Abstract
Species-specific recognition between egg and sperm, a crucial event that marks the beginning of fertilization in multicellular organisms, mirrors the binding between haploid cells of opposite mating type in unicellular eukaryotes such as yeast. However, as implied by the lack of sequence similarity between sperm-binding regions of invertebrate and vertebrate egg coat proteins, these interactions are thought to rely on completely different molecular entities. Here, we argue that these recognition systems are, in fact, related: despite being separated by 0.6–1 billion years of evolution, functionally essential domains of a mollusc sperm receptor and a yeast mating protein adopt the same 3D fold as egg zona pellucida proteins mediating the binding between gametes in humans.
Keywords: fertilization, egg–sperm interaction, egg coat, zona pellucida domain, yeast mating, protein structure
Introduction
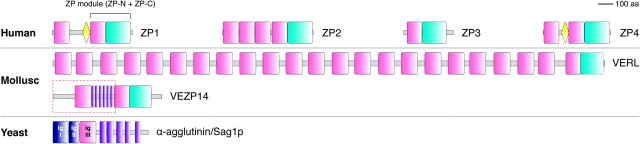
Like their counterparts in the vitelline (egg) envelope (VE) of other vertebrates as well as invertebrates such as the mollusc abalone (Aagaard et al. 2006), mammalian zona pellucida (ZP) subunits ZP1–4 assemble into the nascent egg coat using a common C-terminal “ZP domain” (Bork and Sander 1992; Jovine et al. 2002). This conserved polymerization module consists of two domains, ZP-N and ZP-C (Jovine et al. 2004; Wassarman and Litscher 2008) (fig. 1). Recent crystallographic studies of sperm receptor ZP3 revealed that the ZP-N domain defines a new subtype of the immunoglobulin (Ig) superfamily of proteins, characterized by two disulfide bonds with invariant 1–4, 2–3 connectivity, a unique E' strand implicated in polymerization, and a conserved Tyr residue in strand F (Monné et al. 2008). Moreover, they showed that—despite having a very different sequence—ZP-C also adopts a β-sandwich fold with the same basic topology as ZP-N, suggesting that the two moieties of the ZP module might have originated by duplication of a single ancestral Ig-like domain (Han et al. 2010). Additional copies of ZP-N are found within the N-terminal region of some vertebrate ZP/VE components (Callebaut et al. 2007; Monné et al. 2008) (fig. 1), where—as in the case of mammalian ZP2—they can bind sperm (Tsubamoto et al. 1999) and regulate gamete recognition (Bleil et al. 1981; Gahlay et al. 2010). Notably, repeated sequences located within the N-terminal region of abalone VE subunits VERL and VEZP14 (fig. 1) are also thought to bind sperm (Swanson and Vacquier 1997; Aagaard et al. 2010), but because of very low-sequence similarity, no connection could be made between molluscan and mammalian repeats.
FIG. 1.
Domain architecture of human ZP subunits, mollusc VERL and VEZP14, and yeast α-agglutinin/Sag1p. Pink: ZP-N domain; cyan: ZP-C domain; yellow: trefoil domain; violet: S/T-rich sequence repeat; dark blue: Sag1p Ig-like domains I, II; and dashed red box: SMART Pfam:Candida_ALS match in VEZP14.
Molluscan Egg Coat Protein Repeats Adopt the ZP-N Fold of Mammalian ZP Proteins
Because rapid sequence divergence could mask potential relationships between reproductive proteins from evolutionary distant species (Swanson and Vacquier 2002), we performed a fold recognition analysis using sequence–structure comparison in FUGUE (Shi et al. 2001). Molluscan repeat sequences were threaded against a local copy of the HOMSTRAD database of structural profiles (de Bakker et al. 2001) that included an entry for the canonical ZP-N domain of VERL (Galindo et al. 2002), generated on the basis of the crystal structure of ZP3 ZP-N (Monné et al. 2008; Han et al. 2010). A high-confidence match was found between the sequence of VERL repeat 10 and the Ig-like fold variant specific to ZP-N (supplementary fig. S1, Supplementary Material online). An homology model of repeat 10 created on the basis of this sequence–structure alignment is structurally sound and exposes Asn side chains expected to be glycosylated (Swanson and Vacquier 1997) (fig. 2). Moreover, it can be readily extended to all other VERL repeats, as well as the VERL-like repeat of VEZP14 (Aagaard et al. 2010), because of significant sequence similarity (supplementary figs. S1 and S2a–b, Supplementary Material online). This suggests that all Cys within the repeat array of VERL are engaged in ZP-N-specific Cys1–4, Cys2–3 disulfide bonds, with the exception of C201 and C294 (supplementary fig. S3a, Supplementary Material online). These additional Cys, located in repeat 2, may therefore be responsible for forming the intermolecular disulfides that have been shown to mediate homodimerization of VERL (Swanson and Vacquier 1997). This prediction was experimentally confirmed by loss of VERL dimerization upon introduction of C201D, C294S substitutions within a repeat 1–4 fragment secreted by insect cells (supplementary fig. S3b, Supplementary Material online). Considering that all other abalone VE subunits also contain a ZP module (Aagaard et al. 2010), this data collectively suggest that, as in mammals, the ZP-N domain accounts for the majority of the structure of the molluscan egg coat.
