Abstract
Nuclear receptors are ligand-modulated transcription factors that respond to steroids, retinoids, and thyroid hormones to control development and body physiology. Orphan nuclear receptors, which lack identified ligands, provide a unique, and largely untapped, resource to discover new principles of physiologic homeostasis. We describe the isolation and characterization of the vertebrate orphan receptor, BXR, which heterodimerizes with RXR and binds high-affinity DNA sites composed of a variant thyroid hormone response element. A bioactivity-guided screen of embryonic extracts revealed that BXR is activatable by low-molecular-weight molecules with spectral patterns distinct from known nuclear receptor ligands. Mass spectrometry and 1H NMR analysis identified alkyl esters of amino and hydroxy benzoic acids as potent, stereoselective activators. In vitro cofactor association studies, along with competable binding of radiolabeled compounds, establish these molecules as bona fide ligands. Benzoates comprise a new molecular class of nuclear receptor ligand and their activity suggests that BXR may control a previously unsuspected vertebrate signaling pathway.
Keywords: Xenopus Orphan nuclear receptor, BXR, benzoates, vertebrate signaling pathway
A search for nuclear receptors expressed early in Xenopus laevis development identified cDNAs encoding-transcripts from nine different genes, including the retinoic acid receptors xRARα and xRARγ, the retinoid x receptors xRXRα and xRXRγ, and five different orphan receptors termed XOR-1, XOR-2, XOR-4, XOR-6, and XOR-7 (Xenopus Orphan Receptor; Blumberg et al. 1992). The presence of this diversity of receptors early in development suggests the existence of potential new embryonic ligands. Because all nonsteroid nuclear receptors employ the 9-cis retinoic acid receptor RXR as a common heterodimeric partner, we used this as a preliminary screen for potential hormone inducibility. As described here, we demonstrate that XOR-6 heterodimerizes with RXR both in vitro and in vivo, and preferentially binds a variant DR-4 response element found in the promoter of the rat Pit-1 gene (Rhodes et al. 1993). This enabled the development of a bioactivity-guided screening approach that led to the identification of alkyl amino benzoates as potent and selective agonists and to the renaming of XOR-6 as BXR (for benzoate X receptor). Benzoates represent a new molecular class of nuclear receptor activators. These activators specifically bind to purified BXR protein and appropriately modulate interactions between BXR and nuclear receptor cofactors in vitro. The identification of this activatable receptor, its heterodimerization, DNA, and ligand-binding properties support the existence of a novel vertebrate hormonal signaling pathway and highlight the importance of orphan nuclear receptors in establishing new directions to endocrine physiology.
Results
BXR defines a novel class of orphan nuclear receptor
A new member of the steroid receptor superfamily was isolated from a screen to identify developmental regulatory factors present in a X. laevis embryonic cDNA library (Fig. 1a). The 2191-bp cDNA encodes a predicted protein of 386 amino acids that is related to the vitamin D3 receptor (VDR) (Fig. 1b). The proteins are 73% identical in the DNA-binding domains and 42% in the ligand-binding domain. Among orphan receptors, BXR shows the greatest similarity to the recently identified steroid “X” receptor (B. Blumberg, H. Juguilon, J. Bolado Jr., E.S. Ong, and R.M. Evans, in prep.), the pregnane “X” receptor, PXR (Kliewer et al. 1998), and CAR (previously MB67) (Fig. 1b) (Baes et al. 1994). All of these receptors require heterodimerization with RXR for high-affinity DNA binding. SXR is activatable by a variety of steroids (B. Blumberg, H. Juguilon, J. Bolado Jr., E.S. Ong, and R.M. Evans, in prep.) whereas CAR constitutively activates transcription from βRARE (Baes et al. 1994). Aside from VDR, SXR, PXR, and CAR, BXR shows no more similarity to other nuclear receptors than the different receptor subfamilies do to each other. Following the original isolation of BXR (XOR-6 in Blumberg et al. 1992), a related Xenopus cDNA, ONR-1, was reported (Smith et al. 1994). Although BXR and ONR-1 are more than 99% identical, ONR-1 was reported to differ in its DNA-binding preferences (Smith et al. 1994; see below). Genomic Southern blotting suggests that BXR-related genes occur in a variety of vertebrate species, including human, mouse, and zebrafish (data not shown), therefore BXR appears to define a new subfamily of nuclear receptors. Together, BXR, SXR, PXR, CAR, and VDR appear to constitute a distinct branch of the nuclear receptor family tree.
