Abstract
With the discovery that the Saccharomyces cerevisiae Rad51 protein is both structurally and functionally similar to the Escherichia coli RecA protein, the RecA paradigm for homologous recombination was extended to the Eucarya. The ubiquitous presence of RecA and Rad51 protein homologs raises the question of whether this archetypal protein exists within the third domain of life, the Archaea. Here we present the isolation of a Rad51/RecA protein homolog from the archaeon Sulfolobus solfataricus, and show that this protein, RadA, possesses the characteristics of a DNA strand exchange protein: The RadA protein is a DNA-dependent ATPase, forms a nucleoprotein filament on DNA, and catalyzes DNA pairing and strand exchange.
Keywords: Archaea, RadA, recombination, RecA, DNA strand exchange
Homologous recombination is a process whereby two DNA duplexes interact to transfer genetic information, thus creating new genetic linkages and rearranging segments of their DNA. Genes that are involved in homologous recombination are important for DNA repair, regulation of gene expression, and generation of genetic diversity (Kowalczykowski et al. 1994). A central part of this process is the need for an enzyme to bring two homologous DNA molecules together and to exchange the DNA strands. This crucial function is carried out by the RecA protein in bacteria (Kowalczykowski et al. 1994) and the more recently discovered Rad51 protein in eucarya (Ogawa et al. 1993a). Both of these proteins display striking structural and functional similarities (Story et al. 1993): Upon ATP binding, they nucleate onto DNA to produce a highly structured nucleoprotein filament, in which the DNA is stretched extensively, making it available for homologous pairing (Ogawa et al. 1993b).
The identification of RAD51 homologs in every eukaryote examined has supported the notion that a RecA protein-like mechanism for DNA strand exchange is ubiquitous among the Eucarya (Ogawa et al. 1993a; Heyer 1994) and raised the question of whether this paradigm could be true for the third domain of life, the Archaea. Sandler et al. (1996) reported the first identification and cloning of archaeal genes with significant homology to recA from three different archaeons—Sulfolobus solfataricus, Methanococcus jannaschii, and Haloferax volcanii. These proteins share 42%–49% identity with one another (Sandler et al. 1996) and, interestingly, they are more similar to the eucaryotic Rad51 protein (40% amino acid identity) than to the bacterial RecA protein (20%). Deletion of the radA open reading frame in H. volcanii results in a recombination-deficient archaeon (Woods and Dyall-Smith 1997), providing evidence that this gene bears functional as well as structural resemblance to the RecA and Rad51 proteins. Additionally, radA homologs have been found in each of the three archaeons that have been completely sequenced to date (Bult et al. 1996; Klenk et al. 1997; Smith et al. 1997), showing that this gene is ubiquitous among the Archaea.
Here we present the purification of a Rad51/RecA protein homolog from the hyperthermophilic, sulfur-oxidizing archaeon S. solfataricus. We show that this protein, RadA, possesses the characteristics of a DNA strand exchange protein: It is a DNA-dependent ATPase, it forms a nucleoprotein filament on DNA, it can promote the formation of joint molecules, and can catalyze homologous DNA pairing and strand exchange. All of these reactions occur at elevated temperatures similar to the conditions in which S. solfataricus lives. Thus, RadA protein defines the first DNA strand exchange protein from the Archaea.
Results
RadA protein is a DNA-dependent ATPase
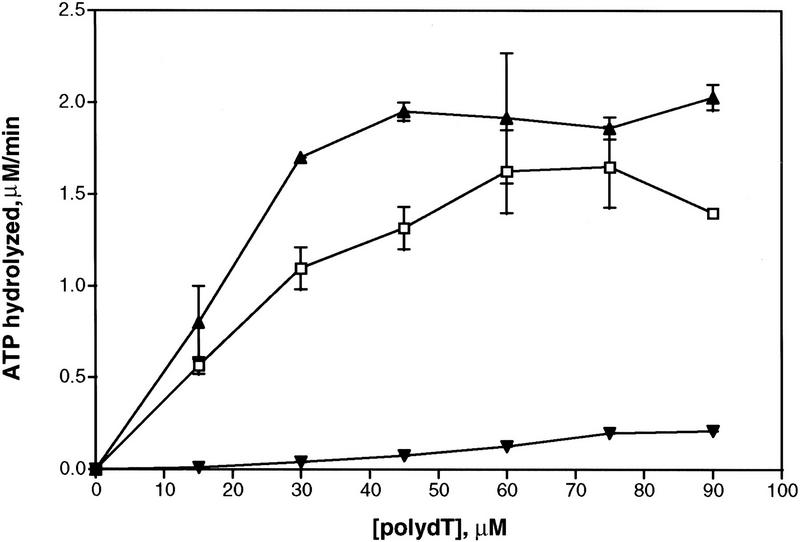
Following purification to near homogeneity (∼90%), the RadA protein was tested for single-stranded DNA (ssDNA)-dependent ATP hydrolysis activity. Similar to that observed for RecA and Rad51 proteins (Cox 1990; Sung 1994), the RadA protein hydrolyzed ATP to ADP only in the presence of both magnesium and ssDNA (Table 1). ATP hydrolysis was less efficient in the presence of double-stranded DNA (dsDNA), and almost no hydrolysis occurred in the absence of DNA. DNA-dependent ATP hydrolysis occurred with a catalytic rate constant (kcat) of 0.10–0.20/min exclusively at elevated temperatures (65°C, 75°C, 85°C), and very little hydrolysis occurred at 37°C. The ATPase activity is comparable to that observed for the Saccharomyces cerevisiae Rad51 protein (kcat of 0.7/min) (Sung 1994) but is substantially lower than that of the Escherichia coli RecA protein (kcat of 30/min) (Kowalczykowski et al 1994). RadA protein also hydrolyzed dATP with a similar efficiency and with the same DNA cofactor and temperature requirements (data not shown). The ATPase activity of RadA protein increased linearly with ssDNA concentration, until a saturation plateau was attained (Fig. 1). At the saturation point, the apparent binding stoichiometry is 1 RadA monomer per 3 nucleotides, as observed for both RecA protein (Lauder and Kowalczykowski 1991) and Rad51 protein (Ogawa et al. 1993b; Sung 1994).