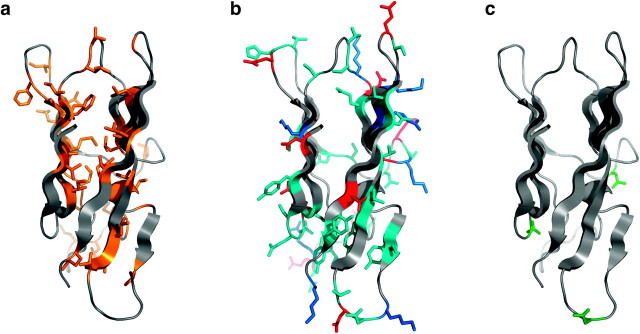
FIG. 2.
Homology model of abalone VERL repeat 10 ZP-N domain, shown in side view using a cartoon representation with relevant residues depicted as sticks. The model is consistent with burial of hydrophobic residues (a; brown), exposure of positively charged, negatively charged, and polar side chains (b; blue, red, and cyan, respectively) and exposure of consensus sites for N-glycosylation (c; green).
A Protein Domain Essential for Mating in Yeast Also Shares ZP-N-Specific Features
Domain analysis with SMART (Letunic et al. 2009) indicates that the N-terminal region of VEZP14, which contains the protein's VERL-like ZP-N repeat (Aagaard et al. 2010) (fig. 1), is in turn related to yeast agglutinin-like proteins (supplementary fig. S1, Supplementary Material online). These highly glycosylated adhesion molecules mediate extracellular interactions, such as mating in Saccharomyces cerevisiae and host invasion in Candida albicans, mainly using the last of three N-terminal Ig domains (Ig III; fig. 1) (Dranginis et al. 2007). Although Ig III was initially modeled on the basis of Ig Kol—the best template available at the time—(de Nobel et al. 1996), FUGUE threading of Ig III sequences against the current protein fold database identifies the ZP-N Ig subtype as the top hit for this domain (supplementary fig. S1, Supplementary Material online), a prediction supported by I-TASSER (Roy et al. 2010). Most importantly, our ZP-N-based model of S. cerevisiae mating protein α-agglutinin/Sag1p Ig III is not only physically realistic (supplementary fig. S4, Supplementary Material online) but also completely consistent with a large amount of available biochemical data (fig. 3). Specifically, the model agrees with circular dichroism spectroscopy studies of the N-terminal half of α-agglutinin (Chen et al. 1995); accounts for the experimentally determined disulfide bond between C202 and C300 (Chen et al. 1995), which corresponds to the canonical Cys1–4 disulfide of the ZP-N fold; predicts burial of C227 and C256 (Cys2,3) (Chen et al. 1995); and is consistent with exposure of residues that were shown experimentally to be accessible to proteases (Chen et al. 1995), glycosylated (Chen et al. 1995), or involved in binding to a-agglutinin (Cappellaro et al. 1991; de Nobel et al. 1996). Moreover, Y270 of α-agglutinin is positioned in correspondence of the conserved F-strand Tyr that lies next to invariant Cys4 within the E'-F-G extension of the ZP-N fold (Monné et al. 2008; Han et al. 2010). Taken together, these considerations suggest that this particular type of Ig-like domain may not be restricted to multicellular eukaryotes as previously thought but also exist in specialized extracellular proteins of yeast that play key roles in mating (S. cerevisiae Sag1p) or adhesion to human tissues and biofilm formation (C. albicans Als1p and Als3p).
