Abstract
The environmental bacterium Stenotrophomonas maltophilia is increasingly described as a multidrug-resistant pathogen of humans, being associated with pneumonia, among other diseases. But the degree to which S. maltophilia is capable of replicating in a mammalian host has been an issue of controversy. Using a model of intranasal inoculation into adult A/J mice, we now document that S. maltophilia strain K279a, the clinical isolate of S. maltophilia whose complete genome sequence was recently determined, is in fact capable of replicating in lungs, displaying as much as a 10-fold increase in c.f.u. in the first 8 h of infection. Importantly, as few as 104 c.f.u. deposited into the A/J lung was sufficient to promote bacterial outgrowth. Bacterial replication in the lungs of the A/J mice was followed by elevations in pro-inflammatory cytokines and also promoted resistance to subsequent challenge. We also found that DBA/2 mice were permissive for S. maltophilia K279a replication, although the level of growth and persistence in these animals was less than it was in the A/J mice. In contrast, the BALB/c and C57BL/6 mouse strains were non-permissive for S. maltophilia K279a growth. Interestingly, when five additional clinical isolates were introduced into the A/J lung, marked differences in survival were observed, with some strains being much less infective than K279a and others being appreciably more infective. These data suggest that the presence of major virulence determinants is variable among clinical isolates. Overall, this study confirms the infectivity of S. maltophilia for the mammalian host, and illustrates how both host and bacterial factors affect the outcome of Stenotrophomonas infection.
INTRODUCTION
Stenotrophomonas maltophilia is an environmental, Gram-negative bacterium that is increasingly identified as an opportunistic and nosocomial pathogen (Falagas et al., 2009; Looney et al., 2009). Originally classified as Pseudomonas maltophilia and then Xanthomonas maltophilia, S. maltophilia belongs to the γ-Proteobacteria and is the best known of the 11 species that form the genus Stenotrophomonas (Kim et al., 2010; Lee et al., 2011; Ryan et al., 2009; Yi et al., 2010). In nature, S. maltophilia exists in a wide variety of environments, being found in water, soil and plant material (Denton & Kerr, 1998; Ryan et al., 2009). But, S. maltophilia also occurs in man-made water systems, and most problematic is its presence in devices and reagents in hospital settings (Denton & Kerr, 1998; Looney et al., 2009). Increasingly isolated by clinical microbiology laboratories, S. maltophilia is associated with a spectrum of diseases, including infections of the lung, blood, heart, urinary tract, eyes, CNS, skin and soft tissue (Denton & Kerr, 1998; Falagas et al., 2009; Looney et al., 2009). The respiratory tract is the most common locale for S. maltophilia infection, with surveys indicating that ~5 % of nosocomial pneumonias are associated with this organism (Denton & Kerr, 1998; Looney et al., 2009). Some of the factors that predispose patients to S. maltophilia are immunosuppression, cancer, indwelling devices, mechanical ventilation, and broad-spectrum antimicrobial therapy (Ansari et al., 2007; Denton & Kerr, 1998; Falagas et al., 2009; Paez et al., 2008; Looney et al., 2009; Safdar & Rolston, 2007). Investigators throughout the world also report an increasing prevalence of S. maltophilia in cystic fibrosis (CF) patients; in the US, S. maltophilia is linked to ~10 % of CF patients, and in Europe, the linkage is as high as 30 % (Davies & Rubin, 2007; Lambiase et al., 2006; Nicoletti et al., 2011; Paschoal et al., 2007; Spicuzza et al., 2009; Waters et al., 2011). A recent study has also determined that chronic S. maltophilia infection is an independent risk factor for lung exacerbations in CF patients (Waters et al., 2011). The prevalence of S. maltophilia in patients is partly explained by the marked resistance of the organism to antibiotics, which involves β-lactamases, enzymes that act on aminoglycosides, and multidrug efflux pumps (Alonso & Martínez, 2000; Avison et al., 2002; Gordon & Wareham, 2010; Gould & Avison, 2006; Hernández et al., 2009; Huang et al., 2010; Looney et al., 2009; Okazaki & Avison, 2007; Sánchez et al., 2008, 2009; Sánchez & Martínez, 2010; Shimizu et al., 2008).
Despite the increasing association of S. maltophilia with disease, our understanding of the pathogenicity and virulence of this bacterium is still quite minimal. Limited phenotypic analysis of S. maltophilia strains in vitro and the recent sequencing of the S. maltophilia genome both suggest that the organism has traits that have been linked to the virulence of other bacteria (Crossman et al., 2008; Denton & Kerr, 1998; Looney et al., 2009; Nicoletti et al., 2011; Rocco et al., 2009; Ryan et al., 2009). Also, inoculation of S. maltophilia into the lungs of mice results in a marked inflammatory response within 24 h that involves, among other things, elevated cytokines (Di Bonaventura et al., 2010; Waters et al., 2007; Zgair & Chhibber, 2010a). However, the degree to which S. maltophilia is capable of replicating on its own within a mammalian host has remained a point of some controversy for several reasons. First, S. maltophilia is often isolated from patients along with other bacteria, especially in cases of pneumonia (Ryan et al., 2009). Second, several studies which used a pure culture of S. maltophilia to infect the lungs of mice or rats did not document significant increases in bacterial numbers post-inoculation; rather, bacterial numbers were only reported as declining between days 1 and 7 post-inoculation (Di Bonaventura et al., 2010; George et al., 1993; McKay et al., 2003; Waters et al., 2007). Thus, we sought to more thoroughly address the issue of S. maltophilia infectivity by examining six different strains of S. maltophilia for their ability to replicate and persist in the lungs of four different strains of inbred mice, utilizing various inoculating doses and assaying bacterial numbers at earlier times post-inoculation. We now document that S. maltophilia strains can indeed replicate as much as 10-fold following inoculation into the lungs of some but not all strains of mice. We also report that clinical isolates of S. maltophilia differ significantly in their ability to grow and persist in the murine lung.
METHODS
Bacterial strains and media.
The primary S. maltophilia strain used in this study was K279a, obtained from Matthew Avison at the University of Bristol. K279a is a multidrug-resistant strain that was originally isolated from the blood of a cancer patient (Avison et al., 2000). The five additional strains of S. maltophilia that were included in the study were obtained from Paul Edelstein at the University of Pennsylvania. These isolates, which were obtained from the respiratory tracts of patients at the University of Pennsylvania Hospital, were designated UPSm1, UPSm2, UPSm3, UPSm4 and UPSm5. Strains of S. maltophilia were routinely cultured on standard Luria–Bertani (LB) agar (Ausubel et al., 1989). Growth of these strains in bacteriological media was also assessed by incubating LB broth cultures (25 ml in a 125 ml flask) at 37 °C with agitation and then monitoring the OD600 using a Beckman Coulter DU 720 spectrophotometer. Inocula for these cultures were derived from exponential-phase pre-cultures.