Table 1.
RadA protein possesses DNA-dependent ATPase activity
| Addition or omission
|
Percent conversion of ATP or ADP
|
kcat (/min)
|
|---|---|---|
| −DNA | 2.0 | 0.01 ± 0.02 |
| +poly(dT)(37°C) | 2.7 | 0.02 ± 0.02 |
| +poly(dT)(65°C) | 14 | 0.12 ± 0.04 |
| +poly(dT)(75°C) | 17.5 | 0.14 ± 0.04 |
| +poly(dT)(85°C) | 20 | 0.17 ± 0.03 |
| +poly(dT) −Mg2+ | 0 | 0 |
| +M13 ssDNA | 8.7 | 0.06 ± 0.01 |
| +M13 ssDNA −Mg2+ | 0 | 0 |
| +M13 dsDNA | 4.3 | 0.03 ± 0.01 |
| +poly(dT) +dATP −ATP | 23 | 0.17 ± 0.03 |
The assays were conducted at 65°C, unless otherwise indicated, for 90 min. kcat values were determined from three different sets of data and calculated from the observed rate of ATP hydrolysis (μm ATP hydrolyzed/min) divided by the total concentration of RadA protein, as the concentration of ssDNA was in excess of the protein concentration.
Figure 1.
RadA protein is a DNA-dependent ATPase. This activity saturates at an apparent stoichiometry of 1 RadA monomer per 3 nucleotides at 75°C. The error bars indicate the s.d. for three sets of experiments. (▴) 15 μm RadA at 75°C; (□) 15μm RadA at 65°C; (▾) 15 μm RadA at 37°C.
RadA forms a nucleoprotein filament on DNA
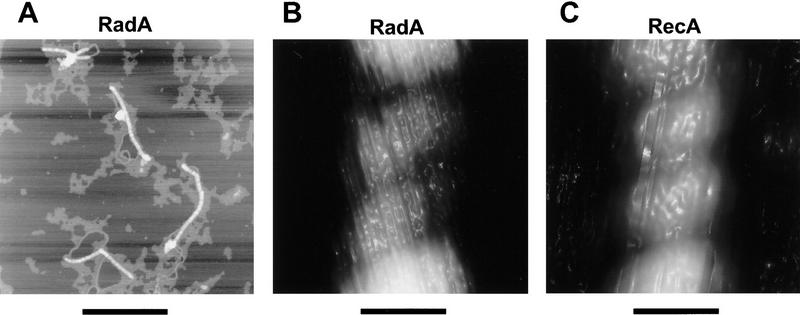
The extended filamentous structure of RecA and Rad51 nucleoprotein filaments is a unique hallmark of this family of proteins. These protein–DNA complexes, either with ssDNA or dsDNA, show striking similarities in helicity, pitch, diameter, and extension of the axial rise per base pair of DNA involved in the complex (Ogawa et al. 1993a; Yu and Egelman 1993; Yu et al. 1995). Complexes of purified RadA protein and pBR322 plasmid DNA were imaged by atomic force microscopy (Fig. 2A). The complexes have the striated appearance of a helical nucleoprotein filament. Although binding is not complete (some free DNA is visible), RadA protein binds to DNA in clusters, suggesting that it binds cooperatively to DNA, as do RecA and Rad51 proteins.
Figure 2.
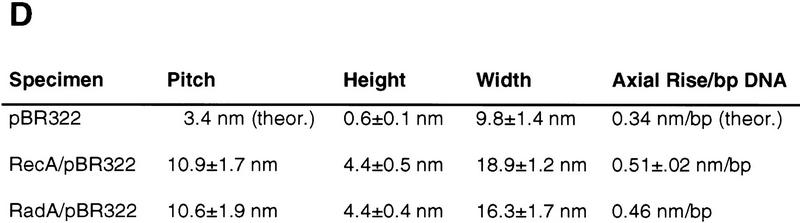
(A) Complexes of RadA protein and pBR322 plasmid DNA imaged by AFM. Bar, 500 nm. (B) The RadA protein-dsDNA filament is a right-handed helical structure that is similar to the RecA protein–dsDNA filament shown in C. Bars (B and C), 10 nm. (D) Physical dimensions of the RecA and RadA nucleoprotein filaments. The axial rise per bp for RadA/pBR322 is based on one measurement. The value reported for RecA/pBR322 is based on three measurements. Reported values for pitch, height, and width are each based on at least 10 measurements.
