1. Introduction
The sequencing of the human genetic code (DNA base pair sequence) has led to the discovery of genes involved with various human diseases [1]. However, for most of the complex common diseases, the genes identified account for only a small effect in the vulnerability of developing them (i.e., addiction, heart disease, diabetes, depression, cancer, Parkinson’s disease). This is likely to reflect the importance of the interactions between environmental and genetic factors in the development of complex human diseases.
It is now widely accepted that epigenetics (regulation of gene activity that is not dependent on DNA sequence) is just as critical as DNA to the healthy development of organisms. Epigenetic processes, such as DNA methylation and histone modifications (e.g. acetylation, methylation, phosphorylation and ubiquitination) play a crucial role in gene expression across species [2]. The epigenetic modifications of the histone proteins of chromatin can have significant impact on most major cell functions including DNA repair, cell growth, differentiation and apoptosis. Of these epigenetic processes, the pattern of histone acetylation, regulated by the enzymes histone deacetylases (HDACs) and histone acetyl transferases (HATs), has been most intensively studied [3–5]. Indeed epigenetic research in cancer has led to new therapeutic approaches (i.e histone deacetylase inhibitors) and to biomarkers to predict treatment response [6]. Though current interest in HDAC as therapeutic targets in cancers, neurodegenerative diseases and inflammation is great, much is not known about the mechanisms that underlie these processes and how they are affected by aging, disease, drugs or other environmental exposures.
The 18 identified isoforms of HDAC have been broadly divided into two groups, the Zn2+ dependent protease and the NAD+ dependent (class III) [7–9]. The Zn2+ dependent protease group has been further divided into three classes: class I (HDAC -1, -2, -3 and -8), class II (HDAC -4, -5, -6, -7, -9 and -10) and class IV (HDAC -11). The class I and II groups are therapeutic targets for treatment of cancer and other diseases [10–14]. Class I HDACs are localized mainly in the nucleus while class II enzymes are larger in size and shuttle between the nucleus and cytoplasm. Tissue expression of class I and II HDACs is reported to be generally low, with significant variation within tissue types but with little differences in expression pattern between normal and malignant tissue [15]. More recently an atlas of HDACs 1–11 throughout more than 50 regions of the rat brain was reported [16].
In addition to histone proteins endogenous substrates for HDACs include several non-histone proteins such as transcription factors (eg p53, E2F1, GATA1, Re1A, YY1, and hormone receptors), α-tubulin, nuclear import protein importin-α7, signal transduction protein β-catein, DNA repair enzymes Ku70 and WRN and the heat shock protein 90 (hsp90) [17–18]. The deacetylation of these non-histone proteins affects their activity, suggesting that HDACs exert a much broader range of activities within the cell than control of gene transcription [19].
Much effort has been focused on identifying new HDAC inhibitors (HDIs), but few isoform-specific ligands have been identified thus far [20–22]. Studies on several nonspecific HDIs have advanced to clinical trials with one inhibitor, suberanilohydroxamic acid (SAHA) already approved for treatment of cutaneous T-cell lymphoma [23]. In addition to inhibiting class I and II HDACs, SAHA was recently shown to possess no inhibitory activity to a number of kinases and phosphatases but shown to inhibit the zinc dependent metalloproteases neprilysin and ACE (angiotensin-converting enzyme), albeit with lower potency [24].
A greater understanding of the biology and pharmacology of the different HDAC isoforms is needed to better guide preclinical and clinical investigations. In addition to insufficient pharmacological knowledge there are also few tools for accessing the efficacy of these inhibitors in vivo. Imaging the expression/availability/activity of HDAC, using positron emission tomography (PET), may provide a complementary method for determining inhibitor action, monitoring therapeutic efficacy and may also provide insight on the pharmacokinetics (PK) and biodistribution of some of the available HDAC inhibitors. PET imaging may also provide the opportunity to track and quantify the levels of HDAC non-invasively in human tissues as a function of age, disease, drug use or other environmental exposures.
Recently 6-([18F]fluoroacetamido)-1-hexanoicanilide ([18F]FAHA) was reported as a potential PET imaging agent and substrate for HDAC with utility for imaging whole body HDAC expression and tumor activity [25]. In PET studies using rats implanted with carcinoma cells, uptake in tumor xenografts was reported to peak at 30 min at 0.43%ID/g [26]. High HDAC activity was observed in tumors as indicated by tumor to muscle ratios of 2.2–2.4 between 30–60 min post-injection. The same group also reported HDAC activity in the rat brain, using [18F]FAHA, as evidenced by a peak uptake of 0.44 % ID/g [27]. In both rodent studies SAHA was shown to inhibit radiotracer uptake.
[18F]FAHA is the first reported PET radiotracer with potential for monitoring an epigenetic process in vivo. The present study attempts to assess the feasibility of using [18F]FAHA for imaging HDAC in the baboon brain and to determine its distribution and pharmacokinetics in peripheral organs. While [18F]FAHA appears promising in rodent PET studies, no characterization of its in vitro binding and specificity has been reported. Given its similarity to the acetylated side chain of lysine, we anticipated that FAHA may be a substrate for HDACs. If so, [18F]FAHA in vivo would be cleaved resulting in a labeled metabolite, [18F]fluoroacetate ([18F]FAC), which has the been shown to cross the blood brain barrier (BBB) and have selective uptake in glial cells [28,29]. The metabolism of [18F]FAHA to [18F]FAC in the brain and periphery could complicate interpretation of PET data depending on the rate of decetylation of [18F]FAHA and the uptake and distribution of its metabolite [18F]FAC.
In order to determine the contribution of [18F]FAC to [18F]FAHA imaging studies, we performed the following supporting measurements: 1. the rate of peripheral metabolism of [18F]FAHA was assessed by analysis of arterial plasma in baboon studies; 2. the biodistribution of [18F]FAC was determined to access the degree of brain uptake; 3. rodent/microPET studies of [18F]FAHA were carried out to determine the PK in the rodent brain and the chemical form of F-18 in the brain evaluated by ex vivo analysis of brain tissue; 4. in vitro whole blood and plasma incubation studies were used to investigate [18F]FAHA metabolism ex vivo and 5. in vitro distribution and stability studies were also performed in two cell lines to determine if [18F]FAC is formed intracellularly and whether it diffuses out of the cell.
2. Experimental procedures
2.1. Reagents and instrumentation
Commercially available reagents and solvents were used and, where necessary, solvents were further dried according to procedures outlined in Purification of Laboratory Chemicals [30]. BNL’s EBCO cyclotron was used to produce [18F]fluoride, by proton irradiation of 95% enriched [18O]water. A Capintec CRC-712MV radioisotope calibrator (Capintec Inc., Ramsey, NJ) was used for larger radioactivity measurements while small radioactive samples were measured in a Packard MINAXI γ 5000 automated gamma counter. Semipreparative and analytical high performance liquid chromatography (HPLC) was performed using a Knauer HPLC system equipped with a model K-1001 pump, a NaI radioactivity detector, a Hewlett Packard 3390A integrator, and a SRI Peak Simple Integration System. For semipreparative HPLC a model 87 variable wavelength monitor was used while a Knauer model K2501 UV detector was used for analytical HPLC. For thin layer chromatography (TLC) analyses, Macherey-Nagel (MN) polygram sil G/UV254 plastic-back TLC plates and a Bioscan System 200 imaging scanner were used.
