Abstract
Although often ignored, human rhinoviruses (HRVs) are the most frequent causes of respiratory tract infections (RTIs). A group of closely related novel rhinoviruses have recently been discovered. Based on their unique phylogenetic position and distinct genomic features, they are classified as a separate species, HRV-C. After their discovery, HRV-C viruses have been detected in patients worldwide, with a reported prevalence of 1.4–30.9% among tested specimens. This suggests that the species contribute to a significant proportion of RTIs that were unrecognized in the past. HRV-C is also the predominant HRV species, often with a higher detection rate than that of the two previously known species, HRV-A and HRV-B. HRV-C infections appear to peak in fall or winter in most temperate or subtropical countries, but may predominate in the rainy season in the tropics. In children, HRV-C is often associated with upper RTIs, with asthma exacerbation and wheezing episodes being common complications. The virus has also been detected in children with bronchitis, bronchiolitis, pneumonia, otitis media, sinusitis and systemic infections complicated by pericarditis. As for adults, HRV-C has been associated with more severe disease such as pneumonia and exacerbation of chronic obstructive pulmonary disease. However, larger clinical studies with asymptomatic controls are required to better define the significance of HRV-C infection in the adult population. On the basis of VP4 sequence analysis, a potential distinct subgroup within HRV-C has also been identified, although more complete genome sequences are needed to better define the genetic diversity of HRV-C.
Introduction
Human rhinoviruses (HRVs) are the most frequent causes of acute respiratory tract infections (RTIs). They are small, nonenveloped, single-stranded, positive-sense, RNA viruses that are now classified within the genus Enterovirus of the eight genera belonging to the family Picornaviridae. Traditionally, they have been associated only with mild upper respiratory tract infections (URTIs). Most ‘classical’ HRVs have a relatively low optimal temperature for growth (33°c), reflecting their adaptation to the human nasopharynx and association with URTIs. However, recent studies have shown that they are increasingly associated with more severe diseases such as pneumonia—especially in infants, the elderly and immunocompromised patients.1–3 ‘Classical’ HRVs consist of more than 100 distinct serotypes, which appear to correlate with VP1 gene sequences.4 The different serotypes can be further classified according to receptor specificity, antiviral susceptibility and nucleotide sequence identities.4 On the basis of VP4/VP2 gene sequence analysis, all serotypes except HRV87 were classified into two species, HRV-A with 74 serotypes, and HRV-B with 25 serotypes.5–8
Because the etiological agents of a significant proportion of RTIs remain unknown,9, 10 intensive research efforts were carried out in the past few years to identify novel respiratory pathogens that could be responsible. Apart from the recent discovery of human metapneumovirus,11 severe acute respiratory syndrome coronavirus,12 human coronavirus NL63 (HCoV-NL63),13, 14 human coronavirus HKU1 (HCoV-HKU1)15–18 and human bocavirus (HBoV),18–20 several research groups have previously reported the detection of novel HRV genotypes in respiratory tract specimens from patients in the United States, Australia and China.21–25 Analysis of the complete genome sequences suggested that these newly identified HRV genotypes belonged to a potentially novel HRV species, human rhinovirus C (HRV-C), with genome features distinct from HRV-A and HRV-B.24, 26 HRVs closely related to HRV-C have subsequently been found in patients from various countries, suggesting that these viruses are circulating worldwide and represent an important cause of respiratory disease.25, 27–41 In this review, we summarize current knowledge of the epidemiology, clinical features, genome features, molecular diagnosis and genetic diversity of this novel HRV species.
Methods
Keywords including ‘rhinovirus’, ‘new’, ‘novel’, ‘HRV’ and ‘HRV-C’ were used in MEDLINE searches. The search results were then manually screened to include literature on the newly described HRV-C species or related strains.
Human rhinovirus C: discovery of a novel HRV species
In 2006, new rhinovirus genotypes were identified in respiratory samples collected from patients in Queensland and New York City.21, 22 Because only partial VP4/VP2 sequences were available, the phylogenetic positions of these new genotypes could not be ascertained then, and the strains found in New York were designated HRV-NY. After complete polyprotein gene sequencing a Queensland strain, designated HRV-QPM, was classified as a subgroup of HRV-A, HRV-A2.23 In a previous study of HBoV infections in children admitted to hospital,24 we also identified the presence of HRV sequences that did not cluster with either HRV-A or HRV-B species. Upon VP4 sequence analysis, these HRV strains from New York, Queensland and Hong Kong fell into a distinct cluster away from HRV-A and HRV-B, suggesting that they represent a novel clade of HRVs. To better ascertain their phylogenetic position and genome structure, we carried out complete genome sequence analysis on three of strains found by us, which revealed distinct genome features supporting the classification of HRV-C as a separate species.24 As a result of limited sequence data available in earlier studies, it was uncertain whether HRV-Cs represent only novel genotypes or a sublineage within HRV-A, so these virus strains were given different names (Table 1). However, based on the currently available sequence data, HRV-C is now proposed by the International Committee on Taxonomy of Viruses (http://talk.ictvonline.org/media/p/ 1201.aspx) as a new HRV species in the genus Enterovirus, family Picornaviridae, order Picornavirales.
Table 1.
