Abstract
Salmonella enteritidis has emerged as one of the most important food-borne pathogens for humans, and the formation of biofilms by this species may improve its resistance to disadvantageous conditions. The spiA gene of Salmonella typhimurium is essential for its virulence in host cells. However, the roles of the spiA gene in biofilm formation and virulence of S. enteritidis remain unclear. In this study we constructed a spiA gene mutant with a suicide plasmid. Phenotypic and biological analysis revealed that the mutant was similar to the wild-type strain in growth rate, morphology, and adherence to and invasion of epithelial cells. However, the mutant showed reduced biofilm formation in a quantitative microtitre assay and by scanning electron microscopy, and significantly decreased curli production and intracellular proliferation of macrophages during the biofilm phase. In addition, the spiA mutant was attenuated in a mouse model in both the exponential growth and biofilm phases. These data indicate that the spiA gene is involved in both biofilm formation and virulence of S. enteritidis.
Introduction
Salmonella enterica serovar Enteritidis (Salmonella enteritidis) has emerged as one of the most important food-borne pathogens for humans; it is mainly associated with consumption of contaminated poultry meat and eggs (Patrick et al., 2004). A number of studies have demonstrated that Salmonella bacteria are capable of forming biofilms on a wide variety of contact surfaces (Korber et al., 1997; Römling et al., 2003), and the formation of biofilms may improve the ability of these organisms to resist stresses such as desiccation, extreme temperatures, antibiotics and antiseptics (Marin et al., 2009; Scher et al., 2005). Biofilm formation allows Salmonella to survive long term in the poultry farm environment and to contaminate poultry meat and eggs, which remain the leading vehicles of food-borne salmonellosis outbreaks (Joseph et al., 2001).
In recent years, many groups have dedicated great effort to identifying the factors involved in biofilm development. Curli and cellulose are the major components of biofilm formation in Salmonella (Römling et al., 1998b; White et al., 2003; Zogaj et al., 2001); capsular polysaccharide (de Rezende et al., 2005), other polysaccharides such as LPS (Anriany et al., 2006) and a large secreted protein, BapA, also contribute to biofilm formation (Latasa et al., 2005). Several regulatory genes are involved in biofilm formation. For example, the csgD gene encodes a transcriptional regulator of the LuxR superfamily and has been shown to critically control expression of curli and cellulose (Gerstel & Römling, 2003), which are positively regulated by the ompR and rpoS genes (Römling et al., 1998a). Hamilton et al. (2009) determined the transcriptomic and proteomic profiles of Salmonella typhimurium biofilms and found that a functional SPI2 secretion system regulator (ssrA), an STM0341 gene of unknown function, and a trpE gene that is involved in tryptophan biosynthesis and transport were needed for biofilm growth. Recently, we identified several genes associated with biofilm formation by transposon mutagenesis in Salmonella (Dong et al., 2008). However, the detailed roles of some of the genes identified to be involved in biofilm formation remain unclear. In this study, the roles of the spiA gene in biofilm formation and virulence were investigated.
Methods
Strains, plasmids and growth conditions.
The bacterial strains and plasmids used in this study are listed in Table 1. Bacteria were grown in rich liquid or solid (15 g agar l−1) Luria broth (LB) medium with antibiotic supplementation as needed, such as ampicillin (Amp, 100 µg ml−1), kanamycin (Km, 25 µg ml−1) and streptomycin (Sm, 100 µg ml−1). Nutrient agar (NA, solid LB medium without NaCl) and nutrient broth (NB, liquid LB medium without NaCl) with 10 % sucrose were used during the allelic-exchange experiments. Growth rates of the wild-type strain and the mutant were measured as follows. An overnight culture was inoculated in 10 ml LB medium (adjusted to OD600 0.1) and shaken at 200 r.p.m. at 37 °C. The bacterial cultures were monitored spectrophotometrically at 600 nm every hour for 7 h. For preparation of bacteria in the exponential phase of growth, the bacteria were streaked on LB agar, incubated at 37 °C for 4–5 h, washed twice with PBS and suspended in PBS. For preparation of bacteria in biofilm growth, overnight broth cultures were diluted 1 : 10 in cell culture flasks and incubated at 28 °C for 48 h. The biofilm cells that formed on flasks were dislodged by vigorous vortexing with glass beads in PBS and then resuspended in PBS.
Table 1. Summary of the strains, plasmids and primers used.