A highly ordered section is shown in Figure 2B at high magnification. For comparison, a section of a RecA protein–dsDNA complex at the same magnification is shown in Figure 2C. The complexes formed by RadA and RecA proteins in Figure 2 are clearly right-handed helical structures.
The structural characteristics of the RadA protein–DNA complexes were compared to an equivalent RecA protein complex (Fig. 2D). The average pitch of 60 turns from RadA protein–dsDNA filaments is 10.6 ± 1.9 nm. This is in good agreement with the pitch of the RecA nucleoprotein complexes prepared in a similar manner and imaged by atomic force microscopy (AFM), which is 10.9 ± 1.7 nm. Using electron microscopy or neutron scattering (Egelman and Stasiak 1986; DiCapua et al. 1990), other groups have measured a pitch of 9.0–10.0 nm for RecA protein–dsDNA filaments; however, pitches of 10.8 and 11.4 nm were also observed (Yu and Egelman 1992). The average apparent width and height of the RadA protein–dsDNA complexes were determined to be 16.3 ± 1.7 nm and 4.4 ± 0.4 nm, respectively. These dimensions, although consistent with measurements of complexes formed by RecA protein using this technique, are different from the 10 nm diameter measured by others (Egelman and Stasiak 1986; DiCapua et al. 1990). The larger apparent width results from a contribution of the AFM probe diameter to the observed sample width measurement (Bustamante et al. 1992), and the shorter height is most likely due to compression of the sample by the tapping force of the probe tip.
RadA protein also increases the apparent contour length of the bound DNA. The contour length of the pBR322 plasmid DNA alone measured by AFM is consistent with the expected length based on the number of base pairs and the axial rise per base pair of B-form DNA (3.4 Å/bp). Although we observed no complexes covered completely by RadA protein, we could measure the length of the protein-bound region and the protein-free DNA. From these measurements, we estimate that RadA protein extends the axial rise per base pair to 4.6 Å/bp. For comparison, the axial rise per bp of DNA in RecA protein–dsDNA filaments was measured by AFM as 5.1 Å/bp.
RadA protein promotes DNA pairing and strand exchange
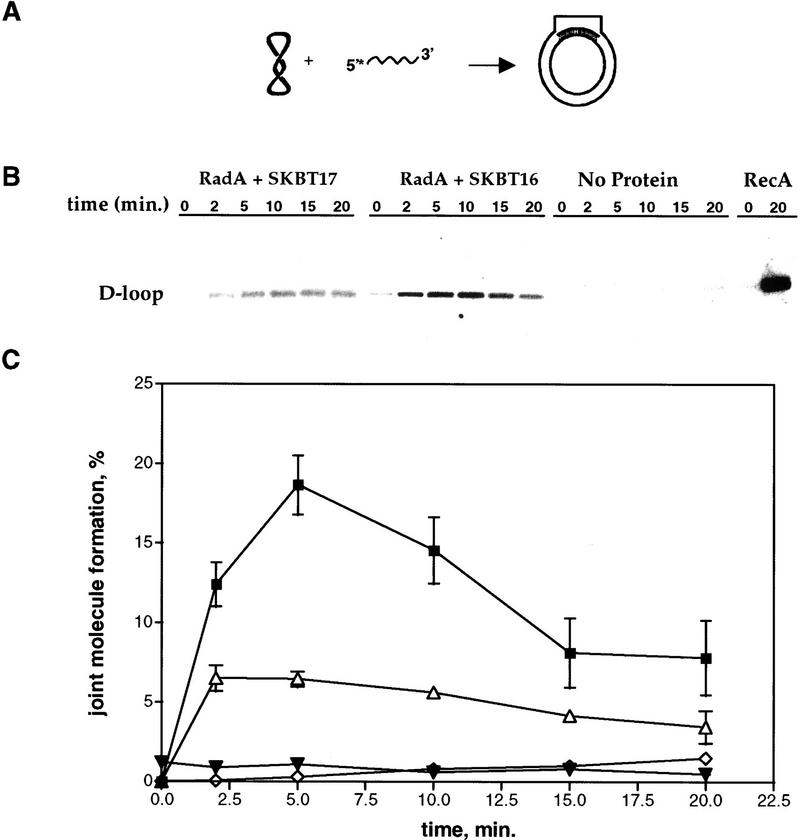
Both the ATPase activity and the filamentous structure suggest that RadA protein is a DNA strand exchange protein. To test this possibility, we employed two different assays. The first (Fig. 3) tested whether RadA protein could catalyze the invasion of supercoiled DNA by homologous ssDNA to form joint molecules. We examined joint molecule formation by RadA protein using a supercoiled plasmid, pBT54CN1 (a derivative of pUC19) (Tracy and Kowalczykowski 1996), and two complementary 54-mer ssDNA oligonucleotides, SKBT16 and its complement SKBT17. Figure 3 shows that the RadA protein can promote the formation of joint molecules with both SKBT16 (rate, 4.0%/min; maximum extent, 16%) and SKBT17 (rate, 1.9%/min; maximum extent, 6%), although the reaction is significantly enhanced with SKBT16; a similar behavior was observed for RecA protein (Tracy and Kowalczykowski 1996) and for Rad51 protein (Tracy et al. 1997). This reaction is homology dependent (pBR322 plasmid DNA was used as the heterologous control), requires RadA protein, and is the most efficient at 65°C (data not shown).
