Abstract
Amyotrophic lateral sclerosis (ALS) and frontotemporal lobar degeneration (FTLD) are neurodegenerative diseases with clinical and pathological overlap. Landmark discoveries of mutations in the transactive response DNA-binding protein (TDP-43) and fused in sarcoma/translocated in liposarcoma (FUS/TLS) as causative of ALS and FTLD, combined with the abnormal aggregation of these proteins, have initiated a shifting paradigm for the underlying pathogenesis of multiple neurodegenerative diseases. TDP-43 and FUS/TLS are both RNA/DNA-binding proteins with striking structural and functional similarities. Their association with ALS and other neurodegenerative diseases is redirecting research efforts toward understanding the role of RNA processing regulation in neurodegeneration.
TDP-43: A COMMON PATHOLOGICAL AND GENETIC DETERMINANT OF AMYOTROPHIC LATERAL SCLEROSIS AND FRONTOTEMPORAL LOBAR DEGENERATION
Amyotrophic lateral sclerosis (ALS) is an adult-onset neurodegenerative disorder characterized by the premature loss of upper and lower motor neurons. The progression of ALS is marked by fatal paralysis and respiratory failure with a typical disease course of 1–5 years. Most incidences of ALS are sporadic but ∼10% of patients have a familial history. Dominant mutations in the copper/zinc superoxide dismutase 1 (SOD1) gene (1) account for ∼20% of the familial ALS forms and ∼1% of sporadic cases. Other rare familial cases with atypical forms of ALS are due to mutations in multiple genes; however, a large majority of ALS cases do not have a known genetic component (reviewed in 2–4). The identification of SOD1 mutations initiated the molecular era of ALS research and significant insights into ALS pathogenesis were provided through the identification of pathways directly affected by the toxicity of mutant SOD1 (reviewed in 2,3,5).
A major shift in our understanding of ALS pathogenesis started in 2006 (6,7) with the identification of the 43 kDa transactive response (TAR) DNA-binding protein (TDP-43) as the main component of ubiquitinated protein aggregates found in sporadic ALS patients and in patients with frontotemporal lobar degeneration (FTLD), a neurodegenerative disease characterized by behavioral and language disorders (8). Motor neuron disease and cognitive deficits of variable severity can be concomitant in patients or within families (9–12), and the identification of a common pathological hallmark defined by TDP-43- and ubiquitin-positive, α-synuclein- and tau-negative, cytoplasmic inclusions (6,7) suggested that ALS and FTLD were part of a broad disease continuum, a point now proven by mutations in TDP-43 as causative of either (see below).
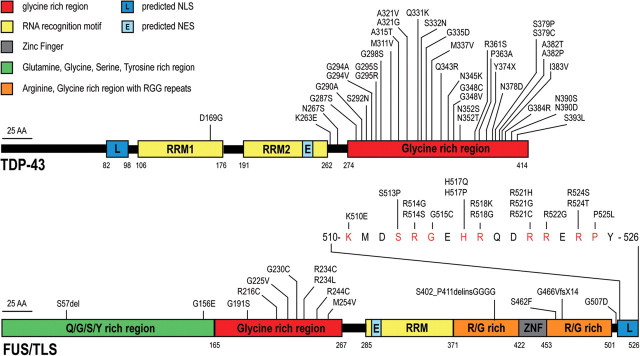
Mislocalization of TDP-43 observed by pathology motivated direct sequencing of the gene encoding it (TARDBP) in cohorts of patients with motor neuron disease and/or frontotemporal dementia. In early 2008, the successful identification of dominant mutations as a primary cause of ALS provided evidence that aberrant TDP-43 can directly trigger neurodegeneration (13–17). A total of 38 mutations (Fig. 1) have now been described in ALS patients with or without apparent family history, corresponding to ∼4% of familial ALS (and <1% of sporadic ALS) (13–33). Most patients with TDP-43 mutation develop a classical ALS phenotype without cognitive deficit, with some variability within families in the site and age of onset. Although initial studies failed to find TARDBP mutations in FTLD patients, rare TDP-43 mutations in patients displaying FTLD, with or without motor neuron disease, were reported in 2009 (34–37).
Figure 1.
TDP-43 and FUS/TLS mutations in ALS and FTLD patients. Thirty-eight dominant mutations have been identified in TDP-43 in sporadic and familial ALS patients and in rare FTLD patients, with most lying in the C-terminal glycine-rich region. All are missense mutations, except for the truncating mutation TDP-43Y374X (upper panel). Thirty mutations have been identified in FUS/TLS in familial and sporadic ALS cases and in rare FTLD patients (R514S and G515S were found in cis). Most mutations are clustered in the last 17 amino acids and in the glycine-rich region (lower panel). NLS, nuclear localization signal; NES, nuclear export signal. Domains have been defined according to http://www.uniprot.org and http://www.cbs.dtu.dk/services/NetNES.
TDP-43 is a 414 amino acid protein encoded by six exons and containing two RNA recognition motifs (RRM1 and 2) and a C-terminal glycine-rich region (38–40). Most of the mutations identified are localized in the glycine-rich region encoded by exon 6 (Fig. 1). All the mutations are dominant missense changes with the exception of a truncating mutation at the extreme C-terminal of the protein (Y374X) (20). Several variants lying in the non-coding regions of the TARDBP gene have been identified in patients but further studies are necessary to prove their pathogenic effect (29,35).
Postmortem analysis of patients with TDP-43 mutations found a pattern similar to the TDP-43 pathology described in sporadic ALS and FTLD patients. TDP-43 inclusions are not restricted to motor neurons but can be widespread in brain in ALS patients with or without dementia (16,17,31,41,42). Under normal conditions, TDP-43 is mainly localized within the nucleus, but abnormal TDP-43 distribution such as neuronal cytoplasmic or intranuclear inclusions and dystrophic neurites (6,7), as well as glial cytoplasmic inclusions (7,31,43–45) have been reported. A very curious, and mechanistically unexplained, aspect of TDP-43 pathology is a significant TDP-43 nuclear clearance in a proportion of neurons containing cytoplasmic aggregates, suggesting that pathogenesis may be driven, at least in part, by loss of one or more nuclear TDP-43 functions (6,17,43,46,47). Some ‘pre-inclusions’ have been proposed to arise from diffuse granular cytoplasmic staining and nuclear clearing (17,31,42,43,47–52). Although a plethora of early reports did not establish how prominent this nuclear clearing was and whether it escalates in frequency during disease progression, further efforts, especially the work of Giordana et al. (42), have established that the cytoplasmic redistribution of TDP-43 appears to be an early event in ALS. The pre-inclusions and the larger TDP-43 inclusions are only partially labeled with anti-ubiquitin antibodies (42,52), whereas intranuclear and cytoplasmic inclusions are strongly recognized by phosphorylation-specific antibodies to TDP-43 (33,53–57).
Immunoblotting of detergent-insoluble protein extracts from affected brain and spinal cord has defined a biochemical signature of disease that includes hyperphosphorylation and ubiquitination of TDP-43, and the production of several C-terminal fragments (CTFs) around 25 kDa (6,7). The extent of this pathologic signature correlates with the density of TDP-43 inclusions detected by immunohistochemistry (58,59). Interestingly, the composition of TDP-43 inclusions seems to differ between cortical brain and spinal cord in ALS patients, as inclusions from cortical regions are preferentially labeled by C-terminal antibodies, whereas spinal cord inclusions display equivalent immunoreactivity between N- and C-terminal-specific antibodies (46). In accordance with this, spinal cord extracts showed an absence or only weak accumulation of ∼25 kDa CTFs compared with brain extracts (54).
FUS/TLS: A NEW ACTOR IN ALS AND FTLD
The identification of TDP-43 mutations in ALS was rapidly followed by the discovery of mutations in another RNA/DNA-binding protein, FUS/TLS (fused in sarcoma/translocated in liposarcoma), as a primary cause of familial ALS (60,61). Thirty mutations (Fig. 1) have now been reported in ∼4% of familial ALS and in rare sporadic patients with no apparent familial history (60–74). The inheritance pattern is dominant except for one recessive mutation (H517Q) found in a family of Cape Verdean origin (60). Most are missense mutations with a few exceptions. Indeed, a single amino acid deletion (S57del) (64), a complex 29 bp deletion and 11 bp insertion mutation in exon 12 (p.S402_P411delinsGGGG) and a de novo splice-site mutation leading to the skipping of exon 14 and the production of a truncated FUS/TLS protein (p.G466VfsX14) (74) were identified in sporadic ALS patients. It is noteworthy that in-frame deletions and insertions in poly-glycine tracts initially identified in ALS patients (60) were subsequently found in several control individuals, challenging their pathogenic effect (62,64,70). Additionally, further studies are necessary to test whether rare synonymous changes and variants lying in the non-coding regions of FUS/TLS gene (63,65,66,71,72) are contributors to pathogenesis.
The site and age of disease onset are variable within families, and incomplete penetrance has been documented for several FUS/TLS mutations (60,63,67,69,72), which may account, at least in part, for the absence of family history in sporadic patients (62,64,66,70,71). Interestingly, several patients harboring the same R521C mutation developed an unusual presentation including an early-onset drop-head syndrome (62,65,67,69). This is an atypical phenotype, as only ∼1% of ALS patients present with severe weakness of neck extensor muscles in the early stage of the disease course (75).
Most patients with FUS/TLS mutations develop a classical ALS phenotype without cognitive defect; however, there is one report of a patient who developed FTLD concurrently with motor neuron disease (65) and two patients presented with FTLD in the absence of motor neuron deficit (67,70). At a minimum, these most recent reports provide further evidence that ALS and FTLD have clinical, pathological and genetic overlaps.
FUS/TLS is a 526 amino acid protein encoded by 15 exons (76) and characterized by an N-terminal domain enriched in glutamine, glycine, serine and tyrosine residues (QGSY region), a glycine-rich region, an RRM, multiple arginine/glycine/glycine (RGG) repeats in an arginine- and glycine-rich region and a C-terminal zinc finger motif (Fig. 1) (77–80). Most of the mutations are clustered in the glycine-rich region and in the extreme C-terminal part of the protein with evidence for mutations in each of the five arginine residues present in this region. The FUS/TLS nuclear localization signal (NLS) is likely to reside in this conserved C-terminal region since the NLS of the Ewing sarcoma (EWS) protein, a member of the TET protein family that includes FUS/TLS (79), has been identified in the last 18 amino acid residues which are highly conserved between EWS and FUS/TLS (81). Future studies are necessary to determine if the disease-related mutations clustering in the C-terminal region are responsible for a disruption of the NLS and the abnormal FUS/TLS cytoplasmic redistribution observed in patients.
Like TDP-43, FUS/TLS is mainly nuclear, with lower levels of cytoplasmic accumulation detected in most cell types (82). Postmortem analysis of brain and spinal cord from patients with FUS/TLS mutations found abnormal FUS/TLS cytoplasmic inclusions in neurons and glial cells (60,61,69). These inclusions were reported to be immunoreactive for FUS/TLS, GRP78/BiP, p62 and ubiquitin, but strikingly not for TDP-43, implying that neurodegenerative processes driven by FUS/TLS mutations are independent of TDP-43 mislocalization (61,69,73). Similar FUS-positive cytoplasmic and intranuclear inclusions were subsequently identified in brain and spinal cord of patients presenting different FTLD subtypes with TDP-43- and tau-negative aggregates (54,83–85). In both ALS and FTLD patients, FUS/TLS nuclear staining was occasionally reduced in neurons bearing cytoplasmic inclusions, but this pattern was less obvious than in TDP-43 proteinopathies and at least some of the normal FUS/TLS nuclear and cytoplasmic staining are retained (61,69,83). Again like TDP-43, ubiquitination of FUS/TLS inclusions has not always been detected, but these inclusions do label strongly with antibodies recognizing the N-terminus, mid-region or C-terminus of the FUS/TLS protein (54,69,83). Immunoblots confirmed the increased levels of full-length FUS/TLS protein in cytoplasmic and insoluble fractions, but no other evidence of biochemical abnormality such as hyperphosphorylation or ubiquitination was found (60,61,83).
TDP-43 AND FUS/TLS PROTEINOPATHIES: A BROADENING SPECTRUM OF DISEASES
TDP-43 and FUS/TLS mislocalizations have now been observed in a large number of disorders (Table 1), leading to the emergence of a new nomenclature for the set of such diseases: TDP-43 and FUS/TLS proteinopathies (43,86–91). Hallmark TDP-43 misaccumulation is observed in both ALS patients with TARDBP mutation and in the vast majority of ALS patients, with the striking exception of familial forms caused by SOD1 mutations and mutant SOD1 transgenic mouse models (45,92–96). TDP-43 proteinopathy is also found in rare cases of FTLD with TDP-43 mutation (36) and in most incidences of sporadic FTLD as well as familial FTLD cases due to mutations in the progranulin (GRN) gene, the valosine-containing protein (VCP) gene and the unidentified gene on chromosome 9p (6,49,97).
Table 1.
Reported TDP-43 and FUS/TLS inclusions in various diseases
 |
Inclusions of TDP-43 and FUS/TLS have been identified in a growing list of pathological conditions. In most instances, only a subset of patients present with aggregations of TDP-43 or FUS/TLS (e.g. around 30% of Alzheimer's disease patients), leading to inconsistency between different reports. Yes, some patients with the corresponding disease were reported to present with aggregations; No, none of the patients tested displayed abnormal inclusions of TDP-43 or FUS/TLS. Yellow, TDP-43 inclusions; Blue, FUS/TLS inclusions; Green, both TDP-43 and FUS/TLS inclusions; Gray, not tested; MND, motor neuron disease; SCA, spinocerebellar ataxia; DRPLA, dentatorubral-pallidoluysian atrophy; FXTAS, fragile X tremor/ataxia syndrome.
*FTD with inclusion body myopathy and Paget's disease.
**Mainly neuronal intranuclear inclusions (NII).
***Sarcoplasmic inclusions.
TDP-43 inclusions have also been reported to occur in various forms of dementia, including ∼30% of Alzheimer's patients (58,98–103) and various forms of Parkinson's disease (101,103–110). In these instances, TDP-43 inclusions co-exist, but only partially co-localize with, tau or α-synuclein aggregates (58,98,101–104,111,112). In such ‘combined TDP-43 proteinopathies’ (57), the mechanism(s) leading to the co-occurrence of aggregations is unknown (58,103). Attempts to correlate the presence or the absence of TDP-43 aggregation with the clinical presentation or severity have produced divergent outcomes (58,103,111–113).
Variability in the extent of TDP-43 pathology among different central nervous system (CNS) regions is observed in the different TDP-43 proteinopathies. The reasons for selective vulnerability are not known nor are TDP-43 misaccumulations restricted to the nervous system. Although TDP-43 localization was reported to be normal in muscle biopsies from ALS patients (114), TDP-43-positive sarcoplasmic inclusions have been described in various forms of myopathies with inclusion bodies and rimmed vacuoles including those due to VCP mutation (115–118). As we go to press, a very provocative doubling (to nearly 100%) of the proportion of all cells in the skin that accumulate TDP-43 has been reported in 15 examples of sporadic ALS (119).
The extent of FUS/TLS proteinopathies is less well defined. However, soon after mutant FUS/TLS was found to aggregate in ALS and FTLD patients, inclusions containing wild-type FUS/TLS protein were reported in three different forms of sporadic FTLD, namely atypical FTLD-U (83), neuronal intermediate filament inclusion disease (120) and basophilic inclusion body disease (84). Moreover, an unbiased proteomic approach identified FUS/TLS as a major component of the intranuclear polyQ aggregates in cellular models for the Huntington and spinocerebellar ataxia type 3 (SCA3) diseases (121). Postmortem analysis of patients with different polyQ diseases (Huntington's disease, SCA1, SCA2 and SCA3 and dentatorubral-pallidoluysian atrophy) confirmed the association of FUS/TLS mainly with neuronal intranuclear inclusions and in rare occasions with small cytoplasmic inclusions (83,85,121,122). This led to the provocative proposal that FUS/TLS directly binds to polyQ aggregates in an early stage of amyloid formation (121).
In the vast majority of cases, the occurrence of TDP-43- or FUS/TLS-positive inclusions appears to be mutually exclusive. One exception may be the polyglutamine diseases, since besides FUS/TLS-positive intranuclear inclusions, two groups have reported the presence of TDP-43-positive cytoplasmic inclusions in Huntington's and SCA3 patients (123,124). Further studies using phospho-specific antibodies to TDP-43 are needed to establish if co-occurrence of TDP-43 and FUS/TLS misaccumulation is a consistent feature of these diseases.
TDP-43 AND FUS/TLS ARE INVOLVED IN MULTIPLE STEPS OF RNA PROCESSING
The precise roles of TDP-43 and FUS/TLS are not fully elucidated. Although there is no evidence that TDP-43 and FUS/TLS act together, they are both structurally close to the family of heterogeneous ribonucleoproteins (hnRNPs) and have been involved in multiple levels of RNA processing including transcription, splicing, transport and translation (Fig. 2A). Such multifunctional proteins could have roles in coupling transcription with splicing and other RNA processes (82,125–129).
Figure 2.
Proposed physiological roles of TDP-43 and FUS/TLS. (A) Summary of major steps in RNA processing from transcription to translation or degradation. (B) TDP-43 binds single-stranded TG-rich elements in promoter regions thereby blocking transcription of the downstream gene [shown for TAR DNA of HIV (38) and mouse SP-10 gene (132,150)]. (C) FUS/TLS associates with TBP within the TFIID complex suggesting that it participates in the general transcriptional machinery (156–159). (D) In response to DNA damage, FUS/TLS is recruited in the promoter region of cyclin D1 (CCND1) by sense and antisense non-coding RNAs (ncRNAs) and represses CCND1 transcription (145). (E) TDP-43 binds a UG track in intronic regions preceding alternatively spliced exons and enhances their exclusion [shown for CFTR (38,40,96,130,131,160) and apolipoprotein A-II (161)]. (F) FUS/TLS was identified as a part of the spliceosome (168–170) and (G) was shown to promote exon inclusion in H-ras mRNA, through indirect binding to structural regulatory elements located on the downstream intron (177). (H) Both proteins were found in a complex with Drosha, suggesting that they may be involved in miRNA processing (183). (I) Both TDP-43 and FUS/TLS shuttle between the nucleus and the cytosol (134,142) and (J) are incorporated in SGs where they form complexes with mRNAs and other RNA binding proteins (82,166,190,201–204). (K) TDP-43 and FUS/TLS are both involved in the transport of mRNAs to dendritic spines and/or the axonal terminal where they may facilitate local translation (187–189,191). Examples of such cargo transcripts are the low molecular weight NFL for TDP-43 (51,208) and the actin-stabilizing protein Nd1-L for FUS/TLS (147).
Structure and nucleic acid properties of TDP-43 and FUS/TLS
TDP-43 contains two RRMs (RRM1 and 2) and a C-terminal glycine-rich region (38–40). The RRM1 of TDP-43 is indispensable for binding to single-stranded RNA with a minimum of five UG repeats (40,130), and its affinity increases with repeat length (40). Competition assays with synthetic oligonucleotides have suggested that TDP-43 also binds single-stranded, but not double-stranded, DNA with TG repeats (131,132). However, TDP-43 can bind to both single- and double-stranded TG-repeat DNA in nitrocellulose filter-binding assays, albeit with slightly higher affinity for single-stranded DNA (133).
The function of RRM2 is still obscure, since it is dispensable for RNA binding (130), but has been proposed to play a role in chromatin organization (134). RRM2 may mediate interaction with DNA since this region can co-crystallize with a TG-rich single-stranded DNA so as to form highly thermal-stable dimeric assemblies (133). A proportion of TDP-43 in cells forms a dimer (135), suggesting a possible functional relevance of the RRM2-mediated dimerization of TDP-43.
The C-terminal glycine-rich region of TDP-43 is essential for interactions with other proteins (39,40,136). In the native protein state, this domain lacks secondary or tertiary structure, suggesting that it might undergo structural transitions upon binding to its protein partners in vivo (137).