2.2. Chemistry
FAHA and its precursor 6-(bromoacetamido)-1-hexanoicanilide were synthesized following literature procedures [25]. The precursor molecule for the [18F]FAC synthesis, ethyl O-mesylglycolate, and the HDAC inhibitor, SAHA, were also prepared using reported procedures [29,31].
2.3. Radiosynthesis
The radiosynthesis of [18F]FAHA was achieved using procedures similar to those reported in the literature [25]: [18F]fluoride was prepared and isolated according to published procedures [32]. The isolated [18F]fluoride was delivered in an acetonitrile (3 ml)/aqueous K2CO3(1 ml: 1 mg K2CO3 in 1 ml water) mixture to a reaction vessel containing kryptofix 2.2.2. (1 mg) and aqueous K2CO3 (50 μl of a 10 mg/ml aqueous K2CO3 solution). The resulting mixture was evaporated azetropically with dry acetonitrile (ACN) at 120 °C under a stream of argon. A solution of 6-(bromoacetamido)-1-hexanoicanilide (3 mg) in dry ACN (0.5 ml) was added to the dry kryptofix mixture. The reaction vessel was sealed and then heated to 115 °C for 25 min. The reaction mixture was then cooled to room temperature and applied to a Waters C18 light Sep-Pak cartridge. The cartridge was then eluted with 30% methanol in dichloromethane (2 × 2.5 ml) and the eluent evaporated under a stream of argon at 100 °C. The resulting residue was diluted with ACN (0.4 ml) followed by the HPLC eluent mixture (1 ml). The resulting mixture was then purified on a Luna C18 semipreparative column with an HPLC eluent mixture of 30% ACN/70% aqueous ammonium formate solution (0.1 M) at a flow rate of 5 ml/min. The fraction containing the labeled tracer was collected over 3 min (7 min–10 min) and found to be contaminated with unlabeled side products. The impure labeled product solution was further purified by first removing the solvent on a rotary evaporator, redissolving the residue in HPLC solvent (1 ml) and further purifying on the same column using the same conditions. The fraction containing the labeled tracer was then collected (over 1 min, 9 min–10 min) and the solvent removed on a rotary evaporator. The residue was dissolved in saline (4 ml) and the solution filtered through an Acrodisc syringe filter (0.2 μm, supor membrane) and collected in a sterile vial for imaging studies.
[18F]FAC was prepared using modified literature procedures [29]:[18F]fluoride delivered from the cyclotron, as outlined above, was added to a reaction vessel containing kryptofix 2.2.2. (6 mg) and aqueous K2CO3 (1 mg). The resulting mixture was evaporated azetropically with dry acetonitrile (ACN) at 120 °C under a stream of argon. A solution of ethyl O-mesylglycolate (1 mg) in dry ACN (0.5 ml) was added to the dry kryptofix mixture, the reaction vessel sealed and the resulting mixture heated at 100 °C for 5 min. The reaction mixture was then cooled to room temperature, diluted with water (7 ml) and applied to an Oasis HLB-6cc cartridge. The cartridge was then rinsed with water (4 × 7 ml) to remove any unreacted fluoride. The radiolabeled intermediate was then eluted from the Oasis cartridge with ethanol (7 ml). Aqueous sodium hydroxide (0.1 M, 200 μl) was added to the eluent and the mixture concentrated under vacuum at 40 °C. The resulting mixture was neutralized with dilute HCl (0.05 M, 200 μl) then purified on a Synergi hydroRP semipreparative column with an eluent mixture of ACN/water solution (2/98) at a flow rate of 1.5 ml/min. The fraction containing the labeled tracer was then collected (over 1 min) and the solvent removed on a rotary evaporator. The residue was redissolved in saline (4 ml) and the solution filtered through an Acrodisc syringe filter (0.2 μm, supor membrane) and collected in a sterile vial.
Specific activities decay corrected back to start of drydown (SOD), were determined from the tracer activity obtained from Capintec readings and the mass of tracer (determined by the UV absorbance of the radiotracer and the use of calibration curves of unlabeled reference compound).
2.4. Quality control
The radiochemical purities of labeled FAHA and FAC were assessed by radio-TLC and radio-HPLC methods. For analysis of [18F]FAHA samples by radio-TLC, MN plates were developed using EtOAc (100%) or an ACN/H2O/NH4OH (90/9/1) mixture and scanned to give Rf’s of 0.42 or 0.77 respectively, which coincided with the unlabeled standard compound. Similarly for radio-TLC analysis of [18F]FAC MN plates were developed using an ACN/H2O/NH4OH (90/9/1) mixture and gave an Rf of 0.26. Coelution of unlabeled FAHA with the radioactive product on a Luna C18 analytical column (254 nm) was used to confirm the identity of [18F]FAHA by radio-HPLC. To confirm the identity of [18F]FAC by radio-HPLC, coelution of the unlabeled fluoroacetate with the radioactive product on a Synergi hydroRP analytical column (210 nm) was used.
2.5. Log D and plasma protein binding
Detailed procedures, used in this study, for measuring partition coefficients and the portion of radiotracer that is not bound to plasma proteins are outlined in an earlier publication [33].
2.6. Baboon PET study
2.6.1. Baboon PET imaging
All animal studies were conducted in compliance with the NIH Guide for the Care and Use of Laboratory Animals and approved by The Brookhaven National Laboratory Animal Care and Use Committee. The protocol used for brain and torso imaging was previously reported [33]. Arterial blood samples, obtained every 5 s for the first 2 min and then at longer time intervals until the end of the study, were centrifuged and the plasma counted. Selected samples were subjected to HPLC analysis to determine the fraction of [18F]FAHA (plasma analysis below). To visualize the entire torso (head to bladder) whole body scans were done at the end of brain and torso studies. For whole body scans, emission data were collected in 2D mode for 5–6 segments (depending on the length of the animal). For each segment a 12 min scanning sequence consisting of an 8 min emission scan followed by a 4 min transmission scan was used.
Two baboons were used for brain studies. For each animal, five brain studies were conducted: one baseline study with [18F]FAHA, one baseline study with [18F]FAC and three inhibition studies. For inhibition studies, SAHA was administered iv in doses of 1 mg/kg, 0.5 mg/kg or 0.25 mg/kg, 1 hour before [18F]FAHA administration. To avoid inter-animal variability, the same animal was used to compare the effects of different doses of SAHA to baseline studies. A third baboon was used for torso studies of [18F]FAHA and [18F]FAC. Animals were given at least two weeks between studies to allow recovery from anesthesia and blood sampling.
2.6.2. Plasma analysis
The % unmetabolized radiotracer in the baboon’s plasma was determined using the following protocol. Baboon plasma (~0.2 ml) sampled at various time points during the PET study was counted, added to a solution of unlabeled standard (20 μl of a 1 mg/ml solution) in acetonitrile (0.3 ml) and the resulting mixture vortexed and centrifuged. The radioactivity trapped in the precipitated protein ranged from 5% to 20% (average (85% ± 2) but the nature of this trapped radioactivity was not investigated. The supernatant was mixed with 0.3 ml water and then analyzed by HPLC (Waters bondapak C18 column with eluents acetonitrile/0.1 M ammonium formate (30/70) at 1.0 ml/min using UV (254 nm) and radio-detection. The fraction of radioactivity coeluting with the unlabeled standard, relative to the total radioactivity from the HPLC column was measured as the % of unmetabolized radiotracer. The total radioactivity eluting the column was between 92 and 102% of the total activity added to the column indicating that there was not significantly retention of radioactivity on the HPLC column. In some cases 102% recovery was obtained due to the counting efficiency of the Picker counter used.
