Abstract
Members of the Ets family of transcription factors mediate transcriptional responses of multiple signaling pathways in diverse cell types and organisms. Targeted deletion of the conserved DNA binding domain of the Ets2 transcription factor results in the retardation and death of homozygous mouse embryos before 8.5 days of embryonic development. Defects in extraembryonic tissue gene expression and function include deficient expression of matrix metalloproteinase-9 (MMP-9, gelatinase B), persistent extracellular matrix, and failure of ectoplacental cone proliferation. Mutant embryos were rescued by aggregation with tetraploid mouse embryos, which complement the developmental defects by providing functional extraembryonic tissues. Rescued Ets2-deficient mice are viable and fertile but have wavy hair, curly whiskers, and abnormal hair follicle shape and arrangement, resembling mice with mutations of the EGF receptor or its ligands. However, these mice are not deficient in the production of TGFα or the EGF receptor. Homozygous mutant cell lines respond mitogenically to TGFα, EGF, FGF1, and FGF2. However, FGF fails to induce MMP-13 (collagenase-3) and MMP-3 (stromelysin-1) in the Ets2-deficient fibroblasts. Ectopic expression of Ets2 in the deficient fibroblasts restores expression of both matrix metalloproteinases. Therefore, Ets2 is essential for placental function, mediating growth factor signaling to key target genes including MMP-3, MMP-9, and MMP-13 in different cell types, and for regulating hair development.
Keywords: Ets2, trophoblast, proteinase, FGF, TGFα, transcription
The Ets family of transcription factors is composed of over 30 members, characterized by the presence of a variant winged helix–turn–helix motif used for DNA binding (Karim et al. 1990; Donaldson et al. 1994; Liang et al. 1994; Kodandapani et al. 1996; Ghysdael and Boureux 1997). A subset of Ets proteins contain a region of 65–100 residues called the pointed or B domain, which has been implicated in signal transduction. In Drosophila, two Ets factors, the PNT-P2 product of the pointed gene and YAN/Pokkuri, mediate signaling by the sevenless/Ras/RAF/MAP kinase pathway, essential for the R7 photoreceptor cell development (Brunner et al. 1994; O’Neill et al. 1994). Similarly, in Caenorhabditis elegans, the Ets transcription factor LIN-1 acts downstream of an EGF/EGF-R/MAP kinase signaling pathway to negatively regulate vulval cell fate (Beitel et al. 1995). The vertebrate Ets1 and Ets2 are most similar to PNT-P2 in both the Ets and pointed domains. Activation of signaling by a mutant neu (Erb-B2), a member of the EGF receptor family, or by v-src, ras, or raf, stimulates the transcriptional activity of both Ets1 and Ets2. This stimulation is dependent upon the phosphorylation of specific threonine residues within the pointed domains (Rabault et al. 1996; Yang et al. 1996). Ets1 and Ets2 cooperate with other transcription factors including AP-1, MafB, SP-1, Pax5, Pit1, NFkB, E2F-1, and GATA-1 to stimulate transcription of a wide variety of genes (Ghysdael and Boureux 1997). Ets1 is less widely expressed than Ets2 in adult tissues and is important for T-cell survival and function (Muthusamy et al. 1995). Ets2 is expressed in many cell types in developing mouse embryos, including limb buds and the embryonic skin, in a pattern overlapping but also largely distinct from Ets1 (Maroulakou et al. 1994).
The number of genes known to utilize Ets binding sites is large and include those coding for transcription factors (Ets1, JunB, TBP, NFκB, c-Fos, GATA-1, Myc), matrix-degrading proteinases (stromelysin-1, collagenase-1, urokinase-type plasminogen activator, gelatinase B), keratins Endo A (mouse K8) and Endo B (mouse K18), cell cycle regulators (p53, cyclin D1), extracellular matrix receptors and ligands (integrins αV, α2, β2, and osteopontin), and growth factors (HB-EGF). However, the similar DNA-binding specificity of multiple members of the Ets family, and the extensive cooperation with other transcription factors has made it difficult to identify specific target genes with certainty. To evaluate the biological importance of Ets2 on gene expression and mammalian development, we have mutated the gene by gene targeting, and have generated cells and mice with no wild-type Ets2 product. Ets2 is a critical regulator of extraembryonic tissue function and hair development.
Results
Targeted mutagenesis of ets2
The mouse ets2 gene was targeted by homologous recombination in D3 ES cells with a vector that replaced all or part of three exons of the gene coding for the Ets DNA binding domain with the pMC1NeoA selectable gene (Henkel et al. 1996). The mutant ets2 gene was expected to either be null or code for a protein that lacks DNA binding activity and the signals necessary for nuclear localization (Boulukos et al. 1989). Six targeted ES cell clones were identified from a total of 109 screened colonies by Southern blot analysis, with probes flanking the sequence included in the targeting vector, and a third probe within the neo gene. (Supplemental data not shown are available at http://www.burnham-inst.org/papers/ets2.) The results confirmed the expected arrangement of the targeted gene, which was designated ets2db1 for DNA-binding domain mutant 1. An ES cell line carrying two targeted alleles was generated by cultivation of a targeted ES cell clone in increasing concentrations of G418 (Henkel et al. 1996). The homozygous clone was used for the generation of teratocarcinoma tumors and the isolation of cell lines.
ets2db1 lacks the DNA-binding domain and is located in the cytoplasm
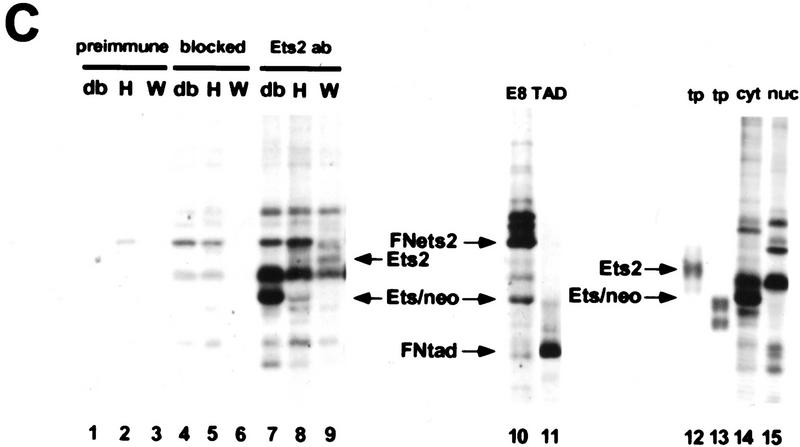
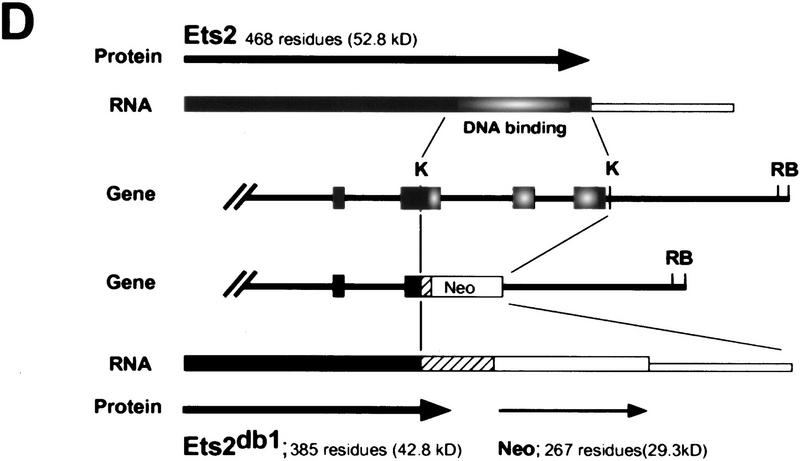
RNase protection analysis confirmed the absence of wild-type RNA in ES cells and fibroblastic cells containing two targeted alleles (Fig. 1A). However, ets2db1/db1 cells expressed normal levels of a slightly larger RNA, which hybridized with a probe for the 5′ end of the ets2 mRNA (Fig. 1B, lane 3). This fusion transcript appears to arise from transcription through the pMC1neo promoter into the large 3′-untranslated portion of ets2 RNA (Watson et al. 1990) because it contained neo coding sequences (Fig. 1B, lanes 5,6). The fusion mRNA from the targeted ets2 allele is predicted to have an open reading frame that extends 77 codons downstream of the fusion site within exon 8 (codon 308) (Fig. 1D). This product could be active as an inhibitor. However, both the preferential cytoplasmic localization and the deletion of the Ets domain would hinder the transcriptional activation potential of the ets2 gene product. The targeting of the ets2 gene results in a deletion of a critical portion of the gene but does not result in a null allele.
Figure 1.
