Abstract
Ribosomal protein S4 binds and stabilizes a five-helix junction in the 5’ domain of the 16S rRNA, and is one of two proteins responsible for nucleating 30S ribosome assembly. Upon binding, both protein S4 and the five-helix junction reorganize their structures. We show that labile S4 complexes rearrange to stable complexes within a few minutes at 42°C, with longer coincubation leading to an increased population of stable complexes. In contrast, prefolding the rRNA has a smaller effect on stable S4 binding. Experiments with minimal rRNA fragments show this structural change depends only on 16S residues within the S4 binding site. SHAPE chemical-probing experiments showed that S4 strongly stabilizes the five-helix junction and helix 18 pseudoknot, which become tightly folded within the first minute of S4 binding. However, a kink in helix 16 that makes specific contacts with the S4 N-terminal extension, and a right angle motif between helices 3, 4 and 18, require a minute or more to become fully structured. Surprisingly, S4 structurally reorganizes the 530-loop and increases the flexibility of helix 3, which is proposed to undergo a conformational switch during 30S assembly. These elements of the S4 binding site may require other 30S proteins to reach a stable conformation.
Keywords: RNA-protein interaction, stable complex, SHAPE, conformation change, RNA folding
INTRODUCTION
Pre-ribosomal rRNA must fold extensively as it complexes with ribosomal proteins during subunit biogenesis. How protein interactions induce specific conformational changes in the rRNA is not yet understood. The association of 5’ domain binding protein S4 with rRNA is a critical step in ribosome biogenesis. S4 is one of the first proteins in vivo to contact the nascent rRNA1, 2, and together with protein S7, is required for nucleation of subunit assembly3. As a primary binding protein, S4 is also required for the association of other proteins such as S16 with nascent subunits4, 5.
Induced-fit is thought to be a common mechanism of RNA–protein recognition,6–8 and previous data suggest that S4 and its rRNA binding site change structure during the formation of stable rRNA-protein complexes. S4 binds to the junction between helices 3, 4, 16, 17, and 18 in the 16S rRNA (Figure 1b) and makes additional interactions to helix (H) 1 and the tip of central domain H219–15. A minimal rRNA consisting of the five-helix junction (5-way-junction or 5WJ) binds S4 with similar affinity as the 16S rRNA (Figure 1b)16.
Figure 1. Chemical probing of protein S4-5WJ rRNA.
(a) Structure of S4 in the E. coli ribosome highlighting the globular domain (cyan) and N-terminal extension (tan). (PDB: 2AVY)56 (b) Secondary structure of 5WJ RNA containing the S4 binding site; residues numbered according to E. coli 16S rRNA. Circles indicate equilibrium protection from hydroxyl-radical cleavage, using Bst S4 and the 5WJ16. Dotted lines summarize time-resolved hydroxyl-radical footprinting experiments with native E. coli 16S and 30S proteins34. Squares and triangles indicate protection and exposure to chemical base modification at 0°C and 42°C20.
Although the 16S 5’ domain can fold without proteins, S4 stabilizes the native conformation of the 5WJ under physiological conditions14, 17 S4 also stabilizes a pseudoknot between nucleotides 505–507 and 524–526 in H18, which correctly positions G530 to interact with substrates in the decoding center of the mature ribosome12, 14, 15, 18. S4 binding depends on temperature, a further indication that binding requires rearrangements in both S4 and the 16S rRNA19, 20. Owing to the large conformational changes that take place during assembly, temperature strongly influences the rRNA interactions of many ribosomal proteins. Temperature-dependent effects tend to be the most significant in large, multi-domain proteins20–22.
Structures of free and bound S4 show that the protein also changes conformation as it is incorporated into the ribosome15, 23–29. S4 has a stably folded C-terminal region and a long, flexible N-terminal extension that is conserved in bacteria but missing in eukaryotes (Figure 1a)30, 31. The C-terminal domains of S4 (cyan; Figure 1a) do not change conformation by binding rRNA27, 32. In contrast, NMR experiments show that the N-terminal extension of S4 (tan; Figure 1a) is largely unstructured in the free protein, yet becomes highly ordered upon rRNA binding27. The N-terminal extension makes extensive conserved contacts along H16 of the 5’ domain rRNA33. That some thermophilic bacteria use zinc to further stabilize the S4 N-terminal arm-H16 interaction highlights the critical nature of these contacts31.
Base-modification experiments on S4-16S complexes formed at different temperatures indicated that some structural changes in the 16S rRNA require coincubation with S4 at 42°C20, suggesting S4 was needed to induce these changes and not just to trap the rRNA in its native conformation. These changes mapped to the pseudoknot and 530-loop in H18, and the junction between H3 and H4 (squares and triangles; Figure 1b). Subsequent time-resolved hydroxyl-radical footprinting experiments showed that contacts between S4 and the H16–H17 and H3–H16 junctions saturate within 0.1 s, while contacts with H18 and H3 formed more slowly34 (dashed lines; Figure 1b).
In this paper, we use kinetic competition assays to show that S4 forms both stable and short-lived complexes at physiological temperatures with minimal rRNA substrates. Prefolding of the rRNA has only a modest effect on formation of the stable complex. Increased S4-rRNA coincubation time, however, greatly increases the population of stable complexes over a range of temperatures. SHAPE chemical modification experiments on populations enriched for either the stable or labile 5WJ – S4 complexes reveal precise structural changes to the rRNA that correlate with the slow emergence of stable complexes. Although the five helix junction and the pseudoknot in H18 of the 5WJ reach the native conformation within the first minute of S4 binding, restructuring of H16, H3 and the 530-loop requires more than a minute to complete.
RESULTS
Competition experiments reveal two distinct S4 - rRNA complexes
Stable RNA-protein recognition frequently requires changes in structure to the RNA, the protein, or both6, 7, 35. How S4 arrives at its native complex, however, is not known20, 27, 34. We used Geobacillus stearothermophilus (Bst) S4 in these studies because Bst S4 is more stable than E. coli S4 in solution19. Bst S4 binds the E. coli 16S rRNA with affinity and specificity equal to that of E. coli S416, 19, because the secondary and tertiary structure of the S4 binding site is strongly conserved among different organisms36, 37. The structure of S4 is also conserved in bacteria, particularly at positions that directly contact the rRNA23, 31.
We previously showed that Bst S4 forms specific, native-like complexes with the E. coli 16S 5’ domain and a minimal 5WJ fragment 16. The advantages of the minimal rRNAs for the current experiments are that S4 binds tightly in the absence of other proteins, non-specific binding is minimized, and native-like complexes form in 4 mM Mg2+, compared to 20 mM generally used in ribosome reconstitution.
Native gel mobility shift assays were used to measure stable S4 – rRNA binding and to define conditions that promote or inhibit the formation of stable S4 complexes. First, we observed that total S4 binding with 32P-labeled 5’ domain and 5WJ RNAs approached equilibrium within 1 min (Figure 2), in agreement with binding rates estimated from time-resolved hydroxyl-radical footprinting and pulse-chase mass spectrometry2, 34, 38. The lower limit for the association rate constant was 2.7 × 106 M−1 sec−1, similar to an earlier estimate of 107 M−1 sec−1 from filter binding assays19.
