Abstract
Sex determination is regulated by diverse pathways. Although upstream signals vary, a cysteine-rich DNA-binding domain (the DM motif) is conserved within downstream transcription factors of Drosophila melanogaster (Doublesex) and Caenorhabditis elegans (MAB-3). Vertebrate DM genes have likewise been identified and, remarkably, are associated with human sex reversal (46, XY gonadal dysgenesis). Here we demonstrate that the structure of the Doublesex domain contains a novel zinc module and disordered tail. The module consists of intertwined CCHC and HCCC Zn2+-binding sites; the tail functions as a nascent recognition α-helix. Mutations in either Zn2+-binding site or tail can lead to an intersex phenotype. The motif binds in the DNA minor groove without sharp DNA bending. These molecular features, unusual among zinc fingers and zinc modules, underlie the organization of a Drosophila enhancer that integrates sex- and tissue-specific signals. The structure provides a foundation for analysis of DM mutations affecting sexual dimorphism and courtship behavior.
Keywords: Drosophila melanogaster, DNA-binding protein, gene regulation, transcription factor, NMR
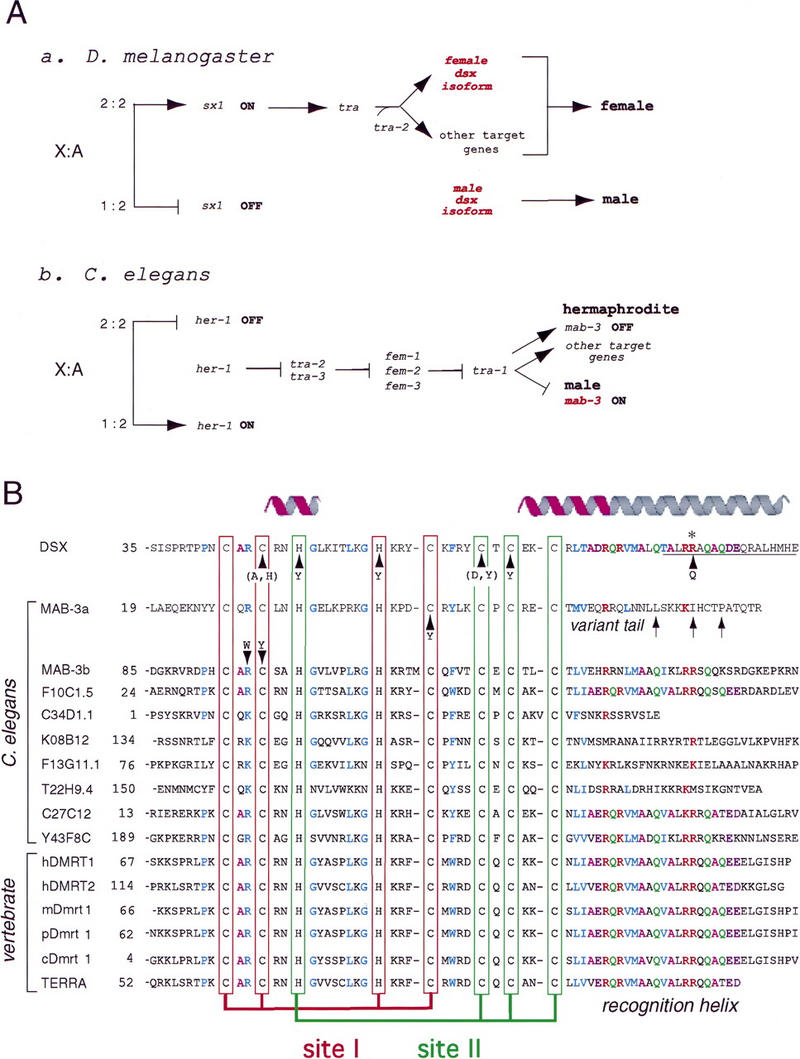
Sexual dimorphism exhibits a diversity of molecular mechanisms. In mammals formation of the testis and subsequent male development is initiated by a gene on the Y chromosome (Page et al. 1987). Studies of human sex reversal led to the identification of SRY (Gubbay et al. 1990; Sinclair et al. 1990), the testis-determining factor (Berta et al. 1990; Koopman et al. 1990). SRY contains a high-mobility group (HMG) box, a minor groove DNA-binding domain (DBD) shared by a family of architectural transcription factors. Regulation of organogenesis requires sharp DNA bending (Giese et al. 1992; van Houte et al. 1993; Pontiggia et al. 1994; Bewley et al. 1998). Alternative genetic mechanisms operate in Drosophila melanogaster (Nagoshi et al. 1988; Burtis et al. 1991; An et al. 1996; Cho and Wensink 1997, 1998; Li and Baker 1998; Marin and Baker 1998) and Caenorhabditis elegans (Villeneuve and Meyer 1990; Hodgkin 1997) wherein sex is determined by the X:autosome ratio, a process linked to X dosage compensation (Cline and Meyer 1996; Dawes et al. 1999). The pathways of the fly and worm (Fig. 1A) seem otherwise unrelated. Whereas somatic sexual differentiation in C. elegans requires cell–cell signaling, analogous decisions in D. melanogaster are cell autonomous (Cline and Meyer 1996). D. melanogaster (but not C. elegans) uses a hierarchy of sex-specific RNA splicing events.
Figure 1.
(A) Genetic pathways of sex determination in D. melanogaster (a) and C. elegans (b) are initiated by X:autosome (A) ratio but otherwise different (Cline and Meyer 1996). (A,a) In the fly a high X:A ratio (2:2) activates sxl, which encodes a splicing factor. In turn, Sxl allows female-specific expression of the splicing factor tra, which together with tra-2, permits expression of female isoform DSXF. The pathway then ramifies. Intersex (ix), hermaphrodite (her), and fruitless (not shown) are other downstream elements. When the X:A ratio is low, male-specific isoform DSXM occurs by default; isoforms differ in carboxy-terminal domain (see Fig. 2A). (A,b) In nematode a high X:A ratio (2:2) is associated with absent her-1 expression, in turn enabling unrepressed expression of tra-2 and tra-3 (unrelated to Drosophila genes of similar nomenclature). Hermaphrodite-specific expression of tra-2 and tra-3 represses fem-1, fem-2, and fem-3. In their absence, tra-1 is expressed in hermaphrodite and turns off male-determinant mab-3 (encoding a DM transcription factor). TRA-1 has MAB-3 independent targets as pathway ramifies. When the X:A ratio is low (1:2), HER-1 (involved in cell–cell signaling) represses tra-2 and tra-3. Unrepressed expression of female genes represses tra-1 and therefore default expression of mab-3 directs male development and turns off hermaphroditic program. Nematode genes (b) that enumerate X:A ratio and control X dosage compensation (including xol-1 and sdc-2; Dawes et al. 1999) are omitted for clarity. (B) Alignment of metazoan DM sequence motifs. Cysteines and histidines that coordinate Zn2+ are aligned as two intertwined binding sites (boxes): site I (red) and site II (green). Conserved residues in zinc module are otherwise shown in blue; conserved residues in tail are color coded (R, red; Q, green; A, magenta; D/E, violet; L/V/T/M, blue). Stable α-helical elements are highlighted by magenta ribbons above DSX sequence. Nascent carboxy-terminal α-helix is indicated by gray extension; it is not meant to convey a continuous α-helix (see text). (DSX) DM domain in D. melanogaster (accession no. M25292). (MAB-3a and MAB-3b) The first and the second DM domains in C. elegans protein (accession no. Z99278). Other C. elegans DM sequences: F10C1.5, cosmid F10C1 (accession no. U49831); C34D1.1, cosmid C34D1 (accession no. Z78060); K08B12, cosmid K08B12 (accession no. U97001); F13G11, cosmid F13G11 (accession no. Z83317); T22H9.4: cosmid T22H9 (accession no. AF101315); C27C12 (accession no. Z69883); and Y43F8C (accession no. AL032637.1). Vertebrate DM sequences: DMRT1 and DMRT2, human homologs on the short arm of chromosome 9 (DMRT1: accession no. AF130728 and DMRT2: accession no. AF130729). Other homologs: mDmrt1 (murine, accession no. AF202778), pDmrt1 (porcine, accession no. AF216651), and cDmrt1 (chicken, accession no. AF123456). TERRA: DM domain of zebrafish terra (accession no. AF080622). Sequences shown include genes (such as TERRA; Meng et al. 1999) not known to be involved in sex determination. Arrows without parentheses indicate sites of point mutations in dsx or mab-3 associated with intersex development; substitutions in parenthesis indicate variants characterized only by biochemical assays (Erdman and Burtis 1993). Three additional putative DM genes of unknown function have recently been found in the genome of D. melanogaster (accession nos. AAF56919, AAF55843, and AAF48261; sequences not shown).
Despite diverse upstream mechanisms, downstream sex-determining genes may be conserved in part (Raymond et al. 1998). An example is provided by homologs doublesex (dsx; Hildreth 1965) and mab-3 (Shen and Hodgkin 1988). In C. elegans MAB-3 is expressed in the male and repressed in the hermaphrodite, whereas the RNA-splicing pathway of D. melanogaster regulates male- and female-specific DSX isoforms (DSXM and DSXF; Baker and Wolfner 1988; Burtis and Baker 1989). Despite these differences, dsx and mab-3 each function downstream of “master” regulatory factors to define one branch of a ramifying pathway (Fig. 1A). Furthermore, dsx and mab-3 each encode transcription factors that in part regulate analogous dimorphic tissues (Bownes et al. 1983; Baker and Wolfner 1988; Burtis and Baker 1989; Coschigano and Wensinck 1993). Functional analogies in downstream pathways exceed the extent of molecular or histologic conservation: for example, DSX and MAB-3 each regulate expression of yolk–protein genes (yp and vit families, respectively), although these yolk proteins are unrelated in sequence (Bownes et al. 1983; Coschigano and Wensink 1993; An and Wensink 1995a,b; Li and Baker 1998; Yi and Zarkower 1999). Similarly, DSX-regulated differentiation of male sensory bristles in D. melanogaster and MAB-3-regulated formation of male sensory rays in C. elegans seem analogous even as the organs themselves differ in histology. Strikingly, DSXM functions in a mab-3− XO worm (ordinarily a chromosomal male with an intersex phenotype; Shen and Hodgkin 1988) to rescue such male features (Raymond et al. 1998). Complementation across phyla demonstrates that downstream mechanisms of gene regulation are in part conserved.
DSX and MAB-3 domains exhibit zinc-dependent DNA binding (Erdman and Burtis 1993) with distinct but overlapping specificities (Erdman et al. 1996; Yi and Zarkower 1999). The proteins contain homologous DBDs (the DM motif) remarkable for a novel pattern of cysteines and histidines (Fig. 1B; Erdman and Burtis 1993; Raymond et al. 1998). Mutations in the motif are associated with intersex development (arrowheads in Fig. 1B; Hildreth 1965; Hodgkin 1983; Erdman et al. 1996). Phenotypes correlate with loss of DNA-binding activity (Erdman and Burtis 1993; Raymond et al. 1998). DSX mutations in the invariant cysteines or histidines impair both Zn2+ coordination and DNA recognition; an intersex mutation in the motif's carboxy-terminal segment disrupts DNA binding without affecting Zn2+ coordination (R91Q; Erdman and Burtis 1993). DSXM and DSXF share the same amino-terminal DBD but distinct carboxy-terminal extensions (Fig. 2A; Burtis and Baker 1989). Unlike DSX, MAB-3 contains two DM domains (Fig. 2A), each required in vivo (Raymond et al. 1998) and proposed to function coordinately in DNA binding (Yi and Zarkower 1999). Multiple homologs of unknown function are encoded in the C. elegans genome (see Fig. 1B; C. elegans Sequence Consortium 1998).
