Abstract
Motivation: Splice junction microarrays and RNA-seq are two popular ways of quantifying splice variants within a cell. Unfortunately, isoform expressions cannot always be determined from the expressions of individual exons and splice junctions. While this issue has been noted before, the extent of the problem on various platforms has not yet been explored, nor have potential remedies been presented.
Results: We propose criteria that will guarantee identifiability of an isoform deconvolution model on exon and splice junction arrays and in RNA-Seq. We show that up to 97% of 2256 alternatively spliced human genes selected from the RefSeq database lead to identifiable gene models in RNA-seq, with similar results in mouse. However, in the Human Exon array only 26% of these genes lead to identifiable models, and even in the most comprehensive splice junction array only 69% lead to identifiable models.
Contact: whwong@stanford.edu
Supplementary information: Supplementary data are available at Bioinformatics online.
1 INTRODUCTION
Alternative splicing is a common mode of gene regulation within cells, being used by 90–95% of human genes (Pan et al., 2008; Wang et al., 2008). Alternative splicing can drastically alter the function of a gene in different tissue types or environmental conditions, or even inactivate the gene completely. Therefore, it is not surprising that alternative splicing is implicated in many diseases (Le et al., 2005; Wang et al., 2003). Precise modeling of tissue- or cell- dependent alternative splicing is therefore of utmost importance.
Alternative splicing can be studied by microarrays containing probes targeting individual exons or junctions. Common array designs include the Affymetrix Exon 1.0 ST array, which contains four probes targeting observed and predicted exons, and the HJAY array from Affymetrix, which contains eight probes targeting observed and predicted exons and splice junctions. Except where noted, we will restrict our attention to probes targeting RefSeq transcripts.
Current arrays are not guaranteed to produce identifiable estimates for isoform-specific expression. For some genes the isoform expressions are non-identifiable in the sense that the expressions of the different isoforms are confounded with each other and also with the probe-specific effects so that they cannot be estimated separately no matter how many replicate experiments are performed to reduce noise. As we will see below, non-identifiability can be substantially reduced by the use of RNA-Seq. However, even in this case the sheer complexity of some isoform sets may still render the estimation problem non-identifiable based on current RNA-Seq protocols. In view of these difficulties, it is important to have a method to detect all isoform sets that are identifiable by a given array design or a given RNA-Seq protocol. Such a method will be useful for understanding the extent of non-identifiability in current transcriptome analysis methods, and for finding ways in which this problem can be abated.
2 METHODS
To derive a characterization of identifiable isoform sets, we start with a popular model for the analysis of exon and junction arrays (Anton et al., 2008; Le et al., 2005; Pan et al., 2004; Wang et al., 2003), which was an extension of the model originally proposed for oligonucleotide gene expression arrays (Li and Wong, 2001).
| (1) |
where yij is the (known) intensity of probe j in array i, ωik is the (unknown) concentration of isoform k on array i, ϕj is the (unknown) affinity of probe j, δkj is the (known) preference of probe j for isoform k and ϵij is random error. Here, we assume δkj=1 if the probe is expected to bind to the transcript, and 0 otherwise, although setting 0 < δkj < 1 to model cross-hybridization is possible.
For any k-mer that belongs to an isoform of a gene, there is a maximum set of isoforms that share this k-mer. All k-mers with the same maximum set of isoforms are said to form a unique probe class. Here and in the sequel, we only consider a k-mer that is unique in the sense that it is mapped to a unique genomic locus (possibly to splice junctions produced from this locus). For the purpose of identifiability, it is convenient to combine the probes into unique probe sets, where a unique probe set is the set of all probes on an array coming from a unique probe class. For example, all non-cross-hybridizing constitutive probes targeting a particular gene would constitute a unique probe set; also, all probes targeting a certain exon-skipping junction constitute a unique probe set because these probes uniquely target the set of isoforms in which this exon is skipped. We note that the concept of a unique probe set is different from the concept of a probe set on an array, since a unique probe set can contain probes from several array probe sets. We let j index the unique probe sets, and yij be the average intensity for all the probes in the unique probe set.