| Strain, plasmid or primer | Description/purpose | Reference or source |
| E. coli strains | ||
| C50041 | Wild-type strain | Qiuchun et al. (2009) |
| χ7213 | For cloning pGMB151 | Kang et al. (2002) |
| S17-λpir (SmR) | For cloning pGMB151 | Huang et al. (2004) |
| ΔspiA | spiA mutant | This study |
| ΔspiAR | Complementation of ΔspiA | This study |
| Plasmids | ||
| pUC18(Ampr) | Plasmid for cloning | Promega |
| pMD18-T | Plasmid for cloning | Takara |
| pGMB151(Ampr Smr) | Suicide plasmid | Huang et al. (2004) |
| pBluescript II SK(−) | Plasmid for cloning | Fermentas |
| pUC4K | Plasmid for kanamycin-resistance gene | Amersham |
| pGEX-6P-1 | Plasmid for spiA gene restoration and csgA gene expression | Amersham |
| Primers | ||
| 12 | 5′-CTCGGATCCCACTATGTACTTGAGTCGTATCATTGCG-3′ (spiA sense, BamHI site underlined) | This study |
| 13 | 5′-CTCGTCGACCCCTTTATGCCAGACAAATGCCA-3′ (spiA antisense, SalI site underlined) | This study |
| 14 | 5′-CTCGAATTCGCGTAAGTAATGAATGAACCGTCC-3′ [pBluescript II SK(−)+spiA1 sense, EcoRI site underlined] | This study |
| 15 | 5′-CTCGAATTCCGGGTTCTTTGTTGCGCGTT-3′ [pBluescript II SK(−)+spiA2 antisense, EcoRI site underlined] | This study |
| 16 | 5′-CAGTGACAGGTTACCTTCATTCAGC-3′ (spiA sense, spiA design primer) | This study |
| 17 | 5′-CTCGGATCCCCCTTTATGCCAGACAAATGCCA-3′ (spiA antisense, BamHI site underlined) | This study |
| 18 | 5′-GAATTCTCAAAGCCACGTTGTGTCTCAAA-3′ (kanamycin sense, EcoRI site underlined) | This study |
| 19 | 5′-GAATTCCGCTGAGGTCTGCCTCGT-3′ (kanamycin antisense, EcoRI site underlined) | This study |
| 20 | 5′-TGCAAGTTAAAGCCAGGTG-3′ (spiC sense) | This study |
| 21 | 5′-CCGAAGGTAATAGCCGATCC-3′ (spiC antisense) | This study |
| 22 | 5′-GAGGTATCGCTGATTACCGTTG-3′ (spiA sense) | This study |
| 23 | 5′-CGTGAGTGTATTGCGTAGGATG-3′ (spiA antisense) | This study |
| 24 | 5′-CACTGGCGCAGGCAGATAC-3′ (ssaD sense) | This study |
| 25 | 5′-GATCAGCAATGGCGTAAGGTTC-3′ (ssaD antisense) | This study |
| 26 | 5′-CTCGGATCCATGGTAGTAAATAAACGTTTAATC-3′ (spiA complementation sense, BamHI site underlined) | This study |
| 27 | 5′-CTCCTCGAGTTAACCATGAGATATGCCATTATTTAC-3′ (spiA complementation antisense, XhoI site underlined) | This study |
Construction of the suicide plasmid, pGMB151-spiA-Kan.
The spiA gene was amplified from the chromosomal DNA from S. enteritidis strain C50041 by using primers 12/13 (see Table 1 for sequences of all primers used). The PCR product was purified and cloned into a pSK(−) vector to obtain pSK-spiA. The SK-spiA fragment was amplified from pSK-spiA by using reverse PCR with primers 14/15. The fragment was then ligated to a kanamycin-resistance cassette (Kan) amplified from pUC4K with primers 18/19 to produce pSK-spiA-Kan, which resulted in insertion of the kanamycin gene into the spiA gene with a 767 bp deletion. The PCR product of spiA-Kan, amplified from pSK-spiA-Kan with primers 12 /17, was digested with BamHI and subsequently inserted into the BamHI site of the suicide plasmid, pGMB151, to form pGMB151-spiA-Kan. After verification by sequence analysis, the resulting plasmid was transferred to Escherichia coli S17-λpir (Smr) and further transferred to E. coli χ7213 as donor strain E. coli χ7213(pGMB151-spiA-Kan).
Construction of a spiA mutant by allelic exchange and complementation of spiA.
The donor strain, E. coli χ7213(pGMB151-spiA-Kan), and the recipient strain, C50041, were grown in LB containing 1 % diaminopimelic acid with shaking at 37 °C to an OD600 of 0.6–0.7. The donor strain and the heat-treated recipient strain were mixed in 10 mM MgSO4, the mixture was immobilized on a 0.45 µm membrane filter and the filter was incubated on LB agar at 30 °C for 18 h. The transconjugants were spread onto LB agar containing Amp, Sm and Km. Colonies were then streaked onto NA with Amp, Sm, Km and sucrose to select bacteria that were sensitive to sucrose (Geng et al., 2009). A single bacterial colony that was sensitive to sucrose was subcultured 8–10 times on NB containing 10 mM MgSO4, 10 % sucrose and Km. The Kmr mutants without Ampr and Smr were screened on LB plates containing Km. Finally, the spiA mutant was confirmed by primers 13/16 and named ΔspiA. For gene complementation, spiA was amplified from the C50041 strain with primers 26/27. The PCR product was cloned into pGEX-6P-1, and the ligation mixture was transformed into E. coli BL21. Plasmid p6P-spiA was isolated from the Ampr transformants, and the presence of an insert was verified by restriction analysis and sequence analysis. Plasmid p6P-spiA was transformed into ΔspiA to create a spiA-complemented strain, ΔspiAR.
RT-PCR.
Total RNA was isolated from exponential phase bacteria of strains C50041 and ΔspiA by the hot phenol method. The RNA was treated with DNase I (Takara) and used as a template for reverse transcription (RT) with random primers. The primer sets for cDNA amplification of target genes spiC, spiA and ssaD are given in Table 1. The PCR products were resolved on 1 % agarose gels and visualized by ethidium bromide staining.
SDS-PAGE.
A single colony each of ΔspiA and C50041 was inoculated into 10 ml LB and shaken at 37 °C for 14 h. The supernatant was collected by centrifugation at 12 000 r.p.m. for 1 min and put on ice for approximately 30 min after the addition of 1 ml trichloroacetic acid. The proteins were harvested by centrifugation at 12 000 r.p.m. for 10 min, washed twice with 5 ml acetone, dried at 4 °C, dissolved in 50 µl PBS and examined by SDS-PAGE on a 10 % separating gel with 40 µg protein.
Biofilm formation.