Published reports on detection of HRV-C from clinical samples
| References | Country/region | Age range of study population (median) | Clinical syndromes | Method of detection | Target(s) for species classification | Type of sample | No. of tested samples | No. (%) of samples positive for HRV-C | Designated virus strain/clade name |
|---|---|---|---|---|---|---|---|---|---|
| Arden et al.21 | Australia | 1 day to 80.3 years (1.2 years) | Acute RTI | RT–PCR | VP4/VP2 | 92.4% NPA | 315 | 9 (2.9) | HRV-A2 |
| Lamson et al. 22 | USA | 4 months to 98 years (25 years) | Influenza-like illness | MassTag PCR | VP4 | Oropharyngeal, NP, nasal swabs | 151 | 8 (5.3) | HRV-NY |
| McErlean et al. 23 | Australia | 1 day to 80 years (1.3 years) | Acute RTI | Real-time RT-PCR | VP4, VP2, VP1 | 92% NPA | 1244 | 17(1.4) | HRV-QPM |
| Lau et al. 24 | Hong Kong | <18 years | Acute RTI | RT-PCR | VP4 | NPA | 203 | 21 (10.3) | HRV-C |
| Kistler et al. 25 | USA | Adults | RTI±asthma | Virochip | VP4/VP2 | Nasal lavage | 82 | 5 (6.1) | HRV 'X' |
| Renwick et al. 27 | Cermany | 14 days to 5 years (10 months) | Acute RTI | Microarray MassTag PCR | VP4/VP2 | NPA | 97 | 30 (30.9) | HRV X |
| Lee et al. 28 | USA | =1 year | Frequent respiratory illnesses | Respiratory Multicode Assay | 5′-NCR | Nasal lavage | 181 | 9 (5.0) | HRVC |
| Briese et al. 29 | South África, Côte d'lvoire, Nepal, India, Australia, Denmark, Spain | 0.4 months to 56 years | Acute RTI | MassTag PCR | VP4/VP2 | Respiratory specimens | 326 | 23 (7.1) | Novel genotype |
| Dominguez et al.30 | USA | Mean 40.1 ±33.4 months | Respiratory symptoms | MassTag PCR | VP4/VP2 | NP washes | 44 | 9 (20.5) | HRV-CO |
| Savolainen-Kopra et al.31 | Finland | <2 years | Acute otitis media | RT–PCR | VP4/VP2 | NPA, middle ear fluid | 92 in 38 AOM events | 1 6 (1 7.4) | HRV-C PNC |
| Kiang et al. 32 | USA | Pediatric and adult patients | Acute RTI | RT–PCR | 5′-NCR | NPS, NPA, ETA, BAL, Pleural fluid | 24 positive for HRV | 5 (20.8) | Genogroup C |
| Xiang et al.33 | China | 1 month to 15 years (10 months) | LRTI | RT–PCR | VP4/VP2 | NPA | 258 | 14 (5.4) | HRV-C BCH |
| Miller et al. 34 | USA | <5 years | Acute RTI or fever | RT–PCR | VP4/VP2 | Nasal/throat swab | 1052 | 77 (7.3) | HRVC |
| Khetsuriani et al. 35 | USA | =2 years | Asthma | Seminested RT-PCR | VP1 | NPS | 142 (65 cases+77 controls) | 8 (5.6) | Genogroup C |
| Tapparel et al. 36 (case report) | Switzerland | 14 months | LRTI with pericarditis | Real-time RT-PCR | VP1 | Plasma, BAL, pericardial effusion, stool | NA | NA | HRV-C |
| Han et al. 37 | South Korea | 1 month to 158 months (14 months) | LRTI | RT–PCR | 5′-NCR | NPA | 470 | 17(3.6) | HRV-C KR |
| Piralla et al. 38 | Italy | Pediatric and adult patients | Acute RTI | Real-time RT-PCR | 5′-NCR, VP4/VP2 | NPA | 301 | 28 (9.3) | HRV-C PV |
| Linsuwanon et al. 39 | Thailand | Pediatric patients | Acute RTI | RT–PCR | VP4/2 | NPA | 289 | 50 (17.3) | HRV-C |
| Lau et al. 40 | Hong Kong | Pediatric and adult patients | Acute RTI | RT–PCR | VP4 | NPA | 1200 | 91 (7.6) | HRV-C |
| Huang et al.41 | Shanghai | Pediatric patients | LRTI | RT–PCR | 5′-NCR, VP4/VP2 | NPS | 827 | 34 (4.1) | HRV-C |
Abbreviations: HRV-C, human rhinovirus C; RTI, respiratory tract infections; LRTI, lower respiratory tract infections; NA, not applicable; NPA, nasopharyngeal aspirate; NPS, nasopharyngeal swab, ETA, endotracheal aspirate; BAL, bronchoalveolar lavage; RT–PCR, reverse transcription–PCR.
Epidemiology of HRV-C
Within three years since its first descriptions up to July 2009, 20 reports have been published describing the detection of HRV-C in clinical samples of patients from countries in Africa, Asia, Australia, Europe and America.21–25, 27–41 Most reports were based on respiratory samples taken from patients with respiratory illness, with the reported prevalence of HRV-C ranging from 1.4 to 30.9% among tested specimens. The difference in reported prevalence among available studies is likely due to differences among patient groups, specimen types and detection methods. Nevertheless, in most studies, HRV-C was detected in >5% of tested specimens, suggesting an important role for the virus in RTIs worldwide. Apart from sporadic infections, HRV-C probably causes frequent outbreaks of respiratory illness in the community and institutions. In our recent study, 12% of cases of HRV-C infections were acquired during institutionalization or admission to hospital.40 In incidents of community-acquired illness, clusters of cases were linked to the same strains of HRV-C within a short period of time and there were frequent reports of contact among patients who suffered from a similar illness.40
HRV-C appears to dominate among HRVs implicated in human RTIs. Among the 26 nasopharyngeal aspirates (NPAs) tested positive for HRV in our earlier study, 21 (81%) belonged to HRV-C whereas five (19%) belonged to HRV-A; HRV-B was not detected.24 In a recent study that included 220 NPAs positive for HRV, HRV-A was the most prevalent species (50%), whereas HRV-C accounted for 41% and HRV-B accounted for 8% of respiratory samples, suggesting that HRV-A and HRV-C are more prevalent than HRV-B in Hong Kong.40 In a number of studies in other countries, HRV-C was found to be the predominant HRV species as it was identified in 50–73% of tested samples, whereas HRV-B was the least common species.21, 22, 27, 39 However, the sensitivity of the various assays used to spot different HRV species was uncertain in many of the studies. In a study from the United States, HRV-C was suspected as the cause of almost half of all HRV-associated hospitalizations.34 On the basis of these data, it can be concluded that this newly described HRV species likely contributes to a significant proportion of HRV infections that were unrecognized in the past.
