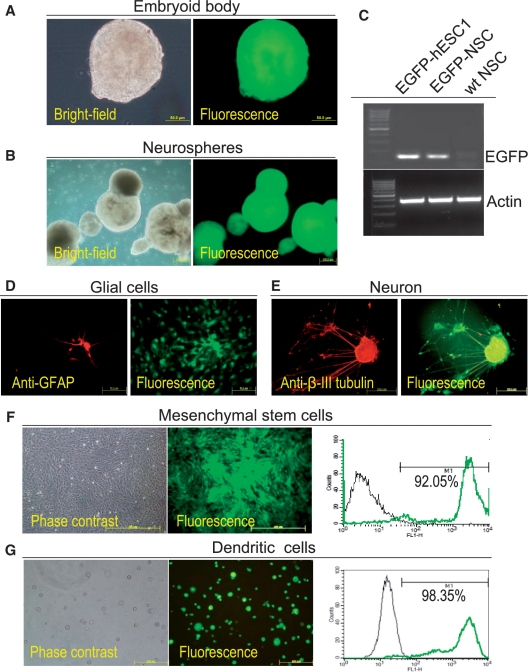
Maintenance of phenotype and pluripotency in genetically modified EGFP-hESCs. (A) RT–PCR analysis of pluripotent markers and stem cell lineage markers in EGFP-hESC1 and derived embryoid bodies. Note that EGFP-hESC1 expresses pluripotent markers Oct-3/4 and Nanog while EGFP-embryoid bodies express ectoderm markers Pax6 and NeuroD, mesoderm marker Brachyury and endoderm marker AFP. (B) Immunostaining to detect the expression of pluripotent markers Oct-3/4, Sox2, Nanog and SSEA-4 in EGFP-hESC1. (C) Molecular characterization of ectoderm-derived cells. Neurospheres derived from EGFP-hESC1 and wild-type hESCs were used for RT–PCR analysis. Neurospheres derived from both hESC lines are positive for SOX2 and nestin as well as display an upregulation of neural progenitor marker SOX1 when compared with EGFP-hESC1. (D) Flow cytometric characterization of mesoderm-derived cells. MSCs derived from EGFP-hESC1 are positive for all five MSC markers CD44, CD73, CD90, CD105 and CD166 as well as for HLA-ABC. They are negative for hematopoietic progenitor markers CD24, CD34 and CD45 as well as for HLA-D(RQP). (E) Flow cytometric characterization of dendritic cells derived from EGFP-hESC1. Cells are positive for DC markers CD11c, CD40, CD86 and CD209 as well as for hematopoietic lineage marker CD45.
To investigate whether EGFP-hESC1 retain their ‘stemness’ property, embryoid bodies were derived from these cells (Figure 5A) and the expression of germ layer markers was analyzed through RT–PCR. We observed the expression of ectoderm, mesoderm and endoderm markers, but not pluripotent markers in the embryoid bodies (Figure 4A). We further demonstrated that neurospheres could be derived from EGFP-hESC1 (Figure 5B) and the derived neurospheres displayed up-regulated expression of neuro-progenitor markers when compared with hESCs (Figure 4C). These neurospheres could further differentiate into β-III tubulin-positive neurons and GFAP-positive glial cells (Figure 5D and E). Cells generated from differentiation of EGFP-hESC1 toward mesenchymal lineage (Figure 5F) exhibited a surface marker profile characteristic of MSCs, including CD44, CD73, CD90, CD105 and CD166, but did not express markers characteristic of hematopoietic progenitor cells, including CD24, CD34 and CD45 (Figure 4D). Through Co-culturing of EGFP-hESC1 with OP9 stromal cells, we generate hematopoietic progenitor cells from EGFP-hESC1, which were further differentiated into dendritic cells (Figure 5G). Flow cytometric analysis indicated that these cells expressed CD86, CD40, CD11c and CD209, the markers characteristic of dendritic cells, and also CD45, a marker characteristic of hematopoietic lineage (Figure 4E). These results confirm that the differentiation potential of EGFP-hESC1 is not compromised by transgene integration into AAVS1.
Figure 5.