Figure 2.
(A) Domain organization of DSXM, DSXF, and MAB-3. Amino-terminal black rectangles indicate DM domains; the carboxy-terminal DSX dimerization domain and sex-specific extension (cross-hatched or gray; not present in MAB-3) are also indicated. (B,top) Three transcription factors (AEF1, DSX, and bZIP1) bind fbe enhancer (An and Wensink 1995a,b) at overlapping target sites (aef1 is shown in black, dsxA in red, and bzip1 in blue); (bottom) consensus sequences. Boxed sites are based on DNase protection; DSX footprint spans 21 bp, whereas DmC/EBP footprint spans 19 bp. DNase footprints overestimate DNA target sites due to steric occlusion between enzyme and protein–DNA complex. (C) Model of sex- and tissue-specific regulation of yolk–protein expression by fbe (An and Wensink 1995a,b). DSX binds as dimer; only monomer is shown (ribbon model). In model dsxA and bzip1 are occupied simultaneously to activate promoter in female. Binding of either DSXM or DSXF displaces AEF1 from its target site aef1. In female, fat body expression of DSXF is higher than that of AEF1, and therefore in the presence of bZIP1, yolk proteins are expressed. Expression is repressed in ovary with higher levels of AEF1, which displace DSXF. Male-specific repression of yolk proteins occurs as binding of DSXM, which is 122 residues longer than DSXF (Burtis and Baker 1989), occludes bzip1 or inactivates bound bZIP1, and therefore is repressed in male (C,bottom; Coschigano and Wensink 1993). It is not known which Drosophila bZIP family member is active in fat body. (D) Overlapping DSX DM and bZIP phosphate contacts are inferred based on methylphosphonate interference studies (red circles); sites of bZIP contact are as predicted by GCN4 cocrystal structure (blue circles; Keller et al. 1995). Filled or half-filled red circles indicate sites of strong or weak DM interference, respectively.
The DM motif is conserved within vertebrates (Fig. 1B, bottom). These include DMRT1 and DMRT2, adjacent genes on human chromosome 9 (9p24; Raymond et al. 1999a). Remarkably, deletions spanning DMRT1 and DMRT2 are associated with XY sex reversal in the absence of abnormalities in SRY (Fryns et al. 1986; Crocker et al. 1988; Magenis et al. 1990; Bennet et al. 1993; McDonald et al. 1997; Flejter et al. 1998; Guioli et al. 1998; Veitia et al. 1998). This clinical correlation suggests broad conservation of DM genes in metazoan sex determination, a hypothesis supported by the embryologic expression of murine Dmrt1 in the differentiating gonadal ridge (Raymond et al. 1999b). Point mutations in DMRT1 or DMRT2 (unlike deletions) are apparently rare in XY humans with unexplained sex reversal or intersex phenotypes, presumably due to functional redundancy (Raymond et al. 1999a). Zebrafish and mammals, like C. elegans, contain additional DM genes, one of which (TERRA) appears to function in mesodermal patterning in both sexes (Meng et al. 1999).
We present here the solution structure of the DSX DM domain and define its overall mode of DNA recognition. The structure reveals a novel zinc module containing intertwined CCHC and HCCC Zn2+-binding sites. A carboxy-terminal tail, disordered in the free domain, is stabilized on DNA binding as a recognition α-helix. Unlike classic zinc fingers and zinc modules (Klug and Schwabe 1995), the DM domain binds in DNA's minor groove. Unlike SRY and other architectural motifs targeted to the minor groove (Giese et al. 1992; van Houte et al. 1993; Haqq et al. 1994), the DM motif does not induce sharp DNA bending. These unusual features rationalize the architecture of a DSX response element active in a dimorphic lineage (the female fat body; Burtis et al. 1991; Coschigano and Wensink 1993). The fat body enhancer (fbe; Fig. 2B) contains tightly overlapping minor and major groove DNA control sites, which function coordinately to integrate sex- and tissue-specific signals (Fig. 2C; An and Wensink 1995a,b). The structure of the DSX DM domain provides a foundation for analysis of intersex mutations in diverse metazoans (Raymond et al. 1998). Because in the fly such mutations lead to allele-specific changes in sexual preference (Villella and Hall 1996), their characterization may provide insight into the genetic encoding of a complex behavior.
Results
Seventy-one-residue DSX- and DMRT1-derived DM polypeptides were obtained by total synthesis. The synthetic strategy exploited the native chemical ligation reaction (Dawson et al. 1994) and used an invariant cysteine residue as a central ligation site. Studies focused on DSX, which was found to be more stable to air oxidation than DMRT1. The DSX polypeptide was shown by mass spectrometry to bind two Zn2+ ions; the (Zn2+)2–polypeptide complex was found by analytical ultracentrifugation to be monomeric in the concentration range 60–140 μm. The synthetic domain's affinity for the fbe (Kd 8 nm as inferred by gel mobility-shift assay; GMSA) is similar to published values (An et al. 1996; Erdman et al. 1996). Zinc-dependent protein folding was demonstrated by circular dichroism (CD) and proton nuclear magnetic resonance (1H-NMR). The visible absorption spectrum of a (Co2+)2–peptide complex is consistent with tetrahedral coordination. Addition of two equivalents of Zn2+ leads to attenuation of Co2+ d–d transitions, indicating (as in the zinc finger; Frankel et al. 1987; Lee et al. 1989) preferential binding of Zn2+.
The DM domain binds cooperatively in the minor groove without DNA bending
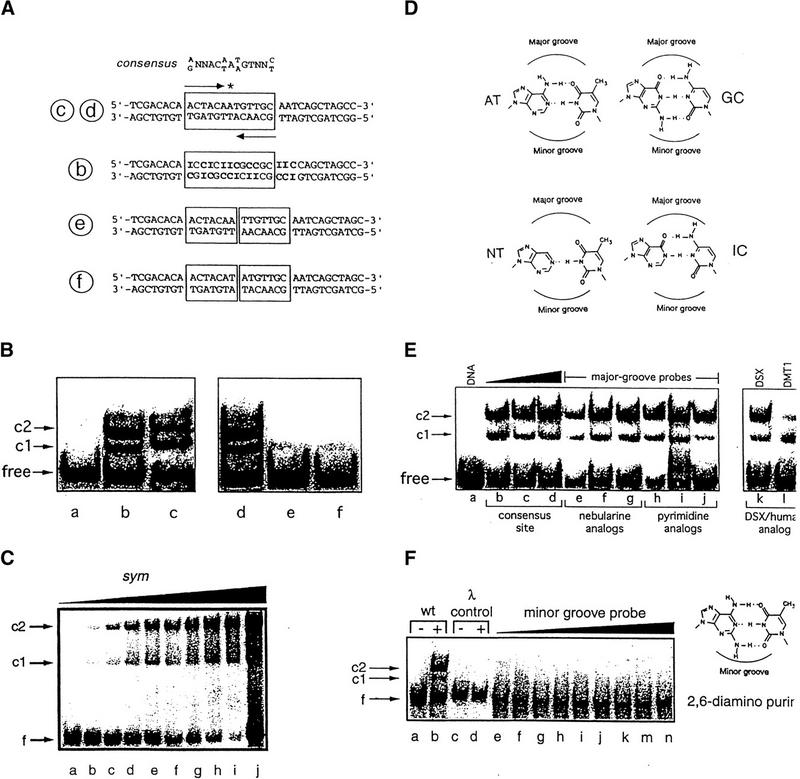
Binding of the DSX DM domain to dsxA (Fig. 3A,B) leads to discrete complexes (c1 and c2). Protein:DNA stoichiometries were verified to be 1:1 and 2:1 by GMSA studies of equimolar and 2:1 solutions at concentrations 500-fold higher than the apparent dissociation constant. The predominance of the 2:1 complex under conditions containing appreciable free DNA demonstrates its cooperative assembly. The domain's sequence specificity is highlighted by its 100-fold decreased affinity for palindromic dsxA analogs containing an additional central AT or TA base pair (Fig. 3A–C); cooperativity is nevertheless retained (Fig. 3C). The human DMRT1 domain also recognizes dsxA (Fig. 3E, lane l). Minor groove binding was established by GMSA studies of DNA sites containing base analogs. Systematic AT → IC substitutions (boldface type in b in Fig. 3A; Lee et al. 1991; Starr and Hawley 1991) alter the global pattern of functional groups in the DNA major groove but preserve a native minor groove (Fig. 3D). Multiple IC substitutions have no effect on specific DNA binding (Fig. 3B, lane b). Base substitutions by uridine, 5-methylcytosine, or nebularine—complementary probes in the major groove—likewise have no effect (Fig. 3E). Robustness of DM–dsxA recognition to major groove chemical modification stands in contrast to its disruption by insertion of a palindromic central base pair (discussed above). An additional negative control is provided by systematic substitution of adenine by diaminopurine, which introduce exocyclic amino groups in the minor groove (right-hand panel of Fig. 3F). As expected, such bulky modifications prevent protein binding (Fig. 3F, lanes e–n). Methylphosphonate interference (Labeots and Weiss 1997) was used to obtain a footprint of DM–DNA contacts (red circles in Fig. 2D). Nine sites of interference were observed. These sites, potential contacts between the DM domain and the DNA backbone, are almost symmetric in distribution and restricted to the core consensus sequence (5′-NACAATGTN-3′ and complement; Erdman et al. 1996). The staggered pattern of half-site contacts is consistent with protein binding to the minor groove of B-DNA. Despite minor groove binding, DSX-induced DNA bending is negligible as inferred from permutation gel electrophoresis: relative mobilities of either fbe complex c1 or c2 exhibit only slight dependence on site position (Fig. 4A,B). Such results are in contrast to the marked electrophoretic anomalies induced by binding of the SRY HMG box (Fig. 4B, control lanes 6,7; Giese et al. 1992). Insubstantial DNA bending was corroborated by analysis of protein-induced changes in fluorescence resonance energy transfer (FRET) between probes (fluoroscein and tetramethylrhodamine) linked to opposite 5′ ends of a 15-bp duplex (Parkhurst et al. 1996). Whereas binding of the SRY HMG box significantly enhances FRET efficiency, consistent with the decreased end-to-end distance of a sharply bent DNA, the effect of DM binding is minor (supplemental information).
Figure 3.