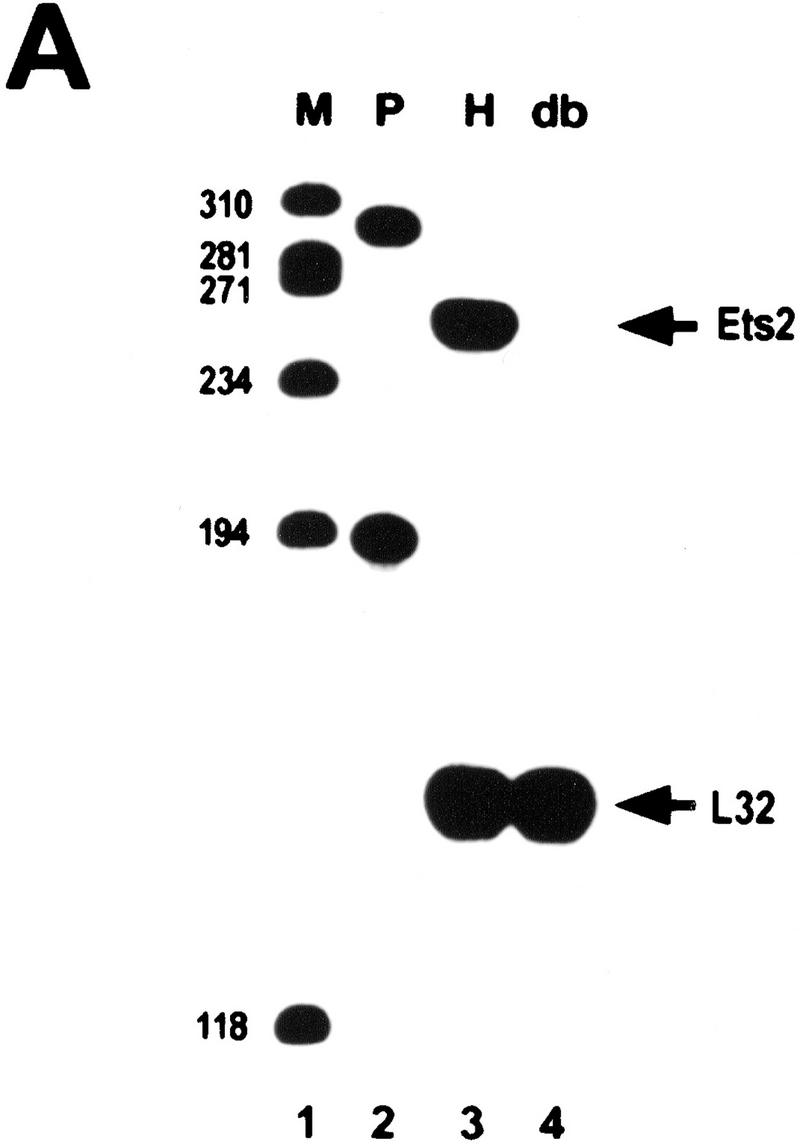
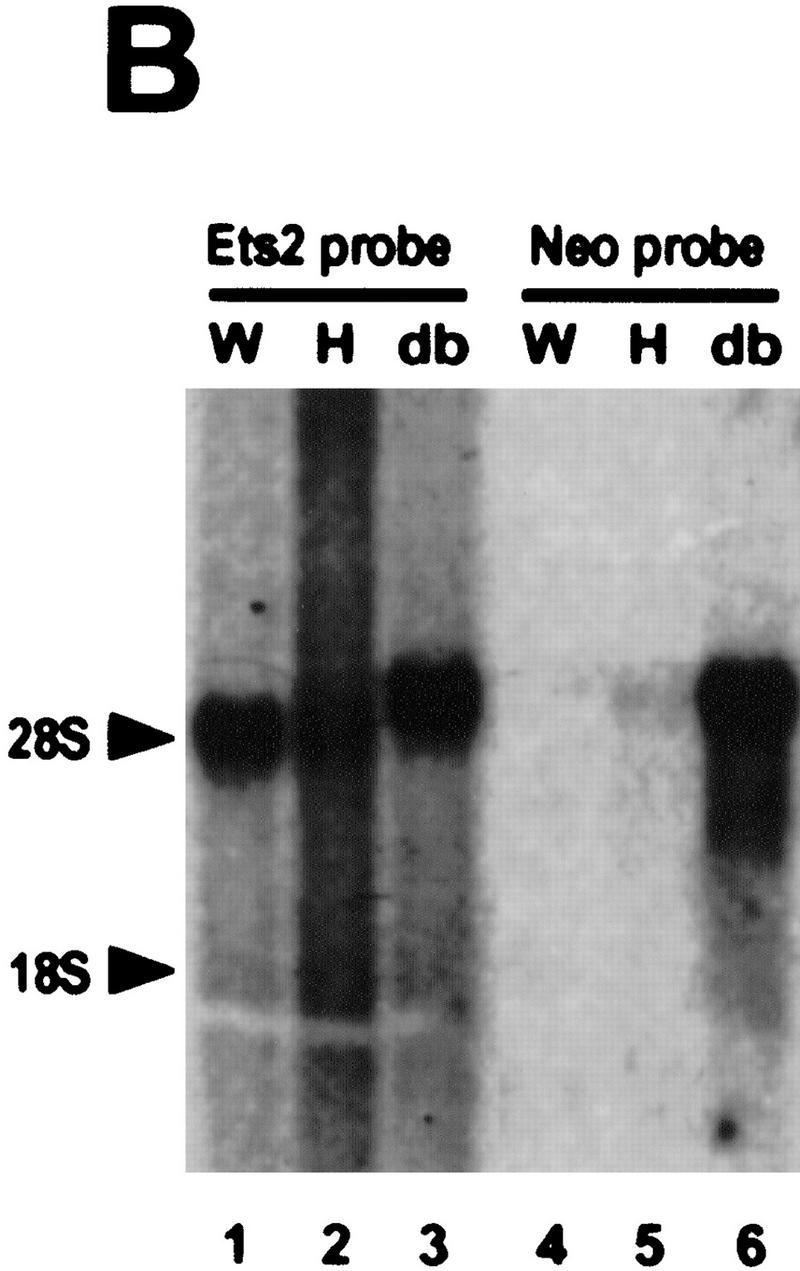
RNA and protein expression from the ets2db1 targeted allele. (A) RNase protection analysis of RNA derived from ets2db1/+ and ets2db1/db1 fibroblast lines utilizing a DNA binding domain probe. (Lane 1) size markers (M); (lane 2) probes (P) for Ets2 (upper) and the L32 ribosomal protein RNA (lower); (lane 3) ets2db1/+ (H) RNA; (lane 4) ets2db1/db1 homozygous (db) RNA. (B) Northern blot analysis of 5 μg of poly(A)+ RNA from the same cell lines. A probe from the 5′ end of the ets2 cDNA detected ets2-related RNA in wild-type (W), heterozygous (H), and homozygous (db) cells. A neo probe detects RNA only in heterozygous ets2db1/+ (lane 5) and ets2db1/db1 RNA (lane 6). (C) Immunoprecipitation analysis of Ets2 and Ets2db1 proteins. Ets2 antiserum (McCarthy et al. 1997) detects the wild-type Ets2 protein (lane 9, Ets2), the product of the ets2db1 allele (lanes 7,10,14, Ets/neo), an epitope-tagged form of Ets2 (lane 10, FNets2) and an epitope-tagged form of the transactivation domain of Ets2 (lane 11, FNtad). No specific signal was detected with preimmune serum (lanes 1–3) or Ets2 antiserum incubated with excess recombinant Ets2 (lanes 4–6). Translation products of ets2 mRNA (lane 12) and the ets2/neo fusion transcript (lane 13). The supernant fraction (cyt) and solubilized pellet (nuc) fractions of ets2db1/db1 cells were immunoprecipitated with Ets2 antiserum (lanes 14,15). The ets2db1/db1, ets2db1/+, and ets2+/+ cell types are EKO1, EHT1, and 3T3 cells, respectively. E8 is a rescued clone of EKO1 that expressed the FNets2 protein (lane 10). (Lane 11) Control cells expressing FNtad. (D) Schematic representation of ets2 and ets2db1 genes, RNAs, and proteins. The structures of the 3′ end of the wild-type and targeted genes are shown in the middle with solid boxes representing exons. The regions coding for the DNA-binding domain are shaded. The position of the pMC1neoA gene is labeled Neo with promoter/enhancer region represented as crosshatched. The predicted RNAs for the two genes are shown at the top and bottom. The expected 3′ noncoding region of both mRNAs are represented by the smaller open rectangles at the right. The proteins coded for by the RNAs are indicted by the arrows with the expected size indicated.
Ets2 protein was expressed at very low levels in wild-type cells, as expected from the reported rapid turnover of the protein (Fig. 1C, lane 9) (Fujiwara et al. 1988). However, its identity was confirmed by its absence from ets2db1/db1 cells (lanes 7,10), its comigration with in vitro synthesized Ets2 (lane 12; data not shown) and its sensitivity to preabsorption with recombinant Ets2. In both ets2db1/db1 and ets2db1/+ cells, we identified a faster migrating protein that was specifically precipitated by the Ets2 antiserum (McCarthy et al. 1997) but not by blocked antibody (Fig. 1C, lanes 7,8, ets/neo). This protein comigrated with a cell-free translation product of a synthetic RNA representing the expected fusion of the 3′ truncated ets2 cDNA and the pMC1neo gene (lanes 12,13). Fractionation of the ets2db1/db1 cells into nuclear and cytoplasmic fractions confirmed that the ets2db1 protein was found only in the cytoplasmic compartment (Fig. 1C, lanes 14,15).
Ets2 is required for embryonic development
Chimeric animals were generated by the injection of the ets2db1/+ ES cells into C57/Bl6 blastocysts. Transmission of the targeted allele to subsequent progeny of both Swiss Bl and 129/Sv strain of mice was confirmed by both Southern blot analysis and PCR. Heterozygous animals in both genetic background appear healthy, fertile, and normal. However, no ets2db1/db1 homozygous animals were detected from 449 progeny of ets2db1/+ heterozygote parents (Table 1). Thus, homozygosity for the ets2db1 allele results in embryonic lethality. Heterozygote adults represented 59% of the total. When heterozygote crosses were analyzed after varying times of development to determine the time of embryonic death, no ets2db1/db1 embryos were recovered at either E11.5 or E8.5 (Table 1). Instead, residual, empty, and smaller decidual swellings were seen. The same result was found in the 129/Sv background and after five generations in the Swiss/Bl background.
Table 1.
Embryonic lethality of ets2db1 mice
|
|
Total no.
|
ets2+/+
|
ets2db1/+
|
ets2db1/db1
|
Empty decidua
|
|---|---|---|---|---|---|
| Adult | 449 | 184 | 265 | 0 | — |
| 11.5 day | 27 | 10 | 17 | 0 | 7 (26%) |
| 8.5 day | 41 | 19 | 22 | 0 | 8 (20%) |
| 7.5 day | 57 | 15 | 28 | 14 | 0 (0%) |
| Blastocyst | 133 | 22 | 82 | 29 | — |
Expression of ets2 in early embryos
In situ hybridization of ets2 RNA revealed very high levels of the RNA in the trophoblastic tissues of E6.0–E7.5 embryos and lower levels of expression in the extraembryonic ectoderm and trophectoderm, but not in the overlying extraembryonic endoderm or primary embryonic ectoderm (Fig. 2A–C). Brachyury (T) expression at E7.5 was used as a control to show that the ets2 expression seen in the trophectoderm and extraembryonic ectoderm was not due to probe trapping (Fig. 2D).
Figure 2.
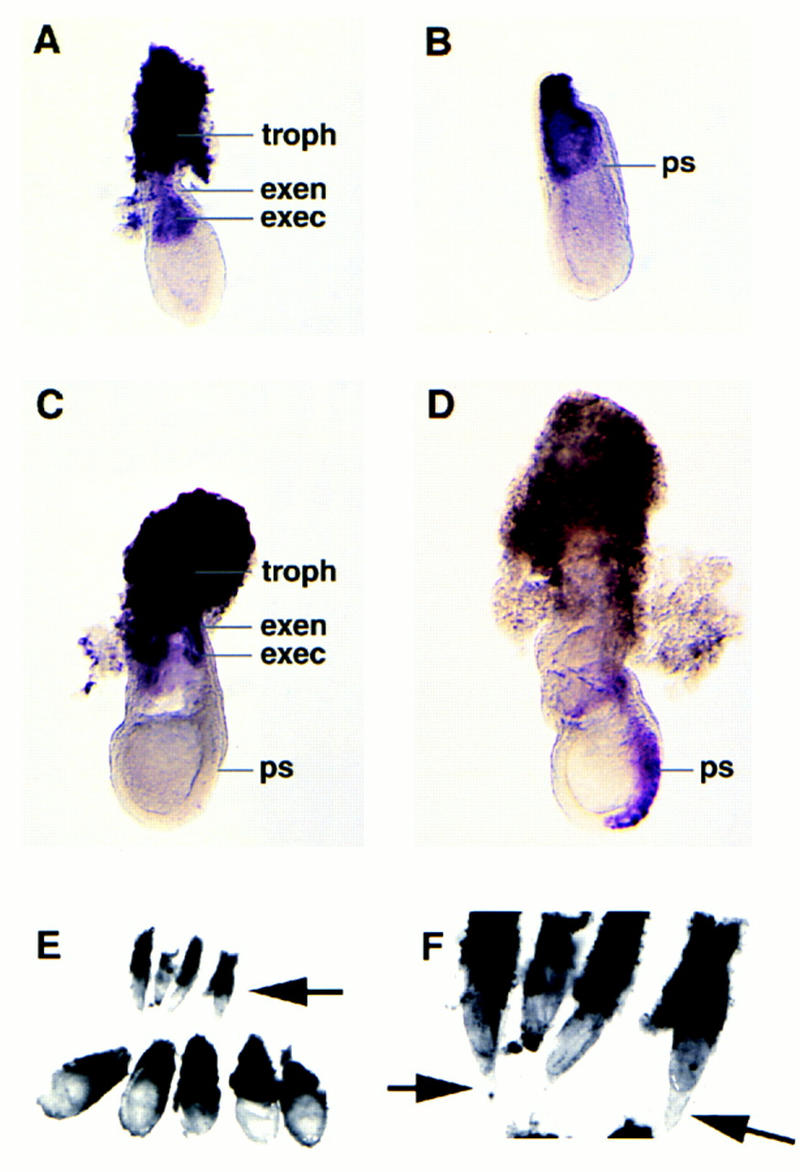
In situ localization of ets2 RNA and retarded growth of ets2db1/db1 embryos. Whole mount in situ analysis of ets2 expression in E6–E7.5 embryos. (A) At E6, prior to primitive streak (ps) formation, expression is seen in the extraembryonic ectoderm (exec) and trophectoderm (troph), but not in the overlying extraembryonic endoderm (exen). (B) Primitive streak stage at E6.5 and elongated at E7.5 (C). (D) Brachyury (T) expression at E7.5, used as a control. (E) Embryos resulting from a mating of ets2db1/+ parents were dissected at E7.5 days. The four small embryos were subsequently identified as ets2db1/db1. The trophoblastic tissue was removed from the normal sized embryos. (F) A higher magnification showing the unusual cone-shaped yolk sac of the arrested ets2db1/db1 embryos.