Maintained transgene expression in differentiated progenies of EGFP-hESCs after AAVS1 integration of a transgene. EGFP expression is retained in EGFP-hESC1-derived embryoid bodies (A), neurospheres (B and C), GFAP-positive glial cells (D), β-III tubulin-positive neurons (E), MSCs (F) and dendritic cells (G). Results from RT–PCR analysis of EGFP-hESC1, neurospheres derived from EGFP-hESC1 (EGFP-NSC) and NSCs derived from wild-type hESCs (wt NSC) are shown in (C). Results from quantitative flow cytometric analysis of EGFP-hESC1-derived MSCs and dendritic cells are shown on the right in (F) and (G), respectively.
Persistent transgene expression maintained after hESC differentiation
While examining whether EGFP-hESC1 can differentiate into multiple cell lineages, we also observed persistent expression of EGFP in differentiated progenies of the genetically modified hESCs. As shown in Figure 5, there was no decline in EGFP expression during the formation of embryoid bodies and their maintenance (Figure 5A). Neurospheres derived from EGFP-hESC1 and cultured for 4 weeks displayed no loss of EGFP fluorescence (Figure 5B). Through RT–PCR analysis, expression of EGFP mRNA was confirmed in neurospheres derived from EGFP-hESC1, but not from wt hESCs (Figure 5C). EGFP-hESC1-derived, terminally differentiated glial cells and neurons continued to express the transgene (Figure 5D and E). EGFP-hESC1-derived MSCs and dendritic cells were cultured for 2 weeks after differentiation and still displayed bright EGFP expression. Quantification by flow cytometric analysis indicated that 92–98% of these differentiated cells were EGFP positive (Figure 5F and G).
Glioma gene therapy potential of hESC-derived neural stem cells after a transgene integrated into AAVS1 in hESCs
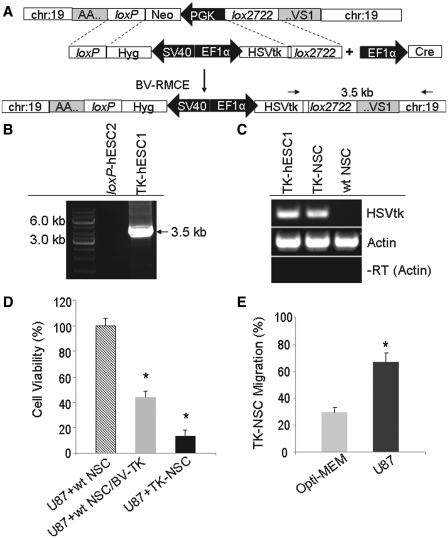
To demonstrate the potential clinical application of BV-RMCE, we generated a hESC line expressing the herpes simplex virus thymidine kinase (HSVtk) suicide gene from the master loxP-hESC2 line (Figure 6A). The HSVtk/ganciclovir system, a commonly used cancer gene therapy regime, uses HSVtk-phosphorylated ganciclovir to inhibit DNA replication, thus inducing tumor cell death (34). HSVtk-phosphorylated GCV molecules may further display bystander killing effect by diffusing to nearby tumor cells. Four colonies were propagated through hygromycin B selection. The DNA of these clones was subjected to PCR genotyping using a primer specific for chromosome 19 and a primer specific for the HSVtk gene present on the baculovirus carrying the EF1α-HSVtk-hyg-lox cassette (BV-TK). The amplification of 3.5 kb fragment indicated the integration of the HSVtk gene within AAVS1. Through this analysis, all four clones were identified as having undergone site-specific integration (Figure 6B and Table 1). TK-hESC Clone #1 (TK-hESC1) was selected randomly and subjected to neural differentiation. RT–PCR analysis confirmed the expression of HSVtk in TK-hESC1 and derived NSCs (TK-NSCs), but not in NSCs derived from wt hESCs (wt NSCs, Figure 6C).
Figure 6.
