Abstract
Homologous recombination between dispersed DNA repeats creates chromosomal rearrangements that are deleterious to the genome. The methylation associated with DNA repeats in many eukaryotes might serve to inhibit homologous recombination and play a role in preserving genome integrity. We have tested the hypothesis that DNA methylation suppresses meiotic recombination in the fungus Ascobolus immersus. The natural process of methylation-induced premeiotically (MIP) was used to methylate the b2 spore color gene, a 7.5-kb chromosomal recombination hot spot. The frequency of crossing-over between two markers flanking b2 was reduced several hundredfold when b2 was methylated on the two homologs. This demonstrates that DNA methylation strongly inhibits homologous recombination. When b2 was methylated on one homolog only, crossing-over was still reduced 50-fold, indicating that the effect of methylation cannot be limited to the blocking of initiation of recombination on the methylated homolog. On the basis of these and other observations, we propose that DNA methylation perturbs pairing between the two intact homologs before recombination initiation and/or impairs the normal processing of recombination intermediates.
Keywords: Ascobolus immersus, chromosome rearrangements, crossing-over, DNA methylation, meiotic recombination, repeat DNA sequences
Crossing-over between dispersed DNA repeats results in chromosomal rearrangements. In eukaryotes, the destructive potential of dispersed repeats through ectopic homologous recombination is well documented (Rouyer et al. 1987; Montgomery et al. 1991; Small et al. 1997). In yeast, artificial duplications placed in ectopic position can interact and generate chromosomal rearrangements through homologous recombination at high frequency during meiosis (Lichten et al. 1987). In higher eukaryotes, the number of repeats per genome is often so high that no single cell would escape genomic rearrangements if ectopic recombination were to occur at high frequencies. Therefore, factors must exist that limit recombination between dispersed repeats. Thuriaux (1977) pointed out that the frequency of crossing-over per unit of physical DNA length decreases with increasing genome size, and proposed that recombination is confined to genes. According to this hypothesis, satellite DNA sequences and interspersed DNA repeats, which constitute the bulk of the intergenic regions, must be poor substrates for meiotic recombination even when in allelic positions. Nucleotidic divergence (Rayssiguier et al. 1989; Radman and Wagner 1993) and an insufficient length of sequence identity (Shen and Huang 1986; Jinks-Robertson et al. 1993) are two factors known to reduce drastically homologous recombination. Nevertheless, other factors that suppress homologous recombination are required to protect genomes against the threat generated by families of long DNA repeats that have diverged little or not at all (e.g., after recent duplication events).
One way to identify such factors would be to determine whether DNA repeats share any feature that might suggest a role in preventing recombination. There is increasing evidence that cytosine methylation is one such common property in the eukaryotic genome that displays this form of DNA modification. In plants, the repeat-rich intergenic regions are often methylated (Bennetzen et al. 1994). In mammals the bulk of DNA methylation involves DNA repeats (Yoder et al. 1997). In the fungus Ascobolus immersus, DNA methylation is typically restricted to DNA repeats (Goyon et al. 1996), most likely as a result of a process called methylation induced premeiotically (MIP) which detects DNA repeats efficiently before meiosis and leads to their methylation (Rhounim et al. 1992).
Some observations can be explained by the hypothesis that methylation serves as a genome stabilizer through the suppression of homologous recombination between dispersed sequences (Rossignol and Faugeron 1994; Yoder et al. 1997). In plants with large genomes, such as maize, recombination appears restricted to hypomethylated gene-containing regions (Whitkus et al. 1992). In humans, genomic regions subjected to parental imprinting, a sex-specific process in which methylation is likely to play a causal role (Barlow 1993; Razin and Cedar 1994), display significant differences in their frequencies of recombination during male and female meioses (Paldi et al. 1995). This last observation is in line with the hypothesis of Thomas and Rothstein (1991), who proposed that differences between male and female meiotic recombination frequencies would result from imprinting. Chromosomal translocations resulting from exchanges between decondensed and demethylated satellites are frequently associated with the immunodeficiency, centromeric heterochromatin instability and facial abnormalities (ICF) syndrome, and are also observed in culture cells treated by the demethylating agent 5-azacytidine (Miniou et al. 1994; Ji et al. 1997).
These observations, however, do not actually prove that DNA methylation suppresses homologous recombination. The only clear demonstration of an effect of methylation on recombination concerns V(D)J joining, which is decreased >100 times when tested on a methylated minichromosome substrate (Hsieh and Lieber 1992). Although abnormal V(D)J joining may generate chromosome rearrangements (Lieber 1992), it is not likely to play a generalized role in such events as it is a specialized recombination process restricted to very specific DNA motifs, and only occurs in specialized somatic cells of vertebrates. As for homologous recombination, a general process that takes place in any living organism, no direct evidence for an inhibitory effect of methylation has been established so far. Attempts to demonstrate such an effect, using extrachromosomal plasmids, have been unsuccessful (Puchta et al. 1992; Liang and Jasin 1995).
By using MIP to methylate a 7.5-kb chromosomal interval encompassing a meiotic recombination hot spot of A. immersus, we show here that DNA methylation reduces by several hundredfold the frequency of crossing-over within this interval. We provide additional experimental evidence indicating that this effect cannot be simply accounted for by an increased resistance of methylated DNA to meiotic endonucleases.
Results
Introduction of markers flanking the b2 recombination hot spot and in vivo methylation of this hot spot
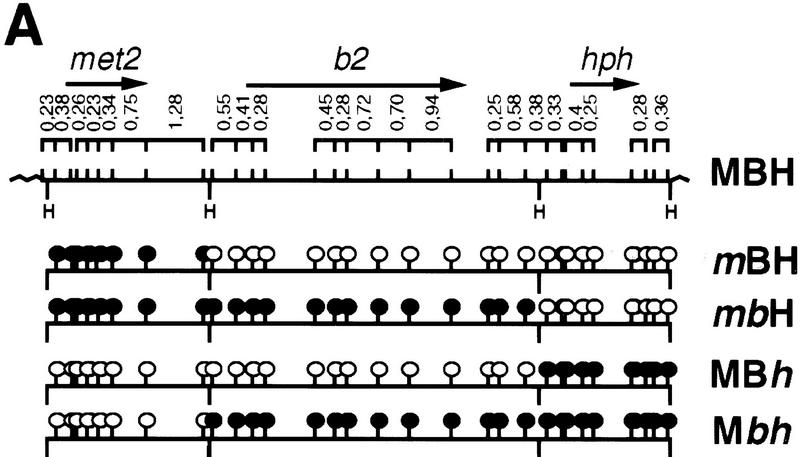
The possible inhibitory role of DNA methylation on crossing-over could be tested in Ascobolus thanks to the MIP process, which enables the in vivo methylation at will of gene-size sequences, and to the existence of a well-characterized recombination hot spot, the b2 spore color gene (Nicolas and Rossignol 1989). The b2 gene used in this study was flanked on a plasmid by an upstream marker, the met2 gene from Ascobolus, and by a downstream marker that confers hygromycin resistance and is based on the hph gene of Escherichia coli. This plasmid was used to transform a haploid Ascobolus strain deleted for the resident copies of the met2 and b2 genes. Correspondingly, this strain exhibited methionine auxotrophy (Met−) and gave white ascospores in crosses. We selected one transgenic strain that was hygromycin resistant (HygR), restored methionine prototrophy (Met+), and displayed the wild-type brown ascospore phenotype. Southern blot analysis (data not shown) showed that the selected transgenic strain resulted from the integration of a single unrearranged copy of the plasmid construct (strain MBH: Met+, Brown spores, HygR; Fig. 1). The shortened designation of strains, in the following paragraphs, will use M and m for Met+ and Met−, respectively, B and b from brown spores and white spores, respectively, and H and h for HygR and HygS, respectively.