ets2db1/db1 conceptuses have extraembryonic defects
Blastocysts recovered from heterozygote matings at E3.5 days appeared normal. In culture, the blastocysts attached to either plastic- or extracellular-matrix-coated surfaces and generated trophoblast outgrowths that were not distinguishable between genotypes (data not shown). At E7.5, 14 of 57 decidual swellings contained much smaller embryos (Fig. 2E) with an unusual cone-shaped yolk sac/trophoblast envelope (Fig. 2F) and a much smaller amount of attached trophoblastic tissue. PCR analysis of the embryonic portion of these embryos confirmed they were all ets2db1/db1. Histological analysis of embryos at E6.5 and E7.5 days revealed multiple defects in ets2db1/db1 embryos (Fig. 3). At E6.5, mutant embryos were distinguished by a small ectoplacental cone region, apparently caused by a failure of trophoblast migration (Fig. 3B). A membrane appeared to cover the ectoplacental cone, which may prevent trophoblast migration. At E7.5 days ets2db1/db1 embryos were much smaller than normal embryos (Fig. 3E; note scale). Neither amnion nor chorion membranes formed, resulting in embryos with only one, rather than three cavities. The trophoblastic tissue in the EPC failed to proliferate, and the primary ectoderm began to die by apoptosis, as indicated by pynotic nuclei (Fig. 3F). Many cells in the primary ectoderm were positive by TUNEL staining of DNA breaks (data not shown). The absence of the chorion and amnion membrane divisions suggests that the embryo failed before or early in gastrulation, which generates both extraembryonic and embryonic mesoderm. By E8.5, no ets2db1/db1 embryos were identified; instead, bloody and largely empty implantation sites showed that the mutant embryos had been resorbed.
Figure 3.
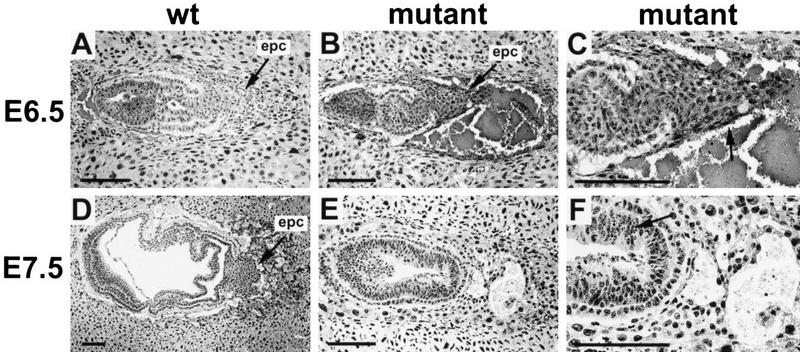
Abnormal implantation in Ets2 mutant embryos. (A,D), normal implantation in E6.5 and E7.5 wild-type embryos in which the embryonic trophoblast cells migrate out from the ectoplacental cone (epc) to form a hybrid vasculature in contact with maternal blood. (B) The trophoblast cells of Ets2 mutant embryos fail to migrate and differentiate, resulting in the characteristic bloody implantation site. (C) A higher magnification of the E6.5 mutant embryo shows a membrane (arrow) covering the ectoplacental cone. (E) At E7.5, the mutant embryo is small and beginning to die. (F) High magnification of the E7.5 mutant shows apoptotic nuclei in embryonic ectoderm (arrow). Scale bars, 125 μm. Hematoxylin and eosin-stained glycol methacrylate sections.
The growth retardation of E7.5 ets2db1/db1 embryos might be attributed to defects in trophoblastic cells in establishing appropriate interaction with the maternal circulation. MMP-9 (gelatinase B), a matrix metalloproteinase and marker of trophoblastic cells at this stage, was dramatically reduced in mutant embryos (Fig. 4A,B). The ets2db1/db1 embryos showed much decreased immunohistochemical staining of PECAM-1, an endothelial marker also expressed in trophoblast cells directly connected to maternal endothelium (Vecchi et al. 1994) (Fig. 4G,H). However, not all trophoblastic gene expression was inhibited. Expression of placental lactogen-1, a hormone secreted by trophoblast cells, was upregulated in mutant embryos (Fig. 4C,D). Laminin, a basement membrane component of Reichert’s membrane, was found in ets2db1/db1 embryos (Fig. 4E,F) and extended into and over the trophoblast. The unusual continuity of the presumptive Reichert’s membrane may reflect the low expression of metalloproteinases such as MMP-9 and likely contributes to the unusual trophoblast/yolk sac envelopes noted in dissected embryos (Fig. 2E,F). These results indicate that the ets2db1/db1 embryos had abnormalities of trophoblast gene expression, poor connection with the maternal circulation, and possible abnormalities in basement membrane metabolism that, together, could contribute to embryonic death.
Figure 4.
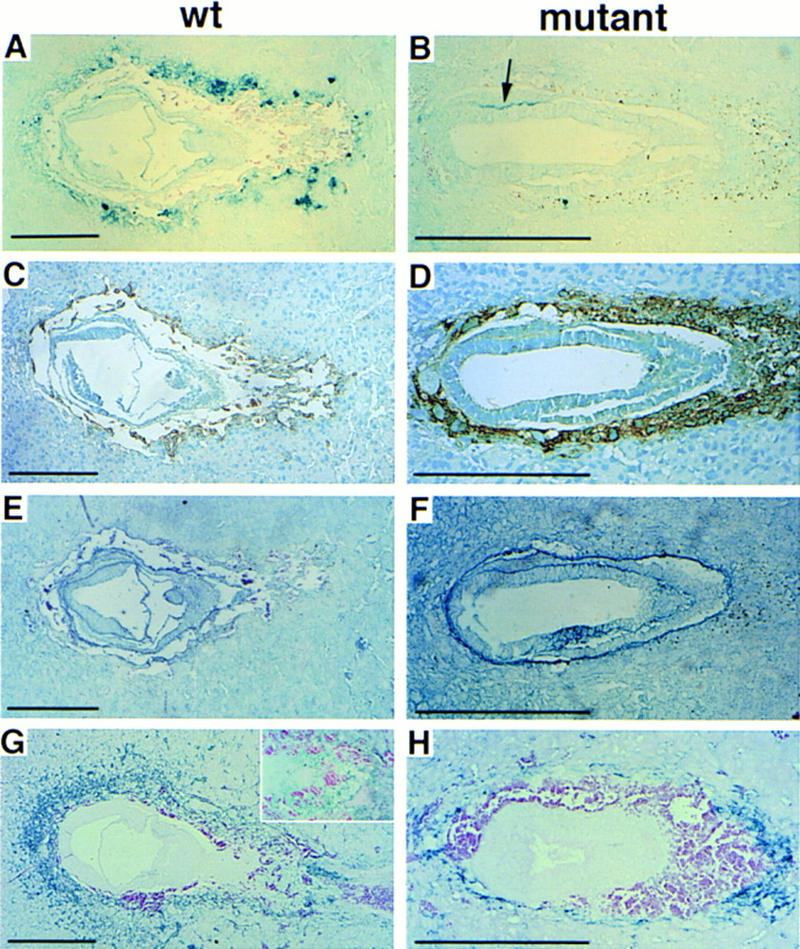
Abnormal protein expression in ets2db1/db1 embryos at E7.5. (A,B) Tissue-specific regulation of MMP-9 in a mutant embryo. Note the absence of immunoreactive MMP-9 in the surrounding trophoblast cells of the mutant (B), as compared to wild-type (A), but the area where primitive streak should form shows a positive stain (arrow) with anti-MMP-9. (C,D) Placental lactogen-1 staining is normal in wild-type (C) (brown color), but is substantially upregulated in the mutant embryo (D). (E,F) Anti-laminin staining for basement membranes (blue color) shows extended expression of laminin around the ectoplacental cone. (G,H) Anti-PECAM antibody to detect endothelial cells of blood vessels (blue color) shows that vascular connections to maternal circulation are severely compromised in the mutant embryo (H), compared to the wild type (G). The higher magnification inset in G shows the staining of ectoplacental cone cells. Primary antibodies were detected with HRP-labeled secondary antibodies, and then histochemical detection for peroxidase. A, B, E, F, G, and H are True Blue substrate with eosin counterstain. C and D are DAB (brown) substrate with methyl green counterstain.
ets2db1/db1 ES cells can contribute to aggregation chimeras with normal embryos
The histological examination of ets2db1/db1 embryos and the in vitro differentiation of ets2db1/db1 ES cells (data not shown) implicated defects in extraembryonic tissue. To evaluate whether the embryonic tissues were also affected, we performed two types of cellular rescue experiments. First, ets2db1/db1 ES cells were injected into C57/Bl6 mouse blastocysts, which were then transferred to foster mothers and sacrificed at E18.5. Morphologically normal chimeric embryos formed. The ES cells contributed to the brain, lung, heart, liver, spleen, gut, and kidney, as assessed by the presence of the B isoform of glucose-6-phosphate isomerase from the strain 129 ES, in addition to the C57/Bl6 A isoform. The contribution of the B isoform provided by the ets2db1/db1 ES cells ranged from 5% to 20% of the total GPI enzyme activity (data not shown). Contribution to heart was higher with values from 15% to 45% of the total activity. Thus the ets2db1/db1 ES cells were capable of contributing to normal development of major organ systems.