DNA-binding studies of DSX domain demonstrate minor groove binding mode. (A,top) Consensus sequence of DSX-binding sites, an imperfect palindrome containing an odd number of base pairs. (Second from top) DSX target site in fbe11–13. (Third from top) Analog containing multiple AT → IC substitutions (boldface type). (Bottom two sequences) Variants containing additional AT or TA base pairs in center. Labels b, c, d, e, f (circled) refer to lanes of gels in B. (B) 33P-detected GMSA at 4°C of DSX domain binding to sites defined in A. Bands c1 and c2 designate 1:1 and 2:1 protein–DNA complexes. Protein binds equally well to native and IC-containing sites. Insertion of central AT or TA base pairs reduces protein binding by about 100-fold. Protein concentration was 8 nm and DNA concentration was 0.5 nm. (C) Cooperative low-affinity binding of the DSX DM domain to a palindromic (sym) DNA site (5′-ACTACAATTGTTGCA-3′; central base pair in boldface type) using fluorescein (at 5′ of top strand) as probe. DNA concentration was 500 nm in lanes a–i, with respective protein concentrations 0, 250, 500, 600, 700, 800, 900, and 1000 nm. Lane j contains an equimolar solution in which concentrations of DNA and protein were each 5 μm; predominance of 2:1 complex and free DNA indicates cooperativity is retained. (D,left) Structures of AT and variant nebularine NT base pair. (D,right) Structures of GC and inosine IC base pair. Positions of major and minor grooves are indicated. (E,right gel) Major groove base modifications do not perturb binding of DSX domain to 15 base-pair DNA probe (5′-CACTACAATGTTGCA-3′ and complement): (lanes e–g) single nebularine substitutions at positions 5, 7, and 8 of upper strand and (lanes h–j) single 5-methylcytosine or uridine substituitions at positions 6, 9, and 11 of upper strand. (E,left gel) DMRT1 DM domain (25 nm) recognizes DSX target site with weaker apparent affinity; cooperative 2:1 binding is maintained. (F) 33P-detected GMSA of fbe analog in which the 8 AT bp in the DSX-binding site (boxed in Fig. 3A, sequence c,d) are substituted by 2,6-diaminopurine (structure at right). No binding is observed to λ operator site OL1 (lanes c,d) or to the diaminopurine fbe analog (lanes e–n) at successive protein concentrations of 0, 1, 2, 4, 8, 16, 32, 64, and 128 nm. DNA concentration was ∼1 nm. Protein concentrations were 8 nm for lanes b and d. Sequence of λ probe is 5′-TACCACTGGCGGTCATA-3′ and complement.
Figure 4.
(A,B) Analysis of protein-directed DNA bending by permutation gel electrophoresis. (A) Probes (each 140 bp) contain fbe site at positions shown; probes 3 and 4 contain SRY target site 5′-TTTGTT-3′. GMSA in B demonstrates very weak dependence of DM–fbe complexes c1 and c2 on fbe position in probe (lanes 1–4). Lanes 6 and 7 illustrate SRY control using probes 3 and 4 (Geise et al. 1992). The weak band under major SRY–DNA complex arises from proteolytic fragment; more slowly migrating species represent higher order complexes. Free probe 3 is shown in lane 5. (C) Tail mutant R91Q impairs specific DNA binding but not cooperativity. GMSA used 33P-labeled 33-bp fbe probe (1.5 nm). (Lanes 2–7) Native DSX domain (respective protein concentrations 4, 8, 12, 18, 24, and 48 nm); (lanes 8–16) R91Q variant domain (respective protein concentrations 12, 24, 48, 96, 120, 156, 240, 480, and 560 nm); the free probe is shown in lane 1. At 48 nm concentrations the native domain is 52% shifted, whereas shift of variant domain is negligible (<1%). Complexes c1 and c2 represent 1:1 and 2:1 binding of domain to dsxA; c3 represents a higher order complex observed at high protein concentrations (>100 nm).
The DM motif encodes a novel intertwined zinc module and disordered tail
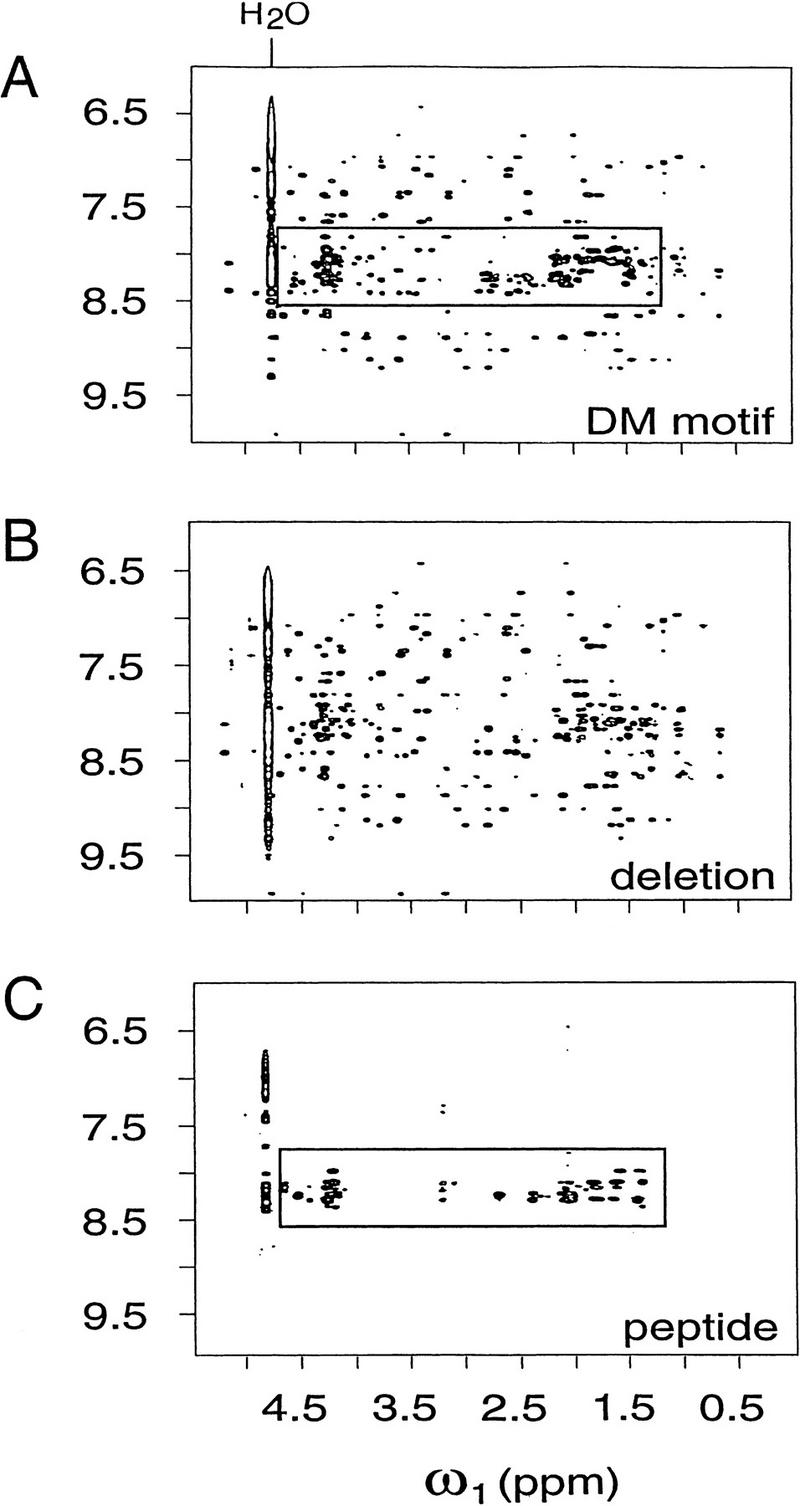
The 1H-NMR spectrum of the DSX DM domain provides evidence of an ordered moiety and disordered tail. The ordered moiety (DSX residues 36–82, including invariant cysteines and histidines; see Fig. 1B) exhibits marked dispersion of chemical shifts, whereas the tail is manifest by an envelope of poorly resolved spin systems at near-random coil chemical shifts. Line widths and chemical shifts are similar in the protein concentration range 60 μm–1.4 mm. Sequential assignment of the ordered moiety, obtained by homonuclear two-dimensional NMR, was facilitated by comparative study of a fragment lacking most of the tail (DSX residues 35–86; DMΔ). The specific DNA-binding activity of DMΔ is reduced by at least 100-fold (not shown). With the exception of tail resonances, 1H-NMR spectra of the intact and truncated domains are essentially identical (Fig. 5A,B). The fragment's structure thus corresponds to the ordered moiety of the intact domain, demonstrating that folding of the zinc module is independent of the tail. The envelope of carboxy-terminal resonances in the spectrum of the intact domain (boxed in Fig. 5A) is similar to that of an isolated tail peptide (19 residues; Fig. 5C).
Figure 5.
NOEsY spectra demonstrate “peptide dissection” of intact DM motif (A) into major and minor fragments: Zn-binding module (DMΔ; B) and carboxy-terminal peptide (peptide DM-p; C). Shown are respective spectra in H2O at 25°C and 600 MHz; mixing times were in each case 175 msec. Cross peaks from amide and aromatic protons (vertical scale, θ2) and aliphatic protons (horizontal scale, θ1) are shown. Boxed regions highlight unresolved envelope in intact DM domain (A) assigned to nascent carboxy-terminal helix (C); this feature is absent in spectrum of DMΔ (B). Peptides were made 1.5 mm in 50 mm deuterated Tris-HCl (pH 6.5) in 90% H2O and 10% D2O. Spectra A and B also were obtained in presence of 4 mm ZnCl2.
A network of 346 interproton nuclear Overhauser effects (NOEs) was observed in the ordered moiety. Of particular interest are 92 long-range NOEs, which include contacts within putative metal-binding sites. An example is provided by NOEs linking the side chains of residues C44, C47, H59, and C63 and in turn linking the side chain of C44 to aliphatic protons of I54 and L56. Contacts likewise occur from C47 to L52 and I54. Analogous NOEs are observed among H50, C68, C70, and C73. Notably, the aromatic ring of F65 contacts both sets of ligands (side chains of C47, H60, and C68). Preliminary distance geometry/simulated annealing (DG/SA) calculations were performed in the absence of explicit Zn2+ coordination. Two potential zinc-binding sites were well defined; each exhibited dimensions consistent with tetrahedral coordination. Such interim models were refined by imposition of zinc-related distance restraints (see Materials and Methods; Qian et al. 1993). An extensive series of control DG/SA calculations was performed imposing alternative ligation schemes; each was excluded based on significant NOE restraint violations. The native model predicted a specific pattern of seven amide–sulfur hydrogen bonds involving four of the six cysteate Sγ atoms. These bonds rationalize a corresponding pattern of protected amide protons in D2O, not otherwise accounted for by secondary structure. The final ensemble was refined by inclusion of amide–sulfur hydrogen bond restraints (Qian et al. 1993). A summary of restraints, violations, and empirical energy terms is provided in Table 1. Two glycines (G51 and G58) exhibit positive φ values. No restraints were obtained for the carboxy-terminal tail (residues 87–105); therefore, they were omitted from the model.
Table 1.