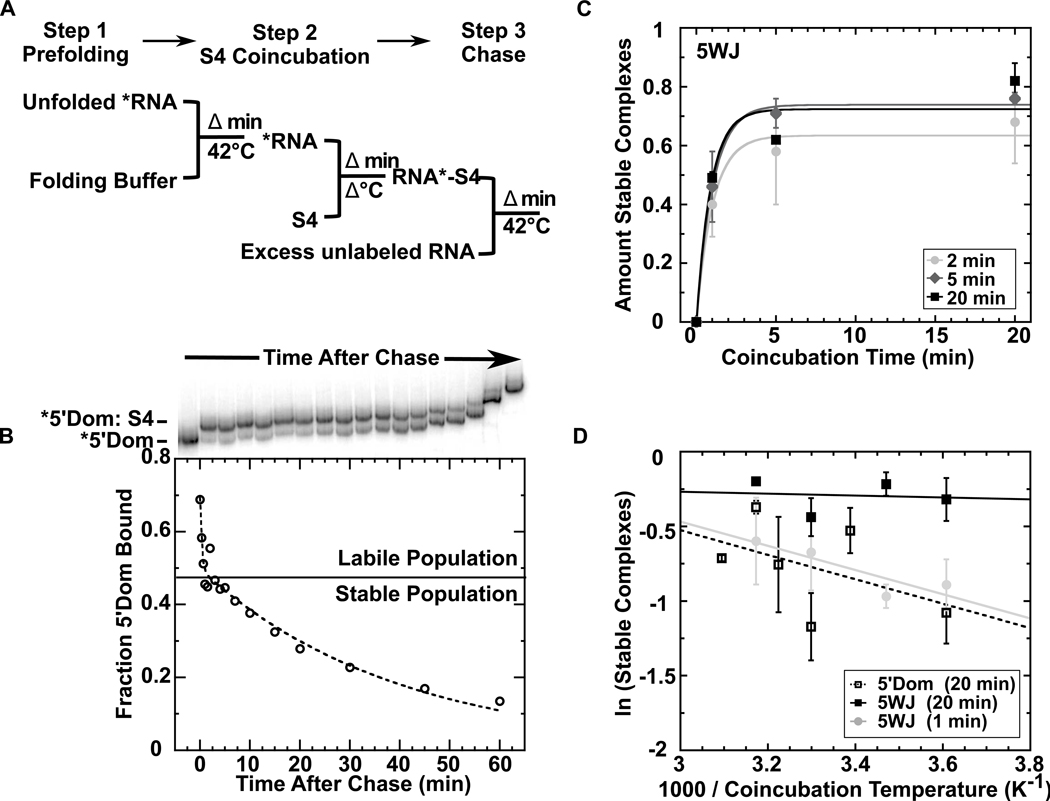
Figure 2. Association kinetics of Bst S4.
(a) Representative gel mobility shift of 5’ domain RNA. Excess S4 was added at 42°C and aliquots loaded directly onto a native polyacrylamide gel at specific time intervals. The gel was run continuously during the experiment. (b) 32P-labeled 5’ domain (open circles, dashed line) or 5WJ (closed circles, solid line) were prefolded in HKM4 buffer 20 min at 42°C before S4 was added. The fraction of bound RNA was fit to fB = fmax(1 − exp(−kobst)); kobs was 7 ± 2 min−1 (5WJ) and 6 ± 1 min−1 (5’ dom).
Next, we used competition experiments to measure the half-life of the S4-rRNA complex (Figure 3). Sub-nanomolar concentrations of 32P-labeled 5’ domain were prefolded and then allowed to bind excess S4 protein. The complexes were chased with unlabeled competitor rRNA to trap free protein. When the loss of the 32P-labeled S4 complex was plotted versus time after addition of the competitor, we observed at least two populations of complexes with different off-rates. About half the complexes dissociated rapidly (koff ≈ 3 min−1), while the remaining population decayed slowly at 42°C (koff = 0.02 min−1) (Figure 3b).
Figure 3. Stable complexes require prolonged coincubation.
(a) Scheme of kinetic competition assay to measure S4 dissociation (see Methods). After prefolding (42°C) and coincubation steps (variable temperature), 500 nM unlabeled 5WJ was added at 42°C to sequester unbound S4 (chase). (b) Representative gel and corresponding plot of fraction bound 5’ domain (open circles) versus time after chase (42°C 20 min prefold, 37 °C 10 min S4 coincubation). The S4 dissociation kinetics from the 5’ domain RNA were fit to fB = A1exp(−k1t) + A2exp(−k2t). Stable and labile complexes are indicated. (c) Amount of stable complex versus S4 coincubation time (42°C). Data are shown for 5WJ RNA, prefolded 2, 5, or 20 min; see Fig. S2 for 5’ domain data. Symbols and error bars represent the mean and standard deviation of 3 trials. Data were normalized to a “no chase” control. Maximum total binding is 80% under these conditions. (d) Dependence on coincubation temperature. Amount of stable complex of 5WJ (filled shapes) and 5’ domain (open squares) after 1 min (gray circles) or 20 min (black squares) coincubation with S4.
The minimal 5WJ rRNA also formed a mixture of stable and unstable complexes, with similar lifetimes as S4 complexes with the entire 16S 5’ domain (Figure S1). Under all conditions, the proportion of stable complexes formed by the 5WJ rRNA was equivalent to or higher than the proportion of stable complexes with the 5’ domain. Thus, the ability to form unstable and stable complexes depends only on interactions within the 5WJ, and not on interactions elsewhere in the 16S rRNA. The slow off-rate likely represents irreversible S4 dissociation, because this rate was the same with or without competitor rRNA (Figure S1).
Stable complex increases with S4 coincubation time and temperature
If the stable complexes represent native interactions between S4 and the rRNA, and the labile complexes represent binding intermediates, manipulating rRNA folding or protein binding conditions should affect how well S4 binds. To test this hypothesis, we varied the incubation time and temperature for the 5WJ and 5’ domain and measured the amount of stable complex.
Prefolding the 5’ domain or the 5WJ in buffer containing MgCl2 at 42°C for various lengths of time had little effect on the formation of stable complexes. Only a modest increase in the stable fraction was seen when the RNA was prefolded 5 min instead of 2 min, and no further increase was seen after 5 min (Figure 3c and S2). That prefolding the RNA had little effect on the ability of S4 to bind tightly is not surprising since the 5’ domain RNA compacts within 15 s at 42°C and folds completely within 2 min39 (Figure S3). By contrast, incubating S4 and the RNA together for a longer time had a greater effect on the amount of stable complex, as can be seen by comparing the proportion of stable complexes formed after 1 min coincubation to the proportion of stable complexes formed after 20 min coincubation for any prefolding condition (Figure 3c and Figure S2).
In contrast to previous base-probing experiments on the full 16S rRNA by Powers and Noller20, the binding temperature didn’t change the final amount of stable complex, but did change the rate at which stable complexes were formed. The proportion of stable complexes formed by the minimal 5WJ rRNA varied less with temperature with increased S4 coincubation time (Figures 3d and S4), and was virtually independent of temperature after 20 min S4 coincubation (Figure 3d). Temperature had a slightly more pronounced, although variable, effect on the proportion of stable complexes formed with the 5’ domain (Figure 3d), possibly because the energy barriers for refolding the longer RNA are higher.