Figure 1.

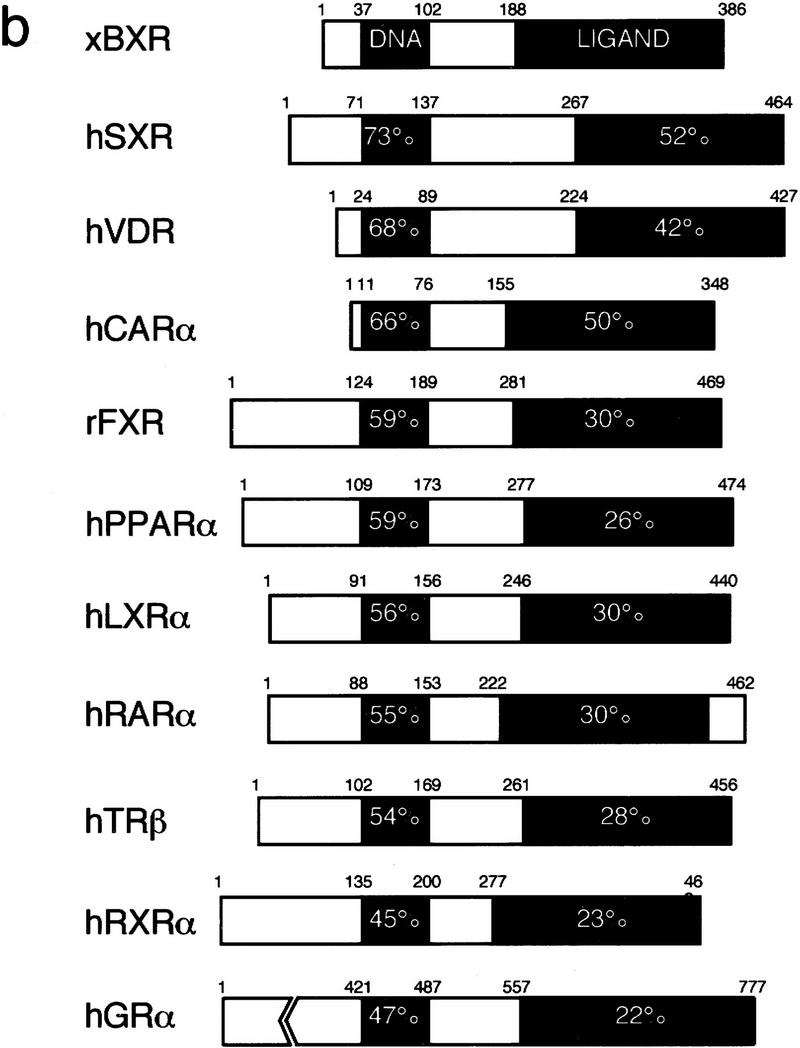
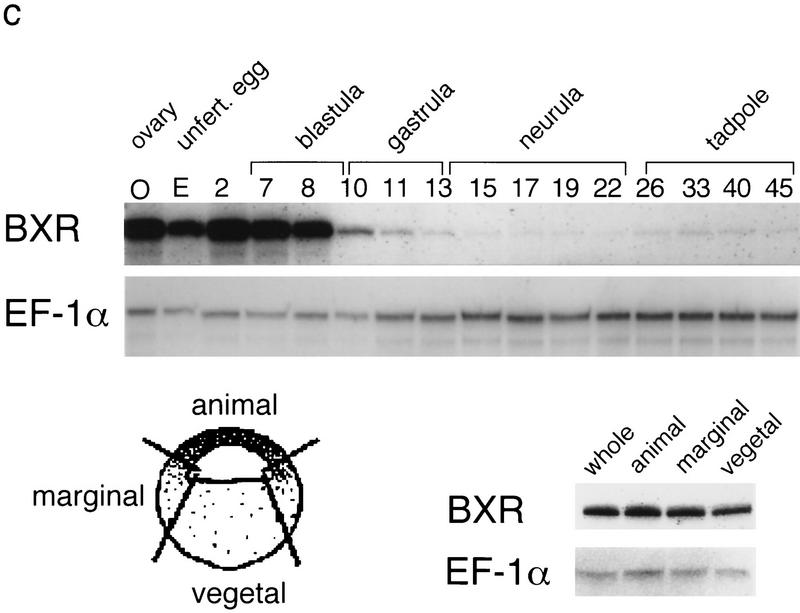
BXR is a novel member of the steroid receptor superfamily. (a) Sequence of the longest BXR cDNA clone. The DNA and ligand-binding domains are boxed. In-frame termination codons are indicated by asterisks. The sequence probably represents an incomplete cDNA, as the canonical AAUAAA polyadenylation signal is not present upstream of the terminal A residues. (b) Schematic comparison between BXR, other known RXR heterodimeric partners, and the human glucocorticoid receptor. Amino acid sequences were aligned using the program GAP (Devereaux et al. 1984). The similarity between the BXR and other receptors is expressed as percent amino-acid identity. (c) Analysis of BXR mRNA expression. The RNase protection probes used are the following: EF-1a (nucleotides 790–1167); BXR (nucleotides 1314–1560). RNase protection was performed with total RNA from the indicated stages and are abbreviated as follows: (O) total ovary (10 μg); (E) unfertilized egg (40 μg); (2) two-cell (40 μg); (7 and 8) blastula (40 μg); (10 and 11) gastrula (stage 10, 10 μg; stage 11, 8 μg); (13–22) neurula (4 μg); (26 and 33) tailbud (4 μg); (40 and 45) swimming tadpole (4 μg). RNase protection was performed with 20 μg of total RNA from whole embryos or dissected animal caps, marginal zone, and vegetal pole.
To determine which embryonic stages might be influenced by BXR activation, we employed RNase protection assays to measure steady-state mRNA levels over a developmental time sequence. BXR mRNA is present in the unfertilized egg and remains at a relatively constant level until gastrulation (Fig. 1c). It persists thereafter at a much reduced level until at least stage 45. In addition, there is no obvious localization of the maternally encoded BXR mRNA to the animal cap, marginal zone or endoderm of early blastula embryos (Fig. 1).
In vitro DNA binding
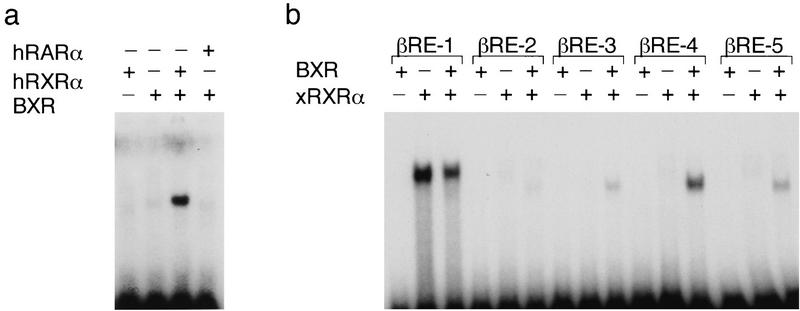
To create a suitable reporter gene for the ligand-screening experiments, in vitro DNA-binding studies were used to determine the mode and specificity of BXR DNA binding. Figure 2a shows that BXR and hRXRα heterodimerize to bind DNA in a cocktail of natural and synthetic hormone response elements. Binding is strongly cooperative, as neither receptor alone showed DNA binding at the protein concentrations used in the assay, and specific to hRXRα (or xRXRα), because hRARα did not enhance BXR DNA binding. A finer analysis of BXR:xRXRα-binding specificity revealed selectivity for a subset of the known response elements in the cocktail. The preferred target is a βDR-4 motif, although some binding to βDR-3 and βDR-5 sites is also seen (Fig. 2b). A scan of naturally occuring targets revealed that the RDE in the mouse Pit-1 gene (RDE, AGTTCATGAGAGTTCA) is a high-affinity element (Rhodes et al. 1993; see below) that was used in a reporter construct [tk-(RDE)2-luc] to screen for BXR activation.
Figure 2.
BXR heterodimerizes with RXR to enable DNA binding. Gel mobility shift analyses of BXR DNA-binding specificity. (a) In vitro-transcribed and -translated proteins were mixed with a cocktail of hormone response elements containing DR0, DR1, PPRE, DR2, MLV–TRE, SPP1, βRARE, GRE, and ERE. (b) BXR and xRXRα proteins were mixed and incubated with the indicated response elements. βRE-1 through βRE-5 are direct repeats of the sequence AGTTCA, separated by 1–5 nucleotides. Reaction conditions and gel electrophoresis were as described (Perlmann et al. 1993). We note also that xRXRα alone binds strongly to the DR-1 motif in this assay and migrates in a position appropriate for xRXRα homodimers. (c) BXR and hRXRα interact in vivo. The indicated plasmids were cotransfected into CV-1 cells along with the reporter tk(galp)3–luc and CMX–βgal. Note the strong suppression of basal transcription when GAL–BXR was added (right). This is characteristic of many known ligand-dependent RXR heterodimeric partners.