Baculoviral transduction to introduce the HSVtk suicide gene into AAVS1 in loxP-hESCs and the use of derived NSCs for cancer therapy. (A) Schematic representation of the generation of a TK-hESC line through BV-RMCE. loxP-hESC2 with a modified AAVS1 was transduced with two baculoviral vectors for site-specific transgene integration. (B) PCR analysis for detection of the integration of the HSVtk gene into AAVS1. TK-hESC clones that underwent site-specific integration are identified through amplification of 3.5 kb fragment. (C) RT–PCR analysis to detect HSVtk expression in TK-hESC1 cells and dervied NSCs. (D) Cell viability analysis following in vitro tumor killing. Each group consisted of 10 repeats and values are expressed as mean ± SD. *P < 0.05 versus the U87+wt NSC group by an unpaired t-test. (E) Migration of TK-NSCs toward U87 glioma cells. Each group consisted of seven repeats and values are expressed as mean ± SD. *P < 0.05 versus the migration toward Opti-MEM by an unpaired t-test.
We then tested whether TK-NSCs had the ability to kill cancer cells through bystander effects by mixing U87 human glioma cells with TK-NSCs or wt NSCs transduced with BV-TK at 1:1 ratio in a co-culture system. We observed that wt NSCs, after transient transduction with BV-TK, killed 55.93 ± 4.51% of the mixed cells in the presence of ganciclovir. TK-NSCs, however, had an ability to kill 86.74 ± 4.64%. The enhanced killing efficiency of TK-NSCs was attributed to the stable expression of HSVtk, as opposed to the transient expression found in baculoviral transduced wt NSCs (Figure 6D).
NSCs are capable of migrating toward tumors and thus can be used in the Trojan horse approach as cellular vehicles for targeted delivery of therapeutic genes to distant tumor sites (35,36). To test whether TK-NSCs derived from the genetically modified hESCs display any tumor tropism, we conducted an in vitro migration assay using Boyden chambers. TK-NSCs displayed a high migration capacity toward U87 glioma cells: the percentage of migrated cells was up to ∼70%, while the percentage of cells migrating toward plain Opti-MEM cell culture medium was <30% (Figure 6E). The strong tropism of TK-NSCs toward glioma cells suggests that these cells are still functionally adequate for tumor targeting after site-specific genetic modification at AAVS1.
DISCUSSION
Previous studies have demonstrated gene targeting at AAVS1 in hESCs using AAV2 and ZFN technologies (6,17). To circumvent some potential limitations associated with the two methods, we have in the current study developed the third method for such genetic modification using a two-step process combining homologous recombination with BV-RMCE. Although two time-consuming steps were used in our approach at the beginning, the generated master loxP-hESC line provides rich opportunities for readily introducing transgenes into AAVS1 through efficient Cre-RMCE. The targeting efficiency in the master hESC line by our BV-RMCE technique could be close to 100%, which is much higher than that provided by the AAV2 technology. The method does not use an enzyme to introduce DNA double-strand breaks and thus could be less complicated in being translated into clinical applications. Furthermore, by integrating the HSVtk suicide gene within AAVS1, we obtained a genetically modified functional NSC population possessing tumor migratory properties as well as the ability to induce gap-junction-mediated bystander killing effects in glioma cells, thereby demonstrating the clinical application potential of the genetic modification approach developed in this study.
Gene targeting through homologous recombination is difficult to achieve in hESCs, primarily due to low clonal efficiency and low gene transfer efficiency (37). So far, only several genes, including POU5F1, HPRT1, ROSA26, Fezf2 and Olig2, have been successfully targeted in hESCs by natural homologous recombination, with varying targeting efficiencies ranging from as low as 1.5% to as high as 40% (19–25). We demonstrated here for the first time gene targeting at AAVS1 in hESCs by conventional homologous recombination without introduction of DNA double-strand breaks. A homologous recombination frequency of 2 × 10−6 was achieved in hESCs, with a targeting efficiency of 30.56 ± 12.11% when our AAVS1 targeting vector pBS-PGK-neo-lox was used. Before these experiments in hESCs, HeLa cells were tested with pBS-PGK-neo-lox to generate G418 resistant colonies. After picking 50 clones, we identified 18 clones (36%) comprising of a modified AAVS1 (C. J. A. Ramachandra et al. unpublished observations). However, when using a long construct, pBS-EF1α-EGFP-neo-lox with the identical homology arms, for integration of the EGFP gene and the neomycin resistance gene within AAVS1 simultaneously through homologous recombination, the frequency decreased by >3-fold (1 out of 11 clones). This result indicated that apart from the homology arms, targeting efficiency is also influenced by the sequence of the integrated gene and its regulatory elements present in the targeting construct. It is likely that using homologous recombination to integrate a large transgene together with a cellular promoter into the AAVS1 locus would be a challenging task, even when the homologous arms are available.