Model (1) translates in a fairly straightforward manner to a model for RNA-Seq data. Let j index the probe classes. Then, yij is the number of reads in run i which belong to class j, and ϕj is a sampling rate for feature j, which is generally assumed to be proportional to the total number of k-mers that belong to class j. As before, ωik are the isoform concentrations and δkj are indicators of whether probe class j is contained in isoform k. Errors can be modeled via the Poisson distribution (Jiang and Wong, 2009).
As noted in (Wang et al., 2003) this model can suffer from identifiability issues. However, by adding appropriate junctions, this problem may become identifiable (Fig. 1). Below we present a sufficient condition to guarantee identifiability of the model, and use it to analyze existing and future designs for exon and splice junction arrays. This condition is general and can apply to any type of alternative splicing event, no matter how complex. See (Fig. 2) for examples. An R script for checking this condition on a set of isoforms is available at http://biogibbs.stanford.edu/~djhiller/nonid_test.R .
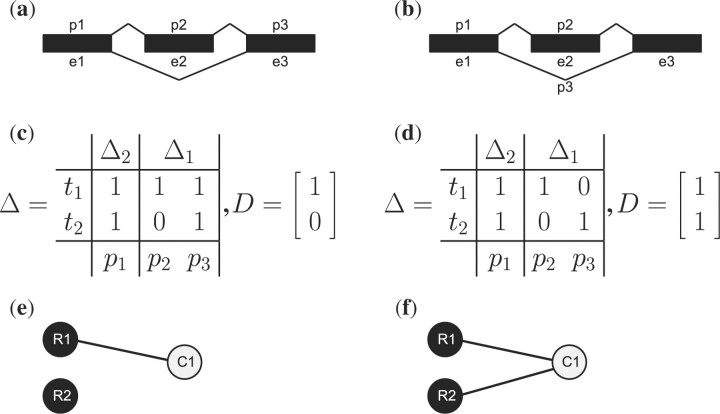
Fig. 1.
(a) Suppose e1 and e3 are constitutive exons and e2 is alternative. Let t1 denote the exon-including isoform and t2 denote the exon-skipping isoform. Let probes p1, p2 and p3 target e1, e2 and e3, respectively. Suppose p1 has intensity (200, 400) in two arrays, and p2 and p3 each have intensity (100, 100). The isoform that includes e2 must be expressed the same in both arrays. The expression ratio of the skipped isoform cannot be determined, however. If ϕ1 = ϕ2 = ϕ3, the skipped isoform is in a ratio of 1:3 in the two arrays. If, however, ϕ1 = 2ϕ2 = 2ϕ3, the ratio becomes 0:1. In other words, we do not even know whether the skipped isoform was present in the first array. (b) If we let p3 target the exon skipping junction, with intensities (0, 200), then the isoform abundances can be deconvolved up to a constant. The skipped isoform ratio is 0:1 (c) The matrices Δ and D corresponding to the case in (a). (d) The matrices Δ and D corresponding to the case in (b). (e) The graph corresponding to D in (c) is disconnected, indicating non-identifiability. R1 and R2 are the rows, C1 is the column. (f) The graph corresponding to D in (d) is connected, indicating identifiability.
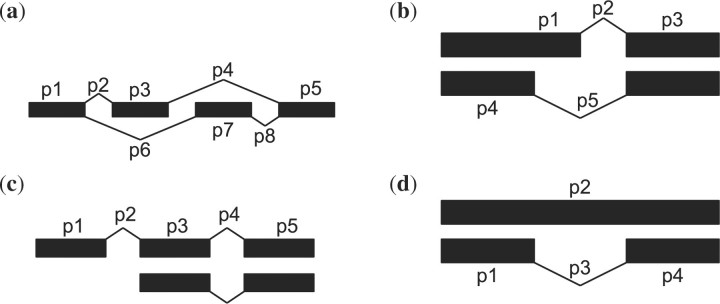
Fig. 2.
