Abstract
Nuclear receptors can function as ligand-inducible transregulators in both mammalian and yeast cells, indicating that important features of control of transcription have been conserved throughout evolution. Here, we report the isolation and characterization of a yeast protein that exhibits properties expected for a coactivator/mediator of the ligand-dependent activation function AF-2 present in the ligand-binding domain (LBD, region E) of the retinoid X (RXRα) and estrogen (ERα) receptors. This protein is identical to Ada3, a component of the yeast Ada coactivator complex. We demonstrate that: (1) the region encompassing residues 347–702 of Ada3 interacts with the LBD of RXRα and ERα in a ligand-dependent manner in yeast; (2) this interaction corresponds to a direct binding and requires the integrity of the core of the AF-2 activating domain (AF-2 AD) of both RXRα and ERα; (3) Ada3 as well as Ada2 and Gcn5, two other components of the Ada complex, are required for maximal AF-2 activity in yeast; and (4) Ada3 is able to enhance the AF-2 activity of RXRα and ERα when overexpressed in yeast and mammalian cells. Taken together, these data indicate that ligand-dependent transactivation by RXRα and ERα in yeast is mediated at least in part by the Ada complex, in which the Ada3 subunit directly binds to the holoreceptor LBD.
Keywords: Transcriptional control, Ada2/Ada3/Gcn5, Saccharomyces cerevisiae, nuclear receptors, RXRα/ERα, transcriptional intermediary factor, coactivator
Nuclear receptors (NRs) represent a large family of ligand-inducible transcriptional regulators that control complex developmental and homeostatic events in vertebrates by binding as homodimers or heterodimers to cognate DNA response elements present in target genes. NRs display a modular structure, with five to six distinct regions (denoted A–E/F; see Fig. 1A). The amino-terminal A/B region contains an autonomous activation function (AF-1), the highly conserved region C belongs to the DNA-binding domain (DBD), and the carboxy-terminal E region contains the ligand-binding domain (LBD), a dimerization surface and a ligand-dependent transcriptional activation function, AF-2 (for reviews and refs, see Mangelsdorf et al. 1995; Chambon 1996; Perlman and Evans 1997). The core of the AF-2 activating domain (AF-2 AD core; see Fig. 1A) has been characterized in the carboxy-terminal part of the E region and corresponds to the conserved amphipathic α helix 12 of the LBD. Upon ligand binding, this helix is thought to fold back over the LBD to generate interacting surface(s) for the binding of transcriptional intermediary factors (TIFs, also denoted coactivators or mediators), whose effects ultimately result in the remodeling of the structure of the chromatin template and/or in the stimulation of initiation of RNA synthesis by the general transcription machinery (for reviews and references, see Chambon 1996; Wurtz et al. 1996; Glass et al. 1997).
Figure 1.

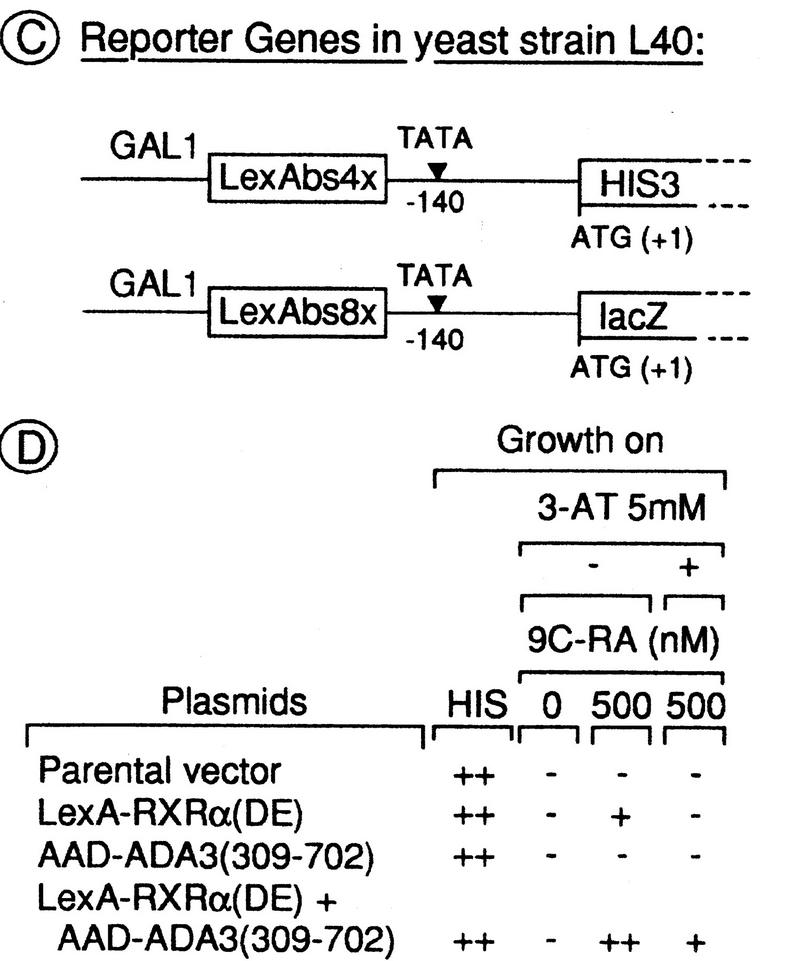
Identification of Ada3 by a two-hybrid screening for yeast proteins that interact with RXRα. (A) Schematic representation of mouse RXRα. Indicated are the various regions of the receptor (denoted A–E) that are conserved among members of the NR family. Transactivation domains (AF-1 and AF-2), DBD, and LBD are indicated. (Filled bar) Core motif of the AF-2 AD. (Numbers) Amino-acid positions. (B) Schematic representation of the LexA protein unfused or fused to the DE region of RXRα. The VP16 AAD-tagged S. cerevisiae genomic DNA library is represented below. The AAD tag also includes codons specifying the nuclear localization signal (NLS) of the yeast ribosomal protein L29. (C) Transcription of the integrated HIS3 and lacZ reporter genes in the reporter strain L40 is driven by a chimeric GAL1 promoter containing four and eight LexA-binding sites (LexAbs), respectively. (D) Growth complementation by interaction of RXRα(DE) with Ada3(309–702). Yeast L40 cells expressing the indicated fusion proteins were plated on medium containing histidine (His) and on His-negative medium +/− 9C-RA and +/− 3-AT as indicated. Plates were incubated at 30°C for 2 days. (++) Wild-type growth; (+) weak gowth; (−) no growth.
Although the yeast Saccharomyces cerevisiae does not possess endogenous NRs, it has been shown that a number of NRs, including the estrogen receptor (ER, now designated ERα), the glucocorticoid receptor (GR), the retinoic acid (RA) receptors (RARs and RXRs), and the thyroid hormone receptor (TRs) can function as ligand-dependent transactivators in yeast (Metzger et al. 1988, 1992; Schena and Yamamoto 1988; Hall et al. 1993; Heery et al. 1993; and references therein). As in vertebrates (Chambon 1996 and references therein), the AF-1 and AF-2 of ERα, RARα, and RXRα can activate transcription independently and synergistically in yeast (Metzger et al. 1988; White et al. 1988; Pierrat et al. 1992; Heery et al.1993; and our unpublished results). Similarly, the transactivation potentials of ERα AF-1 and AF-2 are promoter-context-dependent in both animal and yeast cells (Tora et al. 1989; Berry et al. 1990; Metzger et al. 1992). Thus, some important features of the mechanism(s) by which these AFs stimulate transcription appear to have been conserved during evolution.
Several yeast proteins implicated in control of transcription have been shown to functionally interact with NRs. These factors include the Swi/Snf protein complex (Yoshinaga et al. 1992), Ssn6 (McDonnell et al. 1992), Sin3 (Nawaz et al. 1994), Spt6 (Baniahmad et al. 1995), Rsp5 and Spt3 (Imhof and McDonnell 1996). Mammalian homologs have been identified for Swi/Snf genes (Muchardt and Yaniv 1993; Chiba et al. 1994; Muchardt et al. 1995), Sin3 (Ayer et al. 1995), Spt6 (Segre et al. 1995), and Rsp5 (Imhof and McDonnell 1996). However, a physical interaction between yeast factors and NRs was observed only in the case of the Swi/Snf complex and Spt6, which have been reported to interact with the DBD of GR and the LBD of ERα, respectively (Yoshinaga et al. 1992; Baniahmad et al. 1995).
From the results of a two-hybrid screen for yeast proteins that can interact with the LBD/AF-2 of RXRα in a 9-cis-RA (9C-RA)-dependent manner, we report the identification of such a protein and show that it is involved in the mediation of transactivation by the AF-2 of RXRα and ERα in yeast. This protein is identical to the Ada3 subunit of the Ada coactivator complex that is important for transcriptional activation by specific yeast activators (Piña et al. 1993; Horiuchi et al. 1995; Grant et al. 1997 and references therein). We show that two other subunits of the Ada complex, Ada2 and Gcn5, are required in addition to Ada3 for full activation by the AF-2 of RXRα and ERα in yeast. Interestingly, human homologs of the Ada2 and Gcn5 proteins have recently been isolated, suggesting an evolutionary conservation of this complex (Candau et al. 1996). Thus, the Ada complex may play an important role in mediating the ligand-dependent activation function AF-2 of certain NRs in yeast and higher eukaryotes.
Results
Yeast Ada3 interacts with nuclear receptors
The two-hybrid system (Fields and Sternglanz 1994) was used to identify yeast proteins that interact with the LBD/AF-2-containing region DE of RXRα in the presence of 9C-RA. The bacterial LexA DBD, fused amino-terminally to the DE region of RXRα [LexA–RXRα(DE); Fig. 1B] was expressed in the L40 yeast strain, which contains a HIS3 reporter gene under the control of four LexA-binding sites (Fig. 1C; Vojtek et al. 1993). Addition of 500 nm 9C-RA allowed growth as a result of transactivation of the HIS3 gene by LexA–RXRα(DE) on its own, whereas no growth was observed in the absence of ligand on plates lacking histidine (Fig. 1D). Addition of 5 mm 3-amino-1,2,4-triazole (3-AT), a competitive inhibitor of the HIS3 gene product, suppressed growth in the presence of ligand (Fig. 1D).