RNA structural changes induced by S4
We used selective 2’-hydroxyl acylation analyzed by primer extension, or SHAPE, to compare the rRNA structure in labile and stable S4 complexes at the single nucleotide level. SHAPE reports on the relative flexibility of the rRNA backbone40–43, and can also reveal protein - rRNA contacts that affect relative disorder44. This fine-grained picture of secondary and local tertiary structure complements the read-out of backbone solvent accessibility provided by hydroxyl radical footprinting.
SHAPE experiments were performed on the 5WJ RNA under the same conditions as the mobility shift experiments (Figure 4a). The 5WJ was allowed to fold 2 or 20 min at 42°C (“pre” in Figure 4b), then incubated with S4 1 or 10 min before modification (“S4” in Figure 4b). These conditions produced the greatest differences in the fraction stable complex (Figure 3c). To gather data on the entire 5WJ, the 3’ end of the 5WJ rRNA was extended (see Methods). The extension changed neither the rRNA secondary structure as evaluated by the SHAPE reactivity, nor S4 binding affinity (data not shown).
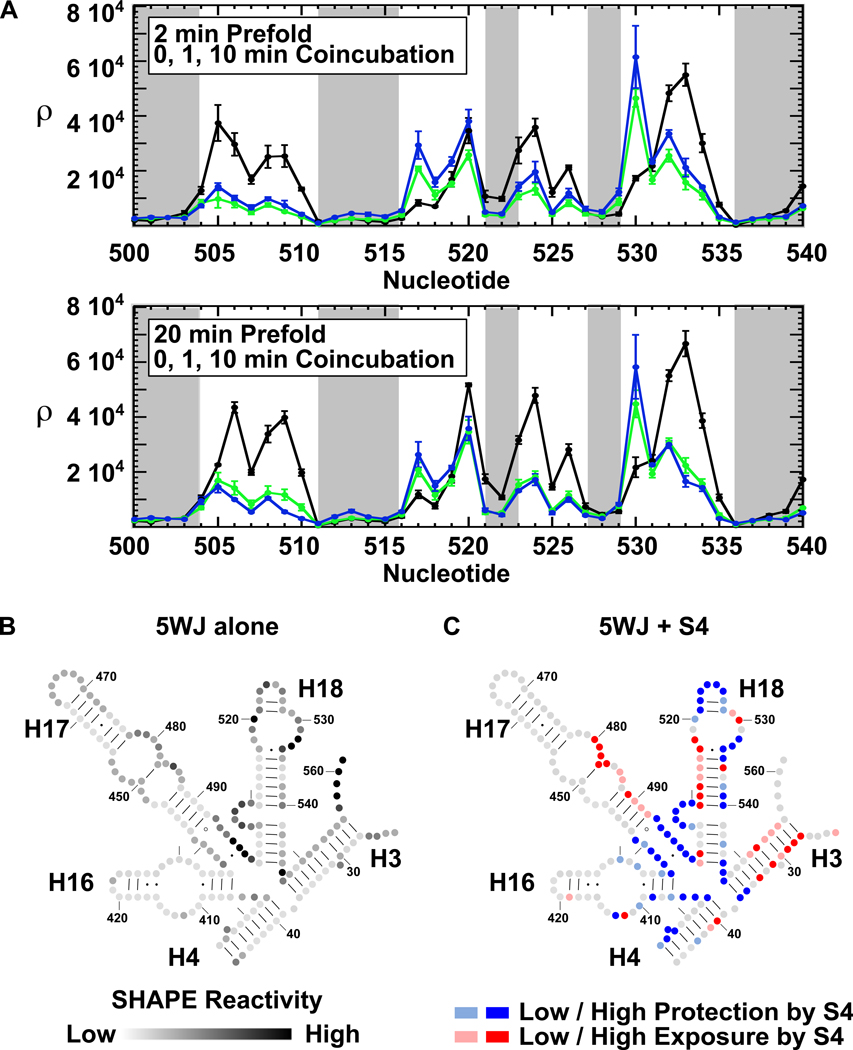
Figure 4. SHAPE chemical probing of S4 – 5WJ complexes.
(a) Chemical probing scheme. Extended 5WJ RNA was prefolded at 42°C in 4 mM MgCl2, mixed with excess Bst S4, and incubated 1 or 10 min at 42°C before modification with 1m7. (b) Representative primer extension of modified RNA. Lanes 1–3: 2 min prefold, no S4. Lanes 4–6: 2 min prefold, 1 min S4 coincubation. Lanes 7–9: 2 min prefold, 10 min S4 coincubation. Lanes 10–12: 20 min prefold, no S4. Lanes 13–15: 20 min prefold, 1 min S4 coincubation. Lanes 16–18: 20 min prefold, 10 min S4 coincubation. Lane 19: No treatment control. Nucleotide positions are indicated to the left.
SHAPE chemical-probing of the 5WJ in the absence of S4 agreed with previously published SHAPE results on the native 16S in the absence of proteins42 and the majority of the 5WJ residues expected to be double-stranded (gray bars, Figure 5a) or single-stranded were protected or exposed as expected (Figure 5b and S5a). Many tertiary interactions were missing, however. Notably, the modification pattern of H18 showed that the pseudoknot is not formed and the 530-loop is improperly folded in the absence of S4. The internal loop of H16 and the junction between H17 and H18 also lacked the native structure.
Figure 5. S4 reduces the flexibility of the 5WJ.
(a) Average adjusted SHAPE intensities (ρ) as a function of nucleotide position. Error bars indicate standard deviations between 3 repeats. RNA was prefolded 2 min (top) or 20 min (bottom). Black, no S4; green, 1 min S4 coincubation; blue, 10 min S4 coincubation. Gray shading indicates double-stranded nucleotides in the ribosome crystal structure (PDB: 2AVY)56. (b) Reactivity of 5WJ nucleotides without S4 after 20 min prefolding. Band intensities were divided by the maximum reactivity (ρ/ρmax). Black > 60%; dark gray > 40%; medium-dark gray > 20%; medium-light gray > 5%. (c) Effect of S4 on 5WJ reactivity. The log ratio of the SHAPE intensities for RNA with and without S4 (log10(ρ10min/ρ0min)) was computed for each residue; dark blue, strongly protected (≤ −0.25); pale blue, weakly protected (≤ −0.10); dark red, strongly enhanced (≥ 0.25); pale red, weakly enhanced (≥ 0.10). RNA was prefolded for 20 min at 42°C before S4 binding.
