Abstract
Staphylococcus aureus possesses 16 two-component systems (TCSs), two of which (GraRS and NsaRS) belong to the intramembrane-sensing histidine kinase (IM-HK) family, which is conserved within the firmicutes. NsaRS has recently been documented as being important for nisin resistance in S. aureus. In this study, we present a characterization of NsaRS and reveal that, as with other IM-HK TCSs, it responds to disruptions in the cell envelope. Analysis using a lacZ reporter–gene fusion demonstrated that nsaRS expression is upregulated by a variety of cell-envelope-damaging antibiotics, including phosphomycin, ampicillin, nisin, gramicidin, carbonyl cyanide m-chlorophenylhydrazone and penicillin G. Additionally, we reveal that NsaRS regulates a downstream transporter NsaAB during nisin-induced stress. NsaS mutants also display a 200-fold decreased ability to develop resistance to the cell-wall-targeting antibiotic bacitracin. Microarray analysis reveals that the transcription of 245 genes is altered in an nsaS mutant, with the vast majority being downregulated. Included within this list are genes involved in transport, drug resistance, cell envelope synthesis, transcriptional regulation, amino acid metabolism and virulence. Using inductively coupled plasma-MS we observed a decrease in intracellular divalent metal ions in an nsaS mutant when grown under low abundance conditions. Characterization of cells using electron microscopy reveals that nsaS mutants have alterations in cell envelope structure. Finally, a variety of virulence-related phenotypes are impaired in nsaS mutants, including biofilm formation, resistance to killing by human macrophages and survival in whole human blood. Thus, NsaRS is important in sensing cell damage in S. aureus and functions to reprogram gene expression to modify cell envelope architecture, facilitating adaptation and survival.
Introduction
Staphylococcus aureus is a highly successful and dangerous human pathogen that is a leading agent of both nosocomial and community-associated infections. It has an extremely diverse pathogenesis, causing infection in a plethora of ecological niches within the host. Typically these proceed from a localized site of infection (e.g. wound infection), through to systemic dissemination (bacteraemia) leading to metastatic disease (e.g. osteomyelitis, endocarditis and septic arthritis). The pathogenic success of S. aureus is largely the result of its arsenal of virulence determinants, which include haemolysins, toxins, adhesins and other exoproteins, such as proteases, staphylokinase and protein A (Lowy 1998; Novick, 2006). These damaging virulence factors are subject to multi-level and multi-factorial regulation, both temporally and spatially, in response to the environments encountered during growth. Accordingly, S. aureus has a diverse and wide-reaching network of regulatory elements that serve to modulate gene expression (Cheung et al., 1992; Fournier et al., 2001; Giraudo et al., 1994; McNamara et al., 2000; Novick et al., 1995) and bring about infection. These include all of the classical regulators of microbial gene expression, including DNA-binding proteins, regulatory RNAs, alternative sigma factors and two-component systems (TCSs). This latter class is an elegant mechanism of modulating gene expression in response to external conditions. As the name suggests, they are composed of two elements: a membrane-associated histidine kinase and a cytoplasmically located response regulator. These systems function to sense change in the external environment and reprogram gene expression accordingly, to circumnavigate stress.
In S. aureus, there are 16 conserved two-component regulators encoded within the various genomes sequenced thus far. Of this number, there are those that control autolysis (ArlRS, LytRS), virulence (SaeRS, AgrCA) and cell wall synthesis/drug resistance (WalKR, GraRS, VraSR) (Brunskill & Bayles, 1996; Delaune et al., 2011; Fournier et al., 2001; Giraudo et al., 1994; Kuroda et al., 2003; Meehl et al., 2007; Novick et al., 1995). There are yet others that sense external iron (HssRS), nitrogen (NreBC) or oxygen (SrrRS) conditions (Schlag et al., 2008; Torres et al., 2007; Yarwood et al., 2001). Recent phylogenetic analysis of bacterial TCSs revealed a conserved group of regulators termed intramembrane-sensing histidine kinases (IM-HKs), which are largely confined to the firmicutes (Joseph et al., 2002; Mascher, 2006). These unique TCSs are characterized as having very small histidine kinase proteins (less than 400 aa) with limited extracellular spanning domains (no more than 25 aa). The IM-HKs can be further divided into two major classes, those that are encoded upstream of a two-component ABC-type transport system, and those that are genetically linked to a conserved membrane protein. The latter class is only found in the low G+C Gram-positive bacteria and bears a HPK7 subfamily protein with multiple membrane-spanning domains. The best studied example of this class of regulators is VraRS from S. aureus, which is involved in resistance to cell-wall-targeting agents, and has been shown to protect against cell damage by modulating components of the peptidoglycan biosynthesis pathway (Gardete et al., 2006; Kuroda et al., 2003).
Quite distinct from the VraRS family is the second, and by far the most numerate, class of IM-HKs, which are defined by an HPK3i subfamily protein, containing only two membrane-spanning domains (Mascher, 2006). This family of regulators functions by sensing cell envelope damage, and upregulating adaptation and survival pathways. This is mediated by a variety of processes, including remodelling of cell architecture, and detoxification via transport systems (Mascher, 2006). In S. aureus there are two members of this latter family, the relatively well-studied GraRS, and the recently named NsaRS (Blake et al., 2011). GraRS has been described as being important in the resistance of S. aureus to antimicrobial peptides by a variety of mechanisms which remodel the cell wall (Herbert et al., 2007; Li et al., 2007). Reference to NsaRS has only recently appeared in the literature, where it was demonstrated to be important for nisin resistance in S. aureus (Blake et al., 2011). In this study we present a characterization of the NsaRS system and reveal that, as with other IM-HK TCSs, it responds to disruption of the cell envelope and redirects gene expression to mediate resistance. Additionally, we demonstrate that NsaRS is required for full biofilm formation across a variety of S. aureus strains, and contributes to survival during challenge by components of the human innate immune system.
Methods
Bacterial strains, plasmids and growth conditions.
S. aureus and Escherichia coli strains, along with the plasmids used in this study, are listed in Table 1. The strains were grown as described previously (Shaw et al., 2008). Cultures for growth experiments were obtained by using the following protocol. Overnight S. aureus cultures (1 ml) were used to inoculate fresh medium and allowed to grow for 3 h. These exponentially growing cultures were used to seed new medium at OD600 0.05. Test cultures were allowed to grow for the time period required for the experiment. Where used, CLR medium consisted of CL (containing 400 µM magnesium) with the following metals at a final concentration of 0.2 µM: calcium chloride, copper sulfate, ferrous sulfate, manganese chloride, nickel sulfate and zinc sulfate. We deviated from published protocols (Horsburgh et al., 2001a) by omitting molybdenum sulfate and including 0.2 µM cobalt sulfate.
Table 1. Strains, plasmids and primers used in this study.
Restriction sites are underlined. Primers are given in 5′–3′ orientation.
| Strain, plasmid or primer | Genotype or description | Reference or source |
| Strains | ||
| E. coli | ||
| DH5α | ϕ80 Δ(lacZ)M15 Δ(argF-lac)U169 endA1 recA1 hsdR17 () deoR thi-1 supE44 gyrA96 relA1 | Lab stocks |
| S. aureus | ||
| RN4220 | Restriction-deficient transformation recipient | Lab stocks |
| 8325-4 | Wild-type laboratory strain rsbU mutant | Lab stocks |
| SH1000 | Wild-type laboratory strain rsbU functional | Horsburgh et al. (2002) |
| Newman | Wild-type laboratory strain | Lab stocks |
| UAMS-1790 | Sequenced USA300-HOU-MRSA isolate cured of pUSA300-HOU-MRSA | Highlander et al. (2007); from M. Smeltzer |
| SLK1 | SH1000 nsaRS–lacZ fusion | This study |
| SLK2 | SH1000 nsaS mutant | This study |
| SLK3 | Newman nsaS mutant | This study |
| SLK4 | USA300 nsaS mutant | This study |
| SLK5 | SH1000 nsaS mutant complemented with pSLK3 | This study |
| SLK6 | Newman nsaS mutant complemented with pSLK3 | This study |
| SLK7 | USA300 nsaS mutant complemented with pSLK3 | This study |
| Plasmids | ||
| pAZ106 | Promoterless lacZ erm insertion vector | Kemp et al. (1991) |
| pMK4 | cm shuttle vector | Sullivan et al. (1984) |
| pSLK1 | pAZ106 containing a 1.5 kb nsaRS promoter fragment | This study |
| pSLK2 | pAZ106 containing an 800 bp mutagenic nsaS fragment | This study |
| pSLK3 | pMK4 containing a 2.5 kb nsaRS complementation fragment | This study |
| Primers | ||
| OL802 | ATGGGATCCGTCACGTGTGCAGCATATGC | |
| OL803 | ATGGAATTCCAGGCGCGTCATGTTAACAGC | |
| OL837 | ATGGGATCCCATTGGTGTCAAGAAATCCGAA | |
| OL838 | ATGGAATTCCAGTTGTGCTGCAGTAATGGGTG | |
| OL931 | ATGGAATTCGTGCTGTCGTAGCATTCAAAC | |
| OL1036 | CCGCGCACATTTCCCCGAAA | |
| OL1180 | TGGTTACGCAAGGTGTTG | |
| OL1181 | TCAACTGGTGAAGGACTG | |
| OL1222 | GAAGCACAACATGGTGGT | |
| OL1223 | TTGCTGCTACTCCACCA | |
| OL1224 | AGATGAACTCGTCCA | |
| OL1225 | GCACATCTGAAGGCG | |
| OL1226 | CGTCATTGATGAGTGGTG | |
| OL1227 | GGTACACTCCAAACATGC | |
| OL1333 | CGGTGTTATTGTCGTTG | |
| OL1334 | ACCATT0TAACGTTGGCA | |
| OL1335 | TGCATGCCATGTTGCT | |
| OL1336 | TTCACCAGCTCCAACT |
Construction of an nsaRS reporter fusion strain.