BXR and RXR form functional heterodimers in vivo
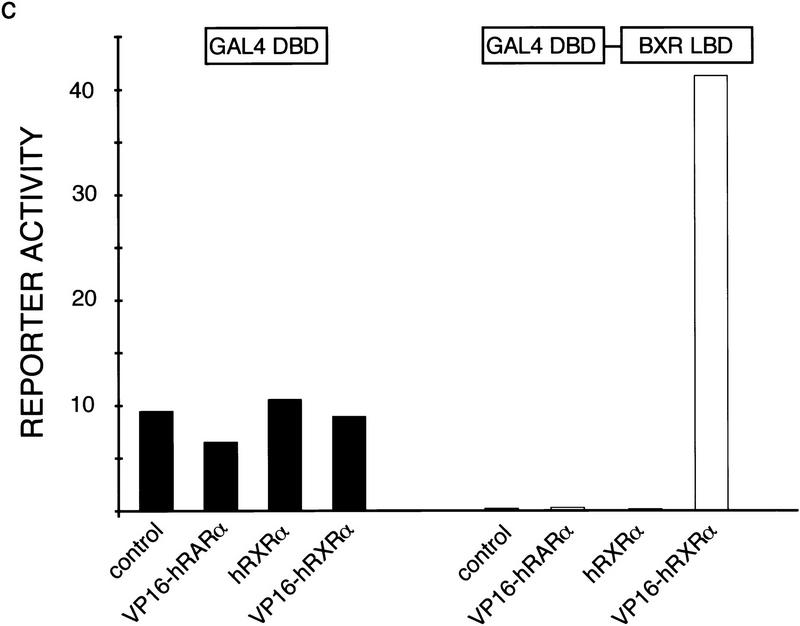
To show that BXR and RXR interact in vivo, we employed a modification of the two-hybrid system for analysis in mammalian cells (Forman et al. 1995a; Perlmann and Jansson 1995; Willy et al. 1995). This system relies on functional dimeric interactions between two fusion proteins, one carrying the GAL4 DNA-binding domain fusion with the ability to bind to a GAL4UAS-containing reporter gene. The other carries the VP16 transactivation domain fusion. When expressed together, DNA binding and transcriptional activation is reconstituted in a single complex. Functional interaction between GAL–BXR and VP16–hRXRα should lead to constitutive activation of the tk(GAL)–luc reporter. As shown in Figure 2c, activation of the reporter occurred only when GAL–BXR and VP16–hRXRα were cotransfected. Moreover, GAL–BXR showed strong suppression of reporter gene basal activity (Fig. 2c, right), which parallels effects elicited by GAL–hRARα, GAL–hTRβ, and GAL–hVDR. We conclude that BXR and RXRα form heterodimers in vivo, that GAL–BXR is unable to activate target genes in the absence of a putative ligand, and that unliganded GAL–BXR, like TR, RAR, and VDR suppresses basal activity of reporter constructs to which it can bind.
BXR is ligand dependent
As BXR apparently defines a novel RXR-dependent, orphan nuclear receptor, we initiated a systematic attempt to identify candidate activators using a bioactivity-guided fractionation approach (Blumberg et al. 1996). The BXR ligand search was based on the presumption that it would be co-localized with its receptor during development. Organic extracts from a mixture of receptor-positive Xenopus embryonic stages were prepared, fractionated by reversed phase HPLC, and tested for BXR activation in transfected CV-1 cells. An activity was identified and purified to homogeneity by reversed-phase HPLC using several solvent systems. A comparison between the activity profile of one purified agonist and its UV absorbance at 254 nm is shown in Figure 3a. The electron impact mass spectrum fragmentation pattern of this embryonic agonist (Fig. 3b) matched that of the library compound, 3-amino ethyl benzoate (3-AEB) (Fig. 3c). Synthetic 3-AEB co-eluted with the purified agonist and showed an identical UV spectrum (data not shown). Chemically pure 3-AEB specifically activates BXR alone among known RXR partners and other vertebrate nuclear receptors (Fig. 3d; data not shown).
Figure 3.
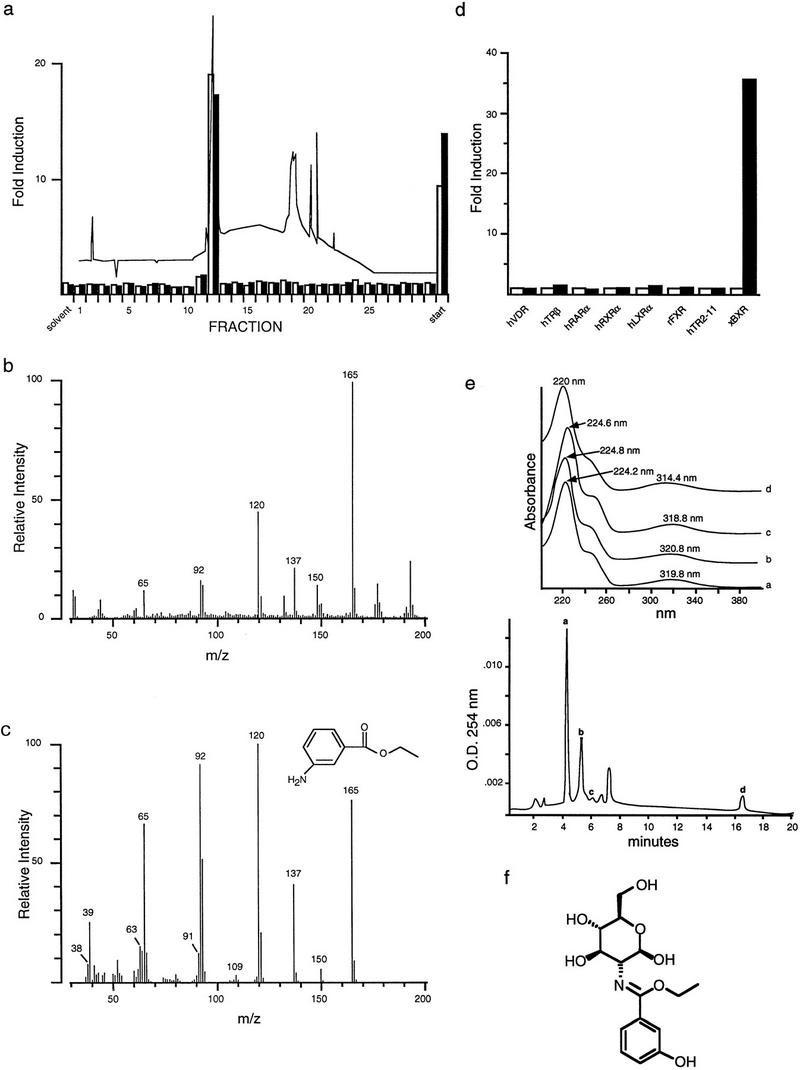
Identification of a BXR agonist. (a) Activation of GAL–BXR (□) and full-length BXR (▪) by fractions from the final purification of BXR agonist. CV-1 cells were transfected with 1 μg of receptor expression plasmid, 5 μg of reporter, and 4 μg of CMX–βgal control plasmid per 5 × 105 cells. 00 254 nm. HPLC fractions were dried, resuspended in 25 μl of methanol, and added in triplicate to cells. (b) Electron impact mass spectrum of the purified BXR agonist. (c) Electron impact mass spectrum of 3-amino-ethyl benzoate. (d) Activation analysis of 10−4 m 3-AEB (▪) on a variety of GAL4–DNA-binding domain/receptor ligand (□) binding domain chimeras. (e, bottom panel) Chomatogram of polar benzoates from mixed embryonic extracts. Apparent benzoate-containing peaks are indicated as a–d; (top panel) UV spectra of endogenous benzoates in active HPLC fractions from embryonic extracts. (f) Structure of ethyl-3-hydroxyl benzoate glucosamine derived from peak a.