Mouse lung infection.
Female A/J, DBA/2, BALB/c and C57BL6 mice were obtained from Jackson Laboratories. After their arrival, mice were acclimated for 1 week and fed a standard diet. At the age of 6–7 weeks, mice were infected with S. maltophilia administered via standard intranasal inoculation. Just prior to their infection, the animals were anaesthetized by intraperitoneal injection of ketamine and xylazine as per veterinary guidelines. The bacteria used for the inoculum were grown for 15 h on LB agar and then suspended and diluted in sterile PBS. Differing amounts of bacteria (as detailed below) were inoculated in a 20 µl aliquot, half of which was delivered into each nostril. At various times post-inoculation, total lung homogenates were obtained as previously described (DebRoy et al., 2006; Rossier et al., 2004), and then the c.f.u. of bacteria within the lungs were determined by plating on LB agar. The c.f.u. recovered immediately after inoculation (t = 0) represented the initial lung deposition. The amount deposited in the lungs was ~30 % of the bacteria administered into the nose. In one set of experiments, animals were also inoculated with heat-killed bacteria, which had been obtained by heating the bacterial suspension for 10 min at 65 °C. All animal experiments were approved by the Animal Care and Use Committee of Northwestern University.
Cytokine assays.
To monitor cytokine levels in infected lungs, A/J mice were infected as described above. At various times post-inoculation, lung homogenates, which had been cleared by centrifugation at 1000 g for 10 min to remove cellular debris, were examined for tumour necrosis factor-α (TNF-α), interleukin (IL)-1β and IL-6 using murine ELISA kits (Ready-SET-Go! kits, eBioscience) according to the manufacturer's instructions.
RESULTS AND DISCUSSION
S. maltophilia K279a replicates within the lungs of A/J mice
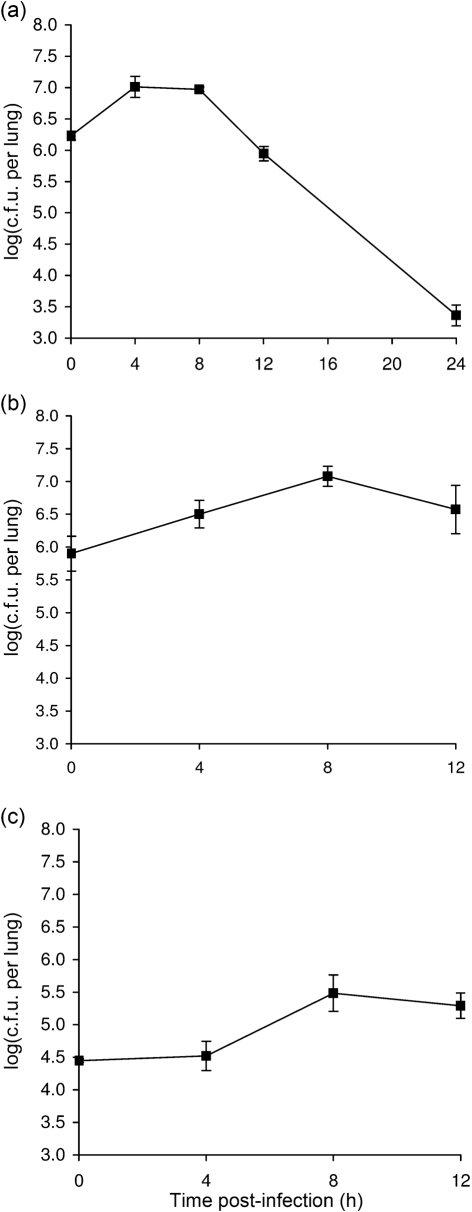
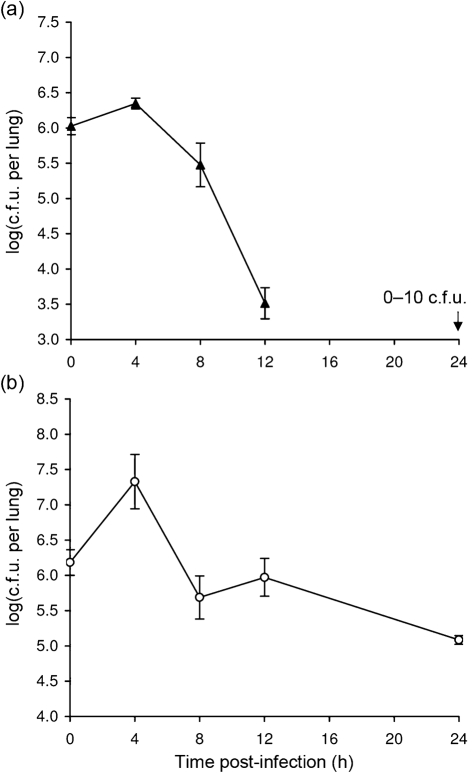
As a first step toward assessing in vivo growth of S. maltophilia, we examined strain K279a for its ability to replicate in the lungs of 6–7 week-old female A/J mice following intranasal inoculation. Strain K279a is the clinical isolate of S. maltophilia for which the genome was recently entirely sequenced (Crossman et al., 2008). We and others have used inbred A/J mice to examine lung infection by Legionella pneumophila (Brieland et al., 1994; DebRoy et al., 2006; Rossier et al., 2004). This inbred strain of mouse has also been used to study infection by a variety of other microbial pathogens, including Acinetobacter baumannii, Bacillus anthracis, Candida albicans, Listeria monocytogenes, Mycobacterium tuberculosis, Pseudomonas aeruginosa and Staphylococcus aureus, some of which are also major causes of lung infection (Ahn et al., 2010; Glomski et al., 2008; Qiu et al., 2009). In an initial infection experiment which began with a K279a deposition count in the lungs of 1.6×106 c.f.u., we observed a fivefold increase in c.f.u. at 4 and 8 h post-inoculation (Student's t test, P<0.05), and then a relatively modest reduction in bacterial recovery at 12 h followed by a larger drop in c.f.u. at 24 h (Fig. 1a). Significant increases in c.f.u. at the two early times points were also observed in four more experiments that utilized the same inoculation dose (data not shown). That 1×106 c.f.u. of K279a deposited into the lungs represents a sublethal dose was confirmed in another trial, when all 20 animals inoculated with this dose survived over the next 21 days of observation. When the assay was next performed with a lower deposition of 8.0×105 c.f.u., significant growth was still observed at 4 and 8 h (P<0.05), although the peak in c.f.u. recovered was shifted to the later time point (Fig. 1b). With this inoculating dose, the overall increase in c.f.u. above the deposition count was now 10-fold. In two additional experiments, when the deposition count was lowered even further to 2.0×104, we still observed a significant increase in c.f.u. (P<0.05) at 8 and 12 h post-inoculation (Fig. 1c and data not shown).