The putative promoter region of the nsaRS locus was amplified as a ~1.5 kb fragment (OL802/OL803) from S. aureus SH1000 genomic DNA. The reporter fusion was then constructed as described previously (Shaw et al., 2004). Briefly, the PCR fragment was cloned into the Gram-positive suicide vector pAZ106 creating plasmid pSLK1. S. aureus RN4220 was transformed with the resulting plasmid, and clones were selected for using TSA containing erythromycin (plasmid-encoded). The resulting strains were confirmed by genomic DNA extraction and PCR analysis. Transformants were used to generate 80α phage lysates for transduction of S. aureus SH1000, with clones again confirmed by PCR analysis. This created strain SLK1.
Construction of the nsaS mutant strain.
We generated an nsaS histidine kinase mutation to probe the role of this system as described previously (Shaw et al., 2004). An 800 bp PCR fragment within the nsaRS locus was amplified (primers OL837/OL838) and cloned into the pAZ106 suicide vector, generating pSLK2, which was used to transform S. aureus RN4220 cells. A representative clone was used to generate a phage lysate for transduction of S. aureus SH1000, Newman and UAMS-1790 [USA300-HOU-MRSA (Highlander et al., 2007) cured of pUSA300-HOU-MRSA, a gift from Dr Mark Smeltzer] using 80α. This created strains SLK2, SLK3 and SLK4, respectively. In this mutation, the forward primer (OL837) is located 191 nt downstream of the ATG of nsaR, whilst the reverse primer (OL838) is located approximately one-third of the way into the nsaS coding region (~350 nt into the 942 nt gene). Accordingly, the recombination event results in a full-length nsaR along with approximately 350 nt of nsaS connected to the natural promoter. The remainder of the nsaS gene, including the histidine kinase region, is disrupted by the insertion event. To further verify the mutation, we conducted reverse-transcription studies, which confirmed, as predicted, that only ~350 bp of nsaS is transcribed with nsaR, and that no mRNA containing the last two-thirds of nsaS (including the histidine kinase domain) is produced (data not shown). To determine if mutation of nsaS affected nsaR, we conducted real-time PCR analysis for nsaR in both the SH1000 wild-type and its nsaS mutant. We observed no alteration in expression of nsaR between the two strains (data not shown), indicating that nsaS mutation produced no unintended effects, such as compensatory nsaR overexpression.
Construction of nsaRS complemented strains.
In order to construct a complementation vector the entire nsaRS locus was amplified (OL802/OL931) as a 2.5 kb fragment. In silico analysis reveals an additional gene (SACOL2647) upstream of nsaR that likely forms the first coding unit of this operon. Accordingly, the forward primer is located 703 nt upstream of SACOL2647, and the reverse primer terminates downstream of nsaS. This fragment was cloned into the Gram-positive shuttle vector pMK4, creating pSLK3. S. aureus RN4220 was transformed with this construct, and clones were confirmed by genomic DNA extraction and PCR analysis, using a combination of gene- and vector-specific primers (OL802/OL1036). A representative isolate was selected to generate an 80α phage lysate, to transduce the SH1000, Newman and UAMS-1790 nsaS mutants. Clones were again confirmed by PCR analysis, creating strains SLK5, SLK6 and SLK7, respectively.
Plate-based assay to determine alterations in transcription resulting from external stress.
This assay was performed as described previously (Shaw et al., 2008). Briefly, TSA plates were overlaid with TSB top agar (0.7 % w/v) containing X-Gal, and seeded with exponentially growing SH1000 nsaRS–lacZ reporter-fusion cells. Sterile filter discs were overlaid onto these plates (three per plate) which were then inoculated with 10 µl of the following stress inducing chemicals: 6 M HCl, 85 % phosphoric acid, 100 % TCA, 88 % formic acid, 0.2 M acetic acid, 6 M sulphuric acid, 6 M nitric acid, 6 M sodium hydroxide, 2 M NaCl, 1 M glucose, 95 % ethanol, 100 % methanol, 100 % 2-propanol, 10 % SDS, 10 % Triton X-100, 10 % Tween-20, 1 M N-lauroyl sarcosine, 30 % hydrogen peroxide, 1 M methyl viologen, 1 % menadione, 2 mg pyrogallol ml−1, 1 M sodium nitroprusside, 1 M methyl-methane sulfonate, 5 mg penicillin G ml−1, 5 mg vancomycin ml−1, 2 mg phosphomycin ml−1, 5 mg spectinomycin ml−1, 100 mg ampicillin ml−1, 5 mg nisin ml−1, 5 mg gramicidin ml−1, 5 mg tetracycline ml−1, 50 mg kanamycin ml−1, 50 mg neomycin ml−1, 10 mg chloramphenicol ml−1, 20 mg puromycin ml−1, 2 mg bacitracin ml−1, 1 mg oxacillin ml−1, 1 mg cefotaxime ml−1, 5 mg d-cycloserine ml−1, 1 mg teicoplanin ml−1, 5 mg polymyxin B ml−1, 5 mg chlorpromazine ml−1, 0.1 mg carbonyl cyanide m-chlorophenylhydrazone (CCCP) ml−1, 1 mg valinomycin ml−1, 2 mg mupirocin ml−1, 500 mM diamide, 12.8 mg berberine chloride ml−1, 4.21 M peracetic acid, 0.1 M EDTA, 1 M DTT. Plates were incubated for 24 h at 37 °C and screened for a blue halo around the perimeter of the filter discs, indicating expression.
Kirby–Bauer sensitivity profiling of cell-wall-damaging agents.
Overnight broth cultures of the SH1000 wild-type and its nsaS mutant were diluted 1 : 1000 and added to 5 ml top agar; this was mixed and used to overlay TSA plates. Lawns were allowed to dry for 15 min and sterile filter disks were added to each plate. Penicillin G, oxacillin, ampicillin, cefotaxime, d-cycloserine, phosphomycin, gramicidin, teicoplanin, bacitracin or polymyxin B (all 10 µl) was added to the filter disks from a 5 mg ml−1 stock solution. Plates were incubated at 37 °C overnight and zones of inhibition were recorded.
β-Galactosidase assays.
Levels of β-galactosidase activity were measured as described previously (Shaw et al., 2006, 2007). Fluorescence was measured using a BioTek Synergy 2 plate reader and calibrated with standard concentrations of 4-methyl umbelliferone (MU). One unit of β-galactosidase activity was defined as the amount of enzyme that catalysed the production of 1 pmol MU min−1 (OD600 unit)−1. Assays were performed on duplicate samples and the mean value was determined. The results presented here are representative of three independent experiments that showed less than 10 % variability.
Transcriptomic studies.
The SH1000 wild-type and nsaS mutant were grown for 3 h, which corresponds to a window of maximal nsaRS expression. After this time RNA was extracted, converted to fluorescently labelled cDNA and hybridized to S. aureus COL genome microarrays version 6.0, as described previously (Delgado et al., 2008). Four hybridizations were performed for this study, including a biological replicate and a dye-swap experiment for each replicate to account for dye bias. Spots flagged as empty or bad were excluded and the raw data from each slide were normalized using the lowess method, with background correction. Data from the replicates were combined (using the median value) and a one-sample t-test was performed. A volcano plot was used with a fold change cut-off of ≥2 and a P-value of <0.05 to filter the genes that were differentially expressed. ORF IDs in the list of differentially expressed genes were then mapped to S. aureus COL IDs where possible. The entire dataset for this study has been deposited in the NCBI GEO database under accession number GSE27061.
Real-time PCR.
Quantitative real-time PCR analysis was conducted as described previously (Livak & Schmittgen, 2001; Riordan et al., 2010) using primers specific for alanine dehydrogenase (OL1222/OL1223), Rex (OL1224/OL1225), NarI (OL1226/OL1227), l-lactate dehydrogenase (OL1180/OL1181), nsaR (OL1333/OL1334) and nsaA (OL1335/OL1336). The control primers were for the 16s rRNA gene, as described previously (Koprivnjak et al., 2006).
Thin-section electron microscopy.
Aliquots of bacteria grown in TSB or 10 % NaCl for 3 and 6 h were processed and fixed as described previously (Shaw et al., 2005). Samples were observed and photographed in an FEI Morgagni 268D (FEI)transmission electron microscope with an Olympus SIS MegaView III (ResAlta Research Technologies) camera at the Integrative Biology Electron Microscopy Core Facility at the University of South Florida.
Negative staining electron microscopy.
A method of fixing bacteria in osmium tetroxide prior to pelleting was employed to stabilize cell walls. Osmium does not cross-link any protein in culture medium to bacterial cells, and allows preservation of the cell wall structure throughout centrifugation. This permits rinsing to remove culture media proteins prior to preparing cells for negative stain preparation and transmission electron microscopy (TEM) analysis. This method imparts electron density similar to that of uranyl acetate or other similar negative stains commonly used to observe bacteria via electron microscopy. Aliquots of cells grown in TSB for 3 and 6 h were initially fixed in an equal volume of 2 % osmium tetroxide in distilled water for 1 h at 4 °C. Following fixation, cells were rinsed in distilled water and pelleted three times at 5000 r.p.m. for 10 min. Dilutions were performed to obtain approximately 2000–3000 cells per drop, before one drop of each sample was applied to a carbon–Formvar-coated copper grid. Grids were allowed to air dry and were visualized by electron microscopy.