Figure 1.
HpaII restriction map of the met2–b2–hph transgenic locus and DNA methylation profiles. (A) The MBH strain is the initial transformant carrying the transgenic locus. The three marker genes are each contained within a HindIII fragment (H). ORFs are indicated by arrows. Only HpaII sites bordering restriction fragments >200 bp are indicated (bars). The methylation status of the HpaII sites (sequence CCGG) present in the transgene is indicated [○ (unmethylated) or • (methylated)] for the four different strains derived from the MBH initial strain. (B) HpaII restriction profiles revealed by Southern blot analyses using a met2 probe (lane 1), a b2 probe (lane 2), and a hph probe (lane 3) are shown for the four progeny strains. HpaII sites are not cut by the enzyme when methylated.
To detect crossing-over in the b2 interval, an epimutated allele of each of the two markers flanking b2 was created using MIP and its gene silencing effect (Rhounim et al. 1992). Methylation and silencing of the entire b2 interval separating the two markers was also achieved by MIP (see Materials and Methods). Strains in which met2 alone, or both met2 and b2, had been methylated and silenced by MIP were termed mBH and mbH, respectively (Fig. 1). Strains in which either hph alone or both hph and b2 had been methylated and silenced were termed MBh and Mbh, respectively.
Methylation of the b2 recombination hot spot leads to a several hundredfold decrease of crossing-over frequency
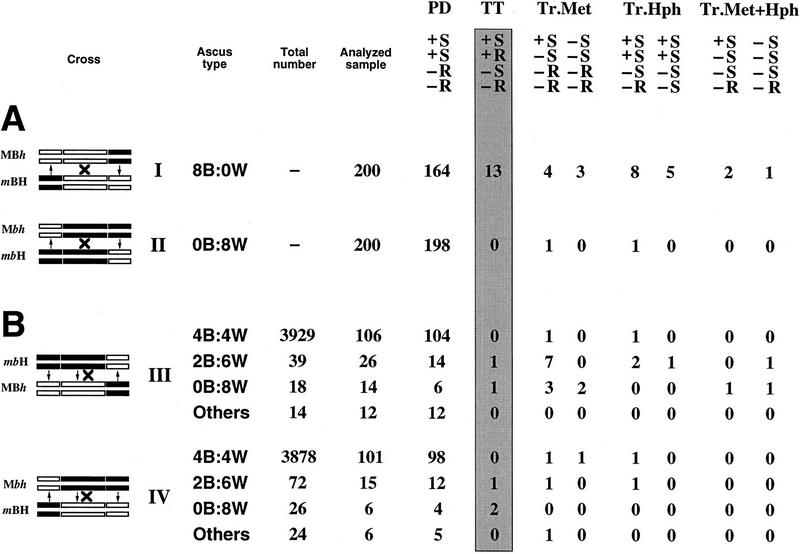
Crossing-over frequency between the met2 and hph markers in the presence or absence of methylation of the b2 interval was measured by performing two crosses (Fig. 2A). The two parental b2 alleles were unmethylated in cross I (MBh × mBH), and methylated in cross II (Mbh × mbH). The frequency of crossing-over in these crosses was first assessed by ascus analysis. A total of 13 crossovers (tetratype asci) were detected in 200 asci analyzed in cross I. In the six tetratype asci that were analyzed at the molecular level by Southern blot analysis, crossing-over was confirmed with full methylation of both flanking markers in one recombinant product and no methylation of these in the reciprocal product (data not shown). In marked contrast, no crossover was detected in 200 asci from cross II. To estimate precisely the decrease in crossing-over frequency between crosses I and II, a random spore analysis was conducted. The frequency of Met+, HygR progeny was 15 × 10−3 among 26,500 germinated spores scored in cross I, and 0.4 × 10−3 among 114,000 germinated spores scored in cross II. Although the incidence of Met+, HygR progeny should reflect crossing-over in the b2 interval (Fig. 2A), it is also possible to obtain such progeny by reversion of the met2 flanking marker. Indeed, unlike the silencing of hph, which never reverts spontaneously, the met2 silencing undergoes reversion at a low frequency. Because the reversion of met silencing is never accompanied by a complete loss of methylation (Rhounim et al. 1992), it is possible to distinguish between reversion and crossing-over by analyzing the methylation status of the met2 flanking marker. This was assessed using the methylation-sensitive restriction enzyme HpaII on a fraction of the Met+, HygR strains recovered from random spore analysis from crosses I and II (Fig. 3). Whereas true recombinants are expected to show no methylation of the flanking markers, revertant strains should show instead some residual methylation of the met2 marker at all restriction sites tested. All 32 progeny strains analyzed from cross I showed methylation patterns indicative of crossing-over (Fig. 3). This indicates that the 15 × 10−3 Met+, HygR progeny does reflect the frequency of true recombinants in cross I. Therefore, we can calculate the frequency of tetratype asci resulting from meiotic crossing-over (see Fig. 2A) in this cross. In these tetratype asci, only one meiotic product out of four is Met+, HygR. Therefore, the frequency of crossing-over per meiosis in cross I is equal to four times the frequency of Met+, HygR (15 × 10−3 × 4). The estimated value (60 × 10−3) is close to that obtained after analysis of individual asci (65 × 10−3). Conversely, only 3 of 20 progeny strains analyzed from cross II showed no methylation of met2 and therefore, were true recombinants. The 17 other progeny strains showed partial methylation of every site indicating the reversion of silencing of the met2 marker (profile D in Fig. 3). As expected from the absence of spontaneous reversion of hph silencing, no methylation was ever found at the hph marker in all Met+, HygR progeny tested (data not shown). On the basis of these results, the frequency of true recombinant Met+, HygR strains was estimated to 0.06 × 10−3 (i.e., 0.4 × 10−3 × 3/20) corresponding to a frequency of crossing-over per meiosis in cross II of 0.24 × 10−3 (i.e., 0.06 × 10−3 × 4). Thus, in these experiments, methylation of b2 reduced crossing-over 250-fold within that interval.