Although analysis of extracts from multiple embryonic stages and other tissues, such as ovary, revealed several additional activities, HPLC analysis showed that all possessed UV absorbtion spectra similar to 3-AEB, suggesting that they are related compounds (Fig. 3e). Sufficient material from one of these peaks (peak a) was obtained for 1H NMR and tandem mass spectrometry and it was shown to be a glucosamine conjugate of 3-hydroxy ethyl benzoate (Fig. 3f). Taken together, these data suggest that a family of structurally related benzoate compounds naturally occurring in Xenopus embryos are BXR activators.
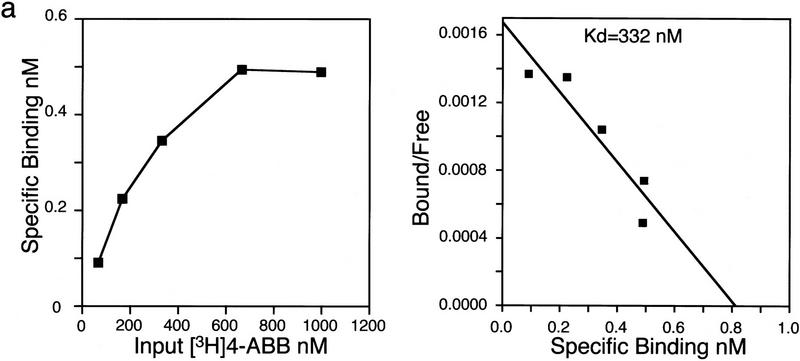
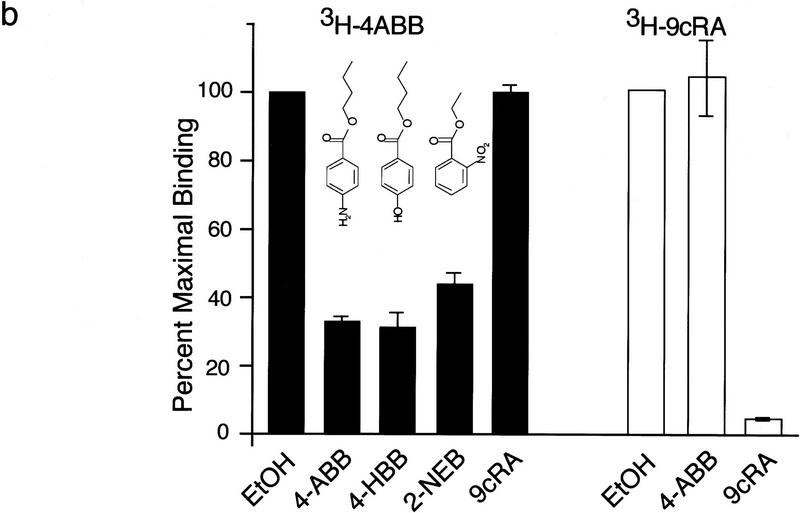
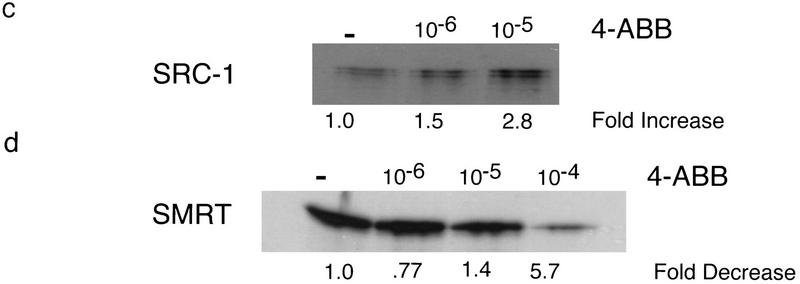
Whether benzoate activators function as bona fide ligands for BXR was addressed using direct receptor binding assays and in vitro cofactor association assays. Purified, baculovirus-expressed full-length BXR:hRXRα heterodimers were tested for binding using 3H-labeled 4-amino butyl benzoate (4-ABB), the most potent activator (see below). 4-ABB binding by BXR:hRXRα heterodimers is saturable as would be expected for specific ligand binding (Fig. 4a). Nonlinear regression and Scatchard analysis establish a Kd of 332 nm for 4-ABB binding by BXR. This binding is specifically competable by benzoates but not by RXR ligands, such as 9-cis-RA (Fig. 4b). RXR is able to bind 9-cis-RA in this heterodimer and this binding is competable by 9-cis-RA but not by 4-ABB (Fig. 4b). 4-ABB induces association between the receptor heterodimer and the nuclear receptor coactivator SRC-1 (Fig. 4c) and dissociation of the complex between BXR, hRXRα, and the transcriptional corepressor SMRT (Fig. 4d; Chen and Evans 1995). The ability of 4-ABB to bind to purified BXR protein and appropriately modulate interactions between the BXR:hRXRα heterodimer and nuclear receptor cofactors, in vitro, establishes it as a bona fide BXR ligand.
Figure 4.