The total radioactivity in arterial plasma for each baboon for each scan was corrected for the presence of labeled metabolites, as determined by the HPLC analysis, and used to obtain an input function for data analysis.
2.6.3. Image analysis
Emission data from the dynamic scans were corrected for attenuation and reconstructed using filtered back projection. All time frames from dynamic images taken over the 90 min scan period were summed and planes then summed in groups of two for placement of regions of interest (ROIs) on the baboon brain. ROIs (global, cerebellum, thalamus, cortical regions etc) were drawn on the summed images then projected to the dynamic images to obtain time activity curves (TACs) expressed as %injected dose/cc (decay corrected) versus time. For the baboon torso, all time frames from dynamic images taken over the 90 min scan period were summed, ROIs drawn on the summed transaxial images and projected to dynamic images to obtain decay corrected TACs.
2.6.4. Data analysis
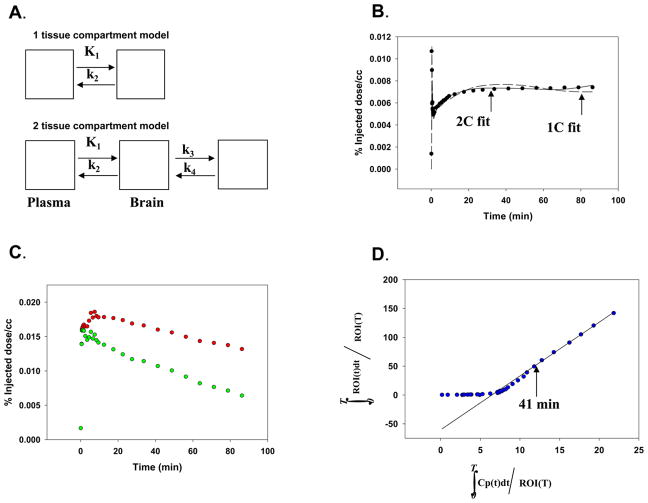
[18F]FAHA in plasma is rapidly converted to [18F]FAC which also crosses the BBB, although to a lesser extent than [18F]FAHA. [18F]FAC is a substrate for acetyl coenzyme A synthase, although the specificity is lower than for acetate [28]. In order to estimate tissue uptake of [18F]FAHA it is necessary to take into account the contribution of the labeled metabolite that originates in plasma and crosses the BBB and contributes to the total [18F] signal as measured by PET. A compartment model was used to fit the uptake and binding (and other transformations) of [18F]FAC from the studies with injected [18F]FAC. Both a 1 compartmental and a 2 compartmental model (illustrated in Fig 2a) were tested to determine which model gave the best fit.
Figure 2. Quantification of HDAC activity.
A. Models tested for uptake (both a 1 and 2 compartment model (1C) and (2C) were used to fit uptake of [18F]FAC).
B. Comparison of model fits to uptake [18F]FAC data using the 1C and 2C models.
C. Uptake in thalamus with (upper curve) and without (lower curve) [18F]FAC from plasma for one animal.
The upper curve is the radioactivity in the thalamus ROI in a baseline [18F]FAHA study. The lower curve is the radioactivity remaining after the estimated contribution from plasma [18F]FAC has been subtracted using the model parameters from the 2C fit illustrated in Fig 2b.
D. Graphical analysis of lower curve in Fig 2C.
The total distribution volume of the corrected thalamus ROI (lower curve in Fig 2c) is the slope of the line from the point labeled 41 min. Vd = 10.9 ml/min/cc.
Using the model parameters determined from these studies, the brain uptake of [18F]FAC from plasma was estimated for all other ([18F]FAHA) studies. In the [18F]FAHA studies the plasma input corresponding to [18F]FAC was assumed to be the metabolite fraction of the measured arterial plasma input function. In the [18F]FAHA studies the estimated [18F]FAC uptake from plasma was subtracted from the brain TACs. Since there appeared to be little regional variation of [18F]FAC uptake, the same [18F]FAC model parameters were used to correct all TACs in the [18F]FAHA studies. The corrected [18F]FAHA TACs and the measured arterial input function of unchanged [18F]FAHA were used to estimate a (model independent) total distribution volume, Vd, using a standard graphical method [34]. This parameter was used to compare regional uptake and uptake after dosing with SAHA.
2.7. Rodent PET study
2.7.1. Small animal imaging
Rodent brain imaging was performed using a commercial small animal PET camera (microPET R4, Concorde Microsystems, Knoxville, TN). Two Sprague-Dawley imaging experiments were performed with [18F]FAHA using the following protocol. Anesthesia was induced using a mixture of ketamine and xylazine (both from Fort Dodge, IA, USA) by intraperitoneal injections of 50 mg/kg ketamine (with 5 mg/kg xylazine) and maintained by subsequent intraperitoneal injections of the same solution at 20% of the initial dose (approximately 15 min apart). After positioning the animal in the camera (with the brain in the center of the field of view), [18F]FAHA (study 1: 0.302 mCi in 0.6 ml saline; study 2: 0.430 mCi in 0.7 ml saline) was injected through a tail vein catheter and a 3D data set was collected as a list mode file from the time of injection for 90 min. Three dimensional (3D) sinograms were converted into 2D sinograms using Fourier rebinning before image reconstruction. Data were corrected for photon scatter using the method of tailfitting of the projections [35]. Attenuation correction was not applied. Scatter-corrected sinograms were reconstructed using an iterative maximum likelihood expectation maximization (MLEM) algorithm, which with the 20 iterations employed here yields an image resolution of ~1.5 mm FWHM (Full Width at Half Maximum) at the center of the field of view. The image pixel size in MLEM reconstructed images was 0.4 mm transaxially with a 1.21 mm slice thickness. Image analysis was accomplished using AMIDE® software and manually drawn 3D ROIs.
2.7.2. Rat biodistribution
At 90 min post injection of [18F]FAHA (301.8 μCi), an anesthetized rat was killed by decapitation. Organs were carefully dissected and weighed. Ex vivo quantification of radioactivity was accomplished using a Packard MINAXI γ 5000 automated gamma counter (Packard Instrument, Meriden, CT) and is reported as %injected dose/g (% ID/g) of sample, decay corrected to the time of injection.
2.7.3. Brain homogenate analysis
Two Sprague Dawley rats were anesthetized with ketamine/xylzine (as detailed in the small animal imaging section). Each animal was catheterized in the tail vein and administered [18F]FAHA (rodent 1, 0.73 mCi; rodent 2, 0.35 mCi). After a given time (rodent 1, 7 min; rodent 2, 30 min), the animal was killed by decapitation. The brain was removed, homogenized with 1 ml of water and an aliquot of this mixture (300 μL) was diluted to 1 ml with cold (4 °C) acetonitrile. The sample was mixed by vortex for 1 min and then separated by centrifugation (2000 rcf, 2 min). The supernatant was analyzed by HPLC and TLC.
2.8. In vitro experiments
2.8.1. Plasma and whole blood assays
Arterial blood and plasma samples (0.5 ml) were obtained from one baboon immediately before pretreatment with SAHA (0.25 mg/kg) and one hour later, just prior to radiotracer administration. [18F]FAHA was added to the blood and plasma samples at room temperature and the stability of the radiotracer was monitored by radio-TLC after 10, 20 and 30 mins.