A library of yeast genomic DNA fragments was constructed in pASV3, a yeast multicopy expression vector that directs the synthesis of polypeptides fused to the acidic activation domain (AAD) of the VP16 protein (Fig. 1B; Le Douarin et al. 1995b). L40 yeast cells expressing LexA–RXRα(DE) were transformed with this library. Approximately 4 × 106 yeast transformants (covering the library four times) were spread directly on histidine-negative plates containing 500 nm 9C-RA and 5 mm 3-AT. Colonies (232) were isolated and retested for activation of a second reporter gene, the Escherichia coli lacZ gene driven by a GAL1 promoter containing eight LexA-binding sites (Fig. 1C). Plasmids were recovered from 110 clones exhibiting strong lacZ expression, amplified, and subjected to restriction analysis. DNA inserts from 15 remaining unique library plasmids were sequenced. Sequence comparison with the NCBI and EMBL databases identified one of these DNA inserts as the sequence encoding amino acids 309–702 of Ada3 (Piña et al. 1993).
Residues 347–702 of Ada3 interact in a ligand-dependent manner with RXRα, TRα, ERα, but not RARα
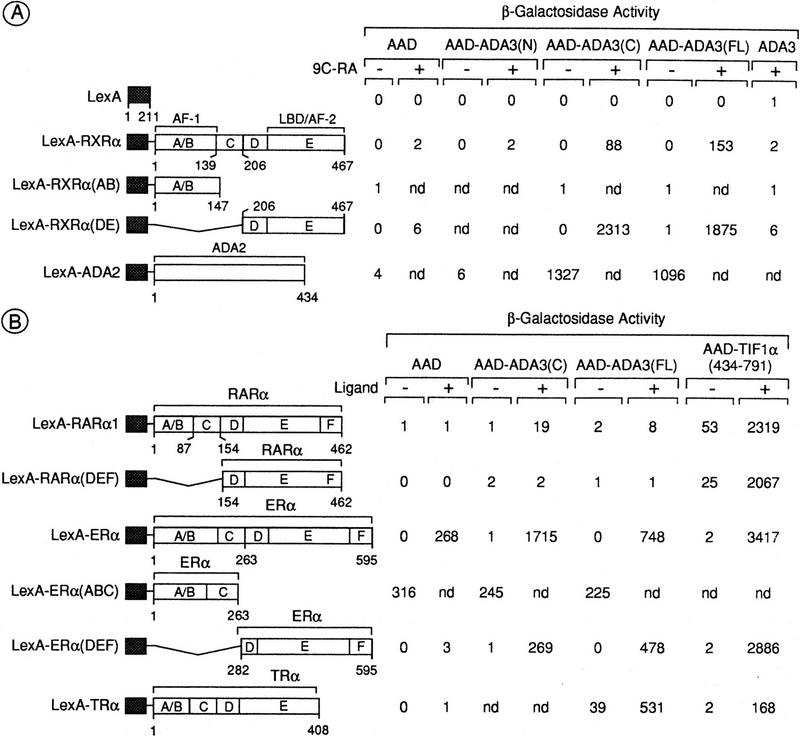
The entire coding sequence of RXRα fused to the LexA protein (LexA–RXRα; Fig. 2A) was expressed in L40 cells with full-length (FL) or truncated Ada3, containing either the amino-terminal (N, amino acids 1–346) or the carboxy-terminal (C, amino acids 347–702) region of Ada3, fused to VP16 AAD [AAD–Ada3(FL), AAD–Ada3(N), and AAD–Ada3(C), Fig. 2A]. The coexpression of these Ada3 halves is known to complement a Δada3 strain as efficiently as full-length Ada3 (Horiuchi et al. 1995). In control assays, the LexA–RXRα fusion was assayed for activation with VP16 AAD or unfused Ada3. For comparison, assays were also performed with LexA–Ada2, which interacts with the carboxy-terminal region of Ada3 (Horiuchi et al. 1995). No increase in reporter activity was detected when LexA–RXRα was coexpressed with Ada3 or AAD–Ada3(N) (Fig. 2A). In contrast, 44- and 75-fold 9C-RA-dependent enhancements were observed in the presence of AAD–Ada3(C) and AAD–Ada3(FL), respectively (Fig. 2A), indicating that the carboxy-terminal moiety of Ada3 can functionally interact with RXRα in a 9C-RA-dependent manner. This interaction was specific to the LBD/AF-2 of RXRα, since no β-galactosidase increase was obtained with LexA–RXRα(AB) containing the amino-terminal AB region of RXRα, which harbors the transcriptional activation function AF-1 (Fig. 2A), whereas coexpression of LexA–RXRα(DE) with AAD–Ada3(FL) or AAD–Ada3(C) resulted in a >300-fold 9C-RA-dependent increase in β-galactosidase activity (Fig. 2A). This increase was similar to that resulting from the interaction of Ada3 and Ada2 [see LexA–Ada2 and AAD–Ada3(FL) or AAD–Ada3(C); Fig. 2A].
Figure 2.
Two-hybrid interaction between Ada3 and various nuclear receptors. (A) Residues 347–702 of Ada3 are sufficient for mediating a ligand-dependent interaction with the LBD/AF-2 of RXRα. (Left) Schematic representations of the LexA–RXRα fusions. These chimera were expressed in the yeast reporter strain L40 together with either the VP16 AAD or the VP16 AAD fused to the amino-terminal residues 1–346 of Ada3 [AAD–Ada3(N)] or the VP16 AAD fused to the carboxy-terminal residues 347–702 of Ada3 [AAD–Ada3(C)] or unfused Ada3. Transformants were grown in liquid medium in the presence (+) or absence (−) of 500 nm 9C-RA. β-Galactosidase activities are expressed in nmoles of substrate/min/mg. (B) Ada3 interacts with the LBD/AF-2 of ERα and full-length TRα, but not with the LBD/AF-2 of RARα. The indicated LexA and AAD fusions were assayed for interaction in the yeast reporter strain L40 grown in the presence (+) or absence (−) of the cognate ligand (500 nm T-RA for RARα, 500 nm E2 for ERα, 5 μm T3 for TRα). β-Galactosidase activities are expressed as in A. In all panels, the values (±20%) are the average of at least three independent transformants.
LexA-fusion proteins containing sequences of RARα, ERα, and TRα or deletion derivatives of these receptors (Fig. 2B) were also coexpressed with either AAD or AAD–Ada3 in the presence or absence of cognate ligands. For comparison, assays were also performed with AAD–TIF1α(434–791), which contains the NR-interacting domain of TIF1α, a putative mouse mediator of the ligand-dependent AF-2 of several NRs (Le Douarin et al. 1995a). A very weak increase in β-galactosidase was observed by coexpression of LexA–RARα with either AAD–Ada3(FL) or AAD–Ada3(C) in the presence of T-RA (Fig. 2B), and no significant increase was observed in cells coexpressing LexA–RARα(DEF) [Fig. 2B; under the same conditions, a high T-RA-stimulated activation was detected with AAD–TIF1α(434–791)]. Thus, in contrast to RXRα, the LBD/AF-2 of RARα cannot efficiently interact with Ada3. In the presence of estradiol (E2), LexA–ERα on its own activated the chimeric GAL1 promoter (Fig. 2B). As expected (Metzger et al. 1992), the activation function AF-1 was mostly responsible for this activation [compare LexA–ERα(ABC) to LexA–ERα(DEF); Fig. 2B]. Although poorly active on its own, the DEF region of ERα exhibited a significant E2-dependent interaction with the full-length and carboxy-terminal moiety of Ada3 (Fig. 2B), indicating that, like RXRα, the LBD of ERα can interact with Ada3. However, β-galactosidase activity resulting from this interaction was 6- to 10-fold lower than that corresponding to the LexA–ERα(DEF)/AAD–TIF1α(434–791) interaction (Fig. 2B). In the case of LexA–TRα, the weak constitutive activation observed with Ada3(FL) was stimulated 16-fold on addition of thyroid hormone (T3) (Fig. 2B).
Mutations in the AF-2 activation domain core motif of RXRα and ERα impair interaction with Ada3 in vivo and in vitro
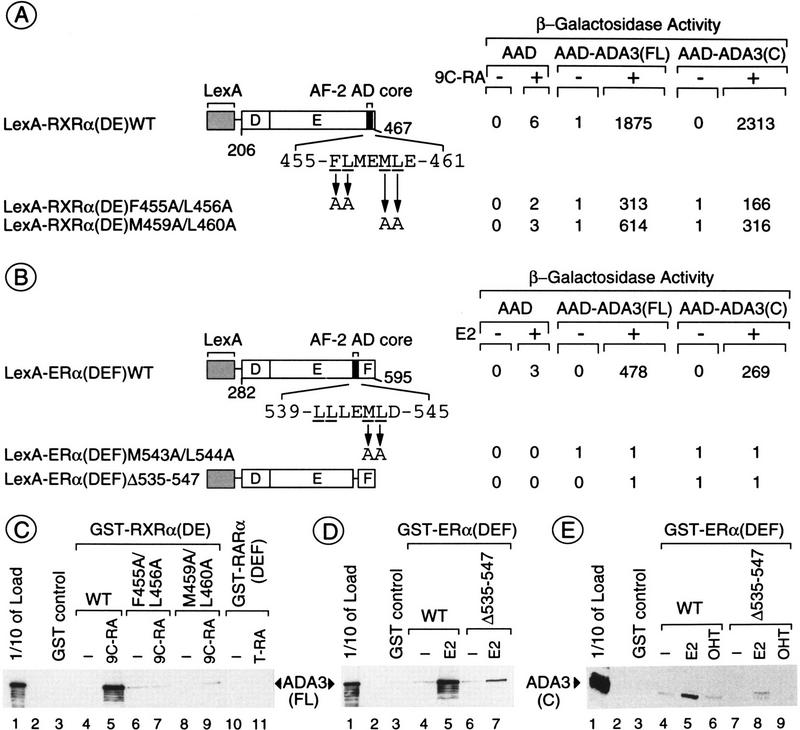
If Ada3 were involved in the mediation of transcriptional activation by NR AF-2, its ligand-dependent interaction with the corresponding LBDs should be affected by mutations in the core motif of the AF-2 activation domain (AD; helix 12 of the LBD), which have been shown to reduce AF-2 activity (but not ligand binding) in mammalian and yeast cells (Danielian et al. 1992; Ince et al. 1993; Durand et al. 1994; Heery et al. 1994) as well as in vitro interaction between NRs and putative mammalian coactivators (e.g., CBP/p300, SRC-1 and TIF2/GRIP1; for review, see Glass et al. 1997). Several RXRα and ERα mutants carrying mutations in the AF-2 AD core motif (see Fig. 3) were tested for their ability to interact with Ada3 in the yeast two-hybrid assay. When compared with wild-type RXRα(DE), RXRα(DE)F455A/L456A and RXRα(DE)M459A/L460A were severely impaired in their ability to interact with Ada3(FL) and Ada3(C) (Fig. 3A). Similarly, a point mutant [ERα(DEF)M543A/L544A] as well as a mutant bearing an internal deletion of the core motif of ERα [ERα(DEF)Δ535–547] showed no interaction with Ada3 (Fig. 3B). Thus, both RXRα and ERα require an intact AF-2 AD core motif to efficiently interact with Ada3 in the yeast two-hybrid system.