A quantitative microtitre assay was performed as described by Pratt & Kolter (1998). The morphology of the biofilm formed was also determined by scanning electron microscopy as described by Anriany et al. (2001), with minor modifications. Strains ΔspiA and C50041 were inoculated in 3 ml LB in a six-well cell-culture plate containing polystyrene coverslips (0.5 cm×0.5 cm) and incubated at 28 °C for 48 h without shaking. The slips with bacterial cells were removed, fixed in 3 % glutaraldehyde-PBS (pH 7.0) for 2 h, gently washed three times with PBS (pH 7.0), and dehydrated through graded ethanol/water mixtures with 30 % (15 min), 50 % (15 min), 70 % (15 min), 80 % (15 min), 90 % (15 min) and 100 % (15 min×3) ethanol. Dried chips were then immersed in ethanol/isoamyl acetate (1 : 1 and 2 : 1) for 30 min and then transferred to isoamyl acetate for 30 min. They were dried by critical-point drying, coated with gold–palladium alloy and observed with a scanning electron microscope (S4800; Hitachi).
Determination of curli and cellulose.
To determine whether ΔspiA produced curli and/or cellulose, 10 µl samples of ΔspiA and the wild-type strain, C50041, cultured overnight, were dropped on LB agar lacking NaCl, supplemented with Congo red (40 µg ml−1; Sigma) and Coomassie brillliant blue G (20 µg ml−1; Sigma), and incubated at 28 °C for 48 h. The morphology was then examined. In a separate experiment, production of cellulose by ΔspiA and C50041 was determined by inoculating 10 µl samples on NA supplemented with 200 mg Calcofluor ml−1 (fluorescent brightener 28; Sigma) at 28 °C for 48 h, and comparing the fluorescence of ΔspiA and C50041 under UV light (366 nm). Production of curli protein was determined as described by Anriany et al. (2006). One millilitre of suspension with ΔspiA, C50041 and ΔspiAR in the biofilm phase (OD600 3.0) was centrifuged, and the cell pellets were resuspended in 100 µl SDS sample buffer [62.5 mM Tris/HCl (pH 6.8), 10 % glycerol, 2 % SDS] and boiled for 10 min. The cell lysate was centrifuged, and the pellet was washed twice with sterile water, dissolved in 100 µl 97 % formic acid (Sigma), frozen at −70 °C and lyophilized. The samples were resuspended in 100 µl SDS sample buffer, sonicated for 5 s and examined by SDS-PAGE. Separated proteins were transferred onto a nitrocellulose membrane. Western blotting was performed using mouse antisera specific against the curli major subunit, CsgA, which was prepared in mice immunized with an expression product of pGEX-6P-1-csgA.
Adherence and invasion assay.
The A549 human epithelial cell line was cultured in RPMI 1640 with 10 % fetal calf serum at 37 °C in 5 % CO2 (Monack et al., 1996). Briefly, a 1 ml portion with 1×105 cells was seeded into each well of 24-well plates and incubated for 16 h. Cell cultures of strains ΔspiA and C50041 in the exponential and biofilm phase of growth were suspended in PBS and adjusted to OD600 0.4. The mean concentrations of strains C50041 and ΔspiA in exponential phase growth were 3.6×108 and 3.8×108 c.f.u. ml−1, respectively, while the concentrations in the biofilm phase were approximately 4.5×108 and 4.8×108 c.f.u. ml−1, respectively, according to viable bacteria counting. Suspensions with an m.o.i. of 100 were inoculated in duplicate in a 24-well plate. This plate was centrifuged for 5 min at 100 g and then incubated at 37 °C for 2 h, the wells were rinsed three times with PBS, and the cells were released from the plate by adding 0.05 % trypsin. For the invasion assay, the cells were further cultured with 100 µg gentamicin ml−1 for 1 h and lysed with 1 % Triton X-100. Both cell suspensions were serially diluted in PBS and spread onto LB plates to determine the number of viable bacteria. Adherence and invasion were expressed as the percentage of bacteria that were attached and that invaded the cells relative to the original number of bacteria added to the well. The experiments were repeated in three independent assays.
Intracellular growth rate assay.
For this assay, a mouse macrophage cell line, RAW264.7, was used and the method was the same as that used for the adherence assay. Cell cultures of strains ΔspiA and C50041 in the exponential and biofilm phases were diluted in PBS to allow for infection at an m.o.i. of 10. Infection cultures were incubated for 2 h, rinsed three times in PBS and incubated with 100 µg gentamicin ml−1 for 1 h. The intracellular bacteria were counted as above. For determination of intracellular bacterial proliferation, cells were incubated for an additional 20 h with 10 µg gentamicin ml−1. The experiments were repeated in three independent assays.
Determination of LD50 in mice.
BALB/c mice (6 weeks of age) were obtained from the experimental animal centre of Yangzhou University. The mice were housed in an animal facility under a standard animal study protocol. Four separate groups of mice (each containing 30 mice) were infected with ΔspiA and C50041 in the exponential and biofilm phases. Mice in each group were further subdivided into five subgroups, each containing six mice. Based on the results of a pilot study, each mouse in the ΔspiA groups was injected intraperitoneally with 0.2 ml of 2×108, 2×107, 2×106, 2×105 and 2×104 c.f.u., respectively, and each mouse in the C50041 groups was injected intraperitoneally with 0.2 ml of 2×104, 2×103, 2×102, 2×101 and 2×100 c.f.u., respectively. Deaths were recorded up to day 14, and the LD50 of each strain was calculated by the method of Reed & Muench (1938).
Distribution of ΔspiA and C50041 in mice.
Strains ΔspiA and C50041 in the exponential and biofilm phases were suspended in PBS and adjusted to 107 c.f.u. ml−1. Six-week-old BALB/c mice were divided into four groups (five per group) and each mouse was administered intraperitoneally with a 0.2 ml bacterial suspension. The numbers of bacteria present in the blood, liver, spleen and lungs of the mice were measured 6 and 48 h post-challenge.
Statistical analysis.
