Abstract
BCL-6 encodes a POZ/zinc finger transcriptional repressor that is required for germinal center formation and may influence apoptosis. Aberrant expression of BCL-6 due to chromosomal translocations is implicated in certain subtypes of non-Hodgkin's lymphoma. The POZ domains of BCL-6 and several other POZ proteins interact with corepressors N-CoR and SMRT. Here we identify and characterize a novel corepressor BCoR (BCL-6 interacting corepressor), which is expressed ubiquitously in human tissues. BCoR can function as a corepressor when tethered to DNA and, when overexpressed, can potentiate BCL-6 repression. Specific class I and II histone deacetylases (HDACs) interact in vivo with BCoR, suggesting that BCoR may functionally link these two classes of HDACs. Strikingly, BCoR interacts selectively with the POZ domain of BCL-6 but not with eight other POZ proteins tested, including PLZF. Additionally, interactions between the BCL-6 POZ domain and SMRT, N-CoR, and BCoR are mutually exclusive. The specificity of the BCL-6/BCoR interaction suggests that BCoR may have a role in BCL-6-associated lymphomas.
Keywords: BCL-6, POZ, BTB, HDAC, transcriptional repression
The BCL-6 gene was originally identified because of its involvement in chromosomal translocations associated with non-Hodgkin's lymphoma (NHL) (Baron et al. 1993; Kerckaert et al. 1993; Ye et al. 1993; Miki et al. 1994). These translocations, which juxtapose the intact coding sequences of BCL-6 to one of a number of different loci, are believed to result in deregulated BCL-6 expression (Ye et al. 1995; Chen et al. 1998). This, in turn, is thought to have a role in lymphomagenesis. BCL-6 rearrangements are found primarily in two common subtypes of NHL, diffuse large cell and follicular lymphomas, where rearrangements occur at a frequency of ∼40% and 10%, respectively (Bastard et al. 1994; Lo Coco et al. 1994; Otsuki et al. 1995).
BCL-6 has an important role in normal immune responses and can either promote or inhibit apoptosis, depending on the situation and cell type (Albagli et al. 1999; Kumagai et al. 1999; Yamochi et al. 1999). BCL-6 is normally expressed in mature B cells and some CD4+ T cells of the germinal centers, as well as at lower levels in several other tissues (Cattoretti et al. 1995; Onizuka et al. 1995; Allman et al. 1996; Bajalica-Lagercrantz et al. 1998). BCL-6-associated lymphomas are thought to have arisen from these germinal center B cells (Stein and Dallenbach 1992; Alizadeh et al. 2000). Targeted gene disruption in mice revealed that BCL-6 is required for germinal center formation and T-cell-dependent antibody responses (Dent et al. 1997; Fukuda et al. 1997; Ye et al. 1997). In addition, BCL-6-deficient mice are prone to inflammatory diseases that are thought to result from Th2-mediated hyperimmune responses (Dent et al. 1997; Ye et al. 1997).
The BCL-6 gene encodes a 706-amino-acid protein that functions as a sequence-specific transcriptional repressor (Deweindt et al. 1995; Chang et al. 1996; Seyfert et al. 1996). It belongs to a subclass of zinc finger proteins that contain a POZ domain (also called BTB or ZIN domain) at the amino terminus and Cys2–His2 zinc fingers at the carboxyl terminus (Numoto et al. 1993; Bardwell and Treisman 1994; Zollman et al. 1994; Albagli et al. 1995). The 120-amino-acid POZ domain is an evolutionarily conserved protein–protein interaction domain that can mediate both homomeric as well as heteromeric POZ–POZ interactions (Bardwell and Treisman 1994; Okabe et al. 1998). The consensus recognition site for BCL-6 has been determined and resembles that of STAT proteins (Dent et al. 1997; Ye et al. 1997; Huynh and Bardwell 1998). Consistent with this, the STAT6-responsive immunoglobulin germ line ε promoter recently was reported to be down-regulated by BCL-6 (Harris et al. 1999).
The POZ domain of BCL-6 is responsible for a large portion of the repressive function of the protein, although an intervening region between the POZ domain and the zinc finger domain can also mediate repression independently (Albagli et al. 1996; Chang et al. 1996; Seyfert et al. 1996). We and others have reported that the BCL-6 POZ domain interacts with corepressors N-CoR and SMRT, which are associated with a large transcriptional regulatory complex that includes mSIN3 and the histone deacetylase HDAC1 (Alland et al. 1997; Dhordain et al. 1997, 1998; Heinzel et al. 1997; Nagy et al. 1997; Huynh and Bardwell 1998). This HDAC1 complex is recruited by several different classes of transcriptional repressors (for review, see Knoepfler and Eisenman 1999). In the case of BCL-6, self-association of the POZ domain is required for interactions with N-CoR and SMRT (Huynh and Bardwell 1998). These corepressors are capable of interacting with several other POZ domain-containing proteins, including PLZF, a transcriptional repressor that is linked to promyelocytic leukemia (Hong et al. 1997; Huynh and Bardwell 1998; Wong and Privalsky 1998). Taken together, these results support a model in which the POZ domain of BCL-6, PLZF, and some but not all (Deltour et al. 1999) other POZ-containing repressors recruit N-CoR/SMRT corepressors and the HDAC1 complex to promoters of their respective target genes, thereby inducing a repressive chromatin state through the functions of HDAC1 (David et al. 1998; Dhordain et al. 1998; Grignani et al. 1998; Lin et al. 1998; Wong and Privalsky 1998).
To date, multiple mammalian HDACs have been identified and characterized, and are grouped into two classes on the basis of sequence homology with yeast HDACs. The class I HDACs (HDAC1–HDAC3) are related to yeast RPD3 (Taunton et al. 1996; Yang et al. 1996, 1997), and the class II HDACs (HDAC4–HDAC7) share homology with yeast HDA1 (Fischle et al. 1999; Grozinger et al. 1999; Miska et al. 1999; Verdel and Khochbin 1999; Kao et al. 2000). In yeast, RPD3 and HDA1 are functionally distinct; they are found in different complexes and regulate different sets of genes (Rundlett et al. 1996, 1998). Two recent reports revealed that the corepressors N-CoR and SMRT, which have hitherto been characterized as functioning with the class I HDAC1/HDAC2 complex, can also associate with class II HDAC4, HDAC5, and HDAC7 (Huang et al. 2000; Kao et al. 2000). These results suggest that the two mammalian HDAC classes are functionally linked, unlike the yeast HDACs.
We report here the identification of a novel corepressor, BCoR (BCL-6 corepressor), which interacts specifically with the POZ domain of BCL-6. The BCoR gene encodes at least two proteins, a long form containing 1721 amino acids (BCoR) and a short form containing 1004 amino acids BCoR-short (BCoR-S). We find that BCoR, but not BCoR-S, can repress transcription when tethered to a promoter and can potentiate transcriptional repression by BCL-6 in a specific manner, which suggests that BCoR is involved in BCL-6 repression. Additionally, both forms of BCoR can associate with specific HDACs of both class I and class II HDACs, which suggests that histone/protein deacetylation is a mechanism for BCoR-mediated repression. Finally, we show that the interactions between the BCL-6 POZ domain and SMRT, N-CoR, and BCoR are mutually exclusive.
Results
Identification of the novel BCL-6-interacting protein BCoR in a yeast two-hybrid screen
The yeast two-hybrid approach was used to identify proteins that can interact with the amino-terminal two-thirds of BCL-6, codons 1–418 [POZ long (POZ-L)]. In addition to corepressors N-CoR and SMRT, described previously, we also identified several other candidate BCL-6-interacting proteins that retested positive for interaction with the original bait and not with irrelevant test baits in yeast (Huynh and Bardwell 1998). In this study we focused on one of these, A3, as its interaction with POZ-L was confirmed in multiple protein–protein interaction assays (see below).
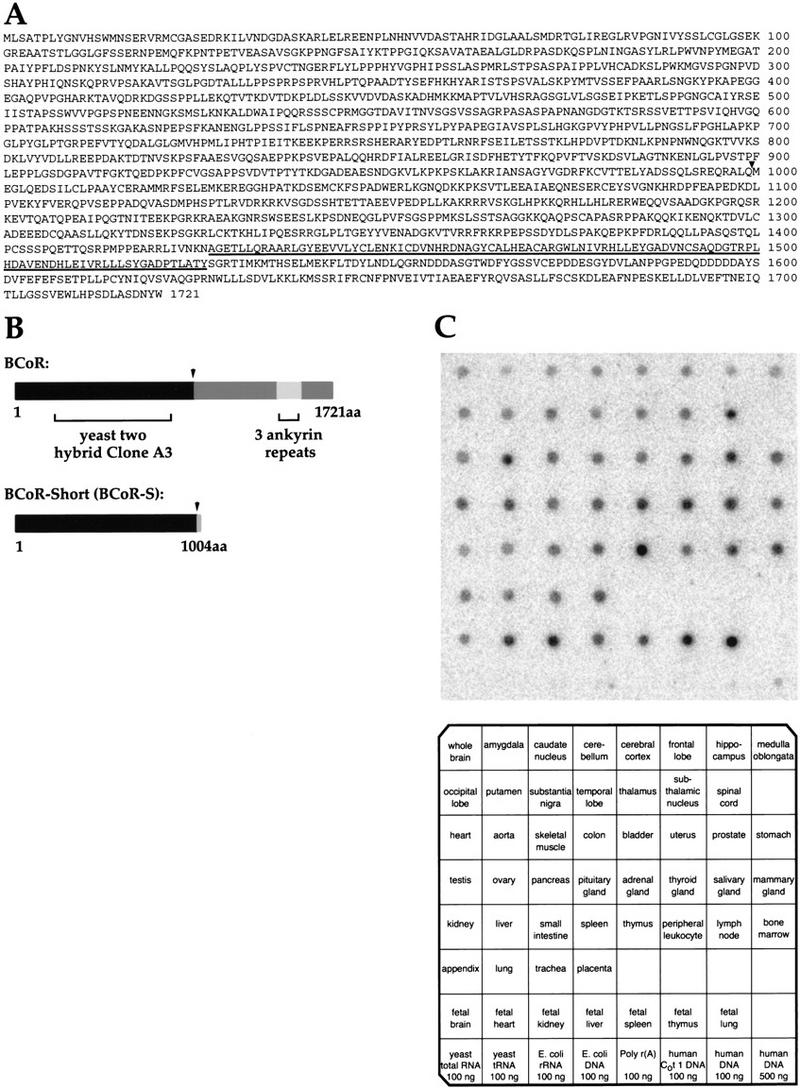
The 1928-bp sequence of the A3 clone contains a single continuous ORF, indicating that it is a partial cDNA. Using conventional cloning methodologies (see Materials and Methods) we obtained two overlapping cDNA clones that extended A3. These two cDNA clones generate a 5708 bp sequence encoding a complete 1721-amino-acid ORF, which we call BCoR. We also isolated cDNAs representing an alternatively processed variant of BCoR; the predicted ORF encodes a shorter protein of 1004 amino acids, designated BCoR-S. BCoR-S shares an amino-terminal sequence with BCoR that ends at amino acid 999 (Fig. 1A,B). The sequence of BCoR-S continues beyond this junction with five more amino acids: VSPPT. Searches of the available databases reveal that BCoR is a novel protein, sharing no significant homology with any known proteins, aside from three tandem ankyrin repeats (residues 1428–1527), that are not contained in BCoR-S (Fig. 1A,B). A related pseudogene is present in the database, AC012062 (A. Keating, K.D. Huynh, and V.J. Bardwell, unpubl.).