Figure 3.
The AF-2 AD core motif is important for interaction between NRs and Ada3. (A) Point mutations in the conserved hydrophobic residues of the AF-2 AD core of RXRα impair interaction with Ada3. The indicated mutants of RXRα were fused to LexA and assayed for interaction with AAD–Ada3(FL) and AAD–Ada3(C) in the yeast reporter strain L40 grown in the presence or absence of 500 nm 9C-RA. β-Galactosidase activities are expressed as in Fig. 2A. (B) Deletion or point mutations in the AF-2 AD core of ERα abolish interaction with Ada3. L40 transformants expressing the indicated LexA and AAD fusions in the presence or absence of 500 nm E2 were treated as described in A. (C) Ada3 binds directly to the LBD of RXRα, but not of RARα, in a ligand- and AF-2-integrity-dependent manner. Purified His-epitope B10-tagged Ada3 was incubated in a batch assay with control GST (lane 3), GST–RXRα(DE) wild type (WT; lanes 4,5), GST–RXRα(DE) mutated in the conserved hydrophobic residues of the AF-2 AD core motif (lanes 6–9), or GST–RARα(DEF) (lanes 10,11) bound to glutathione–Sepharose beads, in the presence or absence of ligands (1 μm 9C-RA for RXRα and 1 μm T-RA for RARα). Bound Ada3 protein was detected by Western blotting with the B10 antibody. (Lane 1) 1/10 the amount of input His–Ada3 fusion (arrow). (D) Ada3 binds directly to the LBD of ERα in a ligand- and AF-2-integrity-dependent manner. Binding assays were done as in C in the presence or absence of 500 nm E2. (E) E2 (500 nm; lane 5), but not OHT (1 μm; lane 6), induces interaction between the ERα LBD and the carboxy-terminal moiety of Ada3.
Binding assays between Ada3 and the LBD/AF-2s of RXRα, RARα, and ERα were then performed in vitro with purified recombinant proteins. Purified E. coli-expressed histidine-epitope B10-tagged Ada3 was mixed with GST–receptor fusion proteins attached to glutathione–Sepharose beads (Fig. 3C–E). The matrix-associated Ada3 protein was revealed by Western blotting. In agreement with the two-hybrid data, a 9C-RA-dependent interaction was observed between the LBD/AF-2 of RXRα and Ada3 (cf. lanes 4 and 5 in Fig. 3C). As expected, the AF-2 AD core mutants GST–RXRα(DE)F455A/L456A (lanes 6,7) and GST–RXRα(DE)M459A/L460A (lanes 8,9) associated very poorly with Ada3. In contrast, as observed in the two-hybrid assays, no significant interaction was detected between Ada3 and GST–RARα(DEF) (lanes 10,11, Fig. 3C). Similarly to RXRα, agarose beads loaded with the LBD/AF-2 region of ERα retained Ada3 in a ligand-dependent manner [see GST–ERα(DEF)WT; lanes 4,5 in Fig. 3D], and this interaction was dependent on the integrity of the AF-2 AD core (cf. lanes 5 and 7, Fig. 3D). Essentially, the same results were obtained for interaction between the ERα DEF region and the carboxy-terminal moiety of Ada3 (Fig. 3E). Moreover, the anti-estrogen hydroxytamoxifen (OHT), which does not induce ER AF-2 activity in animal and yeast cells (Metzger et al. 1988, 1992; White et al. 1988; Berry et al. 1990), did not promote the binding of ERα to either Ada3(FL) or Ada3(C) (Fig. 3E, lanes 6,9 and data not shown), thus showing that both transactivation by AF-2 and receptor–Ada3 interaction require the same agonist-induced conformational changes in the LBD of ERα.
Ada3 acts as a coactivator for the AF-2 of RXRα and ERα in yeast
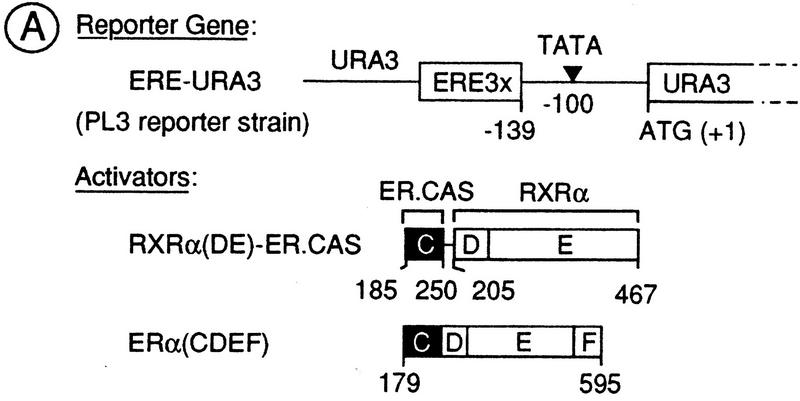
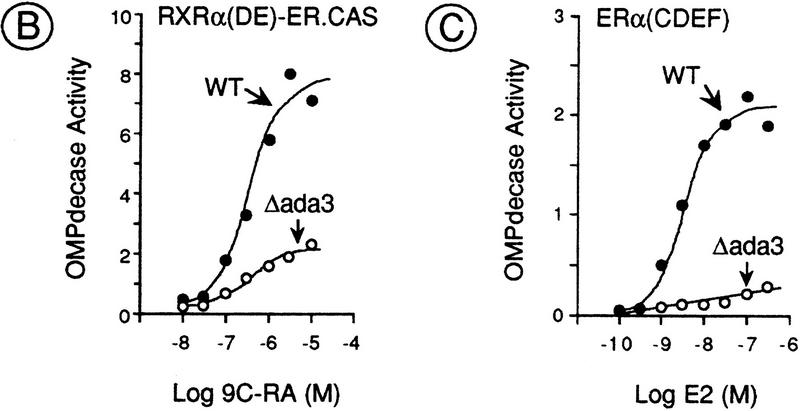
To determine whether Ada3 functions as a yeast mediator for the AF-2 of RXRα and ERα, the transcriptional activity of these AF-2s was tested in the absence of Ada3. The ADA3 gene was disrupted in the PL3 reporter strain, which contains a URA3 reporter gene controlled by three estrogen response elements (EREs, Fig. 4A; Pierrat et al. 1992). Wild-type and Δada3 PL3 strains were transformed with a high-copy-number plasmid YEp90 expressing [under the control of the phosphoglycerate kinase (PGK) promoter] either a chimeric receptor consisting of the DBD of ERα (ER.CAS, ERα amino acid residues 185–250) fused to the LBD/AF-2 of RXRα [RXRα(DE)–ER.CAS; Heery et al. 1993] or an amino-terminally truncated ERα derivative [ERα(CDEF), previously called HEG19; Berry et al. 1990] (Fig. 4A). The transcriptional activity of these proteins, as estimated by OMPdecase activity (the URA3 product) in extracts from cells grown in the presence of increasing amounts of ligand, was reduced ∼3- and 10-fold respectively, in the Δada3 mutant when compared with wild-type cells (Fig. 4B,C). Thus, the AF-2 ligand-dependent activation functions of RXRα and ERα require Ada3 for full activity in yeast.
Figure 4.
