Abstract
With the improvements in diagnostic techniques, Bartonella henselae (B. henselae) infection has recently been recognized to cause a widening spectrum of diseases. Cats are the natural reservoir hosts of B. henselae. The current study aims to investigate the prevalence of B. henselae infection in the cat populations in China. Polymerase chain reaction (PCR) and bacterial cultures confirm that 12.7% of the tested cats were positive for the infection. Old age and outdoor exposure were statistically associated with the infection. Multilocus sequence typing and eBURST analysis of the cat isolates collected in the present study show that 65.4% of the isolates belong to sequence type 1 (ST1). Three new STs (ST16–18) were identified in Midwestern China. These results may aid our understanding of the population structure of B. henselae in China and the relationship between human and cat strains in subsequent studies.
Author Summary
Bartonella infection (Bartonella henselae in paticular) is responsible for a widening spectrum of human diseases. Cats are thought as the major source of human infections. In most of the developing countries, however, this infection lacks recognition especially in clinicians. Some well known bartonelloses, like cat-scratch disease, bacillary angiomatosis, and infective endocarditis had not been clinically noticed in these countries. In the present study, the prevalence of Bartonella infection in cat populations was investigated in mainland China. As a result, 12.7% of cats were confirmed positive with Bartonella infection. Factors of age, gender, outdoor experience, and geographic location were analyzed in association with infection. The authors found old cats and stray cats are susceptible populations. Some new molecular types are prevalent in Mid-Western China. Inclusion of these types also changed the characteristics of original molecular structure. This research provides the first molecular investigation of Bartonella infection in cats in China, and promotes our understanding of the relationship between human and cat strains in subsequent studies.
Introduction
Bartonella is a small, fastidious, intracellular Gram-negative bacterium that has been recently identified in a wide range of domestic and wild mammals. The genus Bartonella has expanded from Bartonella bacilliformis to at least 20 recognized species during the past century, of which at least 10 species and subspecies are known or suspected to be pathogenic to humans [1], [2]. Of these, B. henselae is the most common Bartonella infection worldwide, resulting in cat-scratch disease (CSD), bacillary angiomatosis (BA), endocarditis, and neuroretinitis [3], [4]. Cats are the major reservoir hosts of B. henselae. Two main B. henselae 16S rRNA genotypes have been identified and designated as Houston-1 and Marseille. Marseille is the dominant type in cats in the United States, Europe, and Australia, whereas Houston-1 is dominant in Asia [5], [6]. In China, only rodent-associated bartonellosis has been well documented [7], [8]. However, little is known about the prevalence of B. henselae infection in the cat populations in China. Thus, the current study aims to evaluate B. henselae infection in Chinese cats using bacterial cultures, PCR detection, and sequence typing.
Materials and Methods
Ethics statement
This study was performed in accordance with the recommendations in the Guide for the Care and Use of Laboratory Animals of the Ministry of Health, China. The current study was approved by the Shanghai Animal Management Committee. (Permit Number:SYXK2007-0025).
Sampling
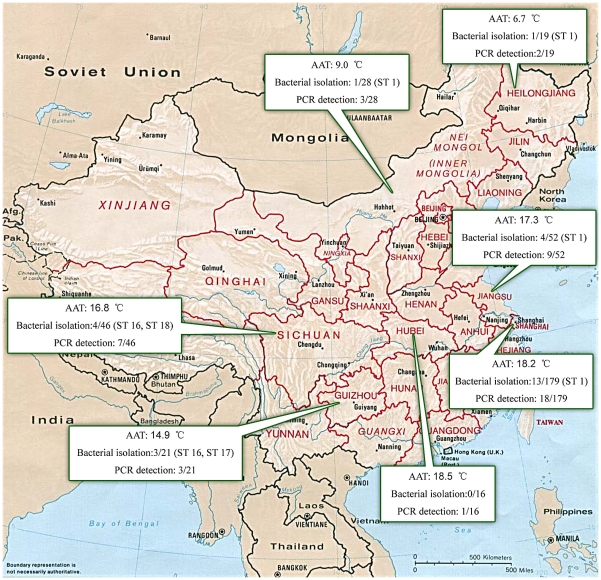
Three hundred fifteen blood samples from pet cats and 46 samples from stray cats were collected from March to September in 2010. The geographic locations are specified in Figure 1. The mean age of the pet cats was 3.1 years (in the range of 4 months to 9 years). The age of the stray cats was unavailable. Of the 361 cats, 209 were male and 152 were female. About 2 ml of EDTA-anticoagulated blood was collected from each cat by venipuncture. One ml was stored at −80°C for subsequent culture. The remaining blood was centrifuged and the cell pellet was collected for PCR detection.