Figure 2.
Ascus analysis in crosses I to IV. (A) Crosses I (MBh × mBH) and II (Mbh × mbH). For each cross, the three marker genes are symbolized on the four chromatids at meiosis I by white rectangles when unmodified or black rectangles when methylated and silenced. Asci of the progeny are indicated by their spore color phenotype (B, brown; W, white). In crosses I and II, they were composed exclusively (several thousand asci scored) of 8B:0W and 0B:8W asci, respectively. Because meiosis is followed by one cell division before ascospore formation, asci contain four pairs of spores that correspond to the four meiotic products. Segregation of the two flanking markers met2 and hph in the different types of asci is shown (+: Met+; −: Met−; S: HygS; R: HygR). Parental ditype asci (PD), which reflect an absence of events, are the most frequently observed. Crossing-over, symbolized by crosses (left), are detected because they lead to tetratype asci (TT, shaded rectangle). Three additional classes of asci were observed as the result of methylation transfer (↑, ↓ at left) from the methylated parental allele to the previously active and unmethylated parental met2 allele (Tr.Met) and/or hph allele (Tr.Hph and Tr.Met+Hph). (B) Crosses III (mbH × MBh) and IV (Mbh × mBH). These crosses, which involve one methylated allele and one unmethylated allele of b2, can give rise to asci showing a transfer of methylation at b2 as well as at the two flanking markers. Methylation transfer at b2 leads to three classes of asci with less than four brown spores (2B:6W, 0B:8W, and Others). (Others) Asci with pink spores indicative of partial inactivation (Colot et al. 1996). A total of 4000 asci were scored in each cross.
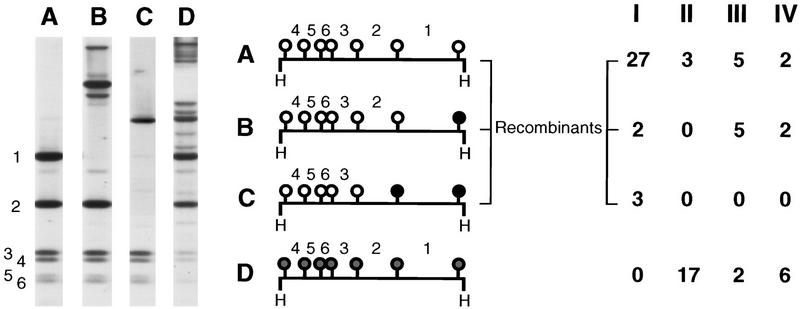
Figure 3.
Methylation pattern of the met2 marker of Met+, HygR strains. DNA was digested with the enzyme HpaII. Southern blots were hybridized with the met2 HindIII fragment. The HpaII sites are indicated by solid circles when fully methylated, shaded circles when partially methylated, and an open circles when unmethylated. HpaII fragments are numbered according to their size. The Roman numerals (I–IV) heading the columns (right), refer to the four genetic crosses in Fig. 2. Four distinct classes of methylation patterns (A,B,C,D) were observed. Within each class, all the strains analyzed exhibited a methylation pattern identical to that examplified in the figure, except for class B. Classes A, B, and C are all indicative of crossing-over. In class A, there was no methylation at any of the met2 restriction sites tested. In class B, there was also no methylation, except at the rightmost HpaII site. Methylation of this site leads to the loss of the HpaII fragment 1. In cross I, where b2 is unmethylated, this fragment was replaced by one fragment 120 bp longer (not shown). In crosses III and IV, it was replaced by various fragments of higher molecular mass, depending on the methylation status of the neighboring HpaII sites within b2 (one example is shown). In class C, the two rightmost HpaII sites were methylated. This led to the loss of HpaII fragments 1 and 2 and to the appearance of a unique fragment of higher molecular mass. Methylation in classes B and C is restricted to the region closest to b2, as expected if they result from events initiated in the b2 interval and propagating in the direction of met2 (see paragraph on methylation transfers; Colot et al. 1996). Class D shows partial methylation of every met2 HpaII site corresponding to the reversion of silencing of the met2 marker.
Methylation of only one parental b2 allele is sufficient to decrease markedly the frequency of crossing-over
Crosses in which only one of the two parental b2 alleles was methylated were also performed. In cross III (MBh × mbH) the two methylated alleles b2 and met2 were associated in the same parent. In the reciprocal cross IV (Mbh × mBH), the methylated b2 allele was associated with the methylated allele of hph (Fig. 2B). The frequencies of Met+, HygR progeny were 0.5 × 10−3 and 0.6 × 10−3, in crosses III and IV, respectively, among ∼94,000 ascospores issued from each of these crosses. The number of true recombinants among these strains was estimated after Southern blot analysis, as before (Fig. 3). The deduced frequencies of crossing-over per meiosis were 1.5 × 10−3 and 1 × 10−3 in crosses III and IV, respectively. These values, which are ∼50-fold lower than those obtained for cross I (60 × 10−3), therefore, indicate that methylation of one parental b2 allele is sufficient to strongly decrease crossing-over. The samples studied do not allow us to decide whether this reduction is really smaller than that found in cross II.
Association of crossing-over with transfers of methylation at b2
Ascus analyses were also performed on the progeny of crosses III and IV. In both cases, a majority of the progeny was composed of asci with four brown and four white spores (4B:4W), indicative of the Mendelian segregation of the active and silenced parental b2 alleles. Nevertheless, a fraction of asci displayed aberrant segregation patterns of spore color, distinguished by a deficit of brown-colored spores (Fig. 2B). These aberrant patterns reflect transfers of methylation and silencing during meiosis from the methylated parental b2 allele to the formerly unmethylated parental b2 allele (Colot et al. 1996; Fig. 4). On the basis of the analysis of a sample of each class of asci (4B:4W, 2B:6W, 0B:8W, and others), the frequency of crossing-over was estimated to be 0.7 × 10−3 in cross III and 3.3 × 10−3 in cross IV (Fig. 2B). Remarkably, all five crossing-over events detected arose in 2B:6W and 0B:8W asci and thus, were associated with a transfer of methylation. To investigate this further, the methylation status of the entire met2–b2–hph transgenic locus was assessed for each of the four meiotic products recovered from the five crossover asci (Fig. 5). Crossing-over was confirmed at the molecular level, with methylation of both flanking markers in one recombinant product and no methylation of these in the reciprocal product. All the crossing-overs were associated with methylation transfers extending to various lengths along the gene. More significant, the methylation transfer tracts always included the site of exchange. Indeed, the two recombinant products always showed a reciprocal exchange between the unmethylated and the methylated parental molecules on either side of the methylation transfer tract. This indicates that crossing-over never occurred outside the region involved in transfer. In asci 1 and 5, methylation transfer observed in the Met+, HygR product included the rightmost met2 HpaII site. This situation accounts for profile B in Figure 3.
Figure 4.