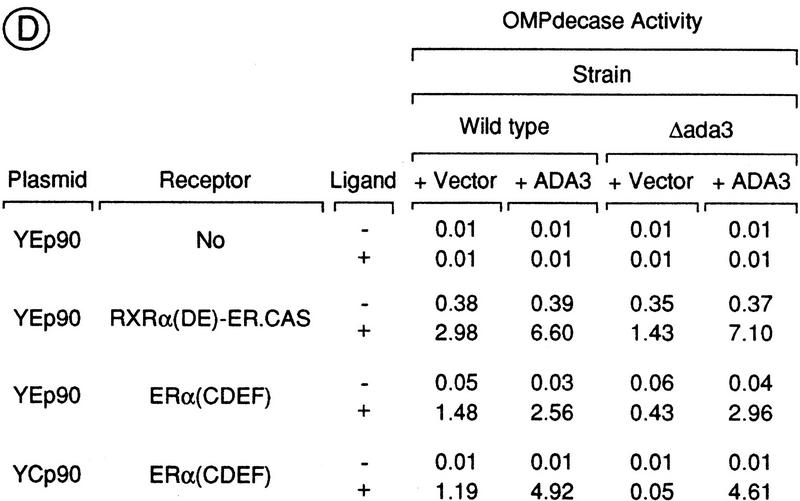
Effect of deletion or overexpression Ada3 on the AF-2 activity of RXRα and ERα. (A) Schematic representation of the reporter gene and activators used in this study. The ERE–URA3 reporter gene (integrated in the yeast strain PL3) was created by replacement of the poly[d(A-T)] sequences and the UASUra site [from position −216 to −139 with respect to the ATG (+1) of the URA3 gene] by three EREs (ERE3x) (Pierrat et al. 1992). (B,C) The ligand-dependent activation function AF-2 of RXRα and ERα requires Ada3 for full activity in yeast. Wild-type (WT) and Δada3 PL3 strains were transformed with high-copy-number (YEp90) plasmids containing the receptor derivatives illustrated in A. Transformants were grown exponentially during five generations on selective medium containing uracil and ligand [9C-RA for RXRα(DE)–ER.CAS and E2 for ERα(CDEF)] at the concentrations indicated. OMPdecase assays were performed on each cell-free extract. Enzyme activity is expressed in nmole of substrate/min/mg protein. (D) Ada3 is a limiting coactivator for the AF-2 of RXRα and ERα. The indicated receptor derivatives were expressed from a high- (YEp90) or low- (YCp90) copy-number plasmid in the wild-type and Δada3 PL3 strains in the presence (+) or absence (−) of ligand [500 nm 9C-RA for RXRα(DE)–ER.CAS and 500 nm E2 for ERα(CDEF)]. Cells were also transformed with an episomal expression vector (YEp10) containing Ada3 (+ Ada3) or no insert (+ vector). OMPdecase activity is expressed as in B. The values (±20%) are the mean of at least three independent experiments.
We also evaluated the effect of Ada3 overexpression on the AF-2 activity of RXRα and ERα. Overexpressing Ada3 in wild-type and Δada3 strains resulted in an approximately two- and fivefold enhancement in the AF-2 activity of RXRα, respectively (Fig. 4D). Similar stimulatory effects were obtained on the AF-2 activity of ERα (Fig. 4D). Interestingly, a more drastic ∼90-fold increase in the AF-2 activity of ERα was observed in the Δada3 cells that express ERα(CDEF) from a low-copy-number plasmid (YCp90) in the absence or presence of Ada3 (Fig. 4D). This effect was strictly ligand dependent (Fig. 4D) and apparently promoter-specific, as no increase in the LexA–GAL1/lacZ reporter gene activity was observed when LexA–RXRα(DE) or LexA–ERα(DEF) was coexpressed with Ada3 in the L40 reporter strain (Fig. 2A and data not shown). To exclude the possibility that Ada3 had an effect on receptor expression levels, Western blot analyses were performed and no significant change in the expression levels of the RXRα and ERα receptor proteins was detected when Ada3 was deleted (not shown), indicating that, as reported (Henriksson et al. 1997), the activity of the PGK promoter is not Ada3-dependent. Taken together, these results indicate that Ada3 is a coactivator for the AF-2 of RXRα and ERα in yeast cells.
Integrity of the Ada complex is required for full activation of transcription by the AF-2 of ERα and RXRα
Ada3 forms a protein complex, the Ada complex, with other yeast proteins, including Ada2 and Gcn5 (see Discussion). We therefore investigated whether Ada3 could mediate the AF-2 activity of ERα through this complex. Deletions were introduced in the ADA2 and GCN5 genes of the yeast reporter strain PL3. Wild-type and mutant strains were transformed with the low-copy-number plasmid YCp90-ERα(CDEF), transformants were grown in the presence of hormone, and OMPdecase activity was determined. The reporter gene activity was approximately six- and fourfold reduced in Δada2 and Δgcn5 strains, respectively (Table 1), indicating that the AF-2 of ERα requires functions of other components of the Ada complex for full activity. However, the AF-2 activity was reduced to a lesser extent in Δada2 and Δgcn5 mutants than in the Δada3 mutant (Table 1). Furthermore, we found that overexpression of Ada3 from a high-copy-number YEp10 vector suppressed the reduction of AF-2 activity in the Δada2 and Δgcn5 strains (Table 1), indicating that Ada3 can function independently of Ada2 and Gcn5, at least when overexpressed. Note also that, in contrast to Ada3, overexpression of Ada2 or Gcn5 in the wild-type strain did not enhance ERα AF-2 activity (Table 1). Thus, Ada3, but not Ada2 and Gcn5, appears to be a limiting cofactor for the AF-2 of ERα in yeast.
Table 1.
Effect of deleting or overexpressing Ada2, Ada3, and Gcn5 on ERα and RXRα AF-2 activity
| Transcriptional activitya
|
||||||
|---|---|---|---|---|---|---|
| Strain
|
YCp90–ERα(CDEF)
|
YCp90–ERα (+ vector)
|
YEp90–RXRα (DE)–ER.CAS (+ vector)
|
|||
| +vector
|
+Ada2
|
+Ada3
|
+Gcn5
|
|||
| Wild type | 1.19 | 0.78 | 4.92 | 0.88 | 10.65 | 2.98 |
| Δada2 | 0.22 | 0.73 | 2.09 | 0.67 | 2.80 | 1.10 |
| Δada3 | 0.05 | 0.05 | 4.61 | 0.17 | 1.30 | 1.43 |
| Δgcn5 | 0.34 | 0.43 | 2.54 | 1.25 | 4.20 | 1.60 |
The indicated receptor constructs were expressed from a low (YCp90) or high (YEp90) copy number plasmid and tested for their ability to activate the ERE–URA3 reporter gene (see Fig. 4A) in the isogenic PL3 strains wild type, Δada2, Δada3, and Δgcn5 grown in the presence of ligand [500 nm E2 for ERα constructs and 500 nm 9C-RA for RXRα(DE)–ER.CAS]. Cells were also transformed with an episomal expression vector (YEp10) containing Ada2, Ada3, Gcn5 or no insert. OMPdecase assays were performed on each cell-free extract. Enzyme activity is expressed in nmoles of substrate/min per mg protein. The values (±20%) are the mean of at least three independent experiments.
Similar to the ERα(CDEF) derivative, full-length ERα expressed from YCp90 exhibited a reduced transcriptional activity in mutant strains when compared to wild-type yeast (Table 1). The transcriptional activity of the chimeric RXRα(DEF)–ER.CAS expressed from the high-copy-plasmid YEp90 was also dependent on Ada2 and Gcn5 (Table 1). Taken together, our results indicate that the transcriptional activities of the AF-2s of ERα and RXRα are maximal when the Ada2, Ada3, and Gcn5 subunits of the ADA complex are all present.
Yeast Ada3 enhances transactivation by RXRα in Cos-1 cells and interacts with human Ada3
To investigate whether Ada3 might affect RXRα-dependent transcription in mammalian cells, RXRα and Ada3 expression vectors were transiently cotransfected into Cos-1 cells, together with a DR1–tk/CAT reporter, which contains a RXR response element (Durand et al. 1994). Cotransfection of increasing amounts of ADA3 stimulated up to approximately fivefold the 9C-RA-dependent transcription of the reporter in a dose-dependent manner, without inducing any significant activation in the absence of ligand (Fig. 5A). Thus, Ada3 can potentiate RXRα activity in cultured mammalian cells. Under similar conditions, a modest (∼1.5- to 2-fold) increase in the E2-dependent transcription of a ERE–TATA–CAT reporter gene was observed when Ada3 was coexpressed with ERα (data not shown). Whether this could be related to differences in the reporter constructs is unknown. Note that similar observations have been made with putative mammalian coactivators, whose overexpression has been reported to enhance the AF-2 activity of only some of the NRs with which they interact (e.g., Voegel et al. 1996).
Figure 5.
