Abstract
Although a variety of cues have been implicated in axonal targeting during embryogenesis and regeneration, the precise mechanisms guiding olfactory axons remain unclear. Appropriate olfactory axon pathfinding is essential for functional chemoreceptive and pheromone receptive systems. Olfactory axon pathfinding is also necessary for establishment of the neuroendocrine LHRH system, cells critical for reproductive function. LHRH cells exhibit neurophilic migration moving from the nasal region along olfactory axons into the brain. Factors involved in the migration of these neuroendocrine cells are as yet unresolved. We report identification of a novel factor termed nasal embryonic LHRH factor (NELF) that was discovered in a differential screen of migrating versus nonmigrating primary LHRH neurons. NELF is expressed in PNS and CNS tissues during embryonic development, including olfactory sensory cells and LHRH cells. NELF antisense experiments indicate that a reduction in NELF expression decreases olfactory axon outgrowth and the number of LHRH neurons that migrate out of the nasal tissue. These results demonstrate that NELF plays a role as a common guidance molecule for olfactory axon projections and subsequently, either directly or indirectly, in the neurophilic migration of LHRH cells.
Keywords: Olfactory, migration, LHRH, GnRH
Appropriate axonal pathfinding and neuronal migration are essential for formation of a normal nervous system. Both of these processes are dependent on spatially and temporally regulated attractants and repellents. Extracellular matrix molecules and cell adhesion molecules have long been known to influence both axon outgrowth and cell movement (for review, see Mori 1993; Mueller 1999). Recently, several families of growth cone guidance molecules [semaphorins (for review, see Pasterkamp et al. 1999) and slits (Wu et al. 1999)] and their receptors [neuropilins/plexins (He and Tessier-Lavigne 1997; Kolodkin et al. 1997; Winberg et al. 1998) and Robo (Nguyen Ba-Charvet et al. 1999)] have been identified. Expression of these molecules throughout the CNS and PNS during development (Adams et al. 1996; Giger et al. 1996; Chen et al. 1997; Kitsukawa et al. 1997; Holmes et al. 1998; Itoh et al. 1998; Brose et al. 1999;) suggests both conservation and redundancy of pathfinding and migrational mechanisms.
The combinatorial effects of these and other molecular signals have been implicated in outgrowth and targeting of olfactory axons during embryogenesis and regeneration. Semaphorins and Slits (as well as their receptors) have been identified in the olfactory system (for review, see Nguyen Ba-Charvet et al. 1999; Pasterkamp et al. 1999) and appear to play a role in the establishment and maintenance of olfactory networks. Proteins such galectin-1, 6B4 proteoglycan, and olfactory receptors have also been proposed to guide olfactory axons from the olfactory epithelium to the olfactory bulb (Nishizuka et al. 1996; Puche et al. 1996; Mombaerts 1999). The targeting of nasal sensory axons, both prenatal and postnatal, is essential for functional chemoreceptive and pheromone receptive systems (Keverne 1999; Mori et al. 1999). In addition, appropriate olfactory axon targeting during development appears to be necessary for the establishment of the neuroendocrine luteinizing hormone-releasing hormone (LHRH) system (Wray et al. 1989). LHRH neurons are critical for reproductive function (for review, see Fink 1988). Disruption of olfactory axon targeting, as occurs in Kallmann's syndrome (Schwanzel-Fukuda et al. 1989; Quinton et al. 1997), results in LHRH cells that do not enter the brain, causing reproductive dysfunction.
LHRH neurons have been documented to migrate along peripherin- and neural cell adhesion molecule (NCAM)-positive axon bundles exiting the vomeronasal organ leading to the brain (Wray et al. 1994). These bundles are believed to be a required substrate for LHRH neuronal migration (Murakami et al. 1991; Schwanzel-Fukuda et al. 1992; Yoshida et al. 1999). The axons from the olfactory pit (vomeronasal organ anlage) and peripherin, NCAM-positive olfactory receptor axons fasciculate into bundles within nasal regions, and the LHRH neurons migrate along these axon bundles through the cribriform plate (nasal–forebrain junction). Once within the brain, a minority of LHRH neurons follow olfactory receptor axons toward the olfactory bulb, but the majority of LHRH cells follow a peripherin-positive, NCAM-negative axonal pathway toward the septal/preoptic area and hypothalamus (Wray et al. 1989, 1994). It is within these locations in the forebrain that the LHRH cells form an essential neuroendocrine component of the reproductive process becoming integral components of the hypothalamic–pituitary–gonadal axis (for review, see Fink 1988).
Cell surface markers expressed on LHRH migrational pathways within the brain include a transient axonal surface glycoprotein (TAG-1) and polysialic acid–NCAM (PSA-NCAM) (Murakami et al. 1991; Yoshida et al. 1995). Experiments have shown PSA-NCAM is required for the proper migration of a subpopulation of LHRH neurons (Yoshida et al. 1999). But a significant percentage of LHRH neurons continued to migrate after removal of PSA-NCAM (Yoshida et al. 1999), and normal LHRH migration was observed in NCAM-null mice (Cremer et al. 1994). In addition, in vitro experiments have documented LHRH migration along NCAM-negative olfactory sensory axons (Fueshko and Wray 1994). Thus, the factors and cues involved in appropriate neurophilic migration of LHRH cells through the nasal regions and CNS have yet to be determined. To gain insight into the molecules involved in this process, a differential screen of primary LHRH neurons was performed comparing a migrating cell and nonmigrating cells. Here, we report identification of a novel factor termed nasal embryonic LHRH factor (NELF) that was expressed in PNS and CNS tissues, including olfactory sensory cells and LHRH cells during embryonic development. Our results indicate that NELF plays a role as a common guidance mechanism for olfactory axon projections and subsequently, either directly or indirectly, in the neurophilic migration of LHRH cells.
Results
To identify molecules involved in LHRH neuronal migration, we used a strategy by which the mRNA complement in a migrating LHRH cell was compared with the mRNA in cells that had ceased migrating. Experimentally, nasal explants were removed from embryonic day (E)11.5 mouse embryos. LHRH neurons have been shown to migrate from the olfactory pit in this explant culture system (Fueshko and Wray 1994) and can be identified in situ (Kusano et al. 1995). Three bipolar neurons were isolated from the nasal explants. One LHRH cell was removed after 3.5 days growth in vitro (div) when cells are migrating, and two cells were removed after 10.5 div (at an age that LHRH neurons cease migrating) (Wray et al. 1989; Fueshko and Wray 1994). A cDNA pool was created from each individual cell, and all cells were shown to be positive for the presence of LHRH mRNA (data not shown). The cDNA library from the 3.5-div LHRH cell was then probed with labeled cDNA from the LHRH cells removed at 10.5 div. Two clones were identified as being unique to the 3.5-div library. These clones were isolated and sequenced. One clone was found to be the transcription factor activator protein-2 (Kramer et al. 2000). The second clone was found to encode a novel gene product we have designated NELF (sequence shown in Fig.1). Southern analysis of the cDNA pools confirmed that the 3.5-div LHRH cell contained the mRNA from NELF, whereas the 10.5-div LHRH cells did not (data not shown).
Characterization of NELF transcript and protein
The cDNA of NELF was isolated by RACE and had an approximate transcript size of 2.7 kb (Fig. 1). Expression of NELF mRNA was present throughout development of the mouse and continued in the adult brain (Fig. 1). In situ hybridization histochemistry and immunohistochemistry (generation of antibody; see Fig. 1 and Materials and Methods) indicated high expression of NELF mRNA (Fig. 2A) and protein (Fig. 2B) in the forebrain, olfactory epithelium, and olfactory pit of developing embryos. The intensity of mRNA signal in cells within the olfactory epithelium and olfactory pit dramatically increased between E12. 5 and E14.5; a maximum level of mRNA expression per area of olfactory tissue appeared to be associated with this embryonic stage, because at E17.5 the signal was again attenuated (data not shown). The relative level of NELF mRNA expression in olfactory epithelial cells thus correlated with the expression level as determined by Northern analysis on whole embryos, peaking around E15.0 (see Fig. 1).
Figure 1.
Sequence of NELF. (Top) Largest ORF. No overt homology was found between NELF and sequences in the gene databases. Underlined regions are phosphorylation consensus elements. Polyclonal antibody 1026 (anti-NELF) was produced against the peptide in the boxed region. (Bottom) Northern analysis indicates NELF mRNA is present throughout mouse development (E7–E17) and in adult mouse brain. The 4.4-Kb product is an unknown cross-hybridizing species. Numbers at right indicate the size in kilobases.
Figure 2.