Coincubation of the rRNA with S4 stiffened the five-helix junction (nt 403–405, 437–440, 494–499, and 545–547), resulting in strong protections of residues at the center of the RNA (blue; Figure 5c and S5b). The junction region interacts directly with the globular C-terminal domain of S4 (cyan, Figure 1a). In addition, the SHAPE results showed that S4 stabilizes the pseudoknot in H18 (nt 505–507, 524–526), and induces a conformational rearrangement in the H18 530-loop that strongly enhances the reactivity of G530 (red; Figure 5c and S5b). Modification of H16 in the presence of S4 was consistent with contacts made to the H16 loop by the N-terminal extension of S4 (Figure 1a, tan)15, 45. Finally, the protections at residues 397–399 (H4) indicated that S4 stabilizes a right angle interaction between H3 and H4, which places the 3’ strand of H4 in the major groove of H3. Surprisingly, S4 enhanced modification of the 5’ and 3’ ends of the 5WJ rRNA, indicating that it destabilizes H3 (red; Figure 5c and S5b).
Overall, the SHAPE protection pattern was consistent with the results of previous chemical-probing studies (Figure 1b)12, 14, 16, 17, 46, 47. Similar structures were attained when the 5WJ rRNA was pre-folded for 2 or 20 min before addition of S4 (Figure 5b,c and S5a,b), reinforcing that the same proportion of stable complexes was obtained after 5 min coincubation with S4, regardless of how the rRNA was pre-folded (Figure 3c).
Most S4-dependent changes to the 5WJ occur within 1 min of coincubation
As the duration of S4 coincubation produced the greatest difference in the amount of stable complex (Figure 3c), we compared the structures of labile and stable complexes by performing SHAPE on S4 - 5WJ complexes after 1 min coincubation (Figure 6). By comparing the reactivity of these complexes to the 5WJ rRNA alone and to the complexes after 10 min coincubation, we identified regions of the rRNA that change conformation slowly in the presence of S4. Nucleotides that achieved >80% of their final protection or exposure within one minute were considered rapidly exposed or protected; the remainder were considered slowly exposed or protected (Figure 6). The rate of change was evaluated separately from the overall extent of protection or exposure (Figure 5c and S5b).
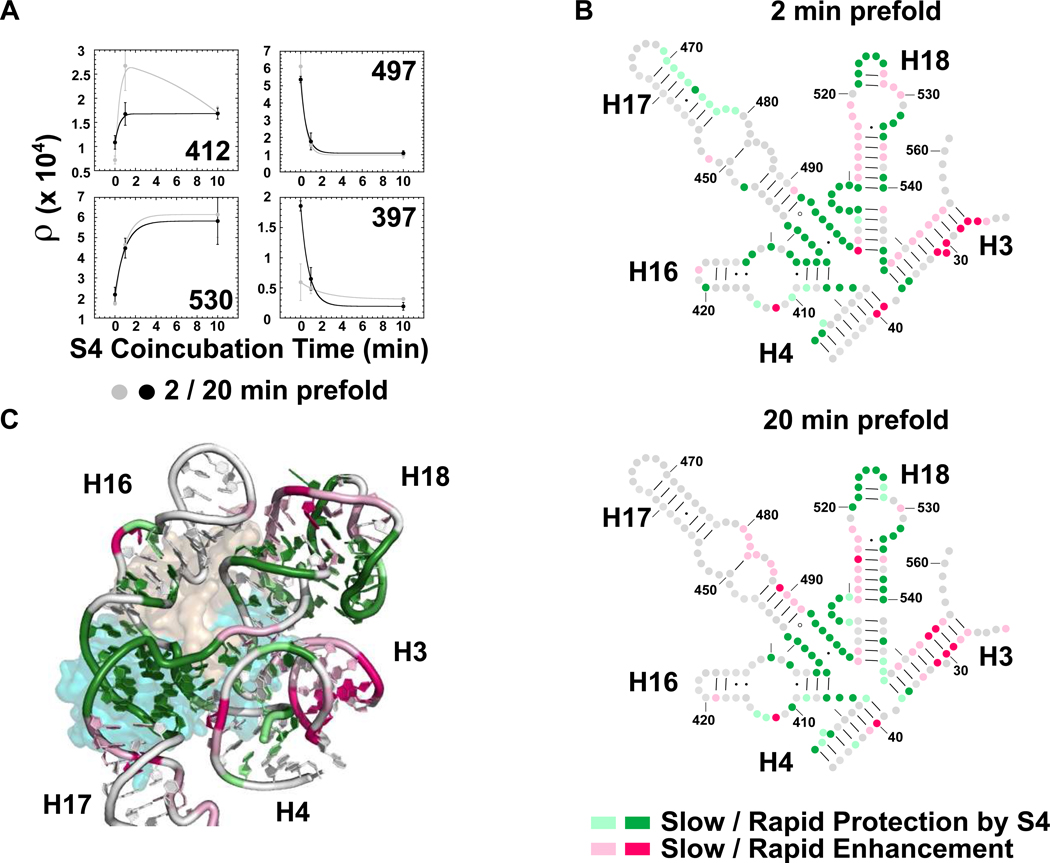
Figure 6. Time-dependence of S4-induced conformational change.
(a) SHAPE reactivity versus S4 coincubation time for four representative 5WJ nucleotides. Gray, 2 min prefold; black, 20 min prefold. Error bars as in Fig. 5a. (b) Secondary structure showing the rate of S4 protection (green) or exposure (pink) of 5WJ complexes prefolded at 42°C for 2 min (top) and 20 min (bottom). Relative differences were used to determine the % change after 1 min S4 coincubation, (ρ1min − ρ0min) / (ρ10min − ρ0min). Gray, no change; dark colors, fast (≥80% at 1 min); pale colors, slow (<80%). (c) Ribbon of E. coli S4 bound to the 5WJ in the ribosome colored as in b for 20 min prefolding (PDB:2AVY)56. S4 is colored as in Figure 1a.
Most residues protected by S4 were fully protected in the first minute of the binding reaction, consistent with rapid association of S4 with the 5WJ rRNA (dark colors; Figure 6 b,c). Nucleotides that form the H18 pseudoknot, the junction between H17 and H18, and the 430 region of the H16 loop were fully structured within this time. By contrast, further changes to H16, and the stacking of H3 upon H4 (right-angle motif) required more than 1 min to complete, suggesting that these slower changes correlate with the transition from labile to stable complexes (light green, Figure 6 b,c). Many of the slowest effects mapped to enhanced modifications, particularly in H18 and H3 (pink, Figure 6b,c). SHAPE modification can arise either from increased backbone flexibility or a distorted conformation that exposes the ribose 2’OH44. Although a distorted backbone can explain the enhanced modification of certain residues, such as G530, our results intriguingly suggest that S4 renders segments of the 16S rRNA more dynamic over time.
When the 5WJ rRNA was prefolded for 2 min prior to adding S4, more residues changed reactivity in the first minute of binding, compared to rRNA prefolded for 20 min (Figure 6b). However, the regions that change slowly after S4 addition were similar in the two experiments. This result suggests that prefolding the rRNA increases the probability of forming productive intermediate complexes with S4, but cannot accelerate conformational changes that require the presence of S4.