The numbers of viable bacteria (c.f.u.) in mice were expressed as the geometric mean±sd. Significance was analysed by using a two-tailed independent Student’s t-test. The adherence, invasion and intracellular growth percentages were analysed by chi-squared tests.
Results
Construction and identification of the ΔspiA mutant
The spiA gene of strain C50041 was amplified with primers 12/13 and cloned into pMD18-T. Nucleotide sequence analysis showed that this DNA fragment contained a single open reading frame of 1494 bp. A ΔspiA mutant was constructed by allelic exchange with a substitution of a kanamycin-resistance cassette and a 767 bp deletion within the spiA coding region. A 1.93 kb fragment amplified with primers 16/13 identified the ΔspiA mutation including the kanamycin-resistance cassette. Sequence analysis of the PCR product confirmed that the kanamycin-resistance cassette had been inserted into spiA of C50041 chromosomal DNA at the predicted position.
To determine whether the insertion had a polar effect on the downstream gene, total RNA isolated from both ΔspiA and C50041 was subjected to RT-PCR analysis using primer sets for spiC (primers 20/21), spiA (primers 22/23), and ssaD (primers 24/25). Compared with the wild-type strain, C50041, insertion of the kanamycin resistance cassette in ΔspiA disrupted only the transcription of the spiA gene and had no effect on transcription of the upstream spiC gene or the downstream ssaD gene (data not shown).
Growth rate and protein pattern of ΔspiA
When grown in LB, the growth rate of ΔspiA was similar to that of C50041 in the exponential phase of growth (P>0.05; Fig. 1a). Comparative analysis of the protein profile indicated that a 45 kDa protein appeared in the supernatant of C50041, while a 47 kDa protein appeared in that of ΔspiA (Fig. 1b). Mass spectrometry analysis (Fitgene, China) showed that both proteins included flagellin peptide residues, which is a similar protein pattern to that described by Ochman et al. (1996).
Fig. 1.
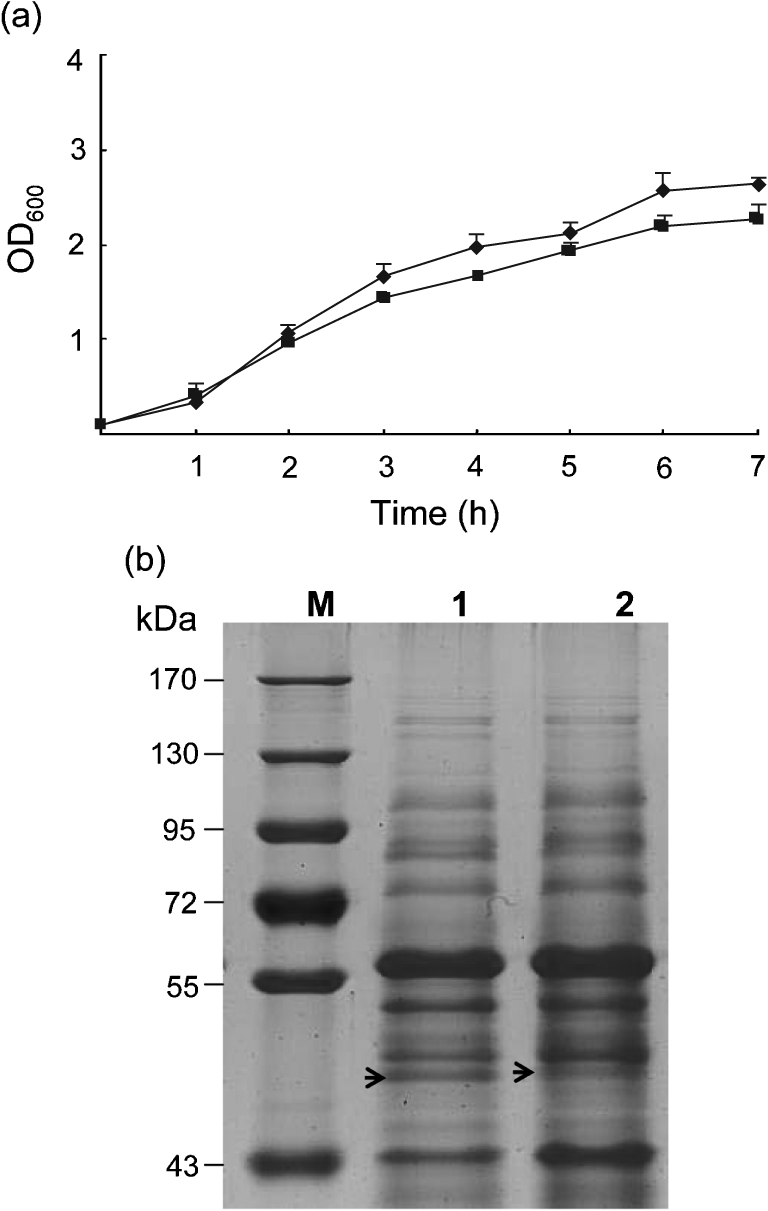
Growth curves and protein patterns of the wild-type strain, C50041, and the spiA mutant, ΔspiA. (a) Cultures of C50041 (⧫) and ΔspiA (▪) were monitored spectrophotometrically at 600 nm hourly for 7 h. (b) Proteins (40 µg samples) prepared from the supernatants of C50041 (lane 1) and ΔspiA (lane 2) cultures were subjected to SDS-PAGE. Prestained protein markers are indicated on the left (lane M).