FUS/TLS is a member of the TET protein family that also includes the EWS protein, the TATA-binding protein (TBP)-associated factor (TAFII68/TAF15) and the Drosophila cabeza/SARF protein (79,125). FUS/TLS, EWS and TAFII68/TAF15 have a similar structure characterized by an N-terminal QGSY-rich region, a highly conserved RRM, multiple RGG repeats, which are extensively dimethylated at arginine residues (138) and a C-terminal zinc finger motif (Fig. 1) (77–80). FUS/TLS was initially identified as a fusion protein caused by chromosomal translocations in human cancers (77,139). In these instances, the promoter and N-terminal part of FUS/TLS is translocated to the C-terminal domain of various DNA-binding transcription factors conferring a strong transcriptional activation domain to the fusion proteins (125,140).
FUS/TLS was independently identified as the hnRNP P2 protein, a subunit of a complex involved in maturation of pre-mRNA (141). Consistently, in vitro studies have shown that FUS/TLS binds RNA, single-stranded DNA and (with lower affinity) double-stranded DNA (77,78,125,142–145). The sequence specificity of FUS/TLS binding to RNA or DNA has not been well established (129); however, using in vitro selection (SELEX), a common GGUG motif has been identified in approximately half of the RNA sequences bound by FUS/TLS (146). A later proposal was that the GGUG motif is recognized by the zinc finger domain and not the RRM (80). Additionally, FUS/TLS has been found to bind a relatively long region in the 3′ untranslated region (UTR) of the actin-stabilizing protein Nd1-L mRNA, suggesting that rather than recognizing specific short sequences, FUS/TLS interacts with multiple RNA-binding motifs or recognizes secondary conformations (147). FUS/TLS has also been proposed to bind human telomeric RNA (UUAGGG)4 and single-stranded human telomeric DNA in vitro (148,149).
Role of TDP-43 and FUS/TLS in transcription regulation
TDP-43 was originally identified as a transcriptional repressor that binds to TAR DNA of the human immunodeficiency virus type 1 (HIV-1) (38), hence its name. TDP-43 was also found to bind the promoter of the mouse SP-10 gene that is required for spermatogenesis (132,150). In both instances, TDP-43 represses transcription by binding these DNA regulatory elements (Fig. 2B)—TAR DNA sequence or mouse SP-10 promoter—but little is known about the mechanisms of this transcriptional repression (reviewed in 96). Consistent with its role in transcription, TDP-43 was found in human brain (151) and in cell culture systems (134,152) to associate with euchromatin—containing actively transcribed genes—and its RRM2 was proposed to mediate this localization (134).
FUS/TLS was found to associate with both general and more specialized factors to influence the initiation of transcription (128). Indeed, FUS/TLS interacts with several nuclear hormone receptors (153) and with gene-specific transcription factors such as Spi-1/PU.1 (154) or NF-κB (155). It also associates with the general transcriptional machinery and may influence transcription initiation and promoter selection by interacting with RNA polymerase II and the TFIID complex (Fig. 2C) (156–158). Recently, FUS/TLS was also shown to repress the transcription of RNAP III genes and to co-immunoprecipitate with TBP and the TFIIIB complex (159).
In addition to its direct interaction with the transcriptional machinery, a recent study provided a direct link between FUS/TLS RNA-binding properties and its role in transcription regulation. Indeed, in response to DNA damage, FUS/TLS is recruited by sense and antisense non-coding RNAs transcribed in the 5′ regulatory region of the cyclin D1 (CCND1) gene. An RNA-dependent allosteric modification of FUS/TLS results in inhibition of CREB-binding protein and p300 histone acetyltransferase activities leading to the repression of cyclin D1 transcription (Fig. 2D) (145).
Role of TDP-43 and FUS/TLS in splicing regulation
Evidence for a role of TDP-43 and FUS/TLS in splicing regulation came from the identification of their association with other splicing factors and that their depletion or overexpression affects the splicing pattern of specific targets. One of the best-characterized examples of alternative splicing regulation for TDP-43 is that of the cystic fibrosis transmembrane regulator (CFTR). The pre-mRNA of CFTR contains an intronic UG tract that is recognized by TDP-43, thereby promoting skipping of exon 9 in CFTR mRNA (Fig. 2E) (39,40,96,130,131,160). TDP-43 was also shown to affect the splicing of apolipoprotein A-II and survival motor neuron (SMN) transcripts (161,162). Alternative splicing coupled with the nonsense-mediated decay (NMD) machinery can sometimes regulate expression levels as in the case of SC35, a serine-/arginine-rich spliceosomal protein. Increased protein levels of SC35 activate splicing events of its own pre-mRNA, resulting in isoforms with premature stop codons that are degraded by the NMD system (163). These splicing events are activated via multiple low-affinity SC35-binding sites and are inhibited by TDP-43 that competes for binding to the same regulatory sequence as SC35 (164).
The association of TDP-43 with a number of proteins involved in splicing (165,166), including SC35 (167), is consistent with its function as a splicing regulator. These interactions are mediated by the C-terminal glycine-rich domain of TDP-43 and truncated TDP-43 proteins that lack this domain do not possess CFTR exon-skipping activity (39,40). Moreover, a TDP-43 interactor protein, hnRNP A2, is a crucial component of splicing regulation, since its knockdown in cells inhibits the exclusion of CFTR exon 9 (136).
Less is known about the role of FUS/TLS in splicing regulation. Proteomic analysis identified this protein as part of the spliceosome machinery (Fig. 2F) (168–170). FUS/TLS associates in vitro with large transcription–splicing complexes that bind the 5′ splice sites of pre-mRNA (127) and have been also proposed to directly bind pre-mRNA 3′ splice site (171). Furthermore, FUS/TLS associates with other splicing factors, including YB-1 (172,173), serine–arginine proteins (SC35 and TASR) (158,174,175), polypyrimidine tract-binding protein (175) or hnRNP A1 and C1/C2 (140). Finally, FUS/TLS overexpression alters the splicing of co-transfected reporter plasmids (146,154,158,172,173,176), and it has recently been proposed to influence alternative splicing of the H-ras mRNA (Fig. 2G) (177).
Despite evidence that TDP-43 and FUS/TLS are involved in RNA splicing (96,128), few of their respective RNA targets have been identified and a comprehensive protein–RNA interaction map still needs to be defined. Recent technologies coupled with high-throughput sequencing have yielded a global insight into RNA regulation (178,179), and such approaches are eagerly anticipated to understand the role of TDP-43 and FUS/TLS in neurodegeneration. Indeed, disrupting the function of an RNA-binding protein can affect many alternatively spliced transcripts, and a growing number of neurological diseases have been linked to this process. Importantly, splicing alterations (180,181) and mRNA-editing errors (182) have been reported in sporadic ALS patients, albeit a role of TDP-43 or FUS/TLS in these modifications has not been explored. The observation of a widespread mRNA splicing defect in TDP-43 and/or FUS/TLS proteinopathies would reinforce the crucial role of splicing regulation for neuronal integrity and potentially identify candidate genes whose altered splicing is central to ALS pathogenesis.
TDP-43 and FUS/TLS are involved in micro-RNA processing
Both TDP-43 and FUS/TLS may play roles in micro-RNA (miRNA) processing. Both have been found (by mass spectrometry) to associate with Drosha (Fig. 2H) (183), the nuclear RNase III-type protein that mediates the first step in miRNA maturation (reviewed in 184). In addition, TDP-43 may be involved in the cytoplasmic cleavage step of miRNA biogenesis—mediated by the Dicer complex—as suggested by its association with proteins known to participate in these functions, such as argonaute 2 and DDX17 (166). Much more can be anticipated soon on the possible involvement of TDP-43 and FUS/TLS in miRNA processing.
Cytosolic roles of TDP-43 and FUS/TLS in regulation of RNA subcellular localization, translation and decay
TDP-43 and FUS/TLS are also present in the cytosol where they are involved in diverse aspects of RNA metabolism, regulating the spatiotemporal fate of mRNA, i.e. subcellular localization, translation or degradation. Indeed, both proteins have been shown by interspecies heterokaryon assays to shuttle between the nucleus and the cytoplasm (Fig. 2I) (134,142). In particular, FUS/TLS was shown to relocalize to the cytoplasm upon inhibition of RNA polymerase II transcription (140,185) and post-translational modifications such as tyrosine phosphorylation by the fibroblast growth factor receptor-1 (FGFR1) kinase (186) or dimethylation of arginine residues may alter its subcellular localization (129,138).
In neurons, TDP-43 and FUS/TLS are found in RNA-transporting granules translocating to dendritic spines upon different neuronal stimuli (187–191). In addition, the loss of TDP-43 reduces dendritic branching as well as synaptic formation in Drosophila neurons (192,193), and cultured hippocampal neurons from FUS/TLS knockout mice (194) displayed abnormal spine morphology and density (190). Collectively, these results suggest that both proteins could play a role in the modulation of neuronal plasticity by altering mRNA transport and local translation in neurons. In particular, FUS/TLS was found to associate with N-methyl-d-aspartate receptor–adhesion protein signaling complexes (189,195,196), suggesting that it participates in the regulation of mRNA translation at excitatory synaptic sites. The accumulation of FUS/TLS in spines is facilitated by myosin-Va (197) and myosin-VI (198) and is dependent on the post-synaptic activation of signaling pathways initiated by the metabotropic glutamate receptor mGluR5 (190). FUS/TLS was also found to bind mRNAs encoding actin-related proteins, such as actin-stabilizing protein Nd1-L, and may be involved in actin reorganization in spines (Fig. 2K) (147). FUS/TLS involvement in the regulation of localized protein synthesis was also suggested by its accumulation with other RNA-binding and ribosomal proteins in the spreading initiation centers of adhering cells (82,199). The role of TDP-43 in the regulation of local translation is less well established; however, it was shown to act as a translational repressor in vitro (200) and has extensive interactions with proteins participating in translation (166).
Increasing evidence suggests that TDP-43 and FUS/TLS are integral components of RNA stress granules (SGs; Fig. 2J) (82,166,200–204), cytoplasmic, microscopically visible foci consisting of mRNA and RNP complexes that stall translation under stress conditions (reviewed in 205). However, in contrast to TIA1 and TIAR, two bona fide components of SGs (206), TDP-43 is neither essential for formation of SGs nor a neuroprotective factor in stress conditions (202). Nonetheless, in axotomized motor neurons in vivo, TDP-43 was found to translocate to the cytoplasm where it formed SGs that dissolved after neuronal recovery (203,207). The role of these TDP-43-positive SGs remains unknown, but they were proposed to mediate the stabilization and transport of the low molecular weight neurofilament (NFL) mRNA to the injury site for local translation of NFL protein required for axonal repair (51,208).
ROLES OF TDP-43 AND FUS/TLS IN DEVELOPMENT AND/OR MAINTENANCE OF GENOME STABILITY
TDP-43 is essential for early mouse embryogenesis as mice with homozygous disruption in the Tardbp gene are embryonically lethal (209–211), a consequence of defective outgrowth of the inner cell mass prior to implantation (210). TDP-43 is developmentally regulated, since the levels of TDP-43 are sustained throughout embryonic development, but in post-natal brains, its accumulation gradually decreases with age (210). TDP-43 expression seems to be tightly regulated, since the heterozygous Tardbp gene disruption mice have apparently unchanged protein levels and are developmentally indistinguishable from control littermates (209–211). In agreement with the above in vivo results, knockdown of TDP-43 induces cell death in cultured tumor cells and differentiated neurons (212,213). TDP-43 disruption in Drosophila leads to lethality at second larvae stage (214), although other groups reported ‘semi-lethal’ phenotypes, with some TDP-43 null flies surviving through adulthood, suggesting additional compensatory mechanisms that might be specific to the fly (192,193).
Two independent sets of mice with disruption of the Fus/Tls gene have been produced by the insertion of gene-trap constructs in exon 8 or 12, respectively (194,215). Recognizing that low levels of a truncated protein are produced, Fus/Tls+/− heterozygous mice do not display overt abnormality, whereas the phenotype of the Fus/Tls−/− gene disruption mice varies depending on the genetic background. Inbred Fus/Tls−/− mice have been reported to be small at birth and die within a few hours with major defects in B-lymphocyte development (194), whereas similar mice in an outbred background survive until adulthood and develop male sterility (215). Murine embryonic fibroblasts derived from the Fus/Tls−/− gene disruption mice display high chromosomal instability and radiation sensitivity (194,215). In fact, several lines of evidence suggest that FUS/TLS is required for the maintenance of genomic integrity and may have a role in DNA double-strand break repair. Indeed, FUS/TLS promotes the annealing of homologous DNA and the formation of DNA D-loops, an essential step in DNA repair by homologous recombination (144,216,217).
FUS/TLS is a key transcriptional regulatory sensor of DNA-damage signals, as exemplified by its recruitment to the cyclin D1 promoter after ionizing radiation exposure where it functions as a transcriptional repressor (145). Moreover, FUS/TLS is phosphorylated by ATM (ataxia-telangiectasia mutated) upon induction of double-strand breaks (218) and mouse models with disruption of FUS/TLS present high levels of chromosomal instability, male sterility due to defects in the formation of autosomal bivalents and increased sensitivity to ionizing irradiation (194,215).
UNDERSTANDING THE PATHOLOGICAL MECHANISMS OF DISEASE: CELLULAR AND ANIMAL MODELS FOR TDP-43
The role of TDP-43, FUS/TLS and mutations thereof in the pathogenesis of the respective proteinopathies has not been established and both a gain of toxic property(ies) and a loss of function via their sequestration in aggregates are plausible. Animal models for the most recently identified FUS/TLS have not yet been reported, whereas for TDP-43, the first outcomes modeling disease in fruit flies or mice have produced a confusing story that has not yet settled the key questions concerning mutant TDP-43-mediated pathogenesis.
Wild-type and mutant TDP-43 in disease
Expression of wild-type human TDP-43 has been found to cause neurodegeneration in Drosophila (192,219,220) and rodent models (221,222). In Drosophila, human TDP-43-mediated neurodegeneration was associated with impaired locomotive activity (219), paralysis and reduced life span (220), whereas in mice it presented as a very rapidly progressive paralysis reminiscent of human ALS in a dose-dependent manner (222). In addition, overexpression of wild-type TDP-43 in primary neurons led to cell death independently of mutation (223).
Although the above observations argue for a pathogenic role of elevated wild-type TDP-43 levels, it remains unclear whether increased levels of TDP-43 are a common finding in human patients with TDP-43 proteinopathies. So far, no copy number variation of the TARDBP gene has been found (19,25,224,225). Brain mRNA levels of TDP-43 were also found unchanged in a handful of patients with TDP-43 proteinopathies, with the exception of a single patient with a 3′ UTR variant of TDP-43 that showed a 2-fold increase in TDP-43 expression levels compared to controls (35). It is noteworthy that elevated levels of TDP-43 mRNA are found in the Wobbler mouse that develops motor neuron disease (226).
Expression of human TDP-43 carrying disease-associated mutations was suggested to exhibit higher toxicity than the wild-type protein in chick embryos (14) and primary rodent neurons (223,227). Mutant TDP-43, but not its wild-type counterpart, caused a motor phenotype in zebrafish associated with shorter and disorganized axons with excessive branching (227). Recently, accumulation to ∼3-fold of the endogenous level of mutant TDP-43 carrying the A315T mutation in mice was reported to trigger a phenotype including gait abnormalities consistent with upper motor neuron disease (228). Since these observations were made in a single transgenic line, it is impossible to exclude integration site artifacts or to determine the role of expression levels or the presence of the point mutation in the induction of the phenotype in this study. These are all crucial points that await independent validation.
Despite the usage of neuronal or more broadly expressed prion protein promoters to drive the expression of transgenes, both wild-type (222) and mutant (228) transgenes have produced loss of cortical layer V pyramidal neurons in the frontal cortex, suggesting a selective vulnerability of this set of neurons to dose-dependent, human TDP-43-mediated toxicity. Moreover, this cortical area exhibited extensive microgliosis and astrocytosis in both mutant and wild-type mouse models (222,228).
Most recently a novel rat model with overexpression of mutant—but not wild-type—human TDP-43 was shown to develop widespread neurodegeneration primarily in the motor system leading to progressive paralysis (229).
Cytoplasmic localization of TDP-43 and intracellular aggregates
Increased cytoplasmic localization of TDP-43 in brains and spinal cords of patients—designated ‘pre-inclusions’—was proposed to be an early event in TDP-43 proteinopathies (42), with the implication of a possible pathogenic role. Consistently, increased cytoplasmic TDP-43 localization was found at presymptomatic stages in mice overexpressing wild-type TDP-43 (222) as well as in an acute rat model with adenovirus-mediated wild-type TDP-43 expression (221). In various cell culture experiments, the expression of TDP-43 proteins carrying mutations that disrupt its NLS (amino acids 78–84) led to localization primarily within the cytoplasm (223,230,231). An elegant study utilized an automated microscopy system for long-term visualization and quantitative correlation between morphologic changes and survival of individual neurons to show that cytoplasmic TDP-43 is toxic for rat primary cortical neurons (223). Although overexpression of wild-type TDP-43 led to increased cytoplasmic localization of TDP-43 and cell death independently of mutation, pathogenic TDP-43 mutations increased the proportion of cytoplasmic TDP-43 (223).
On the other hand, how forced synthesis of high levels of TDP-43 relates to actual pathogenic mechanism for the lower levels of TDP-43 in the physiologically relevant contexts is not established by such approaches. TDP-43 is likely to perform one or more cytoplasmic roles including contribution to neuronal recovery as illustrated by the rapid, transient cytoplasmic translocation of TDP-43 in response to stress (203,207). These observations led to the hypothesis that prominent cytosolic localization in neurons of ALS or FTLD-U patients may actually represent the typical response to stress rather than an initiating event in pathogenesis (203,207). This could provide a partial explanation for the remarkably common incidence of cytoplasmic TDP-43 accumulation in a variety of neurodegenerative conditions of seemingly different origins (Table 1).
Increased cytoplasmic localization of TDP-43 is associated with the formation of intracellular aggregates in the affected areas in patients (6,7,42) and in animal models including rats (221), mice (222) and Drosophila (219). In cell culture systems, increased cytoplasmic localization of TDP-43 was proposed to facilitate the formation of intracellular aggregates (223,230,231), albeit this argument suffers from a ‘chicken versus the egg’ ambiguity. So too do arguments concerning the role of the inclusions in pathogenesis. As in most neurodegenerative diseases where cytoplasmic aggregations are evident, an unresolved controversy is whether inclusions are neurotoxic or neuroprotective (232), the latter presumably through the sequestration of smaller toxic species of misfolded proteins. For example, early studies of hTDP-43 mutants (233,234) in yeast reported that foci of aggregated protein were better determinants of cellular toxicity than diffuse cytosolic staining, but this was not seen for inclusion body formation in the cytoplasm of primary neurons (223) or in Drosophila motor neurons in vivo (220). In any event, the possibility that neurotoxicity of aggregates might stem from the sequestration of essential proteins within the inclusions does not include some known binding partners of TDP-43 (e.g. hnRNP A1, A2/B1 and C and SMN), as these appear to be absent from the inclusions (235).
Phosphorylation and C-terminal fragmentation of TDP-43
The role of phosphorylation of TDP-43 in FTLD-U and ALS patients has been explored with the help of phospho-specific antibodies that strongly bind to nuclear and cytoplasmic TDP-43 inclusions without recognizing normal intranuclear TDP-43 (53–56). Using these, S409/410 have been identified as the major sites of phosphorylation on TDP-43 (55) and were found highly phosphorylated in the proposed pathologic 25 kDa CTFs (see below) (53–56). It is unclear whether phosphorylation of TDP-43 and its CTFs are a primary event, or an epiphenomenon following earlier pathogenic events. Although a correlation between insolubility and phosphorylation of TDP-43 has been reported (236), phosphorylation is not required for C-terminal cleavage, aggregation or toxicity, at least in cellular models (237,238).
Fragments (20–25 kDa) containing the carboxy-proximal portion of TDP-43 accumulate in detergent (sarkosyl)-insoluble fractions derived from patient CNS tissues (6,7). These CTFs originate—at least in part—from proteolytic cleavage at Arg208 (236) and are more prominent in brains of FTLD-U and ALS patients, whereas in the spinal cord of both groups, the predominant species within the inclusions are full-length TDP-43 (46). When expressed in cells, the 25 kDa CTFs recapitulate some of the pathological features of TDP-43 proteinopathy such as increased cytoplasmic accumulation, insolubility, hyperphosphorylation, polyubiquitination and cytotoxicity (46,237,239). In transgenic mice expressing wild-type or mutant TDP-43, the appearance of the 25 kDa CTF (222,228) was shown to increase with disease progression, arguing for a pathogenic role (222). Curiously, in contrast to observations in patients (6,7,46) and in cell culture (204,237,238,240), where 25 kDa CTF is mainly cytoplasmic, the same fragment in transgenic mice was present solely in the nucleus where it formed intranuclear inclusions (222).