DISCUSSION
Previous studies on ribosome assembly have shown how ribosomal protein binding is coupled to folding of the rRNA46, 48. In the specific case of S4, temperature-dependent changes in the conformation of the S4 – 16S complex were found to require the presence of S420. Our data, showing that several minutes of coincubation with S4 is required to form a stable S4-rRNA complex, are consistent with an induced-fit mechanism for S4 recognition. Although the naked rRNA folds into a structure competent to bind S4 within 30–60 s 39 (Figure S3), prefolding the RNA, even for long times, is insufficient for tight binding (Figure 3c). Instead, the proportion of stable complex increases at approximately the same rate for a given temperature (rate42°C = 1.0 ± 0.08 min−1) after S4 is added to the RNA. This change occurs in the minimal 5WJ, indicating it is intrinsic to the S4 binding site. In addition, our data using the Bst S4 protein agree with previous data34,20 obtained with E. coli S4.
Our SHAPE data demonstrate that the H18 pseudoknot, the junction between H17 and H18, and the 430 region of the H16 loop become ordered within the 1 min time window of the experiment. Structural changes that occur more slowly, during the transition to the stable complex, include further rearrangements to H16 and the stacking of H3 at a right angle to H4. This order correlates with the kinetics of S4 binding inferred from time-resolved hydroxyl radical footprinting of 30S assembly 34, as nucleotides near the center of the 5WJ were protected by S4 much more rapidly than residues in H18 and at the top of H16. Since some of the nucleotides that change conformation slowly interact with the N-terminal region of S4, the transition from labile to stable binding is probably linked to refolding of this disordered polypeptide. Pre-ordering of residues 12–15 and 30–33 in the free protein may assist S4-rRNA recognition 27.
The similarity of our results with those of Powers and Noller20 suggest they reflect the same conformational change in the S4-rRNA complex. 16S bases protected from modification by S4 at 4°C 20 were rapidly affected in our SHAPE study (Figure 1b compared to Figure 5c and S5b), while bases requiring 42°C coincubation to reach their native conformation20 map to regions that fold slowly. S4 binding to mature 16S rRNA was more temperature-dependent than binding to minimal rRNA transcripts, perhaps because the other RNA interactions or post-transcriptional modifications in the 530 loop impart greater stability to the mature 16S rRNA 49–51.
Nucleotides that slowly change conformation upon S4 binding are important for 30S assembly and function. First, the pseudoknot in H18 is required for translation accuracy because it positions the universally conserved G530 in the A-site of the 30S decoding center 52–54. Chemical-probing data indicated that the pseudoknot was important for S4 binding 20, and mutations that destabilize the pseudoknot reduced the stability of the S4 – 5WJ complex (−2 kcal/mol)16. Nucleotides that form the pseudoknot base pairs are protected rapidly, while G530 requires a longer S4 coincubation to become exposed to modification (Figure 6). This suggests that the pseudoknot stabilizes further conformational changes in the 530 loop.
Second, the internal loop of H16, which represents a key recognition element for bacterial S4 proteins, rearranges considerably upon S4 coincubation (Figure 5c and S5b). Rapid protection of nt 430–432 and strong exposure of A412 are followed by more subtle changes in the rest of the H16 loop. Molecular dynamics simulations indicate the N-terminus of S4, which directly contacts H16, forms numerous non-native contacts33. These observations suggest S4 initially contacts H16 non-specifically, then rearranges to the specific contacts seen in 30S structures15, 28.
Finally, we previously proposed that H3 is initially displaced from H18 during 30S assembly, and docks with H18 after a conformational switch stabilized by protein S1617. The native right angle between H3 and H4 (right-angle motif), which protects nucleotides 395–398, is one of the later changes in 5WJ structure caused by S4 (Figure 6b, c)34. Unexpectedly, the SHAPE results revealed that S4 increases the reactivity of H3 (red, Figure 5c and S5b), perhaps contributing to the “switch” stabilized by ribosomal protein S16. In the mature ribosome, H3 is presumably stabilized by protein S12 and by interactions with the 16S central domain.
Interestingly, we noticed the reactivity of some nucleotides increased exaggeratedly after 1 min S4 coincubation when the RNA was prefolded only 2 min. These residues were modified to the same level as extensively prefolded rRNA after 10 min of S4 coincubation (Figure 6a). This reproducible pattern mapped to the pseudoknot (nt 505–507, 524–526) and the junction between H16 and H17 (nt 435–440), consistent with reorganization of the RNA interactions during S4 recognition.
Taken together, our binding and SHAPE chemical-probing results are consistent with an induced-fit model of S4 – rRNA interactions. On the one hand, S4 binds and stabilizes the folded structure of the rRNA. On the other hand, S4 also acts as a chaperone to promote structural rearrangements in both the RNA and protein components of the native complex. A mixture of stabilization and refolding was also observed during assembly of the yeast mitochondrial bI3 splicing complex 57. We show that labile S4 complexes rearrange to stable and very long-lived complexes at 42°C, with the longer S4 coincubation times resulting in a greater number of stable complexes. The transformation of labile complexes into stable complexes is linked to specific changes in the 5WJ caused by S4, including remodeling of the H16–H18 interface by the N-terminal domain of S4, formation of the right angle between H3 and H4, and final refolding of the 530-loop in H18.
Although this conformational change can occur in minimal complexes containing only S4, helices extending from the 5WJ interact with proteins S16 and S12 and with the central domain. Thus, changes in the S4 binding site are structurally connected to other regions of the 30S ribosome and can influence other stages of assembly and translation. As S4 complexes are very long-lived once locked in place, disassembly may require protein chaperones or active turnover by proteases and nucleases. Finally, in eukaryotic 40S ribosomes, the S4 homolog (RPS9) lacks an N-terminal extension and the sequence and orientation of H16 varies considerably from bacterial H16, most likely to accommodate the eukaryotic initiation machinery, showing how critical S4-rRNA interactions have coevolved30, 33.
MATERIALS AND METHODS
rRNA Preparation
rRNA containing the E. coli 16S rRNA 5’ domain (nucleotides 21-562) or 5WJ (nucleotides 21–46, 395–562) were transcribed and purified as previously described16, 39. For SHAPE experiments, the 3’ end of the 5WJ rRNA was extended using standard cloning methods to create the plasmid p5WJ-1199. The extended sequence of p5WJ-1199 is 5’-a-a-a-g-c-a-a-t-a-t-a-g-a-c-t-c-g-g-g-a-t-g-a-g-g-a-C-A-T-C-A-T-G-G-C-C-C-T-T-A-C-G-A 3’ and begins after 16S nucleotide 562. The primer-binding site is capitalized. 32P-labeled transcripts were prepared by in vitro transcription as previously described16.
Bst S4 Purification
Geobacillus stearothermophilus (Bst) S4 was purified and stored as described previously16, 19. The final preparation was ≥ 95% full length as judged by silver-stained SDS-PAGE, and properly folded as determined by far UV CD spectroscopy. Bst S4 sediments as a monomer in analytical ultracentrifugation experiments (data not shown). However, we observe irreversible loss of binding activity 1.5–2 hr after stocks are thawed, possibly due to surface adsorption.