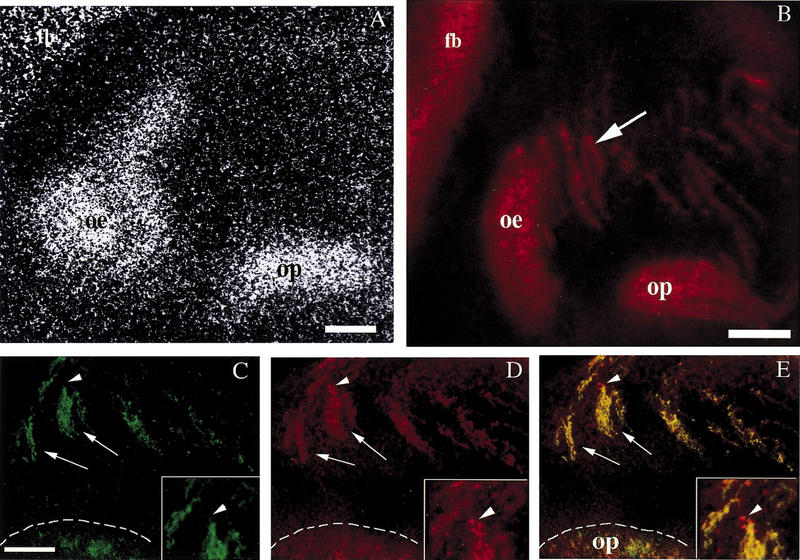
NELF is robustly expressed by nasal epithelia. (A) In situ hybridization histochemistry of an E14.5 embryo indicated high mRNA levels within the forebrain (fb), olfactory epithelium (oe), and olfactory pit (op). (B) Polyclonal 1026 (NELF) antibody staining of an E13.5 embryo revealed protein within these same areas. In addition, protein was also detected within tracks exiting the olfactory pit (arrow). Staining of an E12.5 mouse embryo with peripherin (C) and NELF (D) indicated colocalized staining in the axons (E, arrows) and in the olfactory pit. However, NELF-only staining was observed in cells associated with the axon bundles (arrowhead, and see inset). Bar, 200 μm (A, B) and 100 μm (C–E).
Immunocytochemistry revealed that NELF protein was robustly expressed in both cell soma and processes (Fig. 2B). NELF immunopositive tracks extended from the olfactory epithelium and olfactory pit to the olfactory bulb. Olfactory axons extend from these two structures and are known to express the intermediate filament protein, peripherin (Fueshko and Wray 1994). To establish that the NELF immunopositive tracks were olfactory axons fasciculating into bundles that crossed the nasal septum, double-label immunofluorescence was performed (Fig. 2C–E). Peripherin-positive olfactory axon bundles colabeled with NELF protein. Thus, during development NELF demarcated the olfactory sensory system and the path used for LHRH neuronal migration.
In addition to NELF expression being localized to the LHRH neuronal migratory pathway, LHRH neurons in nasal regions colabeled with NELF (Fig. 3). Immunohistochemical staining for LHRH and 1026 demonstrated that NELF protein was robustly expressed in LHRH neurons from E11.5 to E17.5 but was not observed at PN1 or in adult mice (Fig. 3). Embryonically (E11.5–E17.5), expression of NELF in LHRH neurons was observed as the cells exited the olfactory pit (Fig. 3B–D) and as they migrated along axon bundles through the cribriform plate (nasal–forebrain junction; Fig. 3E–G). However, NELF expression was markedly attenuated once the LHRH cells entered the forebrain (Fig. 3H–J)—a stage when LHRH cells begin to cease migration having reached their adult location. Furthermore, LHRH neurons migrating in the brain toward the olfactory bulb stained for NELF (Fig. 3K–M) in contrast to the absence of NELF staining in LHRH neurons migrating caudally (Fig. 3H–J). Postnatally, LHRH neurons were NELF negative (Fig. 3N, O). These data are consistent with the idea that NELF functions in LHRH neurons while the cells are in the nasal region but not when they enter the forebrain.
Figure 3.
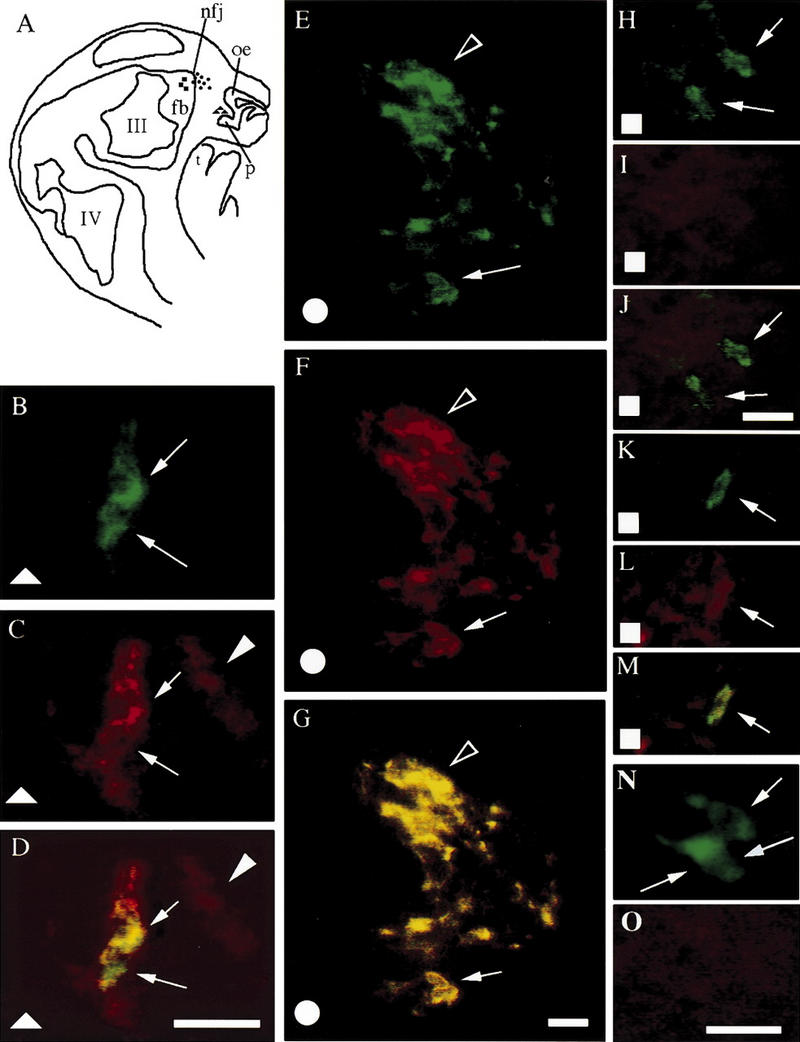
NELF is expressed by LHRH neurons in nasal regions in vivo. (A) Schematic of an E13.5 embryo head, forebrain (fb), nasal forebrain junction (nfj), olfactory epithelium (oe), olfactory pit (op), tongue (t), third (III), and fourth (IV) ventricles are depicted. (B–M) Symbols (squares, circles, triangles) on the schematic indicate location of cells that are from E13.5 embryos. (N,O) cells from postnatal day 1 mouse brain. LHRH staining in shown (B,E,H,K,N). NELF staining is shown (C,F,I,L,O). Simultaneous visualization of both fluorescent wavelengths is shown (D,G,J,M). Cells exiting the olfactory pit (triangles) are positive for both LHRH and NELF (B–D, arrows). In addition to LHRH cells, olfactory axons in this region were also positive for NELF (C,D, arrowhead). LHRH (E) and NELF (F) are also coexpressed (G) in a high percentage of cells at the nasal forebrain junction (circles; arrow points to cluster of three to four cells and arrowhead points to large cluster of cells that converge at nasal forebrain junction, i.e., at the cribriform plate). Upon entering the forebrain (squares), LHRH cells (H,K, arrows) contain little or no signal for NELF (I,L; covisualization J,M). By PN1, LHRH neurons in the brain (N, arrows point to three cells) were NELF negative (O). Bar, 20 μm.
In summary, during development NELF is expressed in LHRH neurons as they migrate from the olfactory pit into the developing forebrain. NELF is also expressed on olfactory axons, including the olfactory axonal pathway on which the LHRH neurons move. These results suggest that NELF may be a molecule that serves as a common guidance mechanism for olfactory axon projections and subsequent neurophilic migration of LHRH cells. As with many other guidance molecules, conservation of function during development is suggested by the fact that NELF was present in many neuronal tissues with migrating cell populations (Table 1).
Table 1.
Expression of NELF in embryonic structures
| Structure
|
mRNAa
|
Proteinb
|
|---|---|---|
| Olfactory epithelium | +++c | +++ |
| Vomeronasal organ | +++ | +++ |
| Cortex anlage | ++ | ++ |
| Forebrain | + | + |
| Spinal cord | + | + |
| Dorsal root ganglia | + | + |
| Heart | − | − |
| Liver | − | − |
| Nasal mesenchyme | − | − |
| Limb bud | − | − |
In situ hybridization of E12.5 and E14.5 embryos.