Figure 1.
Amino acid sequence and tissue distribution of BCoR. (A) Deduced amino acid sequence of BCoR. The point of divergence between BCoR and BCoR-S is indicated (▾); the three tandem ankyrin repeats are underlined. (B) Schematic diagram of BCoR and BCoR-S. The ankyrin repeats (codons 1428–1527) and the region corresponding to the yeast two-hybrid-isolated cDNA clone (A3; codons 112–753) are indicated. (▾) The point of divergence between the two proteins. (C) Expression of BCoR mRNA in various human tissues. A Human RNA Master Blot (Clontech) was probed with a cDNA fragment derived from A3. Tissue identification grid, courtesy of Clontech.
BCoR is expressed ubiquitously
To determine the expression profile of BCoR, its cDNA fragment (A3), which was isolated from the yeast two-hybrid screen, was used as a probe to screen a multitissue mRNA dot blot. Expression was detected in all tissues examined (Fig. 1C), including the spleen and lymph nodes, where BCL-6 expressing mature B cells are found. Two transcripts of ∼6.5 and 7 kb were detected in Northern blot analyses of poly(A)+ RNA from Daudi (a mature B-cell line that expresses BCL-6) and HeLa cells (data not shown). Both transcripts were detected by either a probe for the common region of BCoR and BCoR-S or a probe specific for the unique 3′ portion of BCoR, suggesting that there is additional alternative RNA processing. BCoR-S mRNA was not detected by Northern analysis but was detected in both Daudi and HeLa cells using RT–PCR (data not shown), indicating that BCoR-S mRNA is expressed at significantly lower levels than BCoR mRNA in these cell lines.
Full-length BCL-6 and BCoR interact in vitro and in vivo
A coimmunoprecipitation assay was used to test the interactions of full-length BCL-6 and BCoR. To test for in vitro interactions, myc epitope-tagged BCoR and nontagged BCL-6 full-length proteins were synthesized in vitro and immunoprecipitated with the α-myc antibody. Immunoprecipitated BCoR can coprecipitate full-length BCL-6 (Fig. 2B, bottom, lane 2). This result indicates that full-length BCL-6 and BCoR are capable of interacting in vitro and that the interaction is direct (assuming that a protein in the rabbit reticulocyte lysate is not bridging the interaction). To test the interaction of full-length BCL-6 and BCoR in vivo, total cell extracts from 293 cells transiently transfected with constructs encoding myc-tagged BCoR and nontagged BCL-6 were immunoprecipitated using α-myc antibody. Coprecipitation of the nontagged BCL-6 was observed when constructs for both BCL-6 and BCoR were cotransfected (Fig. 2C, lane 3) and not when BCL-6 was transfected alone (Fig. 2C, lane 2). We conclude from these results that full-length BCL-6 and BCoR are capable of interacting both in vitro and in vivo.
Figure 2.
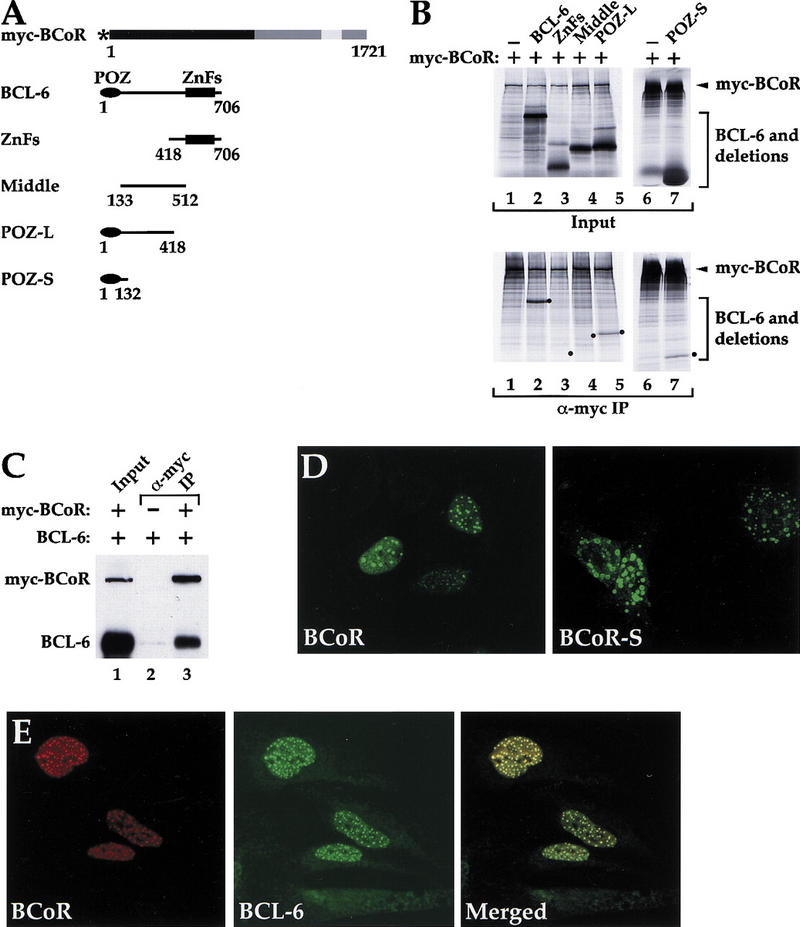
Full-length BCL-6 and BCoR interact in vitro and in vivo, the POZ domain of BCL-6 is both necessary and sufficient for interaction with BCoR in vitro, and BCL-6 and BCoR colocalize in the nucleus. (A) Structure of the proteins used in B and C. (Solid oval) The POZ domain; (solid rectangle) zinc fingers. (*) The myc epitope tag. (B) Coimmunoprecipitation of BCL-6 and BCoR in vitro. Pair-wise combinations of [35S]methionine-labeled nontagged BCL-6 derivatives and myc-tagged BCoR were produced by cotranslation and immunoprecipitated with the α-myc antibody. The immune complexes were recovered and analyzed by SDS-PAGE (10% for lanes 1–5; 17% for lanes 6,7). The intensity of myc–BCoR in lanes 6 and 7 appears stronger due to compression on the 17% gel of non-full-length myc-BCoR proteins. Input represents 20% of the total used. (●) The expected migration of the nontagged BCL-6 derivatives. (C) Coimmunoprecipitation of BCL-6 and BCoR in vivo. 293 cells were transfected with 3 μg of BCL-6 expression plasmid alone (lane 2) or cotransfected with 3 μg each of BCL-6 and myc-tagged BCoR expression plasmids (lanes 1,3). Cell lysates were immunoprecipitated with α-myc antibody (lanes 2,3) and the recovered proteins were detected by Western blot analysis using N-3 α-BCL-6 and α-myc antibodies. The input (lane 1) corresponds to 1/50 of the lysate used in the immunoprecipitation (lane 3). (D) Subcellular localization of BCoR and BCoR-S. Immunofluorescence was performed on HeLa cells transfected with 1.2 μg of either myc-tagged BCoR or BCoR-S expression plasmids. Multiple Z series confocal microscopy images of the given fields were compiled for each image of BCoR and BCoR-S. (E) Colocalization of BCL-6 and BCoR. Coimmunofluorescence was performed on HeLa cells cotransfected with 0.6 μg each of BCL-6 and myc-tagged BCoR, and the data collected as in D. The compiled images of BCoR and BCL-6 were merged to detect colocalization.
BCL-6 interacts with BCoR via the POZ domain
To delineate the minimal region in BCL-6 that is important for interaction with BCoR, we made various BCL-6 deletions (Fig. 2A) and tested them for interaction with BCoR in the in vitro coimmunoprecipitation assay. These BCL-6 deletions include: zinc fingers (ZnFs codons 418–706), middle (codons 133–512), POZ-L (codons 1–418), and POZ-S (POZ short; codons 1–132). To conduct the experiment, pair-wise combinations of myc-tagged BCoR and the indicated BCL-6 deletions were synthesized in vitro and immunoprecipitated with α-myc antibody. Among the BCL-6 deletions tested, only the POZ domain-containing ones were coprecipitated with BCoR, with the minimal POZ (POZ-S) being sufficient for coprecipitation (Fig. 2B, bottom, lanes 2–5,7). Coimmunoprecipitation of the POZ domain was dependent on inclusion of myc–BCoR (data not shown). Identical results were obtained with BCoR-S (data not shown). We conclude that the POZ domain of BCL-6 is both necessary and sufficient for the interaction with BCoR.