Caspase-3, which is activated during apoptosis, has been proposed to be the main protease generating the 25 kDa CTF in cells (204,237,238,240), albeit if so then its production would seem to be a late event as caspase-3 is widely considered to be an executioner caspase whose activation initiates imminent cell death. Recognizing this, a further proposal has emerged that partial caspase-3 activation and subsequent proteolytic processing of TDP-43 occur not only upon induction of apoptosis, but also when progranulin, a secreted growth factor, is downregulated (204,237,240), albeit this was not replicated by others (203,238,241). Despite the above discrepancy, there are independent clues that support a link between progranulin downregulation and TDP-43 cytoplasmic accumulation and possibly also TDP-43 fragmentation. In familial FTLD-U patients with progranulin mutations, all of which lead to progranulin loss of function or haploinsufficiency (242–244), TDP-43 aggregation and C-terminal fragmentation are evident (49). Moreover, TDP-43 cytoplasmic accumulations were reported in progranulin knockout mice (245), albeit the proteolytic processing of TDP-43 or the caspase-3 activation status in these mice was not studied.
Accumulation of another fragment with apparent molecular weight of ∼35 kDa was also found in lymphoblastoid cell lines derived from patients with TDP-43 mutations (15,19), but in CNS tissues of patients, this 35 kDa fragment is either low or undetectable (6,54,55), suggesting that this fragment is not pathologic. Indeed, although a soluble 35 kDa fragment was detected in transgenic mice with wild-type TDP-43 overexpression at early stages, this fragment decreased with disease progression (222). The 35 kDa fragment may well represent a normal TDP-43 isoform generated from an alternative translation start site (204) and may play a physiological role in the formation of SGs (166,200,202,204).
Ubiquitination, the ubiquitin-proteasome system and the autophagic system
The role of ubiquitination of TDP-43 in disease pathogenesis remains unknown, although it is likely to be a late event, since in patients the pre-inclusions and most TDP-43 inclusions are either weakly or not at all ubiquitinated (42,51,52). Nevertheless, extensive ubiquitination of the pathologic CTFs in cells (236) suggests that cellular degradation machineries such as the ubiquitin-proteasome system (UPS) and/or autophagy (reviewed in 246) may be involved in removing TDP-43 aggregates. Indeed, in cell culture systems, inhibition of either UPS or the autophagic system was shown to increase cytoplasmic TDP-43 accumulation and the formation of intracellular aggregates (202,231,239,247–249). Recently, TDP-43 was proposed to associate with ubiquilin 1 (UBQLN) (250), a protein that interacts with ubiquitinated proteins and was previously shown to play a role in Alzheimer's disease (reviewed in 251). UBQLN was found to potentiate TDP-43 autophagosomal degradation in cultured cells (250), and in Drosophila, the co-expression of UBQLN reduced TDP-43 levels but worsened the TDP-43-dependent phenotypes (220).
TDP-43 was also shown to affect the expression of histone deacetylase 6 (HDAC6), another protein associated with autophagosomal degradation (214). HDAC6 promotes the aggregation of polyubiquitinated proteins (252,253) and their autophagic degradation (254,255). A reduction in TDP-43 led to decreased HDAC6 levels resulting in reduced aggregate formation and cell death in a cellular model of spinocerebellar ataxia (214). Genetic ablation of HDAC6 resulted in ubiquitinated protein aggregate accumulation and neurodegeneration in Drosophila and in mice (256). Interestingly, VCP shown to antagonize the HDAC6-dependent aggregation of ubiquitinated proteins (253) is mutated in patients with familial forms of FTD associated with inclusion body myopathy and Paget's disease (257). Moreover, ubiquitinated inclusions in brain and muscle of patients with VCP mutations (6,49,97,115) (Table 1) and of transgenic mice expressing mutant human VCP (258) contain TDP-43. VCP is implicated in multiple steps of degradation of misfolded proteins (reviewed in 259) including a critical role in autophagy, and pathological mutant VCP expression results in autophagic dysfunction in cells and in tissues from patients and transgenic mice (247,260–263), deficits which can be rescued by HDAC6 overexpression in cell culture (260).
Although the interconnections of the different players described above are yet to be established, the autophagic system is likely to play a crucial role in the pathogenesis of neurodegenerative disorders, as suggested by the discovery that the neuronal-specific deletion of genes essential for autophagy leads to accumulation of ubiquitinated protein aggregates and neurodegeneration in mice (264,265).
Loss of TDP-43 nuclear function
The observation that the majority of inclusion-bearing cells in patients display nuclei devoid of TDP-43 led to the hypothesis that some of the deleterious effects of abnormal TDP-43 metabolism may reflect a loss of TDP-43 nuclear function (6,46). Indeed, expression of predominantly cytoplasmic TDP-43—with mutations that disrupt its NLS—led to sequestration of endogenous nuclear TDP-43 in cell culture (230). Furthermore, in transgenic mice expressing mutant human TDP-43, affected neurons displayed nuclei cleared of endogenous TDP-43, despite lack of TDP-43 immunoreactivity of cytoplasmic ubiquitinated inclusions (228). Although the first two sets of heterozygous mice with the Tardbp gene disruption showed no abnormalities (209,210), a most recent study demonstrated motor dysfunctions in aged heterozygous Tardbp+/− mice in the absence of neurodegeneration, despite lack of detectable reduction in TDP-43 protein levels (211). Similarly, TDP-43 loss can drive motor dysfunction in Drosophila (193). The contribution of disease-causing mutations in this possible nuclear loss of function mechanism remains unclear; at least in cell culture, they do not seem to affect the exon-skipping activity of TDP-43 (136,231), or the transcriptional regulation of HDAC6 (214), nor do they alter its protein interaction profile (166).
Although a direct test of an involvement of loss of TDP-43 or FUS/TLS function in disease pathogenesis is still missing, given the known physiological roles of these proteins, it is easy to speculate that their loss of function may have profound effects on RNA processing with detrimental consequences for the cell.
CONCLUSIONS AND OPEN QUESTIONS
In conclusion, the weight of the evidence strongly implicates TDP-43 and FUS/TLS in neurodegeneration through errors in multiple steps of RNA processing. Elucidating the physiological roles of these two proteins within the normal CNS is an essential first step in deciphering disease pathways. The major questions underlying pathogenesis remain unresolved: is disease from TDP-43 or FUS/TLS mutation caused by the loss of normal function, gain of one or more toxic properties or aberrant function or both? Post-translational modifications and cleavage of TDP-43 are markers for the disease state, but a pathogenic role of such alterations has not been established. Going forward, exploitation of advances in high-throughput sequencing is now needed to identify the normal RNA targets and the consequences of mutations on the processing of these RNAs. So too is improved modeling of disease in animal and cell culture systems so as to understand TDP-43- and FUS/TLS-mediated neurodegeneration.
FUNDING
C.L.-T. is the recipient of the Milton-Safenowitz post-doctoral fellowship from the Amytrophic Lateral Sclerosis Association. M.P. is the recipient of a Human Frontier Science Program Long Term Fellowship. D.W.C. receives salary support from the Ludwig Institute for Cancer Research.
ACKNOWLEDGEMENTS
The authors would like to thank the members of the Cleveland Laboratory for fruitful discussions, Dr Dara Ditsworth for comments on the manuscript and Dr Christina Sigurdson for suggestions on Table 1.
Conflict of Interest statement. None declared.
REFERENCES
- 1.Rosen D.R., Siddique T., Patterson D., Figlewicz D.A., Sapp P., Hentati A., Donaldson D., Goto J., O'Regan J.P., Deng H.X., et al. Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature. 1993;362:59–62. doi: 10.1038/362059a0. doi:10.1038/362059a0. [DOI] [PubMed] [Google Scholar]
- 2.Boillee S., Vande Velde C., Cleveland D.W. ALS: a disease of motor neurons and their nonneuronal neighbors. Neuron. 2006;52:39–59. doi: 10.1016/j.neuron.2006.09.018. doi:10.1016/j.neuron.2006.09.018. [DOI] [PubMed] [Google Scholar]
- 3.Pasinelli P., Brown R.H. Molecular biology of amyotrophic lateral sclerosis: insights from genetics. Nat. Rev. Neurosci. 2006;7:710–723. doi: 10.1038/nrn1971. doi:10.1038/nrn1971. [DOI] [PubMed] [Google Scholar]
- 4.Valdmanis P.N., Rouleau G.A. Genetics of familial amyotrophic lateral sclerosis. Neurology. 2008;70:144–152. doi: 10.1212/01.wnl.0000296811.19811.db. doi:10.1212/01.wnl.0000296811.19811.db. [DOI] [PubMed] [Google Scholar]
- 5.Ilieva H., Polymenidou M., Cleveland D.W. Non-cell autonomous toxicity in neurodegenerative disorders: ALS and beyond. J. Cell. Biol. 2009;187:761–772. doi: 10.1083/jcb.200908164. doi:10.1083/jcb.200908164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Neumann M., Sampathu D.M., Kwong L.K., Truax A.C., Micsenyi M.C., Chou T.T., Bruce J., Schuck T., Grossman M., Clark C.M., et al. Ubiquitinated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Science. 2006;314:130–133. doi: 10.1126/science.1134108. doi:10.1126/science.1134108. [DOI] [PubMed] [Google Scholar]
- 7.Arai T., Hasegawa M., Akiyama H., Ikeda K., Nonaka T., Mori H., Mann D., Tsuchiya K., Yoshida M., Hashizume Y., et al. TDP-43 is a component of ubiquitin-positive tau-negative inclusions in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Biochem. Biophys. Res. Commun. 2006;351:602–611. doi: 10.1016/j.bbrc.2006.10.093. doi:10.1016/j.bbrc.2006.10.093. [DOI] [PubMed] [Google Scholar]
- 8.Neary D., Snowden J.S., Gustafson L., Passant U., Stuss D., Black S., Freedman M., Kertesz A., Robert P.H., Albert M., et al. Frontotemporal lobar degeneration: a consensus on clinical diagnostic criteria. Neurology. 1998;51:1546–1554. doi: 10.1212/wnl.51.6.1546. [DOI] [PubMed] [Google Scholar]
- 9.Caselli R.J., Windebank A.J., Petersen R.C., Komori T., Parisi J.E., Okazaki H., Kokmen E., Iverson R., Dinapoli R.P., Graff-Radford N.R., et al. Rapidly progressive aphasic dementia and motor neuron disease. Ann. Neurol. 1993;33:200–207. doi: 10.1002/ana.410330210. doi:10.1002/ana.410330210. [DOI] [PubMed] [Google Scholar]
- 10.Neary D., Snowden J.S., Mann D.M. Cognitive change in motor neurone disease/amyotrophic lateral sclerosis (MND/ALS) J. Neurol. Sci. 2000;180:15–20. doi: 10.1016/s0022-510x(00)00425-1. doi:10.1016/S0022-510X(00)00425-1. [DOI] [PubMed] [Google Scholar]
- 11.Lomen-Hoerth C., Anderson T., Miller B. The overlap of amyotrophic lateral sclerosis and frontotemporal dementia. Neurology. 2002;59:1077–1079. doi: 10.1212/wnl.59.7.1077. [DOI] [PubMed] [Google Scholar]
- 12.Ringholz G.M., Appel S.H., Bradshaw M., Cooke N.A., Mosnik D.M., Schulz P.E. Prevalence and patterns of cognitive impairment in sporadic ALS. Neurology. 2005;65:586–590. doi: 10.1212/01.wnl.0000172911.39167.b6. doi:10.1212/01.wnl.0000172911.39167.b6. [DOI] [PubMed] [Google Scholar]
- 13.Gitcho M.A., Baloh R.H., Chakraverty S., Mayo K., Norton J.B., Levitch D., Hatanpaa K.J., White C.L., 3rd, Bigio E.H., Caselli R., et al. TDP-43 A315T mutation in familial motor neuron disease. Ann. Neurol. 2008;63:535–538. doi: 10.1002/ana.21344. doi:10.1002/ana.21344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sreedharan J., Blair I.P., Tripathi V.B., Hu X., Vance C., Rogelj B., Ackerley S., Durnall J.C., Williams K.L., Buratti E., et al. TDP-43 mutations in familial and sporadic amyotrophic lateral sclerosis. Science. 2008;319:1668–1672. doi: 10.1126/science.1154584. doi:10.1126/science.1154584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kabashi E., Valdmanis P.N., Dion P., Spiegelman D., McConkey B.J., Vande Velde C., Bouchard J.P., Lacomblez L., Pochigaeva K., Salachas F., et al. TARDBP mutations in individuals with sporadic and familial amyotrophic lateral sclerosis. Nat. Genet. 2008;40:572–574. doi: 10.1038/ng.132. doi:10.1038/ng.132. [DOI] [PubMed] [Google Scholar]
- 16.Yokoseki A., Shiga A., Tan C.F., Tagawa A., Kaneko H., Koyama A., Eguchi H., Tsujino A., Ikeuchi T., Kakita A., et al. TDP-43 mutation in familial amyotrophic lateral sclerosis. Ann. Neurol. 2008;63:538–542. doi: 10.1002/ana.21392. doi:10.1002/ana.21392. [DOI] [PubMed] [Google Scholar]
- 17.Van Deerlin V.M., Leverenz J.B., Bekris L.M., Bird T.D., Yuan W., Elman L.B., Clay D., Wood E.M., Chen-Plotkin A.S., Martinez-Lage M., et al. TARDBP mutations in amyotrophic lateral sclerosis with TDP-43 neuropathology: a genetic and histopathological analysis. Lancet Neurol. 2008;7:409–416. doi: 10.1016/S1474-4422(08)70071-1. doi:10.1016/S1474-4422(08)70071-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kuhnlein P., Sperfeld A.D., Vanmassenhove B., Van Deerlin V., Lee V.M., Trojanowski J.Q., Kretzschmar H.A., Ludolph A.C., Neumann M. Two German kindreds with familial amyotrophic lateral sclerosis due to TARDBP mutations. Arch. Neurol. 2008;65:1185–1189. doi: 10.1001/archneur.65.9.1185. doi:10.1001/archneur.65.9.1185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rutherford N.J., Zhang Y.J., Baker M., Gass J.M., Finch N.A., Xu Y.F., Stewart H., Kelley B.J., Kuntz K., Crook R.J., et al. Novel mutations in TARDBP (TDP-43) in patients with familial amyotrophic lateral sclerosis. PLoS Genet. 2008;4:e1000193. doi: 10.1371/journal.pgen.1000193. doi:10.1371/journal.pgen.1000193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Daoud H., Valdmanis P.N., Kabashi E., Dion P., Dupre N., Camu W., Meininger V., Rouleau G.A. Contribution of TARDBP mutations to sporadic amyotrophic lateral sclerosis. J. Med. Genet. 2009;46:112–114. doi: 10.1136/jmg.2008.062463. doi:10.1136/jmg.2008.062463. [DOI] [PubMed] [Google Scholar]
- 21.Lemmens R., Race V., Hersmus N., Matthijs G., Van Den Bosch L., Van Damme P., Dubois B., Boonen S., Goris A., Robberecht W. TDP-43 M311V mutation in familial amyotrophic lateral sclerosis. J. Neurol. Neurosurg. Psychiatry. 2009;80:354–355. doi: 10.1136/jnnp.2008.157677. doi:10.1136/jnnp.2008.157677. [DOI] [PubMed] [Google Scholar]
- 22.Del Bo R., Ghezzi S., Corti S., Pandolfo M., Ranieri M., Santoro D., Ghione I., Prelle A., Orsetti V., Mancuso M., et al. TARDBP (TDP-43) sequence analysis in patients with familial and sporadic ALS: identification of two novel mutations. Eur. J. Neurol. 2009 doi: 10.1111/j.1468-1331.2009.02574.x. [DOI] [PubMed] [Google Scholar]
- 23.Corrado L., Ratti A., Gellera C., Buratti E., Castellotti B., Carlomagno Y., Ticozzi N., Mazzini L., Testa L., Taroni F., et al. High frequency of TARDBP gene mutations in Italian patients with amyotrophic lateral sclerosis. Hum. Mutat. 2009 doi: 10.1002/humu.20950. [DOI] [PubMed] [Google Scholar]
- 24.Williams K.L., Durnall J.C., Thoeng A.D., Warraich S.T., Nicholson G.A., Blair I.P. A novel TARDBP mutation in an Australian amyotrophic lateral sclerosis kindred. J. Neurol. Neurosurg. Psychiatry. 2009;80:1286–1288. doi: 10.1136/jnnp.2008.163261. doi:10.1136/jnnp.2008.163261. [DOI] [PubMed] [Google Scholar]
- 25.Baumer D., Parkinson N., Talbot K. TARDBP in amyotrophic lateral sclerosis: identification of a novel variant but absence of copy number variation. J. Neurol. Neurosurg. Psychiatry. 2009;80:1283–1285. doi: 10.1136/jnnp.2008.166512. doi:10.1136/jnnp.2008.166512. [DOI] [PubMed] [Google Scholar]
- 26.Ticozzi N., Leclerc A.L., van Blitterswijk M., Keagle P., McKenna-Yasek D.M., Sapp P.C., Silani V., Wills A.M., Brown R.H., Jr, Landers J.E. Mutational analysis of TARDBP in neurodegenerative diseases. Neurobiol. Aging. 2009 doi: 10.1016/j.neurobiolaging.2009.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kirby J., Goodall E.F., Smith W., Highley J.R., Masanzu R., Hartley J.A., Hibberd R., Hollinger H.C., Wharton S.B., Morrison K.E., et al. Broad clinical phenotypes associated with TAR-DNA binding protein (TARDBP) mutations in amyotrophic lateral sclerosis. Neurogenetics. 2009 doi: 10.1007/s10048-009-0218-9. [DOI] [PubMed] [Google Scholar]
- 28.Origone P., Caponnetto C., Bandettini di Poggio M., Ghiglione E., Bellone E., Ferrandes G., Mancardi G.L., Mandich P. Enlarging clinical spectrum of FALS with TARDBP gene mutations: S393L variant in an Italian family showing phenotypic variability and relevance for genetic counselling. Amyotroph. Lateral Scler. 2009 doi: 10.3109/17482960903165039. 10.3109/17482960903165039. [DOI] [PubMed] [Google Scholar]