BXR is a benzoate receptor. (a, left panel) Saturable binding of 3H-labeled-4-ABB to purified BXR:hRXRα heterodimers. Purified heterodimeric BXR (2.5 μg) was incubated with the indicated amount of 3H-labeled 4-ABB. Nonspecific binding was determined by competition with excess unlabeled 4-ABB (10−4 m) and subtracted from total binding. (Right panel) Scatchard transformation of the data in the left panel. Kd was determined by nonlinear regression analysis using GraphPad Prism software. (b) Direct binding of radio-labeled 4-ABB to purified BXR:hRXRα heterodimers. (4-HBB) 4-hydroxyl-butyl benzoate; (2NEB) 2-nitro ethyl benzoate. (c) 4-ABB increases the affinity of BXR:hRXRα heterodimers for the nuclear receptor coactivator SRC-1 in vitro. The total radioactivity in each band was determined by phosphorimager analysis and is reported below the image as fold increase over solvent control. (d) 4-ABB inhibits in vitro interactions between BXR:hRXRα heterodimers and the nuclear receptor corepressor SMRT. Phosphorimager quantitation is shown below the image as fold decrease over solvent control.
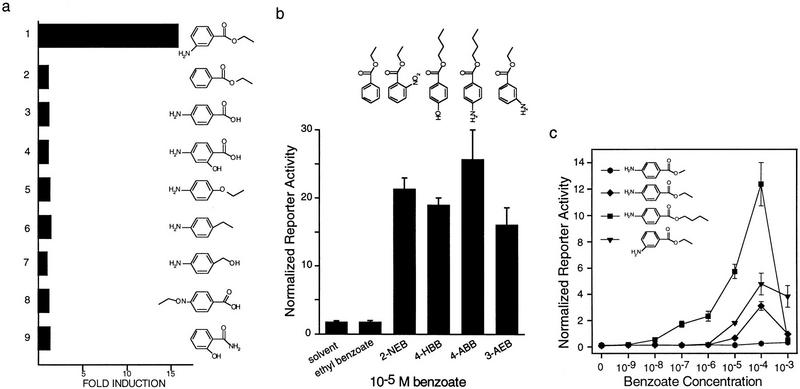
Structure–activity analysis of the benzene ring substituents shows that whereas the ester moiety is required for activation (Fig. 5a), the amino group can be replaced by hydroxyl or nitro but not hydrogen (Fig. 5b). Altered positioning of the amino group showed that there were only moderate differences in the activity of ortho, meta, or para conformers (data not shown).
Figure 5.
Stereochemical requirements for BXR activation. All compounds were tested as dilution series (10−3–10−9 m) for their ability to activate full-length BXR or GAL–BXR using the cotransfection assay as described above. (a) A number of classes of compounds were tested. For simplicity, one representative example from each group is shown. (1) 3-AEB; (2) benzoate esters ranging from methyl to butyl; (3) 2-,3-, or 4-amino benzoic acids; (4) 3- and 4-amino salicylic acids; (5) o-, m-, and p-phenetedines; (6) 2-,3-, and 4-ethyl anilines; (7) 2-,3-, and 4-amino benzyl alcohols; (8) 2-,3-, and 4-acetamidobenzoic acids; (9) salicylamide. Results with 10−4 m of each compound are shown. (b) The effects of substituting the amino group with NO2, OH, or H were tested. (c) The transcriptional effects of increasing the alkyl chain length of the esterified moiety were evaluated using full-length BXR and GAL–BXR in the context of amino benzoates. Results are shown for GAL–BXR.
We then tested the effect of varying the length and shape of the alkyl group esterified to the amino benzoate moiety, reasoning that increasing the overall hydrophobicity of the molecule might improve transport into the cell (Ma et al. 1992, Roy et al. 1993) and therefore activation of BXR. 4-Amino benzoate esters in which the R group was varied from methyl to hexadecyl and including phenyl and geometric isomers of the shorter alkyl groups (isopropyl, isobutyl) were tested (Fig. 5c; data not shown). The more lipophilic alkyl esters showed increased activation with a rank potency of butyl > isobutyl > propyl > isopropyl benzoate ethyl >> methyl. However, increasing the hydrophobicity by extending the alkyl chain to hexyl or hexadecyl, or by substituting phenyl, impaired activation, suggesting that molecular shape is more important than net hydrophobicity for transcriptional activation (data not shown).
Because certain amino benzoic acid esters are local anesthetics that function by blocking sodium channels (Collins 1993), we asked whether BXR activation could be an indirect effect of local anesthesia. Accordingly we tested a variety of common local anesthetics, both similar and different from 3-AEB, for BXR activation. Despite the fact that nearly all of the compounds tested are more potent anesthetics than 3-AEB, none of them activated BXR (data not shown). We also tested the effect of activating protein kinase A or C pathways on BXR activation with, respectively, isobutyl methyl xanthine or phorbol esters. In neither case could we detect receptor-dependent activation of the reporter gene construct (data not shown). Together with the direct ligand-binding data, these pharmacological experiments argue against the possibility that an indirect, membrane-mediated effect is responsible for the activation of BXR by amino benzoates. The specific transcriptional activation (Figs. 3 and 5), binding (Fig. 4a,b), and cofactor association (Fig. 4c,d) suggest that BXR is a novel, ligand-dependent nuclear receptor that can be specifically activated by certain endogenous benzoates.
Discussion
Orphan nuclear receptors represent a diverse and ancient component of the nuclear receptor superfamily, being found in nearly all Metazoan species examined. Although orphan receptors are more numerous (19 families) than receptors with known ligands (11 families), not all orphans (and perhaps only a few) can be expected to represent undisclosed endocrine systems. Our strategy to discover these systems was to first identify embryonic orphan receptors, hypothesizing that one or more of these might be ligand dependent. One of these embryonic orphan receptors, BXR, was identified as an obligate RXR heterodimeric partner, both in vitro and in vivo. BXR:RXR heterodimers bind preferentially to direct repeats of the sequence AGTTCA separated by four nucleotides. BXR and RXR interact in vivo to activate a GAL4-based reporter in the two-hybrid assay.
The demonstration that BXR forms a heterodimer with RXR and the identification of a high-affinity DNA-binding site provided the basis of a bioassay-guided ligand screen. The mRNA expression analysis suggested that a putative ligand would be produced during early embryogenesis and therefore identified a source of endogenous compound. Using these tools, a class of compounds comprised of short-chain alkyl esters of substituted benzoic acids were identified as BXR activators and ligands. This work reveals a novel class of nuclear receptor ligands and simultaneously suggests the existence of a new embryonic signaling pathway.