BCL-6 and BCoR colocalize in the nucleus
To determine the subcellular localization of BCoR and BCoR-S, immunofluorescence was performed on HeLa cells that were transiently transfected with constructs encoding myc-tagged BCoR and BCoR-S. BCoR is localized exclusively in the nucleus in a diffuse pattern and in punctate structures of various sizes (Fig. 2D). Although BCoR-S is also found in punctate structures, in contrast to BCoR, these are not restricted to the nucleus but are found in the cytoplasm as well (Fig. 2D). Because BCL-6 is also found in the nucleus in punctate structures (Cattoretti et al. 1995) and can colocalize with N-CoR (Huynh and Bardwell 1998), we tested whether BCL-6 and BCoR could be found in the same nuclear structures. In HeLa cells cotransfected with constructs encoding BCL-6 and myc-tagged BCoR, BCL-6 and BCoR colocalize to small and large punctate structures (Fig. 2E), indicating that they may function or be stored together in these structures. N-CoR and BCoR also colocalize in similar punctate nuclear structures (data not shown).
Self-association of the BCL-6 POZ domain is required for interaction with BCoR
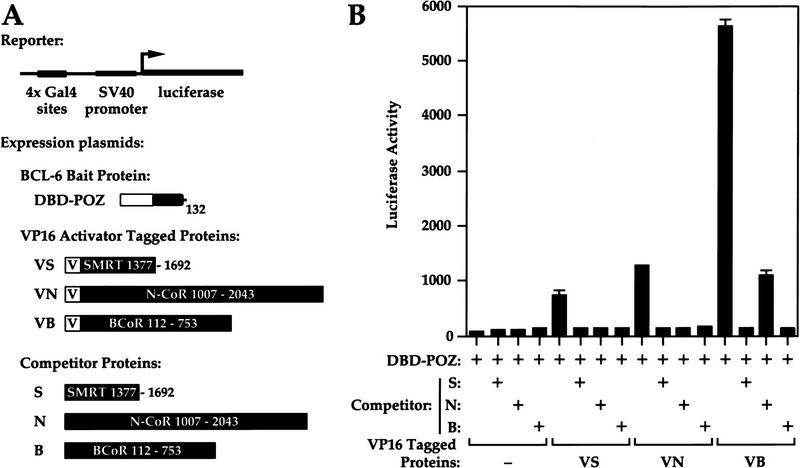
We have shown previously that there is a correlation between the ability of the BCL-6 POZ domain to self-associate, to interact with both N-CoR and SMRT, and to repress transcription (Huynh and Bardwell 1998). To test whether the interaction between BCL-6 and BCoR is also dependent on self-association of the BCL-6 POZ domain, the mammalian two-hybrid assay was used. In this assay HeLa cells were transfected with a GAL4-responsive luciferase reporter, along with pair-wise combinations of the bait and activator constructs. The bait constructs consisted of the GAL4 DNA-binding domain (DBD) alone or DBD fused to either the wild-type POZ domain of BCL-6 (POZ-WT, codons 1–132), or mutants M1, M2, or M3 (Fig. 3A). Mutants M1 and M3 are defective in self-association, interaction with corepressors N-CoR and SMRT, and repression, whereas mutant M2 exhibits wild-type behavior (Huynh and Bardwell 1998). Recent structural studies of the related PLZF POZ domain indicate that M1 and M3 would be predicted to change amino acids critical in the analagous BCL-6 dimerization interface (Ahmad et al. 1998; Li et al. 1999). The activator construct consists of the VP16 activation domain fused to the yeast two-hybrid-isolated cDNA fragment of BCoR (codons 112–753; VB). Cotransfection of the DBD, M1, or M3 bait constructs, along with VB, yielded very little reporter activation. Significant reporter activation was detected, however, with both the POZ-WT and M2 bait constructs (Fig. 3B). These results suggest that BCoR requires self-association of the POZ domain of BCL-6.
Figure 3.
Self association of the POZ domain of BCL-6 is required for interaction with BCoR, and BCoR interacts selectively with the POZ domain of BCL-6. (A) Structures of the reporter construct, bait, and activator proteins used in B. The bait proteins consist of the GAL4 DBD alone, or fused to wild type or the indicated mutant POZ domains. The activator-tagged protein VB consists of the VP16 activation domain fused to the yeast two-hybrid-isolated BCoR clone (codons 112–753). (B) Interactions of BCoR (codons 112–753) with BCL-6 POZ domain point mutants in the mammalian two-hybrid assay. Luciferase and β-gal assays were conducted on cell extracts from HeLa cells transiently transfected with 500 ng of the GAL4-responsive reporter (pAH205), 50 ng of the indicated bait expression plasmids; 0, 50, or 250 ng of the VB expression plasmid; and 25 ng of the pCMVlacZ plasmid, as control for transfection efficiency. Data displayed represent the average of two experiments with the standard deviations depicted. (C) In vitro coimmunoprecipitation of BCoR and BCoR-S with POZ-containing proteins. The indicated pair-wise combinations of [35S]methionine-labeled nontagged POZ proteins (BCL-6, PLZF, BAZF, ZID, TTK, and GAGA) and either myc-tagged BCoR or BCoR-S were produced by cotranslation and immunoprecipitated with α-myc antibody. The proteins were recovered and analyzed by 8% SDS-PAGE. (●) The expected migration of the nontagged BCL-6 derivatives.
BCoR interacts specifically with the POZ domain of BCL-6
Our studies with N-CoR and SMRT revealed that these corepressors can interact with POZ domains from a variety of different proteins other than BCL-6 (Huynh and Bardwell 1998). To determine whether this is also true of BCoR we performed the in vitro coimmunoprecipitation assay again with the same myc-tagged BCoR protein that was used in Figure 2B. The nontagged POZ-containing proteins include full-length forms of human BCL-6, PLZF, and ZID, murine BAZF, and the Drosophila proteins TTK and GAGA. Of the POZ-containing proteins tested, only BCL-6 is coprecipitated with myc-tagged BCoR, suggesting that BCoR interacts specifically with BCL-6 (Fig. 3C). We have confirmed these results using mammalian two-hybrid and/or GST pull-down assays, with which we have also tested human FBI-1, murine ZF5, and a protein derived from the Drosophila E(var)3-93D gene (protein A) and found that they also cannot interact with BCoR (data not shown). Of the nine POZ-containing proteins tested, BCoR interacts selectively with the POZ domain of BCL-6. In addition, we have tested and failed to detect an interaction between BCoR and TRβ (thyroid hormone receptor β) in the in vitro coimmunoprecipitation assay (data not shown), which suggests that BCoR, unlike N-CoR and SMRT, does not interact with nuclear hormone receptors.
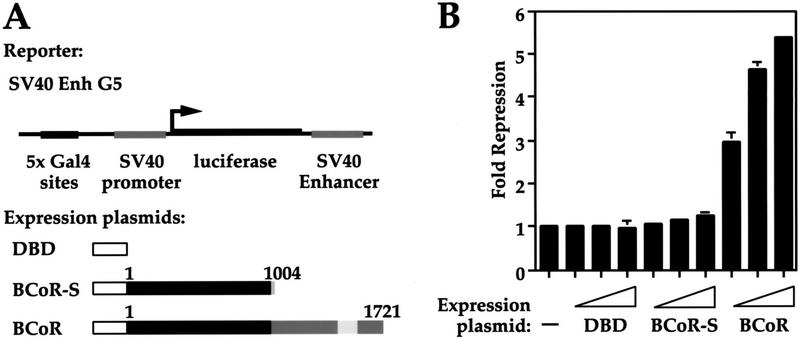
BCoR is a transcriptional corepressor
Having shown that BCL-6 can interact with BCoR and BCoR-S, we wanted to determine the functional role of these proteins. BCL-6 is a transcriptional repressor, and the lack of a recognizable DNA-binding domain in BCoR suggests that BCoR may function as a corepressor. To test this, we fused the GAL4 DBD to BCoR and BCoR-S and determined what transcriptional effects these fusion proteins have on a GAL4-responsive reporter gene. The experiment was performed by cotransfecting HeLa cells with an SV40 promoter- and enhancer-driven, GAL4-responsive luciferase reporter (SV40 Enh G5), along with constructs for DBD alone or fusions with BCoR or BCoR-S (Fig. 4A). We find that BCoR harbors a dose-dependent transcriptional repression activity that is not seen in BCoR-S or the GAL4 DBD alone (Fig. 4B). The DBD BCoR fusion has no effect on the reporter without the GAL4 binding sites (data not shown). At higher concentrations, BCoR-S has a weak repressive effect (data not shown; see Discussion). We conclude from these results that BCoR is a corepressor that can repress transcription when physically tethered to a promoter.
Figure 4.
BCoR is a transcriptional corepressor. (A) Structures of the reporter construct and proteins used in B. (B) Luciferase and β-gal assays were performed on cell extracts from HeLa cells transiently transfected with 50 ng of the GAL4-responsive reporter (SV40 Enh G5), along with 0, 5, 10, or 25 ng of the indicated GAL4 fusion constructs [GAL4 DBD alone (DBD) or DBD fused with BCoR or BCoR-S), and 25 ng of pCMVLacZ plasmid. Data displayed represent the average of two experiments, with the standard deviations depicted. Fold repression was determined by normalizing luciferase/β-gal values of all samples to that of the reporter alone.