Figure 1. Geographic location of tested cats.
The results of infection prevalence and the STs of isolates are specified. AAT means the average annual temperature.
Bacterial culture
The freeze-thawed blood samples (500 µl) were plated in Columbia agar containing 10% defibrinated sheep blood. The plates were incubated at 35°C in a 5% CO2, water-saturated atmosphere. Reference B. henselae strain (Houston-1) was cultured as the control. The plates were examined daily for 5 weeks for evidence of growth. Samples with bacterial contamination were immediately discarded. Positive cultures were further confirmed by PCR detection.
Species-differentiated PCR detection
DNA from the cell pellet of the collected samples was extracted using the QIAamp DNA Blood Mini Kit according to the manufacturer's instructions. PCR primers (5′-CTCTTTCTTCAGATGATGATCC-3′ and 5′-AACCAACTGAGCTACAAGCCCT-3′) resulted in amplified products of approximately 202 (B. bacilliformis), 145 (B. clarridgeiae), 232 (B. elizabethae), 163 (B. henselae), 148 (B. quintana), and 251 bp (B. vinsonii subsp. berkhoffii), which are in accordance with previous study [9]. The DNA fragment was amplified with a 10 min extension step after 36 cycles of 94°C for 1 min, 60°C for 1 min, and 72°C for 1 min,. For better differentiation, 4.5% agarose gel electrophoresis was performed. DNA of B. henselae (Houston-1), B. bacilliformis (KC584), B. quintana (90–268) and B. clarridgeiae (NCSU 94-F40) was used as the control. Amplifications other than B. henselae from tested blood samples were further sequenced.
Multilocus sequence typing (MLST) and data analysis
The B. henselae colonies isolated in the present study were scraped from the plates. DNA was extracted using QIAamp DNA Mini Kit. The PCR procedure and primers used are in accordance with previous studies [10], [11]. Nucleotide sequence data were collected from all B. henselae isolates of nine genetic loci (16SrRNA [rrs], batR, gltA, groEL, ftsZ, nlpD, ribC, rpoB, and eno). The DNA amplifications were purified and sequenced for both strands. All DNA sequences were manually analyzed for polymorphism.
The nucleotide sequences were analyzed using the DNASTAR Lasergene software package 7 (DNASTAR, Madison, USA). New alleles were confirmed by re-sequencing. Alleles and sequence types (STs) were assigned in accordance with the published data [11]. New allelic combinations encountered for the first time in the current study were assigned to new STs in the order of detection. The definition of clonal complexes and the examination of relationships between STs within clonal complexes were carried out using BURST v3 analysis (http://burst.mlst.net).
Statistical analysis
Statistical analysis of the association between B. henselae infection in cats and gender, age, and geographic region was performed using a χ2-test. A value of P<0.05 was considered significant.
Nucleotide sequence accession numbers
New rrs and batR alleles were obtained, and they were designated as rrs alleles 3 and 4, and batR allele 5. The sequences of rrs alleles 3 and 4, as well as that of batR allele 5 were deposited in GenBank under accession numbers JF819177 (rrs allele 3), JF819178 (rrs allele 4), and JF819179 (batR allele 5).
