Abstract
The influenza A virus non-structural protein 1 (NS1) is a multifunctional protein and an important virulence factor. It is composed of two well-characterized domains linked by a short, but not well crystallographically defined, region of unknown function. To study the possible function of this region, we introduced alanine substitutions to replace the two highly conserved leucine residues at amino acid positions 69 and 77. The mutant L69,77A NS1 protein retained wild-type (WT)-comparable binding capabilities to dsRNA, cleavage and polyadenylation specificity factor 30 and the p85β subunit of PI3K. A mutant influenza A virus expressing the L69,77A NS1 protein was generated using reverse genetics. L69,77A NS1 virus infection induced significantly higher levels of beta interferon (IFN-β) expression in Madin–Darby canine kidney (MDCK) cells compared with WT NS1 virus. In addition, the replication rate of the L69,77A NS1 virus was substantially lower in MDCK cells but not in Vero cells compared with the WT virus, suggesting that the L69,77A NS1 protein does not fully antagonize IFN during viral replication. L69,77A NS1 virus infection was not able to activate the PI3K/Akt anti-apoptotic pathway, suggesting that the mutant NS1 protein may not be localized such that it has access to p85β in vivo during infection, which was supported by the altered subcellular localization pattern of the mutant NS1 compared with WT NS1 after transfection or virus infection. Our data demonstrate that this linker region between the two domains is critical for the functions of the NS1 protein during influenza A virus infection, possibly by determining the protein’s correct subcellular localization.
Introduction
Influenza A virus is an important human pathogen. The economic impact of seasonal influenza in terms of loss of life, hospitalizations, direct medical costs and loss of earnings impose a heavy burden on society each year and occasional pandemics have caused great morbidity and mortality worldwide (Bridges et al., 2003). It is therefore of great importance to further our understanding of this virus. The genome of influenza A virus consists of eight ssRNA segments encoding 10–11 viral proteins. Encoded by gene segment 8 of influenza A viruses, the non-structural protein 1 (NS1) is a significant virulence-determining factor that functions as a type I interferon (IFN) antagonist (Kochs et al., 2007; Jackson et al., 2008; Li et al., 2006a, b). The NS1 protein is composed of two well-defined domains with X-ray crystallography structures resolved for both domains independently (Liu et al., 1997; Bornholdt & Prasad, 2006). Although the structure of the full-length NS1 protein has recently been determined using the NS1 protein from an H5N1 virus (A/Vietnam/1203/2004) (Bornholdt & Prasad, 2008), the linker region between the two domains still remains largely unresolved by X-ray crystallography. The N-terminal RNA-binding domain binds to dsRNA, an intermediate product of viral replication, thereby preventing the activation of the protein kinase R (PKR) and 2′-5′-oligoadenylate synthetase/RNase L pathways (Lu et al., 1995; Min & Krug, 2006). The N- and C-terminal domains both participate in the dimerization of the NS1 protein, a function essential for NS1’s dsRNA binding (Bornholdt & Prasad, 2008; Wang et al., 1999). The C-terminal effector domain has a broad range of functions including the binding to cleavage and polyadenylation specificity factor 30 (CPSF30), a cellular factor that is crucial for the 3′-end processing of cellular mRNAs (Nemeroff et al., 1998). Binding of NS1 to CPSF30 during infection inhibits the processing of newly synthesized cellular mRNAs, which include those of alpha interferon (IFN-α)/beta interferon (IFN-β)-independent antiviral genes as well as IFN-α/β itself (Noah et al., 2003). It was recently discovered that this domain of the NS1A protein binds to the p85β subunit of PI3K thereby activating the anti-apoptotic PI3K/Akt pathway and preventing infection-induced cell death so as to maximize viral replication (Hale et al., 2006; Ehrhardt et al., 2007; Shin et al., 2007a, b). In addition to binding CPSF30 and PI3K, the effector domain of NS1 protein also interacts with various other viral and cellular proteins including PKR, retinoic acid inducible gene I (RIG-I), poly(A)-binding protein II (PABII) and ubiquitin ligase TRIM25 (Li et al., 2006b; Chen et al., 1999; Gack et al., 2009). These multiple functions of NS1 protein have been reviewed in detail elsewhere (Hale et al., 2008).
The two domains of the influenza virus NS1 protein are connected by a short linker region of unknown function and unresolved structure. The amino acid sequence of this region is conserved among all influenza A virus strains, suggesting an important viral function. The domain linker regions of other proteins are known to be crucial for their functions, either by providing an interaction site with other proteins, facilitating communications or preventing unfavourable interactions between domains (George & Heringa, 2002; Gonfloni et al., 1997; Mattison et al., 2002). The electron density of NS1 protein amino acid residues 67–74 is relatively weak and the density of amino acid residues 75–79 is not well defined, indicating that this linker region is flexible (Bornholdt & Prasad, 2008). It has also been suggested that the linker region may participate in the modulation of the RNA-binding domain dimerization in a proposed model of dsRNA sequestration (Bornholdt & Prasad, 2008).
In the current study, we investigated the function of the NS1 protein linker region by introducing alanine substitutions to replace the two highly conserved residues (leucine 69 and 77). Using reverse genetics, an influenza A virus expressing this mutant NS1 protein was generated and characterized to show differences in replication, which may alter virulence.
Results
Conservation of amino acid sequence of NS1 protein linker region among influenza strains
The conservation of amino acid sequence within this linker region of the NS1 protein strongly suggests functions in addition to separating the RNA-binding domain and the effector domain (Table 1). To test this hypothesis, we introduced alanine substitutions to replace the highly conserved leucine 69 and leucine 77. Leucine repeats, zippers and leucine-rich domains have been shown to have many various protein interactions. In addition, leucine and alanine both have hydrophobic side chains and are of relatively similar size such that this substitution would be less likely to cause a conformational change that would non-specifically alter the protein function than some of the other options available within this region. For this study, we chose to utilize the NS gene from a more recent human clinical isolate, A/New York/55/04, for these experiments as opposed to using the lab-adapted A/Udorn/72 NS gene.
Table 1. The conservation of amino acid sequence in NS1 protein domain linker.