Biofilm formation by ΔspiA
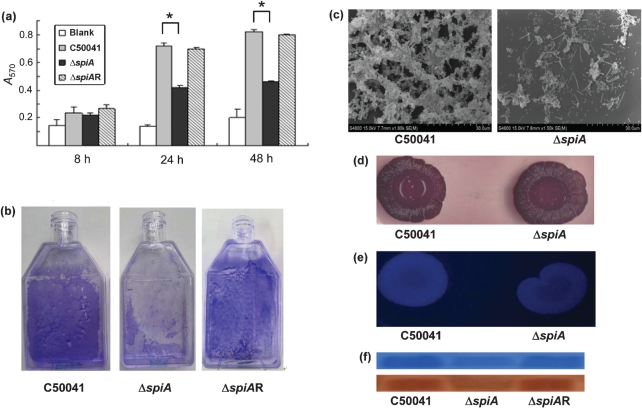
To investigate the effect of the loss of a functional spiA gene on biofilm formation, ΔspiA was tested for biofilm-associated biological activity. In a quantitative microtitre assay, the AOD value of ΔspiA was significantly lower than that of the wild-type strain, C50041, at 24 and 48 h post-inoculation (Fig. 2a). When cultured in a flask, both ΔspiA and C50041 formed biofilms on the flask wall, but the biofilm of ΔspiA was less adherent than that of C50041 (Fig. 2b). In contrast, a spiA complementation strain, ΔspiAR, regained the same biofilm characteristics as the wild-type strain (Fig. 2a, b). Scanning electron microscopy of biofilm formation by the strains showed that ΔspiA produced a loose and thin structure with fewer bacteria, while C50041 produced a very tenacious and thick structure with extracellular substances and fibrils that seemed to bind the cells together (Fig. 2c).
Fig. 2.
Determination of biofilm formation and its components in the wild-type strain (C50041), the spiA mutant (ΔspiA) and the spiA-complemented strain (ΔspiAR). (a) A570 values of crystal-violet-stained C50041, ΔspiA and ΔspiAR biofilms grown in 96-well plates. (b) Overnight broth cultures were diluted 1 : 10 in cell-culture flasks. After incubation at 28 °C for 48 h, the broth was removed and the biofilm cells were stained with crystal violet. (c) Scanning electron micrographs of cells of the wild-type strain C50041 and ΔspiA grown on polystyrene coverslips at 28 °C for 48 h. (d, e) Morphotypes of C50041 and ΔspiA grown on plates of LB agar lacking NaCl and supplemented with Congo red and Coomassie brilliant blue G (d) or with Calcofluor (e). (f) Biofilm cells of strains C50041, ΔspiA and ΔspiAR with the same optical densities (OD600 3.0) were collected and treated. Curli proteins were examined by SDS-PAGE with 15 % separating gel (top) and confirmed by Western blotting (bottom).
Because biofilms of Salmonella are mainly composed of cellulose and curli (Römling et al., 1998b; White et al., 2003; Zogaj et al., 2001), ΔspiA was tested on LB agar lacking NaCl and supplemented with Congo red and Coomassie brilliant blue. Strain ΔspiA formed red, dry and rough colonies, similar to those of the wild-type strain (Fig. 2d). The presence of cellulose was further tested with Calcofluor. The fluorescence of ΔspiA was similar in brightness to that of C50041 (Fig. 2e), indicating that ΔspiA still produced cellulose. Comparative analysis of curli proteins by SDS-PAGE showed that both strains produced a 15.3 kDa band (the major subunit of curli protein; Fig. 2f). However, the intensity of the band produced by ΔspiA was between 47.0 and 60.5 % of that produced by C50041, according to the Quantity One software (Bio-Rad), indicating that ΔspiA produced fewer curli. SDS-PAGE and Western blotting confirmed that the ΔspiAR regained a similar level of expression of the curli protein as the wild-type strain.
Biological activity of ΔspiA
To test the adherence and invasion of ΔspiA of A549 human epithelial cells, bacteria in both the exponential and the biofilm phases of growth were used (Table 2). The percentages of adherence and invasion of A549 cells by ΔspiA in the biofilm phase were higher than those in the exponential growth phase. Lack of expression of the spiA gene caused no reduction in adherence to and invasion of A549 cell lines compared with those of the wild-type strain.
Table 2. Adherence to and invasion of A549 cells by strains C50041 and ΔspiA.
Values are mean±sd.
To test the proliferation capacity of ΔspiA in mouse macrophage-like RAW264.7 cells, both bacteria in the exponential growth and biofilm phases were used (Table 3). For bacteria in the exponential growth phase, the invasion and proliferation ratios of the two strains were similar at 3 and 23 h post-infection. However, for bacteria in the biofilm phase, the proliferation ratio of ΔspiA increased 24.7-fold from 3 to 23 h post-infection, which was significantly lower than that of the wild-type strain (44.29-fold; P<0.05). When comparing the two strains in different conditions, strains in the biofilm phase were more invasive than those in the exponential growth phase.
Table 3. Intramacrophage survival properties of strains C50041 and ΔspiA.
Values are mean±sd.
| Strain | Growth phase | Percentage invasion (3 h) | Percentage proliferation (23 h) | Proliferation ratio (23 h/3 h) |
| C50041 | Exponential | 1.43±0.25 | 35±7.1 | 24.5±0.71 |
| ΔspiA | 1.15±0.21 | 34±4.9 | 29.23±1.09 | |
| C50041 | Biofilm | 29±3.5 | 1273±3.22 | 44.29±5.8* |
| ΔspiA | 46±6 | 1140±3.39 | 24.7±3.98 |
The proliferation rate of strain C50041 was significantly greater than that of ΔspiA in the biofilm phase (P<0.05).