Results
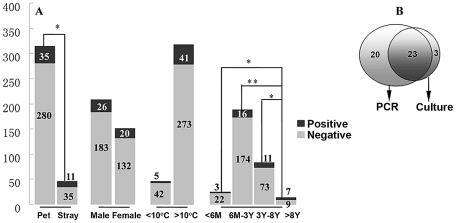
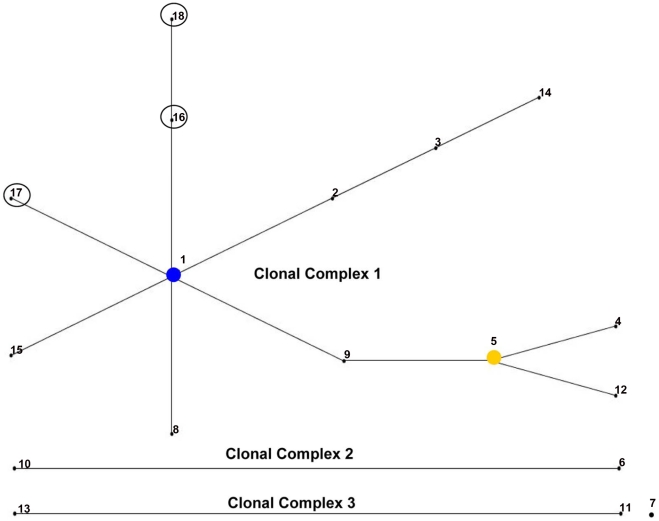
During the five-week bacterial isolation, two samples from pet cats and three samples from stray cats were discarded because of bacterial or fungi contamination. A total of 26 B. henselae strains were successfully isolated from 5.8% of the pet cats (18/313) and 18.6% of the stray cats (8/43), confirming that bacteremia more frequently occurs in stray cats (P<0.05). The average growth time for bacterial isolation was 7.6 days, with the shortest at 4 days and the longest at 23 days. The CFU/ml value of positive samples varied significantly from 4 to 2.4×102. PCR detection using species-differentiated primers confirmed that 10.5% of the pet cats (33/315) and 21.7% of the stray cats (10/46) were PCR positive for B. henselae. Three positive samples with low bacterial loads that were confirmed by culture isolation (<6 CFU/ml) were PCR negative. B. clarridgeiae DNA was detected in four stray cats. Of these, two cats were co-infected with B. henselae. In total, 46 cats were confirmed infected with B. henselae (Figure 2). Statistical analysis shows no significant difference in the infection between male (12.4% positive) and female cats (13.1% positive). However, B. henselae infection was more prevalent in old animals (Figure 2). Based on the average annual temperature, the regions where the tested cats were located are defined as cold and tropical zones. However, climate difference was not statistically associated with infection, although a mild decrease in the prevalence of infection was observed in the colder regions in the current study (Figures 1 and 2). MLST analysis confirmed seventeen of the isolates in current study (65.4%) belong to the ST1 type. Two new rrs alleles (alleles 3 and 4) were obtained from seven and two isolates, respectively. The rrs allele 3 is caused by two nucleotide variations at positions 1414626 (T instead of C) and 1414146 (A instead T) of the B. henselae Houston-1 chromosome, whereas allele 4 consists of a single nucleotide insert based on the allele 3 sequence at position 1414562 of the B. henselae Houston-1 chromosome. A new allele of the batR gene, with a single nucleotide variation at position 85173 (G instead of A) of the B. henselae Houston-1 chromosome, was designated as batR allele 5. Thus, three STs were encountered for the first time in the current study and designated as ST16 (seven isolates), ST 17 (two isolates), and ST18 (one isolate), in the order of detection (Table 1). The ST relationship was analyzed by eBURST. All newly identified STs were assigned to the previously described single clonal complex, Group 1 (Figure 3).
Figure 2. Infection prevalence and association with outdoor frequency, gender, age, and geographic origin.
(A) The risk factors associated with B. henselae infection: “*” P<0.05; “**” P<0.01. (B) The results confirmed by PCR and bacterial isolation.
Table 1. Allele profile of new STs identifed in the present study.
| ST | No of isolates | rrs | batR | gltA | ftsZ | groEL | nlpD | ribC | rpoB | Distribution |
| 16 | 7 | 3 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | SiChuan,Guizhou |
| 17 | 2 | 4 | 1 | 1 | 1 | 1 | 1 | 1 | 1 | Guizhou |
| 18 | 1 | 3 | 5 | 1 | 1 | 1 | 1 | 1 | 1 | SiChuan |
Figure 3. Phylogenetic relationship between the different B. henselae STs as determined by eBURST.
The new STs identified in the present study are closely related to ST1. The inclusion of the new STs altered the structure of the phylogenetic tree: ST1 is defined as the primary founder, whereas ST5 is defined as a subgroup founder.