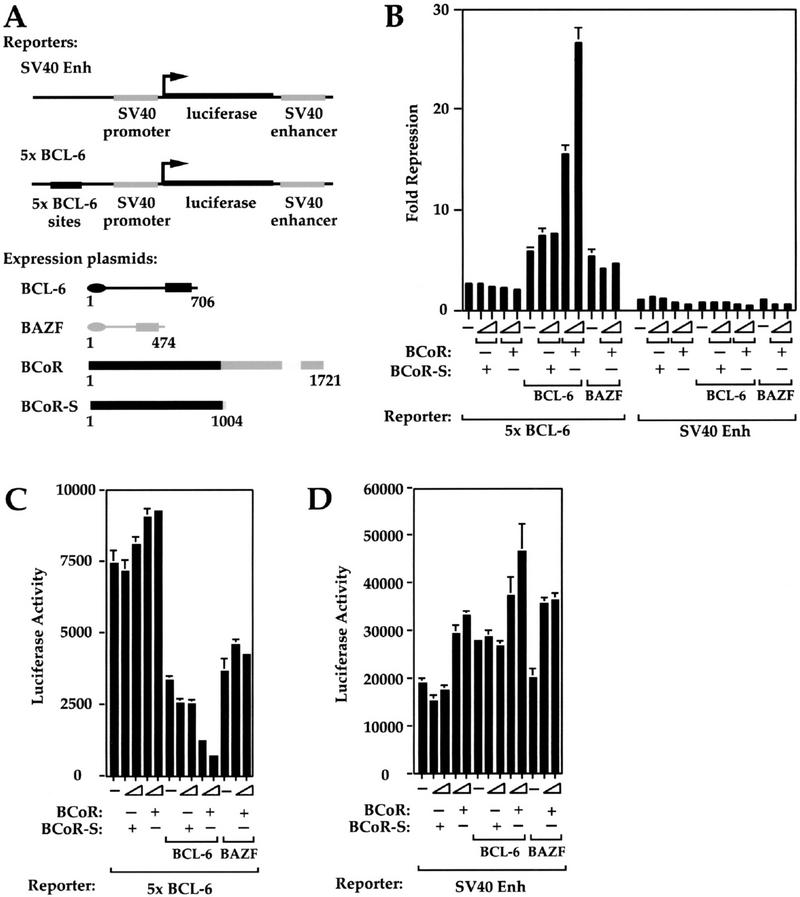
BCoR is involved in BCL-6 repression
Having established that BCoR contains transcriptional repression activity, we sought to determine whether BCoR is involved in BCL-6 repression. If BCoR is a true BCL-6 corepressor, it should potentiate BCL-6 repression when endogenous BCoR is limiting. The experiment was conducted by cotransfecting HeLa cells with an SV40 promoter- and enhancer-driven luciferase reporter containing five BCL-6 binding sites immediately upstream of the SV40 promoter (5× BCL-6), along with various combinations of expression constructs for BCL-6, BCoR, and BCoR-S (Fig. 5A). The parental reporter lacking BCL-6 sites (SV40 Enh) was used as a control. We first transfected HeLa cells with either the 5× BCL-6 or the SV40 Enh reporter construct alone and assayed for luciferase activity of extracts from transfected cells (Fig. 5, B, fold repression, C and D, luciferase data; note different scales in C and D). A two- to threefold repression is observed with the 5× BCL-6 reporter relative to the SV40 Enh reporter (Fig. 5B), presumably because of endogenous BCL-6, which has been reported to be expressed in HeLa cells (Allman et al. 1996). Addition of increasing amounts of BCoR or BCoR-S with either reporter resulted in no change or weak activation. With additional BCL-6 expression, repression of the 5× BCL-6 reporter is increased to a maximum of sixfold. This repression is further potentiated to 28-fold, in a specific and dose-dependent manner when BCoR, but not BCoR-S, is cotransfected along with BCL-6. To confirm the specificity of the potentiation, we tested the effect of BCoR expression on repression by BAZF, a POZ-containing transcriptional repressor that does not interact with BCoR (Fig. 3C). BAZF shares high sequence homology with BCL-6 in its POZ domain and zinc finger region and can bind to the BCL-6 recognition site and repress transcription (Okabe et al. 1998). As expected, we find that overexpression of BCoR failed to potentiate BAZF repression (Fig. 5B), which suggests that potentiation of repression by BCoR is selective for BCL-6. We conclude that BCoR is capable of functioning as a BCL-6 corepressor.
Figure 5.
BCoR can selectively potentiate BCL-6 repression. (A) Structures of the reporter constructs and proteins used in B. (B) Luciferase and β-gal assays were conducted on HeLa cell extracts transiently transfected with 250 ng of either the SV40 Enh or 5× BCL-6 reporter construct, and the indicated combinations of BCL-6 (25 ng), BAZF (25 ng), BCoR (25 and 50 ng), and BCoR-S (25 and 50 ng); pEFLacZ (200 ng) was also included as control for transfection efficiency. Data displayed represent the average of two experiments with the standard deviations depicted. Fold repression was determined by normalizing luciferase/β-gal values (C,D) of all samples to that of the SV40 Enh reporter alone.
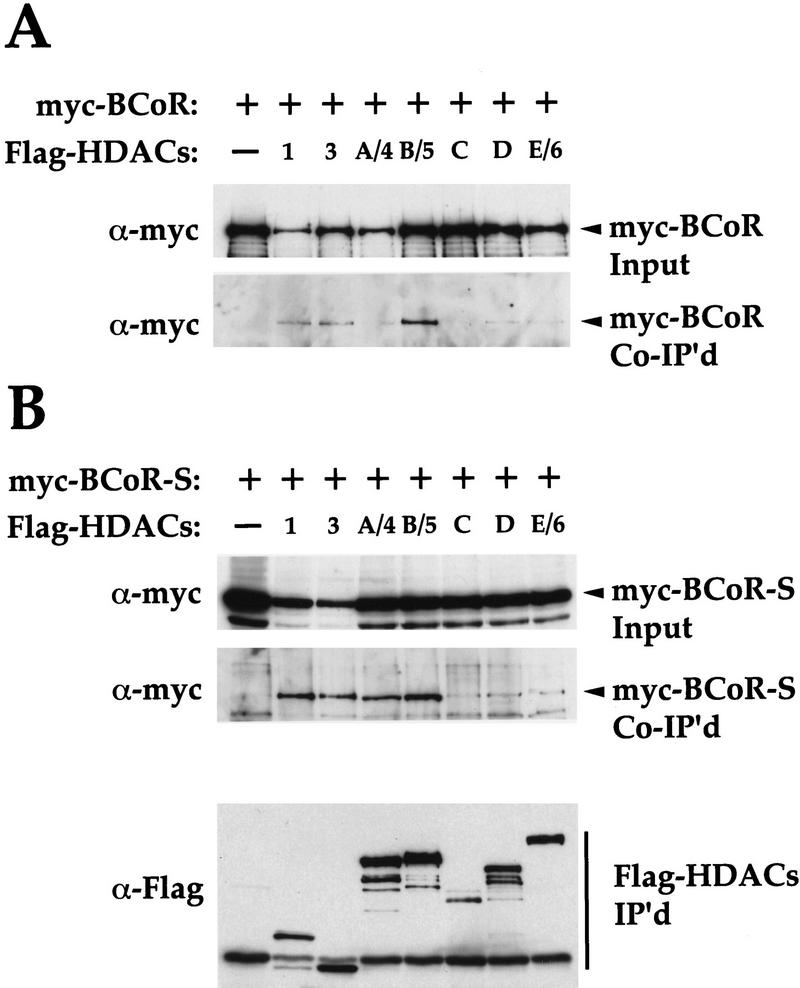
BCoR interacts with specific class I and class II HDACs in vivo
We wanted to address the mechanism of BCoR-mediated repression. Because HDACs appear to be involved in repression by an increasing number of transcriptional repressors, we tested whether BCoR can associate with HDACs. Using the in vivo coimmunoprecipitation assay, we assayed seven of the known human HDACs; the class I HDACs, HDAC1 and HDAC3, and the class II HDACs, HDAC-A/4, HDAC-B/5, HDAC-C, HDAC-D, and HDAC-E/6 (Fischle et al. 1999; Grozinger et al. 1999; W. Fischle and E. Verdin, unpubl.) for interactions with BCoR. We transfected 293 cells with pair-wise combinations of various Flag-tagged HDAC and myc-tagged BCoR expression constructs, and immunoprecipitated the cell extracts with α-Flag antibody. BCoR coprecipitates most strongly with HDAC1, HDAC3, and HDAC-B/5 and more weakly with the other HDACs (Fig. 6A). We also tested BCoR-S for interactions with these HDACs and found that in addition to HDAC1, HDAC3, and HDAC-B/5, it also readily coprecipitates with HDAC-A/4 (Fig. 6B). These results suggest that class I and class II HDACs might be involved in BCoR-mediated repression.
Figure 6.
BCoR and BCoR-S interact with class I and class II HDACs in vivo. 293 cells were transfected with 4 μg of either myc-tagged BCoR (A) or BCoR-S (B) expression plasmids alone (first lane), or cotransfected with pair-wise combinations of 3 μg of the indicated Flag-tagged HDACs and 3 μg of either myc-tagged BCoR (A) or BCoR-S (B) expression plasmids (remaining lanes). Cell lysates were imunoprecipitated with α-Flag antibody, and the recovered proteins were detected by Western blot analysis using α-myc and α-Flag antibodies (shown in B only). The input corresponds to 1/100 of the lysates used for coimmunoprecipitation.