Through the use of ZFN-mediated homologous recombination, the targeting efficiency at AAVS1 is greatly enhanced (17). However, the efficiency of this system is dependent on the type of ZFN pair used as demonstrated with the targeting of the POU5F1 locus (17). Also, the induction of off-target DNA breaks at related sequences throughout the genome could lead to potentially hazardous effects (18). Hence, by generating a loxP-hESC line through natural homologous recombination and developing the BV-RMCE method, we have provided a safer alternative for efficient gene targeting at AAVS1 in hESCs.
The use of baculoviral vectors for the delivery of Cre recombinase and the floxed transgene in hESCs is justified by several factors. Previously, the use of lentiviral vectors was shown to achieve high transient transduction efficiencies as well as induce stable transgene expression in hESCs (38,39). However, these vectors achieve stable transgene expression by integrating randomly within the genome. Although possessing a lower risk of random integration, commonly used adenoviral vectors and AAV2 vectors were shown to achieve low transduction efficiencies in hESCs (40). A study comparing gene transfer efficiencies between the use of viral and non-viral vectors in hESCs showed that the use of electroporation and lipofection were the least effective methods at <3% (41). Baculoviral vectors on the other hand are able to transduce up to 80% of hESCs with no observable cytotoxicity (15,42). Baculoviral vectors derived from Autographa californica multiple nucleopolyhedrovirus (AcMNPV) replicate in insect cells, but become replication incompetent in mammalian cells, providing a property that makes them easy for production and far less harmful in clinical use. When used for genetic modification of human cells, baculoviral vectors, unlike lentiviral vectors, mediate transgene expression without integration into the host genome and the virus DNA degrades quickly within transduced mammalian cells (43), thus suiting the purpose of temporary transgene expression well. These features of baculoviral transduction decrease the probability that baculoviral vectors used ex vivo and transgene products expressed from the vectors will invoke undesired immune responses after the genetically engineered cells are transplanted into the body. This hypothesis, however, needs to be tested in future studies. Nonetheless, the safety of the occluded Heliothis zea baculovirus has been evaluated with feeding tests on volunteer human subjects and clinical tests showed no signs of inflammation, allergy or side effects (44). The use of baculovirus also presents other advantages such as a large cloning capacity to accommodate a DNA insert up to 38 kb (45), rapid construction of recombinant baculovirus using routine cloning methods and easy preparation of high-titer viruses. All these features will contribute to the successful application of baculovirus-mediated RMCE for genetic engineering of hESCs.
The Cre/loxP system has been used previously in attaining genetic modification in hESCs (21,22,25,27,28). Through the use of a cell-permeable Cre recombinase a highly efficient site-specific recombination frequency was achieved in hESCs (27). However, apart from demonstrating the recombination inducing ability of the Cre to delete an integrated transgene, this research did not use any system to integrate transgenes into a specifically defined locus through RMCE. RMCE mediated by the cell-permeable Cre recombinase has later been used for gene insertion after screening to select transgene silencing-resistant hESC lines with one integration copy of a built-in loxP exchange cassette (28). As shown in the study, a site-specific targeting efficiency of ∼33% was achieved and in the rest of the selected clones cell-permeable Cre recombinase mediated either non-specific recombination or random integration. In differentiated somatic cells, the Cre/loxP system has been combined with the use of adenoviral vectors to successfully induce RMCE (26). However, a major drawback with this adenovirus system was that RMCE was only successful in cells that supported adenoviral replication because a high copy number of viral vectors within the cell are required. Obviously, any sort of viral replication within transplantable cells is precarious and hence their use in the clinical setting is undesirable. On the other hand, due to the high transduction efficiency of our baculoviral vectors in hESCs, the use of BV-RMCE in the current study to integrate the EGFP gene and the HSVtk suicide gene within AAVS1 resulted in site-specific targeting efficiencies of 92.59 ± 6.42% and 100%, respectively (Table 1). This result further confirms the limited random integration ability of baculovirus in hESCs.