Figure 3.
RadA protein can promote the formation of joint molecules. (A) DNA substrates and the expected product of the reaction. (B) An agarose gel displaying a time course for formation of joint molecules. The first six lanes show RadA protein-dependent D-loop formation with the 54-mer oligonucleotide SKBT17 at 65°C; the second six lanes show RadA protein-dependent D-loop formation with the 54-mer SKBT16 at 65°C; the third six lanes show the same reaction with SKBT16 in the absence of protein at 65°C; the last two lanes show RecA protein-dependent D-loop formation with SKBT16 at 37°C. (C) Joint molecule formation is dependent on RadA protein and DNA sequence homology. The error bars indicate the s.d. for three sets of experiments. (▪) SKBT16; (▵) SKBT17; (▾) PBR322 heterologous control; (⋄) no RadA.
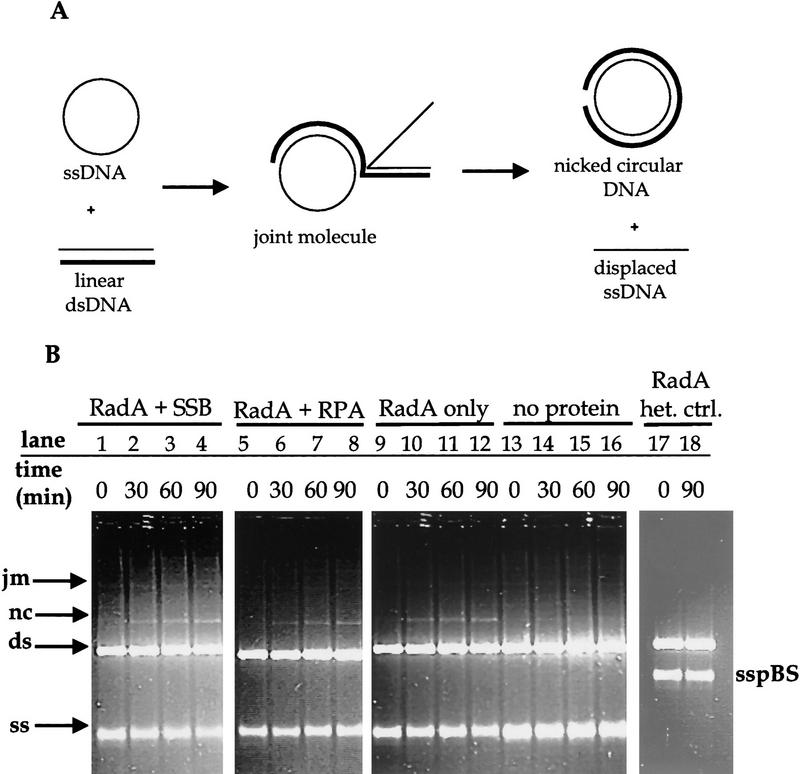
The second pairing assay tested whether RadA protein promotes the complete exchange of DNA strands between ssDNA and homologous linear dsDNA. We used circular φX174 viral ssDNA and linear duplex φX174 DNA—substrates used to test RecA and Rad51 protein function (Fig. 4; Sung 1994). In the RadA protein-promoted reaction, an increasing amount of nicked circular dsDNA product was formed as the reaction progressed, indicating that RadA protein can mediate complete exchange of strands between homologous DNA molecules (Fig. 4B, lanes 1–12). This reaction is not seen at lower temperatures (37°C; data not shown) and is dependent on DNA homology (Fig. 4B, lanes 17,18). E. coli single-stranded DNA binding (SSB) protein slightly increased the amount of nicked circular DNA produced, (17% after 90 min; Fig. 4B, lanes 1–4) but S. cerevisiae replication protein A (RPA) slightly inhibited the reaction (9% after 90 min; Fig. 4B, lanes 4–8) compared to the reaction with RadA protein alone (13% after 90 min; Fig. 4B, lanes 8–12). Because neither SSB nor RPA protein are themselves thermostable, only their effect on presynaptic filament formation was measured, to the extent that this stimulation would persist upon the obligatory temperature shift. The magnitude of stimulation may be enhanced by a thermostable ssDNA-binding protein from the Archaea, which we have provisionally identified (Chédin et al. 1998).
Figure 4.
RadA protein promotes DNA pairing and strand exchange. (A) The DNA substrates as well as the expected intermediates and products of the DNA strand exchange reaction. (B) An agarose gel displaying a time course for formation of these products. Viral φX174 ssDNA (ss) and linear φX174 dsDNA (ds) were incubated with RadA protein, either with or without a ssDNA binding protein in lanes 1–12. Lanes 13–16 show the same reaction without RadA protein; lanes 17 and 18 show the same reaction with heterologous DNA. (jm) Joint molecules; (nc) nicked circular dsDNA; (sspBS) pBluescript ssDNA used as a heterologous control.