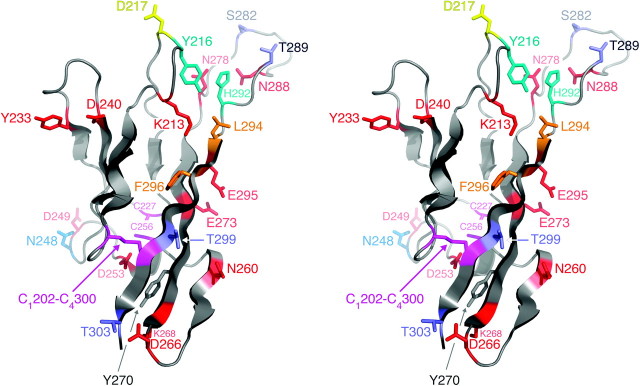
FIG. 3.
Stereograph of the model of Saccharomyces cerevisiae α-agglutinin/Sag1p Ig III. Conserved Cys residues: magenta; protease-accessible residues: red; N-glycosylated residues: light blue; O-glycosylated residues: violet; residues that are essential, important, or play a minor role in binding to a-agglutinin: cyan, orange, and yellow, respectively; and Y270: dark gray.
Conclusions and Functional Implications
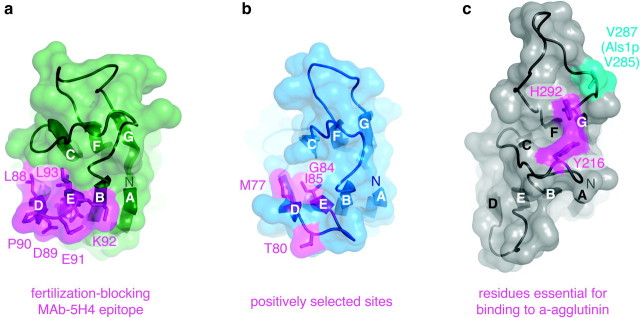
Although the rapid evolution of reproductive proteins makes the direct comparison of their sequences generally uninformative, relationships between these molecules could in principle be recognized by identifying suitably intermediate sequences that connect them (Park et al. 1997) or relying on conservation of common higher-order structural features. In this report, we have combined these approaches to detect unexpected structural similarities between reproductive proteins from both vertebrates and invertebrates, as well as yeast mating proteins. These findings suggest that some of the molecular features that regulate sexual interaction may be much more conserved during evolution than previously appreciated (fig. 1 and supplementary fig. S5, Supplementary Material Online). In this regard, it is particularly remarkable that α-agglutinin amino acids essential for interaction with a-agglutinin (de Nobel et al. 1996) (fig. 4c) are positioned so that they are exposed on the same face of the ZP-N fold as ZP2 and VERL residues implicated in sperm binding (fig. 4a–b). Moreover, specific adherence of Candida to human endothelial and epithelial cells requires Als1p Ig III amino acids centered around V285 (Fu et al. 1998; Loza et al. 2004; Sheppard et al. 2004; Dranginis et al. 2007), a residue that is also predicted to be exposed on the same region of the ZP-N domain (fig. 4c). Because threading per se simply estimates the likeness that a known 3D fold is adopted by a given sequence profile, it does not directly address the point of whether the corresponding proteins share common ancestry or just adopt a similar tertiary structure. Although this is currently unfeasible due to lack of abalone genome sequences and absence of significant conserved syntheny between yeast and human, future identification of related sequences from additional lineages may make it possible to assess whether the similarity that we have uncovered reflects direct homology or is instead the result of convergent evolution. Nevertheless, considering the widespread distribution of the Ig fold, it is striking that reproductive protein sequences from mollusc and yeast specifically match its ZP-N variant, repeats of which had previously only been detected in vertebrate egg coat proteins.
FIG. 4.
Mapping of functionally important residues on the homology models of human ZP2 repeat 1 ZP-N (a), mollusc VERL repeat 1 ZP-N (b), and yeast α-agglutinin/Sag1p Ig III ZP-N (c). Saccharomyces cerevisiae Sag1p Ig III residue V287, corresponding to functionally crucial residue V285 of Candida albicans Als1p, is also indicated in (c). A top view of the proteins is shown, with N termini and Ig fold β-strands marked by uppercase letters.