Application of identifiability criterion to simple alternative splicing events. In all of the following a probe has been placed on every exon and every observed junction. (a) Mutually exclusive exons. Mutually exclusive events are always identifiable. (b) Alternate 3′ or 5′ exon end. These events are identifiable unless the alternative portion is on either end of the transcript. (c) Alternative transcription start site or alternative polyadenylation site. These events are not identifiable in arrays, where the probe effects are unknown. However, they are identifiable in RNA-Seq if we assume that the sampling rates are known. (d) Intron retention. These events are always identifiable.
Definition. —
Suppose that X is a M × N matrix. Let G(X)=(U, V, E) be a bipartite graph corresponding to X, such that U has M nodes, V has N nodes, and there is an edge between ui and vj iff Xij≠0.
Theorem. —
Suppose there are I arrays, J probes and K isoforms. Model (1) can be written in matrix form as
where Ω has elements ωik, Δ has elements δkj, and Φ has diagonal elements ϕj and off-diagonal elements equal to 0. Model (1) is identifiable under the following conditions:
If the probe affinities are known (as in RNA-Seq), we must be able to choose a set S of K probes which are independent in the following sense: Let Δ1 be a K × K matrix with elements equal to δkj with j∈S, then Δ1 is invertible.
If the probe affinities are unknown, we need two other conditions. Let S′ be the set of probes not in S, and let Δ2 have elements equal to δkj with j∈S′. Consider the matrix D=Δ1−1 Δ2. Suppose that Ω is of full rank (which will be true almost surely if the ωik are considered random quantities over a continuous support space), and I≥K. Suppose further that the graph G(D) is connected. Then the model is identifiable. On the other hand, suppose G(D) is not connected, and we do not assume anything about Ω. Then the model is not identifiable. (Fig. 1)
Proof: —
where A represents a perturbation of the transcript abundances that would lead to the same observations yij. Since Φ is invertible, we can simplify:
(2) By assumption, there exists a K × K submatrix of Δ which is invertible. Then Δ has a right inverse ΔR such that Δ ΔR=I, and
(3) Thus given any Ŷ, Δ and Φ there exists a unique estimate for Ω.
(4) where B* is a diagonal matrix with diagonal elements bj. A and B* represent perturbations of the transcript abundances and the probe effects. We will let B=(B*)−1−I, where B is a one-to-one mapping of B*, assuming the diagonal elements are all non-zero. Reducing (5), we have
(5)
(6)
(7) We rewrite (8) as a system of block equations:
(8)
(9) where
(10) We can manipulate (9) into:
(11) Substituting into (10), we get
(12) Since Ω is of full row rank, it follows that Ω is left invertible, so there exists ΩL such that ΩLΩ=I. Canceling, we get:
(13) Premultiplying (14) by (Δ1)−1 gives us:
(14) where D=Δ1−1Δ2. This means that for every i, j:
(15) So Dij ≠ 0 implies that B2j=B1i. Then for each i and j, either B1i=B2j, or Dij=0. But since D is connected, for any i and j there exist probes m1,…, mL such that Dim1≠0, Dmlml+1≠0 and Dmlml−1≠0 for all even l such that 1 ≤ l < L, and DLj≠0. But then for all i and j, B1i=B2j, so B=(k − 1)I for some k. Then substituting B into (8):
(16)
(17) In other words, given Δ and Ŷ there exist estimates of Φ and Ω which are unique up to rescaling by k. Therefore, the model is identifiable.