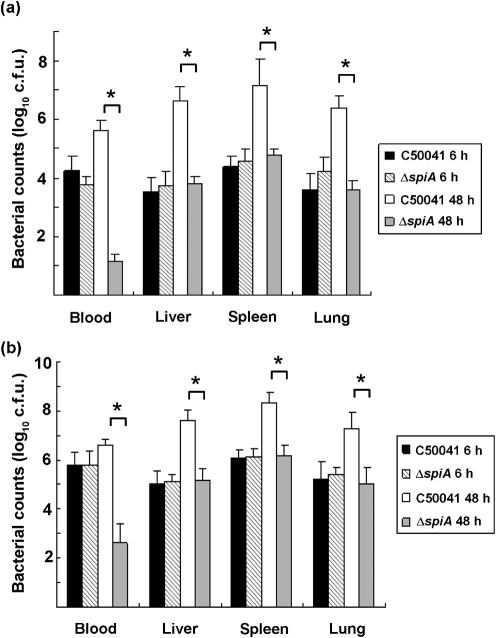
To investigate the effect on the virulence of ΔspiA, mice were injected intraperitoneally with ΔspiA and C50041, and LD50 values were calculated according to the method of Reed & Muench (1938). LD50 values of C50041 in the biofilm and exponential growth phases were 10−0.03 and 100.13, respectively, and those of ΔspiA were 107.47 and 107.13, respectively. To further investigate the distribution of bacteria in vivo, BALB/c mice were injected intraperitoneally with 2×106 c.f.u. and then killed at 6 and 48 h post-infection. At 6 h post-challenge with bacteria in the exponential growth phase, the numbers of ΔspiA present in the blood, liver, spleen and lungs were similar to those of C50041. At 48 h post-challenge, the numbers of ΔspiA in blood, liver, spleen and lungs were significantly lower than those of the wild-type strain, C50041 (Fig. 3a). The distribution pattern in the organs of mice challenged with bacteria in the biofilm phase was similar to that with bacteria in the exponential growth phase. However, bacterial counts in organs challenged with bacteria in the biofilm phase were significantly higher than with bacteria in the exponential growth phase, except for the counts in blood at 6 h post-challenge (Fig. 3b). In addition, the clearance ratio in blood with ΔspiA in the exponential growth phase decreased 412-fold from 6 to 48 h post-infection. By contrast, the clearance ratio in blood with ΔspiA in the biofilm phase decreased 1548-fold from 6 to 48 h post-infection, a 3.75-fold greater clearance rate.
Fig. 3.
Distribution of bacteria in BALB/c mice challenged with exponential-phase cells (a) or biofilm-phase cells (b) of the wild-type strain C50041 and the spiA mutant ΔspiA. Six-week-old BALB/c mice were intraperitoneally injected with 0.2 ml of bacterial suspension. The numbers of bacteria present in the blood, liver, spleen and lungs of the mice were measured 6 and 48 h post-challenge. Significant differences in the bacterial counts of organs were determined by Student’s t-test. *, P<0.05.
Discussion
The spiA gene within S. typhimurium, which encodes an outer-membrane component of the SPI-2 type III secretion system, is essential for virulence in host cells (Ochman et al., 1996). It has also been demonstrated by transposon mutagenesis that the spiA gene may be associated with biofilm formation (Dong et al., 2008). To investigate the effect of the spiA gene on biofilm formation by S. enteritidis, a spiA mutant was constructed, and the biological characteristics of the mutant and wild-type were compared. In a quantitative microtitre assay, the OD600 of ΔspiA was significantly lower than that of the wild-type strain, C50041, which is consistent with previous observations (Dong et al., 2008). Both ΔspiA and C50041 can form biofilms on flask walls, but the biofilm formed by ΔspiA was washed off from the flask wall easily when it was stained by crystal violet. Furthermore, a spiA complementation strain regained the same biofilm characteristics as the wild-type strain. Scanning electron microscopy showed that the ΔspiA strain grown on polystyrene coverslips produced a loose and thin biofilm structure with fewer bacteria, while C50041 produced a very tenacious and thick structure with extracellular substances and fibrils. These results indicated that deletion of the spiA gene resulted in a deficiency in biofilm formation.
The morphotypes of Salmonella grown on Congo red and Coomassie brilliant blue G agar were grouped into five categories: (1) red, dry and rough, indicating curli and cellulose production (RDAR); (2) brown, dry and rough, indicating a lack of cellulose synthesis (BDAR); (3) pink, dry and rough, indicating a defect in curli expression (PDAR); (4) smooth, brown and mucoid, indicating a lack of cellulose synthesis but overproduced capsular polysaccharide (SBAM); and (5) smooth and white, indicating a lack of both curli and cellulose production (SAW) (Malcova et al., 2008; Römling, 2005; Solomon et al., 2005). The ΔspiA strain appeared to be a morphotype of RDAR and had a similarly bright fluorescence to C50041, indicating the presence of curli and cellulose production. However, ΔspiA in the biofilm phase showed a significant decrease in curli production, and a spiA complementation strain in the biofilm phase showed a similar amount of curli production to the wild-type strain. Because curli seem to be more important than cellulose for the formation of cell aggregates (Jonas et al., 2007), the reduced expression of curli in ΔspiA may result in deficient biofilm formation.