Based on the amino acid sequence in the domain linker of influenza A/New York/55/04 NS1 protein, the frequency of the amino acid at each position was calculated by using human H1N1, human H3N2, and all avian influenza strains listed at the National Institute of Allergy and Infectious Diseases (NIAID) Influenza Research Database (IRD) online database at http://www.fludb.org. The leucines at amino acid positions 69 and 77 are in bold.
| Amino acids | Frequency of the linker region amino acids listed | |||
| Human H1N1 | Human H3N2 | All avian strains | ||
| 67 | K | 2423/7233 (33.5 %) | 2377/2545 (93.4 %) | 27/6300 (0.4 %) |
| 68 | I | 7211/7233 (99.7 %) | 2538/2545 (99.7 %) | 6249/6300 (99.2 %) |
| 69 | L | 7216/7233 (99.8 %) | 2537/2545 (99.7 %) | 6260/6300 (99.4 %) |
| 70 | K | 6943/7233 (96.0 %) | 2531/2545 (99.5 %) | 1474/6301 (23.4 %) |
| 71 | E | 6664/7233 (92.1 %) | 2014/2545 (79.1 %) | 4577/6301 (72.6 %) |
| 72 | E | 7217/7233 (99.8 %) | 2538/2545 (99.7 %) | 6272/6301 (99.5 %) |
| 73 | S | 7194/7233 (99.5 %) | 2542/2545 (99.9 %) | 4916/6301 (78.0 %) |
| 74 | D | 4074/7233 (56.3 %) | 2527/2545 (99.3 %) | 6155/6301 (97.7 %) |
| 75 | E | 7144/7233 (98.8 %) | 2535/2545 (99.6 %) | 5789/6301 (91.9 %) |
| 76 | A | 4062/7233 (56.2 %) | 2530/2545 (99.4 %) | 4798/6301 (76.2 %) |
| 77 | L | 6834/7233 (94.5 %) | 2534/2545 (99.6 %) | 6239/6301 (99.0 %) |
| 78 | K | 4079/7233 (56.4 %) | 2513/2545 (98.7 %) | 6073/6301 (96.4 %) |
| 79 | M | 7072/7232 (97.8 %) | 2535/2545 (99.6 %) | 4853/6301 (77.0 %) |
dsRNA and CPSF30 binding affinity of L69,77A and wild-type (WT) NS1 proteins
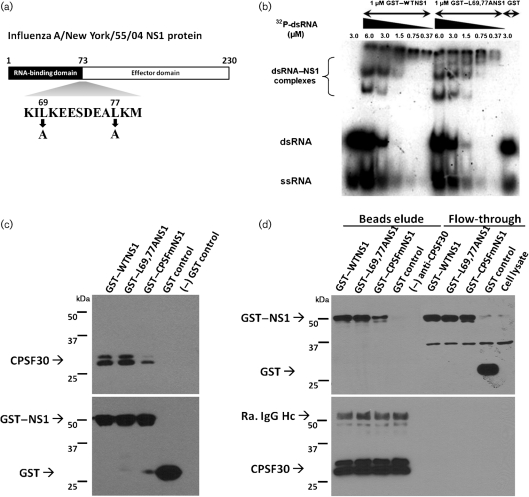
To confirm that the L69,77A mutations (Fig. 1a) within the domain linker region did not destroy known functions of the NS1 protein as a result of misfolding, dsRNA- and CPSF30-binding assays were performed. In a gel shift assay using Escherichia coli-synthesized glutathione S-transferase (GST)-tagged NS1 proteins, the GST–L69,77A NS1 protein had a similar affinity to dsRNA compared to the GST–WT NS1 protein. While the NS1 : dsRNA ratio increased, more NS1 molecules were incorporated into a single NS1–dsRNA complex, as indicated by the increase in the apparent molecular mass of the NS1–dsRNA complexes on the gel (Fig. 1b). Similarly, GST pull-down assays and co-immunoprecipitation assays using E. coli-synthesized GST-tagged NS1 indicated that the L69,77A NS1 protein also bound to CPSF30 as efficiently as the WT NS1 protein while the CPSF30-binding deficient mutant NS1 protein (Noah et al., 2003) that served as a control had a lower affinity to CPSF30 compared with both GST–WT NS1 and GST–L69,77A NS1 (Fig. 1c, d). These results indicate that the L69,77A mutations do not result in misfolding of either the N-terminal RNA-binding domain or the C-terminal effector domain of the NS1 protein.
Fig. 1.
L69,77A NS1 protein has similar dsRNA and CPSF30-binding affinity compared to WT NS1 protein. (a) Schematic representation of influenza A/New York/55/04 NS1 protein and the mutations employed in this study. The domain hinge region is highlighted. (b) Various amounts (0.37–6.0 µM) of 32P-labelled dsRNA (55 bp) were incubated with 1 µM GST–WTNS1 protein or GST–L69,77A NS1 protein in vitro and resolved by 7 % non-denaturing PAGE. (c, d) CPSF30 co-precipitates with both WT NS1 and L69,77A NS1. HeLa cell lysates were mixed with equal amounts of GST-tagged WT NS1, L69,77A NS1, a CPSF30-binding deficient NS1 (CPSFm) and GST control. Proteins were precipitated with glutathione Sepharose 4B beads (c) or with anti-CPSF30 conjugated dynobeads protein A (d). Precipitated proteins and flow-throughs were analysed by Western blot. Molecular masses are indicated in the left.
Comparison of growth and cytopathic effect of L69,77A NS1 virus and WT NS1 virus
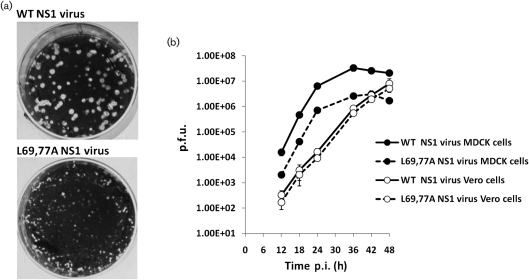
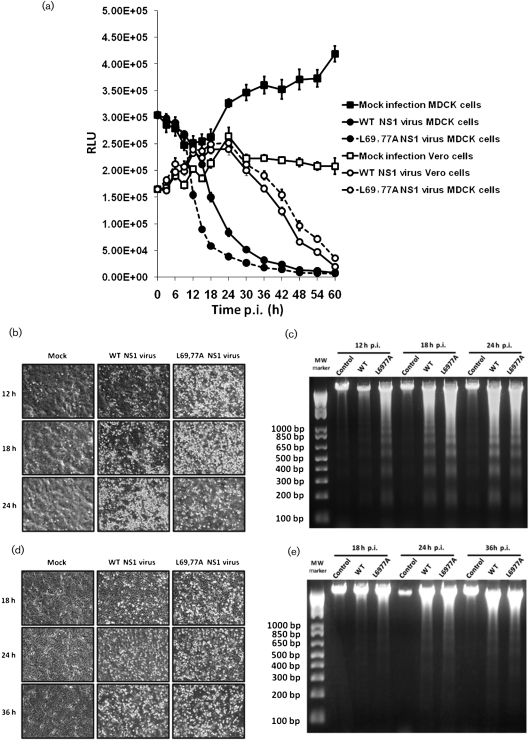
To investigate the effect of the mutations on viral replication, a mutant virus expressing the L69,77A NS1 protein was generated by reverse genetics. This L69,77A NS1 mutant virus was able to replicate in Madin–Darby canine kidney (MDCK) cells and embryonic chicken eggs but did so at an attenuated rate as evidenced by its significantly smaller plaques compared with those formed by the WT NS1 virus (Fig. 2a). The L69,77A NS1 virus also grew to titres ≥10-fold lower compared with those obtained with the WT NS1 virus when MDCK cells were infected at an m.o.i. of 0.001 and virus was allowed to spread. However, the L69,77A NS1 virus was able to grow to titres similar to the WT NS1 virus in Vero cells (Fig. 2b), which lack the ability to make type I IFNs (Emeny & Morgan, 1979). When MDCK cells were infected at an m.o.i. of 5, the L69,77A NS1 mutant virus induced more cell death compared with the WT NS1 virus (Fig. 3a). DNA fragmentation assay confirmed that the cell death was due to apoptosis rather than necrosis (Fig. 3b, c). This difference in virus induced cell death, like that observed in replication, was not significant in Vero cells (Fig. 3d, e).