Biofilm formation assay.
Biofilm formation assays were performed as described previously (Beenken et al., 2003). The absorbance of samples was read using a BioTek Synergy 2 plate reader.
Metal ion profiling using inductively coupled plasma (ICP)-MS.
The SH1000 parental strain, its nsaS mutant and the complemented strain were grown in TSB and metal-limiting media for 4 h at 37 °C. Cells were harvested and their pellets were weighed and dried at 80 °C for 12 h. For hot acid digestion, filter pipette tips were rinsed with 2 % nitric acid in ultrapure water. Dried pellets were allowed to incubate for 1 h at room temperature in 500 µl ultrapure nitric acid OPTIMA. Each sample was then transferred to a borosilicate tube and placed in a silicone oil bath at 140–150 °C until ~200–250 µl each sample remained. To each sample, 250 µl of 30 % hydrogen peroxide ULTREX II was added and incubated at room temperature for 5–10 min. Tubes were placed back into the oil bath at 120 °C and the hydrogen peroxide was allowed to evaporate until ~200–250 µl remained. Tubes were removed from the water bath and ultrapure water was added to each sample up to 5 ml. For ICP-MS analysis, standards of the desired elements (1, 5, 10, 25, 100, 250 and 1000 p.p.b.) in 2 % nitric acid were made to create a standard curve. The ICP mass-spectrometer was tuned with 10 p.p.b. of elements ≥Li for optimization of lenses, and the internal standard used was 10 p.p.b. Indium. A blank (2 % nitric acid) standard curve and the samples were loaded and processed using the Thermo PlasmaLab software. The amount of each metal was determined based on p.p.b. given and the dried cell weights. The data presented are derived from at least five independent cultures.
Macrophage culture and S. aureus intracellular survival assay.
The Newman wild-type and its nsaS mutant were analysed using a human macrophage model of survival and clearance, as described previously (Kubica et al., 2008). The data presented were generated from six independent experiments. The data were analysed using Student’s t-test with a 5 % confidence limit to determine statistical significance.
Whole human blood survival assay.
Exponentially growing cultures of the SH1000 wild-type, nsaS mutant and the complemented strain were washed three times with PBS and resuspended in 1 ml whole human blood. The initial inoculum of each strain was also determined at this time by serial dilution and plating on TSA. Blood cultures were incubated at 37 °C with shaking for 3 h, before the c.f.u. ml−1 of each strain was determined, again by serial dilution and plating. Data are presented as percentage survival of each strain compared with initial inocula. These experiments were performed with two separate blood samples and represent five independent replicates.
Results
Transcriptional profiling of the nsaRS TCS
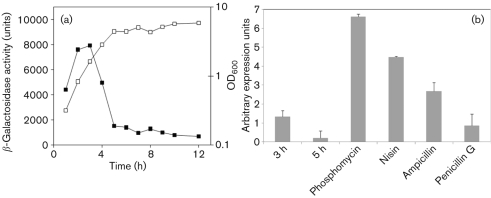
We initially set out to investigate the role of nsaRS by analysing its transcription in the laboratory strain SH1000. A reporter–gene fusion, using a promoterless lacZ cassette, was created and monitored for transcription during growth. Analysis revealed that maximal transcription of nsaRS occurs during the exponential and post-exponential phases of growth (2–5h) and declines into stationary phase (Fig. 1a). Given that TCSs function by sensing external stimuli, we sought to determine if the transcription of nsaRS could be modulated by the presence of chemical stressor compounds. As such, a plate-based assay was employed, as described by Cao et al. (2002) and Shaw et al. (2008), containing a variety of stress-inducing compounds (see Methods) to mimic natural external conditions. What we found was that a variety of cell-envelope-targeting antibiotics upregulated transcription of nsaRS, including ampicillin, phosphomycin, d-cycloserine, gramicidin, nisin, CCCP and penicillin G (data not shown). To quantitatively confirm these observations, we conducted real-time PCR on S. aureus SH1000 grown in either TSB or TSB containing subinhibitory concentrations of ampicillin, phosphomycin, penicillin G and nisin (Fig. 1b). When analysed during a window of peak nsaRS expression (3 h) we found that phosphomycin had the most dramatic effect, increasing expression by approximately fivefold compared with standard conditions. Further to this, analysis using nisin revealed a 3.3-fold upregulation, whilst studies with ampicillin revealed a twofold increase in transcription. Finally, subinhibitory concentrations of penicillin G revealed no alteration in nsaRS expression, perhaps suggesting that greater concentrations of this agent, as used in the plate-based assay, are required to induce expression of this TCS.
Fig. 1.
Transcriptional profiling of nsaRS. (a) A reporter lacZ fusion was created for nsaRS in the SH1000 background, and analysed using 4-MU. Cultures were grown in TSB and OD600 (□) and β-galactosidase activity (▪) was measured every hour throughout growth. Data shown are from at least three independent cultures that demonstrated less than 10 % variability. (b) qRT-PCR was performed for nsaRS under standard conditions (3 and 5 h), and also following the addition of subinhibitory concentrations of cell-wall-damaging chemicals (3 h). The data presented are from at least three independent experiments; error bars, sem.
NsaRS regulates transcription of the downstream transporter NsaAB in response to nisin stress
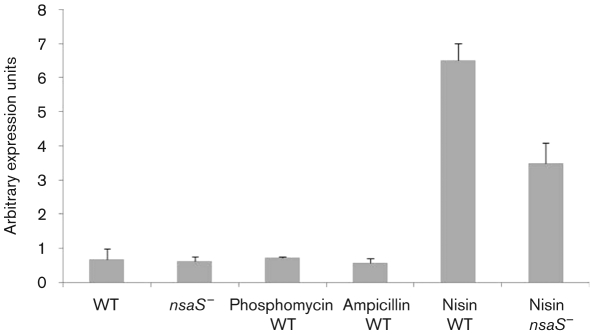
As with other members of the IM-HK TCS family, NsaRS is encoded upstream of a transcriptionally distinct locus that specifies an ABC type transporter (termed NsaAB). The existing paradigm of this arrangement is that the transporter aids in detoxification of the cell in response to cell-envelope-damaging agents, and is commonly regulated by its cognate TCS (Staroń et al., 2011). In order to ascertain if NsaRS regulates NsaAB, we conducted real-time PCR analysis on nsaAB in both the SH1000 wild-type and its nsaS mutant during a window of high NsaRS expression (3 h). Interestingly, we saw no alteration in nsaAB transcription between these two strains under standard conditions (Fig. 2). Given that these ABC transporters commonly function as detoxification units, we repeated nsaAB qRT-PCR analysis with cells grown under conditions shown to induce nsaRS expression. Accordingly, SH1000 was grown for 3 h in the presence of subinhibitory concentrations of phosphomycin, ampicillin and nisin, and analysed for nsaAB expression. Interestingly, whilst phosphomycin and ampicillin did not affect transcription of nsaAB, nisin resulted in an approximately tenfold upregulation of the transporter. To determine if the nisin-dependent stimulation of nsaAB expression was mediated via NsaRS, we repeated these analyses with the nsaS mutant. Compared with the wild-type strain we observed a twofold reduction in nsaAB expression in the nsaS mutant when grown in a subinhibitory concentration of nisin. Interestingly, the expression levels of nsaAB were still increased fivefold above that of the wild-type grown under standard conditions. This suggests that NsaRS plays a major role in regulating nsaAB in response to nisin stress but that other factors may also contribute to this process. These findings are consistent with existing literature on similar systems, which reveals that there is often discrepancy between IM-HK TCS induction and downstream transporter regulation (Staroń et al., 2011).
Fig. 2.
Role of NsaRS in regulating the transporter nsaAB. Transcription of nsaAB was initially determined under standard conditions in SH1000 (WT) and its nsaS mutant grown for 3 h. These analyses were repeated using TSB containing subinhibitory concentrations of phosphomycin, ampicillin and nisin in SH1000 and its nsaS mutant (no change observed for phosphomycin and ampicillin, data not shown). The data presented are from at least three independent experiments; error bars, sem.
NsaRS has a significant role in the ability of S. aureus to develop resistance to bacitracin
In this study, we show that a number of cell-envelope-targeting antimicrobials upregulate nsaRS transcription. Additionally, others have shown the importance of NsaS in the development of resistance to nisin (Blake et al., 2011). Finally, a previous study revealed that an nsaS mutant had a fourfold increase in sensitivity to bacitracin (Matsuo et al., 2010). As such, we set out to assess the sensitivity of the nsaS mutant to a variety of cell-envelope-targeting antimicrobial compounds. Using a modified Kirby–Bauer disk-diffusion approach we were unable to identify any increase in sensitivity of the mutant compared with the wild-type strain (see Methods for list; data not shown). Further qualification of this observation by determining minimum bactericidal concentration (MBC) also revealed no alteration between SH1000 and its nsaS mutant (data not shown). We suggest that these findings are consistent with the existing literature, which demonstrates that mutants of IM-HKs are themselves typically not sensitive to the antibiotics they sense (Staroń et al., 2011). Moreover, it is mutants of the transporters they regulate that demonstrate this type of sensitivity (Staroń et al., 2011). This observation is somewhat surprising, however, given that an nsaS mutant was previously reported as being more sensitive to bacitracin (Matsuo et al., 2010). When performing MIC analyses, we found that the nsaS mutant was not altered for bacitracin in the SH1000 and USA300 backgrounds. The disparity between our findings, and those of Matsuo et al. (2010) is probably explained by bacitracin MICs of the strains chosen; our study used SH1000 and USA300, whilst the previous study used MW2. SH1000 and USA300 have an MIC of 16 µg ml−1 for bacitracin, whilst MW2 has an MIC of 64 µg ml−1. The fact that Matsuo and colleagues reported an MIC of 16 µg ml−1 for the nsaS mutant in MW2, which is that observed for our wild-types, suggests that intrinsic drug resistance mechanisms of MW2 may have some impact on this effect.