Human rhinoviruses are known as a cause of epidemics in early fall and late spring. In some studies, which include clinical specimens collected throughout the year, HRV-C appears to show seasonal patterns of infection. In Hong Kong, a subtropical city, infections caused by HRVs, including HRV-C, occur throughout the year, although a higher incidence has been observed during the fall and winter months.24, 40 In a study from Beijing, HRV-C was also detected in specimens collected in the fall (October–December) but not during summer (July–September).33 A recent study from Thailand has shown that HRV-C can be found throughout the year but predominates in the rainy season,39 whereas in South Korea it has been detected mostly in the spring.37 As for Australia, HRV-C appeared to reach peak prevalence in the winter, but was also detected in spring and summer.23 In a study from the United States, HRV-C prevalence peaked in October (early fall).34 Therefore, it appears that HRV-C shows seasonality, with peaks in fall or winter in most temperate or subtropical countries, but a possible peak incidence during the rainy season in the tropics. As peaks in fall and winter have also been observed in HRV-A and HRV-B infections, infections with these rhinoviruses may show similar seasonality.40 We recently reported an apparent alternating disease activity between HRV-A and HRV-C, although studies of longer duration are required to confirm this phenomenon, which may be due to viral interference or cross-serological protection similar to that seen with human parainfluenza viruses.40, 42
Despite the high prevalence of HRV-C and other HRVs detected in various studies, it is important to note that few data were available on the incidence of co-infections. In our earliest study, HRV-C strains were identified during a search for co-pathogens in HBoV infections.24 In a recent study that involved patients from Thailand, the frequency of coinfection with HRV-C and other respiratory viral pathogens—including HBoV, adenovirus, polyomavirus, influenza virus, parainfluenza virus, respiratory syncytial virus and human metapneumovirus—was as high as 40%.39 In a recent report from Shanghai, co-infection by HRV-A and HRV-B or HRV-C was also observed.41 In fact, co-detection of multiple viruses in respiratory infections has been increasingly frequent as a result of the availability of more accurate detection assays, and the development of microarrays and multiplex detection assays.25, 43–48 The concept of virusassociated RTIs is likely to be revolutionized in the near future, and the role of respiratory viruses will probably be redefined.
Clinical features of HRV-C infections
HRV-C is now known to be associated with both mild URTIs and more severe lower respiratory tract infections (LRTIs) in children and adults. In the earliest studies, HRV-C was detected in patients with influenza-like illness, infants with bronchiolitis and children admitted to hospital for acute respiratory illness.21–24 Although children with HRV-C infections often presented with URTIs, complications were not uncommon and included asthmatic exacerbations or febrile wheeze triggered by the virus infection.23, 24, 34, 35, 39, 40 A study from the United States found that children with HRV-Cs were more likely than those with HRV-As to have a diagnosis of asthma on discharge from hospital.34 In a recent study by us, it was found that wheezing episodes were also more common in patients infected with HRV-C and HRV-A than those who had HRV-B infections.40 HRV-C has been detected frequently in children with LRTIs including bronchitis, bronchiolitis and pneumonia.23, 24, 27, 33, 37, 39, 40 This suggests that these viruses are also associated with more severe respiratory disease. However, a recent report did not find that HRV-C strains had greater clinical impact than HRV-A or HRV-B on respiratory compromise in children with LRTI.41 HRV-C has also been detected in children with acute otitis media and acute sinusitis, in addition to RTIs.24, 40 This fact is supported in a recent study in Finland, which reported the presence of the virus in the middle-ear fluid of young children with acute otitis media.31 In a recent case report from Switzerland, HRV-C was detected in the bronchoalveolar lavage specimen, pericardial fluid, plasma and stool of a 14-month-old boy with an LRTI complicated by severe pericarditis.36 These reports suggest that the new HRV species can cause extrarespiratory and systemic infection, with serious complications such as pericarditis.
The significance of HRV-C infection in the adult population is less well studied. This is probably due to the lower hospitalization and sampling rate for respiratory illness in adults, and the lack of clinical data even in studies where samples from adults are included.21, 22, 29, 32 In reports where a brief clinical diagnosis was provided, HRV-C was detected in adults with exacerbation of chronic obstructive pulmonary disease,23 asthma,25 URTIs and LRTIs.38 In a recent study by us that aimed to better define the role of HRV-C in adult respiratory illness, eight (62%) of the 13 adults with HRV-C infections had pneumonia, a rate significantly higher than that observed with HRV-A infections in the study population.40 The other four patients presented with URTI and one with exacerbation of chronic obstructive pulmonary disease.40 Further clinical studies that involve more adult patients and asymptomatic controls are required to examine whether HRV-C could be more virulent than other HRVs.
Distinct genome features of HRV-C
Genomic studies on HRVs have been limited until recently, with only five HRV-A and one HRV-B genomes published before 2007. Although 97 and 31 complete genome sequences are now available for HRV-A and HRV-B, respectively,49–58 only seven genomes or complete polyprotein coding regions of HRV-C have been sequenced up to the time of writing this article: three from Hong Kong strains (HRV-C 024, 025 and 026), two from California (NAT001 and NAT045), one from New York (NY1078) and one from Australia (HRV-QPM).23–25, 49 The genome organization of HRV-C is typical of Picornaviridae, comprising a 5′-noncoding region (5′-NCR), a single open reading frame encoding a single polyprotein and a 3′-NCR before a polyadenylated tract. However, the genome of HRV-C was the shortest among other reported HRVs and HEVs, a result of several deletions.23, 24 Although the P1 region of HRV-C possessed higher amino-acid identities to that of HRV-A than to the P1 region of HRV-B, the P3 region possessed higher amino-acid identities to that of HRV-B than to the P3 region of HRV-A. The predicted amino-acid sequence of the HRV-C polyprotein shared only 51–52% and 47–48% with that of HRV-A and HRV-B, respectively.23, 24 Phylogenetic analysis of the 5′-NCR region and the predicted proteins revealed that HRV-C strains formed a distinct cluster away from HRV-A and HRV-B, which indicates that they represent a novel HRV species. Although HRV-C is a newly described HRV species, molecular dating analysis using sequences of the VP4/2 coding regions indicated that these viruses have been circulating for at least 250 years, with an estimated evolutionary rate of 6.6×10−4 substitutions per site per year.29
HRV-C shows several distinct genomic features that further support its classification as a separate species. First, a unique putative cleavage site was identified between VP4 and VP2.24 Second, major insertions and deletions were observed in VP1, which is shorter than that of other HRVs, especially in regions that were important for neutralization in HRV-A and HRV-B.23, 24 Third, the conserved amino-acid residues within VP1 that determine receptor binding of HRV-A and HRV-B were found to have frequent substitutions in HRV-C.24 Using structural homology modeling, it was also found the VP1 of HRV-C possesses structural disparities within sites for receptor binding of HRV-A and HRV-B.26 These suggest that HRV-C may use a different cellular receptor, reflected in its failure to grow in traditional cell cultures. In addition, a novel distinct cis-acting replication element located in the VP2 region has been identified in HRV-C, supporting its classification as a separate species.59 By completing the genome sequences for all known HRV serotypes, a recent study also confirmed species-specific sequence and RNA structure elements that differentiate HRV-C from HRV-A and HRV-B.49 Although HRV-A strains are currently susceptible to the antiviral drug pleconaril, analysis of the genome sequence data suggested that HRV-C may be resistant to this drug, in view of the presence of Phe152 and Thr191 in the VP1 of some reported strains.23, 24 However, this would need to be verified by in vitro assays when cell culture systems are established. The use of reverse genetic approaches may overcome the difficulties in isolating HRV-C.