Restraints and restraint violations
| Restraints | |||
| intraresidue | 20 | dihedral angle φ | 16 |
| sequential | 140 | dihedral angle χ1 | 15 |
| medium-range ((i–j) 2–4 residue) | 94 | zinc coordination | 20 |
| long range ((i–j) > 4 residue) | 92 | hydrogen bonds | 16 |
| Average restraint violations | |||
| NOE (Å) | 0.06 | dihedral angle | 0.6 |
| Deviation from ideal covalent geometry | |||
| bond length (Å) | 0.005 | bond angle (θ) | 1.46 |
| Energy function (kcal/mol)a | |||
| NOE restraint energy | 66.2 | van der Waals | 33.7 |
| improper dihedral angles | 22.1 | restrained dihedral angles | 0.04 |
| covalent bonds | 16.2 | bond angles | 158.6 |
NOE and dihedral force constraints were 40 Kcal/Å2 and 40 Kcal/rad2, respectively (1 K = 4.18 kJ).
The structure of the DM domain is remarkable for intertwined CCHC and HCCC zinc-binding sites (sites I and II in Fig. 6A; outlined in schematic form in Fig. 1B). These sites nucleate a unified globular domain containing two α-helices (residues 46–50 and 71–79; Fig. 6B). The first helix, containing C47 and H50, connects Zn2+-binding sites; the second and longer helix extends from site II into the tail. Ordered side chains adjoin each site (Fig. 6C). R46, a site of intersex mutation in C. elegans (motif MAB-3b), is conserved in DMRT1, DMRT2, and other vertebrate homologs (Fig. 1B). Its methylene groups seal one edge of site I, whereas its guanidinium moiety contributes to a positive electrostatic surface. An aromatic ring at position 65 (F65), invariant among DM sequences (Fig. 1B), packs against the imidazole ring of H50 (Fig. 6C). Side chains of N49, K60, and R79 (Fig. 6C) also make long-range contacts and are conserved among a subset of DM sequences (DSX, MAB-3a, DMRT1, and DMRT2; see Fig. 1B). There is no structural relationship between the DM module and classical zinc-binding domains. An unusual distance is observed between Zn2+ ions (9.3 Å), intermediate between that of the C6 binuclear cluster (3.1 Å; Baleja et al. 1992; Kraulis et al. 1992; Marmorstein et al. 1992) and those of C4–C4 nuclear hormone receptors (∼∼13 Å; Hard et al. 1990; Schwabe et al. 1990) or intertwined RING finger (14.2 Å; Borden et al. 1995).
Figure 6.
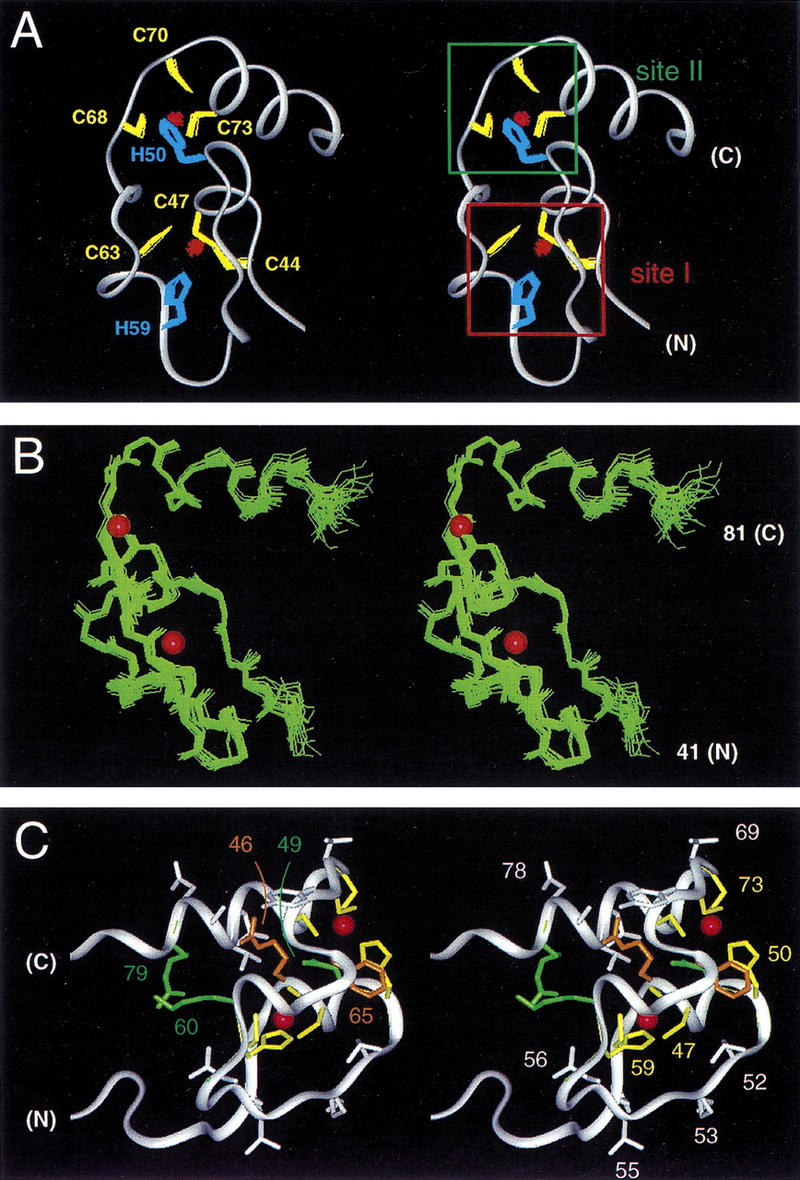
(A) Stereo ribbon representation of the solution structure of the DM domain (DSX residues 41–81). An ensemble of side chains of the coordinating cysteate (yellow) and histidine (blue) side chains and two bound Zn2+ atoms (red) are shown. The two intertwined zinc-binding sites are designated sites I and II (boxes at right). (B) Ensemble of main chain structures (stereo pair) aligned according to the main chain atoms of residues 41–81. The positions of Zn2+ atoms are shown by red spheres (50% of van der Waal radius). (C) Structural relationships among side chains of conserved (green), otherwise ordered (white), or invariant ligand-binding (green) residues. Ligand-binding residues are C44, C47, H59, C63 and H50, C68, C70, C73 (see Fig. 1B). Conserved residues N49 and R79 are shown in green whereas side chains of R46 and F65 are shown in gold to highlight structural environments (see text). Otherwise well-order residues include side chains of N43, A45, R48, L52, K53, T55, L56, Y67, T69, L75, T76, A77, and D78; criterion for inclusion is an ensemble side chain RMSD of <1.0 Å.
The tail contains a nascent DNA recognition helix
Although the carboxy-terminal tail is largely disordered, a strong helical propensity was predicted (Erdman and Burtis 1993). To resolve this apparent discrepancy, the tail's mean conformation was investigated by CD. Comparison of the intact domain and DMΔ (Fig. 7A, a,b) demonstrates that the tail forms a nascent α-helix at low temperature. The inferred helix content of DMΔ (20% or 11 residues) is in accord with its structure (Fig. 7A, b). The intact domain at 4°C (solid line in Fig. 7A, a) exhibits a greater helix content (30% or 21 residues). Their difference spectrum at 4°C exhibits an overall helical signature (Δ in Fig. 7A, c) whose normalization ([Δ] in Fig. 7A, d) predicts that the tail contains at least 10 additional residues of helix (in a static model) or subpopulation of helical structures in an ensemble (dynamic model) as implied by NMR features. The helical character of the tethered tail at 4°C is greater than that of an isolated peptide containing the carboxy-terminal 19 residues (DM-p in Fig. 7A, d), presumably due to nucleation by helix 2.
Figure 7.
The DM tail is a nascent α-helix that folds on DNA binding. (A,B) CD studies of free (A) and bound (B) DSX domain. (A,a) Spectra of intact domain at 4°C (solid line) and 37°C (dotted line); (A,b) corresponding spectra of DMΔ. (A,c) Difference spectra: (solid line a), between DM spectra at 4° and 37°C; (dashed dotted line Δ), between DM and DMΔ at 4°C; (dotted line b), baseline attenuation of stable helices in DMΔ between 4° and 37°C. That difference spectra a and Δ are similar suggests that the labile segment in DM corresponds to the missing element in DMΔ, i.e., a nascent carboxy-terminal helix. (A,d). Difference spectrum [Δ] (dashed dotted line), obtained from Δ normalized relative to 19 deleted residues, exhibits a helical signature. For comparison, spectra of isolated carboxy-terminal peptide DM-p 19 residues; underlined in Fig. 1B) are shown in d at 4° and 37°C (solid and dotted lines). Magnitude of [θ] at 222 nm (P[θ]222) is similar to that of difference spectra Δ and a in A,c. (B) Comparison of spectra of protein–DNA complexes (solid line), free domain (dashed dotted line), and free DNA (dotted or dashed line) at 4°C (B,a) and 37°C (B,b). (B,c) CD melting curves of free DM domain (dashed dotted line) and specific 2:1 complex (solid line) as monitored at 222 nm from 4° to 95°C. Whereas free domain exhibits steep loss in mean residue ellipticity with increasing temperature, the complex exhibits an attenuation rate (d[θ]222/dT) typical of pretransition regime (arrows). (B,d) CD difference spectra at 37°C: (solid line Δ1) between complex and the free DNA; (solid line Δ2), between the spectrum of a complex and the sum of the spectra of its isolated parts. Induced helical feature in far UV is labeled. Arrow in near-UV indicates difference feature arising from change in DNA structure on protein binding. Peptide and DNA concentrations were 10 and 5 μm, respectively. For reference, the spectrum of free DM domain is also shown (dashed dotted line).
Structural independence of the zinc module and tail suggests uncoupled thermal unfolding transitions. Indeed, whereas the CD spectrum of DMΔ changes little between 4° and 37°C, the spectrum of the intact domain undergoes marked thermal attenuation (Fig. 7A, a). The domain's thermal unfolding exhibits an exaggerated and noncooperative overall transition (Fig. 7B, c). The difference spectrum of the intact domain at 4° and 37°C (solid line in Fig. 7A, c) is α-helical in character and similar in magnitude to that between DM and DMΔ at 4°C (dashed dotted line in Fig. 7A, c). Because the zinc module's NMR resonances do not undergo significant changes in this temperature range, we ascribe the domain's attenuation in helix content to thermal fraying of the tail. Similar CD patterns have previously been seen in studies of DNA-binding motifs containing nascent α-helical arms motifs (Weiss et al. 1990; Ferre-D'Amare et al. 1994; Carroll et al. 1997).
To investigate the function of the tail, binding of the DM domain to dsxA was monitored by CD (Fig. 7B). Experiments at 4° and 37°C are shown in Figure 7B, a and b. Difference spectra at 37°C (Fig. 7B, d) indicate a change in structure of both DNA (250–300 nm; arrow in Fig. 7B, d) and protein (200–250 nm; induced α-helix). The α-helical difference feature is more prominent at 37°C than 4°C due to thermal fraying of the free tail; the DNA difference feature is not significantly affected by temperature. DNA-stabilized α-helical structure is maintained in the complex at 4–40°C as indicated by its melting curve (Fig. 7B, c). Negligible CD difference features were observed in control studies using λ operator site OL1 (not shown). Similar temperature- and DNA-dependent α-helical CD difference features accompany the induced fit of nascent α-helical arms in otherwise unrelated DNA-binding motifs (Weiss 1990; Ferre-D'Amare et al. 1994; Carroll et al. 1997). These results demonstrate that the DM tail folds on DNA binding but do not resolve whether the DNA-stabilized structure consists of a single continuous helix or discrete segments. The DM domain's DNA-specific difference feature, unrelated to those of previous CD studies, may reflect the structural accommodation of an α-helix in the minor groove.