Supplementary Material
Supplementary methods and figures S1–S5 are available at Molecular Biology and Evolution online (http://mbe.oxfordjournals.org/).
Acknowledgments
This work was supported by National Institute of Health (NIH) Grants HD057974, HD042563, and HD 054631 (W.J.S.); NIH Grant HD12986 (V.D.V.); the Center for Biosciences, Swedish Research Council grant 2009-5193, an EMBO Young Investigator award, and the European Research Council under the European Union's Seventh Framework Program (FP7/2007–2013)/ERC grant agreement 260759 (L.J.). We thank Tsukasa Matsuda, Stevan Springer, and members of our laboratories for comments and discussions.
References
- Aagaard JE, Vacquier VD, MacCoss MJ, Swanson WJ. ZP domain proteins in the abalone egg coat include a paralog of VERL under positive selection that binds lysin and 18-kDa sperm proteins. Mol Biol Evol. 2010;27:193–203. doi: 10.1093/molbev/msp221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aagaard JE, Yi X, MacCoss MJ, Swanson WJ. Rapidly evolving zona pellucida domain proteins are a major component of the vitelline envelope of abalone eggs. Proc Natl Acad Sci U S A. 2006;103:17302–17307. doi: 10.1073/pnas.0603125103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleil JD, Beall CF, Wassarman PM. Mammalian sperm-egg interaction: fertilization of mouse eggs triggers modification of the major zona pellucida glycoprotein, ZP2. Dev Biol. 1981;86:189–197. doi: 10.1016/0012-1606(81)90329-8. [DOI] [PubMed] [Google Scholar]
- Bork P, Sander C. A large domain common to sperm receptors (Zp2 and Zp3) and TGF-β type III receptor. FEBS Lett. 1992;300:237–240. doi: 10.1016/0014-5793(92)80853-9. [DOI] [PubMed] [Google Scholar]
- Callebaut I, Mornon JP, Monget P. Isolated ZP-N domains constitute the N-terminal extensions of Zona Pellucida proteins. Bioinformatics. 2007;23:1871–1874. doi: 10.1093/bioinformatics/btm265. [DOI] [PubMed] [Google Scholar]
- Cappellaro C, Hauser K, Mrsa V, Watzele M, Watzele G, Gruber C, Tanner W. Saccharomyces cerevisiae a- and α-agglutinin: characterization of their molecular interaction. EMBO J. 1991;10:4081–4088. doi: 10.1002/j.1460-2075.1991.tb04984.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen MH, Shen ZM, Bobin S, Kahn PC, Lipke PN. Structure of Saccharomyces cerevisiae α-agglutinin. Evidence for a yeast cell wall protein with multiple immunoglobulin-like domains with atypical disulfides. J Biol Chem. 1995;270:26168–26177. doi: 10.1074/jbc.270.44.26168. [DOI] [PubMed] [Google Scholar]
- de Bakker PI, Bateman A, Burke DF, Miguel RN, Mizuguchi K, Shi J, Shirai H, Blundell TL. HOMSTRAD: adding sequence information to structure-based alignments of homologous protein families. Bioinformatics. 2001;17:748–749. doi: 10.1093/bioinformatics/17.8.748. [DOI] [PubMed] [Google Scholar]
- de Nobel H, Lipke PN, Kurjan J. Identification of a ligand-binding site in an immunoglobulin fold domain of the Saccharomyces cerevisiae adhesion protein α-agglutinin. Mol Biol Cell. 1996;7:143–153. doi: 10.1091/mbc.7.1.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dranginis AM, Rauceo JM, Coronado JE, Lipke PN. A biochemical guide to yeast adhesins: glycoproteins for social and antisocial occasions. Microbiol Mol Biol Rev. 2007;71:282–294. doi: 10.1128/MMBR.00037-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fu Y, Rieg G, Fonzi WA, Belanger PH, Edwards JEJ, Filler SG. Expression of the Candida albicans gene ALS1 in Saccharomyces cerevisiae induces adherence to endothelial and epithelial cells. Infect Immun. 1998;66:1783–1786. doi: 10.1128/iai.66.4.1783-1786.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gahlay G, Gauthier L, Baibakov B, Epifano O, Dean J. Gamete recognition in mice depends on the cleavage status of an egg's zona pellucida protein. Science. 2010;329:216–219. doi: 10.1126/science.1188178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Galindo BE, Moy GW, Swanson WJ, Vacquier VD. Full-length sequence of VERL, the egg vitelline envelope receptor for abalone sperm lysin. Gene. 2002;288:111–117. doi: 10.1016/s0378-1119(02)00459-6. [DOI] [PubMed] [Google Scholar]