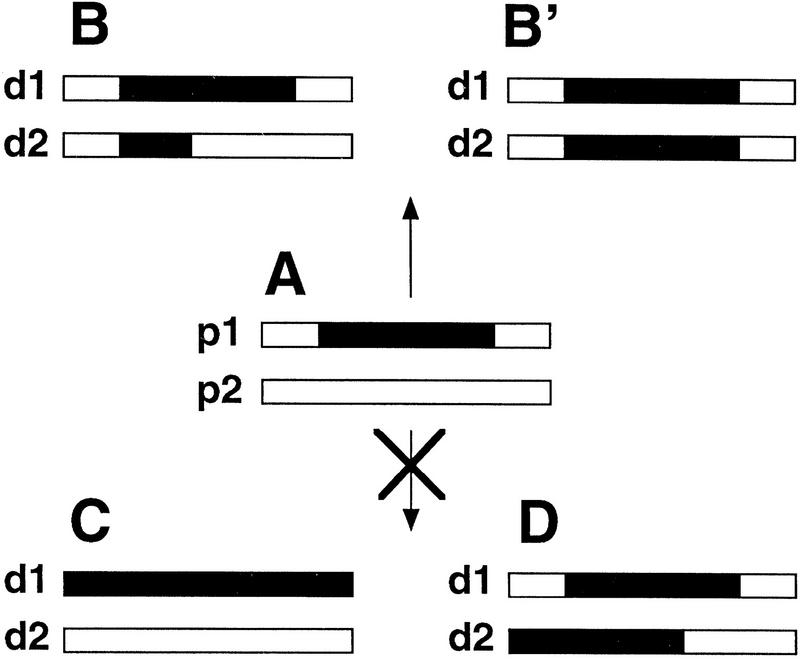
Properties of methylation transfer. Each pair of rectangles represents two allelic regions. Parental DNAs are named p1 and p2. (A) The p1 parental DNA displays methylation (black portion), the p2 parental DNA is unmethylated. The corresponding descendant DNAs after meiosis are named d1 and d2. Methylation transfers are characterized by the appearance of methylated segments of variable length in the d2 DNA issued from p2. This methylated segments always extends within a region allelic to that which was methylated in p1 (B,B′). Methylation never spreads in cis (C), nor is transferred to nonallelic regions (D) (from Colot et al. 1996; this study).
Figure 5.
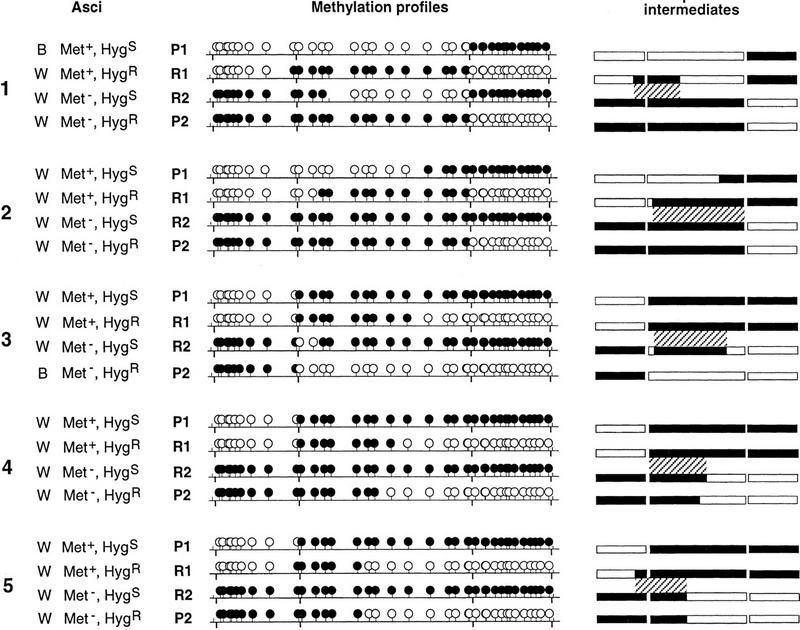
Methylation transfer events associated with crossing-over. The five asci from cross III (asci 1 and 2) and cross IV (asci 3, 4, and 5) showing a crossing-over event (Fig. 2B) are represented (left) with the phenotypes of their four meiotic products. The two parental products are designated P1 and P2; the two recombinant products are designated R1 and R2. The methylation profiles of the four meiotic products of each ascus were determined using the HpaII restriction enzyme. (•) Methylated HpaII sites; (○) unmethylated. The HindIII sites separating the three genes are indicated below the horizontal bar. (Right) Proposed intermediates after methylation transfer. met2, b2, and hph are represented by rectangles; methylated regions are in black and unmethylated regions are in white. For the sake of simplicity, crossing-over was dissociated from transfer of methylation in these drawings. The final products are obtained by performing a reciprocal exchange within the hatched region between the two homologs, which will recombine. This hatched region defines the length of the methylation transfer together with the region where crossing-over must have occurred. Crossing-over outside of this region would have given patched methylation profiles. Methylation transfer involves one (asci 1 and 3) or both (asci 2, 4, and 5) homologous chromatids.
Methylation transfers initiated in b2 show a bidirectional polarity
Previously, we had shown that methylation transfer at the resident b2 locus is polarized 5′ to 3′ (Colot et al. 1996). The same polarity was found in the present study as shown in Figure 6. Eleven of 12 transfer events analyzed in b2 extend rightward from a region located in the left 5′ part. Transfer tracts starting from this same origin extend toward the left and reach the met2 fragment in four cases. Altogether, these transfers display a bidirectional polarity with only one exception.
Figure 6.