Fig. 2.
Replication of the L69,77A NS1 virus is attenuated in MDCK cells but not in Vero cells. (a) Plaques formed by WT NS1 virus and L69,77A NS1 virus on MDCK cell monolayers. (b) Multi-step growth curve of WT NS1 virus and L69,77A NS1 virus in MDCK and Vero cells. MDCK cells and Vero cells were infected with WT NS1 or L6977A NS1 virus at an m.o.i. of 0.001. Media from infected cultures were titrated by plaque assay at 12, 18, 24, 36, 42 and 48 h post-infection (p.i.).
Fig. 3.
L69,77A NS1 virus infection induces more apoptosis in MDCK cells, but not in Vero cells, compared with the WT NS1 virus. (a) MDCK and Vero cells grown on 96-well plates were mock infected or infected with WT NS1 or L69,77A NS1 viruses at an m.o.i. of 5. Cell viability after infection, as reflected by relative luminescence unit (RLU), was determined by CellTiter-Glo assay at every 3 h until 18 h and every 6 h afterwards until 60 h. (b–e) MDCK and Vero cells grown on six-well plates were mock infected or infected with viruses at an m.o.i. of 5. At various time points (12, 18 and 24 h for MDCK cells and 18, 24 and 32 h for Vero cells) after infection, images were collected by a phase-contrast microscope (b and d in MDCK and Vero cells, respectively) and DNA fragmentation was analysed by 1.5 % agarose gels (c and e in MDCK and Vero cells, respectively).
L69,77A NS1 protein is not an efficient type I IFN antagonist
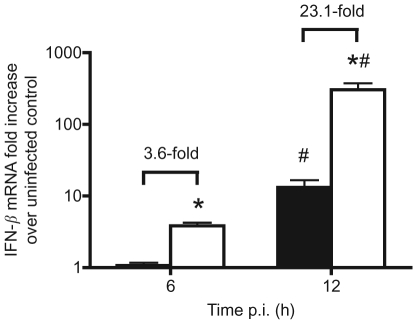
Since Vero cells are unable to produce type I IFNs (Emeny & Morgan, 1979), the observation that the differences in viral growth rate and apoptosis-inducing effects between the L69,77A NS1 and the WT NS1 viruses were not significant in Vero cells indicated that the L69,77A NS1 protein may not be an efficient type I IFN antagonist. To test this hypothesis, MDCK cells were infected with the L69,77A NS1 virus or the WT NS1 virus at m.o.i. of 5 and IFN-β induction was measured by quantitative real-time PCR. In MDCK cells, infection with the L69,77A NS1 virus induced significantly higher levels of IFN-β expression (3.6-fold and 23.1-fold, respectively) at 6 and 12 h after infection compared with infection with the WT NS1 virus (Fig. 4). This indicates that despite the ability of L69,77A NS1 to bind to dsRNA and CPSF30 in vitro, it cannot suppress the induction of type I IFNs during viral infection.
Fig. 4.
L69,77A NS1 virus infection induces higher levels of IFN-β expression compared with WT NS1 virus infection. MDCK cells were mock infected or infected with WT NS1 or L69,77A NS1 viruses at an m.o.i. of 5. Total RNA was extracted and IFN-β expression was determined by quantitative real-time PCR following reverse transcription. Data are expressed as folds of induction over the mock-infected control. The folds of increased IFN-β expression in L69,77A NS1 virus-infected cells (empty bars) over WT NS1 virus-infected cells (filled bars) are also indicated. *, Significant difference relative to the WT NS1 virus within the same time point; #, significant difference between 6 and 12 h.
PI3K/Akt pathway is not activated by L69,77A NS1 virus infection but L69,77A can bind p85β in vitro
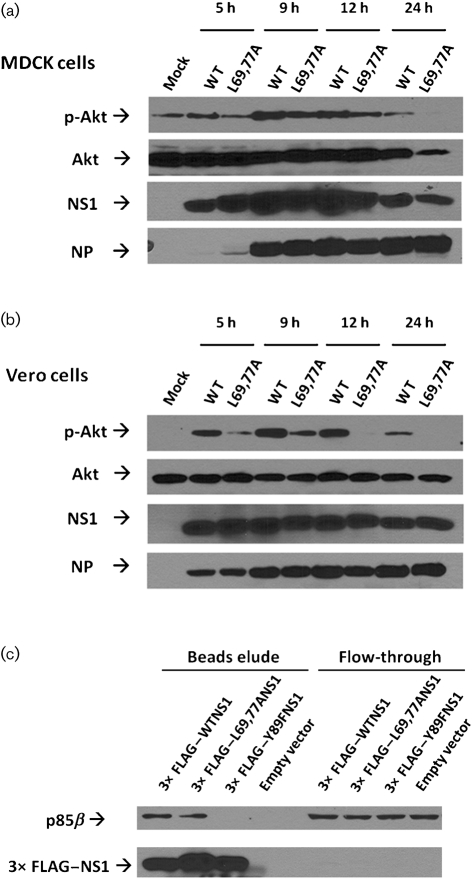
A previous study demonstrated that an NS1-protein-deficient influenza virus (del NS1 virus) induced a greater amount of apoptosis during infection compared with the WT control virus and type I IFNs were thought to play a major role in that difference (García-Sastre et al., 1998). However, since then it has also been discovered that the NS1 protein also directly inhibits apoptosis by binding to the PI3K regulatory subunit p85β and activating the anti-apoptotic/pro-survival PI3K/Akt pathway (Hale et al., 2006; Ehrhardt et al., 2007; Shin et al., 2007a, b). To test whether L69,77A NS1 virus infection would activate the PI3K/Akt pathway, MDCK and Vero cells were infected at an m.o.i. of 5 and the amount of phospho-Akt (p-Akt) in whole-cell lysates was measured by Western blot. The L69,77A NS1-virus-infected cells had less p-Akt but an equal amount of total Akt compared to the WT NS1-virus-infected cells at 5, 9 and 12 h after infection, respectively. At 24 h after infection, there was less total Akt in the L69,77A NS1-virus-infected MDCK cells compared with the WT NS1-virus-infected MDCK cells, and this was probably due to the greater apoptosis caused by L69,77A NS1 virus infection (Fig. 5a, b). Interestingly, despite the inability of the L69,77A NS1 virus to activate the PI3K/Akt pathway during infection, the L69,77A NS1 protein retained p85β-binding capacity in vitro (Fig. 5c), suggesting that L69,77A NS1 protein may have lost access to p85β during infection.