S4 Association Rate
S4 binding reactions were performed in RNAse-free low-retention microcentrifuge tubes (Ambion). 32P-labeled 5’ domain or 5WJ RNAs (0.5 nM) were prefolded 20 min at 42°C in HKM4 Buffer (80 mM K-Hepes pH 7.5, 330 mM KCl, 4 mM MgCl2 20 ng / µL poly dI-dC (Sigma)). Previous work has shown that 4 mM MgCl2 is sufficient for maximum S4 binding16. Bst S4 (25 nM for 5WJ or 36 nM for 5’ domain) was added to the prefolded rRNA at 42°C. The total reaction volume was 20 µL. Aliquots were removed at specific time intervals and loaded directly onto a native polyacrylamide gel. 5’ domain – S4 complexes were resolved on 8% (29:1 mono:bis) polyacrylamide gels in TBE; 5WJ - S4 complexes were resolved on 6% polyacrylamide gels in TKM216. Gels were run 6 h at 15 W at 10 °C. The fractions of bound and free rRNA were obtained by quantifying the counts in each band and fitting to an exponential rate equation, fB = fmax(1 − exp(−kobst). fmax is the maximum fraction of rRNA bound at equilibrium and kobs is the observed rate constant.
Kinetic Competition Assays
For kinetic competition assays 0.5 nM of 32P-labeled 5’ domain was prefolded in HKM4 buffer16, 19. The folded rRNA was then co-incubated with 25 nM Bst S4 (5WJ) or 45–90 nM Bst S4 (5’ domain) up to 20 min at 4°C to 50°C. Competitor rRNA (500 nM – 1 µM unlabeled 5WJ) was added at 42°C, and reaction aliquots loaded directly on a running native gel at various times after the addition of competitor.
The S4 dissociation kinetics from the 5’ domain rRNA were fit to fB = A1exp(−k1t) + A2exp(−k2t), in which A1 and A2 represent the amplitudes of each phase, and k1 and k2 represent the observed decay rates of the labile and stable complexes. The value of k1 was set to 3 min−1. Changing the value of k1 did not affect the values of A1, A2 and k2. As the 5WJ dissociated from S4 too rapidly to estimate the lifetime of the labile complex, the proportion of stable and labile complexes was estimated by comparing experimental reactions to a control reaction without competitor rRNA, which was used to determine the initial fraction of 5WJ bound to S4 (Figure S1). The stated amplitude of the stable phase is the difference between the initial rRNA fraction bound in the control reaction (time “zero”) and the fraction bound at the first point of the experimental reaction (~ 15 s). For all conditions, the experimentally determined value of k2 was 0.02 ± 0.013 min−1, which controls showed represents irreversible denaturation of the stable complex. In all S4 EMSA experiments, maximally ~80% of the RNA was bound, likely due to non-specific complexes that dissociate during gel electrophoresis, but possibly because of some stable misfolding.
SHAPE chemical-probing
Extended 5WJ rRNA (2 pmol) was prefolded 2 or 20 min at 42°C in HKM4 buffer and incubated with 10 pmol of Bst S4 1 or 10 min at 42°C. After S4 incubation, 1-methyl-7-nitroisatoic anhydride (1m7)40, 41 was added to a final concentration of 6.5 mM and incubated 70 s at 42°C. Total reaction volume was 10 µL after 1m7 addition. The modified rRNA was extracted once with phenol and chloroform, precipitated with ethanol, and resuspended in TE (10 mM Tris-HCl pH 8, 1 mM EDTA).
Primer Extension
Primer extension reactions were performed according to the SuperScript III Reverse Transcriptase protocol (Invitrogen) with minor modifications. Briefly, 1 pmol rRNA was incubated with 1 pmol 32P end-labeled 1199 primer47 at 65 °C for 5 min, and then cooled on ice for 1 min. The supplied First Strand buffer was added along with DTT (final concentration 5 mM), 1 mM dNTPs (final), and 100 U SuperScript III reverse transcriptase. The mixture was incubated at 55 °C for 30 min before precipitation with ethanol and analysis on an 8% polyacrylamide sequencing gel.
Analysis of SHAPE Data
Gels were quantified using SAFA55 to obtain the intensity of modification at each nucleotide position. The results were adjusted for loading variations by 1) selecting a candidate band with the least variation between lanes, 2) calculating the degree the experimental band varied from the reference band, and 3) adjusting each lane by a scaling factor to make the reference band intensity the same across all conditions. Reported adjusted intensities (ρ) are averages from an experiment done in triplicate.
To determine the SHAPE reactivity pattern of the 5WJ in the absence of S4 (Figure 5b and S5a) the most reactive nucleotide (ρmax) was set to 100%. All other nucleotide intensities were divided by the maximum possible reactivity (ρ/ρmax). To determine the how S4 coincubation affects 5WJ SHAPE reactivity (Figure 5c and S5b) for each nucleotide, a histogram of relative intensities, a = log10(ρ10min/ρ0min), was evaluated. Residues with |a| ≥ 0.25 were considered significantly enhanced or protected by S4 (13% and 23% of all values), and those with 0.25 > |a| ≥ 0.10 were considered weakly enhanced or protected (14% and 11%). The rate of S4-induced change was estimated from the relative change in ρ after 1 min coincubation (Figure 6), ρrel = (ρ1min − ρ0min) / (ρ10min − ρ0min). Nucleotides with ρrel ≥ 80% at 1 min were considered rapidly changing, whereas those with ρrel < 80% were considered slow. Residues showing an extremely small total change in modification over 10 min of S4 coincubation (ρ10min − ρ0min < 500) were excluded from the analysis.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank D. Draper for the gift of Bst S4 plasmid and helpful advice, and A. Z. E. Tan for assistance with analytical ultracentrifugation experiments. This work was supported by a grant from NIGMS (GM60819).