Discussion
B. henselae has been found responsible for a widening range of clinical syndromes, particularly those of CSD, BA, endocarditis, and neuroretinitis [12]. Cats are the major reservoir host of B. henselae. In China, several studies confirmed a 9.6%–19.6% seropositivity to B. henselae infection in humans using immunofluorescence assays [13], [14]. However, little is known about the status of infections in cats in China. The homemade ELISA using whole bacterial antigens was not used in the present epidemiological study because of the inadequate number of control serum samples for accurate evaluation. Moreover, the high seroprevalence in cats renders the serologic testing impractical for infection diagnosis. Therefore, PCR combined with bacterial culture, was utilized in the present investigation.
Overall, 46 cats were confirmed PCR and/or culture positive. Of these, 26 bacteremic cats (7.3%) were determined by culture isolation. Previous studies have confirmed that Bartonella seroprevalences are higher in older cats than in younger animals, whereas the prevalence of bacteremia is higher in younger animals [15]. However, bacteremia was more frequently identified in cats older than 8 years in the present study. Since the sample size of old cats was relative small in the current study, the association of high prevalence with old age need further confirmation. The seroprevalence of B. henselae infections in cats varies considerably, from none in cold climates (0% in Norway) to high in warm and humid climates (68% in the Philippines) [16]. However, no difference in the prevalence of infection was found between the cold and tropical regions in China. Pet trade may be responsible for the spread of infection in cold regions.
To date, molecular information on Chinese isolates of B. henselae is still unavailable. Therefore, a MLST analysis was performed on the cat isolates collected in the present study. Hitherto, 26 STs were identified; of these, ST16–26 was defined in a recent study [17]. However, the sequence information of these STs has not been deposited in any public database, which enabled us to assign new STs in the order of their detection. Thus, the new allele combinations in the present study were designed from ST15 in the order of detection. In the present study, 65.4% Chinese isolates belonging to ST1 are associated with human infection. Interestingly, all isolates collected from Midwestern China (Sichuan and Guizhou Provinces) have new allele combinations that resulted in ST16–18. The eBURST analysis confirmed that ST16–18 were phylogenetically related to ST1. Although these new STs belonged to a single clonal complex, the inclusion of the new STs changed the characteristics of the original clonal complex 1, resulting in the designation of ST1 as the primary founder; the former primary founder ST5 was consequently designated as a subgroup founder.
In conclusion, the current study confirms the high prevalence of B. henselae infection in the cat populations in China. Molecular information from cat isolates in China was first reported, which shows a genetic diversity in the different regions. These results will facilitate an understanding of the population structure and the relationship between humans and cat strains in China in subsequent studies.
Acknowledgments
We thank Wang Yixiao (DVM) and Li Jingjing (DVM) at Shuangyi Pet hospital for help with sample collection.
Footnotes
The authors have declared that no competing interests exist.
This study is supported by the Science and Technology Commission of Shanghai Municipality (10JC1406500) and in part by the Open Foundation of State Key Laboratory of Veterinary Etiological Biology (SKLVEB2010KFKT011). Dr. Yuan is supported through the “Chen Guang” project supported by the Shanghai Municipal Education Commission and Shanghai Education Development Foundation, and the “Raising Star” project supported by SMG. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Birtles JB, Harrison TG, Saunders NA, Molyneux DH. Proposal to unify the genera Granhamella and Bartonella, with descriptions of Bartonella talpae comb.nov., Bartonella peromysci comb.nov., and three species, Bartonella grahamii sp.nov., Bartonella taylorii sp.nov. and Bartonella doshiae sp.nov. Int J Syst Bacteriol. 1995;45:1–8. doi: 10.1099/00207713-45-1-1. [DOI] [PubMed] [Google Scholar]