Given the recent documentation of the role of NsaS in mediating resistance to nisin, we next determined if the nsaS mutant was impaired in its ability to mediate resistance to other cell-wall-targeting compounds. As such, we analysed spontaneous mutation frequencies for d-cycloserine, phosphomycin, vancomycin, cefotaxime and bacitracin. When performing 13 replicates from five separate cultures, we were unable to find alterations in the spontaneous mutation frequency for d-cycloserine, phosphomycin, vancomycin or cefotaxime. We did, however, find a major discrepancy between the wild-type strain and nsaS mutant for bacitracin. Specifically, we obtained 886 mutants from a combined inocula of 5.77×1010 for SH1000 when plated on bacitracin agar plates containing 3× MIC. In contrast, we were only able to obtain three mutants from a combined inocula of 3.84×1010 for the nsaS mutant strain. This results in a spontaneous mutation frequency of 1.53×10−8 for SH1000 and 7.81×10−11 for the nsaS mutant, representing a 196-fold decrease in spontaneous mutation frequency for the mutant strain.
Profiling the NsaRS transcriptome
Given that TCSs function via modulation of gene expression, we set out to analyse those genes that fall either directly or indirectly under the control of NsaRS. As such, we analysed an nsaS histidine kinase mutant via microarray analysis. Cultures of both SH1000 and its nsaS mutant were grown in TSB for 3 h, which corresponds to a window of maximal nsaRS expression. Total RNA was converted to cDNA and analysed using S. aureus COL genome microarrays v. 6.0 as described previously (Delgado et al., 2008). A total of 245 genes were found to be altered in transcription at a level of twofold or greater. Of these genes, 34 are negatively controlled by NsaRS (see Supplementary Table S1, available with the online version of this paper), whilst 211 are positively regulated (Supplementary Table S2), indicating that this TCS functions overwhelmingly as a positive regulator of gene expression. Of those genes found to be repressed, a large number were components that had been previously implicated in virulence, including β-haemolysin, hyaluronate lyase, fibronectin binding protein A, seven of the genes required for capsule formation and six of the ten extracellular proteases. This last category of enzymes was amongst the most upregulated in the nsaS mutant, with members of the spl operon transcribed at levels more than sixfold higher than in the parental strain. Perhaps unsurprisingly, of the 211 genes positively controlled by NsaRS, a large number specify proteins whose function is associated with the cell envelope. These include components involved in cell wall biosynthesis, surface proteins, cell-wall-anchoring enzymes and, most extensively, those classified as encoding transporters. Indeed, 55 such elements were identified of this latter class, encompassing those either previously shown or implicated in the transport of metal ions, amino acids, peptides, sugars and antimicrobial compounds. In addition to this, there were alterations in genes that function in the biosynthesis and metabolism of amino acids, including arginine, lysine, threonine, glutamate, aspartate, phenylalanine and methionine. Another major class of genes with altered expression included those involved in transcriptional regulation, with 14 such elements identified, including SarS and Rex. Other ontologies of function identified included metabolic enzymes, DNA-damage-repair-encoding genes and elements involved in nucleotide biosynthesis. Interestingly, there were also a large number of genes contained within the transcriptome that have previously been shown to function in anaerobic growth, including acetoin reductase, l-lactate dehydrogenase and d-lactate dehydrogenase (Pagels et al., 2010).
Confirmation of transcriptome data was performed using qRT-PCR analysis for selected genes identified in the dataset. Specifically, this was performed for alanine dehydrogenase, Rex, narI and l-lactate dehydrogenase, revealing 12.56-, 2.69-, 3.83- and 2.12-fold changes, respectively, in the nsaS mutant compared with the wild-type, akin to that seen in the transcriptome.
ICP-MS profiling reveals that NsaRS is required for metal ion transport under limiting growth conditions
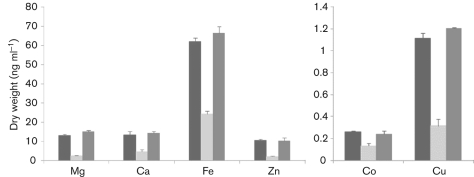
Transcriptomic analysis revealed a total of 55 transporters displaying decreased expression in the nsaS mutant. Of these elements, at least 20 appear to be specific to metal transportation, particularly with respect to divalent metal ions. Accordingly, we set out to analyse by ICP-MS the ability of the wild-type, nsaS mutant and complemented strains to import a variety of these ions. Quintuplet samples of each strain were grown in either TSB or metal-ion-limited media (CLR) supplemented with 400 µM magnesium sulfate and 0.2 µM calcium chloride, copper sulfate, ferrous sulfate, manganese chloride, nickel sulfate, zinc sulfate or cobalt sulfate. Samples were then analysed for the presence of the eight specific ions: Mg, Ca, Fe, Zn, Co, Ni, Cu, and Mn. We found that whilst little variation was observed during growth in replete media, the mutant strain displayed a marked decrease in its ability to import six of the eight metal ions tested when grown in limiting media (Fig. 3). Specifically, we determined a 2-fold decrease in cobalt, a 2.5-fold decrease in iron and calcium, a 3.5-fold decrease in copper and an approximately 5-fold decrease in both zinc and magnesium ions in the nsaS mutant strain. These findings suggest a role for NsaRS in metal ion import, particularly with respect to low-abundance growth conditions.
Fig. 3.
The transport of divalent metal ions under low-abundance conditions requires a functional NsaRS. The SH1000 wild-type (black), its nsaS mutant (light grey) and the complemented strain (dark grey) were grown for 4 h in metal-ion-limiting media (CLR) containing 400 µM magnesium sulfate and 0.2 µM calcium chloride, copper sulfate, ferrous sulfate, manganese chloride, nickel sulfate, zinc sulfate and cobalt sulfate. Cells were harvested and analysed for the presence of the supplementary metal ions using ICP-MS. The data presented are from five independent cultures; error bars, sem. Two graphs are presented because of the widely differing y-axis scales.
Electron microscopy reveals alterations in the cell envelope of nsaS mutant cells
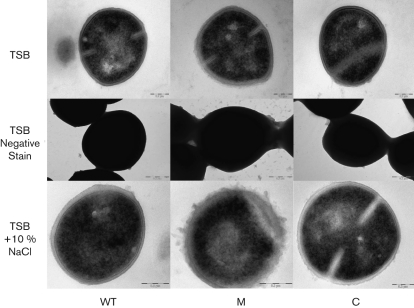
Given that our data thus far suggest a role for the NsaRS TCS in sensing cell envelope stress, we performed TEM analysis of the wild-type, mutant and complemented strains. When cells were grown in TSB for 3 h, we observed that the nsaS mutant had a diffuse and less defined/smooth cell wall compared with the parental and complemented strains (Fig. 4). Additionally, negative stain analysis revealed markedly increased encapsulation of the nsaS mutant after 3 h of growth, which became even more striking at later time points (data shown from 6 h). This finding is perhaps explained by the observed greater than twofold increase in transcription of seven of the ten genes required for capsule biosynthesis in the nsaS mutant. Finally, we also observed decreased transcription of a number of osmoprotectants (cudT and opuCA) in the nsaS mutant, along with a number of solute importers and general permeases. Accordingly, we grew the strains in TSB containing 10 % NaCl to assess whether the nsaS mutant displayed impaired growth. Upon analysis we observed only minor defects in the nsaS mutant when compared with the wild-type strain (1.5-fold maximal decrease in viability at 6 h growth). However when these same cultures (6 h) were analysed by TEM we observed dramatic alterations in cellular appearance. Following the trend of TSB-grown cultures, mutant cells incubated with 10 % NaCl showed highly irregular and diffuse cell walls compared with similarly grown wild-type and complemented cells. Taken together, these data suggest that NsaRS influences cell envelope architecture, particularly during times of stress.
Fig. 4.
NsaRS mediates remodelling of cell envelope architecture. SH1000, its nsaS mutant and the complemented strain were grown in TSB for thin section (3 h cultures) and negative stain (6 h cultures) analyses. Additionally, thin section images were captured for strains grown in TSB containing 10 % NaCl (6 h). WT, Wild-type; M, nsaS mutant; C, nsaS mutant strain complemented in trans within nsaRS. The images presented are representative of more than 10 frames from three independent cultures. Bars, 0.2 µm.