- 29.Luquin N., Yu B., Saunderson R.B., Trent R.J., Pamphlett R. Genetic variants in the promoter of TARDBP in sporadic amyotrophic lateral sclerosis. Neuromuscul. Disord. 2009;19:696–700. doi: 10.1016/j.nmd.2009.07.005. doi:10.1016/j.nmd.2009.07.005. [DOI] [PubMed] [Google Scholar]
- 30.Kamada M., Maruyama H., Tanaka E., Morino H., Wate R., Ito H., Kusaka H., Kawano Y., Miki T., Nodera H., et al. Screening for TARDBP mutations in Japanese familial amyotrophic lateral sclerosis. J. Neurol. Sci. 2009;284:69–71. doi: 10.1016/j.jns.2009.04.017. doi:10.1016/j.jns.2009.04.017. [DOI] [PubMed] [Google Scholar]
- 31.Pamphlett R., Luquin N., McLean C., Jew S.K., Adams L. TDP-43 neuropathology is similar in sporadic amyotrophic lateral sclerosis with or without TDP-43 mutations. Neuropathol. Appl. Neurobiol. 2009;35:222–225. doi: 10.1111/j.1365-2990.2008.00982.x. [DOI] [PubMed] [Google Scholar]
- 32.Xiong H.L., Wang J.Y., Sun Y.M., Wu J.J., Chen Y., Qiao K., Zheng Q.J., Zhao G.X., Wu Z.Y. Association between novel TARDBP mutations and Chinese patients with amyotrophic lateral sclerosis. BMC Med. Genet. 2010;11:8. doi: 10.1186/1471-2350-11-8. doi:10.1186/1471-2350-11-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Tamaoka A., Arai M., Itokawa M., Arai T., Hasegawa M., Tsuchiya K., Takuma H., Tsuji H., Ishii A., Watanabe M., et al. TDP-43 M337V mutation in familial amyotrophic lateral sclerosis in Japan. Intern. Med. 49:331–334. doi: 10.2169/internalmedicine.49.2915. doi:10.2169/internalmedicine.49.2915. [DOI] [PubMed] [Google Scholar]
- 34.Benajiba L., Le Ber I., Camuzat A., Lacoste M., Thomas-Anterion C., Couratier P., Legallic S., Salachas F., Hannequin D., Decousus M., et al. TARDBP mutations in motoneuron disease with frontotemporal lobar degeneration. Ann. Neurol. 2009;65:470–473. doi: 10.1002/ana.21612. doi:10.1002/ana.21612. [DOI] [PubMed] [Google Scholar]
- 35.Gitcho M.A., Bigio E.H., Mishra M., Johnson N., Weintraub S., Mesulam M., Rademakers R., Chakraverty S., Cruchaga C., Morris J.C., et al. TARDBP 3′-UTR variant in autopsy-confirmed frontotemporal lobar degeneration with TDP-43 proteinopathy. Acta Neuropathol. 2009;118:633–645. doi: 10.1007/s00401-009-0571-7. doi:10.1007/s00401-009-0571-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kovacs G.G., Murrell J.R., Horvath S., Haraszti L., Majtenyi K., Molnar M.J., Budka H., Ghetti B., Spina S. TARDBP variation associated with frontotemporal dementia, supranuclear gaze palsy, and chorea. Mov. Disord. 2009;24:1843–1847. doi: 10.1002/mds.22697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Borroni B., Bonvicini C., Alberici A., Buratti E., Agosti C., Archetti S., Papetti A., Stuani C., Di Luca M., Gennarelli M., et al. Mutation within TARDBP leads to frontotemporal dementia without motor neuron disease. Hum. Mutat. 2009;30:E974–E983. doi: 10.1002/humu.21100. doi:10.1002/humu.21100. [DOI] [PubMed] [Google Scholar]
- 38.Ou S.H., Wu F., Harrich D., Garcia-Martinez L.F., Gaynor R.B. Cloning and characterization of a novel cellular protein, TDP-43, that binds to human immunodeficiency virus type 1 TAR DNA sequence motifs. J. Virol. 1995;69:3584–3596. doi: 10.1128/jvi.69.6.3584-3596.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Wang H.Y., Wang I.F., Bose J., Shen C.K. Structural diversity and functional implications of the eukaryotic TDP gene family. Genomics. 2004;83:130–139. doi: 10.1016/s0888-7543(03)00214-3. doi:10.1016/S0888-7543(03)00214-3. [DOI] [PubMed] [Google Scholar]
- 40.Ayala Y.M., Pantano S., D'Ambrogio A., Buratti E., Brindisi A., Marchetti C., Romano M., Baralle F.E. Human, Drosophila, and C. elegans TDP43: nucleic acid binding properties and splicing regulatory function. J. Mol. Biol. 2005;348:575–588. doi: 10.1016/j.jmb.2005.02.038. doi:10.1016/j.jmb.2005.02.038. [DOI] [PubMed] [Google Scholar]
- 41.Geser F., Brandmeir N.J., Kwong L.K., Martinez-Lage M., Elman L., McCluskey L., Xie S.X., Lee V.M., Trojanowski J.Q. Evidence of multisystem disorder in whole-brain map of pathological TDP-43 in amyotrophic lateral sclerosis. Arch. Neurol. 2008;65:636–641. doi: 10.1001/archneur.65.5.636. doi:10.1001/archneur.65.5.636. [DOI] [PubMed] [Google Scholar]
- 42.Giordana M.T., Piccinini M., Grifoni S., De Marco G., Vercellino M., Magistrello M., Pellerino A., Buccinna B., Lupino E., Rinaudo M.T. TDP-43 redistribution is an early event in sporadic amyotrophic lateral sclerosis. Brain Pathol. 2009 doi: 10.1111/j.1750-3639.2009.00284.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Dickson D.W., Josephs K.A., Amador-Ortiz C. TDP-43 in differential diagnosis of motor neuron disorders. Acta Neuropathol. 2007;114:71–79. doi: 10.1007/s00401-007-0234-5. doi:10.1007/s00401-007-0234-5. [DOI] [PubMed] [Google Scholar]
- 44.Neumann M., Kwong L.K., Truax A.C., Vanmassenhove B., Kretzschmar H.A., Van Deerlin V.M., Clark C.M., Grossman M., Miller B.L., Trojanowski J.Q., et al. TDP-43-positive white matter pathology in frontotemporal lobar degeneration with ubiquitin-positive inclusions. J. Neuropathol. Exp. Neurol. 2007;66:177–183. doi: 10.1097/01.jnen.0000248554.45456.58. doi:10.1097/01.jnen.0000248554.45456.58. [DOI] [PubMed] [Google Scholar]
- 45.Mackenzie I.R., Bigio E.H., Ince P.G., Geser F., Neumann M., Cairns N.J., Kwong L.K., Forman M.S., Ravits J., Stewart H., et al. Pathological TDP-43 distinguishes sporadic amyotrophic lateral sclerosis from amyotrophic lateral sclerosis with SOD1 mutations. Ann. Neurol. 2007;61:427–434. doi: 10.1002/ana.21147. doi:10.1002/ana.21147. [DOI] [PubMed] [Google Scholar]
- 46.Igaz L.M., Kwong L.K., Xu Y., Truax A.C., Uryu K., Neumann M., Clark C.M., Elman L.B., Miller B.L., Grossman M., et al. Enrichment of C-terminal fragments in TAR DNA-binding protein-43 cytoplasmic inclusions in brain but not in spinal cord of frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Am. J. Pathol. 2008;173:182–194. doi: 10.2353/ajpath.2008.080003. doi:10.2353/ajpath.2008.080003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Davidson Y., Kelley T., Mackenzie I.R., Pickering-Brown S., Du Plessis D., Neary D., Snowden J.S., Mann D.M. Ubiquitinated pathological lesions in frontotemporal lobar degeneration contain the TAR DNA-binding protein, TDP-43. Acta Neuropathol. 2007;113:521–533. doi: 10.1007/s00401-006-0189-y. doi:10.1007/s00401-006-0189-y. [DOI] [PubMed] [Google Scholar]
- 48.Brandmeir N.J., Geser F., Kwong L.K., Zimmerman E., Qian J., Lee V.M., Trojanowski J.Q. Severe subcortical TDP-43 pathology in sporadic frontotemporal lobar degeneration with motor neuron disease. Acta Neuropathol. 2008;115:123–131. doi: 10.1007/s00401-007-0315-5. doi:10.1007/s00401-007-0315-5. [DOI] [PubMed] [Google Scholar]
- 49.Cairns N.J., Neumann M., Bigio E.H., Holm I.E., Troost D., Hatanpaa K.J., Foong C., White C.L., 3rd, Schneider J.A., Kretzschmar H.A., et al. TDP-43 in familial and sporadic frontotemporal lobar degeneration with ubiquitin inclusions. Am. J. Pathol. 2007;171:227–240. doi: 10.2353/ajpath.2007.070182. doi:10.2353/ajpath.2007.070182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Fujita Y., Mizuno Y., Takatama M., Okamoto K. Anterior horn cells with abnormal TDP-43 immunoreactivities show fragmentation of the Golgi apparatus in ALS. J. Neurol. Sci. 2008;269:30–34. doi: 10.1016/j.jns.2007.12.016. doi:10.1016/j.jns.2007.12.016. [DOI] [PubMed] [Google Scholar]
- 51.Strong M.J., Volkening K., Hammond R., Yang W., Strong W., Leystra-Lantz C., Shoesmith C. TDP43 is a human low molecular weight neurofilament (hNFL) mRNA-binding protein. Mol. Cell. Neurosci. 2007;35:320–327. doi: 10.1016/j.mcn.2007.03.007. doi:10.1016/j.mcn.2007.03.007. [DOI] [PubMed] [Google Scholar]
- 52.Mori F., Tanji K., Zhang H.X., Nishihira Y., Tan C.F., Takahashi H., Wakabayashi K. Maturation process of TDP-43-positive neuronal cytoplasmic inclusions in amyotrophic lateral sclerosis with and without dementia. Acta Neuropathol. 2008;116:193–203. doi: 10.1007/s00401-008-0396-9. doi:10.1007/s00401-008-0396-9. [DOI] [PubMed] [Google Scholar]
- 53.Inukai Y., Nonaka T., Arai T., Yoshida M., Hashizume Y., Beach T.G., Buratti E., Baralle F.E., Akiyama H., Hisanaga S., et al. Abnormal phosphorylation of Ser409/410 of TDP-43 in FTLD-U and ALS. FEBS Lett. 2008;582:2899–2904. doi: 10.1016/j.febslet.2008.07.027. doi:10.1016/j.febslet.2008.07.027. [DOI] [PubMed] [Google Scholar]
- 54.Neumann M., Kwong L.K., Lee E.B., Kremmer E., Flatley A., Xu Y., Forman M.S., Troost D., Kretzschmar H.A., Trojanowski J.Q., et al. Phosphorylation of S409/410 of TDP-43 is a consistent feature in all sporadic and familial forms of TDP-43 proteinopathies. Acta Neuropathol. 2009;117:137–149. doi: 10.1007/s00401-008-0477-9. doi:10.1007/s00401-008-0477-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Hasegawa M., Arai T., Nonaka T., Kametani F., Yoshida M., Hashizume Y., Beach T.G., Buratti E., Baralle F., Morita M., et al. Phosphorylated TDP-43 in frontotemporal lobar degeneration and amyotrophic lateral sclerosis. Ann. Neurol. 2008;64:60–70. doi: 10.1002/ana.21425. doi:10.1002/ana.21425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Kadokura A., Yamazaki T., Kakuda S., Makioka K., Lemere C.A., Fujita Y., Takatama M., Okamoto K. Phosphorylation-dependent TDP-43 antibody detects intraneuronal dot-like structures showing morphological characters of granulovacuolar degeneration. Neurosci. Lett. 2009;463:87–92. doi: 10.1016/j.neulet.2009.06.024. doi:10.1016/j.neulet.2009.06.024. [DOI] [PubMed] [Google Scholar]
- 57.Arai T., Hasegawa M., Nonoka T., Kametani F., Yamashita M., Hosokawa M., Niizato K., Tsuchiya K., Kobayashi Z., Ikeda K., et al. Phosphorylated and cleaved TDP-43 in ALS, FTLD and other neurodegenerative disorders and in cellular models of TDP-43 proteinopathy. Neuropathology. 2010 doi: 10.1111/j.1440-1789.2009.01089.x. [DOI] [PubMed] [Google Scholar]
- 58.Uryu K., Nakashima-Yasuda H., Forman M.S., Kwong L.K., Clark C.M., Grossman M., Miller B.L., Kretzschmar H.A., Lee V.M., Trojanowski J.Q., et al. Concomitant TAR-DNA-binding protein 43 pathology is present in Alzheimer disease and corticobasal degeneration but not in other tauopathies. J. Neuropathol. Exp. Neurol. 2008;67:555–564. doi: 10.1097/NEN.0b013e31817713b5. doi:10.1097/NEN.0b013e31817713b5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Neumann M. Molecular neuropathology of TDP-43 proteinopathies. Int. J. Mol. Sci. 2009;10:232–246. doi: 10.3390/ijms10010232. doi:10.3390/ijms10010232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kwiatkowski T.J., Bosco J.D., LeClerc A.D., Tamrazian E., Van den Berg C.R., Russ C., Davis A., Gilchrist J., Kasarskis E.J., Munsat T., et al. Mutations in the FUS/TLS gene on chromosome 16 cause familial amyotrophic lateral sclerosis. Science. 2009 doi: 10.1126/science.1166066. [DOI] [PubMed] [Google Scholar]
- 61.Vance C., Rogelj B., Hortobagyi T., De Vos K.J., Nishimura A.L., Sreedharan J., Hu X., Smith B., Ruddy D.M., Wright P., et al. Mutations in FUS, an RNA processing protein, cause familial amyotrophic lateral sclerosis type 6. Science. 2009 doi: 10.1126/science.1165942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Corrado L., Del Bo R., Castellotti B., Ratti A., Cereda C., Penco S., Soraru G., Carlomagno Y., Ghezzi S., Pensato V., et al. Mutations of FUS gene in sporadic amyotrophic lateral sclerosis. J. Med. Genet. 2009 doi: 10.1136/jmg.2009.071027. [DOI] [PubMed] [Google Scholar]
- 63.Chio A., Restagno G., Brunetti M., Ossola I., Calvo A., Mora G., Sabatelli M., Monsurro M.R., Battistini S., Mandrioli J., et al. Two Italian kindreds with familial amyotrophic lateral sclerosis due to FUS mutation. Neurobiol. Aging. 2009;30:1272–1275. doi: 10.1016/j.neurobiolaging.2009.05.001. doi:10.1016/j.neurobiolaging.2009.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Belzil V.V., Valdmanis P.N., Dion P.A., Daoud H., Kabashi E., Noreau A., Gauthier J., Hince P., Desjarlais A., Bouchard J.P., et al. Mutations in FUS cause FALS and SALS in French and French Canadian populations. Neurology. 2009;73:1176–1179. doi: 10.1212/WNL.0b013e3181bbfeef. doi:10.1212/WNL.0b013e3181bbfeef. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Ticozzi N., Silani V., LeClerc A.L., Keagle P., Gellera C., Ratti A., Taroni F., Kwiatkowski T.J., Jr, McKenna-Yasek D.M., Sapp P.C., et al. Analysis of FUS gene mutation in familial amyotrophic lateral sclerosis within an Italian cohort. Neurology. 2009;73:1180–1185. doi: 10.1212/WNL.0b013e3181bbff05. doi:10.1212/WNL.0b013e3181bbff05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Drepper C., Herrmann T., Wessig C., Beck M., Sendtner M. C-terminal FUS/TLS mutations in familial and sporadic ALS in Germany. Neurobiol. Aging. 2009 doi: 10.1016/j.neurobiolaging.2009.11.017. [DOI] [PubMed] [Google Scholar]
- 67.Blair I.P., Williams K.L., Warraich S.T., Durnall J.C., Thoeng A.D., Manavis J., Blumbergs P.C., Vucic S., Kiernan M.C., Nicholson G.A. FUS mutations in amyotrophic lateral sclerosis: clinical, pathological, neurophysiological and genetic analysis. J. Neurol. Neurosurg. Psychiatry. 2009 doi: 10.1136/jnnp.2009.194399. [DOI] [PubMed] [Google Scholar]
- 68.Damme P.V., Goris A., Race V., Hersmus N., Dubois B., Bosch L.V., Matthijs G., Robberecht W. The occurrence of mutations in FUS in a Belgian cohort of patients with familial ALS. Eur. J. Neurol. 2009 doi: 10.1111/j.1468-1331.2009.02859.x. [DOI] [PubMed] [Google Scholar]
- 69.Tateishi T., Hokonohara T., Yamasaki R., Miura S., Kikuchi H., Iwaki A., Tashiro H., Furuya H., Nagara Y., Ohyagi Y., et al. Multiple system degeneration with basophilic inclusions in Japanese ALS patients with FUS mutation. Acta Neuropathol. 2009 doi: 10.1007/s00401-009-0621-1. [DOI] [PubMed] [Google Scholar]
- 70.Van Langenhove T., van der Zee J., Sleegers K., Engelborghs S., Vandenberghe R., Gijselinck I., Van den Broeck M., Mattheijssens M., Peeters K., De Deyn P.P., et al. Genetic contribution of FUS to frontotemporal lobar degeneration. Neurology. 2010;74:366–371. doi: 10.1212/WNL.0b013e3181ccc732. doi:10.1212/WNL.0b013e3181ccc732. [DOI] [PubMed] [Google Scholar]
- 71.Lai S.L., Abramzon Y., Schymick J.C., Stephan D.A., Dunckley T., Dillman A., Cookson M., Calvo A., Battistini S., Giannini F., et al. FUS mutations in sporadic amyotrophic lateral sclerosis. Neurobiol. Aging. 2010 doi: 10.1016/j.neurobiolaging.2009.12.020. doi:10.1016/j.neurobiolaging.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Groen E.J., van Es M.A., van Vught P.W., Spliet W.G., van Engelen-Lee J., de Visser M., Wokke J.H., Schelhaas H.J., Ophoff R.A., Fumoto K., et al. FUS mutations in familial amyotrophic lateral sclerosis in the Netherlands. Arch. Neurol. 2010;67:224–230. doi: 10.1001/archneurol.2009.329. doi:10.1001/archneurol.2009.329. [DOI] [PubMed] [Google Scholar]
- 73.Suzuki N., Aoki M., Warita H., Kato M., Mizuno H., Shimakura N., Akiyama T., Furuya H., Hokonohara T., Iwaki A., et al. FALS with FUS mutation in Japan, with early onset, rapid progress and basophilic inclusion. J. Hum. Genet. 2010;55:252–254. doi: 10.1038/jhg.2010.16. [DOI] [PubMed] [Google Scholar]
- 74.Dejesus-Hernandez M., Kocerha J., Finch N., Crook R., Baker M., Desaro P., Johnston A., Rutherford N., Wojtas A., Kennelly K., et al. De novo truncating FUS gene mutation as a cause of sporadic amyotrophic lateral sclerosis. Hum. Mutat. 2010 doi: 10.1002/humu.21241. DOI:10.1002/humu.21241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gourie-Devi M., Nalini A., Sandhya S. Early or late appearance of ‘dropped head syndrome’ in amyotrophic lateral sclerosis. J. Neurol. Neurosurg. Psychiatry. 2003;74:683–686. doi: 10.1136/jnnp.74.5.683. doi:10.1136/jnnp.74.5.683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Aman P., Panagopoulos I., Lassen C., Fioretos T., Mencinger M., Toresson H., Hoglund M., Forster A., Rabbitts T.H., Ron D., et al. Expression patterns of the human sarcoma-associated genes FUS and EWS and the genomic structure of FUS. Genomics. 1996;37:1–8. doi: 10.1006/geno.1996.0513. doi:10.1006/geno.1996.0513. [DOI] [PubMed] [Google Scholar]
- 77.Crozat A., Aman P., Mandahl N., Ron D. Fusion of CHOP to a novel RNA-binding protein in human myxoid liposarcoma. Nature. 1993;363:640–644. doi: 10.1038/363640a0. doi:10.1038/363640a0. [DOI] [PubMed] [Google Scholar]
- 78.Prasad D.D., Ouchida M., Lee L., Rao V.N., Reddy E.S. TLS/FUS fusion domain of TLS/FUS-erg chimeric protein resulting from the t(16;21) chromosomal translocation in human myeloid leukemia functions as a transcriptional activation domain. Oncogene. 1994;9:3717–3729. [PubMed] [Google Scholar]
- 79.Morohoshi F., Ootsuka Y., Arai K., Ichikawa H., Mitani S., Munakata N., Ohki M. Genomic structure of the human RBP56/hTAFII68 and FUS/TLS genes. Gene. 1998;221:191–198. doi: 10.1016/s0378-1119(98)00463-6. doi:10.1016/S0378-1119(98)00463-6. [DOI] [PubMed] [Google Scholar]