Looking ahead, the method developed in the current study has made the repeated transgene insertion at AAVS1 possible, presenting the opportunity for introducing various therapeutic genes safely into the genome of hESCs or human iPS cells for medical applications. Generation of human iPS cells from patients’ cells allows for cell transplantation therapy free from immune rejection. An alternative approach is to establish an iPS cell bank with various human leukocyte antigen (HLA) types. We envision that our technology can be applied to human iPS cells and even be used to establish a small bank of modified iPS cells with common HLA types and heterospecific loxP sites at the AAVS1 locus. Using a cell type- or lineage-specific promoter in our BV-RMCE vectors, it should also be possible to integrate transgenes into AAVS1 in the progenies derived from a master pluripotent cell line with loxP-docking sites at AAVS1. For in vitro biological studies to understand gene function in early human development and cell type differentiation, the master cell line can be used for long-term expression of shRNAs in a uniform chromatin landscape for functional comparison of different genes.
With the AAVS1-modifed hESCs as a starting material for generating transgenic hESC, the time spent on laborious screening of modified hESC clones will be significantly reduced. AAV2 is a common human virus. A recent study using an unbiased genome-wide analysis of wild-type AAV2 integration revealed that the integration sites are scattered throughout the human genome, with only 10% of total integration hotspots being identified within the AAVS1 locus (46). Thus, targeted integration of wild-type AAV2 is not as specific as previously assumed for the AAVS1 locus. Moreover, since genetic modification with homologous recombination and RMCE will interrupt the AAVS1 site in one of two homologous chromosomes 19 (as shown by our Southern blot analysis), the site does not remain intact any longer in the modified hESCs. We speculate that the disrupted site will have significantly reduced functionality as a wild-type AAV2 integration site, further reducing the chance of deleterious interaction of wild-type AAV2 with the integrated transgene.
In conclusion, the key contributions of the current study include confirming the feasibility of homologous recombination at AAVS1, demonstrating the high efficiency of baculovirus-mediated RMCE in hESCs, and developing a two-step process combining homologous recombination with baculovirus-mediated RMCE for site-specific genetic modification of hESCs. The technology holds potential to advance the field of stem cell research.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Institute of Bioengineering and Nanotechnology, Biomedical Research Council, Agency for Science, Technology and Research (A*STAR) in Singapore; National Medical Research Council in Singapore (NMRC/1203/2009). Funding for open access charge: National Medical Research Council in Singapore (NMRC/1203/2009).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank other lab members for helpful discussion and support.
REFERENCES
- 1.Schroder AR, Shinn P, Chen H, Berry C, Ecker JR, Bushman F. HIV-1 integration in the human genome favors active genes and local hotspots. Cell. 2002;110:521–529. doi: 10.1016/s0092-8674(02)00864-4. [DOI] [PubMed] [Google Scholar]
- 2.Hacein-Bey-Abina S, Von Kalle C, Schmidt M, McCormack MP, Wulffraat N, Leboulch P, Lim A, Osborne CS, Pawliuk R, Morillon E, et al. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science. 2003;302:415–419. doi: 10.1126/science.1088547. [DOI] [PubMed] [Google Scholar]
- 3.Yao S, Sukonnik T, Kean T, Bharadwaj RR, Pasceri P, Ellis J. Retrovirus silencing, variegation, extinction and memory are controlled by a dynamic interplay of multiple epigenetic modifications. Mol. Ther. 2004;10:27–36. doi: 10.1016/j.ymthe.2004.04.007. [DOI] [PubMed] [Google Scholar]