(18) Proof of necesarry condition for identifiability when probe effects are unknown: suppose G(D) is disconnected. Starting from (13) we can get
(19) Now suppose G(D) has two unconnected groups. Then we can partition the diagonal elements of B1 and B2 into two groups such that elements in the first group have the value k1, and those in the second group have k2, but it is possible that k1 ≠ k2. Thus, there exist solutions of (15) other than B1 = kI, B2 = kI. But these are also solutions of (13), so there are solutions of (5) other than B = kI. Therefore, the model is not identifiable.
3 RESULTS AND DISCUSSION
We scanned 2256 alternatively spliced human genes (Supplementary Material) for identifiability by the above criterion in four situations: on the Human Exon Array; on the HJAY Array; on the simulated array described below; and in RNA-Seq, in which the sampling rates are known (Table 1). These genes represent a subset of the 4084 genes with multiple isoforms in the RefSeq database [Pruitt et al., 2007, downloaded on April 15, 2008 from UCSC Table Browser (Karolchik et al., 2004) for human genome assembly hg18, NCBI build 36]. The remaining genes were excluded from the analysis either because they could not be reliably mapped to a transcript cluster on both arrays, or because the number of RefSeq transcripts mapping to a transcript cluster was different than the number of transcripts belonging to the original gene, usually indicating that multiple genes mapped to the same cluster.
Table 1.
Summary of the analysis with the Exon, HJAY and simulated arrays and RNA-Seq for 2256 alternatively spliced human genes
| Platform | Unique probesets | Percent identifiable | No. of probes/probeset |
|---|---|---|---|
| Human Exon | 6342 | 26.1 | 4 |
| HJAY | 8339 | 69.0 | 8 |
| Simulated | 9325 | 96.4 | NA |
| RNA-Seq | 9325 | 97.0 | NA |
Column 2 gives the number of probesets which target unique combinations of isoforms. Column 3 gives the fraction of alternatively spliced genes which lead to identifiable models under that platform. Column 4 gives the number of probes per probeset for the arrays. NA, not applicable
The simulated array was constructed as follows. First, we split each gene into probe sets which were either whole exons or, in the case that an exon could have multiple lengths, portions of exons. A simulated probe was assigned to every exon of length ≥25 bp. Additionally, a simulated probe was assigned to every junction observed in the RefSeq database such that the total length of the two exons spanned was ≥25 bp.
In RNA-Seq, 97% of the alternatively spliced genes lead to identifiable gene models. On the simulated array, 96% of the models are identifiable. However, the situation is not so good on actual arrays: in the Exon Array only 26% of gene models were identifiable; and in the HJAY array, which performed significantly better, still only 69% of the gene models were identifiable. These numbers appear to be relatively stable even when we include the unmappable and inconsistent genes eliminated earlier. As a further check, we performed a parallel analysis on 1118 mouse Refseq genes (Supplementary Material; downloaded on August 4, 2009 from UCSC Table Browser for mouse genome assembly mm9, NCBI build 37) using the Mouse Exon array, a mouse simulated array constructed similarly to the human simulated array, and RNA-Seq (Table 2). The mouse genes were chosen based on the same selection criteria as the human genes. The similarity between the mouse and human numbers is striking.
Table 2.
Summary of the analysis with the Exon and simulated arrays and RNA-Seq for 1118 mouse genes
| Platform | Unique probesets | Percent identifiable | No. of probes/probeset |
|---|---|---|---|
| Mouse Exon | 2840 | 29.4 | 4 |
| Simulated | 4176 | 97.0 | NA |
| RNA-Seq | 4176 | 97.9 | NA |
NA, not applicable
While the results for the simulated array and for RNA-Seq seem encouraging, this analysis does not generally take into account practical difficulties in placing probes on particular features. One difficulty which we have attempted to account for is the difficulty in placing probes on short exons. In practise, probes targeting neighboring exon–exon junctions will supply much of the same information as a probe targeting the missing exon would have. A second concern is cross-hybridizing probes, which have not been discarded from the current analysis. A form of cross-hybridization particularly of concern for splice junction arrays is half junction crosstalk (Srinivasan et al., 2005), which happens when a junction probe is bound by a transcript that contains only one of the two exons. However, as long as each unique probe class contains at least one non-cross-hybridizing probe, the identifiability results would not be affected. A third concern is that many probes may have to be discarded due to poor sequence quality, for instance, abnormal GC content. This concern indeed may account for at least some of the discrepancy between the HJAY and simulated arrays. The second and third concerns are at least partially addressed by RNA-Seq: the problem of cross-hybridization is reduced to locations which share high sequence similarity to other regions, and the problem of probe selection is circumvented entirely, although it may be replaced by the problem of low sampling rate on a particular feature.