The DM tail may be stabilized on DNA binding by either tail–DNA contacts or DNA-induced dimerization (or both). To test whether specific binding of a DM monomer to DNA is sufficient to stabilize the tail, CD studies were repeated using an isolated dsxA half site (5′-AGTACATTG-3′ and complement). The domain forms a low affinity 1:1 complex and on binding at 37°C yields a similar α-helical CD difference feature. Furthermore, formation of a 1:1 half-site complex is sufficient to confer thermal stability between 4° and 40°C. DNA-dependent folding can thus occur without dimerization. Direct tail–DNA contacts are also suggested by GMSA studies of an intersex variant (R91Q; see Fig. 1B): DNA recognition is impaired (as previously reported; Erdman and Burtis 1993) but DNA-dependent dimerization is retained (see Fig. 4C). These data strongly suggest that this conserved basic side chain—and hence the tail itself—contributes to the protein–DNA interface; participation of other tail residues in dimerization cannot be excluded. Mutational analyses (Erdman and Burtis 1993; U. Narendra and M.A. Weiss, unpubl.) suggest that additional DNA contacts are made by the Zn module.
Discussion
The present study has defined the structure of the DM motif and its overall mode of DNA recognition. Whereas the genetic regulation of mammalian DM proteins (and in particular their relationship to SRY) is not understood, dsx and mab-3 have been characterized extensively (Fig. 1B; Cline and Meyer 1996). Intersex phenotypes demonstrate that only a subset of dimorphic lineages in either D. melanogaster or C. elegans are affected by DM mutations (Hildreth 1965; Shen and Hodgkin 1988), reflecting the function of dsx and mab-3 in one branch of a ramifying pathway. Downstream pathways are not well defined. A model DSX responsive enhancer (the fbe; Burtis et al. 1991; Coschigano and Wensink 1993) has been isolated in yolk-protein (yp) promoters, active in the fat body of the female adult fly and inactive in the male (Fig. 2B,C; An and Wensink 1995a,b). Although an analogous MAB-3 responsive element has not been characterized, positive regulatory sites have been defined in yolk protein-related vitellogenin (vit) promoters. Mutations that block binding of MAB-3 in vitro also attenuate sex-specific transcriptional regulation in vivo (Yi and Zarkower 1999). Whether the ubiquity of the DM motif indicates conservation of other downstream genes is unknown.
A nascent tail is required for DNA recognition and biological function
The structure of the DM zinc module, like the C4–C4 DBD of the nuclear hormone-receptor superfamily, contains two Zn2+ atoms and adjoining α-helices in a unified core. Although unrelated in detail, these motifs are substantially but not completely folded in the absence of DNA (Hard et al. 1990; Schwabe et al. 1990; Holmbeck et al. 1998). Like the glucocorticoid and estrogen receptor (GR and ER) DBDs in particular, the DM domain recognizes pseudopalindromic DNA sites as an induced homodimer (Luisi et al. 1991; Schwabe et al. 1993). Insertion of a symmetrical central base pair to make a palindromic DNA element in each case leads to a 100-fold reduction in affinity with retention of cooperativity (Luisi et al. 1991; Lundback et al. 1994). These similarities belie fundamental differences. Whereas the nuclear hormone receptor DBD consists of two C4 substructures encoded by separate exons, the DM motif contains intertwined CCHC and HCCC ligands encoded by a single exon (Burtis and Baker 1989; Raymond et al. 1998). Whereas the C4–C4 DBD binds in DNA's major groove, the DM domain binds in the minor groove.
DNA recognition by the DM domain requires a carboxy-terminal basic tail. The present studies demonstrate that the tail undergoes an independent and noncooperative helix–coil equilibrium. These observations are reminiscent of DNA-dependent folding transitions among the basic arms of major groove-binding motifs (Weiss et al. 1990; Ferre-D'Amare et al. 1994; Carroll et al. 1997). Whereas the dimensions of the major groove of B-DNA are commensurate with those of an α-helix, the minor groove is ordinarily too narrow. DM–DNA recognition thus provides an unusual example of the minor groove acting as a template for protein folding (Yang and Steitz 1995). The R91Q intersex mutation in DSX demonstrates that a native tail is necessary for DNA recognition and sex-specific gene regulation in vivo. Tail sequences shared an overall helical propensity. Conservation of tail sequences is more stringent than within the zinc module (exclusive of the invariant cysteines and histidines; Fig. 1B). The DSX tail is similar, for example, to those of MAB-3b and the vertebrate DMRT and TERRA families: the motif L(V/T)X(D/E)RQRVMA(A/L)Q(V/T)-ALRR(Q/A)QA (DSX residues 75–94 including DSX intersex site R91Q; underlined) is invariant. We propose that this segment functions as a DNA recognition helix and therefore, defines a subgroup of DM proteins of related specificity. Such tail–DNA contacts are presumably extended by interactions between the DNA and the Zn module itself.
The tail of MAB-3a is strikingly divergent: nonconservative substitutions are highlighted by arrows in Figure 1B. In particular, MAB-3a contains Ile rather than Arg at DSX intersex site R91Q. Such divergence, retained in C. briggsae (V91; Yi and Zarkower 1999), suggests that MAB-3a recognizes a different DNA sequence or is positioned differently relative to a DNA half site. Indeed, although DSX and MAB-3 exhibit overlapping specificities, random binding-site selection has revealed important differences (Yi and Zarkower 1999). In particular, MAB-3's consensus sequence exhibits marked asymmetry (5′-AATGTTGCGA(T/A)NT-3′ and complement), which contrasts with near-palindromic dsxA half-sites. Because respective DSX and MAB-3 target sites are similar in length, MAB-3 is proposed to bind DNA as an “internal dimer” of DM domains. We imagine that the divergent tails of MAB-3a and MAB-3b contact asymmetric half-sites with different intrinsic specificity. Whereas the genetic function of MAB-3 requires both domains (Raymond et al. 1998), other known DM proteins contain a single domain. We speculate that such proteins undergo DNA-dependent homo- or heterodimerization as an example of combinatorial gene regulation.
DNA-binding properties of the DM motif rationalize the organization of a DSX response element
The fbe, a model DSX response element in a dimorphic tissue, contains three overlapping factor-binding sites (An and Wensink 1995a,b; Fig. 2B). Binding of DSX to the central site (dsxA; red box in Fig. 2B) is proposed to regulate binding of tissue-specific factors to flanking sites (aef1 and bzip1; Fig. 2C). Critical bases in dsxA recognized by DSX are palindromic about a central base pair (Fig. 2B, bottom; An and Wensink 1995b; Erdman et al. 1996). The associated tissue-specific factors (AEF1 and an uncharacterized bZIP transcription factor) contain conserved DNA-binding motifs (classic zinc fingers and basic region-leucine zipper, respectively). In the fat body coordinate binding of DSXF and bZIP1 to the fbe displaces AEF1 and recruits an unknown activator to proximal site ref1, resulting in fat body-specific activation (Fig. 2C, top; other regulatory features are summarized in the caption). Overlapping DNA target sites are thus proposed to provide a mechanism by which sex- and tissue-specific signals are integrated to regulate gene expression (An and Wensink 1995a,b). Although the biochemical basis of fbe regulation remains speculative, functional integration of its discrete factor-binding sites has been demonstrated in transgenic flies. Such features are likely to reflect general principles of transcriptional activation in eukaryotes (Tjian and Maniatis 1994).
Genetic dissection of the fbe has novel and previously unrecognized structural consequences. Because bZIP proteins bind within the major groove (Ellenberger et al. 1992; Glover and Harrison 1995; Keller et al. 1995; Chen et al. 1998), the An-Wensink model (Fig. 2C) predicts that the DM domain—like SRY but unlike classic zinc fingers and other zinc modules—would bind in the minor groove. The model further requires that the DM domain—unlike SRY and other HMG boxes—would not induce sharp DNA bending lest associated compression of the major groove displaces bZIP1. The present studies have tested and confirmed these implicit predictions. Minor groove binding without bending is unusual and may be analogous to that of the T domain–DNA complex of the Brachyury transcription factor (Muller and Herrmann 1997). The putative ternary DSXF–bZIP1–DNA complex would be remarkable for adaptive binding of one nascent α-helix in the DNA major groove (bzip1) and of another nascent α-helix in the overlapping DNA minor groove (dsxA). Methylphosphonate interference experiments suggest intimate juxtaposition of DSXF and bZIP1 complexes. bZIP1–phosphate contacts (blue circles in Fig. 2D), inferred from crystal structures of the GCN4 bZIP–DNA complex (Ellenberger et al. 1992; Keller et al. 1995), overlap sites of DSX–methylphosphonate interference (red circles in Fig. 2D). Such interdigitation of protein–DNA phosphate contacts is consistent with a previous “missing base” interference assay suggesting corecognized bases (An and Wensink 1995b). Simultaneous occupancy of major and minor grooves is likely to be a general feature of enhanceosome assembly (Tjian and Maniatis 1994).
The mechanism of synergistic transcriptional activation between DSXF and bZIP1 (Fig. 2C; An and Wensink 1995a,b) is not well understood. Studies of mammalian liver-specific gene regulation by C/EBPα and NF-Y have demonstrated that synergy can occur in the absence of cooperative promoter binding; binding of C/EBP in fact impairs binding of NF-Y and yet their simultaneous binding leads to synergistic activation of a minimal promoter (Milos and Zaret 1992). Nonetheless, synergy between by CCAAT enhancer-binding factor α (C/EBPα) and nuclear factor Y (NF-Y) results in the formation of a preinitiation complex that is stable through multiple rounds of transcription. Additional biochemical studies will be required to distinguish whether (1) DNA binding by DSXF and bZIP1 is cooperative (a thermodynamic mechanism), (2) the ternary DSXF–DNA–bZIP1 complex dissociates more slowly than either binary complex (a kinetic mechanism), or (3) binding of DSX induces a conformational change in bZIP1 enhancing its potency in transcriptional activation (an allosteric mechanism). The mechanisms by which DSXM represses bzip1 are also not well understood.
Mutations in DSX cause allele-specific changes in courtship behavior
doublesex is representative of a class of genes that not only specify aspects of body plan but also influence behavior (for review, see Yamamoto et al. 1996). Chromosomal female (XX) flies expressing DSXM, although male in external appearance, do not court wild-type females (Villella and Hall 1996). Chromosomal male (haplo-X) flies bearing a deletion in one dsx allele and various point mutations in the other exhibit specific and quantifiable changes in courtship behavior (Hall 1994; Villella and Hall 1996). The haplo-X dsx phenotype differs from that associated with fruitless (male chaining; Ito et al. 1992; Villella et al. 1997). DSX-associated anomalies include an altered courtship song, selective lack of vigor in the pursuit of wild-type females, increased elicitation of courtship from wild-type males, and decreased rejection of subsequent attempted copulation (Villella and Hall 1996). The mechanics of courtship and individual elements of mating behavior are normal with the exception of the extent of abdominal bending during attempted copulation and altered song (Villella and Hall 1996). The latter is of particular interest: the humming component is absent, whereas changes in the rhythmicity of acoustic pulses (as associated with mutations in the period gene; Bargiello et al. 1984; Reddy et al. 1984) are not observed. Unlike fruitless, dsx does not regulate the male-specific induction or female-specific suppression of the abdominal muscle of Lawrence (Gailey et al. 1991), which thus represents a dsx-independent branch of the ramified sex-determining pathway (Fig. 1A).