2.8.2. Cell culture and assays
Two cell lines were used for in vitro experiments: HCT 116 (human colorectal carcinoma) and SH-SY5Y cells (human neuroblastoma). Cells were cultured in medium supplemented with 10% of fetal calf serum. McCoy medium was used for the HCT 116 cells and a mixture of 50% of Dulbecco’s Modified Eagle Medium (DMEM) and 50% of Nutrient Mixture F-12 was used for the SH-5SY cells. About 2×106 of exponentially growing cells were used for each experiment. Cells were carefully detached from the plates by scrapping to ensure plasma membrane integrity, pelleted by centrifugation and resuspended in 2 ml of fresh medium. [18F]FAHA or [18F]fluoroacetate was added to the cell suspension and the mixture incubated for 15 min at 37 °C. Following incubation, cells were pelleted by centrifugation. The supernatant was separated, its radioactivity measured and analyzed by radio-TLC. The cell pellet fraction was used to separate nuclear and cytoplasmic components according to the protocol (http://www.lamondlab.com/f5nucleolarprotocol.htm) with the following modifications: the subcellular fractionation was advanced only to nuclear isolation (step 10) and vigorous vortexing was used instead of a plunger to disrupt the cell membrane. The efficiency of the modifications were confirmed by visual examination of the nuclear morphology under the microscope. The supernatant fractions from the first and the second incubations, containing the cytoplasmic fraction of the cells were combined (cytoplasm fraction). The radioactivity in both the cytoplasm and nuclear fractions was measured and subsequently analyzed by radio-TLC. For inhibition studies, cell cultures were first pretreated with SAHA (1 mg/kg) for 1 h at 37 °C then incubated with [18F]FAHA 15 min at 37 °C. Cell components were then separated and analyzed as above. The amount of the radioactivity retained per the unit of the protein in both the cytoplasm and nuclear fraction was calculated based on the measured protein concentration (determined by NanoDrop).
3. Results and discussion
3.1. Radiosynthesis
[18F]FAHA was produced in 14.7±5.3% (n=6) radiochemical yield (based on total activity at SOD) with 99.7±0.2% (n=8) radiochemical purity. Specific activities ranged from 0.11–1.0 mCi/nmol (4.1–37.0 GBq/μmol) for [18F]FAHA. The incorporation of an additional HPLC purification step in the present study as compared to the single HPLC purification step reported in the literature was required to remove UV active contaminants which were not removed by one HPLC purification step. This resulted in a longer synthesis time of ~105 min (literature time of 67–75 min [25]) for [18F]FAHA. [18F]FAC was produced in 10.5±4.1 (n=4) radiochemical yield (based on total activity at SOD) with 99.3±0.6% (n=4) radiochemical purity. Specific activities for [18F]FAC ranged from 0.06–0.2 mCi/nmol (2.2–7.4 GBq/μmol). Total synthesis time for [18F]FAC was ~75 min as compared to the literature synthesis time of ~35 min due to an added HPLC purification step. Literature procedures utilize only solid phase extraction for [18F]FAC purification [29]. We found that the reaction product after solid phase purification was contaminated by a mixture of UV active compounds thereby necessitating further purification by HPLC.
3.2. LogD and PPB
The measured logD for [18F]FAHA was 1.39±0.1 (n=2) and for [18F]FAC it was 0.70 (n=1). The low logD value for [18F]FAC is not in the optimal range for penetrating the BBB nonetheless, [18F]FAC has been reported to enter the brain [29]. The % of activity unbound to plasma proteins was determined to be 53.93±1.76 (n=6) for [18F]FAHA and 57.2 (n=1) for [18F]FAC. These high unbound percentages suggest that sufficient free radiotracer should be available for entry to the brain.
3.3. Baboon PET study
3.3.1. Baboon plasma analysis
Analysis of arterial plasma samples was undertaken to measure the % of [18F]FAHA available in the blood for entry to the brain during the study. The % unmetabolized [18F]FAHA at four time points in baseline and inhibition studies for two baboons are shown in Table 1.
Table 1.
% Unmetabolized [18F]FAHA in baboon plasma after bolus injection.
| Animal | Study | 1min | 2min | 5min | 10min |
|---|---|---|---|---|---|
| 1 | Baseline | 45 | - | 2 | 1 |
| 0.25mg/kg | 84 | 35 | 10 | 6 | |
| 0.5mg/kg | 78 | 39 | 14 | 7 | |
| 1.0mg/kg | 86 | 34 | 22 | 10 | |
| 2 | Baseline | 52 | 6 | 2 | 1 |
| 0.25mg/kg | 87 | 49 | 11 | 5 | |
| 0.5mg/kg | 78 | 50 | 14 | 6 | |
| 1.0mg/kg | 85 | - | 26 | 20 |
[18F]FAHA was rapidly metabolized in baseline studies, as evidenced by the low parent fraction in the baboon plasma at 1 min and almost complete metabolism by 5 min. Table 1 also shows a decrease the rate of metabolism of [18F]FAHA by SAHA in a dose dependent manner, suggesting that both [18F]FAHA and SAHA may act at the same target enzyme(s). Results were reproducible using different baboons.
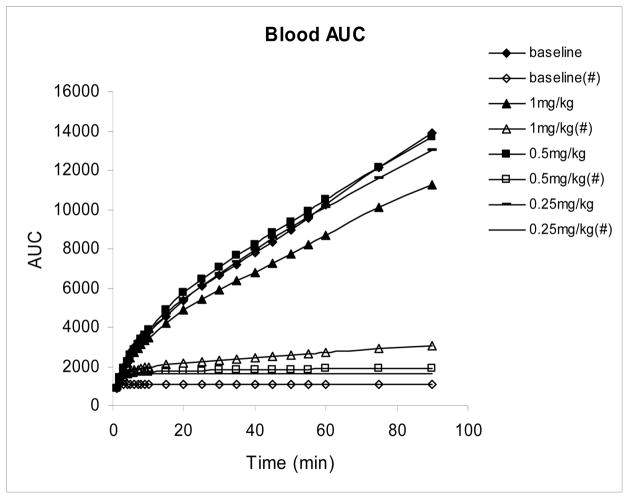
Blood curves showed fast uptake and clearance of total activity and an even more rapid decrease in the fraction of total F-18 which was present as [18F]FAHA. The plasma integral plots (figure 3) also showed the increase in unmetabolized [18F]FAHA in the baboon plasma with increasing SAHA doses again suggesting that SAHA is inhibiting the activity of the enzyme in a dose dependent fashion resulting in slower breakdown of [18F]FAHA and consequently higher blood concentrations of unmetabolized [18F]FAHA.
Figure 3.
Area under the plasma curve (plasma integrals) corrected and uncorrected for metabolites in baseline and SAHA inhibition studies using the same baboon. Plasma integral corrected for metabolites are indicated by #.
3.3.2. Pharmacokinetics
[18F]FAHA pharmacokinetics
For determining pharmacokinetics of [18F]FAHA in the brain of both animals, a baseline study and three inhibition studies, using SAHA in doses of 1.0 mg/kg, 0.5 mg/kg or 0.25 mg/kg were done. Doses of SAHA that were lower than clinical doses (> 5mg/kg well tolerated [36]) were chosen to minimize any of the reported side effects of this non-isoform specific HDAC inhibitor.