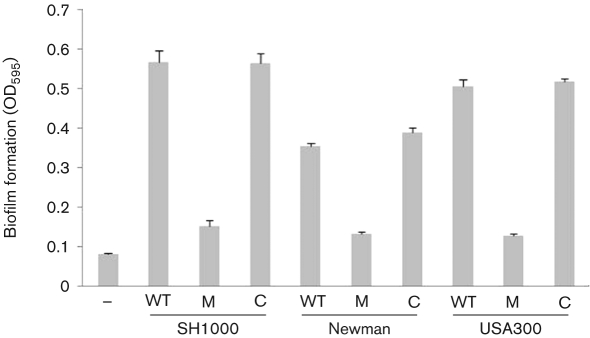
NsaRS contributes to biofilm formation in a variety of diverse S. aureus strains
NsaRS is an orthologue of GraRS in S. aureus, another IM-HK TCS involved in sensing disruption and instability of the cell wall (Li et al., 2007; Kraus et al., 2008). Previous work by other groups has revealed that GraRS mutants are impaired in their ability to form a biofilm (Boles et al., 2010; Herbert et al., 2007). In addition to this, we observed that a number of proteins required for biofilm formation display altered transcription in the nsaS mutant strain. Specifically, proteases are strongly upregulated in the nsaS mutant strain, which, as demonstrated by the recent work by ourselves and others, would be counterproductive to biofilm formation (Beenken et al., 2010; Boles & Horswill, 2008; Tsang et al., 2008). Additionally, the nsaS mutant has reduced expression of a number of anaerobiosis genes, which should also negatively impact biofilm formation (Beenken et al., 2004). Finally, the synthesis of the surface proteins SasG and surface protein A, which have previously been shown to be required for biofilm formation, are also reduced in the mutant (Corrigan et al., 2007; Merino et al., 2009). Analysis of the SH1000 nsaS mutant for biofilm formation (Beenken et al., 2003) revealed a 3.7-fold reduction for the mutant strain compared with the wild-type (Fig. 5). This phenotype was reversed when the nsaRS locus was introduced into the mutant strain in trans. Given the importance of biofilm formation to S. aureus disease progression, we assessed whether this phenotype was reproducible in other strains. Accordingly, we tested nsaS mutants in strain Newman and a community-acquired MRSA USA300 isolate. Again, in each case we were able to demonstrate a decrease in biofilm formation (Newman, 2.7-fold; USA300, 3.9-fold) for nsaS mutant strains, which was restored to wild-type levels upon complementation. Thus, it would appear that NsaRS contributes to biofilm formation across diverse and clonally distinct S. aureus strains.
Fig. 5.
The NsaRS TCS contributes to biofilm formation in S. aureus. nsaS mutants in the SH1000, Newman and USA300 background were assayed for biofilm formation. WT, Wild-type; M, nsaS mutant; C, nsaS mutant strain complemented in trans within nsaRS. The negative control (−) was the non-biofilm-forming strain 8325-4. Data presented are from at least five independent cultures. Error bars, sem.
NsaRS functions in protecting S. aureus cells during interaction with components of the innate immune system
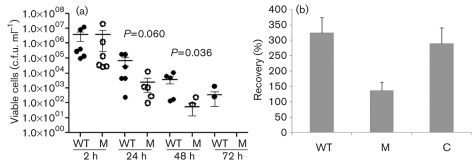
In order to determine the impact of NsaRS on S. aureus virulence, we examined interaction of the nsaS mutant with components of the innate immune system. This was performed using a human macrophage model of survival and clearance previously described by us (Koziel et al., 2009; Kubica et al., 2008). As such, the Newman wild-type and nsaS mutant were challenged with human monocyte-derived macrophages (hMdMs) at an m.o.i. of 1 : 50, and monitored for up to 5 days. Our use of Newman in these studies rather than SH1000, as in other studies, stems from the observation that SH1000 produces variable and inconsistent results in this model, whilst our experience reveals that Newman generates far more reproducible data (Koziel et al., 2009; Kubica et al., 2008). As such, we observed a consistent and significant reduction in the number of nsaS mutant cells recovered from infected hMdMs over the course of the infection period (Fig. 6a). Specifically, we observed a 25-fold reduction in mutant cells after 24 h compared with the parental strain (P = 0.060). Furthermore, by the second day post-infection we observed a further 70-fold reduction in nsaS mutant cells (P = 0.036), and by the third day we were unable to recover any cells from nsaS-infected hMdM cells, whilst the parental strain still returned 100–1000 c.f.u. ml−1 at this time.
Fig. 6.
NsaRS plays an important role during the interaction of S. aureus with components of the human innate immune system. (a) Phagocytosis assays were carried out for the Newman strain (•) and nsaS mutant (○) at an m.o.i. of 1 : 50 (hMdMs). After challenge, cultures were maintained for the desired time before infected cells were lysed by treatment with ice-cold water. Lysates were serially diluted to determine the presence of viable staphylococci remaining within cells. The data presented were generated from six independent experiments and analysed for statistical significance using Student’s t test. (b) Per cent recovery of SH1000 (WT), its nsaS mutant (M) and complemented strain (C) in whole human blood. Data presented are from two separate blood samples and five individual replicates; error bars, sem.
Further to this, we also used a whole human blood survival assay to examine the pathogenic fitness of the nsaS mutant. Accordingly, SH1000, its nsaS mutant and complemented strain were grown to exponential phase in TSB, before being inoculated into whole human blood. Cells were incubated for 3 h, and the survivability of each strain was determined and converted to per cent survival of the inoculum. An analysis with two different blood samples and five different replicates revealed that the wild-type and complemented strains not only were able to survive incubation in whole human blood but also continued to grow. Specifically, we were able to recover 324±43.29 % (sem) and 290±31.29 % of the wild-type and complemented strain inocula, respectively, after 3 h incubation (Fig. 6b). In contrast, the nsaS mutant demonstrated a 2.4-fold decrease in bacterial load compared with the parental strain, returning only 136±48.88 % of the inoculum upon incubation. Thus it would appear that the nsaS mutant strain is not only more rapidly cleared by hMdMs than the wild-type but also impaired in its ability to proliferate in human blood. As such, the NsaRS system seemingly has a significant role during interaction with components of the human innate immune system.
Discussion
In this study, we present a characterization of the TCS NsaRS from S. aureus. This TCS belongs to the IM-HK family of regulators which commonly modulate cell envelope stability by sensing disruption of the cell wall and membrane (Joseph et al., 2002; Mascher, 2006; Staroń et al., 2011). We present evidence here that NsaRS fulfils a similar function in S. aureus, responding to cell envelope damage mediated by a variety of antimicrobial agents. A recent study demonstrated that development of spontaneous resistance to nisin in S. aureus is primarily mediated by NsaS (Blake et al., 2011). Nisin is a polycyclic antibacterial peptide produced by Lactococcus lactis and is particularly effective against other Gram-positive bacteria. It functions by binding to the carbohydrate–phosphate moiety of the cell wall biosynthesis component lipid II (de Kruijff et al., 2008; Schneider & Sahl, 2010). By binding and sequestering lipid II, nisin blocks cell wall biosynthesis and can lead to delocalization of biosynthetic components and aberrant septum formation (Hasper et al., 2006; Hyde et al., 2006). Additionally, nisin is also believed to use this docking event with lipid II to engineer pore formation in the membranes of target cells. In this study we provide corroborative evidence for the observations of Blake et al. (2011) by demonstrating that nisin results in elevated nsaRS expression and upregulation of the downstream NsaAB transporter, which is, at least in part, dependent on NsaRS.
Interestingly, in this study we demonstrate that a number of other antimicrobial agents that target the cell wall also upregulate nsaRS expression. Specifically, both phosphomycin and d-cycloserine, which affect enzymes involved in intracellular peptidoglycan assembly, upregulate expression of nsaRS. The observation with regard to phosphomycin supports that of a previous study, which demonstrates that both nsaR and nsaS are upregulated in S. aureus following phosphomycin challenge (Petek et al., 2010). Furthermore, microarray analysis in this study reveals that NsaRS is responsible for regulation of another TCS, yhcSR. Previous studies with cells depleted for yhcSR reveal elevated sensitivity to this antibiotic (Sun et al., 2005), suggesting interplay between these two systems when sensing phosphomycin-mediated damage. Further to this, using a plate-based screening assay, we demonstrate that ampicillin and penicillin G, both of which are involved in extracellular peptidoglycan cross-linking, results in increased nsaRS expression. This information, in the context of our finding that nsaS mutants have an almost 200-fold decreased capacity for spontaneous bacitracin resistance, which blocks dephosphorylation and recycling of undecaprenyl-pyrophosphate, suggests a major role for NsaRS in sensing and responding to alterations in the cell wall. Indeed this latter finding corroborates a recent study which demonstrates that an NsaS mutant of MW2 has a fourfold increase in sensitivity to bacitracin (Matsuo et al., 2010).
Thus it appears that NsaRS has a role in sensing and responding to perturbation of the cell wall biosynthesis pathway at each step in the cycle, from early, intracellular peptidoglycan subunit assembly (phosphomycin and d-cycloserine), to the transport and anchoring of these moieties via lipid II (nisin), followed by extracellular cross-linking of the peptidoglycan subunits (ampicillin and penicillin G), and finally re-entry of the bactoprenol carrier to continue the cycle (bacitracin). Interestingly, nsaRS does not appear to respond to every cell-wall-targeting antibiotic, as we observe no upregulation of nsaRS or sensitivity of the nsaS mutant to other drugs, including vancomycin. Additionally, we find no upregulation of nsaRS by, or sensitivity of the nsaS mutant to, a variety of antimicrobial peptides (data not shown). We do, however, show that certain cell-membrane-damaging agents, such as gramicidin, nisin and CCCP, upregulate expression of nsaRS. Furthermore, a previous study by Muthaiyan et al. (2008) reveals upregulation of nsaR in response to daptomycin. This indicates that this system responds to perturbation of not only the cell wall but also the cell membrane. Thus, it appears that NsaRS has a general role in sensing and responding to disruption of the cell envelope in S. aureus. The fact that it does not respond to every cell-envelope-targeting compound is consistent with the literature from other IM-HK studies (Staroń et al., 2011) and is perhaps explained in S. aureus by a number of other cell membrane or cell wall stress-responding TCSs, such as GraRS, VraRS and WalKR (Li et al., 2007; Kraus et al., 2008; Pietiäinen et al., 2009; Delaune et al., 2011).