- 80.Iko Y., Kodama T.S., Kasai N., Oyama T., Morita E.H., Muto T., Okumura M., Fujii R., Takumi T., Tate S., et al. Domain architectures and characterization of an RNA-binding protein, TLS. J. Biol. Chem. 2004;279:44834–44840. doi: 10.1074/jbc.M408552200. doi:10.1074/jbc.M408552200. [DOI] [PubMed] [Google Scholar]
- 81.Zakaryan R.P., Gehring H. Identification and characterization of the nuclear localization/retention signal in the EWS proto-oncoprotein. J. Mol. Biol. 2006;363:27–38. doi: 10.1016/j.jmb.2006.08.018. doi:10.1016/j.jmb.2006.08.018. [DOI] [PubMed] [Google Scholar]
- 82.Andersson M.K., Stahlberg A., Arvidsson Y., Olofsson A., Semb H., Stenman G., Nilsson O., Aman P. The multifunctional FUS, EWS and TAF15 proto-oncoproteins show cell type-specific expression patterns and involvement in cell spreading and stress response. BMC Cell. Biol. 2008;9:37. doi: 10.1186/1471-2121-9-37. doi:10.1186/1471-2121-9-37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Neumann M., Rademakers R., Roeber S., Baker M., Kretzschmar H.A., Mackenzie I.R. A new subtype of frontotemporal lobar degeneration with FUS pathology. Brain. 2009;132:2922–2931. doi: 10.1093/brain/awp214. doi:10.1093/brain/awp214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Munoz D.G., Neumann M., Kusaka H., Yokota O., Ishihara K., Terada S., Kuroda S., Mackenzie I.R. FUS pathology in basophilic inclusion body disease. Acta Neuropathol. 2009;118:617–627. doi: 10.1007/s00401-009-0598-9. doi:10.1007/s00401-009-0598-9. [DOI] [PubMed] [Google Scholar]
- 85.Woulfe J., Gray D.A., Mackenzie I.R. FUS-Immunoreactive Intranuclear Inclusions in Neurodegenerative Disease. Brain Pathol. 2009 doi: 10.1111/j.1750-3639.2009.00337.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Cairns N.J., Bigio E.H., Mackenzie I.R., Neumann M., Lee V.M., Hatanpaa K.J., White C.L., 3rd, Schneider J.A., Grinberg L.T., Halliday G., et al. Neuropathologic diagnostic and nosologic criteria for frontotemporal lobar degeneration: consensus of the Consortium for Frontotemporal Lobar Degeneration. Acta Neuropathol. 2007;114:5–22. doi: 10.1007/s00401-007-0237-2. doi:10.1007/s00401-007-0237-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Probst A., Taylor K.I., Tolnay M. Hippocampal sclerosis dementia: a reappraisal. Acta Neuropathol. 2007;114:335–345. doi: 10.1007/s00401-007-0262-1. doi:10.1007/s00401-007-0262-1. [DOI] [PubMed] [Google Scholar]
- 88.Cook C., Zhang Y.J., Xu Y.F., Dickson D.W., Petrucelli L. TDP-43 in neurodegenerative disorders. Expert Opin. Biol. Ther. 2008;8:969–978. doi: 10.1517/14712598.8.7.969. doi:10.1517/14712598.8.7.969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Mackenzie I.R., Neumann M., Bigio E.H., Cairns N.J., Alafuzoff I., Kril J., Kovacs G.G., Ghetti B., Halliday G., Holm I.E., et al. Nomenclature for neuropathologic subtypes of frontotemporal lobar degeneration: consensus recommendations. Acta Neuropathol. 2009;117:15–18. doi: 10.1007/s00401-008-0460-5. doi:10.1007/s00401-008-0460-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Frank S., Tolnay M. Frontotemporal lobar degeneration: toward the end of confusion. Acta Neuropathol. 2009;118:629–631. doi: 10.1007/s00401-009-0602-4. doi:10.1007/s00401-009-0602-4. [DOI] [PubMed] [Google Scholar]
- 91.Mackenzie I.R., Neumann M., Bigio E.H., Cairns N.J., Alafuzoff I., Kril J., Kovacs G.G., Ghetti B., Halliday G., Holm I.E., et al. Nomenclature and nosology for neuropathologic subtypes of frontotemporal lobar degeneration: an update. Acta Neuropathol. 119:1–4. doi: 10.1007/s00401-009-0612-2. doi:10.1007/s00401-009-0612-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Tan C.F., Eguchi H., Tagawa A., Onodera O., Iwasaki T., Tsujino A., Nishizawa M., Kakita A., Takahashi H. TDP-43 immunoreactivity in neuronal inclusions in familial amyotrophic lateral sclerosis with or without SOD1 gene mutation. Acta Neuropathol. 2007;113:535–542. doi: 10.1007/s00401-007-0206-9. doi:10.1007/s00401-007-0206-9. [DOI] [PubMed] [Google Scholar]
- 93.Robertson J., Sanelli T., Xiao S., Yang W., Horne P., Hammond R., Pioro E.P., Strong M.J. Lack of TDP-43 abnormalities in mutant SOD1 transgenic mice shows disparity with ALS. Neurosci. Lett. 2007;420:128–132. doi: 10.1016/j.neulet.2007.03.066. doi:10.1016/j.neulet.2007.03.066. [DOI] [PubMed] [Google Scholar]
- 94.Turner B.J., Baumer D., Parkinson N.J., Scaber J., Ansorge O., Talbot K. TDP-43 expression in mouse models of amyotrophic lateral sclerosis and spinal muscular atrophy. BMC Neurosci. 2008;9:104. doi: 10.1186/1471-2202-9-104. doi:10.1186/1471-2202-9-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Banks G.T., Kuta A., Isaacs A.M., Fisher E.M. TDP-43 is a culprit in human neurodegeneration, and not just an innocent bystander. Mamm. Genome. 2008;19:299–305. doi: 10.1007/s00335-008-9117-x. doi:10.1007/s00335-008-9117-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Buratti E., Baralle F.E. Multiple roles of TDP-43 in gene expression, splicing regulation, and human disease. Front Biosci. 2008;13:867–878. doi: 10.2741/2727. doi:10.2741/2727. [DOI] [PubMed] [Google Scholar]
- 97.Neumann M., Mackenzie I.R., Cairns N.J., Boyer P.J., Markesbery W.R., Smith C.D., Taylor J.P., Kretzschmar H.A., Kimonis V.E., Forman M.S. TDP-43 in the ubiquitin pathology of frontotemporal dementia with VCP gene mutations. J. Neuropathol. Exp. Neurol. 2007;66:152–157. doi: 10.1097/nen.0b013e31803020b9. doi:10.1097/nen.0b013e31803020b9. [DOI] [PubMed] [Google Scholar]
- 98.Amador-Ortiz C., Lin W.L., Ahmed Z., Personett D., Davies P., Duara R., Graff-Radford N.R., Hutton M.L., Dickson D.W. TDP-43 immunoreactivity in hippocampal sclerosis and Alzheimer's disease. Ann. Neurol. 2007;61:435–445. doi: 10.1002/ana.21154. doi:10.1002/ana.21154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Hu W.T., Josephs K.A., Knopman D.S., Boeve B.F., Dickson D.W., Petersen R.C., Parisi J.E. Temporal lobar predominance of TDP-43 neuronal cytoplasmic inclusions in Alzheimer disease. Acta Neuropathol. 2008;116:215–220. doi: 10.1007/s00401-008-0400-4. doi:10.1007/s00401-008-0400-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Freeman S.H., Spires-Jones T., Hyman B.T., Growdon J.H., Frosch M.P. TAR-DNA binding protein 43 in Pick disease. J. Neuropathol. Exp. Neurol. 2008;67:62–67. doi: 10.1097/nen.0b013e3181609361. doi:10.1097/nen.0b013e3181609361. [DOI] [PubMed] [Google Scholar]
- 101.Hasegawa M., Arai T., Akiyama H., Nonaka T., Mori H., Hashimoto T., Yamazaki M., Oyanagi K. TDP-43 is deposited in the Guam parkinsonism-dementia complex brains. Brain. 2007;130:1386–1394. doi: 10.1093/brain/awm065. doi:10.1093/brain/awm065. [DOI] [PubMed] [Google Scholar]
- 102.Higashi S., Iseki E., Yamamoto R., Minegishi M., Hino H., Fujisawa K., Togo T., Katsuse O., Uchikado H., Furukawa Y., et al. Concurrence of TDP-43, tau and alpha-synuclein pathology in brains of Alzheimer's disease and dementia with Lewy bodies. Brain Res. 2007;1184:284–294. doi: 10.1016/j.brainres.2007.09.048. doi:10.1016/j.brainres.2007.09.048. [DOI] [PubMed] [Google Scholar]
- 103.Nakashima-Yasuda H., Uryu K., Robinson J., Xie S.X., Hurtig H., Duda J.E., Arnold S.E., Siderowf A., Grossman M., Leverenz J.B., et al. Co-morbidity of TDP-43 proteinopathy in Lewy body related diseases. Acta Neuropathol. 2007;114:221–229. doi: 10.1007/s00401-007-0261-2. doi:10.1007/s00401-007-0261-2. [DOI] [PubMed] [Google Scholar]
- 104.Geser F., Winton M.J., Kwong L.K., Xu Y., Xie S.X., Igaz L.M., Garruto R.M., Perl D.P., Galasko D., Lee V.M., et al. Pathological TDP-43 in parkinsonism-dementia complex and amyotrophic lateral sclerosis of Guam. Acta Neuropathol. 2008;115:133–145. doi: 10.1007/s00401-007-0257-y. doi:10.1007/s00401-007-0257-y. [DOI] [PubMed] [Google Scholar]
- 105.Miklossy J., Steele J.C., Yu S., McCall S., Sandberg G., McGeer E.G., McGeer P.L. Enduring involvement of tau, beta-amyloid, alpha-synuclein, ubiquitin and TDP-43 pathology in the amyotrophic lateral sclerosis/parkinsonism-dementia complex of Guam (ALS/PDC) Acta Neuropathol. 2008;116:625–637. doi: 10.1007/s00401-008-0439-2. doi:10.1007/s00401-008-0439-2. [DOI] [PubMed] [Google Scholar]
- 106.Maekawa S., Leigh P.N., King A., Jones E., Steele J.C., Bodi I., Shaw C.E., Hortobagyi T., Al-Sarraj S. TDP-43 is consistently co-localized with ubiquitinated inclusions in sporadic and Guam amyotrophic lateral sclerosis but not in familial amyotrophic lateral sclerosis with and without SOD1 mutations. Neuropathology. 2009 doi: 10.1111/j.1440-1789.2009.01029.x. [DOI] [PubMed] [Google Scholar]
- 107.Covy J.P., Yuan W., Waxman E.A., Hurtig H.I., Van Deerlin V.M., Giasson B.I. Clinical and pathological characteristics of patients with leucine-rich repeat kinase-2 mutations. Mov. Disord. 2009;24:32–39. doi: 10.1002/mds.22096. doi:10.1002/mds.22096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Farrer M.J., Hulihan M.M., Kachergus J.M., Dachsel J.C., Stoessl A.J., Grantier L.L., Calne S., Calne D.B., Lechevalier B., Chapon F., et al. DCTN1 mutations in Perry syndrome. Nat. Genet. 2009;41:163–165. doi: 10.1038/ng.293. doi:10.1038/ng.293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Wider C., Dickson D.W., Stoessl A.J., Tsuboi Y., Chapon F., Gutmann L., Lechevalier B., Calne D.B., Personett D.A., Hulihan M., et al. Pallidonigral TDP-43 pathology in Perry syndrome. Parkinsonism Relat. Disord. 2009;15:281–286. doi: 10.1016/j.parkreldis.2008.07.005. doi:10.1016/j.parkreldis.2008.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wider C., Dachsel J.C., Farrer M.J., Dickson D.W., Tsuboi Y., Wszolek Z.K. Elucidating the genetics and pathology of Perry syndrome. J. Neurol. Sci. 2010;289:149–154. doi: 10.1016/j.jns.2009.08.044. doi:10.1016/j.jns.2009.08.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Fujishiro H., Uchikado H., Arai T., Hasegawa M., Akiyama H., Yokota O., Tsuchiya K., Togo T., Iseki E., Hirayasu Y. Accumulation of phosphorylated TDP-43 in brains of patients with argyrophilic grain disease. Acta Neuropathol. 2009;117:151–158. doi: 10.1007/s00401-008-0463-2. doi:10.1007/s00401-008-0463-2. [DOI] [PubMed] [Google Scholar]
- 112.Arai T., Mackenzie I.R., Hasegawa M., Nonoka T., Niizato K., Tsuchiya K., Iritani S., Onaya M., Akiyama H. Phosphorylated TDP-43 in Alzheimer's disease and dementia with Lewy bodies. Acta Neuropathol. 2009;117:125–136. doi: 10.1007/s00401-008-0480-1. doi:10.1007/s00401-008-0480-1. [DOI] [PubMed] [Google Scholar]
- 113.Josephs K.A., Whitwell J.L., Knopman D.S., Hu W.T., Stroh D.A., Baker M., Rademakers R., Boeve B.F., Parisi J.E., Smith G.E., et al. Abnormal TDP-43 immunoreactivity in AD modifies clinicopathologic and radiologic phenotype. Neurology. 2008;70:1850–1857. doi: 10.1212/01.wnl.0000304041.09418.b1. doi:10.1212/01.wnl.0000304041.09418.b1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Soraru G., Orsetti V., Buratti E., Baralle F., Cima V., Volpe M., D'Ascenzo C., Palmieri A., Koutsikos K., Pegoraro E., et al. TDP-43 in skeletal muscle of patients affected with amyotrophic lateral sclerosis. Amyotroph. Lateral Scler. 2009;11:240–243. doi: 10.3109/17482960902810890. doi:10.1080/17482960902810890. [DOI] [PubMed] [Google Scholar]
- 115.Weihl C.C., Temiz P., Miller S.E., Watts G., Smith C., Forman M., Hanson P.I., Kimonis V., Pestronk A. TDP-43 accumulation in inclusion body myopathy muscle suggests a common pathogenic mechanism with frontotemporal dementia. J. Neurol. Neurosurg. Psychiatry. 2008;79:1186–1189. doi: 10.1136/jnnp.2007.131334. doi:10.1136/jnnp.2007.131334. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Kusters B., van Hoeve B.J., Schelhaas H.J., Ter Laak H., van Engelen B.G., Lammens M. TDP-43 accumulation is common in myopathies with rimmed vacuoles. Acta Neuropathol. 2009;117:209–211. doi: 10.1007/s00401-008-0471-2. doi:10.1007/s00401-008-0471-2. [DOI] [PubMed] [Google Scholar]
- 117.Salajegheh M., Pinkus J.L., Taylor J.P., Amato A.A., Nazareno R., Baloh R.H., Greenberg S.A. Sarcoplasmic redistribution of nuclear TDP-43 in inclusion body myositis. Muscle Nerve. 2009;40:19–31. doi: 10.1002/mus.21386. doi:10.1002/mus.21386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Olive M., Janue A., Moreno D., Gamez J., Torrejon-Escribano B., Ferrer I. TAR DNA-binding protein 43 accumulation in protein aggregate myopathies. J. Neuropathol. Exp. Neurol. 2009;68:262–273. doi: 10.1097/NEN.0b013e3181996d8f. doi:10.1097/NEN.0b013e3181996d8f. [DOI] [PubMed] [Google Scholar]
- 119.Suzuki M., Mikami H., Watanabe T., Yamano T., Yamazaki T., Nomura M., Yasui K., Ishikawa H., Ono S. Increased expression of TDP-43 in the skin of amyotrophic lateral sclerosis. Acta Neurol. Scand. 2010 doi: 10.1111/j.1600-0404.2010.01321.x. DOI:10.1111/j.1600-0404.2010.01321.x. [DOI] [PubMed] [Google Scholar]
- 120.Neumann M., Roeber S., Kretzschmar H.A., Rademakers R., Baker M., Mackenzie I.R. Abundant FUS-immunoreactive pathology in neuronal intermediate filament inclusion disease. Acta Neuropathol. 2009;118:605–616. doi: 10.1007/s00401-009-0581-5. doi:10.1007/s00401-009-0581-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Doi H., Okamura K., Bauer P.O., Furukawa Y., Shimizu H., Kurosawa M., Machida Y., Miyazaki H., Mitsui K., Kuroiwa Y., et al. RNA-binding protein TLS is a major nuclear aggregate-interacting protein in huntingtin exon 1 with expanded polyglutamine-expressing cells. J. Biol. Chem. 2008;283:6489–6500. doi: 10.1074/jbc.M705306200. [DOI] [PubMed] [Google Scholar]
- 122.Doi H., Koyano S., Suzuki Y., Nukina N., Kuroiwa Y. The RNA-binding protein FUS/TLS is a common aggregate-interacting protein in polyglutamine diseases. Neurosci. Res. 2010;66:131–133. doi: 10.1016/j.neures.2009.10.004. doi:10.1016/j.neures.2009.10.004. [DOI] [PubMed] [Google Scholar]
- 123.Schwab C., Arai T., Hasegawa M., Yu S., McGeer P.L. Colocalization of transactivation-responsive DNA-binding protein 43 and huntingtin in inclusions of Huntington disease. J. Neuropathol. Exp. Neurol. 2008;67:1159–1165. doi: 10.1097/NEN.0b013e31818e8951. doi:10.1097/NEN.0b013e31818e8951. [DOI] [PubMed] [Google Scholar]
- 124.Tan C.F., Yamada M., Toyoshima Y., Yokoseki A., Miki Y., Hoshi Y., Kaneko H., Ikeuchi T., Onodera O., Kakita A., et al. Selective occurrence of TDP-43-immunoreactive inclusions in the lower motor neurons in Machado-Joseph disease. Acta Neuropathol. 2009;118:553–560. doi: 10.1007/s00401-009-0552-x. doi:10.1007/s00401-009-0552-x. [DOI] [PubMed] [Google Scholar]
- 125.Bertolotti A., Bell B., Tora L. The N-terminal domain of human TAFII68 displays transactivation and oncogenic properties. Oncogene. 1999;18:8000–8010. doi: 10.1038/sj.onc.1203207. doi:10.1038/sj.onc.1203207. [DOI] [PubMed] [Google Scholar]
- 126.Hirose Y., Manley J.L. RNA polymerase II and the integration of nuclear events. Genes Dev. 2000;14:1415–1429. [PubMed] [Google Scholar]
- 127.Kameoka S., Duque P., Konarska M.M. p54(nrb) associates with the 5' splice site within large transcription/splicing complexes. EMBO J. 2004;23:1782–1791. doi: 10.1038/sj.emboj.7600187. doi:10.1038/sj.emboj.7600187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Law W.J., Cann K.L., Hicks G.G. TLS, EWS and TAF15: a model for transcriptional integration of gene expression. Brief Funct. Genomic. Proteomic. 2006;5:8–14. doi: 10.1093/bfgp/ell015. doi:10.1093/bfgp/ell015. [DOI] [PubMed] [Google Scholar]
- 129.Tan A.Y., Manley J.L. The TET family of proteins: functions and roles in disease. J. Mol. Cell. Biol. 2009;1:82–92. doi: 10.1093/jmcb/mjp025. doi:10.1093/jmcb/mjp025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 130.Buratti E., Dork T., Zuccato E., Pagani F., Romano M., Baralle F.E. Nuclear factor TDP-43 and SR proteins promote in vitro and in vivo CFTR exon 9 skipping. EMBO J. 2001;20:1774–1784. doi: 10.1093/emboj/20.7.1774. doi:10.1093/emboj/20.7.1774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Buratti E., Baralle F.E. Characterization and functional implications of the RNA binding properties of nuclear factor TDP-43, a novel splicing regulator of CFTR exon 9. J. Biol. Chem. 2001;276:36337–36343. doi: 10.1074/jbc.M104236200. doi:10.1074/jbc.M104236200. [DOI] [PubMed] [Google Scholar]
- 132.Acharya K.K., Govind C.K., Shore A.N., Stoler M.H., Reddi P.P. cis-requirement for the maintenance of round spermatid-specific transcription. Dev. Biol. 2006;295:781–790. doi: 10.1016/j.ydbio.2006.04.443. doi:10.1016/j.ydbio.2006.04.443. [DOI] [PubMed] [Google Scholar]
- 133.Kuo P.H., Doudeva L.G., Wang Y.T., Shen C.K., Yuan H.S. Structural insights into TDP-43 in nucleic-acid binding and domain interactions. Nucleic Acids Res. 2009;37:1799–1808. doi: 10.1093/nar/gkp013. doi:10.1093/nar/gkp013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Ayala Y.M., Zago P., D'Ambrogio A., Xu Y.F., Petrucelli L., Buratti E., Baralle F.E. Structural determinants of the cellular localization and shuttling of TDP-43. J. Cell. Sci. 2008;121:3778–3785. doi: 10.1242/jcs.038950. doi:10.1242/jcs.038950. [DOI] [PubMed] [Google Scholar]