Fig. 1.
Growth of S. maltophilia strain K279a in the lungs of A/J mice. (a–c) Mice were intranasally inoculated with different amounts of strain K279a (▪), resulting in different levels of deposition (see t = 0, in a–c), and then at 4, 8, 12 and 24 h post-inoculation, the c.f.u. in total lung homogenates were determined by plating. Data are the mean±sd obtained from four or five infected animals. Significant increases in c.f.u. relative to the value at t = 0 were obtained at 4 and 8 h in (a) and (b) and at 8 and 12 h in (c) (Student's t test, P<0.05).
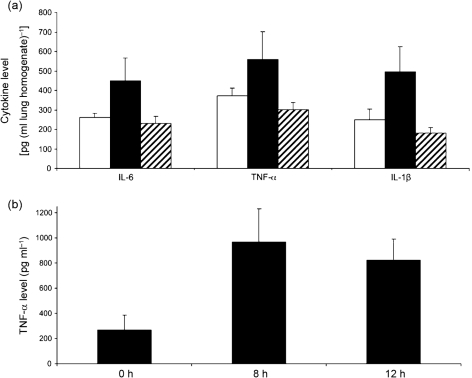
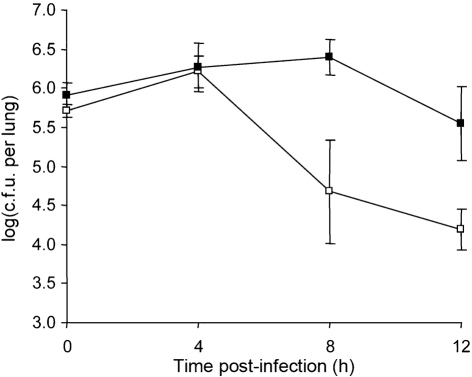
To determine whether infection of the A/J lung with S. maltophilia results in an inflammatory response, we infected the mice with a deposition of ~1×106 c.f.u. of strain K279a as above, and then examined the infected lung for elevations in the pro-inflammatory cytokines IL-6, TNF-α and IL-1β at 24 h post-inoculation. We observed significant increases (P<0.05) in all cytokines compared with uninfected controls (Fig. 2a). An additional infection confirmed the presence of elevated cytokine levels at 24 h in mice infected with viable bacteria compared with uninfected animals (data not shown). In contrast, inoculation with heat-killed K279a did not result in elevated cytokines (Fig. 2a). Further assessments of TNF-α levels indicated that the pro-inflammatory response in the A/J mice was evident as early as 8 h post-inoculation (Fig. 2b). Next, to ascertain whether infection of the A/J lung with strain K279a also results in a functional adaptive immune response, we sought to determine whether infection with strain K279a afforded any protection against a subsequent challenge. To that end, one group of mice (n = 20) was inoculated with a deposition count of 1.1×106 c.f.u. of K279a, and another group (n = 19) was inoculated with PBS alone. Twenty-one days later, we infected the two groups as above and then determined the numbers of c.f.u. in the lungs at 0, 4, 8 and 12 h post-inoculation (Fig. 3). The animals that had not been previously exposed to K279a exhibited a pattern of bacterial growth and survival that was comparable with that of earlier experiments; e.g. compare Fig. 3 with Fig. 1(b). In contrast, those animals that had been previously exposed showed a more rapid clearance (P<0.01) of bacteria that was evident as early as 8 h post-inoculation (Fig. 3).
Fig. 2.
Cytokine production in the lungs of A/J mice infected with S. maltophilia strain K279a. (a) Mice were intranasally inoculated with either PBS alone (white bars), strain K279a (black bars) or an equal amount of heat-killed K279a (hatched bars), and then at 24 h post-inoculation, the levels of IL-6, TNF-α and IL-1β per millilitre of lung homogenate were determined by ELISA. The group infected with viable bacteria consisted of 10 mice, whereas the two control groups had five mice each. The values presented are the mean and sd obtained from each group. The cytokine levels in the infected animals were significantly above those in the two control groups (Student's t test, P<0.05). (b) Mice were intranasally inoculated with strain K279a, and then at 0, 8 and 12 h post-inoculation, the levels of TNF-α were determined. The values presented are the mean and sd obtained from four to five infected animals and are representative of two independent experiments. The cytokine levels observed at 8 and 12 h were significantly above those seen at t = 0 (Student's t test, P<0.05).
Fig. 3.
Effect of prior infection with strain K279a on the susceptibility of A/J mice to S. maltophilia challenge. One group of mice (n = 20) that had been infected with a sublethal dose of K279a 21 days earlier (□) and another group of mice (n = 19) that had previously been inoculated with PBS only (▪) were infected with K279a, and then the numbers of c.f.u. in the lungs were determined at 0, 4, 8 and 12 h post-inoculation. The values presented at each time point are the mean±sd obtained from five infected animals, with the exception of the 0 h point for the PBS control (four animals). Mice that had previous exposure to K279a exhibited a significantly reduced bacterial load compared with the PBS control group at 8 h post-inoculation (Student's t test, P<0.01).
Taken together, these data demonstrated that S. maltophilia is fully capable of replicating at least five- to 10-fold within the A/J mouse lung and without the ‘assistance’ of another bacterial species. Furthermore, Stenotrophomonas replication was observed after an initial lung deposition count as low as 104 c.f.u. Finally, these experiments established that the sequenced clinical isolate of S. maltophilia is capable of both replicating and triggering an innate and adaptive immune response in a mammalian model of disease. The K279a genome sequence reveals a number of potential virulence factors and immune modulators, based upon the in vivo importance of their homologues in other bacteria (Crossman et al., 2008). These factors include type I, II, IV and V protein secretion systems, tissue-degradative exoenzymes, various pili, putative adhesins, flagella, LPS, putative exopolysaccharide, siderophores and quorum sensing (Crossman et al., 2008; Denton & Kerr, 1998; Fouhy et al., 2007; Huang & Wong, 2007; Looney et al., 2009; Minkwitz & Berg, 2001; Rocco et al., 2009; Travassos et al., 2004; Waters et al., 2007; Zgair & Chhibber, 2010b). Thus, the A/J mouse model can be used to compare wild-type K279a with its mutant derivatives in order to formally define the virulence factors of S. maltophilia.