Identifiability issues are critical for quantification of alternate splice forms. A non-identifiable gene model may grossly mis-estimate relative isoform abundances, even declaring a present isoform absent or vice versa (Fig. 1). For this reason, the discrepancy between the proportion of identifiable gene models on the HJAY array and the theoretical optimum is interesting, and it is worth taking a closer look at the possible reasons for this discrepancy. As we noted above, the exclusion of potential probe sets could explain much of the gap. Another observation is that the model is especially sensitive to the number of unique probe sets on the array. In the HJAY array, an 11% reduction in the number of unique probe sets leads to a 28% drop in identifiability. Even more striking, in the Exon Array a 32% drop in the unique probe sets causes a 73% drop in identifiability. This analysis suggests that too stringent of a probe selection criteron may limit the ability to accurately deconvolve isoform concentrations from expression data, particularly when all the probes in a given probe class are eliminated. This analysis also suggests the superiority of RNA-Seq as a tool for alternative splicing analysis, because of its ability to reduce many of the problems inherent in array-based methods.
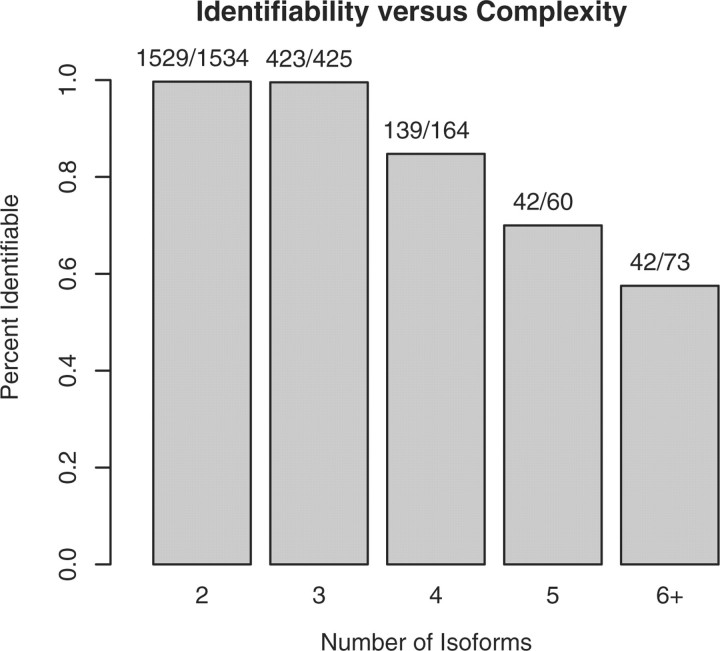
A significant limitation in isoform deconvolution models is that the probability of an identifiable gene model decreases sharply as the number of transcripts increases (Fig. 3). This issue is less significant when using the RefSeq database, because 97% of alternatively spliced genes contain five or fewer transcripts. However, as we include more transcripts, the results quickly deteriorate. In the HJAY array, for example, when considering all 14 800 genes with multiple transcripts in the Refseq and Ensembl databases (58 000 transcripts), the rate drops modestly to 46.7 (Ensembl release 38, April 2006; Hubbard et al., 2007). If we consider all 17 300 genes having multiple predicted transcripts (3 00 000 transcripts), the rate drops to 24.2%. Thus, model (1) is not suitable when we wish to include an arbitrary number of transcripts. Instead, for each gene, we must choose a small set of transcripts which we expect to account for most or all of the transcripts in the cell type being studied. As an alternative to using the RefSeq transcripts, a short list could also be generated from a single run of RNA-Seq on a pooled sample. RNA-Seq is likely to be better suited for novel isoform discovery, due to the digital nature of the measurements and the decreased level of uncertainty in the sampling rate.