Immunohistochemistry of E12.5–E15.5 embryos.
Relative intensity of the signal.
Function of NELF
To test the hypothesis that NELF influences olfactory axon outgrowth and/or LHRH migration, we returned to our in vitro model. Nasal explants show robust and directional outgrowth of olfactory axons and subsequent migration of LHRH neurons (Fig. 4; Fueshko and Wray 1994). NELF protein was robustly expressed in these cultures, with the majority of LHRH neurons and olfactory axons stained for NELF (Fig. 4A–C, n = 12 cultures). As in vivo, olfactory axons labeled by peripherin antibodies coexpressed NELF in vitro (data not shown). NELF staining intensity in LHRH neurons decreased dramatically in older cultures (10.5 div, n = 6), consistent with the mRNA expression pattern of NELF observed earlier (Southern analysis of cDNA pools 3.5 div vs. 10.5 div). In contrast, NELF staining remained high in olfactory axons at 7 and 10.5 div, n = 3 each).
Figure 4.
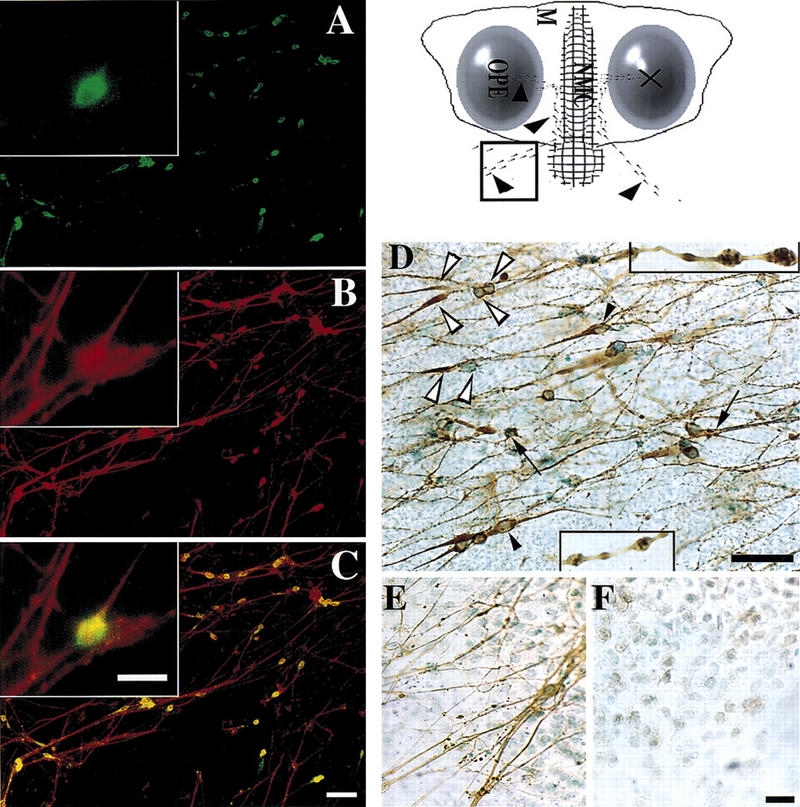
NELF is expressed on the cell surface of LHRH neurons and olfactory axons. Schematic of a nasal explant culture removed from an E11.5 mouse embryo and maintained in serum-free media for 7 days is shown (top, right). Ovals represent the olfactory pit epithelium (OPE), in the center is the nasal midline cartilage (NMC) and the surrounding mesenchyme (M). LHRH neurons (dots) migrate from the OPE to the midline and then off the explant into the periphery (arrowheads). The boxed region within the schematic is the area shown in the remaining panels. (A–C) Double immunofluorescence was performed on explants using antibodies to LHRH and NELF. (A) Many immunopositive LHRH neurons are detected that migrated off the explant into the periphery. (B) Immunostaining for NELF revealed robust staining of fibers as well as cell bodies in this same area. (C) Simultaneous visualization of both fluorescent wavelengths indicated LHRH neurons, both cell soma as well as processes, were positive for NELF. NELF-only positive fibers were also present. (Inset) An enlarged view of the same cell in each panel. Cells and processes are immunopositive for NELF in nasal explants not treated with the detergent Triton X-100 (D, n = 5). In certain cells (D, white arrowheads) the cell soma stained with NELF, but there was a paucity of staining between the soma (right white arrowheads) and distal region of the axon (left white arrowheads). Other bipolar cells (black arrowheads) showed staining for NELF throughout the extent of the cell. Staining with NELF was also intense at fibers terminating in bouton-like structures (arrows) on NELF immunopositive cells. Fibers containing multiple NELF-positive varicosities were detected throughout the periphery of the explant (enlarged views of such fibers are shown, insets). Controls to determine whether membrane permeabilization was necessary for intracellular access of the antibody using our staining procedure included staining for the olfactory intermediate filament protein peripherin with and without the detergent Triton X-100. Explant cultures permeabilized with detergent had positive fibers stained for peripherin (E), but no peripherin staining was detected on explant cultures when detergent was not used (F). Bar, 20 μm (inset, D–F) and 50 μm (A–C).
NELF is localized to membrane surfaces
The pattern of NELF expression is consistent with its proposed function as a migratory factor for LHRH neurons, but migratory factors and/or signals (e.g., PSA-NCAM, bone morphogenetic proteins) are found in, or exposed to, the extracellular milieu. Thus, to determine whether NELF protein was present on the outside of the cellular membrane, immunocytochemistry without detergent (Triton X–100) to permeabilize the tissue was performed (Fig. 4D). In the absence of detergent, robust NELF immunopositive staining was observed. In contrast, no immunostaining was found for peripherin under these same conditions (Fig. 4E,F). NELF protein was present on the outside of the LHRH cell membrane and axons projecting from the olfactory pit and epithelium. In some instances staining was localized to the cell soma and distal axon projections and punctate regions (varicosities) along fibers (Fig. 4D, open arrowheads; enlarged varicosities, see insets). Furthermore, NELF-positive fibers terminated in arborizations that contacted both the cell soma and processes of LHRH neurons (Fig. 4D, arrows). NELF localization on the exterior of both the sensory fiber pathway and LHRH neurons suggests a possible homophilic interaction between the two types of cells. This interaction could function to signal or aid in migration of the LHRH neurons. To further examine this issue, nasal explants were treated with NELF antisense oligo DNA.
NELF protein levels decrease after antisense treatment
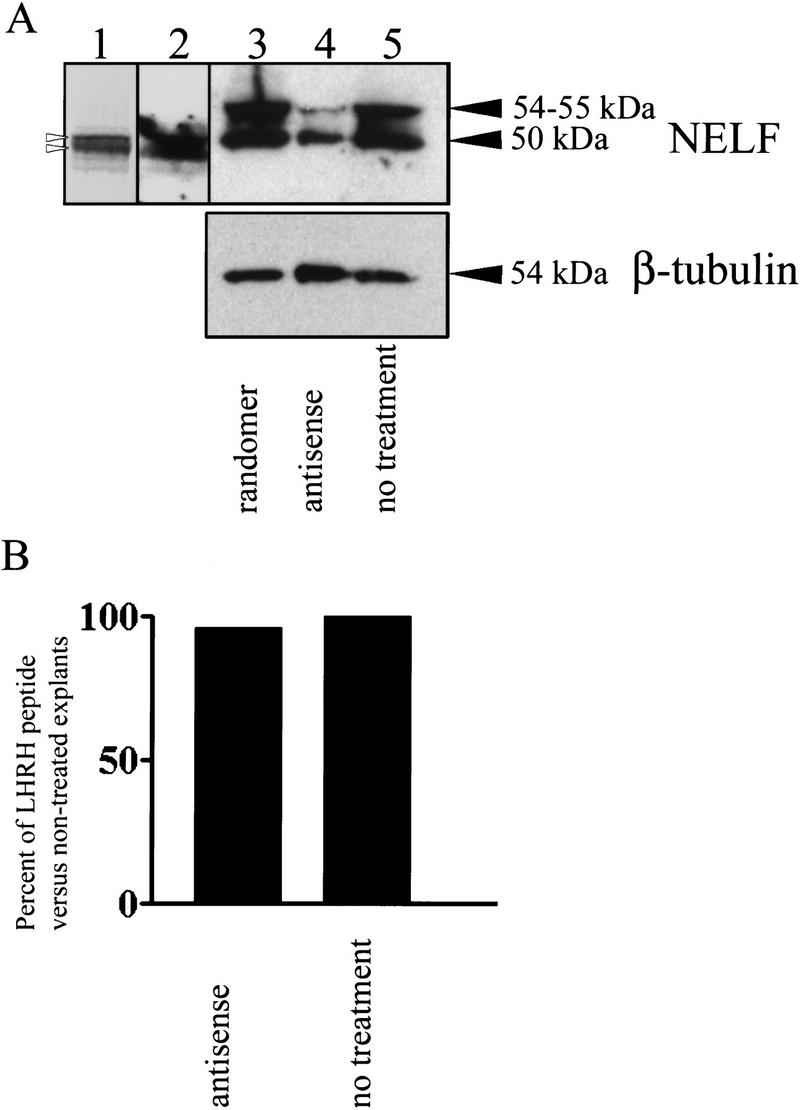
The largest open reading frame (ORF) in the NELF transcript (Fig.1) is predicted to produce a protein with a molecular mass of 49.2 kD. In vitro translation of this ORF produced a product of ∼50 kD (Fig. 5, lane 1 and 2). Multiple in-frame start sites (see Fig.1) result in several smaller translation products (Fig. 5, lane 1). In vivo, Western blot analysis of protein from explant tissue indicated the presence of a 50-kD protein recognized by antibody 1026 consistent with the size of the largest ORF but a 54- to 55-kD positive product was also detected with this antibody (Fig. 5, lanes 3–5). Nasal explant cultures were treated with antisense oligo DNA (n = 5), a randomer (n = 4), or transfection media minus DNA (n = 4). NELF protein was assayed ∼84 hr later. After antisense treatment, the intensity of the 50- and 55-kD bands was reduced (Fig. 5, lane 4) by ∼60% as compared with controls (antisense, 2318 ± 1129; randomer, 5697 ± 840; nontreated, 6421 ± 2479). In contrast, the percent of LHRH peptide in antisense-treated samples did not decrease in comparison with controls (Fig. 5, histogram), as measured by ELISA (Maurer and Wray 1999).