- 2.Brenner DJ, O'Connor S, Winkler HH, Steigerwalt AG. Proposals to unify the genera Bartonella and Rochalimaea, with descriptions of Bartonella quintana comb. nov., Bartonella vinsonii comb.nov. and to remove the family Bartonellaceae from the order Rickettsiales. Int J Syst Bacteriol. 1993;43:777–786. doi: 10.1099/00207713-43-4-777. [DOI] [PubMed] [Google Scholar]
- 3.Karem KL, Paddock CD, Regnery RL. Bartonella henselae, B. quintana, and B. bacilliformis: historical pathogens of emerging significance. Microbes Infect. 2000;2:1193–205. doi: 10.1016/s1286-4579(00)01273-9. [DOI] [PubMed] [Google Scholar]
- 4.Chomel BB, Kasten RW, Williams C, Wey AC, Henn JB, et al. Bartonella endocarditis: a pathology shared by animal reservoirsand patients. Ann NY Acad Sci. 2009;1166:120–126. doi: 10.1111/j.1749-6632.2009.04523.x. [DOI] [PubMed] [Google Scholar]
- 5.La Scola B, Liang Z, Zeaiter Z, Houpikian P, Grimont PA, et al. Genotypic characteristics of two serotypes of Bartonella henselae. J Clin Microbiol. 2002;40:2002–2008. doi: 10.1128/JCM.40.6.2002-2008.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bergmans AM, Schellekens JF, van Embden JD, Schouls LM. Predominance of two Bartonella henselae variants among cat-scratch disease patients in the Netherlands. J Clin Microbiol. 1996;34:254–260. doi: 10.1128/jcm.34.2.254-260.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ying B, Kosoy MY, Maupin GO, Tsuchiya KR, Gage KL. Genetic and ecologic characteristics of Bartonella communities in rodents in southern China. Am J Trop Med Hyg. 2002;66:622–627. doi: 10.4269/ajtmh.2002.66.622. [DOI] [PubMed] [Google Scholar]
- 8.Liu Q, Sun J, Lu L, Fu G, Ding G, et al. Detection of Bartonella species in small mammals from Zhejiang Province, China. J Wild Dis. 2010;46:179–185. doi: 10.7589/0090-3558-46.1.179. [DOI] [PubMed] [Google Scholar]
- 9.Jensen WA, Fall MZ, Rooney J, Kordick DL, Breitschwerdt EB. Rapid Identification and differentiation of Bartonella species using a single-Step PCR assay. J Clin Microbiol. 2000;38:1717–1722. doi: 10.1128/jcm.38.5.1717-1722.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Iredell J, Blanckenberg D, Arvand M, Grauling S, Feil EJ, et al. Characterization of the natural population of Bartonella henselae by multilocus sequence typing. J Clin Microbiol. 2003;41:5071–5079. doi: 10.1128/JCM.41.11.5071-5079.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Arvand M, Feil EJ, Giladi M, Boulouis HJ, Viezens J. Multi-locus sequence typing of Bartonella henselae isolates from three continents reveals hypervirulent and feline-associated clones. PloS ONE. 2007;12:1346. doi: 10.1371/journal.pone.0001346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Florin TA, Zaoutis TE, Zaoutis LB. Beyond Cat Scratch Disease: Widening Spectrum of Bartonella henselae Infection. Pediatrics. 2008;121:1413–1425. doi: 10.1542/peds.2007-1897. [DOI] [PubMed] [Google Scholar]
- 13.Sun J, Fu G, Lin J, Song X, Lu L, Liu Q. Seroprevalence of Bartonella in Eastern China and analysis of risk factors. BMC Infect Dis. 2010;10:121. doi: 10.1186/1471-2334-10-121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhang L, Shan A, Mathew B, Yin J, Fu X, et al. Rickettsial Seroepidemiology among farm workers, Tianjin, People's Republic of China. Emerg Infect Dis. 2008;14:938–940. doi: 10.3201/eid1406.071502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chomel BB, Abbott RC, Kasten RW, Floyd-Hawkins KA, Kass PH, et al. Bartonella henselae prevalence in domestic cats in California: risk factors and association between bacteremia and antibody titers. J Clin Microbiol. 1995;33:2445–50. doi: 10.1128/jcm.33.9.2445-2450.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Boulouis HJ, Chang CC, Henn JB, Kasten RW, Chomel BB. Factors associated with the rapid emergence of zoonotic Bartonella infections. Vet Res. 2005;36:383–410. doi: 10.1051/vetres:2005009. [DOI] [PubMed] [Google Scholar]
- 17.Mietze A, Morick D, Köhler H, Harrus S, Dehio C, et al. Combined MLST and AFLP typing of Bartonella henselae isolated from cats reveals new sequence types and suggests clonal evolution. Vet Microbiol. 2011;148:238–45. doi: 10.1016/j.vetmic.2010.08.012. [DOI] [PubMed] [Google Scholar]