Molecular detection and genetic diversity of HRV-C
5′-NCR and VP4/VP2 regions were the most commonly used targets for detection of HRV-C, although the 5′-NCR region appears to be more sensitive for HRV detection.32, 37 In one study, VP4/VP2 RT-PCR failed to detect all 24 samples with HRVs using cDNA templates identical to those used for 5′-NCR RT-PCR.32 However, 5′-NCR sequence is not suitable for assignment of phylogenetic relationships.22, 60 To better determine the prevalence of HRVs in respiratory specimens, it is important that 5′-NCR regions should be used for detection and VP4/VP2 or VP1 sequences for more accurate species identification. Because VP4 is a highly conserved protein, the use of amino-acid sequences may underestimate the diversity and number of HRV strains as compared with nucleotides sequences. However, it remains to be determined which should be the valid approach for representation of different HRV-C strains.
In addition to the commonly used PCR methodology, other molecular detection methods have also been used to detect HRV-C viruses. In a study from the United States, Virochip was used for virus detection.25 Virochip is a DNA microarray that contains the most conserved sequences of all known viruses identified in humans, animals, plants and microbes.46 This method can detect new members of known virus families by cross-hybridization, which offers a significant advantage compared to PCR-based methods.46, 47 In another study, also from the United States, respiratory multicode assay (RMA) was used to detect viruses.28 RMA is a high-throughput, multiplex PCR-microsphere flow cytometry assay designed for comprehensive detection of common respiratory viruses and consists of five main steps, all of which take place in the same microwell.48 Apart from having the capacity to detect new viruses, these assays also allow simultaneous detection of multiple viruses from the same sample, which may provide better information on rates of co-infection.
Real-time RT-PCR has been used to measure the viral load of HRV-C in clinical specimens. In a study from Italy, the median peak viral load of 28 patients with HRV-C infection was 4.5×106 RNA copies per ml in NPA, which is slightly higher than that seen in 45 patients with HRV-A infection (2.2×106 RNA copies per ml) and in 12 patients with HRV-B infection (1.6×105 RNA copies per ml); however, the difference was not statistically significant.38 In a case report from Switzerland about HRV-C LRTI complicated by pericarditis, the viral load was significantly higher in the bronchoalveolar lavage specimen compared with that in plasma and stool specimens, consistent with the virus’ association with RTIs.36
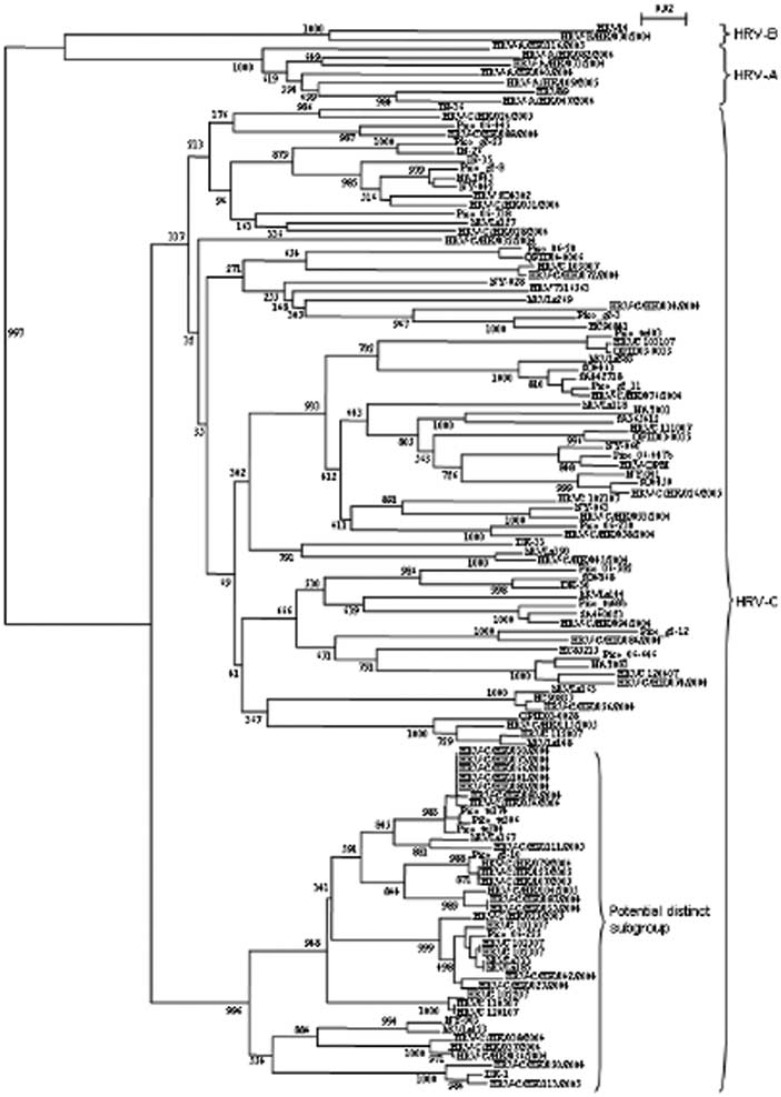
Currently, data on serotype recurrence of HRVs and their seasonal distribution are very limited. In our recent report, recurrent infections by different HRV strains of the same patient were observed within a short period of time.40 Whereas numerous HRV strains belonging to the HRV-C species have been identified, a huge genetic diversity was detected among strains within the species, and this may help the virus evade immune protection. In our study, the VP4 sequences of the identified HRV-C strains showed 67.2–82.1% nucleotide identities to that of our reference HRV-C strain 024.40 By phylogenetic analysis of available VP4 sequences of HRV-C, we also identified a potential distinct subgroup of strains within HRV-C Figure 1.40 Interestingly, HRV-C strains presenting divergent 5′-NCR sequences were also observed in the past,28, 61 and a recent report proposed to subdivide HRV-C in two distinct subspecies based on 5′-NCR regions.41 However, as only 5′-NCR regions were studied for some of the previously reported strains, it remains to be determined if these subspecies correspond to the subgroup we identified based on VP4 sequences. More complete genome sequences of strains from these potential subgroups of HRV-C and successful isolation of different HRV-C genotypes would be required to ascertain the existence of subspecies, and to study their genetic diversity and cross-serological reactivity.