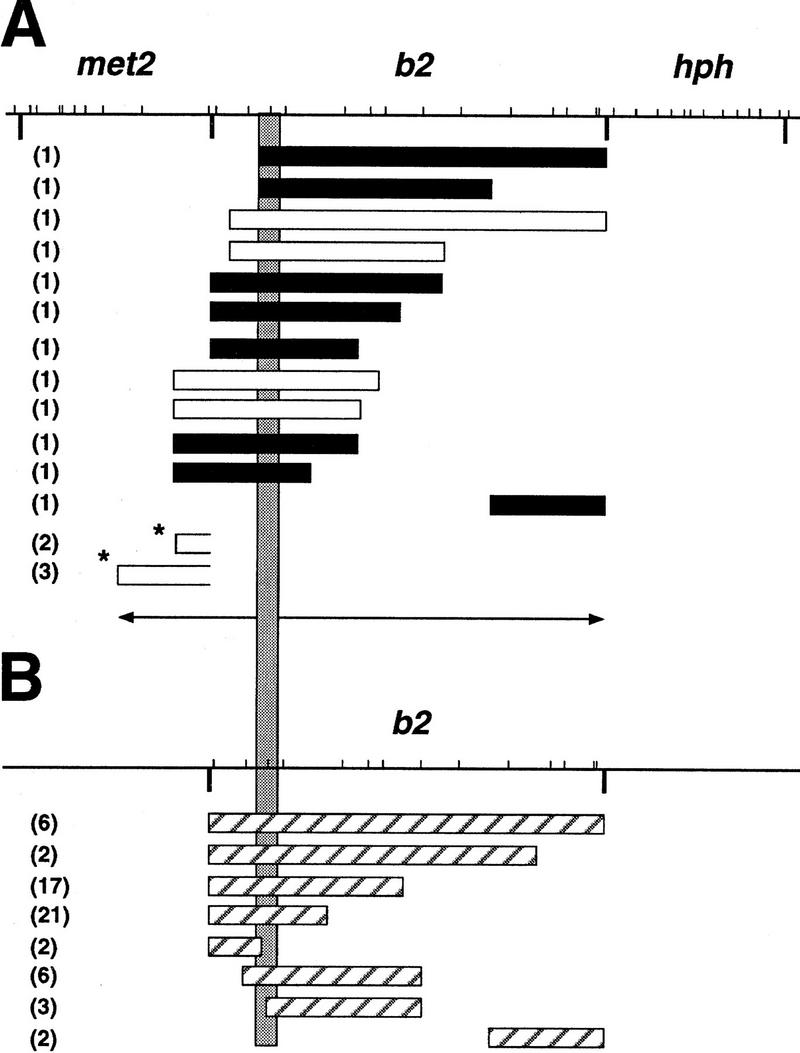
(A) Bidirectional polarity of methylation transfer events. The shaded vertical rectangle defines the region of b2 in which 73 of 76 methylation transfers shown could have been initiated. The bidirectional polarity is suggested by the two opposite arrows. The HpaII restriction map of the met2–b2–hph transgene is at the top (bars). The HindIII sites separating the three genes are indicated below the horizontal bar. Rectangles show the different lengths of methylation transfer observed. Solid rectangles correspond to methylation transfers shown in Fig. 5; open rectangles indicate methylation transfers observed in randomly selected Met+, HygR recombinants (Fig. 3) and correspond to profiles A and B from cross IV. [The extent of the methylation transfer cannot be deduced from the Met+, HygR recombinants in cross III (Fig. 5).] Transfers indicated by asterisks correspond to profiles B and C from cross I and are limited to met2 because b2 was not methylated in this cross. However, they can result from molecular events initiated in b2 and propagating leftward (see Discussion). This is suggested by the right opening of the rectangles. (B) Hatched rectangles show the methylation transfers previously observed at the resident b2 gene, which is devoid of the met2 and hph flanking markers (Colot et al. 1996). For each event, the number of occurrence is given in parenthesis.
The flanking markers met2 and hph also exhibited inactivation transfers in crosses I and II (see Fig. 2A) and in crosses III and IV (see Fig. 2B). As expected, these events correspond to methylation transfers (data not shown). As for b2, inactivation transfers involved either one or the two meiotic products that would otherwise harbor the active allele, leading to asci with either 2Met+:6Met− or 0Met+:8Met−, or either 2HygR:6HygS or 0HygR:8HygS. In crosses III and IV, transfers at met2 and hph were associated frequently with a transfer at b2 (18/20 and 3/6, respectively) (Fig. 2B). Because the process driving the methylation transfer seems to be bidirectional (Fig. 6), we expect that its propagation over longer distances may inactivate b2 together with met2 or hph.
Methylation of b2 leads to a decrease in the frequency of methylation transfers
Remarkably, in crosses I–IV, the frequency of transfers at one or the other of the two flanking markers decreased when the b2 interval was methylated, from 11.5% in cross I to 3% in crosses III and IV and to 1% in cross II. This result can be best explained if most of the methylation transfer events at met2 and hph are initiated within the b2 recombination hot spot. Therefore, DNA methylation affects negatively both homologous recombination and methylation transfer.
Discussion
By using a meiotic recombination hot spot, the b2 gene, that could be methylated in vivo, we have performed a direct test on the effect of methylation on homologous recombination. Our results indicate that methylation of this hot spot in both parents supresses allelic crossing-over efficiently between flanking markers. Methylation of one parental allele also affects drastically homologous recombination, resulting in a 50-fold reduction. In such crosses, methylation transfers to the unmethylated b2 parental allele were observed frequently and the rare crossing-over events that were detected were always physically associated with these transfers. This latter result reinforces the conclusion that meiotic recombination in b2 is mechanistically associated with methylation transfer (Colot et al. 1996) and must be considered in the discussion of the suppressing effect of DNA methylation on recombination.
Molecular insights into meiotic recombination come chiefly from studies in yeast. DNA–DNA pairing interactions are thought to occur at the sites of recombination initiation between the intact duplexes of homologous chromosomes (Weiner and Kleckner 1994). These early interactions have been proposed to play a role in the sensing of DNA homology and in the decision to make double strand breaks (DSBs) (Xu and Kleckner 1995; Rocco and Nicolas 1996). DSBs occur at specific sites that lie close to the high end of gene conversion gradients and are the first chemical disruptions of DNA in meiotic recombination (for review, see Lichten and Goldman 1995). DSBs are followed by the exonucleolytic degradation of their 5′ ends to yield 3′ single strand tails (Sun et al. 1991; Bishop et al. 1992). Invasion of the homologous chromatid by the broken strands occurs subsequently and leads to the formation of joint molecules stabilized by heteroduplex DNA and bordered by double Holliday junctions (Schwacha and Kleckner 1995). DNA repair synthesis from the 3′ ends, together with mismatch correction of the heteroduplex DNA generated by the double Holliday junctions, leads to gene conversion, and resolution of these junctions leads to recombinant products that are either noncrossover or crossover (for other possibilities, see Schwacha and Kleckner 1995). The various steps after the break were predicted by the DSB model of meiotic recombination (Szostak et al. 1983; Sun et al. 1991). Most of the extensive genetic data accumulated on recombination within b2 (Nicolas and Rossignol 1989) and the striking parallel between the bipolarity of gene conversion events in yeast and of methylation transfers in Ascobolus justify the use of the DSB model as a legitimate framework to discuss at which step the recombination process is inhibited by methylation.