The extraembryonic defects of ets2db1/db1 embryos are complemented by tetraploid embryos
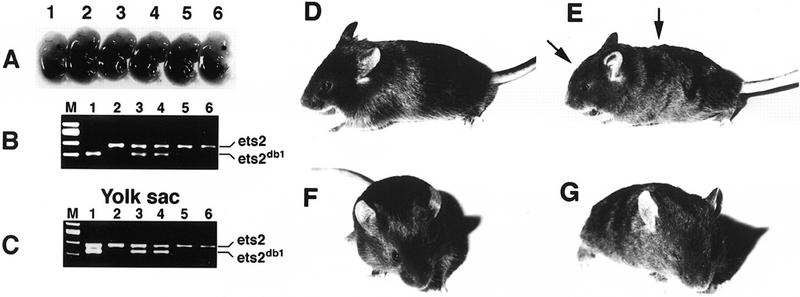
The second type of cellular rescue experiment utilized embryos from ets2db1/+ heterozygotes matings aggregated with tetraploid embryos. Tetraploid embryos are capable of forming normal extraembryonic tissues but do not contribute to the embryonic portion of embryos (Nagy et al. 1990). At E14.5, 2 ets2db1/db1 homozygous embryos were detected by PCR among 31 embryos resulting from the aggregation of tetraploid wild-type embryos and embryos from ets2db1/+ parents. One example is shown in Fig. 5A. Although small, it was still within the variation found for normal embryos (compare embryos 1 and 6, Fig. 5A). The yolk sac of this ets2db1/db1 embryo had both the wild-type and targeted Ets2 alleles because of a mixture of wild-type and ets2db1/db1 cells (Fig. 5C). Both ets2db1/db1 embryos appeared normal upon histological examination (data not shown). The rescue of these embryos indicates that embryo development can proceed in the absence of normal Ets2 product when functional trophoblast and extraembryonic tissue function is provided.
Figure 5.
Rescue of ets2db1/db1 embryos by aggregation with tetraploid embryos. (A) E14.5-day embryos resulting from the aggregation with wild-type tetraploid embryos. (B) PCR analysis of the embryos reveals that embryo 1 is homozygous mutant, whereas embryos 3 and 4 are heterozygous and 2, 5, and 6 are wild-type. (C) PCR analysis of the yolk sac DNA. The equal intensity of the amplified bands of sample 1 indicates approximately equal contribution of wild-type and targeted cells to the yolk sac. (D,F) Three-week-old wild-type mice resulting from a tetraploid embryo aggregation experiment. (E,G) Rescued ets2db1/db1 mouse. Note the wavy hair coat and rounded forehead (arrows).
Adult ets2db1/db1 mice display a hair phenotype resembling that of animals with defective signaling through the EGF receptor.
We next allowed tetraploid embryos aggregated with embryos from ets2dbl/+ heterozygote matings to proceed to term. Of 40 pups, 4 recovered by cesarean section at term were identified as ets2db1/db1 homozygotes. At birth, ets2db1/db1 mice were distinguished by curly whiskers. After ∼2 weeks, the ets2db1/db1 homozygotes were distinguished by curly whiskers, wavy hair, and slightly rounded forehead (Figs. 5 and 6). In adults, all four types of hair (guard hair, awl, auchene, and zigzag) were abnormally curved (Fig. 6F; data not shown). Whole mount analysis of the skin of the ets2db1/db1 animal revealed misaligned hair follicles, resulting in ingrown, curly hairs that failed to penetrate the epidermal surface (Fig. 6H). Sections of the skin confirmed curved and misaligned hair follicles (Fig. 6J). However, the dorsal epidermis of adults was of normal morphology, and the internal morphology of the hair follicles was indistinguishable from normal heterozygote or wild-type littermate controls. These abnormalities of whiskers, hair, and hair follicles closely resemble the phenotype of TGFα-deficient animals (Luetteke et al. 1993; Mann et al. 1993) and of mice with a point mutation of the EGF receptor (EGFR) (Luettke et al. 1994). Two ets2db1/db1 mice developed eye irritations (Fig. 6D) that resolved spontaneously. Eye abnormalities are also characteristic of some, but not all TGFα-null mice (Luetteke et al. 1993; Mann et al. 1993). However, unlike some of the TGFα knockout mice, which were born with open eyes, all ets2db1/db1 newborns had normal closed eyes. The subtle differences in head morphology characteristic of rescued ets2db1/db1 mice were not reported for TGFα-null mice. Whereas the heads of these rescued mice were distinguishable from their littermates, no differences in skull skeletal structure was detected (data not shown). Thus, the differences in appearance of the head are likely caused by muscle or skin differences.
Figure 6.
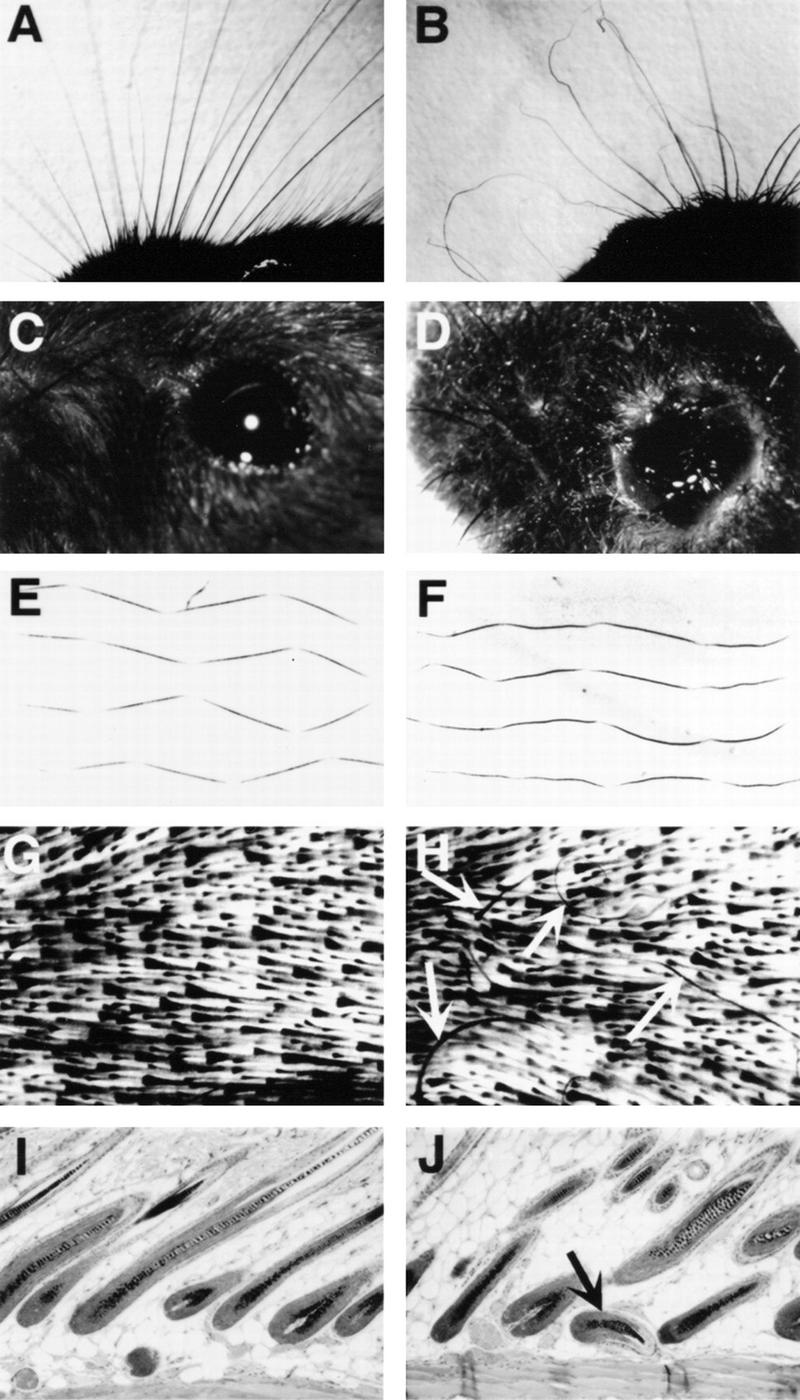
Phenotype in whiskers, hair, and eye alteration of the ets2db1/db1 adult mouse. (A,C,E,G,I) Wild-type mouse whiskers, eye, zigzag hair, whole mount skin, and hair follicle histology respectively; (B,D,F,H,J) from ets2db1/db1 mouse. (D) Eye secretions of the ets2db1/db1 mouse were transient and resolved spontaneously without treatment within a few weeks. (H) Arrows point to ingrown hairs lying wholly within the skin. View is from the lower part of the cleared skin in G and H. (J) Arrow points to abnormally oriented hair follicle. Note the unusual juxtaposition directly against the lower muscle layer of the skin.
One male and one female rescued-ets2db1/db1 mouse generated progeny upon subsequent mating. Thus both sexes are fertile. Other major organs of two adult ets2db1/db1 mice did not reveal abnormalities upon macroscopic and microscopic examination (data not shown). To assess lymphoid and myeloid cell development in the ets2db1/db1 mouse, spleen cells of adult mice were treated with fluorescent conjugates of 16 different monoclonal antibodies (see Materials and Methods) and analyzed by flow cytometry. In all cases, T cell, B cell, and myeloid surface marker profile of the ets2db1/db1 spleen cells was similar to that of normal mice.
The mitogenic responses of T cells, B cells, and macrophages from the ets2db1/db1 mouse were assessed as well. Spleen cells were treated with ConA, anti-CD3, and LPS. No difference in the rates of proliferation were seen between the ets2db1/db1 and normal T and B cells following any of the treatments. Although Ets2 has been implicated as a mediator of mitogenic signals from the M-CSF receptor, we were able to culture and expand bone marrow macrophages from ets2db1/db1 mouse in M-CSF-containing medium. To measure their ability to be activated, these macrophages and macrophages cultured from normal mice were treated with either interferon γ (IFN-γ), LPS or IFN-γ + LPS, and assayed by flow cytometry for the increased expression of IFN-γ receptor, MHC class II antigen I-Ab, and scavenger receptor. In response to these treatments, all assayed antigens in the ets2db1/db1 macrophages were expressed at equivalent levels to that seen in macrophages from normal mice. We conclude that no major defect in either lymphocytes or macrophages occurs in mice that lack wild-type Ets2. With regard to macrophages, these results confirm our earlier analysis of macrophages generated in vitro from the differentiation of doubly targeted ES cells (Henkel et al. 1996).
Both TGFα and EGFR are expressed in ets2db1/db1 mice
Because rescued ets2db1/db1 animals resembled animals with defective EGFR or its ligands, we examined the expression of TGFα and EGFR. TGFα protein, assessed by radioimmuno assay, was not distinguishable from normal adult kidney (wild type, 7 pg/mg total protein; ets2db1/db1, 6.8 pg/mg total protein ) and was 50% or greater of the wild-type level in the skin of the adult ets2db1/db1 mouse (wild type, 3.5 pg/mg total protein; ets2db1/db1, 2 pg/mg total protein). TGFα and the EGFr mRNAs in skin and liver of the ets2db1/db1 mouse were at normal levels (Fig. 7A).
Figure 7.