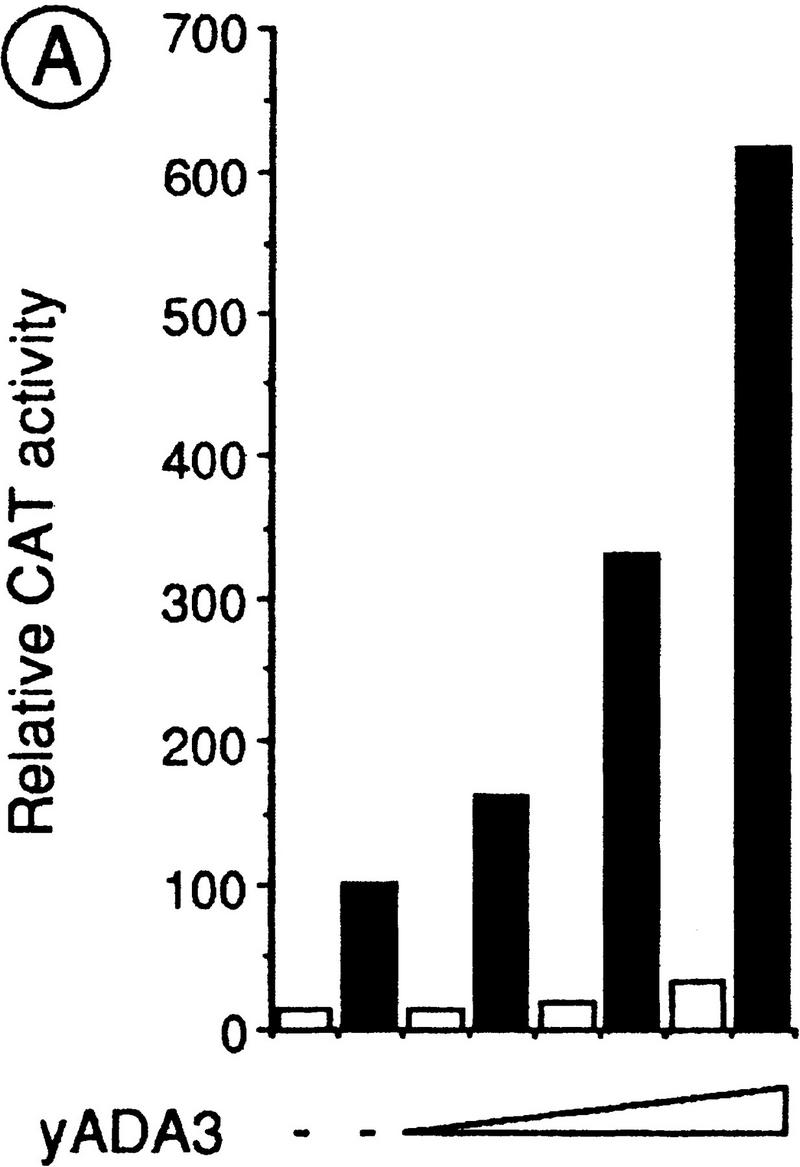
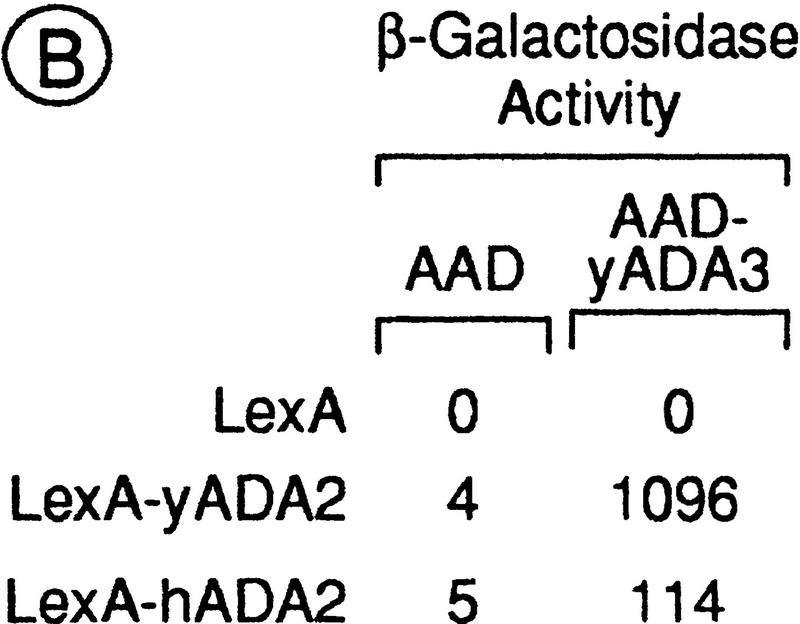
Yeast Ada3 (yAda3) stimulates the RXRα transcriptional activity in mammalian cells and interacts with hAda2 in a yeast two-hybrid assay. (A) Effect of yeast Ada3 on the RXRα transcriptional activity in transfected Cos-1 cells. DR1-tk/CAT reporter (1 μg), RXRα (100 ng), and pCH110 (expressing β-galactosidase; 1 μg) were transiently cotransfected into Cos-1 cells, together with increasing amounts of yeast Ada3 (1, 2, and 5 μg). Cells were treated with control vehicle (□) or 100 nm 9C-RA (▪). Values for CAT activities (+/− 20%) represent the averages of three independent duplicated transfections after normalization for the internal control β-galactosidase activity of pCH110. (B) Two-hybrid interaction between yeast Ada3 and hAda2. The indicated LexA and AAD fusions were introduced into the yeast reporter strain L40. β-Galactosidase activities are expressed as in Fig. 2A.
Interestingly, human homologs of the yeast Ada2 and yeast Gcn5 proteins have recently been identified (hAda2 and hGcn5, respectively; Candau et al. 1996), suggesting an evolutionary conservation of the Ada complex. Because Ada2 is known to interact with Ada3 in the yeast Ada complex (Horiuchi et al. 1995), we investigated whether hAda2 could also interact with yeast Ada3 in the two-hybrid system (Ada3 in Fig. 5B). An ∼20-fold increase in the reporter gene activity above the AAD control was obtained by coexpressing LexA–hAda2 and AAD-yeast Ada3. Although lower than that obtained by coexpression of LexA–Ada2 and AAD–yeast Ada3 (Fig. 5B), this stimulation suggests that yeast Ada3 and hAda2 may form a complex in mammalian cells, like Ada3 and Ada2 in yeast.
Discussion
Ada3, a coactivator/mediator for the ligand-dependent activation function AF-2 of certain nuclear receptors
The ligand-dependent activation function AF-2 of vertebrate nuclear receptors can stimulate transcription in yeast cells in the presence of their cognate ligand, thus suggesting that some of the basic features of the mechanism(s) that mediate these effects have been conserved across eukaryotes (see introductory section for references). Here, we show that, in yeast, the regulatory protein Ada3 (Piña et al. 1993) is a coactivator for the activation function AF-2 of RXRα and ERα. Ada3 interacts functionally in yeast with the AF-2-containing LBD of RXRα and ERα in a ligand-dependent manner. In vitro binding assays demonstrate a direct physical interaction between purified ERα and Ada3 proteins in the presence of E2, but not of the anti-estrogen OHT. Similarly, RXRα interacts in a ligand-dependent manner with Ada3 in vitro. Importantly, both in yeast and in vitro, mutations within the RXRα and ERα AF-2 AD core motifs (helix 12), which inactivate AF-2, also impair Ada3 interaction. Finally, RXRα and ERα AF-2 activity is severely reduced in the absence of Ada3 and enhanced when Ada3 is overexpressed. Thus, Ada3 fulfills the criteria anticipated for a yeast coactivator (mediator) of the AF-2 of RXRα and ERα.
Ada3 can also interact with TRα in a hormone-dependent manner, but not with the AF-2-containing LBD of RARα. This selectivity is consistent with previous studies on putative mammalian mediators, which revealed their differential interaction with the AF-2 ADs of various NRs in spite of the similarity of their respective AF-2 AD core motif (vom Baur et al. 1996; Thénot et al. 1997). Thus, the activity of the RARα AF-2 constructs that can activate transcription in a RA-dependent manner in yeast (Heery et al. 1993) is probably not mediated by Ada3.
Role of the Ada complex(es) in activation of transcription by AF-2 of RXRα and ERα
Ada3 is found in vivo within multisubunit protein complexes of different size (∼0.2, 0.8–0.9, and 1.8–2 MD) and complexity that contain at least three to four additional proteins: Ada1, Ada2, Gcn5 (Ada4), and Ada5 (Grant et al. 1997; Horiuchi et al. 1997; Saleh et al. 1997). Direct and independent interactions have been observed in vitro between Ada2 and both Ada3 and Gcn5 (Horiuchi et al. 1995). In addition, immunoprecipitation assays and biochemical purification of Ada complexes have revealed further association of these three proteins with Ada1 and Ada5 (Marcus et al. 1996; Grant et al. 1997; Horiuchi et al. 1997). Here, we demonstrate that Ada2 and Gcn5, in addition to Ada3, are required for efficient ligand-dependent activation by the AF-2 of RXRα and ERα, indicating that the activity of these AF-2s is most probably mediated by an Ada complex through direct interaction between the liganded LBD and the Ada3 subunit. However, deletion of either the ADA2 or GCN5 gene reduces the transcriptional activity of the AF-2 of ERα to a lesser extent than that of the ADA3 gene. Moreover, Ada3 overexpression results in an increase of AF-2 activity even in Δada2 and Δgcn5 deletion strains. Thus, Ada3 may mediate the ERα AF-2 activity through both Ada2/Gcn5-dependent and independent pathways.
Interestingly, ada5 and ada1 mutants exhibit more severe phenotypic defects and a broader spectrum of transcriptionally affected genes than the ada2 and gcn5 mutants (Marcus et al. 1996; Horiuchi et al. 1997), suggesting that like Ada3, Ada1 and Ada5 possess additional functions that are independent of Ada2 and Gcn5 (see below). Several subunits of the Ada complexes have been shown to interact directly in vitro with the ADs of several activator proteins: Interactions have been reported between Ada2 and either VP16 (Silverman et al. 1994), Gcn4 (Barlev et al. 1995), Adr1 (Chiang et al. 1996), or GR (τ1) (Henriksson et al. 1997), between Gcn5 and Adr1 (Chiang et al. 1996), and between Ada5 and VP16 (Marcus et al. 1996). We show here for the first time that Ada3 can also be the direct target for a transcriptional activator. Thus, it appears that, through interactions with activators, most if not all of the Ada subunits could be involved in the recruitment of Ada complexes to promoter regions. However, the various subunits of the Ada complex may have distinct coactivator functions, as they are not similarly involved in activation of transcription from different promoters. For example, activation through the ADH1 and CYC1 UAS1 yeast promoters is affected in Δada1 and Δada5, but not in Δada2, Δada3, or Δgcn5 strains (Marcus et al. 1996; Horiuchi et al. 1997), whereas our present study indicates that Ada2 and Gcn5 are partially dispensable in the Ada complex recruited to the URA3 promoter through interaction between the liganded LBD of ERα and Ada3.
How do Ada complexes mediate transcriptional activation by NRs?