Fig. 5.
The L69,77A NS1 protein binds to the p85β regulatory subunit of PI3K but L69,77A NS1 virus infection does not activate the PI3K/Akt pathway. MDCK cells (a) and Vero cells (b) were either mock infected, infected with the WT NS1 virus, or the L69,77A NS1 virus at an m.o.i. of 5. The p-Akt, total Akt, viral NS1 and viral NP proteins from whole-cell lysates at 5, 9, 12 and 24 h after infection were detected by Western blot. (c) 293T cells were transfected with plasmids expressing 3× FLAG-tagged WT NS1, L69,77A NS1 or p85β-binding deficient Y89F NS1 proteins. The pCMV10-3× FLAG vector without inserts was also used as a control. Cells were also co-transfected with pCMV6-XL6-p85β plasmid expressing p85β. At 24 h after transfection, 3× FLAG-NS1 and associate proteins were immunoprecipitated with anti-FLAG M2 affinity beads. FLAG-tagged NS1 protein and p85β were detected by Western blot.
L69,77A NS1 protein has altered subcellular localization
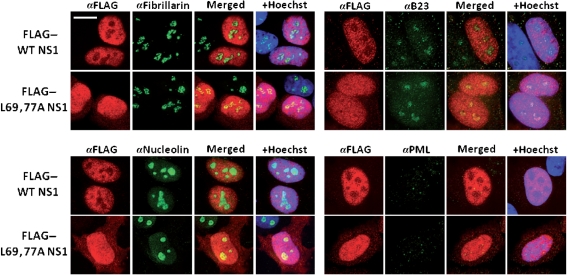
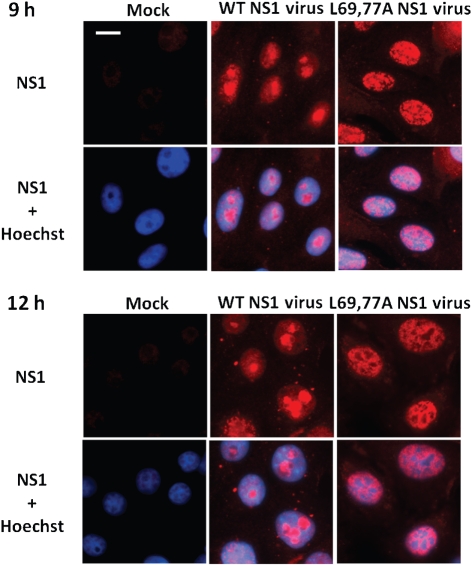
The NS1 protein is mainly localized to the nuclei of transfected or infected cells (Greenspan et al., 1988). A mutant virus expressing an NS1 protein (R38K41A NS1) that mislocalized to the cytoplasm was substantially attenuated (Min & Krug, 2006; Donelan et al., 2003). Infecting MDCK cells with this mutant virus induced significantly higher levels of IFN-β expression compared with the control WT virus (Donelan et al., 2003). Because the L69,77A NS1 virus infection induced higher levels of IFN-β and lower levels of Akt phosphorylation, yet the L69,77A NS1 protein retained in vitro dsRNA-, CPSF30- and p85β-binding functions, we then examined whether the intracellular localization of NS1 was affected by the L69,77A mutations. HeLa cells were transfected with plasmids expressing 3×FLAG-tagged L69,77A NS1 protein or WT NS1 protein. The localization of 3×FLAG–NS1 proteins was illustrated by a FLAG-tag specific antibody. The WT NS1 protein localized to the nuclei of transfected cells and was largely excluded from the nucleoli while the L69,77A NS1 protein distributed homogeneously throughout the nuclei without being excluded from the nucleoli, illustrated by fibrillarin, B23 and nucleolin staining. We also stained promyelocytic leukaemia (PML) nuclear bodies to demonstrate that the nuclear structure was not affected by the accumulation of the NS1 protein in the nuclei (Fig. 6). Next, we examined the subcellular localization of the L69,77A NS1 protein during viral infection. Vero cells were infected at an m.o.i of 5. At 9 and 12 h after infection, cells were fixed with acetone and permeabilized with methanol, as required for use of this particular NS1 antibody, and then stained with NS1 antibody. Like the differential pattern observed in transfected cells, in infected cells the L69,77A NS1 protein clearly had a different staining pattern compared with WT NS1 protein, indicating that the mutations in the domain linker region altered NS1 localization during viral replication (Fig. 7). NS1 staining of infected MDCK cells showed similar patterns to Vero cells (data not shown).
Fig. 6.
L69,77A NS1 protein has altered subcellular localization compared with WT NS1 protein. HeLa cells were transfected with 3× FLAG-tagged NS1 expression plasmids. Cells were fixed and permeablized at 24 h post-transfection. 3× FLAG-tagged NS1, fibrillarin, nucleolin, nucleolar phosphoprotein B23 and PML bodies were detected with antibodies that are specific to FLAG, fibrillarin, nucleolin, B23 and PML, respectively, in combination with Alexa Fluor 568 (red) or Alexa Fluor 488 (green) conjugated secondary antibodies. Bar, 10 µm.
Fig. 7.
L69,77A NS1 protein has altered subcellular localization compared with WT NS1 protein during viral infection. Vero cells were mock infected or infected with WT NS1 or L69,77A NS1 viruses at an m.o.i. of 5. Cells were fixed, permeablized at 9 and 12 h after infection. NS1 protein was detected with NS1 antibody in combination with an Alexa Fluor 568 conjugated secondary antibody. Bar, 10 µm.
Discussion
This study was designed to investigate the possible function(s) of the domain linker region of the influenza A virus NS1 protein, a region that previously had no well defined functions. We found that despite the ability of the L69,77A NS1 protein to bind to dsRNA and CPSF30 in vitro, the recombinant virus expressing L69,77A NS1 replicated to lower titres in MDCK cells but not in Vero cells compared with the control virus carrying WT NS1. Moreover, when infected at multiple m.o.i., the L69,77A NS1 virus induced more apoptosis in MDCK cells compared with the WT NS1 virus. Based on the observation that these differences occurred in MDCK cells but not in type I IFN-deficient Vero cells, we suspected that the L69,77A NS1 protein might not function as an efficient type I IFN antagonist during infection, which was confirmed by quantitative real-time PCR measuring infection-induced IFN-β expression.