It has been reported that a spiA mutant of S. typhimurium displayed wild-type levels of epithelial cell invasion but was defective for intramacrophage survival (Ochman et al., 1996). In our study, regardless of the phase of growth the bacteria were in, ΔspiA showed similar adherence to and invasion of epithelial cells as compared with the wild-type strain. In contrast, ΔspiA showed intracellular proliferation in macrophages in both the exponential growth and the biofilm phases; this may have been due to the use of a different macrophage cell line. Interestingly, ΔspiA and the wild-type strain in the biofilm phase showed much greater adherence to and invasion of epithelial cells or intracellular proliferation in macrophages than those in the exponential growth phase. However, ΔspiA in the biofilm phase showed significantly lower intracellular proliferation than the wild-type strain. Consistent with previous reports (Miki et al., 2004; Ochman et al., 1996), ΔspiA did not kill BALB/c mice when inoculated intraperitoneally at >105 times the median lethal dose of the wild-type strain. In addition, the LD50 in mice with ΔspiA in the biofilm phase was higher than that in mice with ΔspiA in the exponential growth phase; and the LD50 in mice with the wild-type in the biofilm phase was lower than that in mice with the wild-type in the exponential growth phase, indicating that deletion of the spiA gene resulted in a deficiency in biofilm formation and virulence. With regard to the bacterial distribution in mouse organs, ΔspiA showed a similar invasion rate in blood and other organs as the wild-type strain at 6 h post-challenge, but a significantly lower bacterial number in the blood, liver, spleen and lungs at 48 h post-challenge. Consistent with the results of bacterial adherence to and invasion of epithelial cells and bacterial intramacrophage proliferation, bacteria in the biofilms were more invasive in the mouse model than were bacteria in the exponential growth phase. Although the numbers of bacteria of the wild-type strain in both the exponential growth and the biofilm phases showed increases in blood, liver, spleen and lungs from 6 to 48 h post-infection, numbers of ΔspiA bacteria in the biofilm phase showed an accelerated clearance when compared with that of ΔspiA in the exponential growth phase. An additional experiment confirmed that both ΔspiA and the wild-type strain were not sensitive to the normal BALB/c mouse serum (data not shown). The result indicated that deletion of spiA in S. enteritidis had no effect on its adherence and invasion ability, but may result in easy clearance by host cells. Given that the SpiA protein plays a major role in significantly reduced virulence (Ochman et al., 1996), the mechanisms of virulence in bacterial biofilms need further study.
In conclusion, we have constructed a spiA mutant of S. enteritidis and showed that it is deficient in biofilm formation. Although the lack of spiA had no effect on adherence and invasion in epithelial cells, the ΔspiA mutant in both exponential growth and biofilm phases showed reduced adherence and invasion in a mouse model.
Acknowledgements
We thank Dr Wenzhou Hong for his suggestions for the manuscript. This work was supported by the National Natural Science Foundation of China (30871872), the Qing Lan Project, the Program for Changjiang Scholars and Innovative Research Team in University, the High Technology R&D Program of China (2006AA10A206), the National Natural Science Foundation of China for Distinguished Young Scholars (30425031), the National Key Technology R&D Program (2009BADB9B01) and the National Sciences Foundation of Jiangsu Province (BK2008011).
Abbreviations:
- Amp
ampicillin
- Km
kanamycin
- Sm
streptomycin
References
- Anriany Y. A., Weiner R. M., Johnson J. A., De Rezende C. E., Joseph S. W. (2001). Salmonella enterica serovar Typhimurium DT104 displays a rugose phenotype. Appl Environ Microbiol 67, 4048–4056. 10.1128/AEM.67.9.4048-4056.2001.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anriany Y., Sahu S. N., Wessels K. R., McCann L. M., Joseph S. W. (2006). Alteration of the rugose phenotype in waaG and ddhC mutants of Salmonella enterica serovar Typhimurium DT104 is associated with inverse production of curli and cellulose. Appl Environ Microbiol 72, 5002–5012. 10.1128/AEM.02868-05.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Rezende C. E., Anriany Y., Carr L. E., Joseph S. W., Weiner R. M. (2005). Capsular polysaccharide surrounds smooth and rugose types of Salmonella enterica serovar Typhimurium DT104. Appl Environ Microbiol 71, 7345–7351. 10.1128/AEM.71.11.7345-7351.2005.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong H., Zhang X., Pan Z., Peng D., Liu X. (2008). [Identification of genes for biofilm formation in a Salmonella enteritidis strain by transposon mutagenesis]. Wei Sheng Wu Xue Bao 48, 869–873. (in Chinese).. [PubMed] [Google Scholar]
- Geng S. Z., Jiao X. A., Pan Z. M., Chen X. J., Zhang X. M., Chen X. (2009). An improved method to knock out the asd gene of Salmonella enterica serovar Pullorum. J Biomed Biotechnol 2009, 646380.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gerstel U., Römling U. (2003). The csgD promoter, a control unit for biofilm formation in Salmonella typhimurium. Res Microbiol 154, 659–667. 10.1016/j.resmic.2003.08.005.. [DOI] [PubMed] [Google Scholar]
- Hamilton S., Bongaerts R. J., Mulholland F., Cochrane B., Porter J., Lucchini S., Lappin-Scott H. M., Hinton J. C. (2009). The transcriptional programme of Salmonella enterica serovar Typhimurium reveals a key role for tryptophan metabolism in biofilms. BMC Genomics 10, 599. 10.1186/1471-2164-10-599.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X., Phungle V., Dejsirilert S., Tishyadhigama P., Li Y., Liu H., Hirose K., Kawamura Y., Ezaki T. (2004). Cloning and characterization of the gene encoding the z66 antigen of Salmonella enterica serovar Typhi. FEMS Microbiol Lett 234, 239–246. 10.1186/1471-2164-10-599.. [DOI] [PubMed] [Google Scholar]