Figure 1.
Phylogenetic tree of the VP4 region of HRV-C strains detected in different countries, showing the presence of a potential distinct subgroup. 201 nucleotide positions in each VP4 region were included in the analysis. The tree was constructed by a neighbor-joining method and bootstrap values were calculated from 1000 trees. The scale bar indicates the estimated number of substitutions per 50 nucleotides. Strains from Hong Kong are indicated by HRV-C/HK/strain no./year of detection. The GenBank accession numbers of the previously published sequences are as follows: HRV-QPM, EF186077; HRV89, A10937; HRV14, NC_001490; DK-1, EU697851; DK-30, EU697837; DK-33, EU697833; HC85215, EU697846; HC90837, EU697856; HC90841, EU697863; hRVLz118, EU822834; hRVLz123, EU822836; hRVLz127, EU822838; hRVLz144, EU822840; hRVLz148, EU822841; hRVLz163, EU822842; hRVLz167, EU822843; hRVLz185, EU822847; hRVLz269, EU822856; hRVLz333, EU822869; hRVLz383, EU822878; hRVLz390, EU822879; IN-26, EU697865; IN-35, EU697845; IN-36, EU697839; HRV 7316563, EU697850; NY-003, DQ875929; NY-028, DQ875931; NY-041, DQ875921; NY-042, DQ875926; NY-060, DQ875928; NY-063, DQ975924; SA365412, EU697852; SA440023, EU697829; SA442718, EU697828; SO4302, EU697869; SO4450, EU697835; SO4463, EU697854; SO4868, EU697854; NAT001, EF077252; NAT045, EF077253; NAT083, EF077264; QPID03-0035, EU155152; QPID03-0033, EU155153; QPID03-0028, EU155154; QPID04-0006, EU155158; HRVC 102307, EU687518; HRVC 102507, EU687523; HRVC 101507, EU687515; HRVC 120107, EU687527; HRVC102207, EU687516; HRVC 110507, EU687522; HRVC 103007, EU687519; HRVC 112007, EU687525; HRVC 103107, EU687520; HRVC 102107, EU687517; HRVC 111007, EU687524; HRVC 120407, EU687528; Pico tu304, EU081791; Pico tu306, EU081793; Pico tu403, EU081795; Pico 06-445, EU081796; Pico tu68b, EU081797; Pico g2-10, EU081798; Pico g2-11, EU081799; Pico 06-20, EU081800; Pico g2-12, EU081801; Pico g2-25, EU081802; Pico 06-447b, EU081806; Pico 06-230, EU081807; Pico 06-582, EU081809; Pico g2-3, EU081810; Pico 06-646, EU081811; Pico g2-8, EU081812; Pico 06-738, EU081813; Pico tu174, EU081814; Pico 06-225, EU081815.
Concluding remarks
Because HRVs were historically considered to have little health impact and clinical significance, the possible existence of novel species and the relative importance and classification of the different species have been poorly investigated.62 Recent studies have shown that in fact HRVs are more important than respiratory syncytial virus as a cause of respiratory virus-associated hospital admissions, and as predictors of recurrent wheezing in children in large cohorts.2, 63 The discovery of HRV-C as a novel species with diverse genotypes, and its prevalence in human respiratory samples, has ushered a new era in HRV research, by uncovering viruses that account for a significant proportion of previously undiagnosed respiratory illness. However, our understanding of this novel HRV species has been hindered to date by a failure to culture these viruses in cell lines. Therefore, further studies on HRV-C are required to better understand the viruses’ genetic diversity, mechanism of immune evasion, virulence and antiviral susceptibilities. We suggest that these studies should focus on analysis of more HRV-C genome sequences, pathogenesis and the development of culture systems.
Acknowledgements
This work was partly supported by the Research Grant Council Grant (HKU 7687/09M), University Development Fund, Committee on Research and Conference Grants, HKU Special Research Achievement Award and Outstanding Young Researcher Award, The University of Hong Kong; the HKSAR Research Fund for the Control of Infectious Diseases of the Health, Welfare and Food Bureau, and the Consultancy Service for Enhancing Laboratory Surveillance of Emerging Infectious Disease for the Department of Health of the Hong Kong Special Administrative Region of China. We thank Professor York Chow for his support.