The PI3K/Akt pathway is the cellular ‘survival’ pathway that mediates various anti-apoptotic responses (Franke et al., 1997). PI3K exists as a heterodimer in vivo consisting of a p110 catalytic subunit and a p85 (p85α or p85β) regulatory subunit (Yu et al., 1998). The p85 regulatory subunit stabilizes the p110 subunit and inhibits its activity (Yu et al., 1998). Recently, it was demonstrated that during influenza A virus infection, the NS1 protein bound to the p85β (but not p85α) regulatory subunit and induced PI3K activation (Hale et al., 2006; Ehrhardt et al., 2007; Shin et al., 2007a; Ehrhardt et al., 2006). The suppression of premature apoptosis by activating PI3K/Akt during infection is advantageous to the virus. Because the infection-induced apoptosis was greater in MDCK cells infected with the L69,77A NS1 virus compared with those infected with the WT NS1 virus, we examined whether infection with the two viruses would induce different levels of PI3K/Akt activation. Our data indicate that in both MDCK cells and Vero cells, L69,77A NS1 virus infection induces less PI3K/Akt activation compared with WT NS1 virus infection and the difference is more significant in Vero cells. Although in MDCK cells, the lower amount of p-Akt induced by infection may be attributed to the decreased amount of NS1 protein as the result of slower viral replication compared with WT NS1 virus. The results from infected Vero cells clearly demonstrate that the L69,77A NS1 protein did not induce PI3K/Akt activation. In Vero cells, the amount of NS1 did not differ between the L69,77A NS1 virus and the WT NS1 virus-infected cells. However, the amount of p-Akt was lower in the L69,77A NS1 virus-infected cells. Interestingly, a study by Jackson et al. (2010) demonstrated that the loss of PI3K activation during viral infection has no effect on cell viability. This is supported by our data in Vero cells, as L69,77A NS1 virus infection did not result in increased apoptosis compared to WT NS1 virus infection. Since the activation of PI3K/Akt pathway by influenza A viruses is a result of a direct interaction between the viral NS1 protein and the PI3K p85β regulatory subunit (Hale et al., 2006; Ehrhardt et al., 2006; Shin et al., 2007a), we next tested whether the L69,77A NS1 protein was able to bind to p85β, and our co-immunoprecipitation data suggested that the L69,77A NS1 protein bound efficiently to p85β. These data, when taken together, demonstrated that although the L69,77A NS1 protein was able to bind to many known cellular components, including dsRNA, CPSF30 and p85β, as efficiently as the WT NS1 protein in vitro, the mutant NS1 did not appear to efficiently regulate the related cellular functions within the context of infection. Therefore, we speculated that the mutations in the linker region result in mislocalization of NS1 in such a way that the NS1 protein does not have normal access to either dsRNA, CPSF30 or p85β in vivo during infection, a hypothesis that is supported by our transfection experiments in HeLa cells and infection experiments in Vero and MDCK cells. The differences in NS1 localization pattern between transfection and infection were probably a result of the interaction between NS1 and other viral components (e.g. viral RNA, viral proteins). However, because acetone fixation is required to use this particular NS1 antibody, we cannot rule out the possibility that these differences are due to the difference in fixation methods. Regardless of the fixation method, both the WT NS1 and the mutant NS1 were fixed using the same methods, and clearly had different localization patterns during either transfection or infection. The mislocalization could also result in altered or ineffective access or compartmentalization with other known and unknown cellular or viral proteins required for viral replication. It is also possible that the L69,77A NS1 protein binds to but does not activate PI3K, or that the NS1 protein requires interaction with another cellular or viral protein to have these functions, and this domain linker region is essential for that interaction and the mislocalization of the mutant NS1 is a phenotype rather than the cause of the lost function(s). It should be noted that the importance of this linker region to the overall NS1 protein structure is not clear. For instance, this region may be critical in determining the relative positions of the N- and C-terminal domains. Mutations within this region may affect the conformation of the full-length NS1 protein. We are investigating these possibilities.
To our knowledge, our study is the first to demonstrate the importance of the domain linker of the influenza A virus NS1 protein to the virus. Our data demonstrate that this region plays a critical role in the function of influenza A virus NS1 protein during infection and warrant further studies on the functions of this region. Mutations within this region result in mislocalization of the NS1 protein. The increased apoptosis in the L69,77A NS1-infected MDCK cells are probably a result of the combination of both L69,77A NS1’s inability to suppress type I IFN expression and its failure to activate the PI3K/Akt pathway.
Methods
Sequence analysis.
The amino acid sequences of NS1 of human H1N1 and H3N2 influenza subtypes and avian influenza subtypes was obtained from the NIAID IRD online through the website at http://www.fludb.org. The conservation of amino acids at each position in the domain linker region (in reference to the A/New York/55/04 strain) was calculated using the IRD analysis tool at the website.
Cells and viruses.
The HeLa, 293T, Vero and MDCK cell lines were obtained from the ATCC. HeLa cells were maintained in minimum essential medium (Mediatech) supplemented with 0.075 % sodium bicarbonate (Mediatech), 1 mM sodium pyruvate (Mediatech), 0.4× non-essential amino acids (Mediatech) and 10 % FBS (Hyclone). 293T, Vero and MDCK cells were maintained in Dulbecco’s modified Eagle’s medium (Invitrogen) supplemented with 10 % FBS (Hyclone). For the generation of recombinant influenza A/Udorn/72 viruses expressing WT or L69,77A NS1 of influenza A/New York/55/04, viral RNA was extracted from influenza A/New York/55/04 stock using Viral RNA Mini kit (Qiagen) and RT-PCR was performed using the SuperScript III First-strand cDNA Synthesis kit (Invitrogen). Alanine substitutions were performed by using two rounds of PCR with specific primers. NS1 DNA was sequenced and cloned into pHH21 plasmid and co-transfected into 293T cells together with pHH21 plasmids containing the full-length cDNA for each of the other seven genomic RNA segments of influenza A/Udorn/72 and four pcDNA plasmids encoding PB1, PB2, PA and nucleoprotein (NP) proteins of influenza A/Udorn/72. Transfections were performed using the TransIT-LT1 transfection reagent (Mirus). Cells were cultured in Opti-MEM I medium (Invitrogen) containing 3 µg acetylated trypsin (Sigma) ml−1. At 24 h after transfection, MDCK cells were added to the 293T cell culture to sustain viral replication. The supernatants of the 293T/MDCK co-culture were collected after the majority of MDCK cells detached as a result of viral replication. Viruses were titrated by plaque assays on MDCK cell monolayers and individual plaques were amplified and propagated in 6–10 day-old hen eggs (Charles River). Mutations of NS1 gene were confirmed by sequencing analysis.