- 135.Shiina Y., Arima K., Tabunoki H., Satoh J.I. TDP-43 dimerizes in human cells in culture. Cell. Mol. Neurobiol. 2009 doi: 10.1007/s10571-009-9489-9. DOI10.1007/s10571-009-9489-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.D'Ambrogio A., Buratti E., Stuani C., Guarnaccia C., Romano M., Ayala Y.M., Baralle F.E. Functional mapping of the interaction between TDP-43 and hnRNP A2 in vivo. Nucleic Acids Res. 2009;37:4116–4126. doi: 10.1093/nar/gkp342. doi:10.1093/nar/gkp342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Chen A.K., Lin R.Y., Hsieh E.Z., Tu P.H., Chen R.P., Liao T.Y., Chen W., Wang C.H., Huang J.J. Induction of amyloid fibrils by the C-terminal fragments of TDP-43 in amyotrophic lateral sclerosis. J. Am. Chem. Soc. 2010;132:1186–1187. doi: 10.1021/ja9066207. doi:10.1021/ja9066207. [DOI] [PubMed] [Google Scholar]
- 138.Rappsilber J., Friesen W.J., Paushkin S., Dreyfuss G., Mann M. Detection of arginine dimethylated peptides by parallel precursor ion scanning mass spectrometry in positive ion mode. Anal. Chem. 2003;75:3107–3114. doi: 10.1021/ac026283q. doi:10.1021/ac026283q. [DOI] [PubMed] [Google Scholar]
- 139.Rabbitts T.H., Forster A., Larson R., Nathan P. Fusion of the dominant negative transcription regulator CHOP with a novel gene FUS by translocation t(12;16) in malignant liposarcoma. Nat. Genet. 1993;4:175–180. doi: 10.1038/ng0693-175. doi:10.1038/ng0693-175. [DOI] [PubMed] [Google Scholar]
- 140.Zinszner H., Albalat R., Ron D. A novel effector domain from the RNA-binding protein TLS or EWS is required for oncogenic transformation by CHOP. Genes Dev. 1994;8:2513–2526. doi: 10.1101/gad.8.21.2513. doi:10.1101/gad.8.21.2513. [DOI] [PubMed] [Google Scholar]
- 141.Calvio C., Neubauer G., Mann M., Lamond A.I. Identification of hnRNP P2 as TLS/FUS using electrospray mass spectrometry. RNA. 1995;1:724–733. [PMC free article] [PubMed] [Google Scholar]
- 142.Zinszner H., Sok J., Immanuel D., Yin Y., Ron D. TLS (FUS) binds RNA in vivo and engages in nucleo-cytoplasmic shuttling. J. Cell. Sci. 1997;110:1741–1750. doi: 10.1242/jcs.110.15.1741. [DOI] [PubMed] [Google Scholar]
- 143.Perrotti D., Bonatti S., Trotta R., Martinez R., Skorski T., Salomoni P., Grassilli E., Lozzo R.V., Cooper D.R., Calabretta B. TLS/FUS, a pro-oncogene involved in multiple chromosomal translocations, is a novel regulator of BCR/ABL-mediated leukemogenesis. EMBO J. 1998;17:4442–4455. doi: 10.1093/emboj/17.15.4442. doi:10.1093/emboj/17.15.4442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Baechtold H., Kuroda M., Sok J., Ron D., Lopez B.S., Akhmedov A.T. Human 75-kDa DNA-pairing protein is identical to the pro-oncoprotein TLS/FUS and is able to promote D-loop formation. J. Biol. Chem. 1999;274:34337–34342. doi: 10.1074/jbc.274.48.34337. doi:10.1074/jbc.274.48.34337. [DOI] [PubMed] [Google Scholar]
- 145.Wang X., Arai S., Song X., Reichart D., Du K., Pascual G., Tempst P., Rosenfeld M.G., Glass C.K., Kurokawa R. Induced ncRNAs allosterically modify RNA-binding proteins in cis to inhibit transcription. Nature. 2008;454:126–130. doi: 10.1038/nature06992. doi:10.1038/nature06992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Lerga A., Hallier M., Delva L., Orvain C., Gallais I., Marie J., Moreau-Gachelin F. Identification of an RNA binding specificity for the potential splicing factor TLS. J. Biol. Chem. 2001;276:6807–6816. doi: 10.1074/jbc.M008304200. doi:10.1074/jbc.M008304200. [DOI] [PubMed] [Google Scholar]
- 147.Fujii R., Takumi T. TLS facilitates transport of mRNA encoding an actin-stabilizing protein to dendritic spines. J. Cell. Sci. 2005;118:5755–5765. doi: 10.1242/jcs.02692. doi:10.1242/jcs.02692. [DOI] [PubMed] [Google Scholar]
- 148.Takahama K., Kino K., Arai S., Kurokawa R., Oyoshi T. Identification of RNA binding specificity for the TET-family proteins. Nucleic Acids Symp. Ser. (Oxf.) 2008;52:213–214. doi: 10.1093/nass/nrn108. doi:10.1093/nass/nrn108. [DOI] [PubMed] [Google Scholar]
- 149.Takahama K., Arai S., Kurokawa R., Oyoshi T. Identification of DNA binding specificity for TLS. Nucleic Acids Symp. Ser. (Oxf.) 2009;53:247–248. doi: 10.1093/nass/nrp124. doi:10.1093/nass/nrp124. [DOI] [PubMed] [Google Scholar]
- 150.Abhyankar M.M., Urekar C., Reddi P.P. A novel CpG-free vertebrate insulator silences the testis-specific SP-10 gene in somatic tissues: role for TDP-43 in insulator function. J. Biol. Chem. 2007;282:36143–36154. doi: 10.1074/jbc.M705811200. doi:10.1074/jbc.M705811200. [DOI] [PubMed] [Google Scholar]
- 151.Thorpe J.R., Tang H., Atherton J., Cairns N.J. Fine structural analysis of the neuronal inclusions of frontotemporal lobar degeneration with TDP-43 proteinopathy. J. Neural. Transm. 2008;115:1661–1671. doi: 10.1007/s00702-008-0137-1. doi:10.1007/s00702-008-0137-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 152.Casafont I., Bengoechea R., Tapia O., Berciano M.T., Lafarga M. TDP-43 localizes in mRNA transcription and processing sites in mammalian neurons. J. Struct. Biol. 2009;167:235–241. doi: 10.1016/j.jsb.2009.06.006. doi:10.1016/j.jsb.2009.06.006. [DOI] [PubMed] [Google Scholar]
- 153.Powers C.A., Mathur M., Raaka B.M., Ron D., Samuels H.H. TLS (translocated-in-liposarcoma) is a high-affinity interactor for steroid, thyroid hormone, and retinoid receptors. Mol. Endocrinol. 1998;12:4–18. doi: 10.1210/mend.12.1.0043. doi:10.1210/me.12.1.4. [DOI] [PubMed] [Google Scholar]
- 154.Hallier M., Lerga A., Barnache S., Tavitian A., Moreau-Gachelin F. The transcription factor Spi-1/PU.1 interacts with the potential splicing factor TLS. J. Biol. Chem. 1998;273:4838–4842. doi: 10.1074/jbc.273.9.4838. doi:10.1074/jbc.273.9.4838. [DOI] [PubMed] [Google Scholar]
- 155.Uranishi H., Tetsuka T., Yamashita M., Asamitsu K., Shimizu M., Itoh M., Okamoto T. Involvement of the pro-oncoprotein TLS (translocated in liposarcoma) in nuclear factor-kappa B p65-mediated transcription as a coactivator. J. Biol. Chem. 2001;276:13395–13401. doi: 10.1074/jbc.M011176200. doi:10.1074/jbc.M011176200. [DOI] [PubMed] [Google Scholar]
- 156.Bertolotti A., Lutz Y., Heard D.J., Chambon P., Tora L. hTAF(II)68, a novel RNA/ssDNA-binding protein with homology to the pro-oncoproteins TLS/FUS and EWS is associated with both TFIID and RNA polymerase II. EMBO J. 1996;15:5022–5031. [PMC free article] [PubMed] [Google Scholar]
- 157.Bertolotti A., Melot T., Acker J., Vigneron M., Delattre O., Tora L. EWS, but not EWS-FLI-1, is associated with both TFIID and RNA polymerase II: interactions between two members of the TET family, EWS and hTAFII68, and subunits of TFIID and RNA polymerase II complexes. Mol. Cell. Biol. 1998;18:1489–1497. doi: 10.1128/mcb.18.3.1489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Yang L., Embree L.J., Hickstein D.D. TLS-ERG leukemia fusion protein inhibits RNA splicing mediated by serine–arginine proteins. Mol. Cell. Biol. 2000;20:3345–3354. doi: 10.1128/mcb.20.10.3345-3354.2000. doi:10.1128/MCB.20.10.3345-3354.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 159.Tan A.Y., Manley J.L. TLS inhibits RNA polymerase III transcription. Mol. Cell. Biol. 2010;30:186–196. doi: 10.1128/MCB.00884-09. doi:10.1128/MCB.00884-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 160.Ayala Y.M., Pagani F., Baralle F.E. TDP43 depletion rescues aberrant CFTR exon 9 skipping. FEBS Lett. 2006;580:1339–1344. doi: 10.1016/j.febslet.2006.01.052. doi:10.1016/j.febslet.2006.01.052. [DOI] [PubMed] [Google Scholar]
- 161.Mercado P.A., Ayala Y.M., Romano M., Buratti E., Baralle F.E. Depletion of TDP 43 overrides the need for exonic and intronic splicing enhancers in the human apoA-II gene. Nucleic Acids Res. 2005;33:6000–6010. doi: 10.1093/nar/gki897. doi:10.1093/nar/gki897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Bose J.K., Wang I.F., Hung L., Tarn W.Y., Shen C.K. TDP-43 overexpression enhances exon 7 inclusion during the survival of motor neuron pre-mRNA splicing. J. Biol. Chem. 2008;283:28852–28859. doi: 10.1074/jbc.M805376200. doi:10.1074/jbc.M805376200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Sureau A., Gattoni R., Dooghe Y., Stevenin J., Soret J. SC35 autoregulates its expression by promoting splicing events that destabilize its mRNAs. EMBO J. 2001;20:1785–1796. doi: 10.1093/emboj/20.7.1785. doi:10.1093/emboj/20.7.1785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Dreumont N., Hardy S., Behm-Ansmant I., Kister L., Branlant C., Stevenin J., Bourgeois C.F. Antagonistic factors control the unproductive splicing of SC35 terminal intron. Nucleic Acids Res. 2009 doi: 10.1093/nar/gkp1086. doi:10.1093/nar/gkp1086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165.Buratti E., Brindisi A., Giombi M., Tisminetzky S., Ayala Y.M., Baralle F.E. TDP-43 binds heterogeneous nuclear ribonucleoprotein A/B through its C-terminal tail: an important region for the inhibition of cystic fibrosis transmembrane conductance regulator exon 9 splicing. J. Biol. Chem. 2005;280:37572–37584. doi: 10.1074/jbc.M505557200. doi:10.1074/jbc.M505557200. [DOI] [PubMed] [Google Scholar]
- 166.Freibaum B.D., Chitta R.K., High A.A., Taylor J.P. Global analysis of TDP-43 interacting proteins reveals strong association with RNA splicing and translation machinery. J. Proteome Res. 2010;9:1104–1120. doi: 10.1021/pr901076y. doi:10.1021/pr901076y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167.Wang I.F., Reddy N.M., Shen C.K. Higher order arrangement of the eukaryotic nuclear bodies. Proc. Natl Acad. Sci. USA. 2002;99:13583–13588. doi: 10.1073/pnas.212483099. doi:10.1073/pnas.212483099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Zhou Z., Licklider L.J., Gygi S.P., Reed R. Comprehensive proteomic analysis of the human spliceosome. Nature. 2002;419:182–185. doi: 10.1038/nature01031. doi:10.1038/nature01031. [DOI] [PubMed] [Google Scholar]
- 169.Rappsilber J., Ryder U., Lamond A.I., Mann M. Large-scale proteomic analysis of the human spliceosome. Genome Res. 2002;12:1231–1245. doi: 10.1101/gr.473902. doi:10.1101/gr.473902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Hartmuth K., Urlaub H., Vornlocher H.P., Will C.L., Gentzel M., Wilm M., Luhrmann R. Protein composition of human prespliceosomes isolated by a tobramycin affinity-selection method. Proc. Natl Acad. Sci. USA. 2002;99:16719–16724. doi: 10.1073/pnas.262483899. doi:10.1073/pnas.262483899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Wu S., Green M.R. Identification of a human protein that recognizes the 3′ splice site during the second step of pre-mRNA splicing. EMBO J. 1997;16:4421–4432. doi: 10.1093/emboj/16.14.4421. doi:10.1093/emboj/16.14.4421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172.Chansky H.A., Hu M., Hickstein D.D., Yang L. Oncogenic TLS/ERG and EWS/Fli-1 fusion proteins inhibit RNA splicing mediated by YB-1 protein. Cancer Res. 2001;61:3586–3590. [PubMed] [Google Scholar]
- 173.Rapp T.B., Yang L., Conrad E.U., 3rd, Mandahl N., Chansky H.A. RNA splicing mediated by YB-1 is inhibited by TLS/CHOP in human myxoid liposarcoma cells. J. Orthop. Res. 2002;20:723–729. doi: 10.1016/S0736-0266(02)00006-2. doi:10.1016/S0736-0266(02)00006-2. [DOI] [PubMed] [Google Scholar]
- 174.Goransson M., Wedin M., Aman P. Temperature-dependent localization of TLS-CHOP to splicing factor compartments. Exp. Cell. Res. 2002;278:125–132. doi: 10.1006/excr.2002.5566. doi:10.1006/excr.2002.5566. [DOI] [PubMed] [Google Scholar]
- 175.Meissner M., Lopato S., Gotzmann J., Sauermann G., Barta A. Proto-oncoprotein TLS/FUS is associated to the nuclear matrix and complexed with splicing factors PTB, SRm160, and SR proteins. Exp. Cell. Res. 2003;283:184–195. doi: 10.1016/s0014-4827(02)00046-0. doi:10.1016/S0014-4827(02)00046-0. [DOI] [PubMed] [Google Scholar]
- 176.Sato S., Idogawa M., Honda K., Fujii G., Kawashima H., Takekuma K., Hoshika A., Hirohashi S., Yamada T. Beta-catenin interacts with the FUS proto-oncogene product and regulates pre-mRNA splicing. Gastroenterology. 2005;129:1225–1236. doi: 10.1053/j.gastro.2005.07.025. doi:10.1053/j.gastro.2005.07.025. [DOI] [PubMed] [Google Scholar]
- 177.Camats M., Guil S., Kokolo M., Bach-Elias M. P68 RNA helicase (DDX5) alters activity of cis- and trans-acting factors of the alternative splicing of H-Ras. PLoS One. 2008;3:e2926. doi: 10.1371/journal.pone.0002926. doi:10.1371/journal.pone.0002926. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178.Licatalosi D.D., Mele A., Fak J.J., Ule J., Kayikci M., Chi S.W., Clark T.A., Schweitzer A.C., Blume J.E., Wang X., et al. HITS-CLIP yields genome-wide insights into brain alternative RNA processing. Nature. 2008;456:464–469. doi: 10.1038/nature07488. doi:10.1038/nature07488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 179.Yeo G.W., Coufal N.G., Liang T.Y., Peng G.E., Fu X.D., Gage F.H. An RNA code for the FOX2 splicing regulator revealed by mapping RNA-protein interactions in stem cells. Nat. Struct. Mol. Biol. 2009;16:130–137. doi: 10.1038/nsmb.1545. doi:10.1038/nsmb.1545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Lin C.L., Bristol L.A., Jin L., Dykes-Hoberg M., Crawford T., Clawson L., Rothstein J.D. Aberrant RNA processing in a neurodegenerative disease: the cause for absent EAAT2, a glutamate transporter, in amyotrophic lateral sclerosis. Neuron. 1998;20:589–602. doi: 10.1016/s0896-6273(00)80997-6. doi:10.1016/S0896-6273(00)80997-6. [DOI] [PubMed] [Google Scholar]
- 181.Rabin S.J., Kim J.M., Baughn M., Libby R.T., Kim Y.J., Fan Y., La Spada A., Stone B., Ravits J. Sporadic ALS has compartment-specific aberrant exon splicing and altered cell-matrix adhesion biology. Hum. Mol. Genet. 2010;19:313–328. doi: 10.1093/hmg/ddp498. doi:10.1093/hmg/ddp498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 182.Kawahara Y., Ito K., Sun H., Aizawa H., Kanazawa I., Kwak S. Glutamate receptors: RNA editing and death of motor neurons. Nature. 2004;427:801. doi: 10.1038/427801a. doi:10.1038/427801a. [DOI] [PubMed] [Google Scholar]
- 183.Gregory R.I., Yan K.P., Amuthan G., Chendrimada T., Doratotaj B., Cooch N., Shiekhattar R. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–240. doi: 10.1038/nature03120. doi:10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- 184.Kim V.N., Han J., Siomi M.C. Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell. Biol. 2009;10:126–139. doi: 10.1038/nrm2632. doi:10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 185.Zinszner H., Immanuel D., Yin Y., Liang F.X., Ron D. A topogenic role for the oncogenic N-terminus of TLS: nucleolar localization when transcription is inhibited. Oncogene. 1997;14:451–461. doi: 10.1038/sj.onc.1200854. doi:10.1038/sj.onc.1200854. [DOI] [PubMed] [Google Scholar]
- 186.Klint P., Hellman U., Wernstedt C., Aman P., Ron D., Claesson-Welsh L. Translocated in liposarcoma (TLS) is a substrate for fibroblast growth factor receptor-1. Cell. Signal. 2004;16:515–520. doi: 10.1016/j.cellsig.2003.09.007. doi:10.1016/j.cellsig.2003.09.007. [DOI] [PubMed] [Google Scholar]
- 187.Elvira G., Wasiak S., Blandford V., Tong X.K., Serrano A., Fan X., del Rayo Sanchez-Carbente M., Servant F., Bell A.W., Boismenu D., et al. Characterization of an RNA granule from developing brain. Mol. Cell. Proteomics. 2006;5:635–651. doi: 10.1074/mcp.M500255-MCP200. [DOI] [PubMed] [Google Scholar]
- 188.Kanai Y., Dohmae N., Hirokawa N. Kinesin transports RNA: isolation and characterization of an RNA-transporting granule. Neuron. 2004;43:513–525. doi: 10.1016/j.neuron.2004.07.022. doi:10.1016/j.neuron.2004.07.022. [DOI] [PubMed] [Google Scholar]
- 189.Belly A., Moreau-Gachelin F., Sadoul R., Goldberg Y. Delocalization of the multifunctional RNA splicing factor TLS/FUS in hippocampal neurones: exclusion from the nucleus and accumulation in dendritic granules and spine heads. Neurosci. Lett. 2005;379:152–157. doi: 10.1016/j.neulet.2004.12.071. doi:10.1016/j.neulet.2004.12.071. [DOI] [PubMed] [Google Scholar]
- 190.Fujii R., Okabe S., Urushido T., Inoue K., Yoshimura A., Tachibana T., Nishikawa T., Hicks G.G., Takumi T. The RNA binding protein TLS is translocated to dendritic spines by mGluR5 activation and regulates spine morphology. Curr. Biol. 2005;15:587–593. doi: 10.1016/j.cub.2005.01.058. doi:10.1016/j.cub.2005.01.058. [DOI] [PubMed] [Google Scholar]
- 191.Wang I.F., Wu L.S., Shen C.K. TDP-43: an emerging new player in neurodegenerative diseases. Trends Mol. Med. 2008;14:479–485. doi: 10.1016/j.molmed.2008.09.001. doi:10.1016/j.molmed.2008.09.001. [DOI] [PubMed] [Google Scholar]
- 192.Lu Y., Ferris J., Gao F.B. Frontotemporal dementia and amyotrophic lateral sclerosis-associated disease protein TDP-43 promotes dendritic branching. Mol. Brain. 2009;2:30. doi: 10.1186/1756-6606-2-30. doi:10.1186/1756-6606-2-30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193.Feiguin F., Godena V.K., Romano G., D'Ambrogio A., Klima R., Baralle F.E. Depletion of TDP-43 affects Drosophila motoneurons terminal synapsis and locomotive behavior. FEBS Lett. 2009;583:1586–1592. doi: 10.1016/j.febslet.2009.04.019. doi:10.1016/j.febslet.2009.04.019. [DOI] [PubMed] [Google Scholar]