References
- 1.Kaiser L, Aubert JD, Pache JC, Deffernez C, Rochat T, Garbino J, et al. Chronic rhinoviral infection in lung transplant recipients. Am J Respir Crit Care Med. 2006;174:1392–9. doi: 10.1164/rccm.200604-489OC. [DOI] [PubMed] [Google Scholar]
- 2.Miller EK, Lu X, Erdman DD, Poehling KA, Zhu Y, Griffin MR, et al. Rhinovirus-associated hospitalizations in young children. J Infect Dis. 2007;195:773–81. doi: 10.1086/511821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Turner RB. Rhinovirus: more than just a common cold virus. J Infect Dis. 2007;195:765–6. doi: 10.1086/511829. [DOI] [PubMed] [Google Scholar]
- 4.Oberste MS, Maher K, Kilpatrick DR, Pallansch MA. Molecular evolution of the human enteroviruses: correlation of serotype with VP1 sequence and application to picornavirus classification. J Virol. 1999;73:1941–8. doi: 10.1128/jvi.73.3.1941-1948.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Laine P, Savolainen C, Blomqvist S, Hovi T. Phylogenetic analysis of human rhinovirus capsid protein VP1 and 2A protease coding sequences confirms shared genus-like relationships with human enteroviruses. J Gen Virol. 2005;86:697–706. doi: 10.1099/vir.0.80445-0. [DOI] [PubMed] [Google Scholar]
- 6.Ledford RM, Patel NR, Demenczuk TM, Watanyar A, Herbertz T, Collett MS, et al. VP1 sequencing of all human rhinovirus serotypes: insights into genus phylogeny and susceptibility to antiviral capsid-binding compounds. J Virol. 2004;78:3663–74. doi: 10.1128/JVI.78.7.3663-3674.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Savolainen C, Laine P, Mulders MN, Hovi T. Sequence analysis of human rhinoviruses in the RNA-dependent RNA polymerase coding region reveals large within-species variation. J Gen Virol. 2004;85:2271–7. doi: 10.1099/vir.0.79897-0. [DOI] [PubMed] [Google Scholar]
- 8.Savolainen C, Blomqvist S, Mulders MN, Hovi T. Genetic clustering of all 102 human rhinovirus prototype strains: serotype 87 is close to human enterovirus 70. J Gen Virol. 2002;83:333–40. doi: 10.1099/0022-1317-83-2-333. [DOI] [PubMed] [Google Scholar]
- 9.Macfarlane JT, Colville A, Guion A, Macfarlane RM, Rose DH. Prospective study of aetiology and outcome of adult lowerrespiratory- tract infections in the community. Lancet. 1993;341:511–14. doi: 10.1016/0140-6736(93)90275-l. [DOI] [PubMed] [Google Scholar]
- 10.Ruiz M, Ewig S, Marcos MA, Martinez JA, Arancibia F, Mensa J, et al. Etiology of community-acquired pneumonia: impact of age, comorbidity, and severity. Am J Respir Crit Care Med. 1999;160:397–405. doi: 10.1164/ajrccm.160.2.9808045. [DOI] [PubMed] [Google Scholar]
- 11.van den Hoogen BG, de Jong JC, Groen J, Kuiken T, de Groot R, Fouchier RA, et al. A newly discovered human pneumovirus isolated from young children with respiratory tract disease. Nat Med. 2001;7:719–24. doi: 10.1038/89098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Peiris JS, Lai ST, Poon LL, Guan Y, Yam LY, Lim W, et al. Coronavirus as a possible cause of severe acute respiratory syndrome. Lancet. 2003;361:1319–25. doi: 10.1016/S0140-6736(03)13077-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fouchier RA, Hartwig NG, Bestebroer TM, Niemeyer B, de Jong JC, Simon JH, et al. A previously undescribed coronavirus associated with respiratory disease in humans. Proc Natl Acad Sci USA. 2004;101:6212–16. doi: 10.1073/pnas.0400762101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van der Hoek L, Pyrc K, Jebbink MF, Vermeulen-Oost W, Berkhout RJ, Wolthers KC, et al. Identification of a new human coronavirus. Nat Med. 2004;10:368–73. doi: 10.1038/nm1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lau SK, Woo PC, Yip CC, Tse H, Tsoi HW, Cheng VC, et al. Coronavirus HKU1 and other coronavirus infections in Hong Kong. J Clin Microbiol. 2006;44:2063–71. doi: 10.1128/JCM.02614-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Woo PC, Lau SK, Chu CM, Chan KH, Tsoi HW, Huang Y, et al. Characterization and complete genome sequence of a novel coronavirus, coronavirus HKU1, from patients with pneumonia. J Virol. 2005;79:884–95. doi: 10.1128/JVI.79.2.884-895.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Woo PC, Lau SK, Tsoi HW, Huang Y, Poon RW, Chu CM, et al. Clinical and molecular epidemiological features of coronavirus HKU1-associated community-acquired pneumonia. J Infect Dis. 2005;191:1898–907. doi: 10.1086/497151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Sloots TP, McErlean P, Speicher DJ, Arden KE, Nissen MD, Mackay IM. Evidence of human coronavirus HKU1 and human bocavirus in Australian children. J Clin Virol. 2006;35:99–102. doi: 10.1016/j.jcv.2005.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Allander T, Tammi MT, Eriksson M, Bjerkner A, Tiveljung-Lindell A, Andersson B. Cloning of a human parvovirus by molecular screening of respiratory tract samples. Proc Natl Acad Sci USA. 2005;102:12891–6. doi: 10.1073/pnas.0504666102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lau SKP, Yip CCY, Que TL, Lee RA, Au-Yeung RK, Zhou B, et al. Clinical and molecular epidemiology of human bocavirus in respiratory and fecal samples from children in Hong Kong. J Infect Dis. 2007;196:986–93. doi: 10.1086/521310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Arden KE, McErlean P, Nissen MD, Sloots TP, Mackay IM. Frequent detection of human rhinoviruses, paramyxoviruses, coronaviruses, and bocavirus during acute respiratory tract infections. J Med Virol. 2006;78:1232–40. doi: 10.1002/jmv.20689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lamson D, Renwick N, Kapoor V, Liu Z, Palacios G, Ju J, et al. MassTag polymerase-chain-reaction detection of respiratory pathogens, including a new rhinovirus genotype, that caused influenza-like illness in New York State during 2004–2005. J Infect Dis. 2006;194:1398–402. doi: 10.1086/508551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.McErlean P, Shackelton LA, Lambert SB, Nissen MD, Sloots TP, Mackay IM. Characterisation of a newly identified human rhinovirus, HRV-QPM, discovered in infants with bronchiolitis. J Clin Virol. 