- 4.Ellis J. Silencing and variegation of gammaretrovirus and lentivirus vectors. Hum. Gene Ther. 2005;16:1241–1246. doi: 10.1089/hum.2005.16.1241. [DOI] [PubMed] [Google Scholar]
- 5.Xia X, Zhang Y, Zieth CR, Zhang SC. Transgenes delivered by lentiviral vector are suppressed in human embryonic stem cells in a promoter-dependent manner. Stem Cells Dev. 2007;16:167–176. doi: 10.1089/scd.2006.0057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Smith JR, Maguire S, Davis LA, Alexander M, Yang F, Chandran S, ffrench-Constant C, Pedersen RA. Robust, persistent transgene expression in human embryonic stem cells is achieved with AAVS1-targeted integration. Stem Cells. 2008;26:496–504. doi: 10.1634/stemcells.2007-0039. [DOI] [PubMed] [Google Scholar]
- 7.Kotin RM, Linden RM, Berns KI. Characterization of a preferred site on human chromosome 19q for integration of adeno-associated virus DNA by non-homologous recombination. EMBO J. 1992;11:5071–5078. doi: 10.1002/j.1460-2075.1992.tb05614.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lamartina S, Sporeno E, Fattori E, Toniatti C. Characteristics of the adeno-associated virus preintegration site in human chromosome 19: open chromatin conformation and transcription-competent environment. J. Virol. 2000;74:7671–7677. doi: 10.1128/jvi.74.16.7671-7677.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ogata T, Kozuka T, Kanda T. Identification of an insulator in AAVS1, a preferred region for integration of adeno-associated virus DNA. J. Virol. 2003;77:9000–9007. doi: 10.1128/JVI.77.16.9000-9007.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Philpott NJ, Gomos J, Falck-Pedersen E. Transgene expression after rep-mediated site-specific integration into chromosome 19. Hum. Gene Ther. 2004;15:47–61. doi: 10.1089/10430340460732454. [DOI] [PubMed] [Google Scholar]
- 11.DeKelver RC, Choi VM, Moehle EA, Paschon DE, Hockemeyer D, Meijsing SH, Sancak Y, Cui X, Steine EJ, Miller JC, et al. Functional genomics, proteomics and regulatory DNA analysis in isogenic settings using zinc finger nuclease-driven transgenesis into a safe harbor locus in the human genome. Genome Res. 2010;20:1133–1142. doi: 10.1101/gr.106773.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Linden RM, Ward P, Giraud C, Winocour E, Berns KI. Site-specific integration by adeno-associated virus. Proc. Natl Acad. Sci. USA. 1996;93:11288–11294. doi: 10.1073/pnas.93.21.11288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Balague C, Kalla M, Zhang WW. Adeno-associated virus Rep78 protein and terminal repeats enhance integration of DNA sequences into the cellular genome. J. Virol. 1997;71:3299–3306. doi: 10.1128/jvi.71.4.3299-3306.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Howden SE, Voullaire L, Wardan H, Williamson R, Vadolas J. Site-specific, Rep-mediated integration of the intact beta-globin locus in the human erythroleukaemic cell line K562. Gene Ther. 2008;15:1372–1383. doi: 10.1038/gt.2008.84. [DOI] [PubMed] [Google Scholar]
- 15.Zeng J, Du J, Zhao Y, Palanisamy N, Wang S. Baculoviral vector-mediated transient and stable transgene expression in human embryonic stem cells. Stem Cells. 2007;25:1055–1061. doi: 10.1634/stemcells.2006-0616. [DOI] [PubMed] [Google Scholar]
- 16.Durai S, Mani M, Kandavelou K, Wu J, Porteus MH, Chandrasegaran S. Zinc finger nucleases: custom-designed molecular scissors for genome engineering of plant and mammalian cells. Nucleic Acids Res. 2005;33:5978–5990. doi: 10.1093/nar/gki912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hockemeyer D, Soldner F, Beard C, Gao Q, Mitalipova M, DeKelver RC, Katibah GE, Amora R, Boydston EA, Zeitler B, et al. Efficient targeting of expressed and silent genes in human ESCs and iPSCs using zinc-finger nucleases. Nat. Biotechnol. 2009;27:851–857. doi: 10.1038/nbt.1562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cathomen T, Joung JK. Zinc-finger nucleases: the next generation emerges. Mol. Ther. 2008;16:1200–1207. doi: 10.1038/mt.2008.114. [DOI] [PubMed] [Google Scholar]