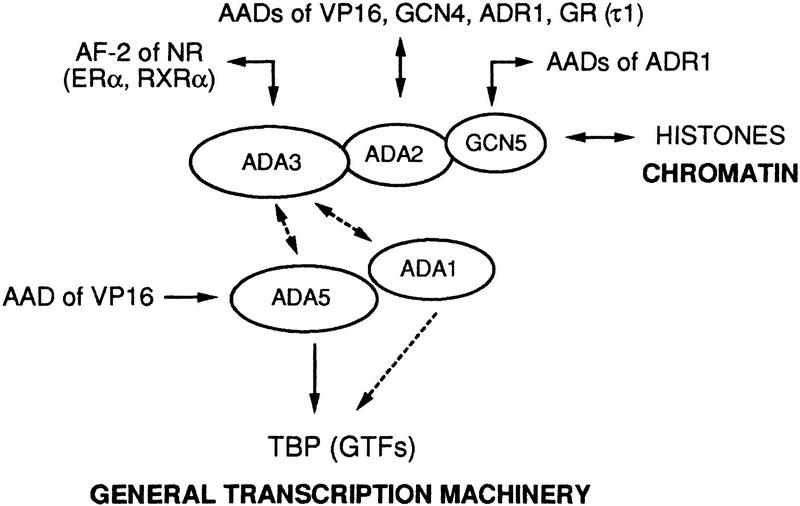
Gcn5 possesses a histone acetyltransferase (HAT) activity (Kuo et al. 1996). Thus, the recruitment of Gcn5-containing Ada complex(es) might result in acetylation of histones, relieving some of their repressive effects on transcription. However, Gcn5 is responsible only for part of the transcriptional activation mediated by the Ada complex recruited by the ER, as the activity of ERα AF-2 is reduced to a lesser extent in a Δgcn5 strain than in a Δada3 strain, while it is enhanced by overexpression of Ada3 in a Δgcn5 strain. Thus, Ada complexes recruited by liganded NRs might also act through additional targets, for instance through general transcription factors (GTFs). This possibility is supported by several recent findings. Interactions between the Ada complex and the GTF TATA-binding protein (TBP) have been reported (Barlev et al. 1995; Saleh et al. 1997), and both ADA5 (SPT20) and ADA1 have been shown to belong to the TBP class of SPT genes (Marcus et al. 1996; Roberts and Winston 1996; Horiuchi et al. 1997 and references therein). This class also includes TBP (SPT15), SPT3, SPT7, and SPT8 (Eisenmann et al. 1992, 1994; Gansheroff et al. 1995). SPT3 has been shown to interact genetically and functionally with TBP and TFIIA (Eisenmann et al. 1992; Madison and Winston 1997). Importantly, a native 1.8-MD Ada complex (named SAGA) has been recently isolated from yeast nuclei; it contains Spt7 and Spt3, and most probably Spt8, in addition to Gcn5, Ada2, Ada3, Ada1, and Ada5 (Grant et al. 1997; Horiuchi et al. 1997). Moreover, a physical interaction has been demonstrated between Ada5/Spt20 and the four other Spt proteins, Spt3, Spt7, Spt8, and TBP (Grant et al. 1997; Roberts and Winston 1997). Thus, at least two mechanisms appear to be involved in the coactivator function of the subunits of Ada multiprotein complexes: remodeling of the chromatin structure through histone acetylation by Gcn5, via Ada2 and recruitment of the trancription machinery to promoter regions through interactions with TBP, and via Ada5 and/or Ada1 (see Fig. 6).
Figure 6.
Schematic representation of the known interactions (double-headed arrows) between the ADs of transactivators and the subunits of the yeast Ada complex. Putative interactions between Ada3 and Ada5 and/or Ada1 are indicated by broken lines. Interactions between Ada5 (and possibly Ada1) and TBP, and between the HAT Gcn5 and the histone tails are also shown. It is postulated that transcriptional activation mediated by the Ada complex involves chromatin remodeling and/or recruitment of the general transcription machinery through interactions with GTFs.
Taken together, these above findings and our present observations indicate that Ada3 could mediate the effect of the AF-2 of NRs to both the HAT Gcn5 and to TBP (and possibly other GTFs; see Fig. 6). This would readily account for our observations that ERα AF-2 activity is not fully suppressed in Δada2 and Δgcn5 strains, and also that increased levels of Ada3 can enhance AF2 activity in the Δada2 and Δgcn5 strains. After completion of this study, Henriksson et al. (1997) reported that the AF-1 amino-terminal activation domain τ1 of GR interacts directly with Ada2 and requires Ada2, Ada3, and Gcn5 for full activity in yeast cells. Thus, at least two components of the Ada complex(es) can serve as coactivator/mediators for AF-1 and AF-2 of some NRs (see Fig. 6). The GR AF-1 (τ1) is an AAD that resembles the AAD of VP16 (Tasset et al. 1990; Henriksson et al. 1997 and references therein). Therefore, like VP16 (Marcus et al. 1996), it may also interact with the Ada5 subunit of the Ada complex (see above). Thus, our present scheme would account for the observation of Henriksson et al. (1997) that an ada3 deletion can cause a greater reduction in τ1 activity than ada2 and gcn5 deletions (Henriksson et al. 1997). Interestingly, ERα and Ada3 could be coimmunoprecipitated from extracts of Δada3 cells coexpressing an amino-terminally FLAG-tagged Ada3 and either full-length ERα or ERα(CDEF). However, this coimmunoprecipitation occurred irrespective of the presence of the ligand (E. vom Baur et al., unpubl.). These observations suggest that, in addition to the ligand-dependent interaction with Ada3, ligand-independent interactions may exist betwen ERα and other components of the Ada complex.
Is there a vertebrate Ada3 homolog?
Human homologs of the yeast Ada2 and Gcn5 proteins have been identified (Candau et al. 1996). Yet, our attempts to isolate a mammalian homolog of Ada3 either by complementing the activation deficiency of the AF-2 of ERα in a Δada3 reporter strain or by using a two-hybrid screening aimed at isolating hAda2 interacting proteins, have been unsuccessful so far (E. vom Baur et al., unpubl.). Nevertheless, several lines of evidence suggest that a homolog of Ada3 might exist in higher eukaryotes. First, we have shown here that overexpression of yeast Ada3 in Cos-1 cells can enhance ligand-activated transcription by RXRα and ERα, whereas overexpression of human Ada2 enhances the τ1 activation function of the GR (Henriksson et al. 1997). Second, we found that, similar to yeast Ada2, hAda2 can interact with yeast Ada3 in a two-hybrid assay, which suggests that Ada2–Ada3 interactions could be conserved in a putative vertebrate Ada complex. Third, a Gcn5-containing protein complex with an apparent size (molecular mass ∼220 kD) similar to that of the smallest yeast Ada complex (Saleh et al. 1997; see above) has been purified from the protozoan Tetrahymena, which might contain the Tetrahymena homologs of the other yeast Ada proteins (Brownell et al. 1996). Interestingly, as in yeast cells (see above), coexpressed ERα or RXRα and FLAG-tagged yeast Ada3 could be coimunoprecipitated from transfected Cos-1 cells, irrespective of the presence of the ligand (E. vom Baur et al., unpubl.), which suggests the possible existence of ligand-independent interactions between these nuclear receptors and other components of a putative vertebrate Ada complex.
Thus, Ada complexes might also exist in higher eukaryotes and, as in yeast, interact with transactivators to mediate their stimulatory effect on initiation of transcription through remodeling of the chromatin structure and/or recruitment of the general transcription machinery. In this respect, the Ada complex exhibits an interesting similarity with the mammalian and Drosophila CBP/p300 coactivator proteins, which do not exist in yeast cells. Indeed, like the Ada complex, CBP/p300 can interact directly with a number of transcriptional activators. CBP/p300 also interacts with the HAT P/CAF, which is related to Gcn5. Moreover, CBP/p300 possesses HAT activity on its own and, as the Ada complex, can interact with components of the general transcription machinery (for review, see Shikama et al. 1997 and references therein). The presence of Ada-like complexes in metazoan organisms, would obviously increase the number of combinatorial possibilities for controlling the multiple gene networks that are required for the realization of complex developmental and homeostatic programs in these organisms.
Materials and methods
Yeast strains, transformations, and media
S. cerevisiae L40 (a) strain [MATa, his3Δ200, trp1-901, leu2-3,112, ade2, LYS::(lexAbs)4–HIS3, URA3::(lexAbs)8–lacZ] was a gift from S.M. Hollenberg (Vojtek et al. 1993). The reporter strain PL3 (α) [MATα, leu2-Δ1, ura3-Δ1, his3-Δ200, trp1::(ERE)3–URA3] was described elsewhere (Pierrat et al. 1992). Δada2, Δada3, and Δgcn5 deletion strains were generated in the PL3 (α) background according to the PCR-based disruption method described by Wach et al. (1994). Deletions were verified by PCR on genomic DNA and functional complementation. For all deletions, the full-length coding sequence of the deleted gene was replaced by the kanamycin-resistance gene. Yeast transformation was carried out by the lithium-acetate procedure and standard media were used for growth (Rose et al. 1990).
Plasmids
Details on individual plasmid constructs, which were all verified by sequencing, are available on request. Receptor cDNAs used in this study correspond to human RARα1 and ERα, mouse RXRα and chicken TRα. Yeast expression plasmids YEp10 (TRP1), YEp90 (His3) and YCp90 (His3), have been described elsewhere (Pierrat et al. 1992; Heery et al. 1994). AAD (VP16) fusion proteins were expressed from the multicopy plasmid pASV3 (Leu2) (Le Douarin et al. 1995b). All these plasmids express inserts under the control of the PGK promoter. LexA-fusion proteins were expressed from a derivative of the episomal plasmid pBTM116 (Trp1) under the control of the alcohol dehydrogenase 1 (ADH1) promoter (Vojtek et al. 1993). Full-length ADA2, GCN5, and ADA3 genes were isolated on the basis of published sequences by PCR on genomic DNA prepared from PL3. For transfection studies in mammalian cells, cDNAs were expressed from pSG5. The reporter genes DR1–tk/CAT and ERE–TATA–CAT have been described previously (Berry et al. 1990; Durand et al. 1994). For in vitro-binding assays, the indicated cDNAs were fused to GST in the pGEX2T plasmid (Pharmacia; vom Baur et al. 1996). Full-length Ada3 and Ada3(347–702) were cloned into the pET15bEpB10 plasmid, which directs the synthesis of 6× His-epitope B10 (region B of human ERα)–tagged fusion proteins in E. coli.
Two-hybrid screening
A yeast genomic VP16 fusion library was constructed by limited Sau3AI digestion of genomic DNA and subsequent insertion of fragments into a modified Leu2 pASV3 vector. The library was introduced by lithium-acetate transformation into the L40 reporter strain expressing LexA–RXRα(DE) from the Trp1 pBTM116m vector. Cells were spread directly on His− Leu− Trp− plates containing 500 nm 9C-RA and 5 mm 3-AT. Two hundred thirty-two clones were isolated and then retested for β-galactosidase activity on permeabilized cells. Library plasmids from the positive isolates were recovered into E. coli HB101 (leu2−). Plasmids were subjected to restriction analysis, and unique inserts were sequenced.