S. maltophilia K279a replicates in the lungs of DBA/2 mice but not BALB/c and C57BL/6 mice
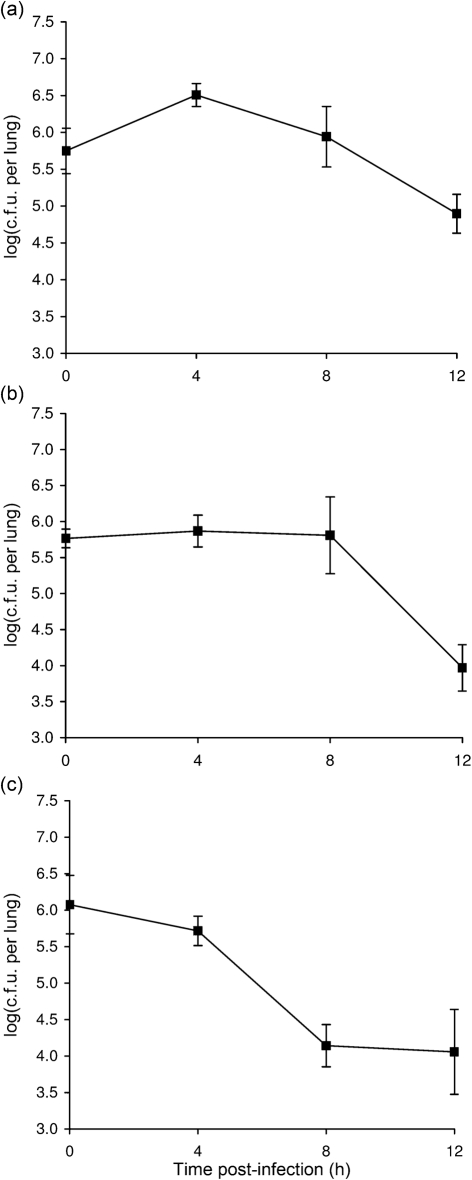
To discern whether or not the growth that we had observed for K279a was specific to A/J mice, we performed intranasal inoculations and lung c.f.u. determinations in three more inbred strains of mice; i.e. 6–7-week-old female DBA/2, BALB/c and C57BL/6 mice. We included the DBA/2 strain in particular because it shares with the A/J mouse a genetic deficiency in complement factor C5 (http://jaxmice.jax.org/strain/000646.html), which could potentially influence susceptibility to S. maltophilia. Although the initial lung deposition of K279a in these strains of mice was comparable with that which had been obtained with the A/J mouse infections, a spectrum of growth and persistence was subsequently observed (Fig. 4). In the DBA/2 host, strain K279a behaved in a manner that was similar but not identical to that which had been observed in the A/J mice; i.e. five- to 10-fold increases (P<0.05) in c.f.u. were detected at 4 h post-inoculation, but then there was a gradual decline in numbers at both 8 and 12 h (Fig. 4a, as compared most closely with Fig. 1b). Late in the course of our study, another group reported infection of 7-week-old female DBA/2 mice with a different strain of S. maltophilia; however, as noted above, this study did not report bacterial replication (Di Bonaventura et al., 2010). Given our findings, the most likely reason that the other study did not observe replication was because it began with lung depositions that were already between 107 and 108 c.f.u. and then first monitored for changes in c.f.u. at 24 h post-inoculation. In the BALB/c mice, strain K279a did not increase its numbers but was able to fully maintain its c.f.u. for the first 8 h before dropping more than 10-fold at 12 h (Fig. 4b). In contrast to this result, a different blood culture isolate of S. maltophilia was very recently reported as being capable of replicating within 12 h following intranasal inoculation into 4–6-week-old male BALB/c mice (Zgair & Chhibber, 2010a). That bacterial replication was observed in that study and not ours could be due to differences in virulence between the bacterial strains used and/or differences in the age, sex or source (lineage) of the BALB/c mice. Additionally, the other study utilized a very large inoculating dose of 109 c.f.u., a dose that is 1000-fold higher than that used in our experiments. In the C57BL/6 mice, K279a appeared unable to replicate, showing a decline in c.f.u. recovery at 4, 8 and 12 h (Fig. 4c). An earlier study reported infection of neonatal (i.e. 7–10-day-old) C57BL/6 mice with different strains of S. maltophilia; but, as noted above, no evidence for bacterial replication was reported after determining lung c.f.u. at 16 h post-inoculation (Waters et al., 2007).
Fig. 4.
Growth of S. maltophilia strain K279a in the lungs of DBA/2, BALB/c and C57BL/6 mice. DBA/2 (a), BALB/c (b) and C57BL/6 (c) mice were intranasally inoculated with strain K279a (▪), resulting in comparable levels of deposition (t = 0), and then at 4, 8 and 12 h post-inoculation, the c.f.u. in lung homogenates were determined by plating. Data are the mean±sd obtained from five infected animals and are representative of at least two independent experiments. Significant increases in c.f.u. relative to the value at t = 0 were obtained at 4 h in (a) (Student's t test, P<0.05).
Taken together, our experiments, which represent the first side-by-side comparison of mouse strains for their susceptibility to S. maltophilia, indicate that the S. maltophilia bacterium replicates in some but not all strains of adult mice. Furthermore, the data indicate that there is a spectrum of susceptibility among commonly used inbred strains of mice, i.e. the A/J strain is the most permissive for S. maltophilia growth and persistence, followed by the DBA/2 strain, then the BALB/c mouse, and finally the non-permissive C57BL/6 strain. It is tempting to hypothesize that the reason that the A/J and DBA/2 mice are most permissive for S. maltophilia is that they both lack the complement factor C5, which is a mediator of neutrophil and macrophage recruitment, and C5 deficiency has been linked to susceptibility to other bacteria (Ahn et al., 2010; Ricklin et al., 2010). However, several points argue that the situation may not be that simple. First, the behaviour of strain K279a in the A/J mice was similar but not identical to that observed in the DBA/2 mice. Second, there are some bacteria for which the primary reason for A/J susceptibility is not linked to the absence of C5 (Qiu et al., 2009); e.g. in S. aureus, the key factors are encoded by the Tnfaip8 and Seh1l loci (Ahn et al., 2010; Qiu et al., 2009), and in L. pneumophila, the critical marker is Naip5/Bircle (Zamboni et al., 2006). Third, the susceptibility of DBA/2 mice to the replication of some bacteria, including P. aeruginosa, is also not linked to the C5 deficiency (Wilson et al., 2007). Based upon our data, genetic crosses between the permissive and non-permissive mice can map and then help define host factors that govern susceptibility to S. maltophilia, with the potential for discovering new attributes of host susceptibility that have implications for explaining human cases of S. maltophilia disease.