- 19.Zwaka TP, Thomson JA. Homologous recombination in human embryonic stem cells. Nat. Biotechnol. 2003;21:319–321. doi: 10.1038/nbt788. [DOI] [PubMed] [Google Scholar]
- 20.Urbach A, Schuldiner M, Benvenisty N. Modeling for Lesch-Nyhan disease by gene targeting in human embryonic stem cells. Stem Cells. 2004;22:635–641. doi: 10.1634/stemcells.22-4-635. [DOI] [PubMed] [Google Scholar]
- 21.Irion S, Luche H, Gadue P, Fehling HJ, Kennedy M, Keller G. Identification and targeting of the ROSA26 locus in human embryonic stem cells. Nat. Biotechnol. 2007;25:1477–1482. doi: 10.1038/nbt1362. [DOI] [PubMed] [Google Scholar]
- 22.Di Domenico AI, Christodoulou I, Pells SC, McWhir J, Thomson AJ. Sequential genetic modification of the hprt locus in human ESCs combining gene targeting and recombinase-mediated cassette exchange. Cloning Stem Cells. 2008;10:217–230. doi: 10.1089/clo.2008.0016. [DOI] [PubMed] [Google Scholar]
- 23.Ruby KM, Zheng B. Gene targeting in a HUES line of human embryonic stem cells via electroporation. Stem Cells. 2009;27:1496–1506. doi: 10.1002/stem.73. [DOI] [PubMed] [Google Scholar]
- 24.Xue H, Wu S, Papadeas ST, Spusta S, Swistowska AM, MacArthur CC, Mattson MP, Maragakis NJ, Capecchi MR, Rao MS, et al. A targeted neuroglial reporter line generated by homologous recombination in human embryonic stem cells. Stem Cells. 2009;27:1836–1846. doi: 10.1002/stem.129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sakurai K, Shimoji M, Tahimic CG, Aiba K, Kawase E, Hasegawa K, Amagai Y, Suemori H, Nakatsuji N. Efficient integration of transgenes into a defined locus in human embryonic stem cells. Nucleic Acids Res. 2010;38:e96. doi: 10.1093/nar/gkp1234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sorrell DA, Robinson CJ, Smith JA, Kolb AF. Recombinase mediated cassette exchange into genomic targets using an adenovirus vector. Nucleic Acids Res. 2010;38:e123. doi: 10.1093/nar/gkq192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Nolden L, Edenhofer F, Haupt S, Koch P, Wunderlich FT, Siemen H, Brustle O. Site-specific recombination in human embryonic stem cells induced by cell-permeant Cre recombinase. Nat. Methods. 2006;3:461–467. doi: 10.1038/nmeth884. [DOI] [PubMed] [Google Scholar]
- 28.Du ZW, Hu BY, Ayala M, Sauer B, Zhang SC. Cre recombination-mediated cassette exchange for building versatile transgenic human embryonic stem cells lines. Stem Cells. 2009;27:1032–1041. doi: 10.1002/stem.38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Balani P, Boulaire J, Zhao Y, Zeng J, Lin J, Wang S. High mobility group box2 promoter-controlled suicide gene expression enables targeted glioblastoma treatment. Mol. Ther. 2009;17:1003–1011. doi: 10.1038/mt.2009.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Swistowski A, Peng J, Han Y, Swistowska AM, Rao MS, Zeng X. Xeno-free defined conditions for culture of human embryonic stem cells, neural stem cells and dopaminergic neurons derived from them. PLoS ONE. 2009;4:e6233. doi: 10.1371/journal.pone.0006233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Reubinoff BE, Itsykson P, Turetsky T, Pera MF, Reinhartz E, Itzik A, Ben-Hur T. Neural progenitors from human embryonic stem cells. Nat. Biotechnol. 2001;19:1134–1140. doi: 10.1038/nbt1201-1134. [DOI] [PubMed] [Google Scholar]
- 32.Slukvin , II, Vodyanik MA, Thomson JA, Gumenyuk ME, Choi KD. Directed differentiation of human embryonic stem cells into functional dendritic cells through the myeloid pathway. J. Immunol. 2006;176:2924–2932. doi: 10.4049/jimmunol.176.5.2924. [DOI] [PubMed] [Google Scholar]