- 194.Hicks G.G., Singh N., Nashabi A., Mai S., Bozek G., Klewes L., Arapovic D., White E.K., Koury M.J., Oltz E.M., et al. Fus deficiency in mice results in defective B-lymphocyte development and activation, high levels of chromosomal instability and perinatal death. Nat. Genet. 2000;24:175–179. doi: 10.1038/72842. doi:10.1038/72842. [DOI] [PubMed] [Google Scholar]
- 195.Husi H., Ward M.A., Choudhary J.S., Blackstock W.P., Grant S.G. Proteomic analysis of NMDA receptor–adhesion protein signaling complexes. Nat. Neurosci. 2000;3:661–669. doi: 10.1038/76615. [DOI] [PubMed] [Google Scholar]
- 196.Selamat W., Jamari I., Wang Y., Takumi T., Wong F., Fujii R. TLS interaction with NMDA R1 splice variant in retinal ganglion cell line RGC-5. Neurosci. Lett. 2009;450:163–166. doi: 10.1016/j.neulet.2008.12.014. doi:10.1016/j.neulet.2008.12.014. [DOI] [PubMed] [Google Scholar]
- 197.Yoshimura A., Fujii R., Watanabe Y., Okabe S., Fukui K., Takumi T. Myosin-Va facilitates the accumulation of mRNA/protein complex in dendritic spines. Curr. Biol. 2006;16:2345–2351. doi: 10.1016/j.cub.2006.10.024. doi:10.1016/j.cub.2006.10.024. [DOI] [PubMed] [Google Scholar]
- 198.Takarada T., Tamaki K., Takumi T., Ogura M., Ito Y., Nakamichi N., Yoneda Y. A protein-protein interaction of stress-responsive myosin VI endowed to inhibit neural progenitor self-replication with RNA binding protein, TLS, in murine hippocampus. J. Neurochem. 2009;110:1457–1468. doi: 10.1111/j.1471-4159.2009.06225.x. doi:10.1111/j.1471-4159.2009.06225.x. [DOI] [PubMed] [Google Scholar]
- 199.de Hoog C.L., Foster L.J., Mann M. RNA and RNA binding proteins participate in early stages of cell spreading through spreading initiation centers. Cell. 2004;117:649–662. doi: 10.1016/s0092-8674(04)00456-8. doi:10.1016/S0092-8674(04)00456-8. [DOI] [PubMed] [Google Scholar]
- 200.Wang I.F., Wu L.S., Chang H.Y., Shen C.K. TDP-43, the signature protein of FTLD-U, is a neuronal activity-responsive factor. J. Neurochem. 2008;105:797–806. doi: 10.1111/j.1471-4159.2007.05190.x. doi:10.1111/j.1471-4159.2007.05190.x. [DOI] [PubMed] [Google Scholar]
- 201.Goodier J.L., Zhang L., Vetter M.R., Kazazian H.H., Jr LINE-1 ORF1 protein localizes in stress granules with other RNA-binding proteins, including components of RNA interference RNA-induced silencing complex. Mol. Cell. Biol. 2007;27:6469–6483. doi: 10.1128/MCB.00332-07. doi:10.1128/MCB.00332-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 202.Colombrita C., Zennaro E., Fallini C., Weber M., Sommacal A., Buratti E., Silani V., Ratti A. TDP-43 is recruited to stress granules in conditions of oxidative insult. J. Neurochem. 2009;111:1051–1061. doi: 10.1111/j.1471-4159.2009.06383.x. doi:10.1111/j.1471-4159.2009.06383.x. [DOI] [PubMed] [Google Scholar]
- 203.Moisse K., Volkening K., Leystra-Lantz C., Welch I., Hill T., Strong M.J. Divergent patterns of cytosolic TDP-43 and neuronal progranulin expression following axotomy: implications for TDP-43 in the physiological response to neuronal injury. Brain Res. 2009;1249:202–211. doi: 10.1016/j.brainres.2008.10.021. doi:10.1016/j.brainres.2008.10.021. [DOI] [PubMed] [Google Scholar]
- 204.Nishimoto Y., Ito D., Yagi T., Nihei Y., Tsunoda Y., Suzuki N. Characterization of alternative isoforms and inclusion body of the TAR DNA-binding protein-43. J. Biol. Chem. 2010;285:608–619. doi: 10.1074/jbc.M109.022012. doi:10.1074/jbc.M109.022012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 205.Anderson P., Kedersha N. RNA granules: post-transcriptional and epigenetic modulators of gene expression. Nat. Rev. Mol. Cell. Biol. 2009;10:430–436. doi: 10.1038/nrm2694. doi:10.1038/nrm2694. [DOI] [PubMed] [Google Scholar]
- 206.Gilks N., Kedersha N., Ayodele M., Shen L., Stoecklin G., Dember L.M., Anderson P. Stress granule assembly is mediated by prion-like aggregation of TIA-1. Mol. Biol. Cell. 2004;15:5383–5398. doi: 10.1091/mbc.E04-08-0715. doi:10.1091/mbc.E04-08-0715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 207.Sato T., Takeuchi S., Saito A., Ding W., Bamba H., Matsuura H., Hisa Y., Tooyama I., Urushitani M. Axonal ligation induces transient redistribution of TDP-43 in brainstem motor neurons. Neuroscience. 2009;164:1565–1578. doi: 10.1016/j.neuroscience.2009.09.050. doi:10.1016/j.neuroscience.2009.09.050. [DOI] [PubMed] [Google Scholar]
- 208.Moisse K., Mepham J., Volkening K., Welch I., Hill T., Strong M.J. Cytosolic TDP-43 expression following axotomy is associated with caspase 3 activation in NFL-/- mice: support for a role for TDP-43 in the physiological response to neuronal injury. Brain Res. 2009;1296:176–186. doi: 10.1016/j.brainres.2009.07.023. doi:10.1016/j.brainres.2009.07.023. [DOI] [PubMed] [Google Scholar]
- 209.Wu L.S., Cheng W.C., Hou S.C., Yan Y.T., Jiang S.T., Shen C.K. TDP-43, a neuro-pathosignature factor, is essential for early mouse embryogenesis. Genesis. 2010;48:56–62. doi: 10.1002/dvg.20584. [DOI] [PubMed] [Google Scholar]
- 210.Sephton C.F., Good S.K., Atkin S., Dewey C.M., Mayer P., 3rd, Herz J., Yu G. TDP-43 is a developmentally regulated protein essential for early embryonic development. J. Biol. Chem. 2010;285:6826–6834. doi: 10.1074/jbc.M109.061846. doi:10.1074/jbc.M109.061846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 211.Kraemer B.C., Schuck T., Wheeler J.M., Robinson L.C., Trojanowski J.Q., Lee V.M., Schellenberg G.D. Loss of murine TDP-43 disrupts motor function and plays an essential role in embryogenesis. Acta Neuropathol. 2010 doi: 10.1007/s00401-010-0659-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 212.Ayala Y.M., Misteli T., Baralle F.E. TDP-43 regulates retinoblastoma protein phosphorylation through the repression of cyclin-dependent kinase 6 expression. Proc. Natl Acad. Sci. USA. 2008;105:3785–3789. doi: 10.1073/pnas.0800546105. doi:10.1073/pnas.0800546105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 213.Iguchi Y., Katsuno M., Niwa J., Yamada S., Sone J., Waza M., Adachi H., Tanaka F., Nagata K., Arimura N., et al. TDP-43 depletion induces neuronal cell damage through dysregulation of Rho family GTPases. J. Biol. Chem. 2009;284:22059–22066. doi: 10.1074/jbc.M109.012195. doi:10.1074/jbc.M109.012195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 214.Fiesel F.C., Voigt A., Weber S.S., Van den Haute C., Waldenmaier A., Gorner K., Walter M., Anderson M.L., Kern J.V., Rasse T.M., et al. Knockdown of transactive response DNA-binding protein (TDP-43) downregulates histone deacetylase 6. Embo J. 2010;29:209–221. doi: 10.1038/emboj.2009.324. doi:10.1038/emboj.2009.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 215.Kuroda M., Sok J., Webb L., Baechtold H., Urano F., Yin Y., Chung P., de Rooij D.G., Akhmedov A., Ashley T., et al. Male sterility and enhanced radiation sensitivity in TLS(-/-) mice. Embo J. 2000;19:453–462. doi: 10.1093/emboj/19.3.453. doi:10.1093/emboj/19.3.453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 216.Bertrand P., Akhmedov A.T., Delacote F., Durrbach A., Lopez B.S. Human POMp75 is identified as the pro-oncoprotein TLS/FUS: both POMp75 and POMp100 DNA homologous pairing activities are associated to cell proliferation. Oncogene. 1999;18:4515–4521. doi: 10.1038/sj.onc.1203048. doi:10.1038/sj.onc.1203048. [DOI] [PubMed] [Google Scholar]
- 217.Guipaud O., Guillonneau F., Labas V., Praseuth D., Rossier J., Lopez B., Bertrand P. An in vitro enzymatic assay coupled to proteomics analysis reveals a new DNA processing activity for Ewing sarcoma and TAF(II)68 proteins. Proteomics. 2006;6:5962–5972. doi: 10.1002/pmic.200600259. doi:10.1002/pmic.200600259. [DOI] [PubMed] [Google Scholar]
- 218.Gardiner M., Toth R., Vandermoere F., Morrice N.A., Rouse J. Identification and characterization of FUS/TLS as a new target of ATM. Biochem. J. 2008;415:297–307. doi: 10.1042/BJ20081135. doi:10.1042/BJ20081135. [DOI] [PubMed] [Google Scholar]
- 219.Li Y., Ray P., Rao E.J., Shi C., Guo W., Chen X., Woodruff E.A., 3rd, Fushimi K., Wu J.Y. A Drosophila model for TDP-43 proteinopathy. Proc. Natl Acad. Sci. USA. 2010;107:3169–3174. doi: 10.1073/pnas.0913602107. doi:10.1073/pnas.0913602107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 220.Hanson K.A., Kim S.H., Wassarman D.A., Tibbetts R.S. Ubiquilin modifies toxicity of the 43 kilodalton TAR-DNA binding protein (TDP-43) in a Drosophila model of amyotrophic lateral sclerosis (ALS) J. Biol. Chem. 2010 doi: 10.1074/jbc.C109.078527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 221.Tatom J.B., Wang D.B., Dayton R.D., Skalli O., Hutton M.L., Dickson D.W., Klein R.L. Mimicking aspects of frontotemporal lobar degeneration and Lou Gehrig's disease in rats via TDP-43 overexpression. Mol. Ther. 2009;17:607–613. doi: 10.1038/mt.2009.3. doi:10.1038/mt.2009.3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 222.Wils H., Kleinberger G., Janssens J., Pereson S., Joris G., Cuijt I., Smits V., Groote C.C., Van Broeckhoven C., Kumar-Singh S. TDP-43 transgenic mice develop spastic paralysis and neuronal inclusions characteristic of ALS and frontotemporal lobar degeneration. Proc. Natl Acad. Sci. USA. 2010 doi: 10.1073/pnas.0912417107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 223.Barmada S.J., Skibinski G., Korb E., Rao E.J., Wu J.Y., Finkbeiner S. Cytoplasmic mislocalization of TDP-43 is toxic to neurons and enhanced by a mutation associated with familial amyotrophic lateral sclerosis. J. Neurosci. 2010;30:639–649. doi: 10.1523/JNEUROSCI.4988-09.2010. doi:10.1523/JNEUROSCI.4988-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 224.Guerreiro R.J., Schymick J.C., Crews C., Singleton A., Hardy J., Traynor B.J. TDP-43 is not a common cause of sporadic amyotrophic lateral sclerosis. PLoS One. 2008;3:e2450. doi: 10.1371/journal.pone.0002450. doi:10.1371/journal.pone.0002450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 225.Gijselinck I., Sleegers K., Engelborghs S., Robberecht W., Martin J.J., Vandenberghe R., Sciot R., Dermaut B., Goossens D., van der Zee J., et al. Neuronal inclusion protein TDP-43 has no primary genetic role in FTD and ALS. Neurobiol. Aging. 2009;30:1329–1331. doi: 10.1016/j.neurobiolaging.2007.11.002. doi:10.1016/j.neurobiolaging.2007.11.002. [DOI] [PubMed] [Google Scholar]
- 226.Dennis J.S., Citron B.A. Wobbler mice modeling motor neuron disease display elevated transactive response DNA binding protein. Neuroscience. 2009;158:745–750. doi: 10.1016/j.neuroscience.2008.10.030. doi:10.1016/j.neuroscience.2008.10.030. [DOI] [PubMed] [Google Scholar]
- 227.Kabashi E., Lin L., Tradewell M.L., Dion P.A., Bercier V., Bourgouin P., Rochefort D., Bel Hadj S., Durham H.D., Vande Velde C., et al. Gain and loss of function of ALS-related mutations of TARDBP (TDP-43) cause motor deficits in vivo. Hum. Mol. Genet. 2010;19:671–683. doi: 10.1093/hmg/ddp534. doi:10.1093/hmg/ddp534. [DOI] [PubMed] [Google Scholar]
- 228.Wegorzewska I., Bell S., Cairns N.J., Miller T.M., Baloh R.H. TDP-43 mutant transgenic mice develop features of ALS and frontotemporal lobar degeneration. Proc. Natl Acad. Sci. USA. 2009;106:18809–18814. doi: 10.1073/pnas.0908767106. doi:10.1073/pnas.0908767106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 229.Zhou H., Huang C., Chen H., Wang D., Landel C.P., Xia P.Y., Bowser R., Liu Y.J., Xia X.G. Transgenic rat model of neurodegeneration caused by mutation in the TDP gene. PLoS Genet. 2010;6:e1000887. doi: 10.1371/journal.pgen.1000887. doi:10.1371/journal.pgen.1000887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 230.Winton M.J., Igaz L.M., Wong M.M., Kwong L.K., Trojanowski J.Q., Lee V.M. Disturbance of nuclear and cytoplasmic TAR DNA-binding protein (TDP-43) induces disease-like redistribution, sequestration, and aggregate formation. J. Biol. Chem. 2008;283:13302–13309. doi: 10.1074/jbc.M800342200. doi:10.1074/jbc.M800342200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 231.Nonaka T., Kametani F., Arai T., Akiyama H., Hasegawa M. Truncation and pathogenic mutations facilitate the formation of intracellular aggregates of TDP-43. Hum. Mol. Genet. 2009;18:3353–3364. doi: 10.1093/hmg/ddp275. doi:10.1093/hmg/ddp275. [DOI] [PubMed] [Google Scholar]
- 232.Caughey B., Lansbury P.T. Protofibrils, pores, fibrils, and neurodegeneration: separating the responsible protein aggregates from the innocent bystanders. Annu. Rev. Neurosci. 2003;26:267–298. doi: 10.1146/annurev.neuro.26.010302.081142. doi:10.1146/annurev.neuro.26.010302.081142. [DOI] [PubMed] [Google Scholar]
- 233.Johnson B.S., McCaffery J.M., Lindquist S., Gitler A.D. A yeast TDP-43 proteinopathy model: exploring the molecular determinants of TDP-43 aggregation and cellular toxicity. Proc. Natl Acad. Sci. USA. 2008;105:6439–6444. doi: 10.1073/pnas.0802082105. doi:10.1073/pnas.0802082105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 234.Johnson B.S., Snead D., Lee J.J., McCaffery J.M., Shorter J., Gitler A.D. TDP-43 is intrinsically aggregation-prone, and amyotrophic lateral sclerosis-linked mutations accelerate aggregation and increase toxicity. J. Biol. Chem. 2009;284:20329–20339. doi: 10.1074/jbc.M109.010264. doi:10.1074/jbc.M109.010264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 235.Neumann M., Igaz L.M., Kwong L.K., Nakashima-Yasuda H., Kolb S.J., Dreyfuss G., Kretzschmar H.A., Trojanowski J.Q., Lee V.M. Absence of heterogeneous nuclear ribonucleoproteins and survival motor neuron protein in TDP-43 positive inclusions in frontotemporal lobar degeneration. Acta Neuropathol. 2007;113:543–548. doi: 10.1007/s00401-007-0221-x. doi:10.1007/s00401-007-0221-x. [DOI] [PubMed] [Google Scholar]
- 236.Igaz L.M., Kwong L.K., Chen-Plotkin A., Winton M.J., Unger T.L., Xu Y., Neumann M., Trojanowski J.Q., Lee V.M. Expression of TDP-43 C-terminal fragments in vitro recapitulates pathological features of TDP-43 proteinopathies. J. Biol. Chem. 2009;284:8516–8524. doi: 10.1074/jbc.M809462200. doi:10.1074/jbc.M809462200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 237.Zhang Y.J., Xu Y.F., Cook C., Gendron T.F., Roettges P., Link C.D., Lin W.L., Tong J., Castanedes-Casey M., Ash P., et al. Aberrant cleavage of TDP-43 enhances aggregation and cellular toxicity. Proc. Natl Acad. Sci. USA. 2009;106:7607–7612. doi: 10.1073/pnas.0900688106. doi:10.1073/pnas.0900688106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 238.Dormann D., Capell A., Carlson A.M., Shankaran S.S., Rodde R., Neumann M., Kremmer E., Matsuwaki T., Yamanouchi K., Nishihara M., et al. Proteolytic processing of TAR DNA binding protein-43 by caspases produces C-terminal fragments with disease defining properties independent of progranulin. J. Neurochem. 2009;110:1082–1094. doi: 10.1111/j.1471-4159.2009.06211.x. doi:10.1111/j.1471-4159.2009.06211.x. [DOI] [PubMed] [Google Scholar]
- 239.Caccamo A., Majumder S., Deng J.J., Bai Y., Thornton F.B., Oddo S. Rapamycin rescues TDP-43 mislocalization and the associated low molecular mass neurofilament instability. J. Biol. Chem. 2009;284:27416–27424. doi: 10.1074/jbc.M109.031278. doi:10.1074/jbc.M109.031278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 240.Zhang Y.J., Xu Y.F., Dickey C.A., Buratti E., Baralle F., Bailey R., Pickering-Brown S., Dickson D., Petrucelli L. Progranulin mediates caspase-dependent cleavage of TAR DNA binding protein-43. J. Neurosci. 2007;27:10530–10534. doi: 10.1523/JNEUROSCI.3421-07.2007. doi:10.1523/JNEUROSCI.3421-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 241.Shankaran S.S., Capell A., Hruscha A.T., Fellerer K., Neumann M., Schmid B., Haass C. Missense mutations in the progranulin gene linked to frontotemporal lobar degeneration with ubiquitin-immunoreactive inclusions reduce progranulin production and secretion. J. Biol. Chem. 2008;283:1744–1753. doi: 10.1074/jbc.M705115200. doi:10.1074/jbc.M705115200. [DOI] [PubMed] [Google Scholar]
- 242.Baker M., Mackenzie I.R., Pickering-Brown S.M., Gass J., Rademakers R., Lindholm C., Snowden J., Adamson J., Sadovnick A.D., Rollinson S., et al. Mutations in progranulin cause tau-negative frontotemporal dementia linked to chromosome 17. Nature. 2006;442:916–919. doi: 10.1038/nature05016. doi:10.1038/nature05016. [DOI] [PubMed] [Google Scholar]
- 243.Cruts M., Gijselinck I., van der Zee J., Engelborghs S., Wils H., Pirici D., Rademakers R., Vandenberghe R., Dermaut B., Martin J.J., et al. Null mutations in progranulin cause ubiquitin-positive frontotemporal dementia linked to chromosome 17q21. Nature. 2006;442:920–924. doi: 10.1038/nature05017. doi:10.1038/nature05017. [DOI] [PubMed] [Google Scholar]
- 244.Cruts M., Kumar-Singh S., Van Broeckhoven C. Progranulin mutations in ubiquitin-positive frontotemporal dementia linked to chromosome 17q21. Curr. Alzheimer Res. 2006;3:485–491. doi: 10.2174/156720506779025251. doi:10.2174/156720506779025251. [DOI] [PubMed] [Google Scholar]