In the present study, the methylation of the b2 gene results in its transcriptional inactivation. Several examples show a link between recombination and transcription (Stewart and Roeder 1989; Nickoloff and Reynolds 1990; Thomas and Rothstein 1992). Meiotic recombination is mainly initiated in promoter regions (Lichten and Goldman 1995). The deletion of these regions prevents the initiation of recombination on the deleted parental strand and leads to a twofold reduction in the frequency of recombination (Nicolas et al. 1989; Schultes and Szostak 1991). Nevertheless, it has been shown that the transcription process itself is not required for recombination. Indeed, although the promoter regions of ARG4 and HIS4 are necessary for homologous recombination in these genes, small deletions in their promoter resulting in a strong decrease of transcription, leave unaltered the rate of recombination (Schultes and Szostak 1991; White et al. 1992). These observations make unlikely the hypothesis that the decrease of recombination when b2 is methylated results simply from a transcriptional effect unless the recombination features in Ascobolus and in yeast are completely different. Independent studies in Saccharomyces cerevisiae (White et al. 1993) and Schizosaccharomyces pombe (Kon et al. 1997) indicate that transcription factors play a role in inducing recombination, but that this is likely to be a consequence of an alteration of chromatin structure necessary for the initial break rather than an effect of these factors on transcription itself. In the present study, methylation preventing or altering the binding of transcription factors necessary for both transcription and the triggering of the initial break could account for the almost total suppression of crossing-over when the two parents are methylated. However, this hypothesis cannot explain the 50-fold reduction of crossing-over observed when only one parent is methylated. Indeed, in this case, the other parent is still transcribed, as attested by the segregation of spore color in meiotic products. This parent should undergo normal recombination initiation, which would result in a twofold reduction of crossing-over only. Therefore, a masking by methylation of the initiation site cannot account for the observed effect. To explain the data, we need to assume either that methylation also inhibits the production of the initial break in trans, on the unmethylated homologous chromatids, or that it acts at a further step.
A way to inhibit the break in trans would be by impairing the process of sensing of homology (Xu and Kleckner 1995; Rocco and Nicolas 1996), which must be the earliest step of recombination and methylation transfer (Colot et al 1996). If methylation were to inhibit recombination by acting at this step only, then the methylation of one parent would make this step 50-fold less efficient, although allowing a frequency of 1.5%–3% methylation transfers. Therefore, this hypothesis im-plies that every meiosis undergoes an efficient sensing of homology at b2 in the unmethylated control cross. It also implies that the sensing of homology events that would escape the inhibitory effect of methylation in crosses with one methylated parent, allow methylation transfer every time. At present, there is no known molecular mechanism that could simply explain directed transfers of methylation in the hypothetical DNA–DNA structure, which is thought to be formed between the two intact DNA duplexes at this step.
Another way to account for the effect of methylation is to assume that it acts at a step after the initial break. Methylation would not prevent the formation of recombination intermediates, but would impair their stability and correspondingly their maturation into recombinant products. This could account for the severely reduced crossing-over frequencies associated with methylation. In this hypothesis, methylation transfer likely occurs through the formation of hemimethylated recombination intermediates (Colot et al. 1996). It is noteworthy, however, that the frequency of methylation transfer at b2 observed in crosses III and IV ranges from 1.5% to 3%, which is between two and four times less than the 6% of crossing-over observed in cross I. This implies in the present hypothesis that 50% or more of the recombination intermediates that are formed in any case are destabilized by methylation before methylation transfer can occur. This conclusion is in agreement with the observation that methylation transfer at the markers flanking b2 is reduced when the b2 interval is methylated. Nevertheless, the latter reduction is approximately fourfold in crosses III and IV, and thus is 10-fold lower than the reduction in crossover frequency observed in the same crosses. Therefore, this would suggest that methylation affects the stability of recombination intermediates in a second step, at least after they become substrates for methylation transfer but before their maturation into crossover products.
The hypothesis that unstable intermediates are formed can also account for the frequent occurrence of asci with transfer of methylation to both unmethylated sister chromatids (0B:8W, 0Met+:8Met−, 0HygR:8HygS). Accordingly, the instability of the recombination intermediates after the initial break would result in unrepaired chromatids, which could then enter a second round of interaction involving different partners. In particular, the broken chromatid may form a new recombination intermediate with the unmethylated chromatid that was not involved in the first round of interaction. This hypothesis assumes that recombination intermediates could be formed between sister chromatids. It has been shown in yeast that meiotic recombination strongly favors intermediates formed between homologous chromatids (Schwacha and Kleckner 1997). Methylation might affect this bias, by favoring the formation of recombination intermediates between sister chromatids.
The way in which methylation might impair recombination intermediates is unknown. DNA methylation is often associated with changes in chromatin conformation (Kass et al. 1997) and triggers such changes in Ascobolus independently of the transcriptional state (J.L. Barra, G. Almouzni, G. Faugeron, and J.-L. Rossignol, unpubl.). DNA methylation, or associated changes, may impair the progress of the recombination process, either by preventing the faithful formation of the molecular complex that constitutes the recombination machinery, or by interfering with one or more activities of this machinery.
Our finding that methylation prevents crossing-over in Ascobolus raises the question of its effect on recombination in other organisms displaying methylation such as plants and vertebrates. In Ascobolus, methylation involves every cytosine, even when they are not part of a symmetrical sequence, which is not the case of vertebrates where methylation mostly involves CpG. An important issue is to know whether methylation in these organisms may contribute to stabilize their genomes by preventing homologous recombination between dispersed DNA repeats. This would provide a major biological significance for the observation that in many eukaryotes, methylation is triggered by DNA repeats (Assaad and Signer 1992; Rossignol and Faugeron 1994) or by foreign DNA (Doerfler 1992; Bestor and Tycko 1996).
Materials and methods
Plasmid construction
The 7.5-kb HindIII fragment, which contains the b2 gene (Colot et al. 1996), was inserted downstream of the Ascobolus met2 gene harbored in PVM1.1, a pBKS-derived plasmid (Colot and Rossignol 1995). This new plasmid was then used to insert downstream of b2 the 3-kb HindIII fragment from plasmid pMP6 (M. Plamann, pers. comm.) that carries a chimeric construct of the E. coli hph gene. This gave plasmid pLmbh, which contains the b2 gene flanked upstream and downstrean by met2 and hph, respectively, all three in the same orientation.
Media and strains
Standard Ascobolus genetic techniques and media were used (referred to in Colot and Rossignol 1995).
MIP triggers the silencing of duplicated genes through cytosine methylation, which is coextensive with the size of the duplications. To methylate and silence the met2 and b2 copies carried by the transgene of the MBH strain (Fig. 1), we first introduced this met2–b2–hph transgene, through an appropriate cross, in a strain that carries the resident met2 and b2 genes. MIP of the met2 and b2 duplications was triggered by crossing the resulting strain with a strain of the opposite mating type. Because this strain carries a deletion of the b2 and met2 resident genes, progeny strains could be isolated that had inherited no resident copy of b2 and met2 and in which the met2 copy alone or both the met2 copy and the b2 copy carried by the met2–b2–hph transgenic locus were methylated and silenced (strains mBH and mbH, respectively; Fig. 1). A similar protocol (using duplications of b2 and hph) was used to isolate strains devoid of b2 and met2 resident copies and in which the met2–b2–hph transgenic locus was methylated and silenced either at hph alone or at both hph and b2 (strains MBh and Mbh, respectively; Fig. 1). These strains exhibited stable inactivated phenotypes, as expected (Rhounim et al. 1992; Colot et al. 1996).