Plasmids and fusion proteins.
To generate N-terminal 3×FLAG-tagged WT NS1, L69,77A NS1 and Y89F NS1 constructs, the ORF encoding NS1 was amplified from influenza A/New York/55/04 cDNA using appropriate primers. Mutations were generated by site-directed mutagenesis as described earlier. The PCR products were cloned into the pCMV10-3×FLAG expression plasmid (Sigma) downstream of the sequence encoding the 3×FLAG tag, using EcoRI and BamHI restriction sites. To prepare GST–NS1 fusion proteins, PCR products encoding A/New York/55/04 WT NS1, L69,77A NS1 and CPSF30-binding deficient NS1 (Noah et al., 2003) were cloned into pGEX-4T-2 plasmid (GE Lifesciences) using EcoRI and BamHI restriction sites. The recombinant constructs were transformed into E. coli strain BL21 and the expressed GST fusion proteins were purified by affinity chromatography using glutathione Sepharose 4B beads (GE Lifesciences). The p85β expression plasmid (pCMV6-XL6-p85β) was purchased commercially (Origene).
Viral growth curve and plaque assay.
For the multi-step growth curves of the viruses, MDCK cells and Vero cells were infected with viruses at 0.001 p.f.u. per cell in Opti-MEM I medium (Invitrogen) containing 3 µg acetylated trypsin (Sigma) ml−1. After 30 min of infection, cells were washed three times with Dulbecco’s PBS (Invitrogen) and resupplied with fresh Opti-MEM I medium containing 3 µg acetylated trypsin ml−1. At various time points, medium samples were collected and viruses were titrated by plaque assay as described previously (Noah et al., 2003).
CellTiter-Glo luminescent cell viability assay.
Infection-induced cell death was measured by the CellTiter-Glo luminescent cell viability assay (Promega) as described previously (Noah et al., 2007).
DNA fragmentation assay for apoptosis.
Low molecular mass cellular DNA from cells was extracted using the high salt extraction method (Hirt, 1967). Briefly, cells grown on six-well tissue culture plates were gently scraped off with bent pipette tips and centrifuged at 1500 g for 10 min at 4 °C. Cell pellets were resuspended in 80 µl PBS (Invitrogen) and mixed with 300 µl buffer containing 10 mM Tris/HCl (pH 7.6), 10 mM EDTA and 0.6 % SDS. The cell suspensions were then mixed with 100 µl 5 M NaCl and incubated at 4 °C overnight. The next day, the mixtures were centrifuged at 13 000 g for 30 min at 4 °C. Supernatants were transferred to a new set of tubes and RNase A (Qiagen) and proteinase K (Promega) were added to each sample to final concentrations of 1 and 0.2 mg ml−1, respectively. Samples were incubated for 20 min in a 37 °C water bath for a complete digestion of RNAs and proteins. Then two volumes of ethanol were mixed with each sample and incubated at −20 °C overnight. Samples were centrifuged at 13 000 g for 30 min to pellet precipitated DNA. DNA pellets were dissolved in 30 µl TE buffer (10 mM Tris/HCl, 1 mM EDTA; pH 7.6) and separated on 1.5 % agarose gels followed by ethidium bromide staining. Molecular masses were indicated by 1 kb plus DNA ladder (Invitrogen).
Quantitative real-time PCR.
Total RNA was extracted from MDCK cells using RNeasy Mini kit (Qiagen) and RT-PCR was performed using the SuperScript III First-strand cDNA Synthesis kit (Invitrogen). Expression of IFN-β gene was determined by quantitative real-time PCR using canine IFN-β specific primers and probe (Applied Biosystems). The amount of 18S rRNA was determined for all samples using pan-species 18S rRNA primers and probe (Applied Biosystems) and used to normalize IFN-β expression.
RNA-binding assay.
The 56 bp 32P-labelled dsRNAs were prepared by annealing the sense and antisense transcripts of pCR3.1 plasmid poly-linker sequence that was synthesized using MEGAscript T7 kit (Ambion). The indicated amounts of GST–NS1 proteins were mixed with this 32P-labelled dsRNA in 10 µl RNA-binding buffer (50 mM Tris/HCl, pH 8.0, 50 mM KCl, 8 % glycerol, 2.5 mM DTT, 50 µg carrier RNA ml−1 and 0.5 U RNasin µl−1) and incubated on ice for 30 min. The GST–NS1–dsRNA complexes were resolved from free RNA by non-denaturing electrophoresis on a 7 % polyacrylamide gel at 4 °C at 100V using a 45 mM TBE buffer.
GST-pull down.
The GST-pull down assay was performed as described previously (Nemeroff et al., 1995) with minor modifications. Briefly, equal amounts of purified GST–WT NS1, GST–L69,77A NS1, GST–CPSFm NS1 or GST control protein were incubated with HeLa cell lysates in an NS1 CPSF30-binding buffer [50 mM Tris/HCl, 150 mM NaCl, 0.25 % NP-40 plus protease inhibitors (Sigma), pH 8.0] together with 30 µl glutathione Sepharose 4B beads (GE Lifesciences) at 4 °C for 3 h with gentle shaking. The beads were then washed three times gently with the NS1 CPSF30-binding buffer and bead-bound proteins were eluded with 30 µl 2× Laemmli sample buffer (Bio-Rad).
Co-immunoprecipitation.
For the CPSF30 GST–NS1 co-immunoprecipitation experiments, HeLa cells were lysed in the NS1 CPSF30-binding buffer as described above and incubated with equal amounts of purified GST–WT NS1, GST–L69,77A NS1, GST–CPSFm NS1 or GST control protein together with rabbit anti-CPSF30 (Bethyl Laboratories) conjugated Dynobeads protein A (Invitrogen) at 4 °C for 3 h with gentle shaking. For the 3× FLAG–NS1 p85β co-immunoprecipitation experiment, 293T cells were co-transfected with the pCMV6-XL6-p85β plasmid expressing p85β or the plasmid expressing 3× FLAG-tagged WT NS1, L69,77A NS1 or Y89F NS1. After 24 h cells were lysed in immunoprecipitation buffer [50 mM Tris/HCl, pH 7.5, 200 mM NaCl, 0.1 % Tween 20 plus protease inhibitors (Sigma)]. The cell lysates were incubated with 30 µl EZview Red Anti-FLAG M2 affinity gel (Sigma) at 4 °C for 3 h with gentle shaking. The beads were then washed three times gently with the immunoprecipitation buffer and bead-bound proteins were eluded with 30 µl 2× Laemmli sample buffer (Bio-Rad).
Electrophoresis and Western blotting.