- 245.Yin F., Banerjee R., Thomas B., Zhou P., Qian L., Jia T., Ma X., Ma Y., Iadecola C., Beal M.F., et al. Exaggerated inflammation, impaired host defense, and neuropathology in progranulin-deficient mice. J. Exp. Med. 207:117–128. doi: 10.1084/jem.20091568. S1–4. doi:10.1084/jem.20091568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 246.Levine B., Kroemer G. Autophagy in the pathogenesis of disease. Cell. 2008;132:27–42. doi: 10.1016/j.cell.2007.12.018. doi:10.1016/j.cell.2007.12.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 247.Ju J.S., Fuentealba R.A., Miller S.E., Jackson E., Piwnica-Worms D., Baloh R.H., Weihl C.C. Valosin-containing protein (VCP) is required for autophagy and is disrupted in VCP disease. J. Cell. Biol. 2009;187:875–888. doi: 10.1083/jcb.200908115. doi:10.1083/jcb.200908115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 248.Wang X., Fan H., Ying Z., Li B., Wang H., Wang G. Degradation of TDP-43 and its pathogenic form by autophagy and the ubiquitin-proteasome system. Neurosci. Lett. 2010;469:112–116. doi: 10.1016/j.neulet.2009.11.055. doi:10.1016/j.neulet.2009.11.055. [DOI] [PubMed] [Google Scholar]
- 249.Urushitani M., Sato T., Bamba H., Hisa Y., Tooyama I. Synergistic effect between proteasome and autophagosome in the clearance of polyubiquitinated TDP-43. J. Neurosci. Res. 2010;88:784–797. doi: 10.1002/jnr.22243. [DOI] [PubMed] [Google Scholar]
- 250.Kim S.H., Shi Y., Hanson K.A., Williams L.M., Sakasai R., Bowler M.J., Tibbetts R.S. Potentiation of amyotrophic lateral sclerosis (ALS)-associated TDP-43 aggregation by the proteasome-targeting factor, ubiquilin 1. J. Biol. Chem. 2009;284:8083–8092. doi: 10.1074/jbc.M808064200. doi:10.1074/jbc.M808064200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 251.Haapasalo A., Viswanathan J., Bertram L., Soininen H., Tanzi R.E., Hiltunen M. Emerging role of Alzheimer's disease-associated ubiquilin-1 in protein aggregation. Biochem. Soc. Trans. 2010;38:150–155. doi: 10.1042/BST0380150. doi:10.1042/BST0380150. [DOI] [PubMed] [Google Scholar]
- 252.Kawaguchi Y., Kovacs J.J., McLaurin A., Vance J.M., Ito A., Yao T.P. The deacetylase HDAC6 regulates aggresome formation and cell viability in response to misfolded protein stress. Cell. 2003;115:727–738. doi: 10.1016/s0092-8674(03)00939-5. doi:10.1016/S0092-8674(03)00939-5. [DOI] [PubMed] [Google Scholar]
- 253.Boyault C., Gilquin B., Zhang Y., Rybin V., Garman E., Meyer-Klaucke W., Matthias P., Muller C.W., Khochbin S. HDAC6-p97/VCP controlled polyubiquitin chain turnover. EMBO J. 2006;25:3357–3366. doi: 10.1038/sj.emboj.7601210. doi:10.1038/sj.emboj.7601210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 254.Boyault C., Zhang Y., Fritah S., Caron C., Gilquin B., Kwon S.H., Garrido C., Yao T.P., Vourc'h C., Matthias P., et al. HDAC6 controls major cell response pathways to cytotoxic accumulation of protein aggregates. Genes Dev. 2007;21:2172–2181. doi: 10.1101/gad.436407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 255.Pandey U.B., Nie Z., Batlevi Y., McCray B.A., Ritson G.P., Nedelsky N.B., Schwartz S.L., DiProspero N.A., Knight M.A., Schuldiner O., et al. HDAC6 rescues neurodegeneration and provides an essential link between autophagy and the UPS. Nature. 2007;447:859–863. doi: 10.1038/nature05853. [DOI] [PubMed] [Google Scholar]
- 256.Lee J.Y., Koga H., Kawaguchi Y., Tang W., Wong E., Gao Y.S., Pandey U.B., Kaushik S., Tresse E., Lu J., et al. HDAC6 controls autophagosome maturation essential for ubiquitin-selective quality-control autophagy. EMBO J. 2010 doi: 10.1038/emboj.2009.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 257.Watts G.D., Wymer J., Kovach M.J., Mehta S.G., Mumm S., Darvish D., Pestronk A., Whyte M.P., Kimonis V.E. Inclusion body myopathy associated with Paget disease of bone and frontotemporal dementia is caused by mutant valosin-containing protein. Nat. Genet. 2004;36:377–381. doi: 10.1038/ng1332. doi:10.1038/ng1332. [DOI] [PubMed] [Google Scholar]
- 258.Custer S.K., Neumann M., Lu H., Wright A.C., Taylor J.P. Transgenic mice expressing mutant forms VCP/p97 recapitulate the full spectrum of IBMPFD including degeneration in muscle, brain and bone. Hum. Mol. Genet. 2010 doi: 10.1093/hmg/ddq050. [DOI] [PubMed] [Google Scholar]
- 259.Halawani D., Latterich M. p97: the cell's molecular purgatory? Mol. Cell. 2006;22:713–717. doi: 10.1016/j.molcel.2006.06.003. doi:10.1016/j.molcel.2006.06.003. [DOI] [PubMed] [Google Scholar]
- 260.Ju J.S., Miller S.E., Hanson P.I., Weihl C.C. Impaired protein aggregate handling and clearance underlie the pathogenesis of p97/VCP-associated disease. J. Biol. Chem. 2008;283:30289–30299. doi: 10.1074/jbc.M805517200. doi:10.1074/jbc.M805517200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 261.Vesa J., Su H., Watts G.D., Krause S., Walter M.C., Martin B., Smith C., Wallace D.C., Kimonis V.E. Valosin containing protein associated inclusion body myopathy: abnormal vacuolization, autophagy and cell fusion in myoblasts. Neuromuscul. Disord. 2009;19:766–772. doi: 10.1016/j.nmd.2009.08.003. doi:10.1016/j.nmd.2009.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 262.Nogalska A., Terracciano C., D'Agostino C., King Engel W., Askanas V. p62/SQSTM1 is overexpressed and prominently accumulated in inclusions of sporadic inclusion-body myositis muscle fibers, and can help differentiating it from polymyositis and dermatomyositis. Acta Neuropathol. 2009;118:407–413. doi: 10.1007/s00401-009-0564-6. doi:10.1007/s00401-009-0564-6. [DOI] [PubMed] [Google Scholar]
- 263.Tresse E., Salomons F.A., Vesa J., Bott L.C., Kimonis V., Yao T.P., Dantuma N.P., Taylor J.P. VCP/p97 is essential for maturation of ubiquitin-containing autophagosomes and this function is impaired by mutations that cause IBMPFD. Autophagy. 2010;6 doi: 10.4161/auto.6.2.11014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 264.Hara T., Nakamura K., Matsui M., Yamamoto A., Nakahara Y., Suzuki-Migishima R., Yokoyama M., Mishima K., Saito I., Okano H., et al. Suppression of basal autophagy in neural cells causes neurodegenerative disease in mice. Nature. 2006;441:885–889. doi: 10.1038/nature04724. doi:10.1038/nature04724. [DOI] [PubMed] [Google Scholar]
- 265.Komatsu M., Waguri S., Chiba T., Murata S., Iwata J., Tanida I., Ueno T., Koike M., Uchiyama Y., Kominami E., et al. Loss of autophagy in the central nervous system causes neurodegeneration in mice. Nature. 2006;441:880–884. doi: 10.1038/nature04723. doi:10.1038/nature04723. [DOI] [PubMed] [Google Scholar]
- 266.Pamphlett R., Kum Jew S. TDP-43 inclusions do not protect motor neurons from sporadic ALS. Acta Neuropathol. 2008;116:221–222. doi: 10.1007/s00401-008-0392-0. doi:10.1007/s00401-008-0392-0. [DOI] [PubMed] [Google Scholar]
- 267.Lin W.L., Dickson D.W. Ultrastructural localization of TDP-43 in filamentous neuronal inclusions in various neurodegenerative diseases. Acta Neuropathol. 2008;116:205–213. doi: 10.1007/s00401-008-0408-9. doi:10.1007/s00401-008-0408-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 268.Davidson Y., Amin H., Kelley T., Shi J., Tian J., Kumaran R., Lashley T., Lees A.J., DuPlessis D., Neary D., et al. TDP-43 in ubiquitinated inclusions in the inferior olives in frontotemporal lobar degeneration and in other neurodegenerative diseases: a degenerative process distinct from normal ageing. Acta Neuropathol. 2009;118:359–369. doi: 10.1007/s00401-009-0526-z. doi:10.1007/s00401-009-0526-z. [DOI] [PubMed] [Google Scholar]
- 269.Nishihira Y., Tan C.F., Toyoshima Y., Yonemochi Y., Kondo H., Nakajima T., Takahashi H. Sporadic amyotrophic lateral sclerosis: widespread multisystem degeneration with TDP-43 pathology in a patient after long-term survival on a respirator. Neuropathology. 2009 doi: 10.1111/j.1440-1789.2008.00999.x. [DOI] [PubMed] [Google Scholar]
- 270.Nishihira Y., Tan C.F., Hoshi Y., Iwanaga K., Yamada M., Kawachi I., Tsujihata M., Hozumi I., Morita T., Onodera O., et al. Sporadic amyotrophic lateral sclerosis of long duration is associated with relatively mild TDP-43 pathology. Acta Neuropathol. 2009;117:45–53. doi: 10.1007/s00401-008-0443-6. doi:10.1007/s00401-008-0443-6. [DOI] [PubMed] [Google Scholar]
- 271.Nishihira Y., Tan C.F., Onodera O., Toyoshima Y., Yamada M., Morita T., Nishizawa M., Kakita A., Takahashi H. Sporadic amyotrophic lateral sclerosis: two pathological patterns shown by analysis of distribution of TDP-43-immunoreactive neuronal and glial cytoplasmic inclusions. Acta Neuropathol. 2008;116:169–182. doi: 10.1007/s00401-008-0385-z. doi:10.1007/s00401-008-0385-z. [DOI] [PubMed] [Google Scholar]
- 272.Seilhean D., Cazeneuve C., Thuries V., Russaouen O., Millecamps S., Salachas F., Meininger V., Leguern E., Duyckaerts C. Accumulation of TDP-43 and alpha-actin in an amyotrophic lateral sclerosis patient with the K17I ANG mutation. Acta Neuropathol. 2009;118:561–573. doi: 10.1007/s00401-009-0545-9. doi:10.1007/s00401-009-0545-9. [DOI] [PubMed] [Google Scholar]
- 273.Sampathu D.M., Neumann M., Kwong L.K., Chou T.T., Micsenyi M., Truax A., Bruce J., Grossman M., Trojanowski J.Q., Lee V.M. Pathological heterogeneity of frontotemporal lobar degeneration with ubiquitin-positive inclusions delineated by ubiquitin immunohistochemistry and novel monoclonal antibodies. Am. J. Pathol. 2006;169:1343–1352. doi: 10.2353/ajpath.2006.060438. doi:10.2353/ajpath.2006.060438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 274.Seelaar H., Schelhaas H.J., Azmani A., Kusters B., Rosso S., Majoor-Krakauer D., de Rijik M.C., Rizzu P., ten Brummelhuis M., van Doorn P.A., et al. TDP-43 pathology in familial frontotemporal dementia and motor neuron disease without Progranulin mutations. Brain. 2007;130:1375–1385. doi: 10.1093/brain/awm024. doi:10.1093/brain/awm024. [DOI] [PubMed] [Google Scholar]
- 275.Lin W.L., Castanedes-Casey M., Dickson D.W. Transactivation response DNA-binding protein 43 microvasculopathy in frontotemporal degeneration and familial Lewy body disease. J. Neuropathol. Exp. Neurol. 2009;68:1167–1176. doi: 10.1097/NEN.0b013e3181baacec. doi:10.1097/NEN.0b013e3181baacec. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 276.Hatanpaa K.J., Bigio E.H., Cairns N.J., Womack K.B., Weintraub S., Morris J.C., Foong C., Xiao G., Hladik C., Mantanona T.Y., et al. TAR DNA-binding protein 43 immunohistochemistry reveals extensive neuritic pathology in FTLD-U: a midwest-southwest consortium for FTLD study. J. Neuropathol. Exp. Neurol. 2008;67:271–279. doi: 10.1097/NEN.0b013e31816a12a6. doi:10.1097/NEN.0b013e31816a12a6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 277.Josephs K.A., Stroh A., Dugger B., Dickson D.W. Evaluation of subcortical pathology and clinical correlations in FTLD-U subtypes. Acta Neuropathol. 2009;118:349–358. doi: 10.1007/s00401-009-0547-7. doi:10.1007/s00401-009-0547-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 278.Yokota O., Tsuchiya K., Arai T., Yagishita S., Matsubara O., Mochizuki A., Tamaoka A., Kawamura M., Yoshida H., Terada S., et al. Clinicopathological characterization of Pick's disease versus frontotemporal lobar degeneration with ubiquitin/TDP-43-positive inclusions. Acta Neuropathol. 2009;117:429–444. doi: 10.1007/s00401-009-0493-4. doi:10.1007/s00401-009-0493-4. [DOI] [PubMed] [Google Scholar]
- 279.Seelaar H., Klijnsma K.Y., de Koning I., van der Lugt A., Chiu W.Z., Azmani A., Rozemuller A.J., van Swieten J.C. Frequency of ubiquitin and FUS-positive, TDP-43-negative frontotemporal lobar degeneration. J. Neurol. 2009 doi: 10.1007/s00415-009-5404-z. 10.1007/s00415-009-5404-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 280.Leverenz J.B., Yu C.E., Montine T.J., Steinbart E., Bekris L.M., Zabetian C., Kwong L.K., Lee V.M., Schellenberg G.D., Bird T.D. A novel progranulin mutation associated with variable clinical presentation and tau, TDP43 and alpha-synuclein pathology. Brain. 2007;130:1360–1374. doi: 10.1093/brain/awm069. doi:10.1093/brain/awm069. [DOI] [PubMed] [Google Scholar]
- 281.Luty A.A., Kwok J.B., Thompson E.M., Blumbergs P., Brooks W.S., Loy C.T., Dobson-Stone C., Panegyres P.K., Hecker J., Nicholson G.A., et al. Pedigree with frontotemporal lobar degeneration–motor neuron disease and Tar DNA binding protein-43 positive neuropathology: genetic linkage to chromosome 9. BMC Neurol. 2008;8:32. doi: 10.1186/1471-2377-8-32. doi:10.1186/1471-2377-8-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 282.Le Ber I., Camuzat A., Berger E., Hannequin D., Laquerriere A., Golfier V., Seilhean D., Viennet G., Couratier P., Verpillat P., et al. Chromosome 9p-linked families with frontotemporal dementia associated with motor neuron disease. Neurology. 2009;72:1669–1676. doi: 10.1212/WNL.0b013e3181a55f1c. doi:10.1212/WNL.0b013e3181a55f1c. [DOI] [PubMed] [Google Scholar]
- 283.Mackenzie I.R., Foti D., Woulfe J., Hurwitz T.A. Atypical frontotemporal lobar degeneration with ubiquitin-positive, TDP-43-negative neuronal inclusions. Brain. 2008;131:1282–1293. doi: 10.1093/brain/awn061. doi:10.1093/brain/awn061. [DOI] [PubMed] [Google Scholar]
- 284.Roeber S., Mackenzie I.R., Kretzschmar H.A., Neumann M. TDP-43-negative FTLD-U is a significant new clinico-pathological subtype of FTLD. Acta Neuropathol. 2008;116:147–157. doi: 10.1007/s00401-008-0395-x. doi:10.1007/s00401-008-0395-x. [DOI] [PubMed] [Google Scholar]
- 285.Josephs K.A., Lin W.L., Ahmed Z., Stroh D.A., Graff-Radford N.R., Dickson D.W. Frontotemporal lobar degeneration with ubiquitin-positive, but TDP-43-negative inclusions. Acta Neuropathol. 2008;116:159–167. doi: 10.1007/s00401-008-0397-8. doi:10.1007/s00401-008-0397-8. [DOI] [PubMed] [Google Scholar]
- 286.Tartaglia M.C., Sidhu M., Laluz V., Racine C., Rabinovici G.D., Creighton K., Karydas A., Rademakers R., Huang E.J., Miller B.L., et al. Sporadic corticobasal syndrome due to FTLD-TDP. Acta Neuropathol. 2009 doi: 10.1007/s00401-009-0605-1. 10.1007/s00401-009-0605-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 287.Holm I.E., Englund E., Mackenzie I.R., Johannsen P., Isaacs A.M. A reassessment of the neuropathology of frontotemporal dementia linked to chromosome 3. J. Neuropathol. Exp. Neurol. 2007;66:884–891. doi: 10.1097/nen.0b013e3181567f02. doi:10.1097/nen.0b013e3181567f02. [DOI] [PubMed] [Google Scholar]
- 288.Holm I.E., Isaacs A.M., Mackenzie I.R. Absence of FUS-immunoreactive pathology in frontotemporal dementia linked to chromosome 3 (FTD-3) caused by mutation in the CHMP2B gene. Acta Neuropathol. 2009;118:719–720. doi: 10.1007/s00401-009-0593-1. doi:10.1007/s00401-009-0593-1. [DOI] [PubMed] [Google Scholar]
- 289.Kadokura A., Yamazaki T., Lemere C.A., Takatama M., Okamoto K. Regional distribution of TDP-43 inclusions in Alzheimer disease (AD) brains: their relation to AD common pathology. Neuropathology. 2009;29:566–573. doi: 10.1111/j.1440-1789.2009.01017.x. doi:10.1111/j.1440-1789.2009.01017.x. [DOI] [PubMed] [Google Scholar]
- 290.Lippa C.F., Rosso A.L., Stutzbach L.D., Neumann M., Lee V.M., Trojanowski J.Q. Transactive response DNA-binding protein 43 burden in familial Alzheimer disease and Down syndrome. Arch. Neurol. 2009;66:1483–1488. doi: 10.1001/archneurol.2009.277. doi:10.1001/archneurol.2009.277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 291.King A., Al-Sarraj S., Shaw C. Frontotemporal lobar degeneration with ubiquitinated tau-negative inclusions and additional alpha-synuclein pathology but also unusual cerebellar ubiquitinated p62-positive, TDP-43-negative inclusions. Neuropathology. 2009;29:466–471. doi: 10.1111/j.1440-1789.2008.00966.x. doi:10.1111/j.1440-1789.2008.00966.x. [DOI] [PubMed] [Google Scholar]
- 292.Schwab C., Arai T., Hasegawa M., Akiyama H., Yu S., McGeer P.L. TDP-43 pathology in familial British dementia. Acta Neuropathol. 2009;118:303–311. doi: 10.1007/s00401-009-0514-3. doi:10.1007/s00401-009-0514-3. [DOI] [PubMed] [Google Scholar]
- 293.Isaacs A.M., Powell C., Webb T.E., Linehan J.M., Collinge J., Brandner S. Lack of TAR-DNA binding protein-43 (TDP-43) pathology in human prion diseases. Neuropathol. Appl. Neurobiol. 2008;34:446–456. doi: 10.1111/j.1365-2990.2008.00963.x. doi:10.1111/j.1365-2990.2008.00963.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 294.Honda M., Arai T., Fukazawa M., Honda Y., Tsuchiya K., Salehi A., Akiyama H., Mignot E. Absence of ubiquitinated inclusions in hypocretin neurons of patients with narcolepsy. Neurology. 2009;73:511–517. doi: 10.1212/WNL.0b013e3181b2a6af. doi:10.1212/WNL.0b013e3181b2a6af. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 295.Mateen F.J., Josephs K.A. TDP-43 is not present in brain tissue of patients with schizophrenia. Schizophr. Res. 2009;108:297–298. doi: 10.1016/j.schres.2008.08.033. doi:10.1016/j.schres.2008.08.033. [DOI] [PubMed] [Google Scholar]