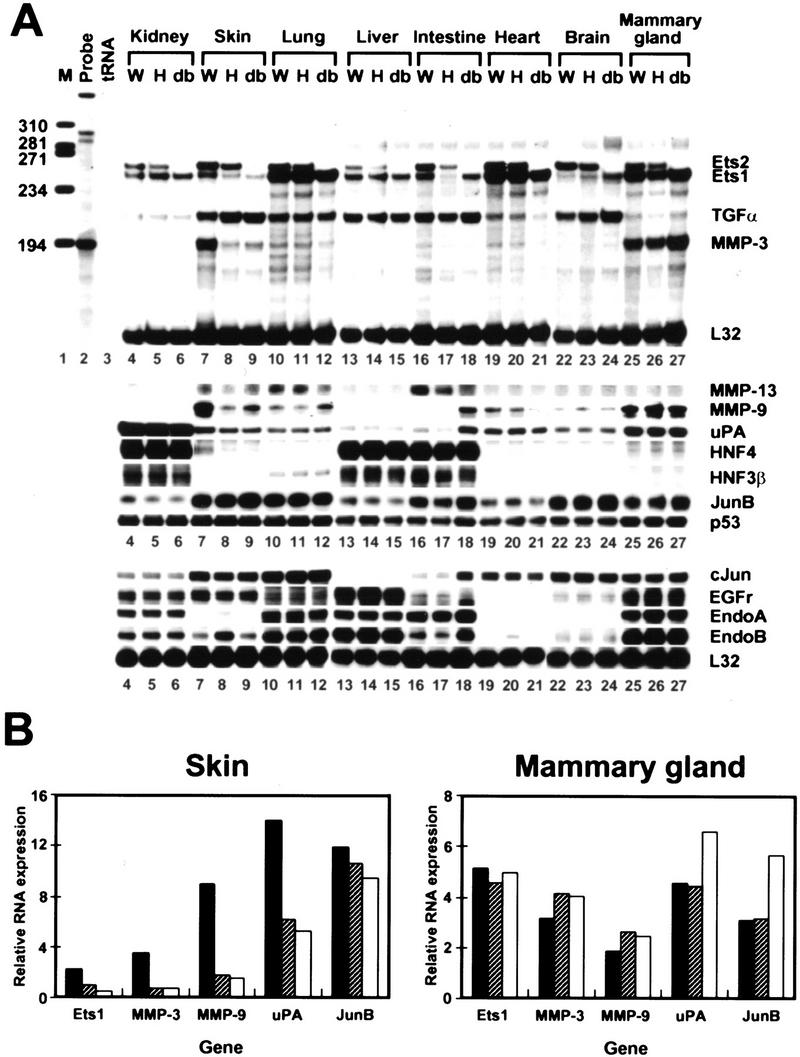
RNase protection analysis of adult organ RNAs. (A) All RNAs were from male 90-day-old tetraploid embryo aggregation littermates except for brain and mammary gland samples of heterozygous and homozygous 50-day littermate females of a second tetraploid embryo aggregation experiment. The wild-type female mammary gland and brain samples were from a 90-day-old adult. Twenty or 10 μg (c-Jun, EGFr, Endo A, and Endo B) of total RNA from the indicated organs was analyzed by RNase protection with probes indicated at right. Protected fragments were resolved by acrylamide gel electrophoresis and detected by autoradiography in the presence of enhancer screens and quantified using a PhosphorImager. Images are from autoradiographic exposures of five different gels, each of which also contained L32 signals (additional data shown). Signals for uPA (lanes 4–9), HNF4, HNF3β (lanes 4–6) and EGFr, endo A, and endo B (lanes 16–18) are 1-day exposures for improved clarity. (B) Quantitation of ets1, MMP-3, MMP-9, uPA, and junB mRNA after normalization to L32 RNA in skin and mammary gland samples.
Altered expression of MMP-3, 9, and 13 in adult ets2db1/db1 tissues
Ets factors regulate many genes. RNA transcripts for the candidate target genes p53, endo A, endo B, junB, HNF-4, and ets1 were not affected by the absence of Ets2 in kidney, lung, liver, heart, and brain of the rescued adult (Fig. 7A). However, MMP-3, MMP-9, uPA, and ets1 were all lower in skin samples for homozygous and heterozygous animals (Fig. 7A), when normalized to L32 RNA levels (Fig. 7B). In contrast, ets1, MMP-3, MMP-9, and uPA RNAs were at similar levels in the mammary glands of wild-type, ets2db1/+, and ets2db1/db1 mice (Fig. 7B). MMP-13 was also lower in ets2db1/db1 lung and intestine (Fig. 7A). Tissue specificity in MMP-9 expression was noted in the E7.5 ets2db1/db1 embryos, which down-regulated this enzyme in trophoblast, yet maintained expression in the primitive streak (Fig. 4B). These data indicate that there is tissue specificity in the effects of wild-type Ets2 on the expression of MMP-3, MMP-9, and MMP-13.
ets2db1/db1 cells are defective in the induction of MMP-13 and MMP-3 RNAs by FGF2
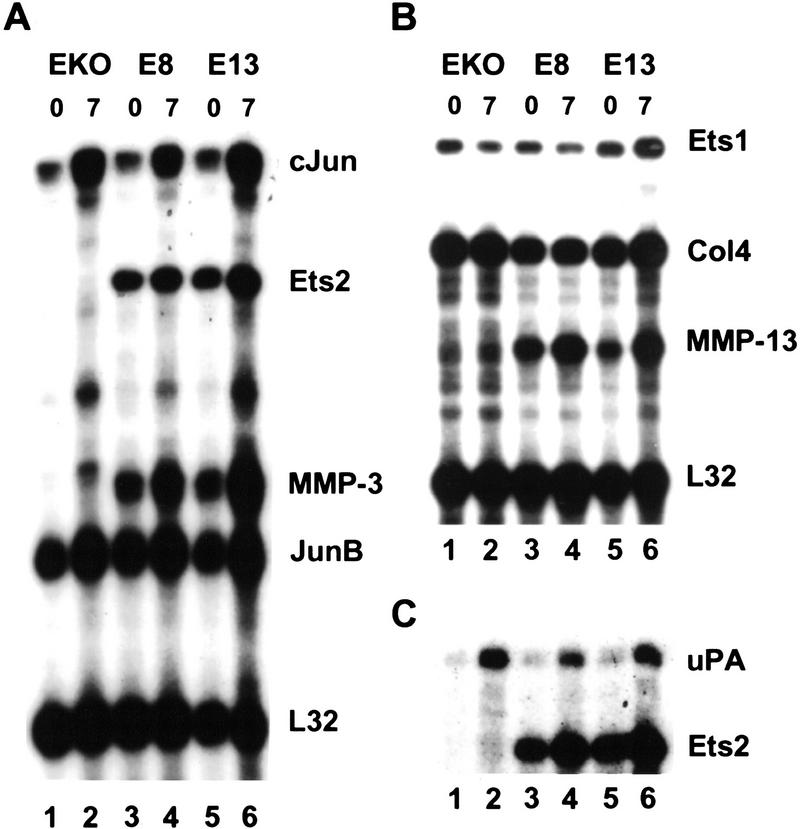
To explore the role of Ets2 in mediating growth factor signaling, we established a fibroblastic cell line from primary cultures of teratocarcinoma tumors formed by the subcutaneous injection of the ets2db1/db1 ES cells. One cell line, which lacked wild-type Ets2 expression, was subsequently transfected with an expression vector for Ets2 or empty expression vector and multiple clones were isolated. Several rescued clones that expressed moderate levels of Ets2 were compared with the parental cell line for gene expression after growth factor exposure. The parental EKO1 line and the E8 and E13 Ets2 rescued clones responded equally to mitogenic stimulation by FGF1, FGF2, EGF, and TGFα, as analyzed by [3H]thymidine incorporation at 24 hr, and grew at comparable rates in media containing fetal bovine serum. However, serum-starved EKO1 cells (Fig. 8) expressed little MMP-3 or MMP-13 RNA, and exposure to FGF2 failed to induce the RNA. Treatment of Ets2 expressing subclones with growth factors increased MMP-13 and MMP-3 RNAs within 7 hr, whereas RNAs for the L32 ribosomal protein, Ets1, type IV and type I collagens (not shown) were relatively insensitive to serum starvation and growth factor treatment. RNAs for cJun and uPA were increased by exposure to FGF2, independent of Ets2 expression. In contrast, TGFα did not induce either MMP-13 or MMP-3 RNAs in any of the three cell lines. These results show that the ets2db1/db1 EKO1 cell line is deficient in the induction of both MMP-13 and MMP-3 in response to FGF2. However, MMP-3 deficiency is probably not the key target gene responsible for either the trophoblastic defects or the hair growth abnormalities, because a MMP-3 knockout mouse is viable and does not have curly hair. However preliminary analyses of MMP-9-targeted embryos suggests that some of the trophoblastic defects of ets2db1/db1 embryos may be a direct consequence of MMP-9 deficiency (Z. Werb, unpubl.).
Figure 8.
Role of Ets2 in FGF2 induction of MMP-13 and MMP-3 in cultured cells. The EKO1 Ets2-deficient fibroblastic cell line and two independent isolates that expressed transfected Ets2 (E8 and E13) were incubated in medium containing 0.5% fetal bovine serum for 2 days. FGF2 (50 ng/ml) was added for 7 hr and RNA was isolated. The indicated RNAs were detected by RNase protection analysis. Collagen IV is indicated by Col4. Shown are autoradiographic exposures of the acrylamide gels used to separate the protected fragments. The time of growth factor exposure is indicated in hours at the top of each lane. Note the increased expression of the MMP-3 (A, lanes 3,5) ) and MMP-13 (B, lanes 3,5) RNAs in the E8 and E13 cell lines and their further increase after 7 hr.
Discussion
Ets2 is essential for ectoplacental cone proliferation and the establishment of vascular interactions with the maternal circulation. The targeting of Ets2 leads to deficient maternal nutrition, impaired growth, and the eventual death of the embryonic ectoderm by apoptosis. The failure of gastrulation contributes to the absence of amnion and chorion membranes but appears to occur after the beginning of the primitive streak (Fig. 4B). Targeted mutagenesis of a large number of genes result in placentation failures (Copp 1995). Among these, tetraploid embryo aggregation has successfully rescued homozygous mutations of Mash-2 (Guillemot et al. 1994), a trophoblast-specific transcription factor, ERR-β, an orphan hormone receptor (Luo et al. 1997) and the AP-1 transcription factor JunB (Erwin Wagner, pers. comm.). The abnormalities of embryonic ectoderm apoptosis and gastrulation of ets2db1/db1 embryos resemble the effects of targeting the HNF4 transcription factor, which are also similarly complemented by aggregation with tetraploid embryos (Chen et al. 1994; Duncan et al. 1997). However, we did not detect alterations in HNF4 expression in either differentiating ets2db1/db1 ES cells or adult rescued mice. The extraembryonic defects seen in the ets2db1/db1 embryos appear different from other placental abnormalities resulting from gene targeting. The failure of ectoplacental cone proliferation, the increased placental lactogen expression, and the complete resorption of the ets2db1/db1 embryos by E8.5 are distinctive.
Both TGFα and FGF5 have been implicated in the regulation of hair growth (Hebert et al. 1994). The abnormalities in hair development, including curly whiskers and hair, misaligned hair follicles and ingrown hairs of the rescued ets2db1/db1 mice, resemble the phenotype of TGFα-null animals (Luetteke et al. 1993; Mann et al. 1993) or the spontaneous waved-1 mutation of the TGFα gene (Berkowitz et al. 1996). However, the hair abnormalities of the ets2db1/db1 mice were not caused by TGFα or EGFR deficiency. Moreover, ets2db1/db1 fibroblast cell lines responded mitogenically to EGF and TGFα.
Ets2 may mediate either TGFα or FGF signaling during embryonic hair follicle development (Hardy 1992) by the regulation of a subset of growth-factor-regulated genes. Clearly, Ets2 does not mediate all such signaling by either type of growth factor because the phenotypes of mice with targeted mutations of different FGFs, EGF, TGFα, and their receptors are distinct from those found in rescued ets2db1/db1 mice (Miettinen et al. 1995; Sibilia and Wagner 1995; Threadgill et al. 1995; Yamaguchi and Rossant 1995). However, Ets family members have been implicated in mediating FGF and EGF signaling through ras signal transduction pathways in Drosophila, C. elegans, and mammals (Ghysdael and Boureux 1997).