Strains of S. maltophilia replicate and persist to differing degrees within the lungs of A/J mice
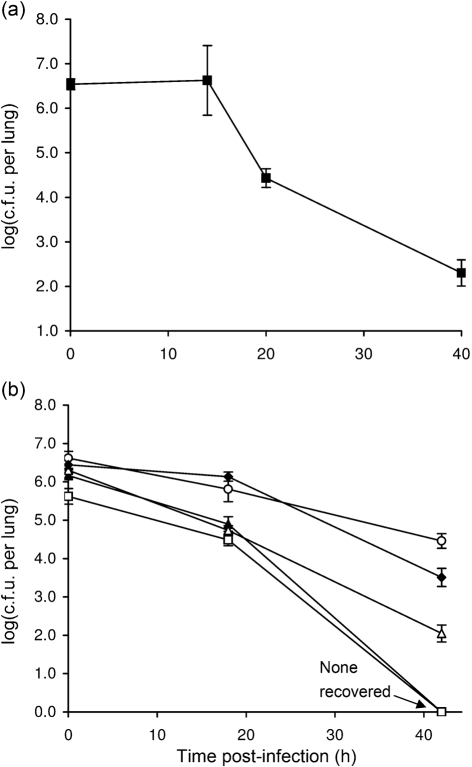
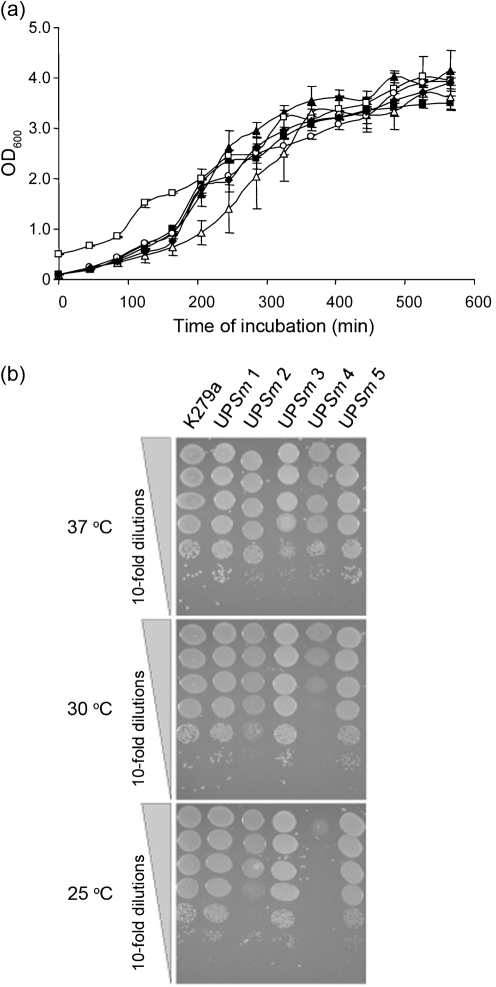
Utilizing the A/J mouse model of infection, we next sought to determine whether strains of S. maltophilia differ in their ability to replicate or persist in the lung. To that end, we employed five clinical isolates obtained from the respiratory tract. The first strain tested, UPSm1, which was obtained from a tracheal aspirate, grew slightly (P<0.05) in the first 4 h and then dropped rapidly over the next 12 h compared with strain K279a (Fig. 5a, compared most closely with Fig. 1a). At the 24 h time point, UPSm1was not recoverable; this was a marked departure from what had been observed for K279a. The second strain tested, UPSm4, obtained from a sputum sample, increased its numbers 10-fold (P<0.05) by 4 h post-inoculation, but then, unlike strain K279a, declined by 8 h (Fig. 5b, as compared with Fig. 1a). However, between 8 and 24 h, UPSm4 was more effective than K279a at maintaining its numbers, e.g. at 24 h, it displayed ~100-fold more c.f.u. than did K279a. Taken together, these experiments indicated that S. maltophilia growth in the lungs is not specific to K279a. However, the data suggested that S. maltophilia strains can differ significantly in their ability to persist in the lungs of A/J mice, with UPSm4 exhibiting strong persistence, K279a intermediate persistence and UPSm1 weak persistence. Thus, we extended the c.f.u. determinations until ~40 h post-inoculation and added three more strains (i.e. UPSm2, obtained from a sputum sample of a CF patient, and UPSm3 and UPSm5, obtained from the respiratory sinus of two other patients) to the analysis. The prototype K279a persisted, showing 100–1000 c.f.u. in the lungs at 40 h post-inoculation (Fig. 6a). The other five could be placed roughly into three groups relative to K279a (Fig. 6b). UPSm1 and UPSm3 were unrecoverable after 40 h, indicative of strains with poor persistence. UPSm2 behaved comparably with K279a, yielding ~105 c.f.u. around the 20 h time point and then 100–1000 c.f.u. after 40 h. Finally, UPSm4 and UPSm5 were more effective at persistence than was K279a. UPSm4 was especially persistent, yielding ~105 c.f.u. after 40 h, which was only about 10-fold less than its deposition. In contrast to these in vivo findings, the strains grew quite similarly when cultured at 37 °C in LB broth or on LB agar (Fig. 7). Incidentally, upon culturing at 30 and 25 °C, UPSm4 and to a lesser degree UPSm2 showed less growth than the other four strains (Fig. 7b). Taken together, these data document that strains of S. maltophilia grow and persist in the lungs to different degrees. That S. maltophilia strains should exhibit different levels of in vivo growth is compatible with past epidemiological analyses which describe high genetic diversity among clinical isolates (Schaumann et al., 2008; Valdezate et al., 2004). The molecular basis for differences in strain virulence may involve the presence or absence of large genomic islands that exist in S. maltophilia genomes but have not been functionally examined. For example, an island in K279a but not in an environmental isolate contains a putative type IV secretion system (Rocco et al., 2009). On the other hand, more subtle genetic or phenotypic differences could explain some of the differences in growth that we have documented. Along those lines, previous studies have made in vitro comparisons among clinical isolates and observed that strains can differ in the expression of putative virulence factors, including proteases, haemolytic activity, and the ability to form biofilms (Di Bonaventura et al., 2007; Figueirêdo et al., 2006; Nicoletti et al., 2011). Thus, future analysis of S. maltophilia virulence in the A/J mouse model should examine not only the sequenced K279a strain but also those strains that have an in vivo phenotype that is significantly different from that of the prototype.
Fig. 5.
Growth of different strains of S. maltophilia in the lungs of A/J mice. Animals were intranasally inoculated with strain UPSm1 (a, ▴) or UPSm4 (b, ○) resulting in comparable levels of deposition (t = 0), and then at 4, 8, 12 and 24 h post-inoculation, the c.f.u. in total lung homogenates were determined by plating. Data are the mean±sd obtained from five infected animals and are representative of at least two independent experiments. Significant increases in c.f.u. relative to the values at t = 0 were obtained only at 4 h in (a) and (b) (Student's t test, P<0.05).