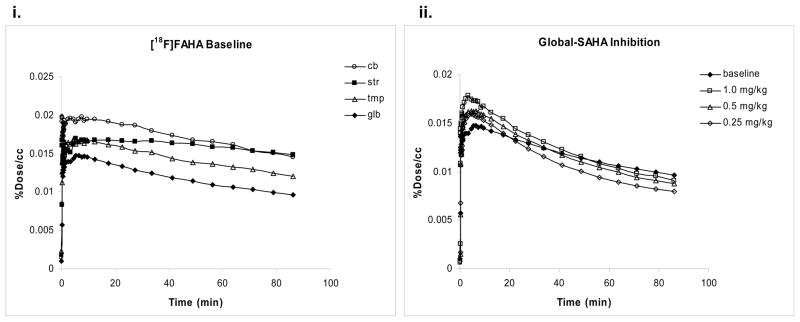
The time course of F-18 radioactivity, normalized to the injected dose, in different regions of the baboon brain is shown in figure 4i.
Figure 4.
(i) Time activity curves for [18F]FAHA in brain regions of the same baboon. (ii) Time activity curves for [18F]FAHA in the brain of the same baboon in baseline and SAHA inhibition studies.
There was similar uptake and clearance in cortical regions. The highest uptake of radioactivity was observed in the cerebellum. The varying kinetics in brain regions suggests that the target molecule(s) that binds [18F]FAHA is not uniformly distributed in the brain.
We also compared the TACs for baseline and inhibition studies in different brain regions (figure 4ii). Higher uptake of radioactivity was observed in SAHA inhibition studies as compared to baseline studies. The higher uptake may be due to higher plasma concentrations.
[18F]FAC Pharmacokinetics
[18F]FAC biodistribution in the brain was measured in the same animals used for [18F]FAHA baseline and inhibition studies. TAC plots of [18F]FAC shows gradual and uniform uptake of radioactivity in brain regions over the time course of the study (data not shown). Uniform uptake of C-14 labeled FAC in brain regions has also previously been reported [28].
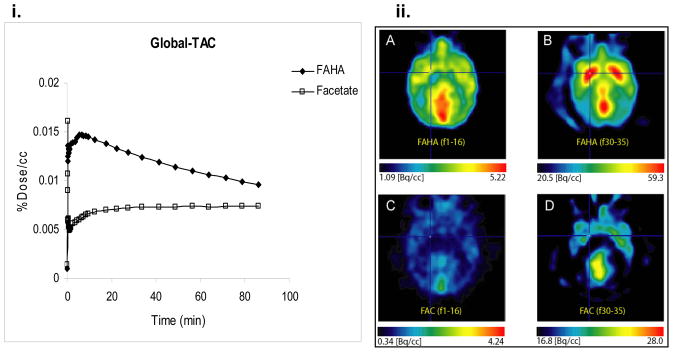
Comparison of [18F]FAHA and [18F]FAC TACs in the same baboon showed that the peak uptake of [18F]FAHA was more than five times greater than its metabolite, [18F]FAC (figure 5i). Notable differences in F-18 biodistribution during the time course of study was also observed in PET images of [18F]FAHA and [18F]FAC in the same baboon (figure 5ii).
Figure 5.
(i) Kinetics of [18F]FAHA and [18F]FAC in the brain of the same baboon. (ii) Transaxial view of PET images, from the same baboon at the level of the striatum (slice 33), after injection of [18F]FAHA (3.45 mCi) and [18F]FAC (2.40 mCi): A. [18F]FAHA PET images summed over the first 3 min of study (frames 1–16). B. [18F]FAHA PET images summed over the last 45 min of study (frames 30–35). C. [18F]FAC PET images summed over the first 3 min of study (frames 1–16). D. [18F]FAC PET images summed over the last 45 min of study (frames 30–35).
3.3.3. Peripheral distribution of [18F]FAHA
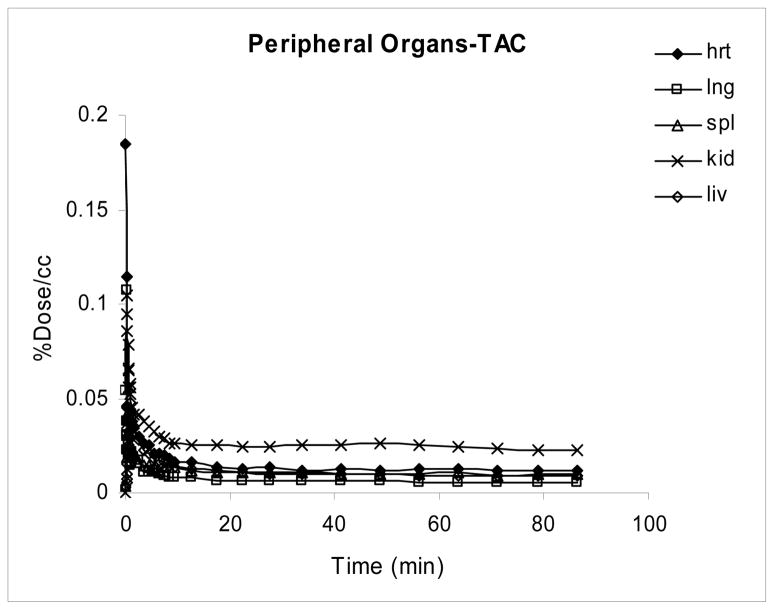
There was rapid uptake and clearance of radioactivity in all peripheral organs studied with higher retention of radioactivity in the kidney as compared to other peripheral organs (figure 6).
Figure 6.
Kinetics of [18F]FAHA in the periphery of the baboon.
A whole body scan (data not shown) showed the highest activity in the kidneys and bladder suggesting that the urinary pathway is the main excretion route. This result is consistent with literature reports, which show highest accumulation of [18F]FAC in the kidney and excretion of metabolic byproducts from the kidney to the bladder [29].
Similar to the peripheral kinetics in [18F]FAHA studies, in [18F]FAC studies there was rapid uptake and clearance of radioactivity in peripheral organs with kidneys showing greatest retention of radioactivity (data not shown).
3.3.4. Modeling results
Parameters for modeling the metabolite ([18F]FAC) uptake were fixed for all four studies with one animal (K1= 0.018 ml/min/cc; k2= 0.04 min−1; k3= 0.008 min−1; k4= .003 min−1). A comparison of a 1 compartment model with the fit from a 2 compartment model (Fig 2a) are shown in Fig 2b. Since the 2 compartment model gave a better fit to the data, it was used to correct the TACs for the presence of plasma metabolites crossing the BBB. Vd results for baseline and blocking studies for one animal (Lulu) are shown in Table 2 (a similar trend was seen for the second animal, Brooke).
Table 2.