ABBREVIATIONS
- 5WJ
five-way-junction
- SHAPE
selective 2’-hydroxyl acylation analyzed by primer extension
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- 1.Powers T, Daubresse G, Noller HF. Dynamics of in vitro assembly of 16 S rRNA into 30 S ribosomal subunits. J. Mol. Biol. 1993;232:362–374. doi: 10.1006/jmbi.1993.1396. [DOI] [PubMed] [Google Scholar]
- 2.Talkington MW, Siuzdak G, Williamson JR. An assembly landscape for the 30S ribosomal subunit. Nature. 2005;438:628–632. doi: 10.1038/nature04261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nowotny V, Nierhaus KH. Assembly of the 30S subunit from Escherichia coli ribosomes occurs via two assembly domains which are initiated by S4 and S7. Biochemistry. 1988;27:7051–7055. doi: 10.1021/bi00418a057. [DOI] [PubMed] [Google Scholar]
- 4.Weitzmann CJ, Cunningham PR, Nurse K, Ofengand J. Chemical evidence for domain assembly of the Escherichia coli 30S ribosome. FASEB J. 1993;7:177–180. doi: 10.1096/fasebj.7.1.7916699. [DOI] [PubMed] [Google Scholar]
- 5.Held WA, Ballou B, Mizushima S, Nomura M. Assembly mapping of 30 S ribosomal proteins from Escherichia coli. Further studies. J. Biol. Chem. 1974;249:3103–3111. [PubMed] [Google Scholar]
- 6.Williamson JR. Induced fit in RNA-protein recognition. Nat. Struct. Biol. 2000;7:834–837. doi: 10.1038/79575. [DOI] [PubMed] [Google Scholar]
- 7.Draper DE, Reynaldo LP. RNA binding strategies of ribosomal proteins. Nucleic Acids Res. 1999;27:381–388. doi: 10.1093/nar/27.2.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Leulliot N, Varani G. Current topics in RNA-protein recognition: control of specificity and biological function through induced fit and conformational capture. Biochemistry. 2001;40:7947–7956. doi: 10.1021/bi010680y. [DOI] [PubMed] [Google Scholar]
- 9.Zimmerman RA, Muto A, Fellner P, Ehresmann C, Branlant C. Location of Ribosomal Protein Binding Sites on 16S Ribosomal RNA. Proc. Natl. Acad. Sci. U. S. A. 1972;69:1282. doi: 10.1073/pnas.69.5.1282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mackie GA, Zimmermann RA. RNA--protein interactions in the ribosome. IV. Structure and properties of binding sites for proteins S4, S16/S17 and S20 in the 16S RNA. J. Mol. Biol. 1978;121:17–39. doi: 10.1016/0022-2836(78)90260-7. [DOI] [PubMed] [Google Scholar]
- 11.Osswald M, Greuer B, Brimacombe R, Stoffler G, Baumert H, Fasold H. RNA-protein cross-linking in Escherichia coli 30S ribosomal subunits; determination of sites on 16S RNA that are cross-linked to proteins S3, S4, S5, S7, S8, S9, S11, S13, S19 and S21 by treatment with methyl p-azidophenyl acetimidate. Nucleic Acids Res. 1987;15:3221–3240. doi: 10.1093/nar/15.8.3221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stern S, Wilson RC, Noller HF. Localization of the binding site for protein S4 on 16 S ribosomal RNA by chemical and enzymatic probing and primer extension. J. Mol. Biol. 1986;192:101–110. doi: 10.1016/0022-2836(86)90467-5. [DOI] [PubMed] [Google Scholar]
- 13.Vartikar JV, Draper DE. S4–16 S ribosomal RNA complex. Binding constant measurements and specific recognition of a 460-nucleotide region. J. Mol. Biol. 1989;209:221–234. doi: 10.1016/0022-2836(89)90274-x. [DOI] [PubMed] [Google Scholar]
- 14.Powers T, Noller HF. Hydroxyl radical footprinting of ribosomal proteins on 16 S rRNA. RNA. 1995;1:194. [PMC free article] [PubMed] [Google Scholar]
- 15.Wimberly BT, Brodersen DE, Clemons WM, Jr, Morgan-Warren RJ, Carter AP, Vonrhein C, Hartsch T, Ramakrishnan V. Structure of the 30S ribosomal subunit. Nature. 2000;407:327–339. doi: 10.1038/35030006. [DOI] [PubMed] [Google Scholar]
- 16.Bellur DL, Woodson SA. A minimized rRNA-binding site for ribosomal protein S4 and its implication for 30S assembly. Nucl. Acids Res. 2009;37:1886. doi: 10.1093/nar/gkp036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ramaswamy P, Woodson SA. Global stabilization of rRNA structure by ribosomal proteins S4, S17, and S20. J. Mol. Biol. 2009;392:666–677. doi: 10.1016/j.jmb.2009.07.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Noller HF. Biochemical characterization of the ribosomal decoding site. Biochimie. 2006;88:935–941. doi: 10.1016/j.biochi.2006.04.006. [DOI] [PubMed] [Google Scholar]
- 19.Gerstner RB, Pak Y, Draper DE. Recognition of 16S rRNA by ribosomal protein S4 from Bacillus stearothermophilus. Biochemistry. 2001;40:7165–7173. doi: 10.1021/bi010026i. [DOI] [PubMed] [Google Scholar]
- 20.Powers T, Noller HF. A temperature-dependent conformational rearrangement in the ribosomal protein S4.16 S rRNA complex. J. Biol. Chem. 1995;270:1238–1242. doi: 10.1074/jbc.270.3.1238. [DOI] [PubMed] [Google Scholar]
- 21.Dutca LM, Jagannathan I, Grondek JF, Culver GM. Temperature-dependent RNP conformational rearrangements: analysis of binary complexes of primary binding proteins with 16 S rRNA. J. Mol. Biol. 2007;368:853–869. doi: 10.1016/j.jmb.2007.02.064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Orr JW, Hagerman PJ, Williamson JR. Protein and Mg(2+)-induced conformational changes in the S15 binding site of 16 S ribosomal RNA. J. Mol. Biol. 1998;275:453–464. doi: 10.1006/jmbi.1997.1489. [DOI] [PubMed] [Google Scholar]
- 23.Davies C, Gerstner RB, Draper DE, Ramakrishnan V, White SW. The crystal structure of ribosomal protein S4 reveals a two-domain molecule with an extensive RNA-binding surface: one domain shows structural homology to the ETS DNA-binding motif. EMBO J. 1998;17:4545–4558. doi: 10.1093/emboj/17.16.4545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Markus MA, Gerstner RB, Draper DE, Torchia DA. Refining the overall structure and subdomain orientation of ribosomal protein S4 delta41 with dipolar couplings measured by NMR in uniaxial liquid crystalline phases. J. Mol. Biol. 1999;292:375–387. doi: 10.1006/jmbi.1999.3061. [DOI] [PubMed] [Google Scholar]
- 25.Markus MA, Gerstner RB, Draper DE, Torchia DA. The solution structure of ribosomal protein S4 delta41 reveals two subdomains and a positively charged surface that may interact with RNA. EMBO J. 1998;17:4559–4571. doi: 10.1093/emboj/17.16.4559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yusupov MM, Yusupova GZ, Baucom A, Lieberman K, Earnest TN, Cate JH, Noller HF. Crystal structure of the ribosome at 5.5 A resolution. Science. 2001;292:883–896. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- 27.Sayers EW, Gerstner RB, Draper DE, Torchia DA. Structural preordering in the N-terminal region of ribosomal protein S4 revealed by heteronuclear NMR spectroscopy. Biochemistry. 2000;39:13602–13613. doi: 10.1021/bi0013391. [DOI] [PubMed] [Google Scholar]