Figure 5.
(A) The largest ORF (Fig. 1) was translated in vitro and produced a doublet of ∼50 kD and a product slightly smaller (lane 1, open arrowheads). The largest ORF has multiple in-frame start sites 3′ of the initial start site that would produce a protein product slightly smaller than the full-length protein (see Fig. 1). Western analysis of the in vitro (lane 2) and in vivo product (explant tissue, lanes 3–5) indicated a band of ∼50 kD, but in vivo, a second 54- to55-kD product is bound by the NELF antibody. Antisense treatment using the oligo 18NPR8.5 decreased the amount of product bound by the polyclonal 1026 antibody by ∼60% [cf. lanes 3 (randomer) and 4 (antisense) with lane 5 (no treatment)]. This Western blot was then stripped and probed with a monoclonal antibody to β-tubulin; comparison of the lanes indicate that there was nearly equal amounts of protein loaded or a slightly higher amount loaded in the antisense lane (lane 4). (B) The histogram depicts the percent of LHRH peptide in the treated and nontreated nasal explants as determined by ELISA analysis and indicates that antisense treatment for NELF had no effect on the level of LHRH peptide. ELISA analysis was completed in duplicate on batches that combined two to six explants (n = 6, nontreated; n = 7, antisense treated).
Olfactory axon outgrowth and LHRH neuronal movement is inhibited after antisense treatment
Nasal explants were treated with NELF antisense oligo as above, fixed, and immunohistochemically stained for NELF (Fig. 6) and LHRH (Fig. 7) using immunofluorescence. Analysis of NELF-positive fibers indicated a reduction of the overall complexity (Fig. 6, cf. A and B) and a significant decrease in the length of the fibers after antisense treatment (Fig. 6, histogram). Antisense treatment also increased the percent of LHRH cells that remained on the main tissue mass (31% to 57%) and significantly reduced the number of LHRH neurons present in the periphery of the explant tissue (Fig. 7, histogram).
Figure 6.
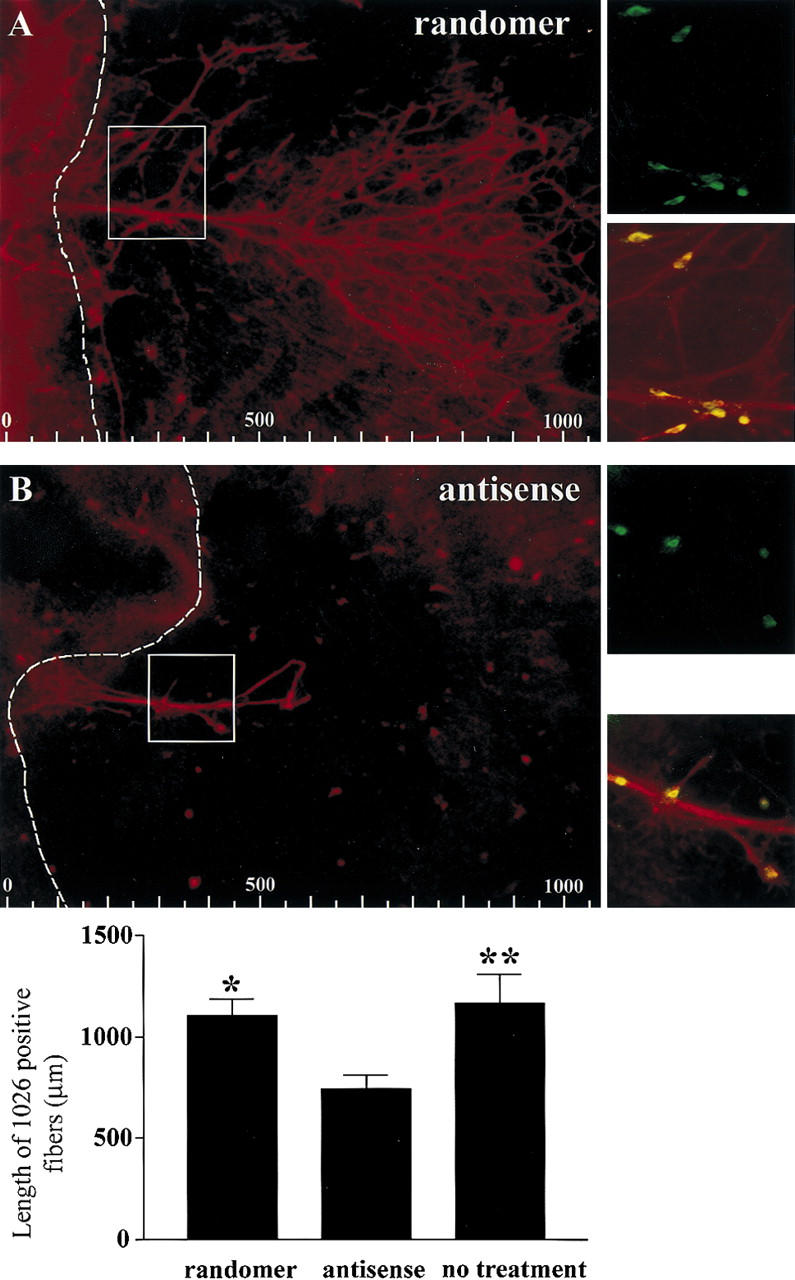
Knockdown experiments using antisense oligo DNA decreased the length of NELF-positive fibers [red fibers; cf. A (randomer) and B (antisense)]. Fiber length measurements were completed averaging the longest two or three NELF-positive fibers present on the culture (note: if a only a single long fiber was present, that value was the only value used). LHRH cells (green) are still present after antisense treatment (red fibers and cells are NELF positive, and yellow indicates colocalization; insets, right are enlarged regions of boxed areas in A and B). Fibers were measured from the edge of the main nasal explant tissue into the periphery (broken line; at left of the broken line is the main tissue of the explant, and at right is the periphery; also see schematic in Fig. 4). Scales at bottom of A and B are in micrometers. The histogram below shows the average length of the NELF positive fibers as a function of treatment (n = 14, randomer; n = 18, antisense; n = 134, no treatment). A significant decrease in the length of NELF-positive fibers was observed in antisense-treated explants as compared with randomer-treated (P < 0.05, *) and nontreated explants (P < 0.01, **).
Figure 7.