Crosses
Five mBH strains and five mbH strains were isolated from the same cross. Five MBh strains and five Mbh strains were isolated from another cross. Crosses I–IV consisted in crossing one individual strain from the first set of strains with one individual strain from the other set. Five different crosses, all involving different strains, were performed for each type of cross I–IV. This allowed to randomize possible genetic background effects.
Ascus and random spore analyses
For ascus analysis an equal number of asci was picked from the progeny of the five crosses constituting each type of cross I–IV, whereas random spore analysis was performed on the progeny of four such crosses. The Met+, HygR strains were detected on selective medium (without methionine and with hygromycin) as germinated spores that showed a normal growth and were clearly distinguishable from the weak residual growth seen with Met− or HygS germinated spores.
DNA methylation analysis
DNA isolation and manipulation were as described in Colot and Rossignol (1995). HpaII digestions were performed on 3 μg of DNA.
Acknowledgments
We thank P. Avner, G. Faugeron, J. Haber, V. Rocco, M. Ronemus, D. Stadler, and P. Thuriaux for discussing the manuscript; I. Cail and S. Ballereau for help in dissecting asci and performing genetic analysis; and J. Delaruelle and V. Haedens for technical assistance. Special thanks to V. Colot for many valuable suggestions and comments. L.M. was a recipient of a Ph. D. studentship from the French Ministère de l’Enseignement Supérieur et de la Recherche. This work was supported by “Groupement de Recherches et d’Etudes sur les Génomes” (contract no. 44/95) and “Association pour la Recherche sur le Cancer” (contract no. 6200).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL rossignol@ijm.jussieu.fr; FAX 33-1-4427-4095.
References
- Assaad FF, Signer ER. Somatic and germinal recombination of a direct repeat in Arabidopsis. Genetics. 1992;132:553–566. doi: 10.1093/genetics/132.2.553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barlow DP. Methylation and imprinting: From host defense to gene regulation? Science. 1993;260:309–310. doi: 10.1126/science.8469984. [DOI] [PubMed] [Google Scholar]
- Bennetzen JL, Schrick KS, Springer PS, Brown WE, SanMiguel P. Active maize genes are unmodified and flanked by diverse classes of modified, highly repetitive DNA. Genome. 1994;37:565–576. doi: 10.1139/g94-081. [DOI] [PubMed] [Google Scholar]
- Bestor TH, Tycko B. Creation of genomic methylation patterns. Nature Genet. 1996;12:363–367. doi: 10.1038/ng0496-363. [DOI] [PubMed] [Google Scholar]
- Bishop DK, Park D, Xu L, Kleckner N. DMC1: A meiosis-specific yeast homolog of E. coli recA required for meiotic recombination, synaptonemal complex formation and cell cycle progression. Cell. 1992;69:439–456. doi: 10.1016/0092-8674(92)90446-j. [DOI] [PubMed] [Google Scholar]
- Colot V, Rossignol J-L. Isolation of the Ascobolus immersus spore color gene b2 and study in single cells of gene silencing by methylation induced premeiotically. Genetics. 1995;141:1299–1314. doi: 10.1093/genetics/141.4.1299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Colot V, Maloisel L, Rossignol J-L. Interchromosomal transfer of epigenetic states in Ascobolus: Transfer of DNA methylation is mechanistically related to homologous recombination. Cell. 1996;86:855–864. doi: 10.1016/s0092-8674(00)80161-0. [DOI] [PubMed] [Google Scholar]
- Doerfler W. DNA methylation: Eukaryotic defense against the transcription of foreign genes? Microb Pathogen. 1992;12:1–8. doi: 10.1016/0882-4010(92)90060-2. [DOI] [PubMed] [Google Scholar]
- Goyon C, Rossignol J-L, Faugeron G. Native DNA repeats and methylation in Ascobolus. Nucleic Acids Res. 1996;24:3348–3356. doi: 10.1093/nar/24.17.3348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsieh C-L, Lieber MR. CpG methylated minichromosomes become inaccessible for V(D)J recombination after undergoing replication. EMBO J. 1992;11:315–325. doi: 10.1002/j.1460-2075.1992.tb05054.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ji W, Hernandez R, Zhang XY, Qu GZ, Frady A, Varela M, Ehrlich M. DNA demethylation and pericentromeric rearrangements of chromosome 1. Mutat Res. 1997;379:33–41. doi: 10.1016/s0027-5107(97)00088-2. [DOI] [PubMed] [Google Scholar]
- Jinks-Robertson S, Michelitch M, Ramcharan S. Substrate length requirements for efficient mitotic recombination in Saccharomyces cerevisiae. Mol Cell Biol. 1993;13:3937–3950. doi: 10.1128/mcb.13.7.3937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kass SU, Landsberger N, Wolffe AP. DNA methylation directs a time-dependent repression of transcription initiation. Curr Biol. 1997;7:157–165. doi: 10.1016/s0960-9822(97)70086-1. [DOI] [PubMed] [Google Scholar]
- Kon N, Krawchuk MD, Warren BG, Smith GR, Wahls WP. Transcription factor Mts1/Mts2 (Atf1/Pcr1, Gad7/Pcr1) activates the M26 meiotic hot spot in S. pombe. Proc Natl Acad Sci. 1997;94:13765–13770. doi: 10.1073/pnas.94.25.13765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang F, Jasin M. Studies on the influence of cytosine methylation on DNA recombination and end-joining in mammalian cells. J Biol Chem. 1995;270:23838–23844. doi: 10.1074/jbc.270.40.23838. [DOI] [PubMed] [Google Scholar]
- Lieber M. The mechanism of V(D)J recombination: A balance of diversity, specificity, and stability. Cell. 1992;70:873–876. doi: 10.1016/0092-8674(92)90237-7. [DOI] [PubMed] [Google Scholar]
- Lichten M, Goldman ASH. Meiotic recombination hotspots. Annu Rev Genet. 1995;29:423–444. doi: 10.1146/annurev.ge.29.120195.002231. [DOI] [PubMed] [Google Scholar]
- Lichten M, Borts RH, Haber JE. Meiotic gene conversion and crossing-over between dispersed homologous sequences occurs frequently in Saccharomyces cerevisiae. Genetics. 1987;115:233–246. doi: 10.1093/genetics/115.2.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miniou P, Jeanpierre M, Blanquet V, Sibella V, Bonneau D, Herbelin C, Fischer A, Niveleau A, Viegas-Péquignot E. Abnormal methylation pattern in constitutive and facultative (X inactive chromosome) heterochromatin of ICF patients. Hum Mol Genet. 1994;3:2093–2102. doi: 10.1093/hmg/3.12.2093. [DOI] [PubMed] [Google Scholar]