Total distribution volume (Vd) in baseline and SAHA inhibition studies.
| Brain region | Baseline | 1mg/kg | 0.50mg/kg | 0.25mg/kg |
|---|---|---|---|---|
| str | 14.2 | 6.90 | 12.34 | 11.5 |
| cb | 13.4 | 5.53 | 9.67 | 9.71 |
| thl | 10.9 | 3.92 | 6.81 | 7.19 ) |
| tmp | 9.0 | 3.87 | 5.42 | 6.60 |
| cing | 6.76 | 3.30 | 4.28 | 5.15 |
| par | 5.4 | 2.88 | 3.74 | 4.14 |
| glb | 4.84 | 2.91 | 3.57 | 3.79 |
The model parameter used for comparison is the total distribution volume, Vd for uptake and conversion of FAHA in brain tissue was determined from graphical analysis. Parameters for modeling the metabolite (FAC) uptake were fixed for all four studies (K1= 0.018 mL/min/cc; k2= 0.04 min−1; k3= 0.008 min−1; k4= .003 min−1.) and were used to estimate the uptake of [18F]FAC from peripheral metabolism of [18F]FAHA. Results for baseline and blocking studies for one animal (similar trends in Vd were observed for the second baboon).
Even though K1 for [18F]FAC was small (~0.018 ml/min/cc), this labeled metabolite makes a significant contribution to the total uptake of radioactivity in brain tissue since [18F]FAHA is rapidly metabolized to [18F]FAC and makes a large contribution to the total plasma radioactivity after 5 min. Figure 2c illustrates the total radioactivity in a ROI from thalamus compared to the ROI with the metabolite contribution arising from plasma removed. Graphical analysis of the corrected ROI is shown in Fig 2d.
The highest activity of HDAC (as measured by Vd for [18F]FAHA) appears to be in striatum and cerebellum followed by thalamus, temporal cortex, cingulate cortex, and parietal cortex. There was significant reduction in Vd with 1 mg/kg for both studies. The average % reduction was 52 ± 8% (Lulu) and 43 ± 7% (Brooke) averaged over all 6 ROIs (excluding global). At 0.25 mg/kg the average reduction was 25 ± 8% (Lulu) and 12 ± 12% (Brooke). There was no significant difference in the average reduction in Vd between 0.5 mg/kg and 0.25 mg/kg (30± 9% and 25± 5% for 0.5 and 0.25 mg). Assumptions used in these calculations are the following: (1) The local concentrations of endogenous substrates are the same between studies, (2) There is no regional variation in [18F]FAC uptake, (3) The same model parameters govern the uptake of [18F]FAC in the presence of the blocking drug and (4) All of the metabolite fraction of [18F]FAHA is due to [18F]FAC.
3.4. Rat biodistribution studies
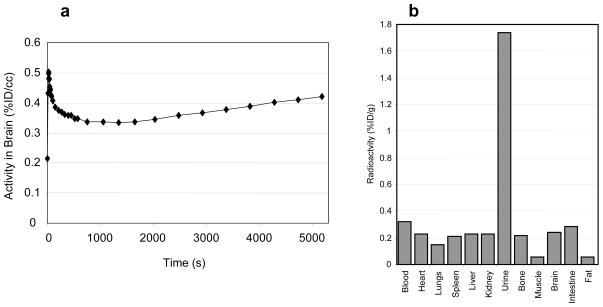
The distribution of radioactivity in the rodent was also studied using [18F]FAHA. The time course of radioactivity in the rodent brain and the distribution of radioactivity in various organs of the body are shown in figure 7.
Figure 7.
(a) Time activity curve for [18F]FAHA in the rodent brain. (b) Biodistribution of radioactivity in Sprague-Dawley rat 90 min after injection of [18F]FAHA.
Radioactivity in the bone suggests that there was some defluorination in vivo. The highest activity was observed in the urine, again indicative of the urinary route being the major excretory pathway.
TLC analysis of brain homogenate samples at 7 min and 60 min determined that no [18F]FAHA was present. Instead one radiopeak with an Rf similar to that of fluoroacetate was observed, suggesting [18F]FAHA is completely converted to [18F]FAC in the rodent brain by 7 min.
3.5. In vitro studies
3.5.1. In vitro blood and plasma analyses
Analysis of whole blood and isolated plasma (minus blood cells) samples incubated with [18F]FAHA were undertaken to determine whether [18F]FAHA was metabolized by plasma proteins or by blood cells and to assess the rate of metabolism of [18F]FAHA in the blood and plasma ex vivo.
Rapid metabolism of [18F]FAHA to [18F]FAC was observed in the blood but not in isolated plasma suggesting that metabolism is mediated by blood cell and not by plasma proteins (Table 3).
Table 3.
% Unmetabolized [18F]FAHA after incubation in baboon plasma in vitro as determined by radio-TLC analysis.
| Time | Blood-pre | Blood-post | Plasma-pre | Plasma-post |
|---|---|---|---|---|
| 10 | 13 | 16 | 98 | 96 |
| 20 | 0 | 0 | 94 | 96 |
| 30 | 0 | 0 | 94 | 94 |
pre: before SAHA pretreatment; post: 1h after SAHA administration
This rapid metabolism in the blood ex vivo implies that the rapid clearance of [18F]FAHA observed in the baboon plasma during PET imaging studies was not solely due to excretion.
3.5.2. In vitro cell studies
In vitro blood and plasma analyses detailed above indicated that blood cells and not plasma proteins were responsible for metabolism of [18F]FAHA in the blood. To corroborate this result and to gain further insights into the metabolism of [18F]FAHA, in vitro cell assays were performed to investigate: 1. whether [18F]FAHA permeates the plasma membrane of cells; 2. whether [18F]FAC formed intracellularly by the metabolism of [18F]FAHA diffuses out of the cell or remains trapped within the cell; 3. how the rate of metabolism of [18F]FAHA within cells is affected by SAHA and 4. the quantitative distribution of radioactivity between cellular compartments.
For these experiments we used two cell lines originating from different tissues: HCT 116 cells (human carcinoma) and SH-SY5Y (neuroblastoma). The HCT 116 cell line was chosen for study as this cell line is widely employed as a model to study the role of HDAC and its modifications in cancer [37]. SH-SY5Y cells were also used as they are representative of cells of neuronal origin and feature distinct morphological characteristics of such cells.
Analysis of the incubation medium and cell components by TLC revealed that [18F]FAHA does indeed enter the cell and is distributed between the cytoplasmic and nuclear compartments. The finding from the animal studies and in vitro blood studies that pretreatment of SAHA slowed the metabolism of [18F]FAHA prompted us to investigate this inhibition in the cell culture assay as well. Pretreatment with SAHA was also shown to retard the rate of [18F]FAHA degradation (Table 4).
Table 4.
[18F]FAHA:[18F]fluoroacetate:[18F]unidentified-polar ratio in the cellular components of HCT and SH-SY5Y cells, untreated and pretreated with SAHA, after 15 min incubation with [18F]-FAHA.
| Cell component | HCT | HCT# | SH-SY5Y | SH-SY5Y# |
|---|---|---|---|---|
| Medium | 83:17:0 | 95:3:2 | 88:12:0 | 95:3:2 |
| Cytoplasm | 64:31:5 | 91:6:3 | 72:17:11 | 96:2:2 |
| Nucleus | 0:80:20 | 22:37:41 | 0:46:54 | 60:30:10 |
-cells pretreated with SAHA 1h before incubation with radioactivity
The detection of [18F]FAC in the medium suggests diffusion of metabolic products out of the cell. To confirm that [18F]FAHA metabolism was occurring inside of the cells and not by some component(s) in the medium we used conditional medium as an additional control. [18F]FAHA was incubated in medium alone and found to be stable up to 2 h with no detection of [18F]FAC. These results indicate that [18F]FAHA enters the cell where it is broken down to [18F]FAC, which subsequently diffuses out of the cell into the medium.