In addition to findings focused on cell-envelope-targeting antibiotics, we also present electron microscopy data revealing that nsaS mutants have a more diffuse and less defined/smooth cell wall. This disparity from the parental and complemented strains is even further enhanced when cells are subject to salt stress, with nsaS mutant cells displaying striking and irregular cell wall appearances. This is perhaps explained by the observation that transcription of a number of osmoprotectants (cudT and opuCA) is repressed in the nsaS mutant, along with a number of solute importers and general permeases. These alterations in cell wall structure can perhaps be further explained by diminished expression in the nsaS mutant strain of genes previously shown to function in cell wall biosynthesis (e.g. tagB, fmhA or scdA). Additionally, we observed decreased expression of glpQ in the nsaS mutant, which is part of the glycerol utilization pathway (Nilsson et al., 1994). Analysis of S. aureus cells that are deprived of their ability to utilize glycerol reveals major remodelling of the cell envelope and an accumulation of fatty acids in the cell resulting from a failure to incorporate them into the phospholipid membrane (Ray et al., 1972). Pilot metabolic profiling conducted in our laboratory reveals that a large number of saturated fatty acids do indeed accumulate in nsaS mutant cells (S. L. Kolar and L. N. Shaw, unpublished observation).
Further electron microscopy studies using negative staining techniques reveal a significant alteration in encapsulation of the nsaS mutant compared with the wild-type and complemented strains. During growth in TSB and high salt concentrations (data not shown) we observed in the mutant strain a significantly increased association of cells in what appears to be an extracellular capsule. This is perhaps explained by microarray analysis, which reveals that nsaS mutant cells demonstrate a greater than twofold increase in transcription of seven of the ten capsule-encoding genes. In addition to this, we also noted a number of other genes in the transcriptome that have functions connected with cell envelope formation and architecture. These include membrane proteins, transporters, cell-wall-anchoring proteins and cell-wall-associated proteins. Thus, it is clear that, in addition to its role in sensing perturbation of the cell wall and membrane, NsaRS has a significant effect on remodelling cell envelope architecture during growth of S. aureus. This is perhaps not surprising given that many other IM-HK TCSs, including the NsaRS orthologue GraRS, have similar roles (Joseph et al., 2002; Mascher, 2006; Li et al., 2007).
In addition to this, and perhaps equally intriguing, is the number of elements identified in the transcriptome that have a role in transport. We identified that 55 such genes have a known or implied role in the import/export of sugars, metal ions, amino acids and antimicrobial compounds. Given this, it is perhaps not surprising that we demonstrate a role for NsaRS in the process of cellular import. When grown during metal-ion-limiting conditions, the levels of intracellular divalent metal ions in the nsaS mutant were significantly reduced. This is in contrast with our finding that the same strain grown in replete media has no such defect. As we have shown that there are alterations in the cell surface of the nsaS mutant, it is possible that these findings result from a decreased association of metal ions with the surface of mutant cells. However, this is perhaps unlikely given that under metal replete conditions we do not observe similar decreases in internal metal concentrations. Thus it would appear that NsaRS mediates internal metal ion pools, not during conditions of abundance but more specifically during times of scarcity. As such, S. aureus cells may utilize NsaRS as a modulator of low abundance metal ion import during times of starvation. It could be speculated that this would be of particular importance during life within the host, which is known to be an environment of famine for metal ion availability. Indeed, such a mechanism might perhaps contribute to the diminished virulence phenotype we observed during ex vivo testing with the nsaS mutant.
Of interest from the microarray studies was the observation that other TCSs are controlled by NsaRS. The positive regulation of SACOL0201/0202 by NsaRS is intriguing because, whilst this regulatory element has not formally been studied thus far, it has been suggested to play a role in the anaerobic response of S. aureus (Fuchs et al., 2007). As such, the finding that a number of components involved in anaerobiosis have reduced expression in the nsaS mutant strain (such as idhA, idh2, acetoin reductase, fdhD and ald1) is reasonable, and could perhaps be mediated, at least in part, via control of SACOL0201/0202 by NsaRS. Further to this, we also identified decreased expression of the anaerobiosis repressor Rex in the nsaS mutant. This finding is somewhat at odds with our other data, as it could be predicted that a strain displaying reduced Rex expression would also demonstrate increased transcription of anaerobically involved loci (Fuchs et al., 2007; Pagels et al., 2010). However, it is tempting to speculate that SACOL0201/0202 and Rex have antagonizing roles in the process of controlling the anaerobic response, and that NsaRS mediates their interaction via regulatory control. It is also equally possible that NsaRS participates in upregulation of the anaerobic response directly, thus proving dominant to the inhibitory capacity of Rex.
We demonstrate that nsaS mutants are impaired in their ability to form biofilms across a variety of S. aureus strains. There are a number of regulatory elements that have been suggested as having roles in this process in S. aureus, including other TCSs and global regulatory loci such as agr, sarA and σB (Beenken et al., 2004). Recent work conducted by ourselves and others (Beenken et al., 2010; Boles & Horswill, 2008; Tsang et al., 2008) has focused not on these regulatory elements but rather on the physiological conditions that facilitate or prevent this aggregate lifestyle. As such, when the NsaRS regulon is analysed, it is perhaps no surprise that a mutant in this system would be impaired in forming and/or maintaining a biofilm. Specifically, the expression of a variety of extracellular proteases (including the V8 protease, staphopain B and a number of Spls), all of which have an important role in the breakdown of extracellular components required for biofilm formation, are increased in the nsaS mutant (Beenken et al., 2010; Boles & Horswill, 2008; Tsang et al., 2008). Further to this, the nsaS mutant has reduced expression of a number of anaerobiosis genes, which would again be counterproductive to this process (Beenken et al., 2004). Finally, the nsaS mutant has diminished synthesis of the surface proteins SasG and surface protein A, which have been shown to be required for biofilm formation (Corrigan et al., 2007; Merino et al., 2009). Collectively, these observations probably explain the role of nsaRS in biofilm formation, a phenotype which is broadly maintained across a number of distinct S. aureus lineages.
Finally, we present data evaluating the role of nsaRS in the virulence of S. aureus. Using a human model of macrophage clearance and survival we observed a 25-fold reduction of mutant cells after 24 h, a 70-fold reduction after 2 days and a lack of mutant cells after this time. Thus it would appear the nsaS mutant is more rapidly cleared than the wild-type upon interaction with human macrophages. Additionally, when performing survival experiments using whole human blood, we observed a reduction in the capacity of the nsaS mutant to proliferate. Whilst the wild-type and complemented strains are able to survive and rapidly proliferate upon challenge, nsaS mutant growth seemingly stalls when faced with these hostile conditions. Human blood is replete with a variety of antimicrobial agents, including leukocytes, antimicrobial peptides/proteins and factors of the complement system (Levy, 2000). As such, whole blood survival assays corroborate the ex vivo macrophage survival and clearance data, and suggest an important role for NsaRS in mediating resistance of S. aureus to the human immune system. Additionally, transcriptome analysis reveals that the expression of a number of elements known to be involved in virulence (such as essABC, sarS, spa, srtB, htrA, clpB and isaB) is decreased in the nsaS mutant. Conversely a number of known virulence factors were also elevated in the transcriptome (such as the V8 protease, staphopain B, FnpB, Hlb, HysA). It is entirely possible that these increases in transcription present a situation akin to that of a sarA mutant, which overexpresses extracellular proteases and has attenuated virulence. Accordingly, this imbalance of virulence determinant expression would likely destabilize the pathogenic response of nsaS mutant cells and perhaps explains the phenotypes observed.
In summary, we present a characterization of the IM-HK family member and GraRS orthologue, NsaRS. NsaRS appears to have parallel roles to its better studied counterpart in a number of processes, including biofilm formation and impaired interaction with components of the innate immune system (Li et al., 2007; Kraus et al., 2008; Boles et al., 2010; Cheung et al., 2010). This is probably explained, in part, by the relatedness of these two systems, and the observation that nsaR has previously been shown to be subject to twofold repression by GraRS (Herbert et al., 2007). Additionally, we demonstrate a number of unique NsaRS phenotypes, indicating its independent contribution to, and role in, sensing and adapting to cell-envelope-mediated stress. We also show that the NsaRS regulatory system appears to have an important role in the transport of metal ions, and is required for full biofilm formation across a number of genetically diverse S. aureus strains. Finally, the defects we present for the nsaS mutant when interacting with components of the innate immune system strongly argue for the importance of this TCS in the in vivo lifestyle of S. aureus.
Acknowledgements
We are grateful to Dr Mark Smeltzer and Dr Karen Beenken for the sharing of bacterial strains and methodologies, and to Dr Stanley Stevens for help with data analysis. We gratefully acknowledge the help of Mr Edward Haller of the Integrative Biology Electron Microscopy Core Facility at the University of South Florida. This study was supported in part the National Institute of Allergies and Infectious Diseases, grant 1R21AI090350-01 (L. N. S.).
Abbreviations:
- CCCP
carbonyl cyanide m-chlorophenylhydrazone
- ICP-MS
inductively coupled plasma-MS
- IM-HK
intramembrane-sensing histidine kinases
- MU
4-methyl umbelliferone
- TCS
two-component system
- TEM
transmission electron microscopy
The dataset for the microarray carried out in this study has been deposited in the NCBI GEO database under accession number GSE27061.