Transactivation assays
Yeast transformants were grown for ∼16 hr to a cell titer of 2–5 × 107 (exponential phase) in minimal medium supplemented with uracil and the required amino acids. When necessary, medium was supplemented with 500 nm of the appropriate ligands. PL3 cell-free extracts were prepared and assayed for OMPdecase activity as described (Pierrat et al. 1992). L40 extracts were prepared and analyzed for β-galactosidase activity according to Rose et al. (1990). Transient transfections of Cos-1 cells, and CAT assays were performed as described (Durand et al. 1994).
Antibodies
Monoclonal antibodies (mAbs) B10 and F3 are directed against the B and F regions of human ERα, respectively, mAb 4RX is directed against the DE region of RXRα, and 2GV4 is directed against VP16 (see Le Douarin et al. 1995a and references therein). A polyclonal rabbit antiserum was raised against LexA expressed in E. coli.
In vitro binding assays
The assay was done as described previously (vom Baur et al. 1996). Briefly, GST or GST-fusion proteins were expressed in E. coli and purified on glutathione–Sepharose (Pharmacia). 6× His-epitope B10-tagged Ada3 fusions were expressed in E. coli and purified on Ni2+-chelating columns (Pharmacia). Purified proteins were quantified by Coomassie staining after SDS-PAGE separation and by Bradford protein assay. Glutathione–Sepharose beads were equilibrated with binding buffer (50 mm Tris-HCl at pH 7.5, 100 mm NaCl, 0.3 mm DTT, 10 mm MgCl2, 8.7% glycerol, 0.1% NP-40), loaded with equimolar amounts of GST or GST fusions and washed. Appropriate ligands and purified His fusions were added to each reaction in the presence of BSA (fraction V, Sigma) to reduce nonspecific binding. Incubation was carried out at room temperature for 30 min. After three washes with binding buffer containing the appropriate ligands and BSA, the beads were dried, resuspended in SDS-loading buffer, and boiled for 10 min, and the proteins were analyzed by SDS-PAGE. Bound Ada3 was detected by immunoblotting with the mAb B10 (see above).
Acknowledgments
We are grateful to S.M. Hollenberg for the generous gifts of yeast strain L40 and plasmid pBTM116. We thank A. Wach and P. Philippsen for KanMX gene-disruption plasmids. We also give special thanks to S. Vicaire and D. Stephan for sequencing all isolated library clones and constructs, to J.-M. Garnier for construction of the yeast genomic VP16 fusion library, to T. Lerouge for construction of many yeast and E. coli expression vectors, to M. Cerviño for technical assistance, and to our colleagues of the yeast and retinoid groups for helpful discussions. This work was supported by the Groupement de Recherches et d’Etudes sur les Génomes (GREG47/95), the CNRS, INSERM, the Centre Hospitalier Universitaire Régional, the Association pour la Recherche sur le Cancer, the Collège de France, the Fondation pour la Recherche Médicale (FRM), the Human Frontier Science Program and Bristol-Myers-Squibb. E.v.B was supported by funds from the Ministère de la Recherche Scientifique and the ULP; M.H. was recipient of a fellowship from the European Economic Community (Human Capital and Mobility Program) and FRM; S.-J.U. was a postdoctoral fellow from INSERM and A.B. is the recipient of a Marie-Curie long-term fellowship from the European Commission (ERBFMBICT961269).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL igbmc@igbmc.u-strasbg.fr; FAX (33) 3 88 65 32 03.
References
- Ayer DE, Lawrence QA, Eisenman RN. Mad–Max transcriptional repression is mediated by ternary complex formation with mammalian homologs of yeast repressor Sin3. Cell. 1995;80:767–776. doi: 10.1016/0092-8674(95)90355-0. [DOI] [PubMed] [Google Scholar]
- Baniahmad C, Nawaz Z, Baniahmad A, Gleeson MA, Tsai MJ, O’Malley BW. Enhancement of human estrogen receptor activity by SPT6: A potential coactivator. Mol Endocrinol. 1995;9:34–43. doi: 10.1210/mend.9.1.7760849. [DOI] [PubMed] [Google Scholar]
- Barlev NA, Candau R, Wang L, Darpino P, Silverman N, Berger SL. Characterization of physical interactions of the putative transcriptional adaptor, ADA2, with acidic activation domains and TATA-binding protein. J Biol Chem. 1995;270:19337–19344. doi: 10.1074/jbc.270.33.19337. [DOI] [PubMed] [Google Scholar]
- Berry M, Metzger D, Chambon P. Role of the two activating domains of the oestrogen receptor in the cell-type and promoter-context dependent agonistic activity of the anti-oestrogen 4-hydroxytamoxifen. EMBO J. 1990;9:2811–2818. doi: 10.1002/j.1460-2075.1990.tb07469.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownell JE, Zhou J, Ranalli T, Kobayashi R, Edmondson DG, Roth SY, Allis CD. Tetrahymena histone acetyltransferase A: A homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell. 1996;84:843–851. doi: 10.1016/s0092-8674(00)81063-6. [DOI] [PubMed] [Google Scholar]
- Candau R, Moore PA, Wang L, Barlev N, Ying CY, Rosen CA, Berger SL. Identification of human proteins functionally conserved with the yeast putative adaptors ADA2 and GCN5. Mol Cell Biol. 1996;16:593–602. doi: 10.1128/mcb.16.2.593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chambon P. A decade of molecular biology of retinoic acid receptors. FASEB J. 1996;10:940–954. [PubMed] [Google Scholar]
- Chiang Y-C, Romarnitsky P, Chase D, Denis CL. ADR1 activation domains contact the histone acetytransferase GCN5 and the core transcriptional factor TFIIB. J Biol Chem. 1996;271:32359–32365. doi: 10.1074/jbc.271.50.32359. [DOI] [PubMed] [Google Scholar]
- Chiba H, Muratmatsu M, Nomoto A, Kato H. Two human homologues of Saccharomyces cerevisiae SWI2/SNF2 and Drosophila brahma are transcriptional coactivators cooperating with the oestrogen receptor and the retinoic acid receptor. Nucleic Acids Res. 1994;22:1815–1820. doi: 10.1093/nar/22.10.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danielian PS, White R, Lees LA, Parker MG. Identification of a conserved region required for hormone dependent transcriptional activation by steroid hormone receptors. EMBO J. 1992;11:1025–1033. doi: 10.1002/j.1460-2075.1992.tb05141.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Durand B, Saunders M, Gaudon C, Roy B, Losson R, Chambon P. Activation function 2 (AF-2) of retinoic acid receptor and 9-cis retinoic acid receptor: Presence of a conserved autonomous constitutive activating domain and influence of the nature of the response element on AF-2 activity. EMBO J. 1994;22:5370–5382. doi: 10.1002/j.1460-2075.1994.tb06872.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eisenmann DM, Arndt KM, Ricupero SL, Rooney JW, Winston F. SPT3 interacts with TFIID to allow normal transcription in Saccharomyces cerevisiae. Genes & Dev. 1992;6:1319–1331. doi: 10.1101/gad.6.7.1319. [DOI] [PubMed] [Google Scholar]
- Eisenmann DM, Chapon C, Roberts SM, Dollard C, Winston F. The Saccharomyces cerevisiae SPT8 gene encodes a very acidic protein that is functionally related to SPT3 and TATA-binding protein. Genetics. 1994;137:647–657. doi: 10.1093/genetics/137.3.647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fields S, Sternglanz R. The two-hybrid system: An assay for protein-protein interactions. Trends Genet. 1994;10:286–292. doi: 10.1016/0168-9525(90)90012-u. [DOI] [PubMed] [Google Scholar]
- Gansheroff LJ, Dollard C, Tan P, Winston F. The Saccharomyces cerevisiae SPT7 gene encodes a very acidic protein important for transcription in vivo. Genetics. 1995;139:523–536. doi: 10.1093/genetics/139.2.523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass CK, Rose DW, Rosenfeld MG. Nuclear receptor coactivators. Curr Opin Cell Biol. 1997;9:222–232. doi: 10.1016/s0955-0674(97)80066-x. [DOI] [PubMed] [Google Scholar]
- Grant PA, Duggan L, Côté J, Roberts SM, Brownell JE, Candau R, Ohba R, Owen-Hughes T, Allis CD, Winston F, Berger SL, Workman JL. Yeast Gcn5 functions in two multisubunit complexes to acetylate nucleosomal histones: Characterization of an Ada complex and the SAGA (Spt/Ada) complex. Genes & Dev. 1997;11:1640–1650. doi: 10.1101/gad.11.13.1640. [DOI] [PubMed] [Google Scholar]
- Hall BL, Smit-McBride Z, Privalsky ML. Reconstitution of retinoid X receptor function and combinatorial regulation of other nuclear hormone receptors in the yeast Saccharomyces cerevisiae. Proc Natl Acad Sci. 1993;90:6929–6933. doi: 10.1073/pnas.90.15.6929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heery DM, Zacharewski T, Pierrat B, Gronemeyer H, Chambon P, Losson R. Efficient transactivation by retinoic acid receptors in yeast requires retinoid X receptors. Proc Natl Acad Sci. 1993;90:4281–4285. doi: 10.1073/pnas.90.9.4281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heery DM, Pierrat B, Gronemeyer H, Chambon P, Losson R. Homo- and heterodimers of the retinoid X receptor (RXR) activated transcription in yeast. Nucleic Acids Res. 1994;22:726–731. doi: 10.1093/nar/22.5.726. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henriksson A, Almhöf T, Ford J, McEwan IJ, Gustafsson J-A, Wright APH. Role of the ADA adaptor complex in gene activation by the glucocorticoid receptor. Mol Cell Biol. 1997;17:3065–3073. doi: 10.1128/mcb.17.6.3065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiuchi J, Silverman N, Marcus GA, Guarente L. ADA3, a putative transcriptional adaptor, consists of two separable domains and interacts with ADA2 and GCN5 in a trimeric complex. Mol Cell Biol. 1995;15:1203–1209. doi: 10.1128/mcb.15.3.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horiuchi J, Sylverman N, Piña B, Marcus GA, Guarente L. ADA1, a novel component of the ADA/GCN5 complex, has broader effects than GCN5, ADA2, or ADA3. Mol Cell Biol. 1997;17:3220–3228. doi: 10.1128/mcb.17.6.3220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imhof MO, McDonnell DP. Yeast RSP5 and its human homolog hRPF1 potentiate hormone-dependent activation of transcription by human progesterone and glucocorticoid receptors. Mol Cell Biol. 1996;16:2594–2605. doi: 10.1128/mcb.16.6.2594. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ince BA, Zhuang Y, Wrenn CK, Shapiro DJ, Katzenellenbogen BS. Powerful dominant negative mutants of the human estrogen receptor. J Biol Chem. 1993;268:14026–14032. [PubMed] [Google Scholar]