In the light of the widespread expression of Ets2 (Maroulakou et al. 1994), the mild effect of targeting Ets2 on the development of rescued mouse embryos was surprising. In contrast to the key roles of the PU.1 and Ets1 members of the Ets family in the immune system (Boulukos et al. 1990; Scott et al. 1994; Muthusamy et al. 1995; McKercher et al. 1996), Ets2 does not appear essential for lymphoid cells. In part, this result confirms the earlier study of Ets2-deficient macrophages generated in vitro from doubly targeted ES cells (Henkel et al. 1996).
Ets family transcription factors have been strongly implicated in the regulation of human MMP-1 (mouse MMP-13) (Gutman and Wasylyk 1990; Logan et al. 1996), rat MMP-3 (Wasylyk et al. 1991), and human MMP-9 (Gum et al. 1996). Thus, the misregulation of these MMPs may be a direct consequence of the absence of wild-type Ets2. Some of the placental and extraembryonic endodermal defects of ets2db1/db1 embryos may reflect a particular importance of Ets2 for proteases that act on extracellular matrix. MMP-9 was poorly expressed in the trophoblast cells of implanting ets2db1/db1 embryos, and the unusual persistence of the Reichert’s membrane may reflect the failure of appropriate remodeling of this extracellular matrix. Similarly, ets2db1/db1 fibroblast cells fail to express both MMP-13 and MMP-3 in response to growth factor exposure unless exogenous Ets2 is supplied. Two tandem, inverted Ets binding sites of the MMP-3 promoter are responsible for Ets factor responsiveness, and a model of this site has been shown to be particularly sensitive for Ets2 transactivation (Galang et al. 1994). However, the dependence of MMP-3 and MMP-9 on Ets2 is cell-type dependent. The levels of MMP-3 RNA were normal in adult ets2db1/db1 mammary gland but markedly reduced in skin. Similarly, MMP-9 was normal in lung and mammary gland but absent in ets2db1/db1 trophoblast of mice. Multiple factors, such as AP-1, are important for optimal expression of each of these genes in particular cell types (McDonnell and Matrisian 1990; Bortner et al. 1993; Wasylyk et al. 1993; Ghysdael and Boureux 1997). Our data indicate that the same gene utilizes different specific transcription factors in different tissues.
Matrix metalloproteinases are important for the remodeling of extracellular matrix, and are involved in normal organ development and the invasive behavior of both normal and tumor cells (McDonnell and Matrisian 1990; Sympson et al. 1994, 1995). A large number of genes have been identified as potential targets of Ets factor regulation (Ghysdael and Boureux 1997). It is now clear that Ets2 is limiting for expression of only a subset of these genes and in particular tissues. The sensitivity and specificity of MMP-13, MMP-3, and MMP-9 gene expression to the levels of Ets2 in certain tissues suggests that targeting specific transcription factors could have very selective effects on the expression of these proteinases in normal tissues and during disease processes. Viable transcription factor knockout mice, coupled with large-scale expression studies, may eventually provide the basis for developing predictive models of tissue-specific, target gene expression.
Materials and methods
Gene targeting of ets2
The construction of the ets2 targeting vector, the isolation of targeted D3 ES cells, and the generation of ES cells with two targeted alleles have been described previously (Henkel et al. 1996). Four independently targeted clones were injected into strain C57/Bl6 mouse blastocysts by standard methods (Stewart and Mintz 1981; Hogan et al. 1986). A single founder chimera transmitted the mutant allele to subsequent generations of Swiss Black and 129/Sv strains of mice.
Cell line isolation
Differentiated ets2db1/db1 cell lines were isolated from teratocarcinomas formed by the injection of ets2+/+, ets2db1/+, and ets2db1/db1 ES cells into strain 129 Sv mice (Oshima et al. 1981). The EKO1 ets2db1/db1 fibroblastic cell line was established by repeated selection for rapid attachment to plastic culture dishes and low-density growth in the absence of LIF. The fibroblastic cell derivative was cloned twice after establishment. This line was subsequently transfected with an expression vector for Ets2 which also contains a FLAG epitope tag and a synthetic nuclear localization signal fused to the amino′ terminus (FNets2) (Galang et al. 1996). FNets2 was expressed from a derivative of the pCIN4 vector (Rees et al. 1996) in which the neo-selectable gene was replaced with the hygromycin gene to generate pCIH–FNets2. Thirteen independent clones with varied Ets2 expression were isolated. The E8 and E13 clones were representative of those with moderate levels of ets2 RNA.
Growth factor treatment
Growth factor induction of DNA synthesis was measured as described (Mohammad et al. 1996) except for the use of medium containing 0.5% fetal bovine serum for starvation. FGF1, FGF2, EGF, or TGFα concentrations ranged from 0.03 to 100 ng/ml. For RNA analysis, 6- or 9-cm dishes of the indicated cell types were subjected to serum starvation (0.5% fetal bovine serum) for 48 hr and then exposed to FGF (50 ng/ml), EGF (10 ng/ml), or TGFα (10 ng/ml) for 7 or 24 hr. RNA was prepared from the cells with the use of acidic phenol (RNAzol) (Chomcznski and Sacchi 1987).
Protein analysis
Cells were labeled by incubation with 500 μCi/ml [35S]methionine in methionine-free medium containing 10% dialyzed fetal bovine serum for 1 hr. Cell lysates were prepared as previously described (Oshima 1981) or in RIPA buffer with similar results. Lysates were precleared with nonimmune rabbit serum and protein A–Sepharose (Pharmacia) and then incubated with 3 μl of rabbit anti-mouse Ets2 antiserum. The antiserum was prepared by immunization of a rabbit with a purified Tryp-E Ets2 fusion protein expressed in bacteria (McCarthy et al. 1997). Reactive proteins were recovered and analyzed as described previously (Oshima 1981) except Protein A–Sepharose instead of formalin-fixed Staphlyococus aureus was used and washes were performed in lysis buffer. For cell fractionation experiments, labeled cells were recovered by scraping in 10 mm Tris-HCl (pH 7.4), 3 mm, MgCl2, 0.1 mm, CaCl2, 50 mm KCl, 0.1% NP-40 detergent, 1% aprotinin solution, 1 mm pepstatin, 1 mm leupeptin. After homogenization or vigorous vortex mixing, the nuclear/cytoskeletal and supernatant (cytoplasmic) fractions were recovered by centrifugation at 5000g for 5 min. The DNA of the nuclear fraction was digested briefly with DNase I, then both fractions were adjusted to 5 mm EDTA, 0.1% SDS, heated to 100°C for 3 min, cooled, and adjusted to 0.5% NP-40. Lysates were stored frozen at −80°C and cleared by centrifugation at 12,000g for 20 min before use.
PCR analysis
Genomic DNA was prepared from ES cells, embryos, and tails by incubation in 0.45% NP-40, 0.45% Tween 20, 10 mm Tris-HCl (pH 8.3), 3 mm MgCl2, and 0.1 mg/ml proteinase K at 56°C (Zeitlin et al. 1995). Lysates were used for PCR after inactivation of the proteinase K at 96°C for 10 min and centrifugation. Alternatively, DNAs were purified by standard protease K digestion in the presence of SDS and phenol–chloroform extraction. Three primers, E163 (CGTCCCTACTGGATGTACAGCGG), E361 (TGCTTTGGTCAAATAGGAGCCACTG), and EN3 (AATGACAAGACGCTGGGCGG) were used in a 20-μl reaction containing 3 mm MgCl2, 1 mm deoxynucleotide triphosphates and 0.5 units of Taq polymerase. The cycling conditions were 94°C for 5 min followed by 35 cycles of 94°C for 30 sec, 70°C for 30 sec, and 72°C for 45 sec. The E163 and E361 primers are specific for ets2 gene. The EN3 primer is specific for the neo gene.
RNA analysis
RNase protection assays were performed as described previously (Neznanov and Oshima 1993). The probes were synthesized from either pGEM1, Bluescript KS, or SP64 vectors containing the indicated fragments of the following mouse cDNAs: ets2, GenBank accession number J04103, bp 1056–1312; Ets1, X53953, bp 620–897 (gift from Barbara Graves, University of Utah, Salt Lake City); collagen IV, J04694, bp 4454–4734; collagen a1(I), K01688, bp 1–335 [gift from Michael Breindl (Rippe et al. 1989)]; stromelysin-1, X63162, bp 1187–1381, (gift of Lynn Matrisian, Vanderbilt University, Nashville, TN), c-jun, X12740, bp 406–713, (gift of Yukio Nishina, Yokohama City University, Japan); junB, J03236, bp 272–455 (gift of Daniel Nathans, Johns Hopkins University, Baltimore, MD); gelatinase B (MMP-9) Z27231, bp 1935–2258, derived from pSP65 92b (Reponen et al. 1994); HNF-4, D29015, bp 628–819 (gift of Shingo Hata, Kyoto University, Japan); HNF3-β, X74937, bp 987–1152 (gift of Kenneth Zaret, Brown University, Providence, RI); urokinase plasminogen activator (uPA), X02389, bp 652–833 (gift of Dominique Belin, Centre Medical Universitaire, Geneva, Switzerland); collagenase-3 (MMP-13), X66473, bp 699–872 (gift of Peter Angel, Kernforschungszentrum Karlsruhe, Karlsruhe, Germany) [previously referred to as collagenase-1 (Gack et al. 1994)]; TGFα, U65016, bp 416–625; and EGFr, X59698, bp 1572–1763. The TGFα and EGFr fragments were obtained by RT–PCR of 2 μg of total mouse kidney RNA.
Whole-mount in situ hybridization
For whole-mount in situ hybridization to analyze plasmids containing 900 bp of 3′ UTR upstream of the ets2 poly(A) tail or Brachyury (Hermann 1991) were linearized. Antisense digoxigenin riboprobes were prepared using digoxigenin-dUTP (Boehringer Mannheim) and the manufacturer’s conditions. These were used to probe E6–E7.5 embryos using a previously described protocol (Henrique et al. 1995). Staining times varied from a few hours to overnight.
Immunocytochemistry
Sections of paraformaldehyde-fixed and paraffin-embedded embryos were incubated with polyclonal rabbit anti-mouse MMP-9/gelatinase B antibody (Behrendtsen et al. 1992), monoclonal rat anti-mouse PECAM-1 antibody (Vecchi et al. 1994), anti-laminin antibody (Collaborative Research Co.), or rabbit anti-mouse placental lactogen-1 (gift of D. Linzer, Northwestern University, Evanston, IL), essentially as described previously (Alexander et al. 1996; Behrendtsen and Werb 1997), by horseradish peroxidase-labeled secondary antibodies and then developed, using the True Blue reagent (KRL) according to the manufacturer’s instructions, or with diaminobenzidine substrate. The sections were counterstained with eosin or methyl green.
Chimera analysis
Ets2db1/db1 ES cells were injected into C57/Bl6 blastocysts and returned to foster mothers. Five of 11 embryos recovered at E18.5 were found to contain ES cell contribution by glucose phosphate isomerase isozyme analysis (Hogan et al. 1986).