Fig. 3.
Bar plot of the percent of gene models which are identifiable against the number of isoforms in the gene model, using data from the human simulated array. Overall, 96% of the gene models were identifiable.
We briefly consider the 3% of genes which are non-identifiable even when using RNA-Seq, to see what are the most difficult situations for current alternative splicing protocols. In 90% of these cases, two alternative splicing events were separated by one or more constitutive exons. Even in the remaining cases, one can always find a subset of transcripts such that this subset contains an exon which is constitutive and which separates two alternative splicing events. Thus, a fundamental limitation of junction arrays and single end RNA-Seq is that they are only able to assess local properties of a transcript. It is possible that paired end sequencing technology will be able to go further in addressing this challenge. In any case, a possible solution for now would be, rather than to quantify the concentration of each transcript, to quantify the rate at which a particular alternative splice event (delineated by constitutive exons) occurs.
ACKNOWLEDGEMENTS
We thank Michael Saunders, Junhee Seok and Wenzhong Xiao for useful discussions.
Funding: National Institute of Health grant (R01-HG004634 to W.H.W.). Ric Weiland Graduate Fellowship (to D.H.). National Institutes of Health grant (U54-GM062119 to W.X.).
Conflict of Interest: none declared.
REFERENCES
- Anton MA, et al. Space: an algorithm to predict and quantify alternatively spliced isoforms using microarrays. Genome Biol. 2008;9:R46. doi: 10.1186/gb-2008-9-2-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hubbard TJP, et al. Ensembl 2007. Nucleic Acids Res. 2007;35:D610–D617. doi: 10.1093/nar/gkl996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jiang H, Wong W. Statistical inferences for isoform expression in RNA-seq. Bioinformatics. 2009;25:1026–1032. doi: 10.1093/bioinformatics/btp113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karolchik D, et al. The UCSC Table Browser data retrieval tool. Nucleic Acids Res. 2004;32:D493–D496. doi: 10.1093/nar/gkh103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le K, et al. Detecting tissue-specific regulation of alternative splicing as a qualitative change in microarray data. Nucleic Acids Res. 2005;32:e180. doi: 10.1093/nar/gnh173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C, Wong W. Model-based analysis of oligonucleotide arrays: Expression index computation and outlier detection. Proc. Natl Acad. Sci. USA. 2001;98:31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pan Q, et al. Revealing global regulatory features of mammalian alternative splicing using a quantitative microarray platform. Mol. Cell. 2004;16:929–941. doi: 10.1016/j.molcel.2004.12.004. [DOI] [PubMed] [Google Scholar]
- Pan Q, et al. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet. 2008;40:1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- Pruitt K, et al. NCBI reference sequences (RefSeq): a curated non-redundant sequence database of genomes, transcripts and proteins. Nucleic Acids Res. 2007;35:D55–D60. doi: 10.1093/nar/gkl842. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srinivasan K, et al. Detection and measurement of alternative splicing using splicing-sensitive microarrays. Methods. 2005;37:345–359. doi: 10.1016/j.ymeth.2005.09.007. [DOI] [PubMed] [Google Scholar]
- Wang ET, et al. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, et al. Gene structure-based splice variant deconvolution using a microarray platform. Bioinformatics. 2003;19:i315–i322. doi: 10.1093/bioinformatics/btg1044. [DOI] [PubMed] [Google Scholar]