Two supplementary tables are available with the online version of this paper.
References
- Beenken K. E., Blevins J. S., Smeltzer M. S. (2003). Mutation of sarA in Staphylococcus aureus limits biofilm formation. Infect Immun 71, 4206–4211. 10.1128/IAI.71.7.4206-4211.2003.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beenken K. E., Dunman P. M., McAleese F., Macapagal D., Murphy E., Projan S. J., Blevins J. S., Smeltzer M. S. (2004). Global gene expression in Staphylococcus aureus biofilms. J Bacteriol 186, 4665–4684. 10.1128/JB.186.14.4665-4684.2004.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beenken K. E., Mrak L. N., Griffin L. M., Zielinska A. K., Shaw L. N., Rice K. C., Horswill A. R., Bayles K. W., Smeltzer M. S. (2010). Epistatic relationships between sarA and agr in Staphylococcus aureus biofilm formation. PLoS ONE 5, e10790. 10.1371/journal.pone.0010790.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blake K. L., Randall C. P., O’Neill A. J. (2011). In vitro studies indicate a high resistance potential for the lantibiotic nisin in Staphylococcus aureus and define a genetic basis for nisin resistance. Antimicrob Agents Chemother 55, 2362–2368. 10.1128/AAC.01077-10.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boles B. R., Horswill A. R. (2008). Agr-mediated dispersal of Staphylococcus aureus biofilms. PLoS Pathog 4, e1000052. 10.1371/journal.ppat.1000052.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boles B. R., Thoendel M., Roth A. J., Horswill A. R. (2010). Identification of genes involved in polysaccharide-independent Staphylococcus aureus biofilm formation. PLoS ONE 5, e10146. 10.1371/journal.pone.0010146.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brunskill E. W., Bayles K. W. (1996). Identification and molecular characterization of a putative regulatory locus that affects autolysis in Staphylococcus aureus. J Bacteriol 178, 611–618.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cao M., Wang T., Ye R., Helmann J. D. (2002). Antibiotics that inhibit cell wall biosynthesis induce expression of the Bacillus subtilis σW and σM regulons. Mol Microbiol 45, 1267–1276. 10.1046/j.1365-2958.2002.03050.x.. [DOI] [PubMed] [Google Scholar]
- Cheung A. L., Koomey J. M., Butler C. A., Projan S. J., Fischetti V. A. (1992). Regulation of exoprotein expression in Staphylococcus aureus by a locus (sar) distinct from agr. Proc Natl Acad Sci U S A 89, 6462–6466. 10.1073/pnas.89.14.6462.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung G. Y., Rigby K., Wang R., Queck S. Y., Braughton K. R., Whitney A. R., Teintze M., DeLeo F. R., Otto M. (2010). Staphylococcus epidermidis strategies to avoid killing by human neutrophils. PLoS Pathog 6, e1001133. 10.1371/journal.ppat.1001133.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Corrigan R. M., Rigby D., Handley P., Foster T. J. (2007). The role of Staphylococcus aureus surface protein SasG in adherence and biofilm formation. Microbiology 153, 2435–2446. 10.1099/mic.0.2007/006676-0.. [DOI] [PubMed] [Google Scholar]
- de Kruijff B., van Dam V., Breukink E. (2008). Lipid II: a central component in bacterial cell wall synthesis and a target for antibiotics. Prostaglandins Leukot Essent Fatty Acids 79, 117–121. 10.1016/j.plefa.2008.09.020.. [DOI] [PubMed] [Google Scholar]
- Delaune A., Poupel O., Mallet A., Coic Y. M., Msadek T., Dubrac S. (2011). Peptidoglycan crosslinking relaxation plays an important role in Staphylococcus aureus WalKR-dependent cell viability. PLoS ONE 6, e17054. 10.1371/journal.pone.0017054.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Delgado A., Zaman S., Muthaiyan A., Nagarajan V., Elasri M. O., Wilkinson B. J., Gustafson J. E. (2008). The fusidic acid stimulon of Staphylococcus aureus. J Antimicrob Chemother 62, 1207–1214. 10.1093/jac/dkn363.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fournier B., Klier A., Rapoport G. (2001). The two-component system ArlS–ArlR is a regulator of virulence gene expression in Staphylococcus aureus. Mol Microbiol 41, 247–261. 10.1046/j.1365-2958.2001.02515.x.. [DOI] [PubMed] [Google Scholar]
- Fuchs S., Pané-Farré J., Kohler C., Hecker M., Engelmann S. (2007). Anaerobic gene expression in Staphylococcus aureus. J Bacteriol 189, 4275–4289. 10.1128/JB.00081-07.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardete S., Wu S. W., Gill S., Tomasz A. (2006). Role of VraSR in antibiotic resistance and antibiotic-induced stress response in Staphylococcus aureus. Antimicrob Agents Chemother 50, 3424–3434. 10.1128/AAC.00356-06.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giraudo A. T., Raspanti C. G., Calzolari A., Nagel R. (1994). Characterization of a Tn551-mutant of Staphylococcus aureus defective in the production of several exoproteins. Can J Microbiol 40, 677–681. 10.1139/m94-107.. [DOI] [PubMed] [Google Scholar]
- Hasper H. E., Kramer N. E., Smith J. L., Hillman J. D., Zachariah C., Kuipers O. P., de Kruijff B., Breukink E. (2006). An alternative bactericidal mechanism of action for lantibiotic peptides that target lipid II. Science 313, 1636–1637. 10.1126/science.1129818.. [DOI] [PubMed] [Google Scholar]
- Herbert S., Bera A., Nerz C., Kraus D., Peschel A., Goerke C., Meehl M., Cheung A., Götz F. (2007). Molecular basis of resistance to muramidase and cationic antimicrobial peptide activity of lysozyme in staphylococci. PLoS Pathog 3, e102. 10.1371/journal.ppat.0030102.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Highlander S. K., Hultén K. G., Qin X., Jiang H., Yerrapragada S., Mason E. O., Jr, Shang Y., Williams T. M., Fortunov R. M., et al. (2007). Subtle genetic changes enhance virulence of methicillin resistant and sensitive Staphylococcus aureus. BMC Microbiol 7, 99. 10.1186/1471-2180-7-99.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsburgh M. J., Clements M. O., Crossley H., Ingham E., Foster S. J. (2001a). PerR controls oxidative stress resistance and iron storage proteins and is required for virulence in Staphylococcus aureus. Infect Immun 69, 3744–3754. 10.1128/IAI.69.6.3744-3754.2001.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsburgh M. J., Ingham E., Foster S. J. (2001b). In Staphylococcus aureus, fur is an interactive regulator with PerR, contributes to virulence, and is necessary for oxidative stress resistance through positive regulation of catalase and iron homeostasis. J Bacteriol 183, 468–475. 10.1128/JB.183.2.468-475.2001.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsburgh M. J., Aish J. L., White I. J., Shaw L., Lithgow J. K., Foster S. J. (2002). σB modulates virulence determinant expression and stress resistance: characterization of a functional rsbU strain derived from Staphylococcus aureus 8325-4. J Bacteriol 184, 5457–5467. 10.1128/JB.183.2.468-475.2001.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyde A. J., Parisot J., McNichol A., Bonev B. B. (2006). Nisin-induced changes in Bacillus morphology suggest a paradigm of antibiotic action. Proc Natl Acad Sci U S A 103, 19896–19901. 10.1073/pnas.0608373104.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph P., Fichant G. &, Quentin Y., Denizot F. (2002). Regulatory relationship of two-component and ABC transport systems and clustering of their genes in the Bacillus/Clostridium group, suggest a functional link between them. J Mol Microbiol Biotechnol 4, 503–513.. [PubMed] [Google Scholar]
- Kemp E. H., Sammons R. L., Moir A., Sun D., Setlow P. (1991). Analysis of transcriptional control of the gerD spore germination gene of Bacillus subtilis 168. J Bacteriol 173, 4646–4652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koprivnjak T., Mlakar V., Swanson L., Fournier B., Peschel A., Weiss J. P. (2006). Cation-induced transcriptional regulation of the dlt operon of Staphylococcus aureus. J Bacteriol 188, 3622–3630. 10.1128/JB.188.10.3622-3630.2006.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koziel J., Maciag-Gudowska A., Mikolajczyk T., Bzowska M., Sturdevant D. E., Whitney A. R., Shaw L. N., DeLeo F. R., Potempa J. (2009). Phagocytosis of Staphylococcus aureus by macrophages exerts cytoprotective effects manifested by the upregulation of antiapoptotic factors. PLoS ONE 4, e5210. 10.1371/journal.pone.0005210.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kraus D., Herbert S., Kristian S. A., Khosravi A., Nizet V., Götz F., Peschel A. (2008). The GraRS regulatory system controls Staphylococcus aureus susceptibility to antimicrobial host defenses. BMC Microbiol 8, 85. 10.1186/1471-2180-8-85.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubica M., Guzik K., Koziel J., Zarebski M., Richter W., Gajkowska B., Golda A., Maciag-Gudowska A., Brix K., et al. (2008). A potential new pathway for Staphylococcus aureus dissemination: the silent survival of S. aureus phagocytosed by human monocyte-derived macrophages. PLoS ONE 3, e1409. 10.1371/journal.pone.0001409.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuroda M., Kuroda H., Oshima T., Takeuchi F., Mori H., Hiramatsu K. (2003). Two-component system VraSR positively modulates the regulation of cell-wall biosynthesis pathway in Staphylococcus aureus. Mol Microbiol 49, 807–821. 10.1046/j.1365-2958.2003.03599.x.. [DOI] [PubMed] [Google Scholar]