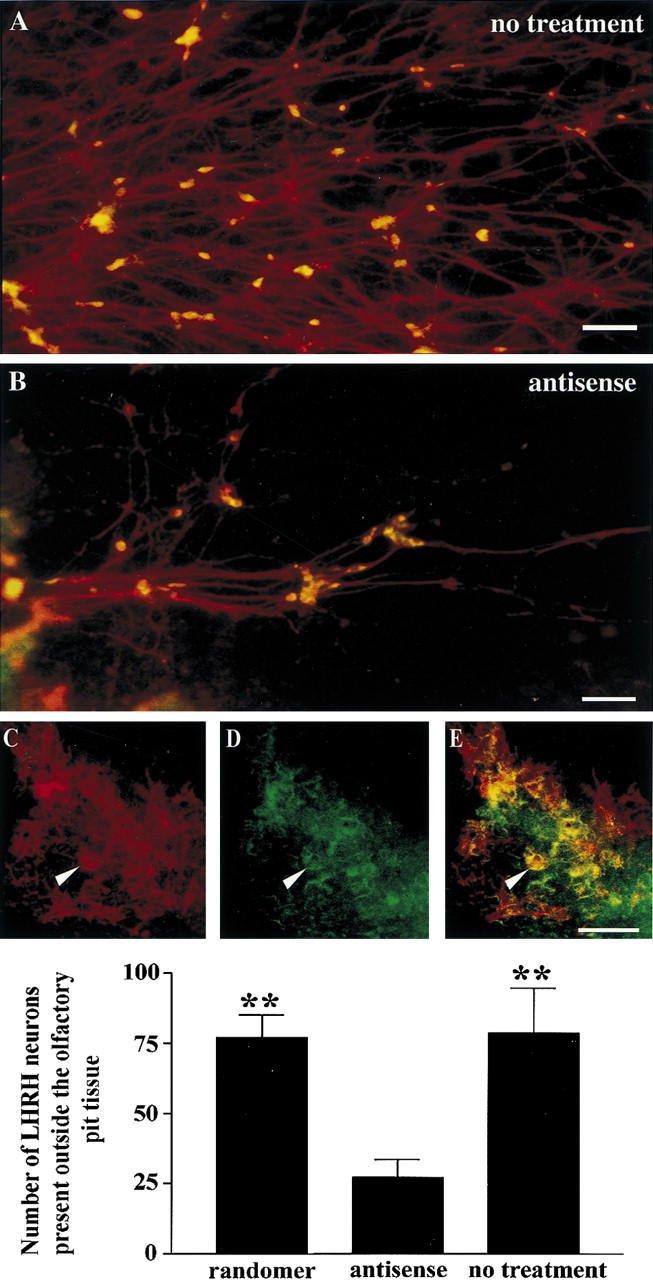
The number of LHRH neurons present within the periphery of nasal explants decreased after treatment with antisense oligo DNA. The complexity of NELF-positive fibers (red) decreased after antisense treatment [cf. A (no treatment) and B (antisense treatment)]. The number of LHRH neurons associated with these fibers in the periphery of the nasal explant also decreased (cf. number of yellow cells in A and B). (Red) NELF; (green) LHRH; (yellow) colocalization. Numerous LHRH neurons are still present within the explant tissue (on the main tissue mass) after antisense treatment (C, NELF; D, LHRH; E, colocalization; arrow points to cell cluster of LHRH neurons). LHRH cell counts of the number of cells that were in the periphery of the nasal explant indicate a significant decrease after treatment with the antisense oligo (n = 18) in comparison with randomer and nontreated explants (P < 0.01, **; n = 11, randomer and no treatment; n = 18, antisense treated). Bar, 100 μm (A–C).
Discussion
A novel gene termed NELF was identified by a cDNA subtraction screen performed on LHRH neurons maintained for 3.5 div versus 10.5 div. We hypothesized that a putative migratory factor would be differentially expressed between LHRH neurons at these ages based on two facts. First, it was shown previously that LHRH neurons in nasal explants cease migrating after 7 div (Fueshko and Wray 1994). Second, the in vitro ages chosen for analysis roughly correspond to LHRH neurons from an E14.5 embryo, when LHRH neurons are still migrating, and a PN1 animal, when LHRH neurons have reached an adult-like distribution b (Schwanzel-Fukuda and Pfaff 1989; Wray et al. 1989). Several lines of evidence suggest that NELF is involved in olfactory axon outgrowth and LHRH neuronal migration.
NELF is a molecule used for neurophilic migration
Spatiotemporal localization
Immunocytochemistry indicated that NELF was more robustly expressed on LHRH neurons in explants grown for 3.5 div in contrast to explants grown for 10.5 div, but NELF staining was present on axons exiting the olfactory pit at both stages. A similar pattern was observed prenatally in vivo. NELF was expressed by olfactory axons, those leaving the main olfactory epithelium (location of olfactory receptor neurons) as well as those leaving the olfactory pit/vomeronasal organ (location of pheromone receptor neurons). In addition, cells exhibiting NELF/LHRH coexpression extended from the olfactory pit to the cribriform plate. In contrast, NELF staining was not observed in LHRH neurons in the forebrain in PN1 animals.
Whether NELF expression in LHRH neurons is dependent on age and/or location signals is difficult to determine because of the development of the LHRH system. At E12.5, LHRH neurons span across the nasal septum into the developing forebrain. At this stage, robust NELF expression was observed in LHRH cells up to the cribriform plate but decreased in cells in the developing CNS. One could interpret these results to indicate that NELF expression in LHRH cells relies on nasal cues and/or is inhibited by CNS cues, that is, location dependent. However, it is likely that the LHRH cells entering the CNS at this time are the earliest LHRH cells to differentiate (LHRH neurons become postmitotic between E10.0 and E11.0; Wray et al. 1989) and thus are developmentally older. Hence, NELF expression in LHRH cells may rely on time-linked events, that is, are age dependent. In vitro, LHRH cells down-regulated NELF expression between 3.5 div and 10.5 div. In the nasal explants, no brain tissue is taken (Fueshko and Wray 1994). Thus, LHRH cells at 10.5 div down-regulated NELF expression in the absence of CNS cues. Location and age, with respect to LHRH cells, are still difficult to separate in this in vitro model. However, both in vivo and in vitro, NELF expression in LHRH cells is correlated with cell movement in nasal regions.
Although the specific cues that regulate NELF expression in olfactory axons and LHRH cells are presently unknown, the spatiotemporal localization of the NELF protein is consistent with NELF acting as a migratory factor or migratory signal within the nasal region. Preliminary studies also show NELF expression at later prenatal stages (E18.5) in the developing cortex and cerebellum, two areas known to exhibit robust neuronal migration. The role of NELF in these areas awaits further investigations but again is consistent with NELF acting as a migratory signal. Furthermore, the down-regulation of NELF on LHRH cells as they cross the cribriform plate and enter the telencephalon may be a critical event, insuring migration of LHRH cells along a different pathway in the CNS (from those leading to the olfactory bulb and accessory nucleus) and thereby facilitate establishment of the appropriate adult-like LHRH distribution.
Homology searches for protein binding domains within the NELF protein were negative. The lack of protein modules in NELF suggested few other binding partners. NELF was expressed on both LHRH neurons and the axons on which they migrate. The fact that no other potential binding partners were identified and that NELF was expressed on both migratory cell and pathway led us to hypothesis a homophilic-like interaction. If NELF acts as a migratory/axonal outgrowth molecule via homophilic interactions, then one would predict that NELF would be localized to the extracellular portion of the cell membrane (Edelman 1985; Cunningham et al. 1987; Rutishauser and Jessell 1988). Visualization of robust, punctate NELF signal in the absence of cell permeabilization supports localization of NELF on the cell surface of both axons and cell soma. Analysis of the NELF protein sequence revealed no strong hydrophobic regions, suggesting a lack of transmembrane domains. In native Western blots, a higher complex of ∼150 kD was occasionally observed, suggesting the presence of a potential cytoplasmic/transmembrane partner by which NELF may be anchored to the membrane proper. Further studies are necessary to examine this issue in more detail. However, the overall expression pattern of NELF suggests a homophilic interaction between NELF on LHRH neurons and NELF on the axon bundles on which they migrate, perhaps being a determining factor in neurophilic migration of LHRH in nasal regions.
Disruption of NELF expression decreases olfactory axon outgrowth and, subsequently, LHRH migration
If NELF is primarily an “ectodomain” protein, the possibility arose that our antibody could be used to determine the functional role of NELF in neuronal migration and/or axonal outgrowth. In a preliminary set of experiments, the 1026 antibody was added (at varying concentrations) to nasal explant cultures. No consistent changes were observed as compared with control nasal explants. A null effect may have occurred as a result of another factor assuming the role of NELF (redundancy) and/or the fact that the antibody generated does not thoroughly cover the functional domain of NELF (Fig. 1). Further investigations into the function of NELF were thus initiated by inhibiting the expression of NELF at the mRNA/protein translation level.