As these compounds are both isolated from embryos and bind BXR in vitro, they are likely to be bona fide endogenous ligands. We estimate that the total concentration of embryonic benzoates is at least 10−5 m at the blastula stage (data not shown). This correlates well with the concentration required for effective receptor activation (Fig. 5c) and suggests that endogenous benzoates function as signaling molecules. The identification of four endogenous benzoates leads one to wonder whether all are authentic receptor ligands or rather if one is the true ligand, whereas the others represent precursors, breakdown products, or storage forms. The compound shown in Figure 3f is likely to be a storage form that can be easily hydrolyzed to 3-hydroxyl-ethyl benzoate. Understanding the relationship among the four molecules however, must await their structural elucidation.
It is intriguing that the endogenous benzoates are related to the nutrient p-amino benzoic acid (PABA). PABA is an integral component of the essential B-vitamin folic acid. Because PABA is generated from folate breakdown, it is impossible to achieve PABA deficiency in mammals without folic acid deficiency. This implies that some symptoms of folate deficiency might result from the absence of folate-derived PABA rather than folate itself. Folate is required for the synthesis of methylcobalamin, which in turn is essential for normal synthesis of purines, and pyrimidines, and therefore DNA. This methyl transfer is also essential for the generation of tetrahydrofolate, which contributes to intermediary metabolism by functioning as a substrate for numerous metabolic reactions. In addition, the folate-cobalamin interaction catalyzes the conversion of methylmalonyl coenzyme A (CoA) to succinyl CoA, which is critical to lipid metabolism and triglyceride synthesis. Therefore, it is possible that BXR may have a previously unexpected role in these metabolic pathways, perhaps by controlling the activity of genes regulating pivotal enzymatic steps. Interestingly, folate lowers blood levels of homocysteine, which in high levels has been linked to heart disease and hypertension. This further suggests a connection betwen folate metabolism and BXR activation. In summary, our discovery of BXR as a ligand-depedent receptor suggests the existence of a new physiologic pathway and provides the initial tools to establish its role in normal development, physiology, and disease.
Materials and methods
Identification of BXR
BXR was identified in a screen for maternally expressed nuclear hormone receptors (Blumberg et al. 1992). Three clones were identified from an egg cDNA library, an additional two were isolated from a dorsal blastopore lip cDNA library. The longest clone was sequenced completely on both strands using a combination of directed subcloning and specific oligonucleotide priming. DNA sequences were compiled and aligned using the programs of Staden (1986), University of Wisconsin Genetics Computer Group (Devereaux et al. 1984), and Feng and Doolittle (1987). Database searching was performed using the BLAST network server at the National Center for Biotechnology Information (Altschul et al. 1990).
RNA preparation and analysis
RNA was prepared from fertilized X. laevis eggs and staged embryos as described (Blumberg et al. 1992). The temporal and spatial patterns of expression were determined using RNase protection (Blumberg et al. 1992). The probe was nucleotides 1314–1560, which represents the last three amino acids of the protein and part of the 3′ untranslated region (UTR). For localization studies, stage 8–9 embryos were dissected into animal, marginal, and vegetal fragments and RNA was prepared using a proteinase K method (Blumberg et al. 1992).
In vitro DNA binding
DNA-binding analysis was performed using in vitro-transcribed, -translated proteins and oligonucleotides as described previously (Umesono et al. 1991; Perlmann et al. 1993).
Cell culture and transfection
A suitable eukaryotic expression vector for these studies was constructed from the commercially available vector pCDNAI–AMP (Invitrogen). This vector allows expression from the strong cytomegalovirus early promoter and bacteriophage T7 and SP6 promoter-driven production of sense and antisense RNA, respectively. The cloning strategy was as follows. The three endogenous NcoI sites were removed by site-directed mutagenesis, the polylinker region between XhoI and XbaI was removed by double digestion, endfilling, and self ligation. A cassette consisting of the Xenopus β-globin leader and trailer derived from the plasmid pSP36T (Amaya et al. 1991), separated by a synthetic polylinker (containing unique sites for NcoI, SphI, EcoRI, SalI, EcoRV, BamHI, and XbaI), was inserted between HindIII and NotI sites in the vector. The resulting plasmid, designated pCDG1, can be linearized with NotI to produce mRNA from the bacteriophage T7 promoter. The BXR protein-coding region was cloned between the NcoI and BamHI sites of pCDG1 and designated pCDG–BXR. pCMX–GAL4–BXR was constructed by cloning nucleotides encoding amino acids 103–386 of BXR protein into the SalI–XbaI sites of pCMX–GAL4 (Umesono et al. 1991; Perlmann et al. 1993). pCMX–VP16 receptor chimeras were constructed by fusing the potent VP16 transactivation domain (Sadowski et al. 1988) to the amino terminus of the full-length hRXRα, hRARα, or VDR protein-coding regions.
CV-1 cells were maintained in Dulbecco’s modified Eagle medium (DMEM) containing 10% resin-charcoal stripped fetal bovine serum and liposome-mediated transfections performed in triplicate as described (Blumberg et al. 1996). Reporter gene expression was normalized to the β-galactosidase transfection control.
3-AEB isolation and identification
Fresh or flash frozen Xenopus embryos were homogenized in a large volume of 50% CH2Cl2 50% CH3OH, typically 10 ml/gram of tissue. Denatured proteins were removed by filtration through diatomaceous earth (Sigma) and the liquid phase recovered and evaporated to dryness with a Buchi rotary evaporator. The resulting material was resuspended in a minimum volume of iso-octane and transferred to a separatory funnel. Nonpolar and polar compounds were separated by partitioning between large volumes of iso-octane and methanol. The BXR agonist partitioned primarily into methanol. The methanol phase was partitioned between ethyl acetate and H2O with the BXR agonist partitioning >95% into ethyl acetate. The ethyl acetate phase was dried and fractionated by reversed-phase HPLC using several solvent systems. Absorbance was monitored between 200–600 nm using a Waters 996 photodiode array detector and fractions were collected, dried, and tested in the cotransfection assay for their ability to activate BXR. Active fractions were pooled and rechromatographed on the same column using a gradient of methanol, 10 mm NH4-acetate at pH 7.5, beginning at 30% methanol, held for 15 min, and increasing to 100% methanol over the next 45 min. Fractions were again tested for bioactivity and active fractions pooled. Final purification was accomplished using a dioxane/water gradient beginning at 20% dioxane and then increasing linearly to 100% dioxane from 15 to 45 min.