Interactions between the BCL-6 POZ domain and N-CoR, SMRT, and BCoR are mutually exclusive
Because SMRT, N-CoR, and BCoR can all interact with the POZ domain of BCL-6 (Figs. 2B and 3B; Dhordain et al. 1997; Huynh and Bardwell 1998; Wong and Privalsky 1998), we wanted to determine whether the interactions are mutually exclusive. We used a variation of the mammalian two-hybrid assay in which we assayed the ability of the corepressors to compete with their VP16-tagged counterparts for interactions with the BCL-6 POZ bait (DBD–POZ, Fig. 7A). If two corepressors can bind simultaneously, they should not compete for DBD–POZ binding. We first determined the amount of VP16-tagged corepressor that would yield 80%–90% maximal reporter activation for a given amount of DBD–POZ bait. In this context, we added the non-VP16-tagged competitor corepressors (Fig. 7A). As expected, each of the corepressors can compete with its respective VP16-tagged corepressor for binding to the BCL-6 POZ bait (Fig. 7B). More importantly, competition is also observed with all combinations of VP16-tagged and nontagged corepressors (SMRT/N-CoR, SMRT/BCoR, and N-CoR/BCoR). This competition is dose dependent (data not shown). The suppression of reporter activation by non-VP16-tagged corepressors is due to competition with VP16-tagged corepressors for binding to the BCL-6 POZ bait and not due to inhibition of VP16 activity, as these competitor proteins had minimal effect on reporter activation by the GAL4 DBD VP16 fusion (data not shown). In addition, in the absence of VP16-tagged corepressors, non-VP16-tagged corepressors had a weak activating (perhaps dominant-negative) effect on DBD–POZ (Fig. 7B). We conclude from these results that the interactions among the POZ domain of BCL-6 and SMRT, N-CoR, and BCoR are mutually exclusive.
Figure 7.
Interactions between the BCL-6 POZ domain and N-CoR, SMRT, and BCoR are mutually exclusive. (A) Structures of the reporter construct and proteins used in B. (B) Luciferase and β-gal assays were performed on HeLa cell extracts transiently transfected with 250 ng of the GAL4-responsive reporter (pAH205), 2.5 ng of the POZ bait, and the indicated combinations of VP16-tagged (VS, 25 ng; VN, 25 ng; VA, 5 ng) and nontagged (S, N, A, at 100 ng each) corepressor expression plasmids, along with 25 ng of the pCMVlacZ plasmid. Data displayed represent the average of two experiments with the standard deviations depicted.
Discussion
In this study our interest in dissecting the mechanisms of repression by BCL-6 led us to the identification and characterization of a novel BCL-6-interacting corepressor, BCoR. Several lines of evidence suggest that the interaction between BCL-6 and BCoR is both specific and functionally important, that it is mutually exclusive with N-CoR and SMRT, and that BCoR is a corepressor that is likely to function by recruiting class I and II HDACs. First, BCoR and BCL-6 interact in vitro and in vivo (Fig. 2B,C) and colocalize in vivo (Fig. 2E). Second, the POZ domain of BCL-6 is both necessary and sufficient for interaction with BCoR (Fig. 2B), whereas the eight other POZ domain-containing proteins tested do not interact with BCoR (Fig. 3C; data not shown). Third, BCoR can compete with N-CoR and SMRT for binding to the BCL-6 POZ domain (Fig. 7). Fourth, BCoR represses transcription when tethered to a promoter (Fig. 4). Fifth, BCoR can potentiate repression by BCL-6 but not by BAZF (Fig. 5), a POZ-containing repressor closely related to BCL-6. Finally, BCoR can interact in vivo with both class I and class II HDACs (Fig. 6).
BCoR is unrelated to other mammalian proteins in the databases and also has no apparent orthologs in yeast or nematodes. This suggests that BCoR, like the corepressors N-CoR and SMRT, may perform functions specific to higher eukaryotes—in contrast to corepressors such as Sin3 that are widely conserved among eukaryotes, from yeast to mammals. The ankyrin repeats of BCoR, which in other proteins are involved in protein–protein interactions, are not required for interaction with BCL-6 or the HDACs, raising the possibility that additional components of a multiprotein complex interact with BCoR.
Our results suggest that self-association of the POZ domain of BCL-6 is required for interaction with BCoR (Fig. 3B). It is possible that the residues mutated are critical for the direct interaction of BCL-6 with BCoR; however, this is unlikely because these same mutations affect the association of BCL-6 with N-CoR and SMRT. POZ domains can associate to form dimers, and there is structural (Li et al. 1999) and functional data indicating that POZ domain dimers also can associate to form higher order multimers. Multimer formation of POZ proteins is likely to have a role in transcriptional regulation, for example, by functionally coupling dispersed transcription factor binding sites. The best example of such a mechanism involves the Drosophila POZ protein GAGA. POZ–POZ interaction of GAGA bound to multiple widely spaced sites in a single target promoter can serve to draw GAGA and its associated DNA sites together (Espinas et al. 1999; Katsani et al. 1999). If BCL-6 also binds its target genes in this way, BCoR might recognize a specific interface generated by either dimerization or higher order multimerization of the BCL-6 POZ domain.
In this study we have shown that BCoR interacts with BCL-6 and potentiates transcriptional repression by BCL-6 with striking specificity. Of the proteins tested, only BCL-6 interacts with BCoR. However, BCL-6 has a more limited tissue distribution than the ubiquitously expressed BCoR (Fig. 1C), suggesting that BCoR may perform additional functions. BCoR is likely to be targeted to DNA by a specific subset of transcriptional repressors, much like the corepressors N-CoR and SMRT, which interact with specific members of multiple transcription factor families, including some POZ-containing proteins (Huynh and Bardwell 1998; Knoepfler and Eisenman 1999).
BCoR can interact with HDAC1, HDAC3, and HDAC-B/5 more strongly than with HDAC-A/4, HDAC-C, HDAC-D, and HDAC-E. Because these represent HDACs from both class I and class II, the two mammalian HDAC classes might function together, unlike their yeast counterparts. This possibility is supported further by two recent reports demonstrating that both SMRT and N-CoR, which previously appeared to be associated only with the class I HDAC1 complex, can also associate with certain class II HDACs (Huang et al. 2000; Kao et al. 2000). Perhaps higher eukaryotes, through corepressors N-CoR, SMRT, and BCoR, have evolved in such a way as to functionally link the two HDAC classes, thus increasing potential regulatory complexity. The tissue distribution of some of the class II HDACs is quite distinct (Fischle et al. 1999; Grozinger et al. 1999; Verdel and Khochbin 1999; Wang et al. 1999). Therefore, BCoR may interact only with a subset of HDACs in a given tissue, thus further increasing the potential regulatory complexity.
We also identified an alternatively processed variant of BCoR, BCoR-S, which is a truncated protein that shares amino-terminal sequence with BCoR. In HeLa and Daudi cells BCoR-S mRNA is expressed at levels significantly lower than BCoR (data not shown), and the transfected protein is found in both the cytoplasm and the nucleus in contrast to BCoR, which is exclusively nuclear (Fig. 2D). BCoR-S is capable of interacting with BCL-6 and HDACs (HDAC1, HDAC3, HDAC-A/4, and HDAC-B/5) but represses transcription only weakly when brought to DNA and does not potentiate BCL-6 repression. Because BCoR-S lacks sequences that are present in BCoR, including the ankyrin repeats, it is likely that these additional sequences are important for generating a multiprotein complex that is fully functional and that interaction with HDACs is not sufficient for full corepressor activity. Alternatively B-CoR-S may be capable of functioning as a corepressor under specific circumstances or with different transcription factors. A third possibility is that BCoR-S, when expressed at higher levels in specific tissues, might act as an antagonist of BCoR function. A final possibility is that the BCoR-S transcript is an incompletely processed BCoR mRNA.
At present, it is unclear whether BCoR is a previously unidentified component of a complex with NCoR/SMRT or whether it is a component of a distinct complex(es). Like N-CoR and SMRT, BCoR interacts with the POZ domain of BCL-6, and self-association of the POZ domain is required for this interaction. Furthermore, the interaction between the BCL-6 POZ domain and each of these corepressors is mutually exclusive (Fig. 7), indicating that they contact the same or overlapping sites on the POZ domain or that binding of one allosterically inhibits binding of the others. This result, coupled with our inability to detect an interaction between BCoR and N-CoR or mSin3 with the in vivo coimmunoprecipitation assay, using either transfected or endogenous proteins (data not shown), suggests the possibility of a distinct complex. However, it is also possible that these corepressors are components of the same complex that is recruited by POZ domain multimers, with each corepressor contacting different POZ dimers. Further studies will be needed to determine of which complex(es) BCoR is a component. If the N-CoR/SMRT and BCoR corepressors occur in distinct complexes, it will be important to ascertain how the selectivity of BCL-6 is generated for these different complexes and whether it is context dependent. Recruitment of SMRT by some nuclear hormone receptors, as well as by the POZ protein PLZF, is dependent on tyrosine kinase signaling (Hong et al. 1998). We speculate that extracellular signaling likewise might regulate the recruitment of complexes by BCL-6. Future elucidation of the functions and regulated assembly of regulatory complexes recruited by BCL-6 might have important benefits for the rational design of therapeutic agents directed against diffuse large cell lymphoma and follicular lymphoma.
Materials and methods
Plasmids
Construction of plasmids was by standard methods; all structures were verified by appropriate restriction digest and/or sequencing.
Yeast two-hybrid assay
The BCL-6 POZ-L bait construct and the Daudi (a mature B-cell line that expresses BCL-6) Activator Library (carboxy-terminal library) were described previously (Huynh and Bardwell 1998).