Discussion
Our results indicate that RadA protein is a member of the ubiquitous family of recombination proteins that includes the E. coli RecA protein and the S. cerevisiae Rad51 protein. RadA protein hydrolyzes ATP and promotes homologous DNA pairing and strand exchange, two properties inherent to both RecA and Rad51 proteins. The RadA protein–DNA filament is also structurally similar to the filament formed by RecA and Rad51 proteins. Based on the slower rates of both ATP hydrolysis and DNA strand exchange, and on the relatively low yield of DNA strand exchange product formed, the RadA protein behaves more similarly to the eucaryal Rad51 protein than to the bacterial RecA protein. This is consistent with the sequence data which show more homology between RadA and Rad51 proteins, and with phylogenetic analyses, which show that the Archaea and Eucarya share a more recent common ancestor than either do with Bacteria (Woese 1987; Brown and Doolittle 1995).
At present, the yield of DNA strand exchange products produced by RadA protein is low. However, this low amount of DNA strand exchange product is likely due to the absence of a thermostable ssDNA-binding protein in our reactions. With eukaryotic proteins, it was shown that in the absence of RPA, Rad51 protein can achieve only a limited amount of pairing and DNA strand exchange (Sung 1994; Sung and Robberson 1995; Sung and Stratton 1996; Sugiyama et al. 1997). RPA, like its prokaryotic analog SSB protein, is thought to act both at the presynaptic and postsynaptic step of DNA strand exchange. Presynaptically it stimulates the DNA strand exchange reaction by removing secondary structure of the ssDNA and allowing the formation of a contiguous presynaptic filament formed by Rad51; postsynaptically, it binds to the displaced ssDNA that is released after the DNA strands have exchanged (Kowalczykowski et al. 1994). Our recent identification of a SSB/RPA homolog in the Archaea will enable us to assess the effects of a thermostable SSB/RPA on RadA protein-promoted DNA strand exchange and will lend further insight as to the recombination machinery in this third domain (Smith et al. 1997; Chédin et al. 1998).
Although the finding that RadA protein is a DNA strand exchange protein solves the question of which protein is involved in the homologous pairing phase during homologous recombination in Archaea, the genes involved in the initiation step, as well as the heteroduplex extension and resolution steps of this pathway, still remains a mystery. There are presently three archaeal genomes that have been completely sequenced (Bult et al. 1996; Klenk et al. 1997; Smith et al. 1997). To date, a putative Schizosaccharomyces pombe rad32 homolog (a homolog of S. cerevisiae MRE11) is present in both Archaeoglobus fulgidus and Methanobacterium thermoautotrophicum (Klenk et al. 1997; Smith et al. 1997), which lends some insight into the initiation step of recombination in the Archaea and further supports the notion that recombination in the Archaea is more eucaryal than bacterial. Interestingly, no other homologs of eucaryal recombination genes were identified in the Archaea, which explicitly suggests an important divergence from the Eucarya, especially as there are as yet no structural homologs for proteins that are important in eucaryal recombination, such as RAD52, RAD54, RAD55, RAD57, or RAD59 (Game 1993).
The genes involved in nucleic acid metabolism reflect the shared history of the Eucarya and Archaea (Bult et al. 1996; Olsen and Woese 1997) in the sense that most of the genes involved in these processes in the archaeal cell are more homologous to eukaryotic genes than to bacterial genes. In light of this knowledge, the Archaea may afford a unique and simpler insight into the much more complex recombination apparatus (Game 1993) of the eukaryotic cell. Further biochemical characterization of RadA protein and genetic analysis of radA in Archaea will provide insight into an evolutionarily “older” recombination process and may provide information about the development of recombination proteins in the Bacteria and Eucarya.
Materials and methods
Expression of the RadA protein in E. coli
The S. solfataricus radA gene was PCR-amplified from pSJS1047, which contains a 7.6-kb SacI–HindIII genomic fragment cloned into pUC118 (Sandler et al. 1996) so that the E. coli translational start signals were fused to the first ATG after the start of the radA gene. The amplified DNA was gel-purified and treated with DNA polymerase I Klenow fragment in the presence of dNTPs to blunt the DNA ends. This DNA and pSJS582 (Sandler and Clark 1993) were restricted with HpaI and AflII. The DNAs were mixed and used to transform DH5α-competent cells at 30°C. One transformant was saved and called pSJS1106.
We cloned the fragment of DNA from pSJS1106 containing the temperature-sensitive cI gene, the λ PL and PR promoters, the translational start signals, and the beginning of the S. solfataricus radA gene to a cloned copy of the radA gene on pSJS1062 (GenBank accession no. U45310). pSJS1062 was restricted completely with BglII, pSJS1106 was partially restricted with BglII, and both DNAs were further restricted with SacI. The appropriate DNA fragments were purified, mixed, treated with DNA ligase, and used to transform DH5α-competent cells at 30°C. One plasmid was saved and called pSJS1109.
The S. solfataricus radA gene has a large proportion of codons that use A or T in the third position [the S. solfataricus genome is ∼30% GC (Gouy and Gautier 1982)]. To avoid problems associated with poor codon usage, we overexpressed the radA gene in E. coli cells bearing the plasmid pRI952, carrying cloned copies of the tRNA genes that encode AGA, AGG arginine, and AUA isoleucine (Del Tito et al. 1995).