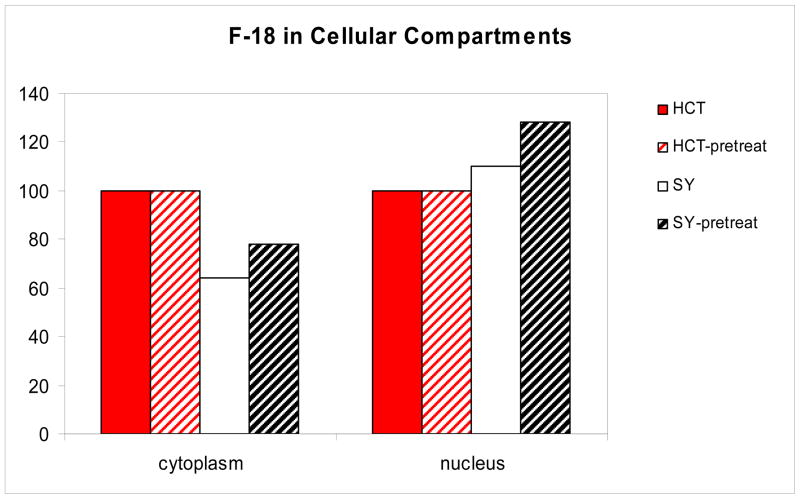
Measurements of the radioactivity in the cytoplasm and nuclear fractions of both cell types revealed higher radioactivity in the cytoplasm as compared to the nucleus but little difference between the cell types. The HCT 116 cells and SH-SY5Y cells have very different morphology: the nuclei of the SH-SY5Y cells are much smaller in comparison to the cytoplasm. To assess whether [18F]FAHA binding is influenced by the types of proteins expressed in cells of different origin, we measured the protein concentration in each fraction and calculated the amount of radioactivity bound per unit of the protein (mg). For comparative purposes the amount of radioactivity in the nuclear fractions of the HCT 116 cells were arbitrarily set to 100 and the amount of radioactivity in the SH-SY5Y nuclear fractions reported relative to this set point (figure 8). The radioactivity distribution in the two cell types suggests that there is greater retention of radioactivity by the proteins of the nucleus of the cells of neuronal origin (SH-SY5Y) as compared to the nucleus of epidermal (HCT 116) cells.
Figure 8.
Distribution of F-18 normalized to protein mass. Radioactivity in the nuclear fractions of the HCT 116 cells were arbitrarily set to 100 and the radioactivity in the SH-SY5Y nuclear fractions reported relative to this set point.
4. Conclusions
The studies described in this article show that [18F]FAHA enters the brain and shows different kinetics in various brain regions. We found no published measurements of the affinity of FAHA for HDAC or its specificity for this class of enzymes. However, inhibition studies using the known HDAC inhibitor SAHA indicate that [18F]FAHA interacts with the same biological target as SAHA. This together with the structural similarity between FAHA and the endogenous HDAC substrate is strong evidence that FAHA may indeed be a substrate for HDAC. However, our studies indicate that the practical utilization of [18F]FAHA for quantitative imaging of HDAC in the baboon brain (and presumably the human brain) may be confounded by its rapid metabolism to [18F]FAC in cells and in blood and the fact that plasma-borne [18F]FAC accumulates in the brain. We have calculated a total distribution volume for [18F]FAHA by estimating the uptake of [18F]FAC and subtracting it from the total tissue uptake. Also our in vitro cell studies indicate that [18F]FAC produced in the brain may not remain localized to the point of deacetylation of [18F]FAHA. We were able to analyze the data using a model independent graphical technique appropriate for nontrapped radioligands, showing a dose-dependent blocking effect with SAHA. These results however depend upon the validity of the assumptions set forth in the modeling results section 3.3.4. Validating the nature of this interaction as HDAC-specific as well as the determination of the affinity of FAHA for HDAC will require additional studies.
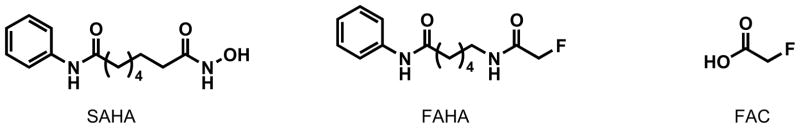
Figure 1.
Chemical structures of SAHA, unlabeled FAHA and FAC.
Acknowledgments
This work was supported by the US Department of Energy’s Office of Biological and Environmental Research and by the National Institutes of Health-National Institute on Alcohol Abuse and Alcoholism Intramural Research Program. Special thanks to Kwesi Amoa and Frank Ogero of Medgar Evers College for help in chemical synthesis and Michael Schueller, Lisa Muench, Pauline Carter, Payton King and Donald Warner of the PET Imaging Group at Brookhaven National Laboratory for technical support.
References
- 1.Peltonen L, McKusick VA. Dissecting human disease in the postgenomic era. Science. 2001;291:1224–1229. doi: 10.1126/science.291.5507.1224. [DOI] [PubMed] [Google Scholar]
- 2.Egger G, Liang G, Aparicio A, Jones P. Epigenetics in human diseases and prospects for epigenetic therapy. Nature. 2004;429:457–463. doi: 10.1038/nature02625. [DOI] [PubMed] [Google Scholar]
- 3.Yang XJ, Seto E. HATs and HDAC: from structure, function and regulation to novel strategies for therapy and prevention. Oncogene. 2007;26:5310–5318. doi: 10.1038/sj.onc.1210599. [DOI] [PubMed] [Google Scholar]
- 4.Lehrmann H, Pritchard L, Harel-Bellan Histone acetyltransferases and deacetylases in the control of cell proliferation and differentiation. Adv Cancer Res. 2002;86:41–65. doi: 10.1016/s0065-230x(02)86002-x. [DOI] [PubMed] [Google Scholar]
- 5.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 6.Weidman JR, Dolinoy DC, Murphy SK, Jirtle RL. Cancer susceptibility: epigenetic manifestation of environmental exposures. Cancer J. 2007;13:9–16. doi: 10.1097/PPO.0b013e31803c71f2. [DOI] [PubMed] [Google Scholar]
- 7.De Ruijter AJ, van Gennip AH, Caron HN, Kemp S, van Kuilenburg AB. Histone deacetylases (HDACs): characterization of the classical HDAC family. Biochem J. 2003;370:737–749. doi: 10.1042/BJ20021321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Gray SG, Ekstrom TJ. The human histone deacetylase family. Exp Cell Res. 2001;262:75–83. doi: 10.1006/excr.2000.5080. [DOI] [PubMed] [Google Scholar]
- 9.Voelter-Mahlknecht S, Ho AD, Mahlknecht U. Chromosomal organization and location of the novel class IV human histone deacetylase 11 gene. Int J Mol Med. 2005;16:589–598. [PubMed] [Google Scholar]
- 10.Pan L, Lu J, Huang B. HDAC Inhibitors: A potential new category of anti-tumor agents. Cell Mol Immun. 2007;4:337–343. [PubMed] [Google Scholar]
- 11.Liu T, Kuljaca S, Tee A, Marshall G. Histone deacetylase inhibitors: Multifunctional anticancer agents. Cancer Treat Rev. 2006;32:157–165. doi: 10.1016/j.ctrv.2005.12.006. [DOI] [PubMed] [Google Scholar]
- 12.Minucci S, Pelicci PG. Histone deacetylase inhibitors and the promise of epigenetic (and more) treatments for cancer. Nature. 2006;6:38–51. doi: 10.1038/nrc1779. [DOI] [PubMed] [Google Scholar]