Treating the nasal culture with antisense oligo DNA had a profound effect on the overall outgrowth of olfactory axons and the number of LHRH neurons present outside the main nasal tissue explant. However, a dramatic decrease in the number of LHRH neurons per length of NELF fibers that had extended from the culture was not observed. These fibers and cells may have been the earliest group to extend/migrate from the culture. As such, treatment at 3 div may not have perturbed their initial development but inhibited subsequent outgrowth and/or cell movement. If this occurred, then a decrease in NELF may have directly disrupted both olfactory axon outgrowth and LHRH cell migration. Alternatively, the reduction in LHRH neurons in the explant periphery may be a consequence of reduction in axon outgrowth and not direct inhibition of the migration of LHRH neurons themselves. The latter hypothesis argues against NELF functioning as the sole neurophilic molecule between LHRH neurons and olfactory axons and implies that NELF aids LHRH cell movement by playing a role in extending the pathway used by LHRH cells during migration through nasal regions.
NELF in the olfactory system
Interestingly, the developing olfactory bulb expresses NELF after olfactory axons begin to make contact in this area, causing bulb induction and evagination (Brunjes and Frazier 1986; Bailey et al. 1999). Robust NELF expression is observed in the outer nerve layer between E14.5 and E17.5. In the mouse, this is a time when axodendritic synapses are starting to form on the dendritic growth cones of the developing mitral cells, directly under the outer nerve layer (Brunjes and Frazier 1986). Thus, the spatiotemporal expression of NELF in olfactory receptor axons is consistent with a molecular cue that signals maturation in the olfactory bulb through olfactory axonal presence and/or innervation (Burd and Sein 1998; Bailey et al. 1999; Treloar et al. 1999).
Possible NELF variants or family genes
Western analysis of tissue samples revealed a 54- to 55-kDA protein cross-reactive with antibody 1026. A larger variant of NELF could be present within the cells using the 4.4-kb mRNA transcript observed upon Northern hybridization of multiple tissues (Fig. 1). This cross-reacting species could be a family member of NELF. Thus far, low-stringency screens of cDNA libraries containing high levels of the 4.4-kb transcript have produced a 2.8-kb cDNA transcript having the exact same sequence homology as NELF except for ∼200 bp 5′ of the sequence. This 5′ region does not contain the translational start site present in the NELF cDNA and suggests that alternatively spliced variants are present for NELF giving rise to larger protein products.
During preparation of this manuscript, a 1560-bp cDNA clone of the human homolog for NELF was submitted to GenBank (accession no. AL117660). Translation of the largest ORF indicated a high degree of conservation in the amino acid sequence, but most importantly, the peptide against which NELF was produced has 100% homology, suggesting the polyclonal antibody 1026 should bind to the human homolog of NELF.
In conclusion, a novel protein designated NELF was differentially expressed in LHRH neurons migrating within the nasal regions in contrast to LHRH neurons in the brain. NELF's expression pattern and extracellular location suggests a role as an axon outgrowth/migration factor, via a homophilic interaction, in nasal regions and specifically while the neuroendocrine LHRH neurons are migrating across the nasal septum into the developing CNS.
Materials and methods
Cell isolation and cDNA construction and anaysis
E11.0–E11.5 embryos were removed from NIH-Swiss mice in accordance with NIH guidelines, and unilateral and bilateral olfactory pits were dissected, trimmed, and grown in vitro for several days under serum-free conditions (Fueshko and Wray 1994). Single cells were isolated using microcapillary pipettes, cDNA was produced, and PCR amplification and library screening were performed as described previously (Dulac and Axel 1995; Kramer et al. 2000). In this procedure, single-cell aliquots were placed into a reaction mixture containing 4 μl of lysis buffer (Kramer et al. 2000) and 10 μm of each dATP, dCTP, dTTP, and dGTP) at 4°C. The cells were lysed, 50 UNITS of Moloney murine leukemia virus and 0.5 UNITS of avian reverse transcriptases (Bethesda Research Laboratories, Beverly, MA) were added, and the mixture was incubated for 15 min at 37°C and then heat inactivated at 65°C for 10 min. Then, a poly(A) addition was performed on the cDNA followed by PCR amplification using an AL1 primer (Kramer et al. 2000). Southern analysis was performed using 1 μg of amplified single-cell cDNA run on a 1.5% agarose gel, blotted on nylon membrane (Genescreenplus: DuPont, Boston, MA). The membranes were probed with LHRH (an ∼360-bp BamHI–EcoRI fragment of the rat LHRH cDNA including exons 1–4, a gift from Dr. J. Adelman; Adelman et al. 1986). The PCR products of LHRH-positive cells were digested with EcoRI and packaged using Gigapack Gold packaging extract (Stratagene, La Jolla, CA) according to manufacturer's directions.
Comparative analysis of LHRH neurons, and PCR amplification and sequencing of inserts
Plaque-lifts using GeneScreenPlus membranes (DuPont, Boston, MA) were completed. Membranes displaying the single-cell cDNA library from a cell grown in vitro for 3.5 days was hybridized with a PCR labeled cDNA library from two other LHRH-positive neurons grown in vitro for 10.5 days (Kramer et al. 2000). Probes were labeled with [α-32P]-dCTP by PCR amplification (Kramer et al. 2000). The membranes were hybridized for >12 hr, washed twice for 45 min at 60°C in a solution of 2× SSC (300 mm NaCl, 30 mm sodium citrate at pH 7.0), 1% SDS, and exposed to film.
Differentially expressed cDNA inserts were PCR-amplified by picking isolated plaques with a pipette tip (Kramer et al. 2000). Southern analysis was performed on duplicate blots. Each blot was hybridized (procedures described above) with a different [32P]-dCTP labeled cDNA library (Kramer et al. 2000); one blot was hybridized with the 3.5 div. library and the other with the 10.5 div. library. The inserts that were differentially expressed in the second screen were purified using Microcon 100 concentrators (Amicon, Beverly, MA), primers were designed, and the insert DNA was sequenced.
RACE analysis
Rapid amplification of cDNA ends (RACE) was perfomed using the Marathon cDNA amplification Kit (Clontech, Palo Alto, CA) and primer 1802R (5′-GCTCCAACTATGTACAAGAGAGCCCAC-3′) along with the Mouse Brain Marathon-Ready cDNA (Clontech) according to manufacturer's protocols. The RACE product was cloned directly into the TA vector pCR 2.1 (Invitrogen, Calsbad, CA).
Northern analysis
Premade mouse embryo multiple tissue and mouse brain Northern blots were obtained from Clontech (Palo Alto, CA) and hybridized according to the manufacturer's directions using a 1600-bp XhoI fragment from the NELF cDNA clone as a probe.
In vitro translation
The NotI cDNA fragment was cloned into pZero-2 (Invitrogen). In vitro translation was performed using the TnT coupled reticulocyte lysate systems (Promega, WI) using T7 polymerase and l-[35S]methionine according to the manufacturer's directions.
In situ hybridization histochemistry
In situ hybridization was performed as described previously (Wray et al. 1991) using a synthetic deoxynucleotide against NELF mRNA. The synthetic 48-base nucleotide probe (5 pmoles), complementary to NELF (5′-CTAAGGGATATCGTACATTGTTCAGTG-CCCAATCCTAACTCCCCTGGGTC-3′), was 3′ end-labeled with 35S-labeled dATP (specific activity 1000–1500 Ci/mmole, DuPont–NEN).
Single- and double-label immunocytochemistry
Fresh, frozen mouse embryos sections or explants were processed as described previously (Fueshko and Wray 1994; Wray et al. 1994). ProLHRH antisera (Wray et al. 1988) was used at 1:2500, 1026 polyclonal antisera was used at 1:4000, peripherin was used at 1:2000 (Chemicon, Temecula, CA). Production of the 1026 antibody was performed using alternating injections of 150–200 μg of the KLH, BSA-conjugated peptide in rabbits (see Fig. 1). Single-label immunocytochemistry was preformed on explants using standard ABC-horseradish peroxidase and diaminobenzidine [brown reaction (Fueshko and Wray 1994)]. Dual fluorescence used directly conjugated fluorescent Cy-3 goat anti-rabbit secondary (1:1000; Jackson ImmunoResearch Laboratories, West Grove, PA) and goat anti-rabbit avidin–FITC complex (Vector, Burlinghame, CA). Positive cells were captured with a Videoscope ICCD-35OF (Sterling, Virginia) camera or a confocal microscope (Zeiss 410 LSM). Controls for double-label immunocytochemical experiments consisted of the replacement of either the first primary or the second primary antibody with a normal goat serum incubation. Control sections revealed no cross-reactivity between the first and second labeling procedures (data not shown).