The electron impact mass spectrum was measured on a JEOL JMS-HX100 double-focusing mass spectrometer using a 70 eV electron impact ionization source, 180°C ion source temperature, about 40°C direct insertion probe temperature, and a resolution of 5000.
Solid-phase extraction was employed to isolate potentially more polar compounds. Mixed embryonic stages (11–34) were homogenized in deionized water (60 embryos/ml) and the extract clarified by centrifugation. The supernatant was extracted by stirring with preconditioned (according to the manufacturer’s recommendation) Amberlite XAD-2 nonionic polymeric resin (Sigma) at a ratio of resin/supernatant =  . The mixture was stirred for 210 min and the resin recovered by filtration. After rinsing with water, the adsorbed material was eluted with methanol and acetone. Both methanol and acetone solubles were combined, clarified by filtration, rotary evaporated and reconstituted in 3 ml of chloroform/methanol (1:2 ratio) under argon. The BXR agonists were purified by reversed-phase HPLC using various solvent systems guided by the bioactivity as described above. The resin extract was fractionated on a 22 × 250 mm C18 column (Vydac 218TP15022) using a linear methanol water gradient of 100% water to 70% methanol/water in 30 min and then increasing further to 100% methanol at 35 min. Active fractions were combined and separated further on a 4.6 × 250 mm C18 column (Vydac 201HS54) by isocratic elution developed at 1.5 ml/min with 75% water in acetonitrile. Major activities were purified to homogeneity uisng the same column, using two different isocratic elutions—85% water/acetonitrile for the benzoate glucosamine and 73% water/acetonitrile for 3-amino ethyl benzoate.
. The mixture was stirred for 210 min and the resin recovered by filtration. After rinsing with water, the adsorbed material was eluted with methanol and acetone. Both methanol and acetone solubles were combined, clarified by filtration, rotary evaporated and reconstituted in 3 ml of chloroform/methanol (1:2 ratio) under argon. The BXR agonists were purified by reversed-phase HPLC using various solvent systems guided by the bioactivity as described above. The resin extract was fractionated on a 22 × 250 mm C18 column (Vydac 218TP15022) using a linear methanol water gradient of 100% water to 70% methanol/water in 30 min and then increasing further to 100% methanol at 35 min. Active fractions were combined and separated further on a 4.6 × 250 mm C18 column (Vydac 201HS54) by isocratic elution developed at 1.5 ml/min with 75% water in acetonitrile. Major activities were purified to homogeneity uisng the same column, using two different isocratic elutions—85% water/acetonitrile for the benzoate glucosamine and 73% water/acetonitrile for 3-amino ethyl benzoate.
Electrospray ionization mass spectrometry and electrospray tandem mass spectrometry for the benzoate glucosamine were performed at the mass spectrometry facility in the Scripps Research Institute.
1H NMR analysis
Pure compounds (∼10 μg) were reconstituted in methanol and then dried under vacuum to remove residual water. All NMR experiments were performed using a 500 MHz Inova 500 (Varian) equipped with a nanoprobe (Varian) in 40 μl of 100% deuterated solvents (Cambridge Isotope Laboratories). 1H-labeled NMR spectra were obtained by using a PRESAT sequence in 100% methanol-d4 and 100% acetonitrile-d3. Decoupling experiments were carried out in 100% methanol-d4. DQCOSY spectra were acquired in 100% methanol-d4.
In vitro ligand binding
The full-length BXR-coding region was cloned into the plasmid pAcHLT (Pharmingen), which yielded a His6 fusion protein. Baculovirus production, infection of SF-9 cells, and purification of GST-hRXRα:His6–xBXR heterodimers were as described (Chen et al. 1997). It was empirically determined that coinfection of a virus producing GST–hRXRα was required to recover soluble BXR protein. Ligand-binding assays used protein prebound to glutathione–Sepharose beads. Assays were performed in 300 μl volume and consisted of 1 μm purified BXR:hRXRα heterodimer, 3H-labeled 4-amino butyl benzoate (50 Ci/mmole, American Radiolabeled Chemicals), 20 mm Tris-HCl at pH 7.5, 50 mm KCl, 1.5% CHAPS, 8% glycerol, 1 μg/ml of pepstatin A, 1 μg/ml of leupeptin, and 1 mm Pefablock (Boehringer Manheim). Binding was carried out for 2 hr at 4°C. Subsequently, three 20-min washes were performed at 4°C in the same buffer except that Tris was replaced by 20 mm HEPES, glycerol was 20%, and 1 mm dithiothreitol and 1 mm EDTA were included. Binding was determined by scintillation counting, all assays were performed in duplicate and replicated in independent experiments. Nonspecific binding was determined by including unlabeled 4-ABB in the binding reactions at 10−4 m and subtracted from total binding. Saturation-binding experiments used the radiolabel concentration indicated in the figure. Nonlinear regression and Scatchard analysis to determine the dissociation constant were performed using GraphPad Prism software. Competition-binding experiments contained 2.5 μCi of 3H-labeled 4-ABB, 10−5 m competitor ligand, and were otherwise performed as described above.
In vitro cofactor association
In vitro-transcribed and -translated 35S-labeled SRC-1 and C-SMRT were prepared using the TNT system (Promega). Binding reactions were as described above except that 1% bovine serum albumin was included and radiolabeled ligand was replaced by 1 μl of SRC-1 protein or 5 μl of C-SMRT protein. Binding was for 30 min at 4°C and three 20-min washes were perfomed as above. Subsequently, the beads were resuspended in 30 μl of SDS-loading buffer, boiled for 10 min, and 20 μl loaded onto 12% SDS-polyacrylamide gels. After fixation and salicylate enhancement, the gels were dried and exposed to x-ray film.
Acknowledgments
We thank Bethany Janowski for advice on ligand binding; Dr. Osman Guner (MDL Information Systems, Inc.) for structural database searching; Drs. William Fenical and Bob Ramos for advice on natural product purification and identification; and Drs. Barry M. Forman and John W.R. Schwabe for comments on the manuscript. We especially thank Dr. William Fenical (Scripps Institute of Oceanography) for his generosity in the use of various equipment and supplies and Dr. Adriana Manzi for access to the nanoprobe-equipped 500 MHz Varian NMR in the University of California, San Diego, Cancer Center Glycobiology Core Facility. B.B. is, and K.U. was, a staff scientist at the Salk Institute for Biological Studies. B.B. gratefully acknowledges the American Cancer Society for financial support in the early stages of this work. T.P. was the recipient of a fellowship from the European Molecular Biology Organization. H.K. acknowledges the La Jolla Interdisciplinary Scholars Training Program and the Burroughs Wellcome fund for fellowship support. Work at the University of California, Los Angeles, was supported by the National Institutes of Health (NIH) and the Lucille Markey Charitable trust (B.B. and E.D.R.). R.M.E. is an Investigator of the Howard Hughes Medical Institute at the Salk Institute for Biological Studies. Work at the Salk Institute was supported by the NIH, G. Harold and Leila Y. Mathers Charitable Foundation (R.M.E.), and in part by the Foundation for Medical Research, Inc. (A.G.C.).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL blumberg@axp1.salk.edu; FAX (619) 455-1349.