- 13.Kouraklis G, Misiakos EP, Theocharis S. Histone deacetylase inhibitors as a potential therapeutic agent for cancer treatment. Targ Oncol. 2006;1:34–41. [Google Scholar]
- 14.Blanchard F, Chipoy C. Histone deacetylase inhibitors: new drugs for the treatment of inflammatory diseases? Drug Discov Today. 2005;10:197–204. doi: 10.1016/S1359-6446(04)03309-4. [DOI] [PubMed] [Google Scholar]
- 15.De Ruijter A, van Gennip A, Caron H, Kemp S, Van Kuilenburg A. Histone deacetylases (HDACs): characterization of the classical HDAC family. Biochem J. 2003;370:737–749. doi: 10.1042/BJ20021321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Broide R, Redwine J, Aftahi N, Young W, Bloom F, Winrow C. Distribution of histone deacetylases 1–11 in the rat brain. J Mol Neurosci. 2007;31:47–58. doi: 10.1007/BF02686117. [DOI] [PubMed] [Google Scholar]
- 17.Bhalla K. Epigenetic and chromatin modifiers as targeted therapy of hematologic malignancies. J clin Onocol. 2005;23:3971–3993. doi: 10.1200/JCO.2005.16.600. [DOI] [PubMed] [Google Scholar]
- 18.Kazantsev A, Thompson L. Therapeutic application of histone deacetylase inhibitors for central nervous system disorders. Nature Rev Drug Discov. 2008;7:854–868. doi: 10.1038/nrd2681. [DOI] [PubMed] [Google Scholar]
- 19.Waltregny D, de Leval, Glenisson W, et al. Expression of histone deacetylase 8, a class 1 histone deacetylase, is restricted to cells showing smooth muscle differentiation in normal human tissues. Am J Path. 2004;165:552–564. doi: 10.1016/S0002-9440(10)63320-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Acharaya MR, Sparreboom A, Venitz J, Figg W. Rational development of histone deacetylase inhibitors as anticancer agents: A review. Mol Pharmacol. 2005;68:917–932. doi: 10.1124/mol.105.014167. [DOI] [PubMed] [Google Scholar]
- 21.Paris M, Porcelloni M, Binaschi M, Fattori D. Histone deacetylase inhibitors: From bench to clinic. J Med Chem. 2008;51:1505–1528. doi: 10.1021/jm7011408. [DOI] [PubMed] [Google Scholar]
- 22.Miller TA, Witter DJ, Belvedere S. Histone deacetylase inhibitors. J Med Chem. 2003;46:5097–5116. doi: 10.1021/jm0303094. [DOI] [PubMed] [Google Scholar]
- 23.Glaser KB. HDAC inhibitors: Clinical update and mechanism-based potential. Biochem Pharmacol. 2007;74:659–671. doi: 10.1016/j.bcp.2007.04.007. [DOI] [PubMed] [Google Scholar]
- 24.Blackwell L, Norris J, Suto CM, Janzen WP. The use of diversity profiling to characterize chemical modulators of the histone deacetylases. Life Sciences. 2008;82:1050–1058. doi: 10.1016/j.lfs.2008.03.004. [DOI] [PubMed] [Google Scholar]
- 25.Mukhopadhyay U, Tong W, Gelovani J, Alauddin M. Radiosynthesis of 6- ([18F]fluoroacetamido)-1-hexanoicanilide ([18F]FAHA) for PET imaging of histone deacetylase (HDAC) J Label Comp Radiopharm. 2006;49:997–1006. [Google Scholar]
- 26.Nishii R, Mukhopadhyay U, Yeh H, Sohomonyan S, Volgin A, Alauddin M, Tong W, Gelovani J. Non-invasive imaging of histone deacetylase activity in human breast carcinoma xenografts in rats using positron emission tomography (PET) with [18F]FAHA. J Nucl Med. 2007;48S2:34. [Google Scholar]
- 27.Nishii R, Mukhopadhyay U, Yeh H, Sohomonyan S, Volgin A, Alauddin M, Tong W, Gelovani J. PET imaging of histone deacetylase activity in rat brain using 6-([18F]fluoroacetamide)-1-hexanoicanilide ([18F]FAHA) J Nucl Med. 2007;48S2:336. [Google Scholar]
- 28.Lear JL, Ackerman RF. Evaluation of radiolabeled acetate and fluoroacetate as potential tracers of cerebral oxidative metabolism. Metab Brain Dis. 1990;5:45–56. doi: 10.1007/BF00996977. [DOI] [PubMed] [Google Scholar]
- 29.Ponde DE, Dence CS, Oyama N, Kim J, Tai YC, Laforest R, Siegel BA, Welch MJ. 18F-Fluoroacetate: A Potential Acetate Anaolg for Prostate tumor Imaging-In vivo evaluation of 18F-Fluoroacetate Versus 11C-Acetate. J Nucl Med. 2007;48:420–428. [PubMed] [Google Scholar]
- 30.Perrin DD, Armarego WL. Purification of Laboratory Chemicals. 3. New York: Pergamon Press Inc; 1988. [Google Scholar]
- 31.Soli ED, Braun MP. Synthesis of [phenyl-U-14C]aryl and [8–14C]carboxy labeled tracers of vorinostst. J Label Comp Radiopharm. 2006;49:437–443. [Google Scholar]
- 32.Schueller M, Ferrieri R, Schlyer D. An automated system for oxygen-18 water recovery and fluorine-18 delivery. Nucl Instrum Methods Phys B. 2005;241:660–664. [Google Scholar]
- 33.Reid AE, Ding YS, Eckelman WC, Logan JJ, Alexoff D, Shea C, Xu Y, Fowler JS. Comparison of the Pharmacokinetics of Different Analogs of 11C-Labeled TZTP for Imaging Muscarinic M2 Receptors with PET. Nucl Med Bio. 2008;35:287–298. doi: 10.1016/j.nucmedbio.2008.01.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Logan J, Fowler JS, Volkow ND, et al. Graphical analysis of reversible radioligand binding from time-activity measurements applied to [N-11C-methyl](-)-cocaine PET studies in human subjects. J Cereb Blood Flow Metab. 1990;10:740–747. doi: 10.1038/jcbfm.1990.127. [DOI] [PubMed] [Google Scholar]
- 35.Alexoff DL, Vaska P, Marstellar D, et al. Reproducibility of 11C-raclopride binding in the rat brain measured with the microPET R4: effects of scatter correction and tracer specific activity. J Nucl Med. 2003;44:815–822. [PubMed] [Google Scholar]
- 36.Kelly WK, O’Connor OA, Krug LM, Chiao JH, et al. Phase I study of an oral histone deacetylase inhibitor, suberoylanilide hydroxamic acid, in patients with advanced cancer. J Clin Oncol. 2005;23:3923–3931. doi: 10.1200/JCO.2005.14.167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ropero S, Ballestar E, Alaminos M, Arango D, et al. Transforming pathways unleashed by a HDAC2 mutation in human cancer. Oncogene. 2008;27:4008–4012. doi: 10.1038/onc.2008.31. [DOI] [PubMed] [Google Scholar]