2007;39:67–75. doi: 10.1016/j.jcv.2007.03.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lau SK, Yip CC, Tsoi HW, Lee RA, So LY, Lau YL, et al. Clinical features and complete genome characterization of a distinct human rhinovirus (HRV) genetic cluster, probably representing a previously undetected HRV species, human rhinovirus-C, associated with acute respiratory illness in children. J Clin Microbiol. 2007;45:3655–64. doi: 10.1128/JCM.01254-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kistler A, Avila PC, Rouskin S, Wang D, Ward T, Yagi S, et al. Pan-viral screening of respiratory tract infections in adults with and without asthma reveals unexpected human coronavirus and human rhinovirus diversity. J Infect Dis. 2007;196:817–25. doi: 10.1086/520816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McErlean P, Shackelton LA, Andrews E, Webster DR, Lambert SB, Nissen MD, et al. Distinguishing molecular features and clinical characteristics of a putative new rhinovirus species, human rhinovirus C (HRV C) PLoS ONE. 2008;3:1847. doi: 10.1371/journal.pone.0001847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Renwick N, Schweiger B, Kapoor V, Liu Z, Villari J, Bullmann R, et al. A recently identified rhinovirus genotype is associated with severe respiratory-tract infection in children in Germany. J Infect Dis. 2007;196:1754–60. doi: 10.1086/524312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lee WM, Kiesner C, Pappas T, Lee I, Grindle K, Jartti T, et al. A diverse group of previously unrecognized human rhinoviruses are common causes of respiratory illnesses in infants. PLoS One. 2007;2:e966. doi: 10.1371/journal.pone.0000966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Briese T, Renwick N, Venter M, Jarman RG, Ghosh D, Köndgen S, et al. Global distribution of novel rhinovirus genotype. Emerg Infect Dis. 2008;14:944–7. doi: 10.3201/eid1406.080271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dominguez SR, Briese T, Palacios G, Hui J, Villari J, Kapoor V, et al. Multiplex MassTag-PCR for respiratory pathogens in pediatric nasopharyngeal washes negative by conventional diagnostic testing shows a high prevalence of viruses belonging to a newly recognized rhinovirus clade. J Clin Virol. 2008;43:219–22. doi: 10.1016/j.jcv.2008.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Savolainen-Kopra C, Blomqvist S, Kilpi T, Roivainen M, Hovi T. Novel species of human rhinoviruses in acute otitis media. Pediatr Infect Dis J. 2009;28:59–61. doi: 10.1097/INF.0b013e318182c90a. [DOI] [PubMed] [Google Scholar]
- 32.Kiang D, Kalra I, Yagi S, Louie JK, Boushey H, Boothby J, et al. Assay for 5′ noncoding region analysis of all human rhinovirus prototype strains. J Clin Microbiol. 2008;46:3736–45. doi: 10.1128/JCM.00674-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Xiang Z, Gonzalez R, Xie Z, Xiao Y, Chen L, Li Y, et al. Human rhinovirus group C infection in children with lower respiratory tract infection. Emerg Infect Dis. 2008;14:1665–7. doi: 10.3201/eid1410.080545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miller EK, Edwards KM, Weinberg GA, Iwane MK, Griffin MR, Hall CB, et al. A novel group of rhinoviruses is associated with asthma hospitalizations. J Allergy Clin Immunol. 2009;123:98–104. doi: 10.1016/j.jaci.2008.10.007. e1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Khetsuriani N, Lu X, Teague WG, Kazerouni N, Anderson LJ, Erdman DD. Novel human rhinoviruses and exacerbation of asthma in children. Emerg Infect Dis. 2008;14:1793–6. doi: 10.3201/eid1411.080386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tapparel C, L'Huillier AG, Rougemont AL, Beghetti M, Barazzone-Argiroffo C, Kaiser L. Pneumonia and pericarditis in a child with HRV-C infection: a case report. J Clin Virol. 2009;45:157–60. doi: 10.1016/j.jcv.2009.03.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Han TH, Chung JY, Hwang ES, Koo JW. Detection of human rhinovirus C in children with acute lower respiratory tract infections in South Korea. Arch Virol. 2009;154:987–91. doi: 10.1007/s00705-009-0383-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Piralla A, Rovida F, Campanini G, Rognoni V, Marchi A, Locatelli F, et al. Clinical severity and molecular typing of human rhinovirus C strains during a fall outbreak affecting hospitalized patients. J Clin Virol. 2009;45:311–17. doi: 10.1016/j.jcv.2009.04.016. [DOI] [PubMed] [Google Scholar]
- 39.Linsuwanon P, Payungporn S, Samransamruajkit R, Posuwan N, Makkoch J, Theanboonlers A, et al. High prevalence of human rhinovirus C infection in Thai children with acute lower respiratory tract disease. J Infect. 2009;59:115–21. doi: 10.1016/j.jinf.2009.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lau SKP, Yip CCY, Lin AWC, Lee RA, So LY, Lau YL, et al. Clinical and molecular epidemiology of a novel rhinovirus species, human rhinovirus C. J Infect Dis. 2009;200:1096–103. doi: 10.1086/605697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Huang T, Wang W, Bessaud M, Ren P, Sheng J, Yan H, et al. Evidence of recombination and genetic diversity in human rhinoviruses in children with acute respiratory infection. PLoS One. 2009;4:6355. doi: 10.1371/journal.pone.0006355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Fry AM, Curns AT, Harbour K, Hutwagner L, Holman RC, Anderson LJ. Seasonal trends of human parainfluenza viral infections: United States, 1990–2004. Clin Infect Dis. 2006;43:1016–22. doi: 10.1086/507638. [DOI] [PubMed] [Google Scholar]