Fig. 6.
Persistence of different strains of S. maltophilia in the lungs of A/J mice. Animals were intranasally inoculated with comparable numbers of strain K279a (a) or strain UPSm1 (▴), UPSm2 (▵), UPSm3 (□), UPSm4 (○) or UPSm5 (⧫) (b), and then at indicated times post-inoculation, the c.f.u. in total lung homogenates were determined by plating. Data are the mean±sd obtained from four infected animals and are representative of at least two independent experiments.
Fig. 7.
Growth of S. maltophilia strains in bacteriological media. (a) Equal numbers of c.f.u. of strains K279a (▪) UPSm1 (▴), UPSm2 (▵), UPSm3 (□), UPSm4 (○) and UPSm5 (⧫) were inoculated into LB broth, and then the growth of the cultures was monitored spectrophotometrically. Data are the mean±sd obtained from replicate samples and are representative of three independent experiments. The slightly reduced optical density readings for the UPSm2 cultures were only significant at the 205 min time point (Student's t test, P<0.05). Although the UPSm3 cultures were started at a slightly higher optical density in order to ensure equal c.f.u. at the outset, they subsequently displayed a growth pattern that was akin to that of the other strains. (b) As indicated, 10-fold dilutions of the five S. maltophilia strains were spotted onto LB agar and then incubated at 37, 30 and 25 °C. After 1 day of incubation, growth was recorded. The patterns of growth depicted here were observed on three independent occasions.
Concluding thoughts
We have demonstrated that S. maltophilia is indeed capable of replicating in the murine lung. This observation lends support to the belief that S. maltophilia is infective and in some cases pathogenic for humans, especially as it relates to pulmonary infection. The different mouse models that we have presented here should prove very useful for future studies aimed at understanding both the bacterial and host factors that influence S. maltophilia infection and disease. Finally, the finding that S. maltophilia strains can differ dramatically in their ability to grow and persist in the lung means that efforts at strain-typing should be intensified in order to better define those strain types or clones that are most likely to trigger the more serious or chronic forms of human disease.
ACKNOWLEDGEMENTS
The authors thank Matthew Avison and Paul Edelstein for sending us bacterial strains. We also acknowledge past and present members of the Cianciotto lab for many helpful discussions, as well as Brendan Mulhern for assistance with figures. This work was supported in part by NIH grants R03 AI082541 and R21 AI076693 awarded to N. P. C.
Abbreviations:
- CF
cystic fibrosis
- IL
interleukin
- TNF-α
tumour necrosis factor-α
REFERENCES
- Ahn S. H., Deshmukh H., Johnson N., Cowell L. G., Rude T. H., Scott W. K., Nelson C. L., Zaas A. K., Marchuk D. A., et al. (2010). Two genes on A/J chromosome 18 are associated with susceptibility to Staphylococcus aureus infection by combined microarray and QTL analyses. PLoS Pathog 6, e1001088. 10.1371/journal.ppat.1001088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alonso A., Martínez J. L. (2000). Cloning and characterization of SmeDEF, a novel multidrug efflux pump from Stenotrophomonas maltophilia. Antimicrob Agents Chemother 44, 3079–3086. 10.1128/AAC.44.11.3079-3086.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ansari S. R., Hanna H., Hachem R., Jiang Y., Rolston K., Raad I. (2007). Risk factors for infections with multidrug-resistant Stenotrophomonas maltophilia in patients with cancer. Cancer 109, 2615–2622. 10.1002/cncr.22705. [DOI] [PubMed] [Google Scholar]
- Ausubel F. M., Brent R., Kingston R. E., Moore D. D., Seidman J. G., Smith J. A., Struhl K. (1989). Current Protocols in Molecular Biology. New York: Wiley. [Google Scholar]
- Avison M. B., von Heldreich C. J., Higgins C. S., Bennett P. M., Walsh T. R. (2000). A TEM-2 β-lactamase encoded on an active Tn1-like transposon in the genome of a clinical isolate of Stenotrophomonas maltophilia. J Antimicrob Chemother 46, 879–884. 10.1093/jac/46.6.879. [DOI] [PubMed] [Google Scholar]
- Avison M. B., Higgins C. S., Ford P. J., von Heldreich C. J., Walsh T. R., Bennett P. M. (2002). Differential regulation of L1 and L2 β-lactamase expression in Stenotrophomonas maltophilia. J Antimicrob Chemother 49, 387–389. 10.1093/jac/49.2.387. [DOI] [PubMed] [Google Scholar]
- Brieland J., Freeman P., Kunkel R., Chrisp C., Hurley M., Fantone J., Engleberg C. (1994). Replicative Legionella pneumophila lung infection in intratracheally inoculated A/J mice. A murine model of human Legionnaires' disease. Am J Pathol 145, 1537–1546. [PMC free article] [PubMed] [Google Scholar]
- Crossman L. C., Gould V. C., Dow J. M., Vernikos G. S., Okazaki A., Sebaihia M., Saunders D., Arrowsmith C., Carver T., et al. (2008). The complete genome, comparative and functional analysis of Stenotrophomonas maltophilia reveals an organism heavily shielded by drug resistance determinants. Genome Biol 9, R74. 10.1186/gb-2008-9-4-r74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies J. C., Rubin B. K. (2007). Emerging and unusual Gram-negative infections in cystic fibrosis. Semin Respir Crit Care Med 28, 312–321. 10.1055/s-2007-981652. [DOI] [PubMed] [Google Scholar]
- DebRoy S., Dao J., Söderberg M., Rossier O., Cianciotto N. P. (2006). Legionella pneumophila type II secretome reveals unique exoproteins and a chitinase that promotes bacterial persistence in the lung. Proc Natl Acad Sci U S A 103, 19146–19151. 10.1073/pnas.0608279103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Denton M., Kerr K. G. (1998). Microbiological and clinical aspects of infection associated with Stenotrophomonas maltophilia. Clin Microbiol Rev 11, 57–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di Bonaventura G., Prosseda G., Del Chierico F., Cannavacciuolo S., Cipriani P., Petrucca A., Superti F., Ammendolia M. G., Concato C., et al. (2007). Molecular characterization of virulence determinants of Stenotrophomonas maltophilia strains isolated from patients affected by cystic fibrosis. Int J Immunopathol Pharmacol 20, 529–537. [DOI] [PubMed] [Google Scholar]