- Montgomery EA, Huang S-M, Langley CH, Judd BH. Chromosome rearrangement by ectopic recombination in Drosophila melanogaster. Genetics. 1991;129:1085–1098. doi: 10.1093/genetics/129.4.1085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicolas A, Rossignol J-L. Intermediates in homologous recombination revealed by marker effects in Ascobolus. Genome. 1989;31:528–535. [Google Scholar]
- Nicolas A, Treco D, Schultes NP, Szostak JW. An initiation site for meiotic gene conversion in the yeast Saccharomyces cerevisiae. Nature. 1989;338:35–39. doi: 10.1038/338035a0. [DOI] [PubMed] [Google Scholar]
- Nickoloff JA, Reynolds RJ. Transcription stimulates homologous recombination in mammalian cells. Mol Cell Biol. 1990;10:4837–4845. doi: 10.1128/mcb.10.9.4837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paldi A, Gyapay G, Jami J. Imprinted chromosomal regions of the human genome display sex-specific meiotic recombination frequencies. Curr Biol. 1995;5:1030–1035. doi: 10.1016/s0960-9822(95)00207-7. [DOI] [PubMed] [Google Scholar]
- Puchta H, Kocher S, Hohn B. Extrachromosomal homologous DNA recombination in plant cells is fast and is not affected by CpG methylation. Mol Cell Biol. 1992;12:3372–3379. doi: 10.1128/mcb.12.8.3372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radman M, Wagner R. Mismatch recognition in chromosomal interactions and speciation. Chromosoma. 1993;102:369–373. doi: 10.1007/BF00360400. [DOI] [PubMed] [Google Scholar]
- Rayssiguier C, Thaler DS, Radman M. The barrier to recombination between Escherichia coli and Salmonella typhimurium is disrupted in mismatch-repair mutants. Nature. 1989;342:396–401. doi: 10.1038/342396a0. [DOI] [PubMed] [Google Scholar]
- Razin A, Cedar H. DNA methylation and genomic imprinting. Cell. 1994;77:473–476. doi: 10.1016/0092-8674(94)90208-9. [DOI] [PubMed] [Google Scholar]
- Rhounim L, Rossignol J-L, Faugeron G. Epimutation of repeated genes in Ascobolus immersus. EMBO J. 1992;11:4451–4457. doi: 10.1002/j.1460-2075.1992.tb05546.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocco V, Nicolas A. Sensing of DNA nonhomology lowers the initiation of meiotic recombination in yeast. Genes in Cells. 1996;1:645–661. doi: 10.1046/j.1365-2443.1996.00256.x. [DOI] [PubMed] [Google Scholar]
- Rossignol J-L, Faugeron G. Gene inactivation triggered by recognition between DNA repeats. Experientia. 1994;50:307–317. doi: 10.1007/BF01924014. [DOI] [PubMed] [Google Scholar]
- Rouyer F, Simmler MC, Page D, Weissenbach J. A sex chromosome rearrangement in a human XX male caused by Alu–Alu recombination. Cell. 1987;51:417–425. doi: 10.1016/0092-8674(87)90637-4. [DOI] [PubMed] [Google Scholar]
- Schultes NP, Szostak JW. A poly(dA.dT) tract is a component of the recombination initiation site at the ARG4 locus in Saccharomyces cerevisiae. Mol Cell Biol. 1991;11:322–328. doi: 10.1128/mcb.11.1.322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwacha A, Kleckner N. Identification of double Holliday junctions as intermediates in meiotic recombination. Cell. 1995;83:783–791. doi: 10.1016/0092-8674(95)90191-4. [DOI] [PubMed] [Google Scholar]
- ————— Interhomolog bias during meiotic recombination: meiotic functions promote a highly differentiated interhomolog-only pathway. Cell. 1997;90:1123–1135. doi: 10.1016/s0092-8674(00)80378-5. [DOI] [PubMed] [Google Scholar]
- Shen P, Huang HV. Homologous recombination in Escherishia coli: Dependence on substrate length and homology. Genetics. 1986;112:441–457. doi: 10.1093/genetics/112.3.441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Small K, Iber J, Warren ST. Emerin deletion reveals a common X chromosome inversion mediated by inverted repeats. Nature Genet. 1997;16:96–99. doi: 10.1038/ng0597-96. [DOI] [PubMed] [Google Scholar]
- Stewart SE, Roeder GS. Transcription by RNA polymerase I stimulates mitotic recombination in Saccharomyces cerevisiae. Mol Cell Biol. 1989;9:3464–3472. doi: 10.1128/mcb.9.8.3464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun H, Treco D, Szostak JW. Extensive 3′-overhanging, single-stranded DNA associated with the meiosis-specific double-strand breaks at the ARG4 recombination initiation site. Cell. 1991;64:1155–1161. doi: 10.1016/0092-8674(91)90270-9. [DOI] [PubMed] [Google Scholar]
- Szostak JW, Orr-Weaver TL, Rothstein RJ, Stahl FW. The double-strand break repair model of recombination. Cell. 1983;33:25–35. doi: 10.1016/0092-8674(83)90331-8. [DOI] [PubMed] [Google Scholar]
- Thomas BJ, Rothstein R. Sex, maps, and imprinting. Cell. 1991;64:1–3. doi: 10.1016/0092-8674(91)90199-9. [DOI] [PubMed] [Google Scholar]
- ————— Elevated recombination rates in transcriptionally active DNA. Cell. 1992;56:619–630. doi: 10.1016/0092-8674(89)90584-9. [DOI] [PubMed] [Google Scholar]
- Thuriaux P. Is recombination confined to structural genes on the eukaryotic genome? Nature. 1977;268:460–462. doi: 10.1038/268460a0. [DOI] [PubMed] [Google Scholar]
- Weiner BM, Kleckner N. Chromosome pairing via multiple interstitial interactions before and during meiosis in yeast. Cell. 1994;77:977–991. doi: 10.1016/0092-8674(94)90438-3. [DOI] [PubMed] [Google Scholar]
- White MA, Detloff P, Strand M, Petes TD. A promoter deletion reduces the rate of mitotic, but not meiotic, recombination at the HIS4 locus in yeast. Curr Genet. 1992;21:109–116. doi: 10.1007/BF00318468. [DOI] [PubMed] [Google Scholar]
- White MA, Dominska M, Petes TD. Transcription factors are required for the meiotic recombination hotspot at the His4 locus in Saccharomyces cerevisiae. Proc Natl Acad Sci. 1993;90:6621–6625. doi: 10.1073/pnas.90.14.6621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitkus R, Doebley J, Lee M. Comparative genome mapping of sorghum and maize. Genetics. 1992;132:1119–1130. doi: 10.1093/genetics/132.4.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu L, Kleckner N. Sequence nonspecific double-strand breaks and interhomolog interactions prior to double-strand break formation at a meiotic recombination hotspot in yeast. EMBO J. 1995;14:5115–5128. doi: 10.1002/j.1460-2075.1995.tb00194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoder JA, Walsh CP, Bestor TB. Cytosine methylation and the ecology of intragenomic parasites. Trends Genet. 1997;13:335–340. doi: 10.1016/s0168-9525(97)01181-5. [DOI] [PubMed] [Google Scholar]