- Han L, Monné M, Okumura H, Schwend T, Cherry AL, Flot D, Matsuda T, Jovine L. Insights into egg coat assembly and egg-sperm interaction from the X-ray structure of full-length ZP3. Cell. 2010;143:404–415. doi: 10.1016/j.cell.2010.09.041. [DOI] [PubMed] [Google Scholar]
- Jovine L, Qi H, Williams Z, Litscher E, Wassarman PM. The ZP domain is a conserved module for polymerization of extracellular proteins. Nat Cell Biol. 2002;4:457–461. doi: 10.1038/ncb802. [DOI] [PubMed] [Google Scholar]
- Jovine L, Qi H, Williams Z, Litscher ES, Wassarman PM. A duplicated motif controls assembly of zona pellucida domain proteins. Proc Natl Acad Sci U S A. 2004;101:5922–5927. doi: 10.1073/pnas.0401600101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Letunic I, Doerks T, Bork P. SMART 6: recent updates and new developments. Nucleic Acids Res. 2009;37:D229–D232. doi: 10.1093/nar/gkn808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loza L, Fu Y, Ibrahim AS, Sheppard DC, Filler SG, Edwards JEJ. Functional analysis of the Candida albicans ALS1 gene product. Yeast. 2004;21:473–482. doi: 10.1002/yea.1111. [DOI] [PubMed] [Google Scholar]
- Monné M, Han L, Schwend T, Burendahl S, Jovine L. Crystal structure of the ZP-N domain of ZP3 reveals the core fold of animal egg coats. Nature. 2008;456:653–657. doi: 10.1038/nature07599. [DOI] [PubMed] [Google Scholar]
- Park J, Teichmann SA, Hubbard T, Chothia C. Intermediate sequences increase the detection of homology between sequences. J Mol Biol. 1997;273:349–354. doi: 10.1006/jmbi.1997.1288. [DOI] [PubMed] [Google Scholar]
- Roy A, Kucukural A, Zhang Y. I-TASSER: a unified platform for automated protein structure and function prediction. Nat Protoc. 2010;5:725–738. doi: 10.1038/nprot.2010.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sheppard DC, Yeaman MR, Welch WH, Phan QT, Fu Y, Ibrahim AS, Filler SG, Zhang M, Waring AJ, Edwards JEJ. Functional and structural diversity in the Als protein family of Candida albicans. J Biol Chem. 2004;279:30480–30489. doi: 10.1074/jbc.M401929200. [DOI] [PubMed] [Google Scholar]
- Shi J, Blundell TL, Mizuguchi K. FUGUE: sequence-structure homology recognition using environment-specific substitution tables and structure-dependent gap penalties. J Mol Biol. 2001;310:243–257. doi: 10.1006/jmbi.2001.4762. [DOI] [PubMed] [Google Scholar]
- Swanson WJ, Vacquier VD. The abalone egg vitelline envelope receptor for sperm lysin is a giant multivalent molecule. Proc Natl Acad Sci U S A. 1997;94:6724–6729. doi: 10.1073/pnas.94.13.6724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swanson WJ, Vacquier VD. The rapid evolution of reproductive proteins. Nat Rev Genet. 2002;3:137–144. doi: 10.1038/nrg733. [DOI] [PubMed] [Google Scholar]
- Tsubamoto H, Hasegawa A, Nakata Y, Naito S, Yamasaki N, Koyama K. Expression of recombinant human zona pellucida protein 2 and its binding capacity to spermatozoa. Biol Reprod. 1999;61:1649–1654. doi: 10.1095/biolreprod61.6.1649. [DOI] [PubMed] [Google Scholar]
- Wassarman PM, Litscher ES. Mammalian fertilization: the egg's multifunctional zona pellucida. Int J Dev Biol. 2008;52:665–676. doi: 10.1387/ijdb.072524pw. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.