References
- Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J Mol Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- Amaya E, Musci TJ, Kirschner MW. Expression of a dominant negative mutant of the FGF reeptor disrupts mesoderm formation in Xenopus embryos. Cell. 1991;66:257–270. doi: 10.1016/0092-8674(91)90616-7. [DOI] [PubMed] [Google Scholar]
- Baes M, Gulick T, Choi HS, Martinoli MG, Simha D, Moore DD. A new orphan member of the nuclear hormone receptor superfamily that interacts with a subset of retinoic acid response elements. Mol Cell Biol. 1994;14:1544–1551. doi: 10.1128/mcb.14.3.1544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumberg B, Mangelsdorf DJ, Dyck J, Bittner DA, Evans RM, De Robertis EM. Multiple retinoid-responsive receptors in a single cell: Families of RXRs and RARs in the Xenopus egg. Proc Natl Acad Sci. 1992;89:2321–2325. doi: 10.1073/pnas.89.6.2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blumberg B, Bolado Jr J, Derguini F, Craig AG, Moreno TA, Charkravarti D, Heyman RA, Buck J, Evans RM. Novel RAR ligands in Xenopus embryos. Proc Natl Acad Sci. 1996;93:4873–4878. doi: 10.1073/pnas.93.10.4873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen H, Lin RJ, Schiltz RL, Chakravarti D, Nash A, Nagy L, Privalsky ML, Nakatani Y, Evans RM. Nuclear receptor coactivator ACTR is a novel histone acetyltransferase and forms a multimeric activation complex with P/CAF and CBP/p300. Cell. 1997;90:569–580. doi: 10.1016/s0092-8674(00)80516-4. [DOI] [PubMed] [Google Scholar]
- Chen JD, Evans RM. A transcriptional co-repressor that interacts with nuclear hormone receptors. Nature. 1995;377:454–457. doi: 10.1038/377454a0. [DOI] [PubMed] [Google Scholar]
- Collins VJ. Principles of anesthesiology: General and regional anesthesia. 3rd ed. Philadelphia, PA: Lea and Febiger; 1993. pp. 1232–1281. [Google Scholar]
- Devereaux J, Haeberli P, Smithies O. A comprehensive set of sequence analysis programs for the VAX. Nucleic Acids Res. 1984;12:387–395. doi: 10.1093/nar/12.1part1.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feng D-F, Doolittle RF. Progressive sequence alignment as a prerequisite to correct phylogenetic trees. J Mol Evol. 1987;25:351–360. doi: 10.1007/BF02603120. [DOI] [PubMed] [Google Scholar]
- Forman BM, Umesono K, Chen J, Evans RM. Unique response pathways are established by allosteric interactions among nuclear hormone receptors. Cell. 1995;81:541–550. doi: 10.1016/0092-8674(95)90075-6. [DOI] [PubMed] [Google Scholar]
- Kliewer SA, Moore JT, Wade L, Staudinger JL, Jones MA, McKee DD, Oliver BM, Willson TM, Zetterstrom RH, Perlmann T, Lehmann J. An orphan nuclear receptor activated by pregnanes defines a novel steroid signaling pathway. Cell. 1998;92:73–82. doi: 10.1016/s0092-8674(00)80900-9. [DOI] [PubMed] [Google Scholar]
- Ma L, Ramachandran C, Weiner ND. Bulk organic solvent-water systems as a possible model to predict alkyl p-aminobenzoate partitioning in liposomes. J Pharmaceut Sci. 1992;81:1104–1108. doi: 10.1002/jps.2600811113. [DOI] [PubMed] [Google Scholar]
- Perlmann T, Jansson L. A novel pathway for vitamin A signaling mediated by RXR heterodimerization with NGFI-B and NURR1. Genes & Dev. 1995;9:769–782. doi: 10.1101/gad.9.7.769. [DOI] [PubMed] [Google Scholar]
- Perlmann T, Rangarajan PN, Umesono K, Evans RM. Determinants for selective RAR and TR recognition of direct repeat HREs. Genes & Dev. 1993;7:1411–1422. doi: 10.1101/gad.7.7b.1411. [DOI] [PubMed] [Google Scholar]
- Rhodes SJ, Chen R, DiMattia GE, Scully KM, Kalla KA, Lin S, Yu VC, Rosenfeld MG. A tissue-specific enhancer confers Pit-1-dependent morphogen inducibility on the pit-1 gene. Genes & Dev. 1993;7:913–932. doi: 10.1101/gad.7.6.913. [DOI] [PubMed] [Google Scholar]
- Roy SD, Fujiki J, Fleitman JS. Permeabilities of alkyl p-aminobenzoates through living skin equivalent and cadaver skin. J Pharmaceut Sci. 1993;82:1266–1268. doi: 10.1002/jps.2600821217. [DOI] [PubMed] [Google Scholar]
- Sadowski I, Ma J, Triezenberg S, Ptashne M. GAL4-VP16 is an unusually potent transcriptional activator. Nature. 1988;335:563–564. doi: 10.1038/335563a0. [DOI] [PubMed] [Google Scholar]
- Smith DP, Mason CS, Jones EA, Old RW. A novel nuclear receptor superfamily member in Xenopus that associates with RXR, and shares extensive sequence similarity to the mammalian vitamin D3 receptor. Nucleic Acids Res. 1994;22:66–71. doi: 10.1093/nar/22.1.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staden R. The current status and portability of our sequence handling software. Nucleic Acids Res. 1986;14:217–231. doi: 10.1093/nar/14.1.217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umesono K, Murakami KK, Thompson CC, Evans RM. Direct repeats as selective response elements for the thyroid hormone, retinoic acid and vitamin D3 receptors. Cell. 1991;65:1255–1266. doi: 10.1016/0092-8674(91)90020-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willy PJ, Umesono K, Ong ES, Evans RM, Heyman RA, Mangelsdorf DJ. LXR, a nuclear receptor that defines a distinct retinoid-response pathway. Genes & Dev. 1995;9:1033–1045. doi: 10.1101/gad.9.9.1033. [DOI] [PubMed] [Google Scholar]