The extent and spectrum of behavioral change associated with the various point mutations in dsx are allele specific. The biochemical bases of these phenotypic differences have not been elucidated. Courtship behavior reflects the complex integration of multiple systems, including pheromone production, elaboration, and sensing; development of the peripheral nervous system and its central ramifications; and function of a putative courtship command center in the brain (Hall 1977; Villella and Hall 1996). Whether and how dsx influences the differentiation of the male brain are not well understood. Although dsx controls the sex-specific pattern of post embryonic proliferation of neuroblasts in an abdominal ganglion (Taylor and Truman 1992), its effect on patterning of the central nervous system (and in particular, on the function of a courtship command center; Villella and Hall 1996) is not well characterized. In light of the allele-specific effects of mutations on extent of courtship anomalies, the DSX pathway promises to provide an attractive model for study of the biochemical basis of a complex behavior.
Materials and methods
Protein synthesis
Solid phase peptide synthesis used an Applied Biosystems 430A synthesizer. t-Boc amino acids were used with the following protection: Arg(Tosyl), Asn(Xanthyl), Asp(OcHxl), Cys(4MeBzl), Glu(OcHxl), His(DNP), Lys(2ClZ), Ser(Bzl), Thr(Bzl), and Tyr(BrZ). Peptide DSX(35–67) was synthesized on an carboxy-terminal thioester-generating resin. The peptides DSX(68–105) and acetyl-DSX(36–105) were synthesized on a Boc–Glu(OcHxl)–OCH2–Pam resin, and DSX(68–86) on a Boc–Gln–OCH2–Pam resin. Full-length polypeptides were prepared by native chemical ligation (Dawson et al. 1994). The amino-terminal peptide consisted of DSX residues 35–67; the carboxy-terminal peptide consisted of residues 68–105 (DM domain) or 68–86 (DMD fragment). The R91Q variant domain was prepared similarly. Ligation products were purified by reverse-phase HPLC and characterized by electrospray mass spectrometry. Residue numbers refer to native sequence.
Analytical ultracentrifugation
Self-association was characterized in 50 mm Tris-HCl (pH 6.5) at 25°C using a 60Tvi rotor and Beckman model XL-A (20,000 rpm) at a concentration of 70 and 140 μm. Although data were inconsistent with a monomer–dimer model, the domain does not behave as an ideal species; fitting yielded an apparent molecular mass (9350 ± 590 Da) larger than that of the monomer (8481 Da), presumably due to hydrodynamic consequences of the disordered tail. The sedimentation equilibrium of DMΔ (DSX residues 35–86) was consistent with an ideal monomer of expected mass.
DNA-binding assays
Specific DNA binding was monitored by GMSA (10% acrylamide) in 0.5x TBE [1x, 89 mm Tris-borate (pH 8.0), and 2 mm ethylene-diamine-tetraacetic acid] at 175 V and 4°C. Sequences are given in Figure 3; binding was studied in 10 mm Tris-HCl (pH 7.4), 50 mm KCl, 0.1 mm ZnCl2, 34 μg/ml bovine serum albumin, and 0.6 ng/μl poly[d(I–C)] DNA (unlabeled). The concentration of 33P-labeled DNA was 1 nm; binding in Figure 4 was at higher salt (150 mm KCl) and nonspecific competitor DNA (0.8 ng/μl poly[d(I–C)] to limit nonspecific binding to the fbe probe. GMSA studies of DNA analogs containing systematic AT → IC substitutions used sequences 5′-TCGACACAICCICICCGCCGCIICCAGCTAGCC-3′ annealed to 3′-AGCTGTGTCGICGCIICIICGCCIGTCGATCGG-5′. Methylphosphonate interference was evaluated in a 15-bp dsxA duplex (Labeots and Weiss 1997). To obtain predicted bZIP contacts (Fig. 2D), bzip1 sequence was aligned with ATF/CREB site of yeast activator GCN4 (5′-GAGATGACGTCATCC-3′; Keller et al. 1995). Underlined bases indicate homology with bzip1 (5′-ACAATGTTGCAATCAGCGG-3′; An and Wensink 1995a,b).
DNA-bending assay
An fbe duplex (5′-GTGCACAACTACAATGTTGCAATCAG-3′ and complement) containing dsxA was cloned in unique EcoR1 site of pBluescript-M13+ (Stratagene Inc., La Jolla, CA). Probes of equal length, with the binding site at varying distance from ends were generated by PCR and 5′-labeled with [33P]ATP and T4 polynucleotide kinase. Binding reactions (25 μl) contained 150 mm KCl, 20 mm Tris-HCl (pH 7.4), 5 mm MgCl2, 20 ng poly[d(I–C)], 0.8 ng bovine serum albumin, 5% glycerol, approximately 1 nm 33P-γ-labeled DNA probe, and 24 nm purified DSX DM domain. Control bending reactions contained the SRY HMG box (King and Weiss 1993) at a concentration of 4 nm in 150 mm KCl, 60 mm Tris-HCl (pH 7.4), 5 mm MgCl2, 33 ng poly[d(I–C)], 0.8 ng of bovine serum albumin, and 5% glycerol.
Circular dichroism
Spectra were obtained using an Aviv spectropolarimeter. Helix contents were estimated by magnitude of mean residue ellipticity at 222 nm, assuming that 100% α-helix corresponds to a value of −33,000 at 4°C. DNA-binding studies used site 5′-CACTACAATGTTGCA-3′ and complement (target site underlined). The protein concentration was 10 μm in a 50 mm Tris-HCl (pH 6.5) buffer containing 50 mm KCl and a 2.5-fold molar excess of ZnCl2. The DNA concentration was 5 μm.
NMR spectroscopy
Samples were prepared as in CD studies using deuterated Tris-HCl buffer and protein concentrations of 60 μm, 460 μm, and 1.4 mm (intact domain) and 2 mm (DMΔ and DM-p). Solutions were purged with N2 before use. Experiments were performed at 25°C on Varian Inova 400, 500, and 600 MHz spectrometers: NOEsY (mixing times, 75, 175, 300 msec), DQF-COSY, and TOCSY (mixing time, 55 msec) in H2O and D2O as described (Qian et al. 1993).
Structural calculations
Models were obtained by DG/SA using X-PLOR (Brünger 1992) and visualized using Insight-II (Molecular Simulations, Inc., San Diego, CA). Distance and torsion angles restraints were derived as described (Qian et al. 1993). Root mean square deviation (RMSD) was calculated using program Superpose (Diamond 1992). RMSD for each residue was calculated using X-PLOR. The SA ensemble exhibits a main-chain RMSD of 0.43 Å. The eight Zn2+-binding residues are characterized with a precision of 0.13 Å (main chain RMSD) and 0.15 Å (side chain RMSD). In refined models respective Zn–S′′γ and Sγ–Sγ distances in metal-binding sites were restrained to be 2.2–2.4 Å and 3.6–3.9 Å; Zn–N and N–SÅ distances were restrained to be 1.9–2.1 Å and 3.4–3.8 Å.
Assignment of hydrogen bond
Protected amide resonances in D2O were observed at 25°C in one-dimensional 1H-NMR and TOCSY spectra obtained immediately after dissolution of Zn–DM powder. Protected amide protons were assigned as putative hydrogen bond donors; acceptors were identified based on preliminary DG/SA models. Two distance restraints were imposed per hydrogen bond. Nine peptide–carbonyl and seven amide–sulfur hydrogen bonds were identified. The former constraints were NH-O (1.8–2.6 Å) and HN-O (2.7–3.3 Å); the latter constraints were NH-Sγ (2.2–2.8 Å) and HN-Sγ (3.1–3.7 Å).
Acknowledgments
We thank P.K. Donahoe, E. Haas, A. Mahowald, and J. Rowley for discussion; K. Hallenga, Q.X. Hua, V. Ittah, D. Jones, L. Milos, M. ter Horst, and B. Wang for advice regarding methods; B. Li for database searches, M. Jeuland for assistance with gel shifts, and W. Jia for assistance with figures. M.A.W. thanks A. Kossiakoff for encouraging the continuation of these studies in Cleveland. This work was supported by a grant from the NIH to M.A.W. and by Gryphon Sciences, Inc. L.-Y.Z. was an NIH Post-Doctoral Fellow in the Cardiovascular Pathophysiology and Biochemistry Training Program at The University of Chicago (5T32HL07237).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL weiss@biochemistry.cwru.edu; FAX (216) 368-3419.