- 28.Tocilj A, Schlunzen F, Janell D, Gluhmann M, Hansen HA, Harms J, Bashan A, Bartels H, Agmon I, Franceschi F, Yonath A. The small ribosomal subunit from Thermus thermophilus at 4.5 A resolution: pattern fittings and the identification of a functional site. Proc. Natl. Acad. Sci. U. S. A. 1999;96:14252–14257. doi: 10.1073/pnas.96.25.14252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schluenzen F, Tocilj A, Zarivach R, Harms J, Gluehmann M, Janell D, Bashan A, Bartels H, Agmon I, Franceschi F, Yonath A. Structure of functionally activated small ribosomal subunit at 3.3 angstroms resolution. Cell. 2000;102:615–623. doi: 10.1016/s0092-8674(00)00084-2. [DOI] [PubMed] [Google Scholar]
- 30.Ben-Shem A, Jenner L, Yusupova G, Yusupov M. Crystal structure of the eukaryotic ribosome. Science. 2010;330:1203–1209. doi: 10.1126/science.1194294. [DOI] [PubMed] [Google Scholar]
- 31.Chen K, Roberts E, Luthey-Schulten Z. Horizontal gene transfer of zinc and non-zinc forms of bacterial ribosomal protein S4. BMC Evol. Biol. 2009;9:179. doi: 10.1186/1471-2148-9-179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Newberry V, Yaguchi M, Garrett RA. A trypsin-resistant fragment from complexes of ribosomal protein S4 with 16-S RNA of Escherichia coli and from the uncomplexed protein. Eur. J. Biochem. 1977;76:51–61. doi: 10.1111/j.1432-1033.1977.tb11569.x. [DOI] [PubMed] [Google Scholar]
- 33.Chen K, Eargle J, Sarkar K, Gruebele M, Luthey-Schulten Z. Functional role of ribosomal signatures. Biophys. J. 2010;99:3930–3940. doi: 10.1016/j.bpj.2010.09.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Adilakshmi T, Bellur DL, Woodson SA. Concurrent nucleation of 16S folding and induced fit in 30S ribosome assembly. Nature. 2008;455:1268–1272. doi: 10.1038/nature07298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brimacombe R. RNA-protein interactions in the Escherichia coli ribosome. Biochimie. 1991;73:927–936. doi: 10.1016/0300-9084(91)90134-m. [DOI] [PubMed] [Google Scholar]
- 36.Woese CR, Fox GE, Zablen L, Uchida T, Bonen L, Pechman K, Lewis BJ, Stahl D. Conservation of primary structure in 16S ribosomal RNA. Nature. 1975;254:83–86. doi: 10.1038/254083a0. [DOI] [PubMed] [Google Scholar]
- 37.Gutell RR, Weiser B, Woese CR, Noller HF. Comparative anatomy of 16-S-like ribosomal RNA. Prog. Nucleic Acid Res. Mol. Biol. 1985;32:155–216. doi: 10.1016/s0079-6603(08)60348-7. [DOI] [PubMed] [Google Scholar]
- 38.Mulder AM, Yoshioka C, Beck AH, Bunner AE, Milligan RA, Potter CS, Carragher B, Williamson JR. Visualizing ribosome biogenesis: parallel assembly pathways for the 30S subunit. Science. 2010;330:673–677. doi: 10.1126/science.1193220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Adilakshmi T, Ramaswamy P, Woodson SA. Protein-independent folding pathway of the 16S rRNA 5' domain. J. Mol. Biol. 2005;351:508–519. doi: 10.1016/j.jmb.2005.06.020. [DOI] [PubMed] [Google Scholar]
- 40.Wilkinson KA, Merino EJ, Weeks KM. Selective 2'-hydroxyl acylation analyzed by primer extension (SHAPE): quantitative RNA structure analysis at single nucleotide resolution. Nat. Protoc. 2006;1:1610–1616. doi: 10.1038/nprot.2006.249. [DOI] [PubMed] [Google Scholar]
- 41.Mortimer SA, Weeks KM. A fast-acting reagent for accurate analysis of RNA secondary and tertiary structure by SHAPE chemistry. J. Am. Chem. Soc. 2007;129:4144–4145. doi: 10.1021/ja0704028. [DOI] [PubMed] [Google Scholar]
- 42.Deigan KE, Li TW, Mathews DH, Weeks KM. Accurate SHAPE-directed RNA structure determination. Proc. Natl. Acad. Sci. U. S. A. 2009;106:97–102. doi: 10.1073/pnas.0806929106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Low JT, Weeks KM. SHAPE-directed RNA secondary structure prediction. Methods. 2010;52:150–158. doi: 10.1016/j.ymeth.2010.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gherghe CM, Shajani Z, Wilkinson KA, Varani G, Weeks KM. Strong correlation between SHAPE chemistry and the generalized NMR order parameter (S2) in RNA. J. Am. Chem. Soc. 2008;130:12244–12245. doi: 10.1021/ja804541s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Brodersen DE, Clemons WM, Jr, Carter AP, Wimberly BT, Ramakrishnan V. Crystal structure of the 30 S ribosomal subunit from Thermus thermophilus: structure of the proteins and their interactions with 16 S RNA. J. Mol. Biol. 2002;316:725–768. doi: 10.1006/jmbi.2001.5359. [DOI] [PubMed] [Google Scholar]
- 46.Stern S, Powers T, Changchien LM, Noller HF. RNA-protein interactions in 30S ribosomal subunits: folding and function of 16S rRNA. Science. 1989;244:783–790. doi: 10.1126/science.2658053. [DOI] [PubMed] [Google Scholar]
- 47.Moazed D, Stern S, Noller HF. Rapid chemical probing of conformation in 16 S ribosomal RNA and 30 S ribosomal subunits using primer extension. J. Mol. Biol. 1986;187:399–416. doi: 10.1016/0022-2836(86)90441-9. [DOI] [PubMed] [Google Scholar]
- 48.Mizushima S, Nomura M. Assembly mapping of 30S ribosomal proteins from E. coli. Nature. 1970;226:1214. doi: 10.1038/2261214a0. [DOI] [PubMed] [Google Scholar]
- 49.Charette M, Gray MW. Pseudouridine in RNA: what, where, how, and why. IUBMB Life. 2000;49:341–351. doi: 10.1080/152165400410182. [DOI] [PubMed] [Google Scholar]
- 50.Helm M. Post-transcriptional nucleotide modification and alternative folding of RNA. Nucleic Acids Res. 2006;34:721–733. doi: 10.1093/nar/gkj471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Chow CS, Lamichhane TN, Mahto SK. Expanding the nucleotide repertoire of the ribosome with post-transcriptional modifications. ACS Chem. Biol. 2007;2:610–619. doi: 10.1021/cb7001494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Thurlow DL, Zimmermann RA. Conservation of ribosomal protein binding sites in prokaryotic 16S RNAs. Proc. Natl. Acad. Sci. U. S. A. 1978;75:2859–2863. doi: 10.1073/pnas.75.6.2859. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Powers T, Noller HF. A functional pseudoknot in 16S ribosomal RNA. EMBO J. 1991;10:2203–2214. doi: 10.1002/j.1460-2075.1991.tb07756.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ogle JM, Brodersen DE, Clemons WM, Jr, Tarry MJ, Carter AP, Ramakrishnan V. Recognition of cognate transfer RNA by the 30S ribosomal subunit. Science. 2001;292:897–902. doi: 10.1126/science.1060612. [DOI] [PubMed] [Google Scholar]
- 55.Das R, Laederach A, Pearlman SM, Herschlag D, Altman RB. SAFA: semi-automated footprinting analysis software for high-throughput quantification of nucleic acid footprinting experiments. RNA. 2005;11:344–354. doi: 10.1261/rna.7214405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Schuwirth BS, Borovinskaya MA, Hau CW, Zhang W, Vila-Sanjurjo A, Holton JM, Cate JH. Structures of the bacterial ribosome at 3.5 A resolution. Science. 2005;310:827–834. doi: 10.1126/science.1117230. [DOI] [PubMed] [Google Scholar]
- 57.Duncan CDS, Weeks KM. Nonhierarchical Ribonucleoprotein Assembly Suggests a Strain-Propagation Model for Protein-Facilitated RNA Folding. Biochemistry. 2010;49:5418–5425. doi: 10.1021/bi100267g. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.