- Di Bonaventura G., Pompilio A., Zappacosta R., Petrucci F., Fiscarelli E., Rossi C., Piccolomini R. (2010). Role of excessive inflammatory response to Stenotrophomonas maltophilia lung infection in DBA/2 mice and implications for cystic fibrosis. Infect Immun 78, 2466–2476. 10.1128/IAI.01391-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falagas M. E., Kastoris A. C., Vouloumanou E. K., Dimopoulos G. (2009). Community-acquired Stenotrophomonas maltophilia infections: a systematic review. Eur J Clin Microbiol Infect Dis 28, 719–730. 10.1007/s10096-009-0709-5. [DOI] [PubMed] [Google Scholar]
- Figueirêdo P. M., Furumura M. T., Santos A. M., Sousa A. C., Kota D. J., Levy C. E., Yano T. (2006). Cytotoxic activity of clinical Stenotrophomonas maltophilia. Lett Appl Microbiol 43, 443–449. 10.1111/j.1472-765X.2006.01965.x. [DOI] [PubMed] [Google Scholar]
- Fouhy Y., Scanlon K., Schouest K., Spillane C., Crossman L., Avison M. B., Ryan R. P., Dow J. M. (2007). Diffusible signal factor-dependent cell-cell signaling and virulence in the nosocomial pathogen Stenotrophomonas maltophilia. J Bacteriol 189, 4964–4968. 10.1128/JB.00310-07. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- George S. E., Kohan M. J., Gilmour M. I., Taylor M. S., Brooks H. G., Creason J. P., Claxton L. D. (1993). Pulmonary clearance and inflammatory response in C3H/HeJ mice after intranasal exposure to Pseudomonas spp. Appl Environ Microbiol 59, 3585–3591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glomski I. J., Dumetz F., Jouvion G., Huerre M. R., Mock M., Goossens P. L. (2008). Inhaled non-capsulated Bacillus anthracis in A/J mice: nasopharynx and alveolar space as dual portals of entry, delayed dissemination, and specific organ targeting. Microbes Infect 10, 1398–1404. 10.1016/j.micinf.2008.07.042. [DOI] [PubMed] [Google Scholar]
- Gordon N. C., Wareham D. W. (2010). Novel variants of the Smqnr family of quinolone resistance genes in clinical isolates of Stenotrophomonas maltophilia. J Antimicrob Chemother 65, 483–489. 10.1093/jac/dkp476. [DOI] [PubMed] [Google Scholar]
- Gould V. C., Avison M. B. (2006). SmeDEF-mediated antimicrobial drug resistance in Stenotrophomonas maltophilia clinical isolates having defined phylogenetic relationships. J Antimicrob Chemother 57, 1070–1076. 10.1093/jac/dkl106. [DOI] [PubMed] [Google Scholar]
- Hernández A., Maté M. J., Sánchez-Díaz P. C., Romero A., Rojo F., Martínez J. L. (2009). Structural and functional analysis of SmeT, the repressor of the Stenotrophomonas maltophilia multidrug efflux pump SmeDEF. J Biol Chem 284, 14428–14438. 10.1074/jbc.M809221200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang T. P., Wong A. C. (2007). A cyclic AMP receptor protein-regulated cell-cell communication system mediates expression of a FecA homologue in Stenotrophomonas maltophilia. Appl Environ Microbiol 73, 5034–5040. 10.1128/AEM.00366-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang Y. W., Lin C. W., Hu R. M., Lin Y. T., Chung T. C., Yang T. C. (2010). AmpN-AmpG operon is essential for expression of L1 and L2 β-lactamases in Stenotrophomonas maltophilia. Antimicrob Agents Chemother 54, 2583–2589. 10.1128/AAC.01283-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim H. B., Srinivasan S., Sathiyaraj G., Quan L. H., Kim S. H., Bui T. P., Liang Z. Q., Kim Y. J., Yang D. C. (2010). Stenotrophomonas ginsengisoli sp. nov., isolated from a ginseng field. Int J Syst Evol Microbiol 60, 1522–1526. 10.1099/ijs.0.014662-0. [DOI] [PubMed] [Google Scholar]
- Lambiase A., Raia V., Del Pezzo M., Sepe A., Carnovale V., Rossano F. (2006). Microbiology of airway disease in a cohort of patients with cystic fibrosis. BMC Infect Dis 6, 4–10. 10.1186/1471-2334-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee M., Woo S. G., Chae M., Shin M. C., Jung H. M., Ten L. N. (2011). Stenotrophomonas daejeonensis sp. nov., isolated from sewage. Int J Syst Evol Microbiol 61, 598–604. 10.1099/ijs.0.017780-0. [DOI] [PubMed] [Google Scholar]
- Looney W. J., Narita M., Mühlemann K. (2009). Stenotrophomonas maltophilia: an emerging opportunist human pathogen. Lancet Infect Dis 9, 312–323. 10.1016/S1473-3099(09)70083-0. [DOI] [PubMed] [Google Scholar]
- McKay G. A., Woods D. E., MacDonald K. L., Poole K. (2003). Role of phosphoglucomutase of Stenotrophomonas maltophilia in lipopolysaccharide biosynthesis, virulence, and antibiotic resistance. Infect Immun 71, 3068–3075. 10.1128/IAI.71.6.3068-3075.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minkwitz A., Berg G. (2001). Comparison of antifungal activities and 16S ribosomal DNA sequences of clinical and environmental isolates of Stenotrophomonas maltophilia. J Clin Microbiol 39, 139–145. 10.1128/JCM.39.1.139-145.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nicoletti M., Iacobino A., Prosseda G., Fiscarelli E., Zarrilli R., De Carolis E., Petrucca A., Nencioni L., Colonna B., Casalino M. (2011). Stenotrophomonas maltophilia strains from cystic fibrosis patients: genomic variability and molecular characterization of some virulence determinants. Int J Med Microbiol 301, 34–43. 10.1016/j.ijmm.2010.07.003. [DOI] [PubMed] [Google Scholar]
- Okazaki A., Avison M. B. (2007). Aph(3′)-IIc, an aminoglycoside resistance determinant from Stenotrophomonas maltophilia. Antimicrob Agents Chemother 51, 359–360. 10.1128/AAC.00795-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paez J. I., Tengan F. M., Barone A. A., Levin A. S., Costa S. F. (2008). Factors associated with mortality in patients with bloodstream infection and pneumonia due to Stenotrophomonas maltophilia. Eur J Clin Microbiol Infect Dis 27, 901–906. 10.1007/s10096-008-0518-2. [DOI] [PubMed] [Google Scholar]