Samples were separated by 10 % (wt : v) SDS-polyacrylamide gels and electrophoretically blotted onto PVDF membranes (Invitrogen) according to standard procedures. The membranes were blocked with 5 % non-fat milk. For GST-pull down and co-immunoprecipitation experiments, membranes were treated with rabbit anti-CPSF30 (Bethyl Laboratories), rabbit anti-GST (Santa Cruz), mouse anti-FLAG M2 (Sigma) and mouse anti-p85β (Serotech). For the p-Akt measurement, cells were lysed in cold M-PER whole-cell lysis buffer (Thermo) with protease inhibitors (Sigma) followed by electrophoresis and Western blot. Membranes were treated with rabbit anti-p-Akt (Cell Signaling Technology), rabbit anti-Akt (Cell Signaling Technology), and an influenza antibody that reacts with NP protein (GeneTex). The anti-influenza NS1 antibody was a kind gift from Dr Yan Zhou (Vaccine and Infectious Diseases Organization, University of Saskatchewan, Canada). All primary antibodies were diluted in PBS containing 5 % BSA and incubated with membranes overnight at 4 °C. Membranes were then incubated with HRP-conjugated secondary antibodies (Santa Cruz) in 5 % non-fat milk for 1 h at room temperature and stained with SuperSignal West Pico Chemiluminescent Substrate (Thermo).
Immunofluorescence.
HeLa cells were transfected with 3×FLAG-tagged NS1 expression plasmids using the TransIT-LT1 transfection reagent (Mirus). Cells were fixed at 24 h post-transfection with 4 % paraformaldehyde and permeablized with 0.1 % Triton-100. Cells were stained with anti-FLAG M2 mAb (Sigma) and Alexa Fluor 568 secondary antibody. Fibrillarin, nucleolin, nucleolar phosphoprotein B23 and PML bodies were detected with anti-fibrillarin rabbit polyclonal antibody (Abcam), anti-nucleolin rabbit polyclonal antibody (Santa Cruz), anti-B23 rabbit polyclonal antibody (Santa Cruz) and anti-PML rabbit polyclonal antibody (Santa Cruz), respectively. An Alexa Fluor 488 conjugated antibody was used as the secondary antibody. Nuclei were counterstained with Hoechst 33258 (Sigma). A Nikon TE2000 U inverted epifluorescence microscope, Coolsnap HQ CCD camera (Photometrics) and Metamorph software (Universal Imaging) were used for image acquisition. For the infection experiment, Vero cells were grown on sterilized microscope coverslips in six-well plates and mock infected, or infected by WT or L69,77A NS1 virus at an m.o.i. of 5. NS1 immunofluorescent staining was performed as previously described with modifications (Shin et al., 2007b). Briefly, at 9 and 12 h after infection, coverslips were taken out of the plates and fixed in acetone/methanol (1 : 1) for 15 min at −20 °C. After rehydration with PBS, cells were stained with an anti-NS1 antibody and an Alexa Fluor 556 secondary antibody. Nuclei were counterstained with Hoechst 33258 (Sigma). A Zeiss Axiovert 200 microscope and AxioVision 4.5 system software (Zeiss) were used for image acquisition.
Statistical analysis.
Real-time PCR data were log-transformed and analysed using two-way ANOVA followed by Tukey HSD test. All analyses were performed with SigmaStat (Jandel Scientific). P<0.001 was considered significant.
Acknowledgements
This research was supported by NIAID grant R21 AI70758 to D. L. N.
Footnotes
Present address: The W. Harry Feinstone Department of Molecular Microbiology and Immunology, The Johns Hopkins Bloomberg School of Public Health, Baltimore, MD 21218, USA.
References
- Bornholdt Z. A., Prasad B. V. (2006). X-ray structure of influenza virus NS1 effector domain. Nat Struct Mol Biol 13, 559–560 10.1038/nsmb1099 [DOI] [PubMed] [Google Scholar]
- Bornholdt Z. A., Prasad B. V. (2008). X-ray structure of NS1 from a highly pathogenic H5N1 influenza virus. Nature 456, 985–988 10.1038/nature07444 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bridges C. B., Harper S. A., Fukuda K., Uyeki T. M., Cox N. J.,, Singleton J. A., Advisory Committee on Immunization Practices (2003). Prevention and control of influenza. Recommendations of the Advisory Committee on Immunization Practices (ACIP). MMWR Recomm Rep 52(RR-8), 1–34 [PubMed] [Google Scholar]
- Chen Z., Li Y., Krug R. M. (1999). Influenza A virus NS1 protein targets poly(A)-binding protein II of the cellular 3′-end processing machinery. EMBO J 18, 2273–2283 10.1093/emboj/18.8.2273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donelan N. R., Basler C. F., García-Sastre A. (2003). A recombinant influenza A virus expressing an RNA-binding-defective NS1 protein induces high levels of beta interferon and is attenuated in mice. J Virol 77, 13257–13266 10.1128/JVI.77.24.13257-13266.2003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehrhardt C., Marjuki H., Wolff T., Nürnberg B., Planz O., Pleschka S., Ludwig S. (2006). Bivalent role of the phosphatidylinositol-3-kinase (PI3K) during influenza virus infection and host cell defence. Cell Microbiol 8, 1336–1348 10.1111/j.1462-5822.2006.00713.x [DOI] [PubMed] [Google Scholar]
- Ehrhardt C., Wolff T., Pleschka S., Planz O., Beermann W., Bode J. G., Schmolke M., Ludwig S. (2007). Influenza A virus NS1 protein activates the PI3K/Akt pathway to mediate antiapoptotic signaling responses. J Virol 81, 3058–3067 10.1128/JVI.02082-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emeny J. M., Morgan M. J. (1979). Regulation of the interferon system: evidence that Vero cells have a genetic defect in interferon production. J Gen Virol 43, 247–252 10.1099/0022-1317-43-1-247 [DOI] [PubMed] [Google Scholar]
- Franke T. F., Kaplan D. R., Cantley L. C. (1997). PI3K: downstream AKTion blocks apoptosis. Cell 88, 435–437 10.1016/S0092-8674(00)81883-8 [DOI] [PubMed] [Google Scholar]
- Gack M. U., Albrecht R. A., Urano T., Inn K. S., Huang I. C., Carnero E., Farzan M., Inoue S., Jung J. U., García-Sastre A. (2009). Influenza A virus NS1 targets the ubiquitin ligase TRIM25 to evade recognition by the host viral RNA sensor RIG-I. Cell Host Microbe 5, 439–449 10.1016/j.chom.2009.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- García-Sastre A., Egorov A., Matassov D., Brandt S., Levy D. E., Durbin J. E., Palese P., Muster T. (1998). Influenza A virus lacking the NS1 gene replicates in interferon-deficient systems. Virology 252, 324–330 10.1006/viro.1998.9508 [DOI] [PubMed] [Google Scholar]