Purification of RadA protein
An extract was prepared from 24 grams of E. coli strain JC10291 harboring pRI952 and pSJS1109. RadA was first precipitated with 0.8% polymin P, eluted with 150 mm ammonium sulfate, precipitated by ammonium sulfate, and redissolved in P1 buffer [200 mm NaCl, 20 mm KH2PO4 (pH 6.5), 10% (vol/vol) glycerol, 0.1 mm DTT, 0.1 mm EDTA]. This was applied to a phosphocellulose column and the RadA-containing fractions were eluted in the flow through. The pool was dialyzed extensively against T1 buffer [25 mm Tris-HC (pH 7.5), 0.1 mm EDTA, 0.1 mm DTT, 10% (vol/vol) glycerol] and applied to a Mono Q column that was developed with a 350-mL, 100–700 mm NaCl gradient. The RadA protein peak (∼275 mm NaCl) was dialyzed against P2 buffer [20 mm KH2PO4 (pH 7.5), 10% (vol/vol) glycerol, 0.1 mm DTT, 10 mm KCl] and applied to a hydroxylapatite column with a 40-ml, 50–500 mm KH2PO4 gradient. RadA protein eluted from hydroxylapatite at 275 mm KH2PO4 was dialyzed against P3 buffer [20 mm KH2PO4 (pH 6.5), 25 mm EDTA, 0.1 mm DTT, 10% (vol/vol) glycerol], and applied onto a ssDNA cellulose column. RadA protein eluted at 250 mm NaCl and was then dialyzed against storage buffer [25 mm Tris-HCl (pH 7.5), 0.1 mm EDTA, 1 mm DTT, 10% (vol/vol) glycerol, 200 mm NaCl] and frozen at −80°C in 50-μl aliquots. RadA protein concentration was determined using an extinction coefficient of 16,100/m per cm at 280 nm.
ATPase assays
Reaction mixtures (20 μl) contained 25 mm Tris-HCl (pH 7.5), 6 mm MgCl2, 0.1 mm DTT, 1 mm ATP, 0.2 μCi [γ-32P]ATP or [α-32P]dATP, 15 μm RadA protein, and 100 μm nucleotides of either poly(dT), M13 ssDNA, or M13 dsDNA. The assays were incubated at 65°C (unless otherwise indicated) for 90 min. In Figure 1, all conditions were the same except that the amount of poly(dT) varied for each reaction. The amount of ATP hydrolyzed was determined by thin layer chromatography using PEI cellulose sheets developed in 1 m formic acid and 0.5 m LiCl (Weinstock et al. 1981).
AFM imaging
The complexes were formed as follows: RadA protein (7.5 μm) was incubated with 30 μm (nucleotides) pBR322 DNA in 25 mM Tris-acetate (pH 7.2) containing 10 mm magnesium acetate and 2.5 mm ATP at 65°C for 1 hr. NaF and Al(NO3)3 were then added to final concentrations of 5 and 0.2 mm, respectively; incubation continued for another 30 min. The reaction mixture was then applied to a Clontech S-1000 gel filtration spin column that was pre-equilibrated with incubation buffer containing NaF and Al(NO3)3, and it was centrifuged at 1400g for 5 min at room temperature. A 20-μl aliquot of the eluate was then applied to a freshly cleaved mica surface, which was rinsed immediately with 1 ml of ultrapure water and dried with a stream of nitrogen gas. The sample surface was imaged by atomic force microscopy with a Digital Instruments Nanoscope III in air using a tapping mode with a TESP cantilever. Images were analyzed using Digital Instruments nanoscope software and NIH image. Widths of complexes were determined by viewing the complex in cross section and measuring the width at half the height.
The RecA protein–dsDNA complexes were formed using the same procedures as for RadA protein but with the following exceptions: 5 μm RecA protein was used; ATPγS was used instead of ATP; and NaF, Al(NO3)3, and the gel filtration of the sample were omitted.
Joint molecule formation
The ssDNA 54-mers SKBT16 and SKBT17 were labeled at their 5′-ends with T4 polynucleotide kinase and [γ-32P]ATP. Reactions (40 μl final volume) containing 25 mm Tris-acetate (pH 7.5), 15 mm magnesium acetate, 1 mm ATP, 1 mm DTT, 1 μm (nucleotides) 5′-end-labeled SKBT16 or SKBT17, and 0.33 μm RadA protein were incubated at 65°C for 5 min, after which 18 μm (nucleotide) of supercoiled pBT54CN1 was added. Aliquots were removed at the indicated time points and deproteinized by the addition of 0.5% SDS and 1 mg/ml proteinase K, with incubation at 37°C for 10 min. Reactions with RecA protein were under the same conditions except for 4 mm magnesium acetate and incubation at 37°C. Finally, the samples were subjected to gel electrophoresis in 1% agarose gels at 1.2 V/cm. The dried gels were visualized and quantitated on a Betagen 603 β-particle counter.
DNA strand exchange
Reactions (60 μl final volume) containing 30 mm Tris-acetate (pH 7.5), 1 mm DTT, 50 μg/ml BSA, 20 mm magnesium acetate, 2.5 mm ATP, and 5 μm RadA protein were preincubated at 65°C for 5 min; then 15 μm (nucleotides) viral φX174 ssDNA was added; and the mixture was incubated for 10 min at 65°C. Next, the temperature was decreased to 37°C, 1.0 μm SSB protein or 0.5 μm RPA was added, and the reaction incubated for an additional 10 min at 37°C. Finally, 30 μm (nucleotides) of PstI-linearized φX174 dsDNA was added, and the reaction proceeded at 65°C for the times indicated. The reactions were stopped by the addition of 0.5% SDS and 1 mg/ml proteinase K, and incubated at 37°C for an additional 10 min before being subjected to electrophoresis in 1% agarose gels at 0.8 V/cm. Exonucleolytic activity was tested using 3′- and 5′-end-labeled heat-denatured or duplex pBR322 DNA. A stoichiometric amount of purified RadA was incubated with the labeled DNA at 65°C for 90 min, and the amount of TCA-soluble counts were measured by a Beckman LS6000IC scintillation counter (Eichler and Lehman 1977). The percentage of TCA-soluble fragments for either 5′- or 3′-end labeled pBR322 ssDNA or dsDNA was <4% after 90 min.