Antisense treatment
Nasal explants grown in defined media (Fueshko and Wray 1994) were treated on day 3 with 200 μl of Lipofectin–oligo complexes for 24 hr. Lipofectin–oligo complexes were made by adding 12 μl of Lipofectin reagent (GIBCO BRL) to 88 μl of OPTIMEM (GIBCO BRL) incubated for 30 min at room temperature and combining with a mixture containing ∼0.2 μmole of oligo in 100 μl of OPTIMEM and incubating 15 min at room temp before addition. After treatment fresh SFM was added, and the cultures were kept at standard conditions (Fueshko and Wray 1994) for an additional 3.5 days. The random sequence oligo RMCNT1 (5′-CTCGCAGTGCACCGTGGAGCG-3′) was chosen by the critera that it had a similar G + C content as the antisense oligo 18NPR8.5 (5′-GCTGCCTGGGCTGGCACGCAC-3′), lacked repeat elements, and lacked sequences that have the potential to form hairpins. After treatment, cultures were either fixed and processed for immunocytochemistry as descibed above (n = 45), processed for LHRH content via ELISA assay methods as reported previously (Maurer and Wray 1999), or processed for protein content by Western analysis (n = 13; see below).
Western analysis
Embryonic cell lysates were homogenized in lysate buffer (100 mm NaCl, 25 mm tris-HCL at pH 7.0, 1 mm EDTA, 1 mm EGTA, 1 mm AEBSF, 0.5% NP-40, 10 mm β-glycerolphosphate), incubated on ice for 30 min, mixed, and centrifuged, and the total protein of the supernatants was quantitated using the BCA protein assay kit (Pierce, Rockford, IL). For each experiment, 2–6 nasal explant cultures were combined, mixed with loading dye (SDS–protein gel loading solution; Quality Biological, Gaithersburg, MD), and heated at 95°C for 5 min. The lysate and translated product was electrophoresed through a Novex (San Diego, CA) 8%–16% Tris-gylcine–acrylamide gel at 125 V and transferred onto PVDF membrane in 25 mm Tris, 192 mm glycine, 0.1% SDS (pH 8.3) at 150 mA for 5 hr at room temperature. The membrane was blocked [20 mm Tris-HCl, 150 mm NaCl, 0.2% Tween 20, 2% dried milk (wt/vol) at pH 7.4] for 1 hr. The polyclonal antibody 1026 was added at 1:50,000 dilution; monoclonal antibody β-tubulin (Amersham) was added at 1:3000 dilution and incubated at 4°C overnight. The membrane was washed, and an HRP–goat anti-rabbit (for 1026) or anti-mouse (for β-tubulin) secondary antibody was added. After a 2-hr incubation and washing at room temperature, the complex was reacted using the Western blot chemiluminescence reagent plus kit (NEN, DuPont, CN) according to the manufacturer's directions and exposed to film. Quantitation of the band's optical density was performed on a scanned image of the film using NIH Image 1.62 (NIH, Bethesda, MD, Wayne Rasband). Values are given as the mean optical density ± standard error of the mean, O.D. ± s.e.m. The optical density of the β-tubulin band was used as a reference to correct for differences in loading between wells [(1/O.D. β-tubulin * 1000) * O.D. 1026] = corrected O.D. of 1026 band.
Analysis of antisense-treated cultures
Statistical signifigance was determined using a one-way Anova. Analysis between groups was performed using Tukey's Multiple Comparison Test or Bonferroni's Multiple Comparison Test.
Acknowledgments
We are indebted to Dr. P. Moore for the generation of nasal explants and ELISA assays for LHRH content after antisense treatment. We thank J. Nagle of the NINDS sequencing facility. NELF accession no. AF266508.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL swray@codon.nih.gov; FAX (301) 496-8578.
References
- Adams RH, Betz H, Puschel AW. A novel class of murine semaphorins with homology to thrombospondin is differentially expressed during early embryogenesis. Mech Dev. 1996;57:33–45. doi: 10.1016/0925-4773(96)00525-4. [DOI] [PubMed] [Google Scholar]
- Adelman JP, Mason AJ, Hayflick JS, Seeburg PH. Isolation of the gene and hypothalamic cDNA for the common precursor of gonadotropin-releasing hormone and prolactin release-inhibiting factor in human and rat. Proc Natl Acad Sci. 1986;83:179–183. doi: 10.1073/pnas.83.1.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey MS, Puche AC, Shipley MT. Development of the olfactory bulb: Evidence for glia-neuron interactions in glomerular formation [see comments] J Comp Neurol. 1999;415:423–448. [PubMed] [Google Scholar]
- Brose K, Bland KS, Wang KH, Arnott D, Henzel W, Goodman CS, Tessier-Lavigne M, Kidd T. Slit proteins bind Robo receptors and have an evolutionarily conserved role in repulsive axon guidance. Cell. 1999;96:795–806. doi: 10.1016/s0092-8674(00)80590-5. [DOI] [PubMed] [Google Scholar]
- Brunjes PC, Frazier LL. Maturation and plasticity in the olfactory system of vertebrates. Brain Res. 1986;396:1–45. doi: 10.1016/s0006-8993(86)80188-3. [DOI] [PubMed] [Google Scholar]
- Burd GD, Sein V. Influence of olfactory innervation on neurogenesis in the developing olfactory bulb of the frog, Xenopus laevis. Ann NY Acad Sci. 1998;855:270–273. doi: 10.1111/j.1749-6632.1998.tb10581.x. [DOI] [PubMed] [Google Scholar]
- Chen H, Chedotal A, He Z, Goodman CS, Tessier-Lavigne M. Neuropilin-2, a novel member of the neuropilin family, is a high affinity receptor for the semaphorins Sema E and Sema IV but not Sema III [published erratum appears in Neuron 1997 Sep;19(3):559] Neuron. 1997;19:547–559. doi: 10.1016/s0896-6273(00)80371-2. [DOI] [PubMed] [Google Scholar]
- Cremer H, Lange R, Christoph A, Plomann M, Vopper G, Roes J, Brown R, Baldwin S, Kraemer P, Scheff S. Inactivation of the N-CAM gene in mice results in size reduction of the olfactory bulb and deficits in spatial learning. Nature. 1994;367:455–459. doi: 10.1038/367455a0. [DOI] [PubMed] [Google Scholar]
- Cunningham BA, Hemperly JJ, Murray BA, Prediger EA, Brackenbury R, Edelman GM. Neural cell adhesion molecule: Structure, immunoglobulin-like domains, cell surface modulation, and alternative RNA splicing. Science. 1987;236:799–806. doi: 10.1126/science.3576199. [DOI] [PubMed] [Google Scholar]
- Dulac C, Axel R. A novel family of genes encoding putative pheromone receptors in mammals. Cell. 1995;83:195–206. doi: 10.1016/0092-8674(95)90161-2. [DOI] [PubMed] [Google Scholar]
- Edelman GM. Cell adhesion and the molecular processes of morphogenesis. Annu Rev Biochem. 1985;54:135–169. doi: 10.1146/annurev.bi.54.070185.001031. [DOI] [PubMed] [Google Scholar]
- Fink G. Gonadotropin secretion and its control. In: Knobil E, Neil J, editors. The physiology of reproduction. New York, NY: Raven Press; 1988. pp. 1349–1377. [Google Scholar]
- Fueshko S, Wray S. LHRH cells migrate on peripherin fibers in embryonic olfactory explant cultures: An in vitro model for neurophilic neuronal migration. Dev Biol. 1994;166:331–348. doi: 10.1006/dbio.1994.1319. [DOI] [PubMed] [Google Scholar]
- Giger RJ, Wolfer DP, De Wit GM, Verhaagen J. Anatomy of rat semaphorin III/collapsin-1 mRNA expression and relationship to developing nerve tracts during neuroembryogenesis. J Comp Neurol. 1996;375:378–392. doi: 10.1002/(SICI)1096-9861(19961118)375:3<378::AID-CNE3>3.0.CO;2-#. [DOI] [PubMed] [Google Scholar]
- He Z, Tessier-Lavigne M. Neuropilin is a receptor for the axonal chemorepellent semaphorin III. Cell. 1997;90:739–751. doi: 10.1016/s0092-8674(00)80534-6. [DOI] [PubMed] [Google Scholar]
- Holmes GP, Negus K, Burridge L, Raman S, Algar E, Yamada T, Little MH. Distinct but overlapping expression patterns of two vertebrate slit homologs implies functional roles in CNS development and organogenesis. Mech Dev. 1998;79:57–72. doi: 10.1016/s0925-4773(98)00174-9. [DOI] [PubMed] [Google Scholar]