Tetraploid embryo rescue
Superovulated CD-1 females were mated with FVB/N males. E1.5 embryos at the two-cell stage were flushed from the oviducts and collected in M2 medium. After equilibration in fusion solution (0.3 m mannitol, 0.1 mm MgSO4, 50 mm CalCl2, 3% bovine serum albumin) (Duncan et al. 1997), the embryos were placed between the electrodes of a BTX 2001 electro cell manipulator (Genetronics, Inc.), aligned in a 0.6-V AC field and treated with a single 100-msec DC pulse of 1.3–1.5 kV/cm. The embryos were transferred to microdrops of KSOM medium under mineral oil and incubated at 37°C for 30–60 min, at which time successfully fused embryos were selected for overnight culture. The afternoon of the next day, four-cell-stage tetraploid embryos were use for aggregation.
Diploid E2.5 embryos at the six- to eight-cell state were isolated just before use from the oviducts and uterine horn of ets2db1/+ females mated with ets2db1/+ males. Zonae pellucida in both diploid and tetraploid embryos were removed in acidic Tyrode’s solution (pH 2.5). Prior to aggregation, the embryos were decompacted by incubation in calcium-free M2 medium at 37°C for 5 min. Two tetraploid embryos were aggregated with one diploid embryo using the “darning needle” technique (Nagy et al. 1990; Duncan et al. 1997; Luo et al. 1997). After 24–26 hr of incubation, successfully aggregated embryos formed single blastocysts that were transferred to the uterine horn of day 2.5 pseudopregnant CD-1 recipients. Some recipients were sacrificed at E14.5 for analysis of embryonic and extraembryonic tissues. Other groups of recipients were subjected to cesarean section at E18.5 (Hogan et al. 1986). Viable fetuses were fostered by females who had delivered litters on the same day.
Flow cytometry
Cell surface fluorescence was determined essentially as described (McKercher et al. 1996). Fluorescein isothyocyanate (FITC)- or phycoerythrin (PE)-labeled antibodies were previously described or purchased from PharMingen (San Diego, CA), BioSource (Camarillo, CA) or Harlan Bioproducts for Science (Indianapolis, IN). T-cell markers were CD4, CD8, CD5 (53-7.3, PharMingen) and CD90 (HIS51, PharMingen). B-cell markers were CD19 (1D3, PharMingen), I-Ab (C3H, PharMingen), IgM (G53-238, PharMingen), and B220. Myeloid markers were sialoadhesin (3D6.112, BioSource), F4/80, CD11b, MOMA-2 (Harlan), Fc γ receptor III/II (CD 32/CD16) (2.4G2, PharMingen), scavenger receptor (2F8, Harlan), and GRI.
Acknowledgments
All embryo aggregation experiments were performed by the Mouse Genetics Shared Service of the Burnham Institute. We thank Jacquiline Avis and Grace Cecena for expert technical assistance, E. Dejana and D. Linzer for antibody gifts, C. Hauser for Ets2 antiserum, recombinant protein, multiple Ets2 constructions, and stimulating discussions. We are also grateful for advice from A. Nagy on the tetraploid embryo aggregation method. This work was supported by grants from the National Institutes of Health (HD 26732 to Z.W., AI 20656 to R.A.M.), the National Cancer Institute, (CA42302 and CA74547 to R.G.O. supporting the targeting of ES cells and subsequent analysis of the mice respectively) and the Burnham Institute Cancer Center support grant (P30 CA30199).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL rgoshima@ljcrf.edu; FAX (619) 646-3193.
References
- Alexander CM, Hansell EJ, Behrendtsen O, Flannery ML, Kishnani NS, Hawkes SP, Werb Z. Expression and function of matrix metalloproteinases and their inhibitors at the maternal-embryonic boundary during mouse embryo implantation. Development. 1996;122:1723–1736. doi: 10.1242/dev.122.6.1723. [DOI] [PubMed] [Google Scholar]
- Behrendtsen O, Werb Z. Metaloproteinases regulate differentiation and migration of parietal endoderm in mouse embryos. Dev Dyn. 1997;208:255–265. doi: 10.1002/(SICI)1097-0177(199702)208:2<255::AID-AJA12>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Behrendtsen O, Alexander CM, Werb Z. Metalloproteinases mediate extracellular matrix degradation by cells from mouse blastocyst outgrowths. Development. 1992;114:447–456. doi: 10.1242/dev.114.2.447. [DOI] [PubMed] [Google Scholar]
- Beitel GJ, Tuck S, Greenwald I, Horvitz HR. The Caenorhabditis elegans gene lin-1 enclodes an ETS-domain protein and defines a branch of the vulval induction pathway. Genes & Dev. 1995;9:3149–3162. doi: 10.1101/gad.9.24.3149. [DOI] [PubMed] [Google Scholar]
- Berkowitz EA, Seroogy KB, Schroder JA, Russell WE, Evans EP, Riedel RF, Phillips HK, Harrison CA, Lee DC, Luetteke NC. Characterization of the mouse tranforming growth factor α gene: Its expression during eyelid development and in Waved 1 tissues. Cell Growth Differ. 1996;7:1271–1282. [PubMed] [Google Scholar]
- Bortner DM, Langer SJ, Ostrowski MC. Non-nuclear oncogenes and the regulation of gene expression in transformed cells. Crit Rev Oncol. 1993;4:137–160. [PubMed] [Google Scholar]
- Boulukos KE, Pognonec P, Rabault B, Begue A, Ghysdael J. Definition of an Ets1 protein domain required for nuclear localization in cells and DNA-binding activity in vitro. Mol Cell Biol. 1989;9:5718–5721. doi: 10.1128/mcb.9.12.5718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boulukos KE, Pognonec P, Sariban E, Bailly M, Lagrou C, Ghysdael J. Rapid and transient expression of Ets2 in mature macrophages following stimulation with CMGF, LPS, and PKC activators. Genes & Dev. 1990;4:401–409. doi: 10.1101/gad.4.3.401. [DOI] [PubMed] [Google Scholar]
- Brunner D, Dücker K, Oellers N, Hafen E, Scholz H, Klämbt C. The ETS domain protein Pointed-P2 is a target of MAP kinase in the Sevenless signal transduction pathway. Nature. 1994;370:386–389. doi: 10.1038/370386a0. [DOI] [PubMed] [Google Scholar]
- Chen WS, Manova K, Weinstein DC, Duncan SA, Plump AS, Prezioso VR, Bachvarova RF, Darnell Jr JE. Disruption of the HNF-4 gene, expressed in visceral endoderm, leads to cell death in embryonic ectoderm and imparied gastrulation of mouse embryos. Genes & Dev. 1994;8:2466–2477. doi: 10.1101/gad.8.20.2466. [DOI] [PubMed] [Google Scholar]
- Chomcznski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Copp AJ. Death before birth: Clues from gene knockouts and mutations. Trends Genet. 1995;11:87–93. doi: 10.1016/S0168-9525(00)89008-3. [DOI] [PubMed] [Google Scholar]
- Donaldson LW, Petersen JM, Graves BJ, McIntosh LP. Secondary structure of the ETS domain places murine Ets-1 in the superfamily of winged helix-turn-helix DNA-binding proteins. Biochemistry. 1994;33:13,509–13,516. doi: 10.1021/bi00250a001. [DOI] [PubMed] [Google Scholar]
- Duncan SA, Nagy A, Chan W. Murine gastrulation requires HNF-4 regulated gene expression in the visceral endoderm: tetraploid rescue of Hnf-4-/- embryos. Development. 1997;124:279–287. doi: 10.1242/dev.124.2.279. [DOI] [PubMed] [Google Scholar]
- Fujiwara S, Fisher RJ, Bhat NK, Espina S, Diazdela M, Papas TS. A short-lived nuclear phosphoprotein encoded by the human ets-2 proto-oncogene is stabilized by activation of protein kinase C. Mol Cell Biol. 1988;8:4700–4706. doi: 10.1128/mcb.8.11.4700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gack S, Vallon R, Schaper J, Ruther U, Angel P. Phenotypic alterations in Fos-transgenic mice correlate with changes in Fos/Jun-dependent collagenase type I expression. J Biol Chem. 1994;269:10363–10369. [PubMed] [Google Scholar]
- Galang CK, Der CJ, Hauser CA. Oncogenic Ras can induce transcriptional activation through a variety of promoter elements, including tandem c-Ets-2 binding sites. Oncogene. 1994;9:2913–2921. [PubMed] [Google Scholar]
- Galang CK, Garcia-Ramirez JJ, Solski PA, Der CJ, Neznanov NN, Oshima RG, Hauser CA. Oncogenic neu/ErbB-2 increases Ets, AP-1, and NF-kB-dependent gene expression, and inhibiting Ets activation blocks Neu-mediated cellular transformation. J Biol Chem. 1996;271:7992–7998. doi: 10.1074/jbc.271.14.7992. [DOI] [PubMed] [Google Scholar]
- Ghysdael J, Boureux A. The ETS family of transcriptional regulators. In: Yaniv B, Ghysdael J, editors. Oncogenes as Transcriptional Regulators. Basel, Switzerland: Birkhauser Verlag; 1997. pp. 29–89. [Google Scholar]
- Guillemot F, Nagy A, Auerbach A, Rossant J, Joyner AL. Essential role of mash-2 in extraembryonic development. Nature. 1994;371:333–336. doi: 10.1038/371333a0. [DOI] [PubMed] [Google Scholar]
- Gum R, Lengyel E, Juarez J, Chen JH, Sato H, Seiki M, Boyd D. Stimulation of 92-kDa gelatinase B promoter activity by ras is mitogen-activated protein kinase kinase 1-independent and requires multiple transcription factor binding sites including closely spaced PEA3/ets and AP-1 sequences. J Biol Chem. 1996;271:10672–10680. doi: 10.1074/jbc.271.18.10672. [DOI] [PubMed] [Google Scholar]
- Gutman A, Wasylyk B. The collagenase gene promoter contains a TPA and oncogene-responsive unit encompassing the PEA3 and AP-1 binding sites. EMBO J. 1990;9:2241–2246. doi: 10.1002/j.1460-2075.1990.tb07394.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hardy MH. The secret life of the hair follicle. Trends Genet. 1992;8:55–61. doi: 10.1016/0168-9525(92)90350-d. [DOI] [PubMed] [Google Scholar]
- Hebert JM, Rosenquist T, Gotz J, Martin GR. FGF5 as a regulator of the hair growth cycle: Evidence from targeted and spontaneous mutations. Cell. 1994;78:1017–1025. doi: 10.1016/0092-8674(94)90276-3. [DOI] [PubMed] [Google Scholar]
- Henkel GW, McKercher SR, Yamamoto H, Anderson KL, Oshima RG, Maki RA. PU.1 but not Ets-2 is essential for macrophage development from ES cells. Blood. 1996;88:2917–2926. [PubMed] [Google Scholar]
- Henrique D, Adam J, Myat A, Chitnis A, Lewis J, Ish-Horowicz D. Expression of a Delta homologue in prospective neurons in the chick. Nature. 1995;375:787–790. doi: 10.1038/375787a0. [DOI] [PubMed] [Google Scholar]
- Hermann BG. Expression pattern of the Brachyury gene in whole-mount TWis/TWis mutant embryos. Development. 1991;113:913–917. doi: 10.1242/dev.113.3.913. [DOI] [PubMed] [Google Scholar]