Coimmunoprecipitations
For in vivo experiments, full-length BCL-6 (codons 1–706), and amino-terminal myc-tagged BCoR (codons 1–1721) and BCoR-S (codons 1–1004) were cloned into pEFplink2 (Huynh and Bardwell 1998). Carboxy-terminal Flag-tagged human HDACs (HDAC1, HDAC3, HDAC-A/4 (codons 118–1085), HDAC-B/5 (codons 132–1123), HDAC-C, HDAC-D, and HDAC-E/6) were inserted into pcDNA3.1 (Invitrogen). For in vitro experiments, all cDNAs used were in T7βplink vector (Dalton and Treisman 1992): amino-terminal myc-tagged BCoR (codons 1–1721) and BCoR-S (codons 1–1004), BCL-6 full-length (codons 1–706), BCL-6 POZ-L (codons 1–418), BCL-6 POZ-S (codons 1–132), BCL-6 middle (codons 133–512), BCL-6 zinc fingers (codons 418–706), PLZF (codons 1–673), BAZF (codons 1–474), ZID (codons 1–424), TTK (codons 1–641), and GAGA (codons 1–519).
Immunofluorescence analyses
Mammalian expression plasmids for BCL-6 and myc-tagged BCoR and BCoR-S were identical to those used for in vivo immunoprecipitation assays.
Mammalian two-hybrid and transcriptional repression analyses
The Gal4-responsive luciferase reporter (pAH205) and the DBD, POZ-WT (POZ-S), M1 (L19S), M2 (N23H), and M3 (L25S, R26L) bait constructs were described previously (Huynh and Bardwell 1998). The yeast two-hybrid-isolated cDNA fragments of B-CoR (codons 112–753), SMRT (codons 1377–1692), and N-CoR (codons 1007–2043) [Huynh and Bardwell 1998, current amino acid numberings are according to NP_006303 (SMRT) and NP_006302.1 (N-CoR)] were cloned into a pEFplink2 expression construct that either contains or lacks a VP16 activation domain, to generate the VP16-tagged (VB, VS, and VN) or nontagged (B, S, and N) proteins, respectively. Both sets of expression constructs contain an in-frame SV40 nuclear localization signal (PKKKRKV). The SV40 Enh (pGL3-Control; Promega) and the SV40 Enh G5 reporters, as well as the DBD and BCL-6 (codons 1–706) mammalian expression constructs were described previously (Huynh and Bardwell 1998). The 5× BCL-6 luciferase reporter was constructed by insertion of five copies of the oligonucleotide 5′-TCGAATTCCTCGAAAGTCGA-3′ into the XhoI site of pGL3-Control vector (oligonucleotide orientation: − + + + +). BCoR (codons 1–1721) and BCoR-S (codons 1–1004) cDNAs were cloned into the GAL4 DBD construct (Wilkinson and Towle 1997) to generate GAL4 DBD fusions. BCoR and BCoR-S expression plasmids for potentiation experiments were the same as those used for in vivo coimmunoprecipitation analyses. Full-length BAZF (codons 1–474) was inserted into pEFplink2. pEFlacZ and pCMVlacZ, used as controls for transfection efficiency, were described previously (Huynh and Bardwell 1998).
Isolation of BCoR cDNAs
The yeast two-hybrid screen using BCL-6 POZ-L bait (codons 1–418) was described previously (Huynh and Bardwell 1998). The yeast two-hybrid isolated clone of BCoR, A3, which was isolated once, contains a 1928-bp cDNA. A 5′ portion of this cDNA (721-bp fragment, from the start of the cDNA to the PstI site) was used as a probe to screen a Lambda-ZAPII human adult frontal cortex cDNA library (a generous gift of M. Macdonald, Massachusetts General Hospital, Charlestown, MA). This resulted in the isolation of a 3205-bp cDNA (designated 1a) that encompasses the entire sequence of A3 but still lacks a translation stop codon in its predicted ORF. The entire sequence of this cDNA is shared between BCoR and BCoR-S. Further hybridization screening yielded an overlapping cDNA (3583 bp) that was combined with 1a to yield the full-length BCoR cDNA. The full-length BCoR-S cDNA was generated by combining 1a with an overlapping expressed sequence tag (EST AA806538) clone obtained from Research Genetics.
Cell culture and transfections
HeLa and 293 cells were maintained in complete (10% fetal calf serum and 0.1 mg/ml penicillin G/streptomycin) RPMI and DMEM media, respectively. Transfections of HeLa cells were done by the liposome-mediated method, using the Tfx-20 reagent (Promega). Cells were plated at 2 × 105 cells per well in six-well plates 1 day prior to transfection. For immunofluorescence analyses, cells were plated on coverslips. Each transfection contained the amounts of expression plasmids as indicated in the figure legends (Fig. 2D,E; 3A,B; 4; 5; 7), along with 3.6 μl of Tfx-20 reagent (yielding a 2:1 lipid to DNA ratio) in 1 ml of serum-free media. The total amount of DNA per transfection was adjusted to 1.2 μg by inclusion of the appropriate amount of empty parental expression plasmids to keep expression promoter concentrations constant, and/or a pBluescript (Stratagene) based filler plasmid. After a 1 hr incubation at 37°C, 2 ml of complete media was added. Transfections of 293 (a human kidney cell line) were done with the calcium phosphate-mediated method, using the Profection Mammalian Transfection System (Promega). Transfections were done in 60-mm plates, according to the manufacturer's instructions, with amounts of expression constructs as indicated in the figure legends (Fig. 2C; 6). For all transfections, cells were harvested 44-48 hr post-transfection.
Coimmunoprecipitations
In vivo coimmunoprecipitation assays were performed by transfecting 293 cells with the amounts of expression constructs as indicated in the figure legends (Fig. 2C; 6). Transfected cells were washed twice with PBS and lysed in 500 μl of cold immunoprecipitation (IP) buffer (PBS, 10% glycerol, 0.5% NP-40, and a complete protease inhibitor cocktail from Boehringer-Mannheim) for 10 min on ice. Lysates were briefly sonicated and centrifuged for 10 min at 4° C. The supernatants were collected and immunoprecipitated with either 1 μg of α-myc (9E10; Santa Cruz Biotechnology) or 2 μg of α-Flag M2 (Sigma) antibodies. After a 1 hr incubation at 4°C with rotation, 20 μl of protein A/G Plus-Agarose slurry (Santa Cruz Biotechnology) was added and reincubated as above. The immune complexes were then collected and washed three times with 500 μl of cold IP buffer. The immunoprecipitated proteins were fractionated on 8% SDS–polyacrylamide gels and transferred to nitrocellulose. Western analyses were performed with primary antibodies as indicated in the figure legends [α-BCL-6 (N-3; Santa Cruz Biotechnology), α-myc (9E10), or α-Flag M2], according to protocols provided by Santa Cruz Biotechnology. Proteins were visualized by ECL (Amersham Pharmacia Biotech). For in vitro coimmunoprecipitation experiments, the indicated combinations of [35S]methionine-labeled proteins were cosynthesized using the TNT T7 Quick Coupled Transcription/Translation System (Promega), according to the manufacturer's instructions. Coimmunoprecipitation experiments were performed as follows: Five microliters of the indicated TNT reaction mixture was added to a 45-μl solution (containing 3 μl of unprogrammed reticulocyte lysate, 10 μl of 50% protein A slurry, and 32 μl of IP buffer) that had been preincubated at 4°C. After a 1 hr incubation at 4°C with rotation, the immune complexes were collected and washed four times with 500 μl of cold IP buffer. The immunoprecipitated proteins were fractionated on 8%, 10%, or 17% SDS–polyacrylamide gels.
Immunofluorescence analyses
Immunofluorescence analyses were performed as described (Skinner et al. 1997). Antibody incubations were as follows: for BCoR and BCoR-S immunofluorescence, (1) 9E10 mouse monoclonal α-myc (1:100), (2) rabbit anti-mouse (1:200), (3) Cy2-conjugated goat anti-rabbit (1:300; Jackson ImmunoResearch); for BCL-6 and BCoR coimmunofluorescence, (1) 9E10 and N-3 rabbit α-BCL-6 (1:100), (2) Cy2-conjugated goat anti-mouse (Jackson ImmunoResearch) and rhodamine-conjugated goat anti-rabbit (Accurate Chemical and Scientific), both at 1:300.
Other methods
Expression analysis for BCoR using the Human RNA Master Blot (Clontech) was performed according to the manufacturer's instructions. For reporter gene assays, transfected HeLa cells were lysed in 100 μl of Reporter Lysis Buffer (Promega). Luciferase and β-gal assays were conducted according to protocols provided by Promega and Clontech, respectively.
Acknowledgments
We thank M. Macdonald for the Lambda-ZAPII human frontal cortex cDNA library, R. Sheaff for 293 cells, T. Lebien for HeLa cells, D. Bernlohr for pAH205 reporter plasmid, R. Marais for pEFplinkz and pEFlacZ expression plasmids, H. Towle for mammalian GAL4 DBD and pCMVlacZ plasmids, T. Tokuhisa for BAZF cDNA, N. Hernandez for FBI-1 cDNA, and A. Zelent for PLZF cDNA. We also thank K. Conklin, D. Largaespada, J. Simon, H. Towle, R. Treisman, B. Van Ness, and D. Zarkower for critical reading of the manuscript. This work was supported by NIH grant 5R29CA71540 (V.B.), the University of Minnesota Graduate School (V.B.), Minnesota Medical Foundation (V.B.), and a University of Minnesota Cancer Center Predoctoral Fellowship (K.D.H.).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL bardwell@lenti.med.umn.edu; FAX (612) 626-6140.