- Itoh A, Miyabayashi T, Ohno M, Sakano S. Cloning and expressions of three mammalian homologues of Drosophila slit suggest possible roles for Slit in the formation and maintenance of the nervous system. Brain Res Mol Brain Res. 1998;62:175–186. doi: 10.1016/s0169-328x(98)00224-1. [DOI] [PubMed] [Google Scholar]
- Keverne EB. The vomeronasal organ. Science. 1999;286:716–720. doi: 10.1126/science.286.5440.716. [DOI] [PubMed] [Google Scholar]
- Kitsukawa T, Shimizu M, Sanbo M, Hirata T, Taniguchi M, Bekku Y, Yagi T, Fujisawa H. Neuropilin-semaphorin III/D-mediated chemorepulsive signals play a crucial role in peripheral nerve projection in mice. Neuron. 1997;19:995–1005. doi: 10.1016/s0896-6273(00)80392-x. [DOI] [PubMed] [Google Scholar]
- Kolodkin AL, Levengood DV, Rowe EG, Tai YT, Giger RJ, Ginty DD. Neuropilin is a semaphorin III receptor. Cell. 1997;90:753–762. doi: 10.1016/s0092-8674(00)80535-8. [DOI] [PubMed] [Google Scholar]
- Kramer PR, Krishnamurthy R, Mitchell PJ, Wray S. Transcription factor activator protein-2 is required for continued luteinizing hormone-releasing hormone expression in the forebrain of developing mice. Endocrinology. 2000;141:1823–1838. doi: 10.1210/endo.141.5.7452. [DOI] [PubMed] [Google Scholar]
- Kusano K, Fueshko S, Gainer H, Wray S. Electrical and synaptic properties of embryonic luteinizing hormone-releasing hormone neurons in explant cultures. Proc Natl Acad Sci. 1995;92:3918–3922. doi: 10.1073/pnas.92.9.3918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurer JA, Wray S. Luteinizing hormone-releasing hormone quantified in tissues and slice explant cultures of postnatal rat hypothalami “In Process Citation”. Endocrinology. 1999;140:791–799. doi: 10.1210/endo.140.2.6534. [DOI] [PubMed] [Google Scholar]
- Mombaerts P. Seven-transmembrane proteins as odorant and chemosensory receptors. Science. 1999;286:707–711. doi: 10.1126/science.286.5440.707. [DOI] [PubMed] [Google Scholar]
- Mori K. Molecular and cellular properties of mammalian primary olfactory axons. Microsc Res Tech. 1993;24:131–141. doi: 10.1002/jemt.1070240205. [DOI] [PubMed] [Google Scholar]
- Mori K, Nagao H, Yoshihara Y. The olfactory bulb: Coding and processing of odor molecule information. Science. 1999;286:711–715. doi: 10.1126/science.286.5440.711. [DOI] [PubMed] [Google Scholar]
- Mueller BK. Growth cone guidance: First steps towards a deeper understanding. Annu Rev Neurosci. 1999;22:351–388. doi: 10.1146/annurev.neuro.22.1.351. [DOI] [PubMed] [Google Scholar]
- Murakami S, Seki T, Wakabayashi K, Arai Y. The ontogeny of luteinizing hormone-releasing hormone (LHRH) producing neurons in the chick embryo: Possible evidence for migrating LHRH neurons from the olfactory epithelium expressing a highly polysialylated neural cell adhesion molecule. Neurosci Res. 1991;12:421–431. doi: 10.1016/0168-0102(91)90073-8. [DOI] [PubMed] [Google Scholar]
- Nguyen Ba-Charvet KT, Brose K, Marillat V, Kidd T, Goodman CS, Tessier-Lavigne M, Sotelo C, Chedotal A. Slit2-mediated chemorepulsion and collapse of developing forebrain axons. Neuron. 1999;22:463–473. doi: 10.1016/s0896-6273(00)80702-3. [DOI] [PubMed] [Google Scholar]
- Nishizuka M, Ikeda S, Arai Y, Maeda N, Noda M. Cell surface-associated extracellular distribution of a neural proteoglycan, 6B4 proteoglycan/phosphacan, in the olfactory epithelium, olfactory nerve, and cells migrating along the olfactory nerve in chick embryos. Neurosci Res. 1996;24:345–355. doi: 10.1016/0168-0102(95)01010-6. [DOI] [PubMed] [Google Scholar]
- Pasterkamp, R.J., Ruitenberg, M.J., and Verhaagen, J. 1999. Semaphorins and their receptors in olfactory axon guidance. Cell Mol. Biol.(Noisy.-le.-grand.) 45: 763–779. [PubMed]
- Puche AC, Poirier F, Hair M, Bartlett PF, Key B. Role of galectin-1 in the developing mouse olfactory system. Dev Biol. 1996;179:274–287. doi: 10.1006/dbio.1996.0257. [DOI] [PubMed] [Google Scholar]
- Quinton R, Hasan W, Grant W, Thrasivoulou C, Quiney RE, Besser GM, Bouloux PM. Gonadotropin-releasing hormone immunoreactivity in the nasal epithelia of adults with Kallmann's syndrome and isolated hypogonadotropic hypogonadism and in the early midtrimester human fetus. J Clin Endocrinol Metab. 1997;82:309–314. doi: 10.1210/jcem.82.1.3673. [DOI] [PubMed] [Google Scholar]
- Rutishauser U, Jessell TM. Cell adhesion molecules in vertebrate neural development. Physiol Rev. 1988;68:819–857. doi: 10.1152/physrev.1988.68.3.819. [DOI] [PubMed] [Google Scholar]
- Schwanzel-Fukuda M, Pfaff DW. Origin of luteinizing hormone-releasing hormone neurons. Nature. 1989;338:161–164. doi: 10.1038/338161a0. [DOI] [PubMed] [Google Scholar]
- Schwanzel-Fukuda M, Bick D, Pfaff DW. Luteinizing hormone-releasing hormone (LHRH)-expressing cells do not migrate normally in an inherited hypogonadal (Kallmann) syndrome. Brain Res Mol Brain Res. 1989;6:311–326. doi: 10.1016/0169-328x(89)90076-4. [DOI] [PubMed] [Google Scholar]
- Schwanzel-Fukuda M, Abraham S, Crossin KL, Edelman GM, Pfaff DW. Immunocytochemical demonstration of neural cell adhesion molecule (NCAM) along the migration route of luteinizing hormone-releasing hormone (LHRH) neurons in mice. J Comp Neurol. 1992;321:1–18. doi: 10.1002/cne.903210102. [DOI] [PubMed] [Google Scholar]
- Treloar HB, Purcell AL, Greer CA. Glomerular formation in the developing rat olfactory bulb. J Comp Neurol. 1999;413:289–304. [PubMed] [Google Scholar]
- Winberg ML, Noordermeer JN, Tamagnone L, Comoglio PM, Spriggs MK, Tessier-Lavigne M, Goodman CS. Plexin A is a neuronal semaphorin receptor that controls axon guidance. Cell. 1998;95:903–916. doi: 10.1016/s0092-8674(00)81715-8. [DOI] [PubMed] [Google Scholar]
- Wray S, Gahwiler BH, Gainer H. Slice cultures of LHRH neurons in the presence and absence of brainstem and pituitary. Peptides. 1988;9:1151–1175. doi: 10.1016/0196-9781(88)90103-9. [DOI] [PubMed] [Google Scholar]
- Wray S, Grant P, Gainer H. Evidence that cells expressing luteinizing hormone-releasing hormone mRNA in the mouse are derived from progenitor cells in the olfactory placode. Proc Natl Acad Sci. 1989;86:8132–8136. doi: 10.1073/pnas.86.20.8132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wray S, Kusano K, Gainer H. Maintenance of LHRH and oxytocin neurons in slice explants cultured in serum-free media: Effects of tetrodotoxin on gene expression. Neuroendocrinology. 1991;54:327–339. doi: 10.1159/000125910. [DOI] [PubMed] [Google Scholar]
- Wray S, Key S, Qualls R, Fueshko SM. A subset of peripherin positive olfactory axons delineates the luteinizing hormone releasing hormone neuronal migratory pathway in developing mouse. Dev Biol. 1994;166:349–354. doi: 10.1006/dbio.1994.1320. [DOI] [PubMed] [Google Scholar]
- Wu W, Wong K, Chen J, Jiang Z, Dupuis S, Wu JY, Rao Y. Directional guidance of neuronal migration in the olfactory system by the protein Slit [see comments] Nature. 1999;400:331–336. doi: 10.1038/22477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K, Tobet SA, Crandall JE, Jimenez TP, Schwarting GA. The migration of luteinizing hormone-releasing hormone neurons in the developing rat is associated with a transient, caudal projection of the vomeronasal nerve. J Neurosci. 1995;15:7769–7777. doi: 10.1523/JNEUROSCI.15-12-07769.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoshida K, Rutishauser U, Crandall JE, Schwarting GA. Polysialic acid facilitates migration of luteinizing hormone-releasing hormone neurons on vomeronasal axons. J Neurosci. 1999;19:794–801. doi: 10.1523/JNEUROSCI.19-02-00794.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]