- George R. A., Heringa J. (2002). An analysis of protein domain linkers: their classification and role in protein folding. Protein Eng 15, 871–879 10.1093/protein/15.11.871 [DOI] [PubMed] [Google Scholar]
- Gonfloni S., Williams J. C., Hattula K., Weijland A., Wierenga R. K., Superti-Furga G. (1997). The role of the linker between the SH2 domain and catalytic domain in the regulation and function of Src. EMBO J 16, 7261–7271 10.1093/emboj/16.24.7261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenspan D., Palese P., Krystal M. (1988). Two nuclear location signals in the influenza virus NS1 nonstructural protein. J Virol 62, 3020–3026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale B. G., Jackson D., Chen Y. H., Lamb R. A., Randall R. E. (2006). Influenza A virus NS1 protein binds p85β and activates phosphatidylinositol-3-kinase signaling. Proc Natl Acad Sci U S A 103, 14194–14199 10.1073/pnas.0606109103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hale B. G., Randall R. E., Ortín J., Jackson D. (2008). The multifunctional NS1 protein of influenza A viruses. J Gen Virol 89, 2359–2376 10.1099/vir.0.2008/004606-0 [DOI] [PubMed] [Google Scholar]
- Hirt B. (1967). Selective extraction of polyoma DNA from infected mouse cell cultures. J Mol Biol 26, 365–369 10.1016/0022-2836(67)90307-5 [DOI] [PubMed] [Google Scholar]
- Jackson D., Hossain M. J., Hickman D., Perez D. R., Lamb R. A. (2008). A new influenza virus virulence determinant: the NS1 protein four C-terminal residues modulate pathogenicity. Proc Natl Acad Sci U S A 105, 4381–4386 10.1073/pnas.0800482105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson D., Killip M. J., Galloway C. S., Russell R. J., Randall R. E. (2010). Loss of function of the influenza A virus NS1 protein promotes apoptosis but this is not due to a failure to activate phosphatidylinositol 3-kinase (PI3K). Virology 396, 94–105 10.1016/j.virol.2009.10.004 [DOI] [PubMed] [Google Scholar]
- Kochs G., García-Sastre A., Martínez-Sobrido L. (2007). Multiple anti-interferon actions of the influenza A virus NS1 protein. J Virol 81, 7011–7021 10.1128/JVI.02581-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z., Jiang Y., Jiao P., Wang A., Zhao F., Tian G., Wang X., Yu K., Bu Z., Chen H. (2006a). The NS1 gene contributes to the virulence of H5N1 avian influenza viruses. J Virol 80, 11115–11123 10.1128/JVI.00993-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S., Min J. Y., Krug R. M., Sen G. C. (2006b). Binding of the influenza A virus NS1 protein to PKR mediates the inhibition of its activation by either PACT or double-stranded RNA. Virology 349, 13–21 10.1016/j.virol.2006.01.005 [DOI] [PubMed] [Google Scholar]
- Liu J., Lynch P. A., Chien C. Y., Montelione G. T., Krug R. M., Berman H. M. (1997). Crystal structure of the unique RNA-binding domain of the influenza virus NS1 protein. Nat Struct Biol 4, 896–899 10.1038/nsb1197-896 [DOI] [PubMed] [Google Scholar]
- Lu Y., Wambach M., Katze M. G., Krug R. M. (1995). Binding of the influenza virus NS1 protein to double-stranded RNA inhibits the activation of the protein kinase that phosphorylates the elF-2 translation initiation factor. Virology 214, 222–228 10.1006/viro.1995.9937 [DOI] [PubMed] [Google Scholar]
- Mattison K., Oropeza R., Kenney L. J. (2002). The linker region plays an important role in the interdomain communication of the response regulator OmpR. J Biol Chem 277, 32714–32721 10.1074/jbc.M204122200 [DOI] [PubMed] [Google Scholar]
- Min J. Y., Krug R. M. (2006). The primary function of RNA binding by the influenza A virus NS1 protein in infected cells: Inhibiting the 2′-5′ oligo (A) synthetase/RNase L pathway. Proc Natl Acad Sci U S A 103, 7100–7105 10.1073/pnas.0602184103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nemeroff M. E., Qian X. Y., Krug R. M. (1995). The influenza virus NS1 protein forms multimers in vitro and in vivo. Virology 212, 422–428 10.1006/viro.1995.1499 [DOI] [PubMed] [Google Scholar]
- Nemeroff M. E., Barabino S. M., Li Y., Keller W., Krug R. M. (1998). Influenza virus NS1 protein interacts with the cellular 30 kDa subunit of CPSF and inhibits 3′ end formation of cellular pre-mRNAs. Mol Cell 1, 991–1000 10.1016/S1097-2765(00)80099-4 [DOI] [PubMed] [Google Scholar]
- Noah D. L., Twu K. Y., Krug R. M. (2003). Cellular antiviral responses against influenza A virus are countered at the posttranscriptional level by the viral NS1A protein via its binding to a cellular protein required for the 3′ end processing of cellular pre-mRNAs. Virology 307, 386–395 10.1016/S0042-6822(02)00127-7 [DOI] [PubMed] [Google Scholar]
- Noah J. W., Severson W., Noah D. L., Rasmussen L., White E. L., Jonsson C. B. (2007). A cell-based luminescence assay is effective for high-throughput screening of potential influenza antivirals. Antiviral Res 73, 50–59 10.1016/j.antiviral.2006.07.006 [DOI] [PubMed] [Google Scholar]
- Shin Y. K., Liu Q., Tikoo S. K., Babiuk L. A., Zhou Y. (2007a). Influenza A virus NS1 protein activates the phosphatidylinositol 3-kinase (PI3K)/Akt pathway by direct interaction with the p85 subunit of PI3K. J Gen Virol 88, 13–18 10.1099/vir.0.82419-0 [DOI] [PubMed] [Google Scholar]
- Shin Y. K., Li Y., Liu Q., Anderson D. H., Babiuk L. A., Zhou Y. (2007b). SH3 binding motif 1 in influenza A virus NS1 protein is essential for PI3K/Akt signaling pathway activation. J Virol 81, 12730–12739 10.1128/JVI.01427-07 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Riedel K., Lynch P., Chien C. Y., Montelione G. T., Krug R. M. (1999). RNA binding by the novel helical domain of the influenza virus NS1 protein requires its dimer structure and a small number of specific basic amino acids. RNA 5, 195–205 10.1017/S1355838299981621 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu J., Zhang Y., McIlroy J., Rordorf-Nikolic T., Orr G. A., Backer J. M. (1998). Regulation of the p85/p110 phosphatidylinositol 3′-kinase: stabilization and inhibition of the p110alpha catalytic subunit by the p85 regulatory subunit. Mol Cell Biol 18, 1379–1387 [DOI] [PMC free article] [PubMed] [Google Scholar]