- Jonas K., Tomenius H., Kader A., Normark S., Römling U., Belova L. M., Melefors O. (2007). Roles of curli, cellulose and BapA in Salmonella biofilm morphology studied by atomic force microscopy. BMC Microbiol 7, 70. 10.1186/1471-2180-7-70.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joseph B., Otta S. K., Karunasagar I., Karunasagar I. (2001). Biofilm formation by Salmonella spp. on food contact surfaces and their sensitivity to sanitizers. Int J Food Microbiol 64, 367–372. 10.1016/S0168-1605(00)00466-9.. [DOI] [PubMed] [Google Scholar]
- Kang H. Y., Srinivasan J., Curtiss R, 3rd. (2001). Immune responses to recombinant pneumococcal PspA antigen delivered by live attenuated Salmonella enterica serovar Typhimurium vaccine. Infect Immun 70, 1739–1749. 10.1016/S0168-1605(00)00466-9.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korber D. R., Choi A., Wolfaardt G. M., Ingham S. C., Caldwell D. E. (1997). Substratum topography influences susceptibility of Salmonella enteritidis biofilms to trisodium phosphate. Appl Environ Microbiol 63, 3352–3358.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latasa C., Roux A., Toledo-Arana A., Ghigo J. M., Gamazo C., Penadés J. R., Lasa I. (2005). BapA, a large secreted protein required for biofilm formation and host colonization of Salmonella enterica serovar Enteritidis. Mol Microbiol 58, 1322–1339. 10.1111/j.1365-2958.2005.04907.x.. [DOI] [PubMed] [Google Scholar]
- Malcova M., Hradecka H., Karpiskova R., Rychlik I. (2008). Biofilm formation in field strains of Salmonella enterica serovar Typhimurium: identification of a new colony morphology type and the role of SGI1 in biofilm formation. Vet Microbiol 129, 360–366. 10.1016/j.vetmic.2007.12.006.. [DOI] [PubMed] [Google Scholar]
- Marin C., Hernandiz A., Lainez M. (2009). Biofilm development capacity of Salmonella strains isolated in poultry: risk factors and their resistance against disinfectants. Poult Sci 88, 424–431. 10.3382/ps.2008-00241.. [DOI] [PubMed] [Google Scholar]
- Miki T., Okada N., Danbara H. (2004). Two periplasmic disulfide oxidoreductases, DsbA and SrgA, target outer membrane protein SpiA, a component of the Salmonella pathogenicity island 2 type III secretion system. J Biol Chem 279, 34631–34642. 10.1074/jbc.M402760200.. [DOI] [PubMed] [Google Scholar]
- Monack D. M., Raupach B., Hromockyj A. E., Falkow S. (1996). Salmonella typhimurium invasion induces apoptosis in infected macrophages. Proc Natl Acad Sci U S A 93, 9833–9838. 10.1073/pnas.93.18.9833.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ochman H., Soncini F. C., Solomon F., Groisman E. A. (1996). Identification of a pathogenicity island required for Salmonella survival in host cells. Proc Natl Acad Sci U S A 93, 7800–7804. 10.1073/pnas.93.15.7800.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patrick M. E., Adcock P. M., Gomez T. M., Altekruse S. F., Holland B. H., Tauxe R. V., Swerdlow D. L. (2004). Salmonella enteritidis infections, United States, 1985–1999. Emerg Infect Dis 10, 1–7.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pratt L. A., Kolter R. (1998). Genetic analysis of Escherichia coli biofilm formation: roles of flagella, motility, chemotaxis and type I pili. Mol Microbiol 30, 285–293. 10.1046/j.1365-2958.1998.01061.x.. [DOI] [PubMed] [Google Scholar]
- Qiuchun L., Xu Y., Jiao X. (2009). Identification of Salmonella pullorum genomic sequences using suppression subtractive hybridization. J Microbiol Biotechnol 19, 898–903. 10.1046/j.1365-2958.1998.01061.x.. [DOI] [PubMed] [Google Scholar]
- Reed L. J., Muench H. (1938). A simple method of estimating fifty percent endpoints. Am J Hyg 27, 493–497. [Google Scholar]
- Römling U. (2005). Characterization of the rdar morphotype, a multicellular behaviour in Enterobacteriaceae. Cell Mol Life Sci 62, 1234–1246. 10.1007/s00018-005-4557-x.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Römling U., Bian Z., Hammar M., Sierralta W. D., Normark S. (1998a). Curli fibers are highly conserved between Salmonella typhimurium and Escherichia coli with respect to operon structure and regulation. J Bacteriol 180, 722–731.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Römling U., Sierralta W. D., Eriksson K., Normark S. (1998b). Multicellular and aggregative behaviour of Salmonella typhimurium strains is controlled by mutations in the agfD promoter. Mol Microbiol 28, 249–264. 10.1046/j.1365-2958.1998.00791.x.. [DOI] [PubMed] [Google Scholar]
- Römling U., Bokranz W., Rabsch W., Zogaj X., Nimtz M., Tschäpe H. (2003). Occurrence and regulation of the multicellular morphotype in Salmonella serovars important in human disease. Int J Med Microbiol 293, 273–285. 10.1078/1438-4221-00268.. [DOI] [PubMed] [Google Scholar]
- Scher K., Romling U., Yaron S. (2005). Effect of heat, acidification, and chlorination on Salmonella enterica serovar Typhimurium cells in a biofilm formed at the air-liquid interface. Appl Environ Microbiol 71, 1163–1168. 10.1128/AEM.71.3.1163-1168.2005.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon E. B., Niemira B. A., Sapers G. M., Annous B. A. (2005). Biofilm formation, cellulose production, and curli biosynthesis by Salmonella originating from produce, animal, and clinical sources. J Food Prot 68, 906–912.. [DOI] [PubMed] [Google Scholar]
- White A. P., Gibson D. L., Collinson S. K., Banser P. A., Kay W. W. (2003). Extracellular polysaccharides associated with thin aggregative fimbriae of Salmonella enterica serovar Enteritidis. J Bacteriol 185, 5398–5407. 10.1128/JB.185.18.5398-5407.2003.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zogaj X., Nimtz M., Rohde M., Bokranz W., Römling U. (2001). The multicellular morphotypes of Salmonella typhimurium and Escherichia coli produce cellulose as the second component of the extracellular matrix. Mol Microbiol 39, 1452–1463. 10.1046/j.1365-2958.2001.02337.x.. [DOI] [PubMed] [Google Scholar]