- Hogan B, Costantini F, Lacy E. Manipulating the mouse embryo: A laboratory manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1986. [Google Scholar]
- Karim FD, Urness LD, Thummel CS, Klemsz MJ, McKercher SR, Celada A, VanBeveren C, Maki RA. The ETS domain: A new DNA binding motif that recognizes a purine-rich core DNA sequence. Genes & Dev. 1990;4:1451–1453. doi: 10.1101/gad.4.9.1451. [DOI] [PubMed] [Google Scholar]
- Kodandapani R, Pio F, Ni C-Z, Piccialli G, Klemsz M, McKercher S, Maki RA, Ely KR. A new pattern for helix-turn-helix recognition revealed by the PU.1 ETS-domain-DNA complex. Nature. 1996;380:456–460. doi: 10.1038/380456a0. [DOI] [PubMed] [Google Scholar]
- Liang H, Olejniczak ET, Mao X, Nettesheim DG, Yu L, Thompson CB, Fesik SW. The secondary structure of the ets domain of human Fli-1 resembles that of the helix-turn-helix DNA-binding motif of the Escherichia coli catabolite gene activator protein. Proc Natl Acad Sci. 1994;91:11655–11659. doi: 10.1073/pnas.91.24.11655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Logan SK, Garabedian MJ, Campbell CE, Werb Z. Synergistic transcriptional activation of the tissue inhibitor of metalloproteinases-1 promoter via functional interaction of AP-1 and Ets-1 transcription factors. J Biol Chem. 1996;271:774–782. doi: 10.1074/jbc.271.2.774. [DOI] [PubMed] [Google Scholar]
- Luetteke NC, Qiu TH, Peiffer RL, Oliver P, Smithies O, Lee DC. TGFα deficiency results in hair follicle and eye abnormalities in targeted and waved-1 mice. Cell. 1993;73:263–278. doi: 10.1016/0092-8674(93)90228-i. [DOI] [PubMed] [Google Scholar]
- Luetteke NC, Phillips HK, Qiu TH, Copeland NG, Earp HS, Jenkins NA, Lee DC. The mouse waved-2 phenotype results from a point mutation in the EGF receptor tyrosine kinase. Genes & Dev. 1994;8:399–413. doi: 10.1101/gad.8.4.399. [DOI] [PubMed] [Google Scholar]
- Luo J, Sladek R, Bader J-A, Matthyssen A, Rossant J, Giguere V. Placental abnormalities in mouse embryos lacking the orphan nuclear receptor ERR-β. Nature. 1997;388:778–782. doi: 10.1038/42022. [DOI] [PubMed] [Google Scholar]
- Mann GB, Fowler KJ, Gabriel A, Nice EC, Williams RL, Dunn AR. Mice with a null mutation of the TGFα gene have abnormal skin architecture, wavy hair, and curly whiskers and often develop corneal inflammation. Cell. 1993;73:249–261. doi: 10.1016/0092-8674(93)90227-h. [DOI] [PubMed] [Google Scholar]
- Maroulakou IG, Papas TS, Green JE. Differential expression of ets-1 and ets-2 proto-oncogenes during murine embryogenesis. Oncogene. 1994;9:1551–1565. [PubMed] [Google Scholar]
- McCarthy SA, Chen D, Yang B-S, Garcia Ramirez JJ, Cherwinski H, Chen X-R, Klagsbrun M, Hauser CA, Ostrowski MC, McMachon M. Rapid phosphorylation of Ets-2 accompanies mitogen-activated protein kinase activation and the induction of heparin-binding epidermal growth factor gene expression by oncogenic Raf-1. Mol Cell Biol. 1997;17:2401–2412. doi: 10.1128/mcb.17.5.2401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonnell S, Matrisian LM. Stromelysin in tumor progression and metastasis. Cancer Metastasis Rev. 1990;9:305–319. doi: 10.1007/BF00049521. [DOI] [PubMed] [Google Scholar]
- McKercher SR, Torbett BE, Anderson KL, Henkel GW, Vestal DJ, Baribault H, Klemsz M, Feeney AJ, Wu GE, Paige CJ, Maki RA. Targeted disruption of the PU.1 gene results in multiple hematopoietic abnormalities. EMBO J. 1996;15:5647–5658. [PMC free article] [PubMed] [Google Scholar]
- Miettinen PJ, Berger JE, Meneses J, Phung Y, Pedersen RA, Werb Z, Derynck R. Epithelial immaturity and multiorgan failure in mice lacking epidermal growth factor receptor. Nature. 1995;376:337–341. doi: 10.1038/376337a0. [DOI] [PubMed] [Google Scholar]
- Mohammad M, Dikic I, Sorokin A, Burgess WH, Jaye M, Schlessinger J. Identification of six novel autophosphorylation sites on fibroblast growth factor receptor 1 and elucidation of their importance in receptor activation and signal transduction. Mol Cell Biol. 1996;16:977–989. doi: 10.1128/mcb.16.3.977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muthusamy N, Barton K, Leiden JM. Defective activation and survival of T cells lacking the Ets-1 transcription factor. Nature. 1995;377:639–642. doi: 10.1038/377639a0. [DOI] [PubMed] [Google Scholar]
- Nagy A, Gocza E, Merentes-Diaz E, Prideaux VR, Ivanyi E, Markkula M, Rossant J. Embryonic stem cells alone are able to support fetal development in the mouse. Development. 1990;110:815–821. doi: 10.1242/dev.110.3.815. [DOI] [PubMed] [Google Scholar]
- Neznanov NS, Oshima RG. Cis regulation of the keratin 18 gene in transgenic mice. Mol Cell Biol. 1993;13:1815–1823. doi: 10.1128/mcb.13.3.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Neill EM, Rebay I, Tijan R, Rubin GM. The activities of two Ets-related transcription factors required for Drosophila eye development are modulated by the Ras/MAPK pathway. Cell. 1994;78:137–147. doi: 10.1016/0092-8674(94)90580-0. [DOI] [PubMed] [Google Scholar]
- Oshima RG. Identification and immunoprecipitation of cytoskeletal proteins from murine extra-embryonic endodermal cells. J Biol Chem. 1981;256:8124–8133. [PubMed] [Google Scholar]
- Oshima RG, McKerrow J, Cox D. Murine embryonal carcinoma hybrids: Decreased abiltiy to spontaneoulsy differentiate as a dominat trait. J Cell Physiol. 1981;109:195–204. doi: 10.1002/jcp.1041090202. [DOI] [PubMed] [Google Scholar]
- Rabault B, Roussel MF, Quang CT, Ghysdael J. Phosphorylation of Ets1 regulates the complementation of a CSF-1 receptor impaired in mitogenesis. Oncogene. 1996;13:877–881. [PubMed] [Google Scholar]
- Rees S, Coote J, Stables J, Goodson S, Harris S, Lee MG. Bicistronic vector for the creation of stable mammalian cell lines that predisposes all antibiotic-resistant cells to express recombinant protein. BioTechniques. 1996;20:102–110. doi: 10.2144/96201st05. [DOI] [PubMed] [Google Scholar]
- Reponen P, Sahlberg C, Munaut C, Thesleff I, Tryggvason K. High expression of 92-kD type IV collagenase (gelatinase B) in the osteoclast lineage during mouse development. J Cell Biol. 1994;124:1091–1102. doi: 10.1083/jcb.124.6.1091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rippe RA, Lorenzen S, Brenner DA, Breindl M. Regulatory elements in the 5’-flanking region and the first intron contribute to the transcriptional control of the mouse alpha 1 type I collagen gene. Mol Cell Biol. 1989;9:2224–2227. doi: 10.1128/mcb.9.5.2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott EW, Simon MC, Anastasi J, Singh H. Requirement of transcription factor PU.1 in the development of multiple hematopoietic lineages. Science. 1994;265:1573–1577. doi: 10.1126/science.8079170. [DOI] [PubMed] [Google Scholar]
- Sibilia M, Wagner EF. Strain-dependent epithelial defects in mice lacking the EGF receptor. Science. 1995;269:234–238. doi: 10.1126/science.7618085. [DOI] [PubMed] [Google Scholar]
- Stewart TA, Mintz B. Successive generations of mice produced from an established culture line of euploid teratocarcinoma cells. Proc Natl Acad Sci. 1981;78:6314–6318. doi: 10.1073/pnas.78.10.6314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sympson CJ, Talhouk RS, Alexander CM, Chin JR, Clift SM, Bissell MJ, Werb Z. Targeted expression of stromelysin-1 in mammary gland provides evidence for a role of proteinases in branching morphogenesis and the requirement for an intact basement membrane for tissue-specific gene expression. J Cell Biol. 1994;125:681–693. doi: 10.1083/jcb.125.3.681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sympson CJ, Talhouk RS, Bissell MJ, Werb Z. The role of metalloproteinases and their inhibitors in regulating mammary epithelial morphology and function in vivo. Perspectives Drug Discovery Design. 1995;2:401–411. [Google Scholar]
- Threadgill DW, Dlugosz AA, Hansen LA, Tennenbaum T, Lichti U, Yee D, LaMantia C, Mourton T, Herrup K, Harris RC, Barnard JA, Yuspa SH, Coffey RJ, Magnuson T. Targeted disruption of mouse EGF receptor: effect of genetic background on mutant phenotype. Science. 1995;269:230–234. doi: 10.1126/science.7618084. [DOI] [PubMed] [Google Scholar]
- Vecchi A, Garlanda C, Lampugnani MG, Resnati M, Matteucci C, Stopparcciaro A, Schnurch H, Risau W, Ruco L, Mantovani A, Dejana E. Monoclonal antibodies specific for endothelial cells of mouse blood vessels. Their application to the identification of adult and embryonic endothelium. Eur J Cell Biol. 1994;63:247–254. [PubMed] [Google Scholar]
- Wasylyk C, Gutman A, Nicholson R, Wasylyk B. The c-Ets oncoprotein activates the stromelysin promoter through the same elements as several non-nuclear oncoproteins. EMBO J. 1991;10:1127–1134. doi: 10.1002/j.1460-2075.1991.tb08053.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wasylyk B, Hahn SL, Giovane A. The Ets family of transcription factors. Eur J Biochem. 1993;211:7–18. doi: 10.1007/978-3-642-78757-7_2. [DOI] [PubMed] [Google Scholar]
- Watson DK, Mavrothalassitis GJ, Jorcyk CL, Smyth FE, Papas TS. Molecular organization and differential polyadenylation sites of the human ETS2 gene. Oncogene. 1990;5:1521–1527. [PubMed] [Google Scholar]
- Yamaguchi TP, Rossant J. Fibroblast growth factors in mammalian development. Curr Opin Genet Dev. 1995;5:485–491. doi: 10.1016/0959-437x(95)90053-j. [DOI] [PubMed] [Google Scholar]
- Yang B-S, Hauser CA, Henkel G, Colman MS, Van Beveren C, Stacey KJ, Hume DA, Maki RA, Ostrowski MC. Ras-mediated phosphorylation of a conserved threonine residue enhances the transactivation activity of c-Ets1 and c-Ets2. Mol Cell Biol. 1996;16:538–547. doi: 10.1128/mcb.16.2.538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeitlin S, Liu J-P, Chapman DL, Papaioannou VE, Efstratiadis A. Increased apoptosis and early embryonic lethality in mice nullizygous for the Huntington’s disease gene homologue. Nature Genet. 1995;11:155–163. doi: 10.1038/ng1095-155. [DOI] [PubMed] [Google Scholar]