References
- An W, Wensink PC. Integrating sex- and tissue-specfic regulation within a single Drosophila enhancer. Genes & Dev. 1995a;9:256–266. doi: 10.1101/gad.9.2.256. [DOI] [PubMed] [Google Scholar]
- ————— Three protein binding sites form an enhancer that regulates sex- and fat body-specific transcription of Drosophila yolk protein genes. EMBO J. 1995b;14:1221–1230. doi: 10.1002/j.1460-2075.1995.tb07105.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- An W, Cho S, Ishii H, Wensink PC. Sex-specific and non-sex-specific oligomerization domains in both of the doublesex transcription factors from Drosophila melanogaster. Mol Cell Biol. 1996;16:3106–3111. doi: 10.1128/mcb.16.6.3106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baker BS, Wolfner MF. A molecular analysis of doublesex, a bifunctional gene that controls both male and female sexual differentiation in Drosophila melanogaster. Genes & Dev. 1988;4:477–489. doi: 10.1101/gad.2.4.477. [DOI] [PubMed] [Google Scholar]
- Baleja JD, Marmorstein R, Harrison SC, Wagner G. Solution structure of the DNA-binding domain of Cd2+-GAL4 from S. cerevisiae. Nature. 1992;356:450–453. doi: 10.1038/356450a0. [DOI] [PubMed] [Google Scholar]
- Bargiello TA, Jackson FR, Young MW. Restoration of circadian behavioural rhythms by gene transfer in Drosophila. Nature. 1984;312:752–754. doi: 10.1038/312752a0. [DOI] [PubMed] [Google Scholar]
- Bennett CP, Docherty Z, Robb SA, Ramani P, Hawkins JR, Grant D. Deletion 9p and sex reversal. J Med Genet. 1993;30:518–520. doi: 10.1136/jmg.30.6.518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berta P, Hawkins JR, Sinclair AH, Taylor A, Griffiths BL, Goodfellow PN, Fellous M. Genetic evidence equating SRY and the testis-determining factor. Nature. 1990;348:448–450. doi: 10.1038/348448A0. [DOI] [PubMed] [Google Scholar]
- Bewley CA, Gronenborn AM, Clore GM. Minor groove-binding architectural proteins: Structure, function, and DNA recognition. Ann Rev Biophys Biomol Struc. 1998;27:105–131. doi: 10.1146/annurev.biophys.27.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borden KL, Boddy MN, Lally J, O'Reilly NJ, Martin S, Howe K, Solomon E, Freemont PS. The solution structure of the RING finger domain from the acute promyelocytic leukaemia proto-oncoprotein PML. EMBO J. 1995;14:1532–1541. doi: 10.1002/j.1460-2075.1995.tb07139.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bownes M, Dempster M, Blair M. Expression of the yolk-protein genes in the mutant doublesex dominant (dsxD) of Drosophila melanogaster. J Embryol Exper Morph. 1983;75:241–257. [PubMed] [Google Scholar]
- Brünger T. X-PLOR version 3.1: A system for crystallography and NMR. New Haven, CT: Yale University; 1992. [Google Scholar]
- Burtis KC, Baker BS. Drosophila doublesex gene controls somatic sexual differentiation by producing alternatively spliced mRNAs encoding related sex-specific polypeptides. Cell. 1989;56:997–1010. doi: 10.1016/0092-8674(89)90633-8. [DOI] [PubMed] [Google Scholar]
- Burtis KC, Coschigano KT, Baker BS, Wensink PC. The doublesex proteins of Drosophila melanogaster bind directly to a sex-specific yolk protein gene enhancer. EMBO J. 1991;10:2577–2582. doi: 10.1002/j.1460-2075.1991.tb07798.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll AS, Gilbert DE, Liu X, Cheung JW, Michnowicz JE, Wagner G, Ellenberger TE, Blackwell TK. SKN-1 domain folding and basic region monomer stabilization upon DNA binding. Genes & Dev. 1997;11:2227–2238. doi: 10.1101/gad.11.17.2227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- C. elegans Sequencing Consortium. Genome sequence of the nematode C. elegans: A platform for investigating biology. Science. 1998;282:2012–2018. doi: 10.1126/science.282.5396.2012. [DOI] [PubMed] [Google Scholar]
- Chen L, Glover JN, Hogan PG, Rao A, Harrison SC. Structure of the DNA-binding domains from NFAT, Fos and Jun bound specifically to DNA. Nature. 1998;392:42–48. doi: 10.1038/32100. [DOI] [PubMed] [Google Scholar]
- Cho S, Wensink PC. DNA binding by the male and female doublesex proteins of Drosophila melanogaster. J Biol Chem. 1997;272:3185–3189. doi: 10.1074/jbc.272.6.3185. [DOI] [PubMed] [Google Scholar]
- ————— Linkage between oligomerization and DNA binding in Drosophila doublesex proteins. Biochemistry. 1998;37:11301–11308. doi: 10.1021/bi972916x. [DOI] [PubMed] [Google Scholar]
- Cline TW, Meyer BJ. VIVE LA DIFFÉRENCE: Males vs females in flies vs worms. Annu Rev Genet. 1996;30:637–702. doi: 10.1146/annurev.genet.30.1.637. [DOI] [PubMed] [Google Scholar]
- Coschigano KT, Wensink PC. Sex-specific transcriptional regulation by the male and female doublesex proteins of Drosophila. Genes & Dev. 1993;7:42–54. doi: 10.1101/gad.7.1.42. [DOI] [PubMed] [Google Scholar]
- Crocker M, Coghill SB, Cortinho R. An unbalanced autosomal translocation (7:9) associated with feminization. Clin Genet. 1988;34:70–73. doi: 10.1111/j.1399-0004.1988.tb02618.x. [DOI] [PubMed] [Google Scholar]
- Dawes HE, Berlin DS, Lapidus DM, Nusbaum C, Davis TL, Meyer BJ. Dosage compensation proteins targeted to X chromosomes by a determinant of hermaphrodite fate. Science. 1999;284:1800–1804. doi: 10.1126/science.284.5421.1800. [DOI] [PubMed] [Google Scholar]
- Dawson PE, Muir TW, Clark-Lewis I, Kent SB. Synthesis of proteins by native chemical ligation. Science. 1994;266:776–779. doi: 10.1126/science.7973629. [DOI] [PubMed] [Google Scholar]
- Diamond R. On the multiple simultaneous superposition of molecular structures by rigid body transformations. Protein Sci. 1992;1:1279–1287. doi: 10.1002/pro.5560011006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellenberger TE, Brand CJ, Struhl K, Harrison SC. The GCN4 basic region leucine zipper binds DNA as a dimer of uninterrupted α-helices: Crystal structure of the protein–DNA complex. Cell. 1992;71:1223–1237. doi: 10.1016/s0092-8674(05)80070-4. [DOI] [PubMed] [Google Scholar]
- Erdman SE, Burtis KC. The Drosophila doublesex proteins share a novel zinc-finger related DNA-binding domain. EMBO J. 1993;12:527–535. doi: 10.1002/j.1460-2075.1993.tb05684.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdman SE, Chen HJ, Burtis KC. Functional and genetic characterization of the oligomerization and DNA binding properties of the Drosophila doublesex proteins. Genetics. 1996;144:1639–1652. doi: 10.1093/genetics/144.4.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferre-D'Amare AR, Pognonec P, Roeder RG, Burley SK. Structure and function of the b/HLH/Z domain of USF. EMBO J. 1994;13:180–189. doi: 10.1002/j.1460-2075.1994.tb06247.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flejter WL, Fergestad J, Gorski J, Varvill T, Chandrasekharappa S. A gene involved in XY sex reversal is located on chromosome 9, distal to marker D9S1779. Am J Hum Genet. 1998;63:794–802. doi: 10.1086/302016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankel AD, Berg JM, Pabo CO. Metal-dependent folding of a single zinc finger from transcription factor IIIA. Proc Natl Acad Sci. 1987;84:4841–4845. doi: 10.1073/pnas.84.14.4841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fryns JP, Kleczkowska A, Casaer P, van den Berghe H. Double autosomal chromosomal aberration (3p trisomy/9p monosomy) and sex-reversal. Ann Genet. 1986;29:49–52. [PubMed] [Google Scholar]
- Gailey DA, Taylor BJ, Hall JC. Elements of the fruitless locus regulate development of the muscle of Lawrence, a male-specific structure in the abdomen of Drosophila melanogaster adults. Development. 1991;113:879–890. doi: 10.1242/dev.113.3.879. [DOI] [PubMed] [Google Scholar]
- Giese K, Cox J, Grosschedl P. The HMG domain of lymphoid enhancer factor 1 bends DNA and facilitates assembly of functional nucleoprotein structures. Cell. 1992;69:185–195. doi: 10.1016/0092-8674(92)90129-z. [DOI] [PubMed] [Google Scholar]
- Glover JN, Harrison SC. Crystal structure of the heterodimeric bZIP transcription factor c-Fos–c-Jun bound to DNA. Nature. 1995;373:257–261. doi: 10.1038/373257a0. [DOI] [PubMed] [Google Scholar]
- Gubbay J, Collignon J, Koopman P, Capel B, Economou A, Munsterberg A, Vivian N, Goodfellow P, Lovell-Badge R. A gene mapping to the sex-determining region of the mouse Y chromosome is a member of a novel family of embryonically expressed genes. Nature. 1990;346:245–250. doi: 10.1038/346245a0. [DOI] [PubMed] [Google Scholar]
- Guioli S, Schmitt K, Critcher R, Bouzyk M, Spurr NK, Ogata T, Hoo JJ, Pinsky L, Gimelli G, Pasztor L, et al. Molecular analysis of 9p deletions associated with XY sex reversal: Refining the localization of a sex-determining gene to the tip of the chromosome. Am J Hum Genet. 1998;63:905–908. doi: 10.1086/302017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall JC. Portions of the central nervous system controlling reproductive behavior in Drosophila melanogaster. Behav Genet. 1977;7:291–312. doi: 10.1007/BF01066800. [DOI] [PubMed] [Google Scholar]
- ————— The mating of a fly. Science. 1994;264:1702–1714. doi: 10.1126/science.8209251. [DOI] [PubMed] [Google Scholar]
- Haqq CM, King CY, Ukiyama E, Falsafi S, Haqq TN, Donahoe PK, Weiss MA. Molecular basis of mammalian sexual determination: Activation of Mullerian inhibiting substance gene expression by SRY. Science. 1994;266:1494–1500. doi: 10.1126/science.7985018. [DOI] [PubMed] [Google Scholar]
- Hard T, Kellenbach E, Boelens R, Maler BA, Dahlman K, Freedman LP, Carlstedt-Duke J, Yamamoto KR, Gustafsson JA, Kaptein R. Solution structure of the glucocorticoid receptor DNA-binding domain. Science. 1990;249:157–160. doi: 10.1126/science.2115209. [DOI] [PubMed] [Google Scholar]
- Hildreth PE. Doublesex, a recessive gene that transforms both males and females of Drosophila into intersexes. Genetics. 1965;51:659–678. doi: 10.1093/genetics/51.4.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hodgkin J. Male phenotypes and mating efficiency in Caenorhabditis elegans. Genetics. 1983;103:43–64. doi: 10.1093/genetics/103.1.43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Sex determination and dosage compensation in Caenorhabditis elegans. Annu Rev Genet. 1997;21:133–154. doi: 10.1146/annurev.ge.21.120187.001025. [DOI] [PubMed] [Google Scholar]
- Holmbeck SMA, Dyson HJ, Wright PE. DNA-induced conformational changes are the basis for cooperative dimerization by the DNA binding domain of the retinoid X receptor. J Mol Biol. 1998;3:533–539. doi: 10.1006/jmbi.1998.2207. [DOI] [PubMed] [Google Scholar]