References
- Ahmad KF, Engel CK, Prive GG. Crystal structure of the BTB domain from PLZF. Proc Natl Acad Sci. 1998;95:12123–12128. doi: 10.1073/pnas.95.21.12123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albagli O, Dhordain P, Deweindt C, Lecocq G, Leprince D. The BTB/POZ domain: A new protein-protein interaction motif common to DNA- and actin-binding proteins. Cell Growth Differ. 1995;6:1193–1198. [PubMed] [Google Scholar]
- Albagli O, Dhordain P, Bernardin F, Quief S, Kerkaert JP, Leprince D. Multiple domains participate in distance-independent LAZ3/BCL6-mediated transcriptional repression. Biochem Biophys Res Commun. 1996;220:911–915. doi: 10.1006/bbrc.1996.0505. [DOI] [PubMed] [Google Scholar]
- Albagli O, Lantoine D, Quief S, Quignon F, Englert C, Kerckaert JP, Montarras D, Pinset C, Lindon C. Overexpressed BCL6 (LAZ3) oncoprotein triggers apoptosis, delays S phase progression and associates with replication foci. Oncogene. 1999;18:5063–5075. doi: 10.1038/sj.onc.1202892. [DOI] [PubMed] [Google Scholar]
- Alizadeh AA, Eisen MB, Davis RE, Ma C, Lossos IS, Rosenwald A, Boldrick JC, Sabet H, Tran T, Yu X, et al. Distinct types of diffuse large B-cell lymphoma identified by gene expression profiling. Nature. 2000;403:503–511. doi: 10.1038/35000501. [DOI] [PubMed] [Google Scholar]
- Alland L, Muhle R, Hou H, Jr, Potes J, Chin L, Schreiber-Agus N, DePinho RA. Role for N-CoR and histone deacetylase in Sin3-mediated transcriptional repression. Nature. 1997;387:49–55. doi: 10.1038/387049a0. [DOI] [PubMed] [Google Scholar]
- Allman D, Jain A, Dent A, Maile RR, Selvaggi T, Kehry MR, Staudt LM. BCL-6 expression during B-cell activation. Blood. 1996;87:5257–5268. [PubMed] [Google Scholar]
- Bajalica-Lagercrantz S, Piehl F, Farnebo F, Larsson C, Lagercrantz J. Expression of the BCL6 gene in the pre- and postnatal mouse. Biochem Biophys Res Commun. 1998;247:357–360. doi: 10.1006/bbrc.1998.8551. [DOI] [PubMed] [Google Scholar]
- Bardwell VJ, Treisman R. The POZ domain: A conserved protein–protein interaction motif. Genes & Dev. 1994;8:1664–1677. doi: 10.1101/gad.8.14.1664. [DOI] [PubMed] [Google Scholar]
- Baron BW, Nucifora G, McCabe N, Espinosa RD, Le Beau MM, McKeithan TW. Identification of the gene associated with the recurring chromosomal translocations t(3;14)(q27;q32) and t(3;22)(q27;q11) in B-cell lymphomas. Proc Natl Acad Sci. 1993;90:5262–5266. doi: 10.1073/pnas.90.11.5262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bastard C, Deweindt C, Kerckaert JP, Lenormand B, Rossi A, Pezzella F, Fruchart C, Duval C, Monconduit M, Tilly H. LAZ3 rearrangements in non-Hodgkin's lymphoma: Correlation with histology, immunophenotype, karyotype, and clinical outcome in 217 patients. Blood. 1994;83:2423–2427. [PubMed] [Google Scholar]
- Cattoretti G, Chang CC, Cechova K, Zhang J, Ye BH, Falini B, Louie DC, Offit K, Chaganti RS, Dalla-Favera R. BCL-6 protein is expressed in germinal-center B cells. Blood. 1995;86:45–53. [PubMed] [Google Scholar]
- Chang CC, Ye BH, Chaganti RS, Dalla-Favera R. BCL-6, a POZ/zinc-finger protein, is a sequence-specific transcriptional repressor. Proc Natl Acad Sci. 1996;93:6947–6952. doi: 10.1073/pnas.93.14.6947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Iida S, Louie DC, Dalla-Favera R, Chaganti RS. Heterologous promoters fused to BCL6 by chromosomal translocations affecting band 3q27 cause its deregulated expression during B-cell differentiation. Blood. 1998;91:603–607. [PubMed] [Google Scholar]
- Dalton S, Treisman R. Characterization of SAP-1, a protein recruited by serum response factor to the c-fos serum response element. Cell. 1992;68:597–612. doi: 10.1016/0092-8674(92)90194-h. [DOI] [PubMed] [Google Scholar]
- David G, Alland L, Hong SH, Wong CW, DePinho RA, Dejean A. Histone deacetylase associated with mSin3A mediates repression by the acute promyelocytic leukemia-associated PLZF protein. Oncogene. 1998;16:2549–2556. doi: 10.1038/sj.onc.1202043. [DOI] [PubMed] [Google Scholar]
- Deltour S, Guerardel C, Leprince D. Recruitment of SMRT/N-CoR-mSin3A-HDAC-repressing complexes is not a general mechanism for BTB/POZ transcriptional repressors: The case of HIC-1 and gammaFBP-B. Proc Natl Acad Sci. 1999;96:14831–14836. doi: 10.1073/pnas.96.26.14831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dent AL, Shaffer AL, Yu X, Allman D, Staudt LM. Control of inflammation, cytokine expression, and germinal center formation by BCL-6. Science. 1997;276:589–592. doi: 10.1126/science.276.5312.589. [DOI] [PubMed] [Google Scholar]
- Deweindt C, Albagli O, Bernardin F, Dhordain P, Quief S, Lantoine D, Kerckaert JP, Leprince D. The LAZ3/BCL6 oncogene encodes a sequence-specific transcriptional inhibitor: A novel function for the BTB/POZ domain as an autonomous repressing domain. Cell Growth Differ. 1995;6:1495–1503. [PubMed] [Google Scholar]
- Dhordain P, Albagli O, Lin RJ, Ansieau S, Quief S, Leutz A, Kerckaert JP, Evans RM, Leprince D. Corepressor SMRT binds the BTB/POZ repressing domain of the LAZ3/BCL6 oncoprotein. Proc Natl Acad Sci. 1997;94:10762–10767. doi: 10.1073/pnas.94.20.10762. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhordain P, Lin RJ, Quief S, Lantoine D, Kerckaert JP, Evans RM, Albagli O. The LAZ3(BCL-6) oncoprotein recruits a SMRT/mSIN3A/histone deacetylase containing complex to mediate transcriptional repression. Nucleic Acids Res. 1998;26:4645–4651. doi: 10.1093/nar/26.20.4645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espinas ML, Jimenez-Garcia E, Vaquero A, Canudas S, Bernues J, Azorin F. The N-terminal POZ domain of GAGA mediates the formation of oligomers that bind DNA with high affinity and specificity. J Biol Chem. 1999;274:16461–16469. doi: 10.1074/jbc.274.23.16461. [DOI] [PubMed] [Google Scholar]
- Fischle W, Emiliani S, Hendzel MJ, Nagase T, Nomura N, Voelter W, Verdin E. A new family of human histone deacetylases related to Saccharomyces cerevisiae HDA1p. J Biol Chem. 1999;274:11713–11720. doi: 10.1074/jbc.274.17.11713. [DOI] [PubMed] [Google Scholar]
- Fukuda T, Yoshida T, Okada S, Hatano M, Miki T, Ishibashi K, Okabe S, Koseki H, Hirosawa S, Taniguchi M, et al. Disruption of the Bcl6 gene results in an impaired germinal center formation. J Exp Med. 1997;186:439–448. doi: 10.1084/jem.186.3.439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grignani F, De Matteis S, Nervi C, Tomassoni L, Gelmetti V, Cioce M, Fanelli M, Ruthardt M, Ferrara FF, Zamir I, et al. Fusion proteins of the retinoic acid receptor-alpha recruit histone deacetylase in promyelocytic leukaemia. Nature. 1998;391:815–818. doi: 10.1038/35901. [DOI] [PubMed] [Google Scholar]
- Grozinger CM, Hassig CA, Schreiber SL. Three proteins define a class of human histone deacetylases related to yeast Hda1p. Proc Natl Acad Sci. 1999;96:4868–4873. doi: 10.1073/pnas.96.9.4868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harris MB, Chang CC, Berton MT, Danial NN, Zhang J, Kuehner D, Ye BH, Kvatyuk M, Pandolfi PP, Cattoretti G, et al. Transcriptional repression of Stat6-dependent interleukin-4-induced genes by BCL-6: Specific regulation of iepsilon transcription and immunoglobulin E switching. Mol Cell Biol. 1999;19:7264–7275. doi: 10.1128/mcb.19.10.7264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heinzel T, Lavinsky RM, Mullen TM, Soderstrom M, Laherty CD, Torchia J, Yang WM, Brard G, Ngo SD, Davie JR, et al. A complex containing N-CoR, mSin3 and histone deacetylase mediates transcriptional repression. Nature. 1997;387:43–48. doi: 10.1038/387043a0. [DOI] [PubMed] [Google Scholar]
- Hong SH, David G, Wong CW, Dejean A, Privalsky ML. SMRT corepressor interacts with PLZF and with the PML-retinoic acid receptor alpha (RARalpha) and PLZF-RARalpha oncoproteins associated with acute promyelocytic leukemia. Proc Natl Acad Sci. 1997;94:9028–9033. doi: 10.1073/pnas.94.17.9028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong SH, Wong CW, Privalsky ML. Signaling by tyrosine kinases negatively regulates the interaction between transcription factors and SMRT (silencing mediator of retinoic acid and thyroid hormone receptor) corepressor. Mol Endocrinol. 1998;12:1161–1171. doi: 10.1210/mend.12.8.0160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang EY, Zhang J, Miska EA, Guenther MG, Kouzarides T, Lazar MA. Nuclear receptor corepressors partner with class II histone deacetylases in a Sin3-independent repression pathway. Genes & Dev. 2000;14:45–54. [PMC free article] [PubMed] [Google Scholar]