- Paschoal I. A., de Oliveira Villalba W., Bertuzzo C. S., Cerqueira E. M., Pereira M. C. (2007). Cystic fibrosis in adults. Lung 185, 81–87. 10.1007/s00408-006-2597-0. [DOI] [PubMed] [Google Scholar]
- Qiu H., KuoLee R., Harris G., Chen W. (2009). High susceptibility to respiratory Acinetobacter baumannii infection in A/J mice is associated with a delay in early pulmonary recruitment of neutrophils. Microbes Infect 11, 946–955. 10.1016/j.micinf.2009.06.003. [DOI] [PubMed] [Google Scholar]
- Ricklin D., Hajishengallis G., Yang K., Lambris J. D. (2010). Complement: a key system for immune surveillance and homeostasis. Nat Immunol 11, 785–797. 10.1038/ni.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rocco F., De Gregorio E., Colonna B., Di Nocera P. P. (2009). Stenotrophomonas maltophilia genomes: a start-up comparison. Int J Med Microbiol 299, 535–546. 10.1016/j.ijmm.2009.05.004. [DOI] [PubMed] [Google Scholar]
- Rossier O., Starkenburg S. R., Cianciotto N. P. (2004). Legionella pneumophila type II protein secretion promotes virulence in the A/J mouse model of Legionnaires' disease pneumonia. Infect Immun 72, 310–321. 10.1128/IAI.72.1.310-321.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan R. P., Monchy S., Cardinale M., Taghavi S., Crossman L., Avison M. B., Berg G., van der Lelie D., Dow J. M. (2009). The versatility and adaptation of bacteria from the genus Stenotrophomonas. Nat Rev Microbiol 7, 514–525. 10.1038/nrmicro2163. [DOI] [PubMed] [Google Scholar]
- Safdar A., Rolston K. V. (2007). Stenotrophomonas maltophilia: changing spectrum of a serious bacterial pathogen in patients with cancer. Clin Infect Dis 45, 1602–1609. 10.1086/522998. [DOI] [PubMed] [Google Scholar]
- Sánchez M. B., Martínez J. L. (2010). SmQnr contributes to intrinsic resistance to quinolones in Stenotrophomonas maltophilia. Antimicrob Agents Chemother 54, 580–581. 10.1128/AAC.00496-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez M. B., Hernández A., Rodríguez-Martínez J. M., Martínez-Martínez L., Martínez J. L. (2008). Predictive analysis of transmissible quinolone resistance indicates Stenotrophomonas maltophilia as a potential source of a novel family of Qnr determinants. BMC Microbiol 8, 148. 10.1186/1471-2180-8-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sánchez M. B., Hernandez A., Martinez J. L. (2009). Stenotrophomonas maltophilia drug resistance. Future Microbiol 4, 655–660. 10.2217/fmb.09.45. [DOI] [PubMed] [Google Scholar]
- Schaumann R., Laurin F., Rodloff A. C. (2008). Molecular typing of clinical isolates of Stenotrophomonas maltophilia by pulsed-field gel electrophoresis and random primer PCR fingerprinting. Int J Hyg Environ Health 211, 292–298. 10.1016/j.ijheh.2007.05.005. [DOI] [PubMed] [Google Scholar]
- Shimizu K., Kikuchi K., Sasaki T., Takahashi N., Ohtsuka M., Ono Y., Hiramatsu K. (2008). Smqnr, a new chromosome-carried quinolone resistance gene in Stenotrophomonas maltophilia. Antimicrob Agents Chemother 52, 3823–3825. 10.1128/AAC.00026-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spicuzza L., Sciuto C., Vitaliti G., Di Dio G., Leonardi S., La Rosa M. (2009). Emerging pathogens in cystic fibrosis: ten years of follow-up in a cohort of patients. Eur J Clin Microbiol Infect Dis 28, 191–195. 10.1007/s10096-008-0605-4. [DOI] [PubMed] [Google Scholar]
- Travassos L. H., Pinheiro M. N., Coelho F. S., Sampaio J. L., Merquior V. L., Marques E. A. (2004). Phenotypic properties, drug susceptibility and genetic relatedness of Stenotrophomonas maltophilia clinical strains from seven hospitals in Rio de Janeiro, Brazil. J Appl Microbiol 96, 1143–1150. 10.1111/j.1365-2672.2004.02248.x. [DOI] [PubMed] [Google Scholar]
- Valdezate S., Vindel A., Martín-Dávila P., Del Saz B. S., Baquero F., Cantón R. (2004). High genetic diversity among Stenotrophomonas maltophilia strains despite their originating at a single hospital. J Clin Microbiol 42, 693–699. 10.1128/JCM.42.2.693-699.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters V. J., Gómez M. I., Soong G., Amin S., Ernst R. K., Prince A. (2007). Immunostimulatory properties of the emerging pathogen Stenotrophomonas maltophilia. Infect Immun 75, 1698–1703. 10.1128/IAI.01469-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters V., Yau Y., Prasad S., Lu A., Atenafu E., Crandall I., Tom S., Tullis E., Ratjen F. (2011). Stenotrophomonas maltophilia in cystic fibrosis: Serologic response and effect on lung disease. Am J Respir Crit Care Med 183, 635–640. 10.1164/rccm.201009-1392OC. [DOI] [PubMed] [Google Scholar]
- Wilson K. R., Napper J. M., Denvir J., Sollars V. E., Yu H. D. (2007). Defect in early lung defence against Pseudomonas aeruginosa in DBA/2 mice is associated with acute inflammatory lung injury and reduced bactericidal activity in naive macrophages. Microbiology 153, 968–979. 10.1099/mic.0.2006/002261-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yi H., Srinivasan S., Kim M. K. (2010). Stenotrophomonas panacihumi sp. nov., isolated from soil of a ginseng field. J Microbiol 48, 30–35. 10.1007/s12275-010-0006-0. [DOI] [PubMed] [Google Scholar]
- Zamboni D. S., Kobayashi K. S., Kohlsdorf T., Ogura Y., Long E. M., Vance R. E., Kuida K., Mariathasan S., Dixit V. M., et al. (2006). The Birc1e cytosolic pattern-recognition receptor contributes to the detection and control of Legionella pneumophila infection. Nat Immunol 7, 318–325. 10.1038/ni1305. [DOI] [PubMed] [Google Scholar]
- Zgair A. K., Chhibber S. (2010a). Immunological and pathological aspects of respiratory tract infection with Stenotrophomonas maltophilia in BALB/c mice. J Microbiol Biotechnol 20, 1585–1591. 10.4014/jmb.1003.03013. [DOI] [PubMed] [Google Scholar]
- Zgair A. K., Chhibber S. (2010b). Stenotrophomonas maltophilia flagellin induces a compartmentalized innate immune response in mouse lung. J Med Microbiol 59, 913–919. 10.1099/jmm.0.020107-0. [DOI] [PubMed] [Google Scholar]