- Ito H, Fujitani K, Usui K, Shimizu-Nishikawa K, Tanaka S, Yamamoto D. Sexual orientation in Drosophila is altered by the satori mutation in the sex-determination gene fruitless that encodes a zinc finger protein with a BTB domain. Proc Natl Acad Sci. 1992;93:9687–9692. doi: 10.1073/pnas.93.18.9687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller W, Konig P, Richmond TJ. Crystal structure of a bZIP/DNA complex at 2.2 Å: Determinants of DNA specific recognition. J Mol Biol. 1995;254:657–667. doi: 10.1006/jmbi.1995.0645. [DOI] [PubMed] [Google Scholar]
- King CY, Weiss MA. The SRY high-mobility-group box recognizes DNA by partial intercalation in the minor groove: A topological mechanism of sequence specificity. Proc Natl Acad Sci. 1993;90:11990–11994. doi: 10.1073/pnas.90.24.11990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klug A, Schwabe JW. Protein motifs 5. Zinc fingers. FASEB J. 1995;8:597–604. [PubMed] [Google Scholar]
- Koopman P, Munsterberg A, Capel B, Vivian N, Lovell-Badge R. Expression of a candidate sex-determining gene during mouse testis differentiation. Nature. 1990;348:450–452. doi: 10.1038/348450a0. [DOI] [PubMed] [Google Scholar]
- Kraulis PJ, Raine AR, Gadhavi PL, Laue ED. Structure of the DNA-binding domain of zinc GAL4. Nature. 1992;356:448–450. doi: 10.1038/356448a0. [DOI] [PubMed] [Google Scholar]
- Labeots LA, Weiss MA. Electrostatics and hydration at the homeodomain–DNA interface: Chemical probes of an interfacial water cavity. J Mol Biol. 1997;269:113–128. doi: 10.1006/jmbi.1997.1026. [DOI] [PubMed] [Google Scholar]
- Lee DK, Horikoshi M, Roeder RG. Interaction of TFIID in the minor groove of the TATA element. Cell. 1991;67:1241–1250. doi: 10.1016/0092-8674(91)90300-n. [DOI] [PubMed] [Google Scholar]
- Lee MS, Gippert GP, Soman KV, Case DA, Wright PE. Three-dimensional solution structure of a single zinc finger DNA-binding domain. Science. 1989;245:635–637. doi: 10.1126/science.2503871. [DOI] [PubMed] [Google Scholar]
- Li H, Baker BS. hermaphrodite and doublesex function both dependently and independently to control various aspects of sexual differentiation in Drosophila. Development. 1998;125:2641–2651. doi: 10.1242/dev.125.14.2641. [DOI] [PubMed] [Google Scholar]
- Luisi BF, Xu WX, Otwinowski Z, Freedman LP, Yamamoto KR, Sigler PB. Crystallographic analysis of the interaction of the glucocorticoid receptor with DNA. Nature. 1991;8:497–505. doi: 10.1038/352497a0. [DOI] [PubMed] [Google Scholar]
- Lundback T, Zilliacus J, Gustafsson JA, Carlstedt-Duke J, Hard T. Thermodynamics of sequence-specific glucocorticoid receptor-DNA interactions. Biochemistry. 1994;33:5955–5965. doi: 10.1021/bi00185a037. [DOI] [PubMed] [Google Scholar]
- Magenis RE, Allen LJ, Brownm MG, Dougherty TM, Black M, Hafits-Borchardt V, Toth-Fejel S, Lawce H, Smith L. 9p monosomy associated with XY gonadal dysgenesis: A contiguous gene syndrome? Am J Hum Genet (Suppl.) 1990;47:A33. [Google Scholar]
- Marin I, Baker BS. The evolutionary dynamics of sex determination. Science. 1998;281:1990–1994. doi: 10.1126/science.281.5385.1990. [DOI] [PubMed] [Google Scholar]
- Marmorstein R, Carey M, Ptashne M, Harrison SC. DNA recognition by GAL4: Structure of a protein–DNA complex. Nature. 1992;356:408–414. doi: 10.1038/356408a0. [DOI] [PubMed] [Google Scholar]
- McDonald MT, Flejter W, Sheldon S, Putzi MJ, Gorski JL. XY sex reversal and gonadal dysgenesis due to 9p24 monosomy. Am J Med Genet. 1997;73:321–326. [PubMed] [Google Scholar]
- Meng A, Morre B, Tang H, Yuan B, Lin S. A Drosophila doublesex-related gene, terra, is involved in somitogenesis in vertebrates. Development. 1999;126:1259–1268. doi: 10.1242/dev.126.6.1259. [DOI] [PubMed] [Google Scholar]
- Milos PM, Zaret KS. A ubiquitous factor is required for C/EBP-related proteins to form stable transcription complexes on an albumin promoter segment in vitro. Genes & Dev. 1992;6:991–1004. doi: 10.1101/gad.6.6.991. [DOI] [PubMed] [Google Scholar]
- Muller CW, Herrmann BG. Crystallographic structure of the T domain–DNA complex of the Brachyury transcription factor. Nature. 1997;389:884–888. doi: 10.1038/39929. [DOI] [PubMed] [Google Scholar]
- Nagoshi PN, McKeown M, Burtis KC, Belote JM, Baker BS. The control of alternative splicing at genes regulating sexual differentiation in D. melanogaster. Cell. 1988;53:229–236. doi: 10.1016/0092-8674(88)90384-4. [DOI] [PubMed] [Google Scholar]
- Page DC, Mosher R, Simpson EM, Fisher EM, Mardon G, Pollack J, McGillivray B, de la Chapelle A, Brown LG. The sex-determining region of the human Y chromosome encodes a finger protein. Cell. 1987;51:1091–1104. doi: 10.1016/0092-8674(87)90595-2. [DOI] [PubMed] [Google Scholar]
- Parkhurst KM, Brenowitz M, Parkhurst LJ. Simultaneous binding and bending of promoter DNA by the TATA binding protein: Real time kinetic measurements. Biochemistry. 1996;35:7459–7465. doi: 10.1021/bi9530301. [DOI] [PubMed] [Google Scholar]
- Pontiggia A, Rimini R, Harley VR, Goodfellow PN, Lovell-Badge R, Bianchi ME. Sex-reversing mutations affect the architecture of SRY-DNA complexes. EMBO J. 1994;13:6115–6124. doi: 10.1002/j.1460-2075.1994.tb06958.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Qian X, Gozani SN, Yoon H, Jeon CJ, Agarwal K, Weiss MA. Novel zinc finger motif in the basal transcriptional machinery: Three-dimensional NMR studies of the nucleic acid binding domain of transcriptional elongation factor TFIIS. Biochemistry. 1993;32:9944–9959. doi: 10.1021/bi00089a010. [DOI] [PubMed] [Google Scholar]
- Raymond CS, Shamu CE, Shen MM, Seifert KJ, Hirsch B, Hodgkin J, Zarkower D. Evidence for evolutionary conservation of sex-determining genes. Nature. 1998;391:691–695. doi: 10.1038/35618. [DOI] [PubMed] [Google Scholar]
- Raymond CS, Parker ED, Kettlewell JR, Brown LG, Page DC, Kusz K, Jaruzelska J, Reinberg Y, Flejter WL, Bardwell VJ, et al. A region of human chromosome 9p required for testis development contains two genes related to known sexual regulators. Hum Mol Genet. 1999a;8:989–996. doi: 10.1093/hmg/8.6.989. [DOI] [PubMed] [Google Scholar]
- Raymond CS, Kettlewell JR, Hirsch B, Bardwell VJ, Zarkower D. Expression of Dmrt1 in the genital ridge of mouse and chicken embryos suggests a role in vertebrate sexual development. Dev Biol. 1999b;215:208–220. doi: 10.1006/dbio.1999.9461. [DOI] [PubMed] [Google Scholar]
- Reddy P, Zehring WA, Wheeler DA, Pirrotta V, Hadfield C, Hall JC, Rosbash M. Molecular analysis of the period locus in Drosophila melanogaster and identification of a transcript involved in biological rhythms. Cell. 1984;38:701–710. doi: 10.1016/0092-8674(84)90265-4. [DOI] [PubMed] [Google Scholar]
- Schwabe JW, Neuhaus D, Rhodes D. Solution structure of the DNA-binding domain of the oestrogen receptor. Nature. 1990;348:458–461. doi: 10.1038/348458a0. [DOI] [PubMed] [Google Scholar]
- Schwabe JW, Chapman L, Finch JT, Rhodes D. The crystal structure of the estrogen receptor DNA-binding domain bound to DNA: How receptors discriminate between their response elements. Cell. 1993;75:567–578. doi: 10.1016/0092-8674(93)90390-c. [DOI] [PubMed] [Google Scholar]
- Shen MM, Hodgkin J. mab-3, a gene required for sex-specific yolk protein expression and a male-specific lineage in C. elegans. Cell. 1988;54:1019–1031. doi: 10.1016/0092-8674(88)90117-1. [DOI] [PubMed] [Google Scholar]
- Sinclair AH, Berta P, Palmer MS, Hawkins JR, Griffiths BL, Smith MJ, Foster JW, Frischauf AM, Lovell-Badge R, Goodfellow PN. A gene from the human sex-determining region encodes a protein with homology to a conserved DNA-binding motif. Nature. 1990;346:240–244. doi: 10.1038/346240a0. [DOI] [PubMed] [Google Scholar]
- Starr DB, Hawley DK. TFIID binds in the minor groove of the TATA box. Cell. 1991;67:1231–1240. doi: 10.1016/0092-8674(91)90299-e. [DOI] [PubMed] [Google Scholar]
- Taylor BJ, Truman JW. Commitment of abdominal neuroblasts in Drosophila to a male or female fate is dependent on genes of the sex-determining hierarchy. Development. 1992;114:625–642. doi: 10.1242/dev.114.3.625. [DOI] [PubMed] [Google Scholar]
- Tjian R, Maniatis T. Transcriptional activation: A complex puzzle with few easy pieces. Cell. 1994;77:5–8. doi: 10.1016/0092-8674(94)90227-5. [DOI] [PubMed] [Google Scholar]
- van Houte L, van Oers A, van de Wetering M, Dooijes D, Kaptein R, Clevers H. The sequence-specific high mobility group 1 box of TCF-1 adopts a predominantly alpha-helical conformation in solution. J Biol Chem. 1993;268:18083–18087. [PubMed] [Google Scholar]
- Veitia RA, Nunes M, Quintana-Murci L, Rappaport R, Thibaud E, Jaubert F, Fellous M, McElreavey K, Goncalves J, Silva M, et al. Swyer syndrome and 46,XY partial gonadal dysgenesis associated with 9p deletions in the absence of monosomy-9p syndrome. Am J Hum Genet. 1998;63:901–905. doi: 10.1086/302023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villella A, Hall JC. Courtship anomalies caused by doublesex mutations in Drosophila melanogaster. Genetics. 1996;143:331–344. doi: 10.1093/genetics/143.1.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villella A, Gailey DA, Berwald B, Ohshima S, Barnes PT, Hall JC. Extended reproductive roles of the fruitless gene in Drosophila melanogaster revealed by behavioral analysis of new fru mutants. Genetics. 1997;147:1107–1130. doi: 10.1093/genetics/147.3.1107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villeneuve AM, Meyer BJ. The regulatory hierarchy controlling sex determination and dosage compensation in C. elegans. Adv Genet. 1990;27:117–188. doi: 10.1016/s0065-2660(08)60025-5. [DOI] [PubMed] [Google Scholar]
- Weiss MA. Thermal unfolding studies of a leucine zipper domain and its specific DNA complex: Implications for scissor's grip recognition. Biochemistry. 1990;29:8020–8024. doi: 10.1021/bi00487a004. [DOI] [PubMed] [Google Scholar]
- Weiss MA, Ellenberger T, Wobbe CR, Lee JP, Harrison SC, Struhl K. Folding transition in the DNA-binding domain of GCN4 on specific binding to DNA. Nature. 1990;347:575–578. doi: 10.1038/347575a0. [DOI] [PubMed] [Google Scholar]
- Yamamoto D, Ito H, Fujitani K. Genetic dissection of sexual orientation: Behavioral, cellular, and molecular approaches in Drosophila melanogaster. Neurosci Res. 1996;26:95–107. doi: 10.1016/s0168-0102(96)01087-5. [DOI] [PubMed] [Google Scholar]
- Yang W, Steitz TA. Crystal structure of the site-specific recombinase γδ resolvase complexed with a 34 bp cleavage site. Cell. 1995;82:193–207. doi: 10.1016/0092-8674(95)90307-0. [DOI] [PubMed] [Google Scholar]
- Yi W, Zarkower D. Similarity of DNA binding and transcriptional regulation by Caenorhabditis elegans MAB-3 and Drosophila melanogaster DSX suggests conservation of sex determining mechanisms. Development. 1999;126:873–881. doi: 10.1242/dev.126.5.873. [DOI] [PubMed] [Google Scholar]