Acknowledgments
We thank Dan Anderson, Deana Arnold, Piero Bianco, Frederic Chedin, Jason Churchill, Frank Harmon, Noriko Kantake, Alex Mazin, James New, Tomohiko Sugiyama, Bob Tracy, and Eugene Zaitsev for critical reading of this manuscript. This work was supported by National Institutes of Health grant AI-18987 to S.C.K. and was conducted under the auspices of the U.S. Department of Energy, supported (in part) by funds provided by the University of California for the conduct of discretionary research by Los Alamos National Laboratory.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL sckowalczykowski@ucdavis.edu; FAX (530) 752-5939.
References
- Brown JR, Doolittle WF. Root of the universal tree of life based on ancient aminoacyl-tRNA synthetase gene duplications. Proc Natl Acad Sci. 1995;92:2441–2445. doi: 10.1073/pnas.92.7.2441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bult CJ, White O, Olsen GJ, Zhou L, Fleischmann RD, Sutton GG, Blake JA, Fitzgerald LM, Clayton RA, Gocayne JD. Complete genome sequence of the methanogenic archaeon, Methanococcus jannaschii. Science. 1996;273:1058–1073. doi: 10.1126/science.273.5278.1058. [DOI] [PubMed] [Google Scholar]
- Bustamante C, Vesenka J, Tang CL, Rees W, Guthold M, Keller R. Circular DNA molecules imaged in air by scanning force microscopy. Biochemistry. 1992;31:22–26. doi: 10.1021/bi00116a005. [DOI] [PubMed] [Google Scholar]
- Chédin, F., E.M. Seitz, and S.C. Kowalczykowski. 1998. A highly conserved DNA-binding motif permits identification of a replication protein-A homologue in Archaea. Trends Biochem. Sci. (in press). [DOI] [PubMed]
- Cox MM. Binding of two DNA molecules at once: The recA protein. In: Revzin A, editor. The biology of nonspecific DNA-protein interactions. Boca Raton, FL: CRC Press; 1990. pp. 171–196. [Google Scholar]
- Del Tito BJ, Jr, Ward JM, Hodgson J, Gershater CJ, Edwards H, Wysocki LA, Watson FA, Sathe G, Kane JF. Effects of a minor isoleucyl tRNA on heterologous protein translation in Escherichia coli. J Bacteriol. 1995;177:7086–7091. doi: 10.1128/jb.177.24.7086-7091.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DiCapua E, Schnarr M, Ruigrok RW, Lindner P, Timmins PA. Complexes of recA protein in solution. A study by small angle neutron scattering. J Mol Biol. 1990;214:557–570. doi: 10.1016/0022-2836(90)90198-U. [DOI] [PubMed] [Google Scholar]
- Egelman EH, Stasiak A. Structure of helical RecA-DNA complexes. Complexes formed in the presence of ATP-γ-S or ATP. J Mol Biol. 1986;191:677–697. doi: 10.1016/0022-2836(86)90453-5. [DOI] [PubMed] [Google Scholar]
- Eichler DC, Lehman IR. On the role of ATP in phosphodiester bond hydrolysis catalyzed by the recBC deoxyribonuclease of Escherichia coli. J Biol Chem. 1977;252:499–503. [PubMed] [Google Scholar]
- Game JC. DNA double-strand breaks and the RAD50-RAD57 genes in Saccharomyces. Semin Cancer Biol. 1993;4:73–83. [PubMed] [Google Scholar]
- Gouy M, Gautier C. Codon usage in bacteria: Correlation with gene expressivity. Nucleic Acids Res. 1982;10:7055–7074. doi: 10.1093/nar/10.22.7055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heyer W-D. The search for the right partner: Homologous pairing and DNA strand exchange proteins in eukaryotes. Experientia. 1994;50:223–233. doi: 10.1007/BF01924005. [DOI] [PubMed] [Google Scholar]
- Klenk HP, Clayton RA, Tomb JF, White O, Nelson KE, Ketchum KA, Dodson RJ, Gwinn M, Hickey EK, Peterson JD, et al. The complete genome sequence of the hyperthermophilic, sulphate-reducing archaeon Archaeoglobus fulgidus. Nature. 1997;390:364–370. doi: 10.1038/37052. [DOI] [PubMed] [Google Scholar]
- Kowalczykowski SC, Dixon DA, Eggleston AK, Lauder SD, Rehrauer WM. Biochemistry of homologous recombination in Escherichia coli. Microbiol Rev. 1994;58:401–465. doi: 10.1128/mr.58.3.401-465.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lauder SD, Kowalczykowski SC. Asymmetry in the recA protein-DNA filament. J Biol Chem. 1991;266:5450–5458. [PubMed] [Google Scholar]
- Ogawa T, Shinohara A, Nabetani A, Ikeya T, Yu X, Egelman EH, Ogawa H. RecA-like recombination proteins in eukaryotes: Function and structures of RAD51 genes. Cold Spring Harb Symp Quant Biol. 1993a;58:567–576. doi: 10.1101/sqb.1993.058.01.063. [DOI] [PubMed] [Google Scholar]