- 43.Chiu CY, Urisman A, Greenhow TL, Rouskin S, Yagi S, Schnurr D, et al. Utility of DNA microarrays for detection of viruses in acute respiratory tract infections in children. J Pediatr. 2008;153:76–83. doi: 10.1016/j.jpeds.2007.12.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bharaj P, Sullender WM, Kabra SK, Mani K, Cherain J, Tyagi V, et al. Respiratory viral infections detected by multiplex PCR among pediatric patients with lower respiratory tract infections seen at an urban hospital in Delhi from 2005 to 2007. Virol J. 2009;6:89. doi: 10.1186/1743-422X-6-89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brittain-Long R, Nord S, Olofsson S, Westin J, Anderson LM, Lindh M. Multiplex real-time PCR for detection of respiratory tract infections. J Clin Virol. 2008;41:53–6. doi: 10.1016/j.jcv.2007.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wang D, Urisman A, Liu YT, Springer M, Ksiazek TG, Erdman DD, et al. Viral discovery and sequence recovery using DNA microarrays. PLoS Biol. 2003;1:E2. doi: 10.1371/journal.pbio.0000002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Chiu CY, Pouskin S, Koshy A, Urisman A, Fischer K, Yagi S, et al. Microarray detection of human parainfluenzavirus 4 infection associated with respiratory failure in an immunocompetent adult. Clin Infect Dis. 2006;43:e71–6. doi: 10.1086/507896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lee WM, Grindle K, Pappas T, Marshall DJ, Moser MJ, Beaty EL, et al. High-throughput, sensitive, and accurate multiplex PCR-microsphere flow cytometry system for large-scale comprehensive detection of respiratory viruses. J Clin Microbiol. 2007;45:2626–34. doi: 10.1128/JCM.02501-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Palmenberg AC, Spiro D, Kuzmickas R, Wang S, Djikeng A, Rathe JA, et al. Sequencing and analyses of all known human rhinovirus genomes reveal structure and evolution. Science. 2009;324:55–9. doi: 10.1126/science.1165557. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Callahan PL, Mizutani S, Colonno RJ. Molecular cloning and complete sequence determination of RNA genome of human rhinovirus type 14. Proc Natl Acad Sci USA. 1985;82:732–6. doi: 10.1073/pnas.82.3.732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Skern T, Sommergruber W, Blaas D, Gruendler P, Fraunorfer F, Pieler C, et al. Human rhinovirus 2: complete nucleotidesequence and proteolytic processing signals in the capsid protein region. Nucleic Acids Res. 1985;13:2111–26. doi: 10.1093/nar/13.6.2111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Duechler M, Skern T, Sommergruber W, Neubauer C, Gruendler P, Fogy I, et al. Evolutionary relationships within the human rhinovirus genus: comparison of serotypes 89, 2, and 14. Proc Natl Acad Sci USA. 1987;84:2605–9. doi: 10.1073/pnas.84.9.2605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Hughes PJ, North C, Jellis CH, Minor PD, Stanway G. The nucleotide sequence of human rhinovirus 1B: molecular relationships with the rhinovirus genus. J Gen Virol. 1988;69((Part 1)):49–58. doi: 10.1099/0022-1317-69-1-49. [DOI] [PubMed] [Google Scholar]
- 54.Lee WM, Monroe SS, Rueckert RR. Role of maturation cleavage in infectivity of picornaviruses: activation of an infectosome. J Virol. 1993;67:2110–22. doi: 10.1128/jvi.67.4.2110-2122.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lee WM, Wang W, Rueckert RR. Complete sequence of the RNA genome of human rhinovirus 16, a clinically useful common cold virus belonging to the ICAM-1 receptor group. Virus Genes. 1995;9:177–81. doi: 10.1007/BF01702661. [DOI] [PubMed] [Google Scholar]
- 56.Harris JR, Racaniello VR. Amino acid changes in proteins 2B and 3A mediate rhinovirus type 39 growth in mouse cells. J Virol. 2005;79:5363–73. doi: 10.1128/JVI.79.9.5363-5373.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Kistler AL, Webster DR, Rouskin S, Magrini V, Credle JJ, Schnurr DP, et al. Genome-wide diversity and selective pressure in the human rhinovirus. Virol J. 2007;4:40. doi: 10.1186/1743-422X-4-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Tapparel C, Junier T, Gerlach D, Cordey S, Van Belle S, Perrin L, et al. New complete genome sequences of human rhinoviruses shed light on their phylogeny and genomic features. BMC Genomics. 2007;8:224. doi: 10.1186/1471-2164-8-224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cordey S, Gerlach D, Junier T, Zdobnov EM, Kaiser L, Tapparel C. The cis-acting replication elements define human enterovirus and rhinovirus species. RNA. 2008;14:1568–78. doi: 10.1261/rna.1031408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Savolainen-Kopra C, Blomqvist S, Smura T, Roivainen M, Hovi T. 5′ Noncoding region alone does not unequivocally determine genetic type of human rhinovirus strains. J Clin Microbiol. 2009;47:1278–80. doi: 10.1128/JCM.02130-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tapparel C, Junier T, Gerlach D, Van-Belle S, Turin L, Cordey S, et al. New respiratory enteroviruses and recombinant rhinoviruses among circulating picornaviruses. Emerg Infect Dis. 2009;15:719–26. doi: 10.3201/eid1505.081286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Stanway G, Brown F, Christian P, Hovi T, Hyypiä T, King AMQ, et al. Family Picornaviridae. In: Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA, editors. Virus Taxonomy: Eighth Report of the International Committee in Taxonomy. San Diego: Elsevier; 2005. pp. 757–78. [Google Scholar]
- 63.Lemanske Jr RF, Jackson DJ, Gangnon RE, Evans MD, Li Z, Shult PA, King AMQ, et al. Rhinovirus illnesses during infancy predict subsequent childhood wheezing. J Allergy Clin Immunol. 2005;116:571–7. doi: 10.1016/j.jaci.2005.06.024. [DOI] [PubMed] [Google Scholar]