- Levy O. (2000). Antimicrobial proteins and peptides of blood: templates for novel antimicrobial agents. Blood 96, 2664–2672.. [PubMed] [Google Scholar]
- Li M., Cha D. J., Lai Y., Villaruz A. E., Sturdevant D. E., Otto M. (2007). The antimicrobial peptide-sensing system aps of Staphylococcus aureus. Mol Microbiol 66, 1136–1147. 10.1111/j.1365-2958.2007.05986.x.. [DOI] [PubMed] [Google Scholar]
- Livak K. J., Schmittgen T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCt method. Methods 25, 402–408. 10.1006/meth.2001.1262.. [DOI] [PubMed] [Google Scholar]
- Lowy F. D. (1998). Staphylococcus aureus infections. N Engl J Med 339, 520–532. 10.1056/NEJM199808203390806.. [DOI] [PubMed] [Google Scholar]
- Mascher T. (2006). Intramembrane-sensing histidine kinases: a new family of cell envelope stress sensors in Firmicutes bacteria. FEMS Microbiol Lett 264, 133–144. 10.1111/j.1574-6968.2006.00444.x.. [DOI] [PubMed] [Google Scholar]
- Matsuo M., Kato F., Oogai Y., Kawai T., Sugai M., Komatsuzawa H. (2010). Distinct two-component systems in methicillin-resistant Staphylococcus aureus can change the susceptibility to antimicrobial agents. J Antimicrob Chemother 65, 1536–1537. 10.1093/jac/dkq141.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McNamara P. J., Milligan-Monroe K. C., Khalili S., Proctor R. A. (2000). Identification, cloning, and initial characterization of rot, a locus encoding a regulator of virulence factor expression in Staphylococcus aureus. J Bacteriol 182, 3197–3203. 10.1128/JB.182.11.3197-3203.2000.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meehl M., Herbert S., Götz F., Cheung A. (2007). Interaction of the GraRS two-component system with the VraFG ABC transporter to support vancomycin-intermediate resistance in Staphylococcus aureus. Antimicrob Agents Chemother 51, 2679–2689. 10.1128/AAC.00209-07.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merino N., Toledo-Arana A., Vergara-Irigaray M., Valle J., Solano C., Calvo E., Lopez J. A., Foster T. J., Penadés J. R., Lasa I. (2009). Protein A-mediated multicellular behavior in Staphylococcus aureus. J Bacteriol 191, 832–843. 10.1128/JB.01222-08.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muthaiyan A., Silverman J. A., Jayaswal R. K., Wilkinson B. J. (2008). Transcriptional profiling reveals that daptomycin induces the Staphylococcus aureus cell wall stress stimulon and genes responsive to membrane depolarization. Antimicrob Agents Chemother 52, 980–990. 10.1128/AAC.01121-07.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nilsson R. P., Beijer L., Rutberg B. (1994). The glpT and glpQ genes of the glycerol regulon in Bacillus subtilis. Microbiology 140, 723–730. 10.1099/00221287-140-4-723.. [DOI] [PubMed] [Google Scholar]
- Novick R. P. (2006). Staphylococcal pathogenesis and pathogenicity factors: genetics and regulation. In Gram-positive Pathogens, pp. 496–516. Edited by Fischetti V. A., Novick R. P., Ferretti J. J., Portnoy D. A., Rood J. I. Washington, DC: American Society for Microbiology. [Google Scholar]
- Novick R. P., Projan S. J., Kornblum J., Ross H. F., Ji G., Kreiswirth B., Vandenesch F., Moghazeh S., Novick R. P. (1995). The agr P2 operon: an autocatalytic sensory transduction system in Staphylococcus aureus. Mol Gen Genet 248, 446–458. 10.1007/BF02191645.. [DOI] [PubMed] [Google Scholar]
- Pagels M., Fuchs S., Pané-Farré J., Kohler C., Menschner L., Hecker M., McNamarra P. J., Bauer M. C., von Wachenfeldt C., et al. (2010). Redox sensing by a Rex-family repressor is involved in the regulation of anaerobic gene expression in Staphylococcus aureus. Mol Microbiol 76, 1142–1161. 10.1111/j.1365-2958.2010.07105.x.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petek M., Baebler S., Kuzman D., Rotter A., Podlesek Z., Gruden K., Ravnikar M., Urleb U. (2010). Revealing fosfomycin primary effect on Staphylococcus aureus transcriptome: modulation of cell envelope biosynthesis and phosphoenolpyruvate induced starvation. BMC Microbiol 10, 159. 10.1186/1471-2180-10-159.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietiäinen M., François P., Hyyryläinen H. L., Tangomo M., Sass V., Sahl H. G., Schrenzel J., Kontinen V. P. (2009). Transcriptome analysis of the responses of Staphylococcus aureus to antimicrobial peptides and characterization of the roles of vraDE and vraSR in antimicrobial resistance. BMC Genomics 10, 429. 10.1186/1471-2164-10-429.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ray P. H., Lillich T. T., White D. C. (1972). Consequences of glycerol deprivation on the synthesis of membrane components in a glycerol auxotroph of Staphylococcus aureus. J Bacteriol 112, 413–420.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riordan J. T., Tietjen J. A., Walsh C. W., Gustafson J. E., Whittam T. S. (2010). Inactivation of alternative sigma factor 54 (RpoN) leads to increased acid resistance, and alters locus of enterocyte effacement (LEE) expression in Escherichia coli O157 : H7. Microbiology 156, 719–730. 10.1099/mic.0.032631-0.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlag S., Fuchs S., Nerz C., Gaupp R., Engelmann S., Liebeke M., Lalk M., Hecker M., Götz F. (2008). Characterization of the oxygen-responsive NreABC regulon of Staphylococcus aureus. J Bacteriol 190, 7847–7858. 10.1128/JB.00905-08.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schneider T., Sahl H. G. (2010). Lipid II and other bactoprenol-bound cell wall precursors as drug targets. Curr Opin Investig Drugs 11, 157–164.. [PubMed] [Google Scholar]
- Shaw L., Golonka E., Potempa J., Foster S. J. (2004). The role and regulation of the extracellular proteases of Staphylococcus aureus. Microbiology 150, 217–228. 10.1099/mic.0.26634-0.. [DOI] [PubMed] [Google Scholar]
- Shaw L. N., Golonka E., Szmyd G., Foster S. J., Travis J., Potempa J. (2005). Cytoplasmic control of premature activation of a secreted protease zymogen: deletion of staphostatin B (SspC) in Staphylococcus aureus 8325-4 yields a profound pleiotropic phenotype. J Bacteriol 187, 1751–1762. 10.1128/JB.187.5.1751-1762.2005.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw L. N., Aish J., Davenport J. E., Brown M. C., Lithgow J. K., Simmonite K., Crossley H., Travis J., Potempa J., Foster S. J. (2006). Investigations into σB-modulated regulatory pathways governing extracellular virulence determinant production in Staphylococcus aureus. J Bacteriol 188, 6070–6080. 10.1128/JB.00551-06.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw L. N., Jonsson I. M., Singh V. K., Tarkowski A., Stewart G. C. (2007). Inactivation of traP has no effect on the agr quorum-sensing system or virulence of Staphylococcus aureus. Infect Immun 75, 4519–4527. 10.1128/IAI.00491-07.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shaw L. N., Lindholm C., Prajsnar T. K., Miller H. K., Brown M. C., Golonka E., Stewart G. C., Tarkowski A., Potempa J. (2008). Identification and characterization of σ, a novel component of the Staphylococcus aureus stress and virulence responses. PLoS ONE 3, e3844. 10.1371/journal.pone.0003844.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staroń A., Finkeisen D. E., Mascher T. (2011). Peptide antibiotic sensing and detoxification modules of Bacillus subtilis. Antimicrob Agents Chemother 55, 515–525. 10.1128/AAC.00352-10.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan M. A., Yasbin R. E., Young F. E. (1984). New shuttle vectors for Bacillus subtilis and Escherichia coli which allow rapid detection of inserted fragments. Gene 29, 21–26. [DOI] [PubMed] [Google Scholar]
- Sun J., Zheng L., Landwehr C., Yang J., Ji Y. (2005). Identification of a novel essential two-component signal transduction system, YhcSR, in Staphylococcus aureus. J Bacteriol 187, 7876–7880. 10.1128/JB.187.22.7876-7880.2005.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres V. J., Stauff D. L., Pishchany G., Bezbradica J. S., Gordy L. E., Iturregui J., Anderson K. L., Dunman P. M., Joyce S., Skaar E. P. (2007). A Staphylococcus aureus regulatory system that responds to host heme and modulates virulence. Cell Host Microbe 1, 109–119. 10.1016/j.chom.2007.03.001.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsang L. H., Cassat J. E., Shaw L. N., Beenken K. E., Smeltzer M. S. (2008). Factors contributing to the biofilm-deficient phenotype of Staphylococcus aureus sarA mutants. PLoS ONE 3, e3361. 10.1371/journal.pone.0003361.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yarwood J. M., McCormick J. K., Schlievert P. M. (2001). Identification of a novel two-component regulatory system that acts in global regulation of virulence factors of Staphylococcus aureus. J Bacteriol 183, 1113–1123. 10.1128/JB.183.4.1113-1123.2001.. [DOI] [PMC free article] [PubMed] [Google Scholar]