- Ogawa T, Yu X, Shinohara A, Egelman EH. Similarity of the yeast RAD51 filament to the bacterial RecA filament. Science. 1993b;259:1896–1899. doi: 10.1126/science.8456314. [DOI] [PubMed] [Google Scholar]
- Olsen GJ, Woese CR. Archaeal genomics: An overview. Cell. 1997;89:991–994. doi: 10.1016/s0092-8674(00)80284-6. [DOI] [PubMed] [Google Scholar]
- Sandler SJ, Clark AJ. Use of high and low level overexpression plasmids to test mutant alleles of the recF gene of Escherichia coli K-12 for partial activity. Genetics. 1993;135:643–654. doi: 10.1093/genetics/135.3.643. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandler SJ, Satin LH, Samra HS, Clark AJ. recA-like genes from three archaean species with putative protein products similar to Rad51 and Dmc1 proteins of the yeast Saccharomyces cerevisiae. Nucleic Acids Res. 1996;24:2125–2132. doi: 10.1093/nar/24.11.2125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith DR, Doucette-Stamm LA, Deloughery C, Lee H, Dubois J, Aldredge T, Bashirzadeh R, Blakely D, Cook R, Gilbert K, et al. Complete genome sequence of Methanobacterium thermoautotrophicum ΔH: Functional analysis and comparative genomics. J Bacteriol. 1997;179:7135–7155. doi: 10.1128/jb.179.22.7135-7155.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Story RM, Bishop DK, Kleckner N, Steitz TA. Structural relationship of bacterial RecA proteins to recombination proteins from bacteriophage T4 and yeast. Science. 1993;259:1892–1896. doi: 10.1126/science.8456313. [DOI] [PubMed] [Google Scholar]
- Sugiyama T, Zaitseva EM, Kowalczykowski SC. A single-stranded DNA-binding protein is needed for efficient presynaptic complex formation by the Saccharomyces cerevisiae Rad51 protein. J Biol Chem. 1997;272:7940–7945. doi: 10.1074/jbc.272.12.7940. [DOI] [PubMed] [Google Scholar]
- Sung P. Catalysis of ATP-dependent homologous DNA pairing and strand exchange by yeast RAD51 protein. Science. 1994;265:1241–1243. doi: 10.1126/science.8066464. [DOI] [PubMed] [Google Scholar]
- Sung P, Robberson DL. DNA strand exchange mediated by a RAD51-ssDNA nucleoprotein filament with polarity opposite to that of RecA. Cell. 1995;82:453–461. doi: 10.1016/0092-8674(95)90434-4. [DOI] [PubMed] [Google Scholar]
- Sung P, Stratton SA. Yeast Rad51 recombinase mediates polar DNA strand exchange in the absence of ATP hydrolysis. J Biol Chem. 1996;271:27983–27986. doi: 10.1074/jbc.271.45.27983. [DOI] [PubMed] [Google Scholar]
- Tracy RB, Kowalczykowski SC. In vitro selection of preferred DNA pairing sequences by the Escherichia coli RecA protein. Genes & Dev. 1996;10:1890–1903. doi: 10.1101/gad.10.15.1890. [DOI] [PubMed] [Google Scholar]
- Tracy RB, Baumohl JK, Kowalczykowski SC. The preference for GT-rich DNA by the yeast Rad51 protein defines a set of universal pairing sequences. Genes & Dev. 1997;11:3423–3431. doi: 10.1101/gad.11.24.3423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinstock GM, McEntee K, Lehman IR. Hydrolysis of nucleoside triphosphates catalyzed by the recA protein of Escherichia coli: Characterization of ATP hydrolysis. J Biol Chem. 1981;256:8829–8834. [PubMed] [Google Scholar]
- Woese CR. Bacterial evolution. Microbiol Rev. 1987;51:221–271. doi: 10.1128/mr.51.2.221-271.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Woods WG, Dyall-Smith ML. Construction and analysis of a recombination-deficient (radA) mutant of Haloferax volcanii. Mol Microbiol. 1997;23:791–797. doi: 10.1046/j.1365-2958.1997.2651626.x. [DOI] [PubMed] [Google Scholar]
- Yu X, Egelman EH. Structural data suggest that the active and inactive forms of the RecA filament are not simply interconvertible. J Mol Biol. 1992;227:334–346. doi: 10.1016/0022-2836(92)90702-l. [DOI] [PubMed] [Google Scholar]
- ————— DNA conformation induced by the bacteriophage T4 UvsX protein appears identical to the conformation induced by the Escherichia coli RecA protein. J Mol Biol. 1993;232:1–4. doi: 10.1006/jmbi.1993.1363. [DOI] [PubMed] [Google Scholar]
- Yu X, Angov E, Camerini-Otero RD, Egelman EH. Structural polymorphism of the RecA protein from the thermophilic bacterium Thermus aquaticus. Biophys J. 1995;69:2728–2738. doi: 10.1016/S0006-3495(95)80144-X. [DOI] [PMC free article] [PubMed] [Google Scholar]