- Huynh KD, Bardwell VJ. The BCL-6 POZ domain and other POZ domains interact with the co-repressors N-CoR and SMRT. Oncogene. 1998;17:2473–2484. doi: 10.1038/sj.onc.1202197. [DOI] [PubMed] [Google Scholar]
- Kao HY, Downes M, Ordentlich P, Evans RM. Isolation of a novel histone deacetylase reveals that class I and class II deacetylases promote SMRT-mediated repression. Genes & Dev. 2000;14:55–66. [PMC free article] [PubMed] [Google Scholar]
- Katsani KR, Hajibagheri MA, Verrijzer CP. Co-operative DNA binding by GAGA transcription factor requires the conserved BTB/POZ domain and reorganizes promoter topology. EMBO J. 1999;18:698–708. doi: 10.1093/emboj/18.3.698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerckaert JP, Deweindt C, Tilly H, Quief S, Lecocq G, Bastard C. LAZ3, a novel zinc-finger encoding gene, is disrupted by recurring chromosome 3q27 translocations in human lymphomas. Nat Genet. 1993;5:66–70. doi: 10.1038/ng0993-66. [DOI] [PubMed] [Google Scholar]
- Knoepfler PS, Eisenman RN. Sin meets NuRD and other tails of repression. Cell. 1999;99:447–450. doi: 10.1016/s0092-8674(00)81531-7. [DOI] [PubMed] [Google Scholar]
- Kumagai T, Miki T, Kikuchi M, Fukuda T, Miyasaka N, Kamiyama R, Hirosawa S. The proto-oncogene Bc16 inhibits apoptotic cell death in differentiation-induced mouse myogenic cells. Oncogene. 1999;18:467–475. doi: 10.1038/sj.onc.1202306. [DOI] [PubMed] [Google Scholar]
- Li X, Peng H, Schultz DC, Lopez-Guisa JM, Rauscher FJ, III, Marmorstein R. Structure-function studies of the BTB/POZ transcriptional repression domain from the promyelocytic leukemia zinc finger oncoprotein. Cancer Res. 1999;59:5275–5282. [PubMed] [Google Scholar]
- Lin RJ, Nagy L, Inoue S, Shao W, Miller WH, Jr, Evans RM. Role of the histone deacetylase complex in acute promyelocytic leukaemia. Nature. 1998;391:811–814. doi: 10.1038/35895. [DOI] [PubMed] [Google Scholar]
- Lo Coco F, Ye BH, Lista F, Corradini P, Offit K, Knowles DM, Chaganti RS, Dalla-Favera R. Rearrangements of the BCL6 gene in diffuse large cell non-Hodgkin's lymphoma. Blood. 1994;83:1757–1759. [PubMed] [Google Scholar]
- Miki T, Kawamata N, Arai A, Ohashi K, Nakamura Y, Kato A, Hirosawa S, Aoki N. Molecular cloning of the breakpoint for 3q27 translocation in B-cell lymphomas and leukemias. Blood. 1994;83:217–222. [PubMed] [Google Scholar]
- Miska EA, Karlsson C, Langley E, Nielsen SJ, Pines J, Kouzarides T. HDAC4 deacetylase associates with and represses the MEF2 transcription factor. EMBO J. 1999;18:5099–5107. doi: 10.1093/emboj/18.18.5099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagy L, Kao HY, Chakravarti D, Lin RJ, Hassig CA, Ayer DE, Schreiber SL, Evans RM. Nuclear receptor repression mediated by a complex containing SMRT, mSin3A, and histone deacetylase. Cell. 1997;89:373–380. doi: 10.1016/s0092-8674(00)80218-4. [DOI] [PubMed] [Google Scholar]
- Numoto M, Niwa O, Kaplan J, Wong KK, Merrell K, Kamiya K, Yanagihara K, Calame K. Transcriptional repressor ZF5 identifies a new conserved domain in zinc finger proteins. Nucleic Acids Res. 1993;21:3767–3775. doi: 10.1093/nar/21.16.3767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okabe S, Fukuda T, Ishibashi K, Kojima S, Okada S, Hatano M, Ebara M, Saisho H, Tokuhisa T. BAZF, a novel Bcl6 homolog, functions as a transcriptional repressor. Mol Cell Biol. 1998;18:4235–4244. doi: 10.1128/mcb.18.7.4235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Onizuka T, Moriyama M, Yamochi T, Kuroda T, Kazama A, Kanazawa N, Sato K, Kato T, Ota H, Mori S. BCL-6 gene product, a 92- to 98-kD nuclear phosphoprotein, is highly expressed in germinal center B cells and their neoplastic counterparts. Blood. 1995;86:28–37. [PubMed] [Google Scholar]
- Otsuki T, Yano T, Clark HM, Bastard C, Kerckaert JP, Jaffe ES, Raffeld M. Analysis of LAZ3 (BCL-6) status in B-cell non-Hodgkin's lymphomas: Results of rearrangement and gene expression studies and a mutational analysis of coding region sequences. Blood. 1995;85:2877–2884. [PubMed] [Google Scholar]
- Rundlett SE, Carmen AA, Kobayashi R, Bavykin S, Turner BM, Grunstein M. HDA1 and RPD3 are members of distinct yeast histone deacetylase complexes that regulate silencing and transcription. Proc Natl Acad Sci. 1996;93:14503–14508. doi: 10.1073/pnas.93.25.14503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rundlett SE, Carmen AA, Suka N, Turner BM, Grunstein m. Transcriptional repression by UME6 involves deacetylation of lysine 5 of histone H4 by RPD3. Nature. 1998;392:831–835. doi: 10.1038/33952. [DOI] [PubMed] [Google Scholar]
- Seyfert VL, Allman D, He Y, Staudt LM. Transcriptional repression by the proto-oncogene BCL-6. Oncogene. 1996;12:2331–2342. [PubMed] [Google Scholar]
- Skinner PJ, Koshy BT, Cummings CJ, Klement IA, Helin K, Servadio A, Zoghbi HY, Orr HT. Ataxin-1 with an expanded glutamine tract alters nuclear matrix-associated structures. Nature. 1997;389:971–974. doi: 10.1038/40153. [DOI] [PubMed] [Google Scholar]
- Stein H, Dallenbach F. In: Neoplastic hematopathology. Knowles DM, editor. Baltimore, MD: Williams and Wilkins; 1992. p. 675. [Google Scholar]
- Taunton J, Hassig CA, Schreiber SL. A mammalian histone deacetylase related to the yeast transcriptional regulator Rpd3p. Science. 1996;272:408–411. doi: 10.1126/science.272.5260.408. [DOI] [PubMed] [Google Scholar]
- Verdel A, Khochbin S. Identification of a new family of higher eukaryotic histone deacetylases. Coordinate expression of differentiation-dependent chromatin modifiers. J Biol Chem. 1999;274:2440–2445. doi: 10.1074/jbc.274.4.2440. [DOI] [PubMed] [Google Scholar]
- Wang AH, Bertos NR, Vezmar M, Pelletier N, Crosato M, Heng HH, Th'ng J, Han J, Yang XJ. HDAC4, a human histone deacetylase related to yeast HDA1, is a transcriptional corepressor. Mol Cell Biol. 1999;19:7816–7827. doi: 10.1128/mcb.19.11.7816. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkinson JR, Towle HC. Identification and characterization of the AF-1 transactivation domain of thyroid hormone receptor beta1. J Biol Chem. 1997;272:23824–23832. doi: 10.1074/jbc.272.38.23824. [DOI] [PubMed] [Google Scholar]
- Wong CW, Privalsky ML. Components of the SMRT corepressor complex exhibit distinctive interactions with the POZ domain oncoproteins PLZF, PLZF-RARalpha, and BCL-6. J Biol Chem. 1998;273:27695–27702. doi: 10.1074/jbc.273.42.27695. [DOI] [PubMed] [Google Scholar]
- Yamochi T, Kaneita Y, Akiyama T, Mori S, Moriyama M. Adenovirus-mediated high expression of BCL-6 in CV-1 cells induces apoptotic cell death accompanied by down-regulation of BCL-2 and BCL- X(L) Oncogene. 1999;18:487–494. doi: 10.1038/sj.onc.1202334. [DOI] [PubMed] [Google Scholar]
- Yang WM, Inouye C, Zeng Y, Bearss D, Seto E. Transcriptional repression by YY1 is mediated by interaction with a mammalian homolog of the yeast global regulator RPD3. Proc Natl Acad Sci. 1996;93:12845–12850. doi: 10.1073/pnas.93.23.12845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang WM, Yao YL, Sun JM, Davie JR, Seto E. Isolation and characterization of cDNAs corresponding to an additional member of the human histone deacetylase gene family. J Biol Chem. 1997;272:28001–28007. doi: 10.1074/jbc.272.44.28001. [DOI] [PubMed] [Google Scholar]
- Ye BH, Rao PH, Chaganti RS, Dalla-Favera R. Cloning of bcl-6, the locus involved in chromosome translocations affecting band 3q27 in B-cell lymphoma. Cancer Res. 1993;53:2732–2735. [PubMed] [Google Scholar]
- Ye BH, Chaganti S, Chang CC, Niu H, Corradini P, Chaganti RS, Dalla-Favera R. Chromosomal translocations cause deregulated BCL6 expression by promoter substitution in B cell lymphoma. EMBO J. 1995;14:6209–6217. doi: 10.1002/j.1460-2075.1995.tb00311.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ye BH, Cattoretti G, Shen Q, Zhang J, Hawe N, de Waard R, Leung C, Nouri-Shirazi M, Orazi A, Chaganti RS, et al. The BCL-6 proto-oncogene controls germinal-centre formation and Th2- type inflammation. Nat Genet. 1997;16:161–170. doi: 10.1038/ng0697-161. [DOI] [PubMed] [Google Scholar]
- Zollman S, Godt D, Prive GG, Couderc JL, Laski FA. The BTB domain, found primarily in zinc finger proteins, defines an evolutionarily conserved family that includes several developmentally regulated genes in Drosophila. Proc Natl Acad Sci. 1994;91:10717–10721. doi: 10.1073/pnas.91.22.10717. [DOI] [PMC free article] [PubMed] [Google Scholar]