- 33.Hwang NS, Varghese S, Lee HJ, Zhang Z, Ye Z, Bae J, Cheng L, Elisseeff J. In vivo commitment and functional tissue regeneration using human embryonic stem cell-derived mesenchymal cells. Proc. Natl Acad. Sci. USA. 2008;105:20641–20646. doi: 10.1073/pnas.0809680106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fillat C, Carrio M, Cascante A, Sangro B. Suicide gene therapy mediated by the Herpes Simplex virus thymidine kinase gene/Ganciclovir system: 15 years of application. Curr. Gene Ther. 2003;3:13–26. doi: 10.2174/1566523033347426. [DOI] [PubMed] [Google Scholar]
- 35.Aboody KS, Brown A, Rainov NG, Bower KA, Liu S, Yang W, Small JE, Herrlinger U, Ourednik V, Black PM, et al. Neural stem cells display extensive tropism for pathology in adult brain: evidence from intracranial gliomas. Proc. Natl Acad. Sci. USA. 2000;97:12846–12851. doi: 10.1073/pnas.97.23.12846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ahmed AU, Alexiades NG, Lesniak MS. The use of neural stem cells in cancer gene therapy: predicting the path to the clinic. Curr. Opin. Mol. Ther. 2010;12:546–552. [PMC free article] [PubMed] [Google Scholar]
- 37.Nakayama M. Homologous recombination in human iPS and ES cells for use in gene correction therapy. Drug Discov. Today. 2010;15:198–202. doi: 10.1016/j.drudis.2010.01.006. [DOI] [PubMed] [Google Scholar]
- 38.Gropp M, Itsykson P, Singer O, Ben-Hur T, Reinhartz E, Galun E, Reubinoff BE. Stable genetic modification of human embryonic stem cells by lentiviral vectors. Mol. Ther. 2003;7:281–287. doi: 10.1016/s1525-0016(02)00047-3. [DOI] [PubMed] [Google Scholar]
- 39.Ma Y, Ramezani A, Lewis R, Hawley RG, Thomson JA. High-level sustained transgene expression in human embryonic stem cells using lentiviral vectors. Stem Cells. 2003;21:111–117. doi: 10.1634/stemcells.21-1-111. [DOI] [PubMed] [Google Scholar]
- 40.Smith-Arica JR, Thomson AJ, Ansell R, Chiorini J, Davidson B, McWhir J. Infection efficiency of human and mouse embryonic stem cells using adenoviral and adeno-associated viral vectors. Cloning Stem Cells. 2003;5:51–62. doi: 10.1089/153623003321512166. [DOI] [PubMed] [Google Scholar]
- 41.Cao F, Xie X, Gollan T, Zhao L, Narsinh K, Lee RJ, Wu JC. Comparison of gene-transfer efficiency in human embryonic stem cells. Mol. Imaging Biol. 2010;12:15–24. doi: 10.1007/s11307-009-0236-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Du J, Zeng J, Zhao Y, Boulaire J, Wang S. The combined use of viral transcriptional and post-transcriptional regulatory elements to improve baculovirus-mediated transient gene expression in human embryonic stem cells. J. Biosci. Bioeng. 2010;109:1–8. doi: 10.1016/j.jbiosc.2009.06.017. [DOI] [PubMed] [Google Scholar]
- 43.Ho YC, Chen HC, Wang KC, Hu YC. Highly efficient baculovirus-mediated gene transfer into rat chondrocytes. Biotechnol. Bioeng. 2004;88:643–651. doi: 10.1002/bit.20239. [DOI] [PubMed] [Google Scholar]
- 44.Heimpel AM, Buchanan LK. Human feeding tests using a nuclear-polyhedrosis virus of Heliothis zea. J. Invertebr. Pathol. 1967;9:55–57. doi: 10.1016/0022-2011(67)90043-2. [DOI] [PubMed] [Google Scholar]
- 45.Cheshenko N, Krougliak N, Eisensmith RC, Krougliak VA. A novel system for the production of fully deleted adenovirus vectors that does not require helper adenovirus. Gene Ther. 2001;8:846–54. doi: 10.1038/sj.gt.3301459. [DOI] [PubMed] [Google Scholar]
- 46.Hüser D, Gogol-Döring A, Lutter T, Weger S, Winter K, Hammer EM, Cathomen T, Reinert K, Heilbronn R. Integration preferences of wild-type AAV-2 for consensus rep-binding sites at numerous loci in the human genome. PLoS Pathog. 2010;6:e1000985. doi: 10.1371/journal.ppat.1000985. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.