- Kuo MH, Brownell JE, Sobel RE, Ranalli TA, Cook RG, Edmondson DG, Roth SY, Allis CD. Transcription-linked acetylation by Gcn5p of histones H3 and H4 at specific lysines. Nature. 1996;383:269–272. doi: 10.1038/383269a0. [DOI] [PubMed] [Google Scholar]
- Le Douarin B, Zechel C, Garnier J-M, Lutz Y, Tora L, Pierrat B, Heery D, Gronemeyer H, Chambon P, Losson R. The amino-terminal part of TIF1, a putative mediator of the ligand-dependent activation function (AF-2) of nuclear receptors, is fused to B-raf in the oncogenic protein T18. EMBO J. 1995a;14:2020–2033. doi: 10.1002/j.1460-2075.1995.tb07194.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Douarin B, Pierrat B, vom Baur E, Chambon P, Losson R. A new version of the two-hybrid assay for detection of protein–protein interactions. Nucleic Acids Res. 1995b;23:876–878. doi: 10.1093/nar/23.5.876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Madison JM, Winston F. Evidence that Spt3 functionally interacts with Mot1, TFIIA, and TATA-binding protein to confer promoter-specific transcriptional control in Saccharomyces cerevisiae. Mol Cell Biol. 1997;17:287–295. doi: 10.1128/mcb.17.1.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mangelsdorf DJ, Thummel C, Beato M, Herrlich P, Schütz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, Evans RM. The nuclear receptor superfamily: The second decade. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus GA, Horiuchi J, Silverman N, Guarente L. ADA5/SPT20 links the ADA and SPT genes, which are involved in yeast transcription. Mol Cell Biol. 1996;16:3197–3205. doi: 10.1128/mcb.16.6.3197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonnell DP, Vegeto E, O’Malley BW. Identification of a negative regulatory function for steroid receptors. Proc Natl Acad Sci. 1992;89:10563–10567. doi: 10.1073/pnas.89.22.10563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Metzger D, White JH, Chambon P. The human oestrogen receptor functions in yeast. Nature. 1988;334:31–36. doi: 10.1038/334031a0. [DOI] [PubMed] [Google Scholar]
- Metzger D, Losson R, Bornert J-M, Lemoine Y, Chambon P. Promoter specificity of the two transcriptional activation functions of the human oestrogen receptor in yeast. Nucleic Acids Res. 1992;20:2813–2817. doi: 10.1093/nar/20.11.2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muchardt C, Yaniv M. A human homologue of Saccharomyces cerevisiae SNF2/SWI2 and Drosophila brm genes potentiates transcriptional activation by the glucocorticoid receptor. EMBO J. 1993;12:4279–4290. doi: 10.1002/j.1460-2075.1993.tb06112.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muchardt C, Sardet C, Bourachot B, Onufryk C, Yaniv M. A human protein with homology to Saccharomyces cerevisae SNF5 interacts with the potential helicase hbrm. Nucleic Acids Res. 1995;23:1127–1132. doi: 10.1093/nar/23.7.1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nawaz Z, Baniahmad C, Burris TP, Stillman DJ, O’Malley BW, Tsai MJ. The yeast SIN3 gene product negatively regulates the activity of the human progesterone receptor and positively regulates the activities of GAL4 and the HAP1 activator. Mol & Gen Genet. 1994;245:724–733. doi: 10.1007/BF00297279. [DOI] [PubMed] [Google Scholar]
- Perlman T, Evans RM. Nuclear receptors in Sicily: All in the famiglia. Cell. 1997;90:391–397. doi: 10.1016/s0092-8674(00)80498-5. [DOI] [PubMed] [Google Scholar]
- Pierrat B, Heery DM, Lemoine Y, Losson R. Functional analysis of the human estrogen receptor using a phenotypic transactivation assay in yeast. Gene. 1992;119:237–245. doi: 10.1016/0378-1119(92)90277-v. [DOI] [PubMed] [Google Scholar]
- Piña B, Berger SL, Marcus G, Silverman N, Agapite J, Guarente L. ADA3: A gene, identified by resistance to GAL4-VP16, with properties similar to and different from those of ADA2. Mol Cell Biol. 1993;13:5981–5989. doi: 10.1128/mcb.13.10.5981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts SM, Winston F. SPT20/ADA5 encodes a novel protein functionally related to the TATA-binding protein and important for transcription in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:3206–3213. doi: 10.1128/mcb.16.6.3206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Essential functional interactions of SAGA, a Saccharomyces cerevisiae complex of Spt, Ada and GCN5 proteins with the Swi/Snf and Srt/mediator complexes. Genetics. 1997;147:451–465. doi: 10.1093/genetics/147.2.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rose MD, Winston F, Hieter P. Methods in yeast genetics: A laboratory course manual. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1990. [Google Scholar]
- Saleh A, Lang V, Cook R, Brandl CJ. Identification of native complexes containing the yeast coactivator/repressor proteins NGG1/ADA3 and ADA2. J Biol Chem. 1997;272:5571–5578. doi: 10.1074/jbc.272.9.5571. [DOI] [PubMed] [Google Scholar]
- Schena M, Yamamoto KR. Mammalian glucocorticoid receptor derivatives enhance transcription in yeast. Science. 1988;241:965–967. doi: 10.1126/science.3043665. [DOI] [PubMed] [Google Scholar]
- Segre JA, Nemhauser JL, Taylor BA, Nadeau JH, Lander ES. Positional cloning of the nude locus: Genetic, physical, and transcription maps of the region and mutations in the mouse and rat. Genomics. 1995;28:549–559. doi: 10.1006/geno.1995.1187. [DOI] [PubMed] [Google Scholar]
- Shikama N, Lyon J, La Thangue NB. The p300/CBP family: Integrating signals with transcription factors and chromatin. Trends Cell Biol. 1997;7:230–236. doi: 10.1016/S0962-8924(97)01048-9. [DOI] [PubMed] [Google Scholar]
- Silverman N, Agapite J, Guarente L. Yeast ADA2 protein binds to the VP16 protein activation domain and activates transcription. Proc Natl Acad Sci. 1994;91:11665–11668. doi: 10.1073/pnas.91.24.11665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tasset D, Tora L, Fromental C, Scheer E, Chambon P. Distinct classes of transcriptional activating domains function by different mechanisms. Cell. 1990;62:1177–1187. doi: 10.1016/0092-8674(90)90394-t. [DOI] [PubMed] [Google Scholar]
- Thénot S, Henriquet C, Rochefort H, Cavailles V. Differential interaction of nuclear receptors with the putative human transcriptional coactivator hTIF1. J Biol Chem. 1997;272:12062–12068. doi: 10.1074/jbc.272.18.12062. [DOI] [PubMed] [Google Scholar]
- Tora L, White J, Brou C, Tasset D, Webster N, Scheer E, Chambon P. The human oestrogen receptor has two independent nonacidic transcriptional activation functions. Cell. 1989;59:447–487. doi: 10.1016/0092-8674(89)90031-7. [DOI] [PubMed] [Google Scholar]
- Voegel JJ, Heine MJS, Zechel C, Chambon P, Gronemeyer H. TIF2, a 160 kDa transcriptional mediator for the ligand-dependent activation function AF-2 of nuclear receptors. EMBO J. 1996;15:3667–3675. [PMC free article] [PubMed] [Google Scholar]
- Vojtek BA, Hollenberg SM, Cooper JA. Mammalian Ras interacts directly with the serine/threonine kinase Raf. Cell. 1993;74:205–214. doi: 10.1016/0092-8674(93)90307-c. [DOI] [PubMed] [Google Scholar]
- vom Baur E, Zechel C, Heery D, Heine MJS, Garnier J-M, Vivat V, Le Douarin B, Gronemeyer H, Chambon P, Losson R. Differential ligand-dependent interactions between the AF-2 activating domain of nuclear receptors and putative transcriptional intermediary factors mSUG1 and TIF1. EMBO J. 1996;15:119–124. [PMC free article] [PubMed] [Google Scholar]
- Wach A, Brachat A, Pohlmann R, Philippsen P. New heterologous modules for classical or PCR-based gene disruptions in Saccharomyces cerevisiae. Yeast. 1994;10:1793–1808. doi: 10.1002/yea.320101310. [DOI] [PubMed] [Google Scholar]
- White JH, Metzger D, Chambon P. Expression and function of the human estrogen receptor in yeast. Cold Spring Harbor Symp Quant Biol. 1988;53:819–828. doi: 10.1101/sqb.1988.053.01.093. [DOI] [PubMed] [Google Scholar]
- Wurtz JM, Bourguet W, Renaud JP, Vivat V, Chambon P, Moras D, Gronemeyer H. A canonical structure for the ligand binding domain of nuclear receptors. Nature Struct Biol. 1996;3:87–94. doi: 10.1038/nsb0196-87. [DOI] [PubMed] [Google Scholar]
- Yoshinaga S, Peterson C, Herskowitz I, Yamamoto K. Roles of SWI1, SWI2, and SWI3 proteins for transcriptional enhancement by steroid receptors. Science. 1992;258:1598–1604. doi: 10.1126/science.1360703. [DOI] [PubMed] [Google Scholar]