Abstract
Messenger RNA export is mediated by the TAP-p15 heterodimer, which belongs to the family of NTF2-like export receptors. TAP-p15 heterodimers also bind to the constitutive transport element (CTE) present in simian type D retroviral RNAs, and mediate export of viral unspliced RNAs to the host cytoplasm. We have solved the crystal structure of the RNA recognition and leucine-rich repeat motifs of TAP bound to one symmetrical-half of CTE RNA. L-shaped conformations of protein and RNA are involved in a mutual molecular embrace on complex formation. We have monitored the impact of structure-guided mutations on binding affinities in vitro and transport assays in vivo. Our studies define the principles by which CTE RNA subverts the mRNA export receptor TAP, thereby facilitating nuclear export of viral genomic RNAs, and more generally, provide insights on cargo RNA recognition by mRNA export receptors.
Keywords: nucleocytoplasmic retroviral RNA transport, mRNA export factor TAP, retroviral constitutive transport element (CTE), RNA recognition motif (RRM), leucine-rich repeat (LRR) motif
Nucleocytoplasmic transport receptors1,2 facilitate the export of processed RNA from the nucleus to the cytoplasm, where the RNA provides the critical information-carrying template that mediates the translation process of protein synthesis. Specific RNA cargoes are recognized by versatile soluble transport receptors (exportins) and exported through the nuclear pore complex (NPC) in a unidirectional manner3–6.
Transport of small nuclear, ribosomal, micro and transfer RNAs through nuclear pore complexes is mediated by the β-karyopherin superfamily of exportins, composed of superhelices of modular HEAT repeats, in conjunction with the regulator Ran, a guanosine triphosphatase. The RNA cargoes are bound by exportin-Ran-GTP complex in the nucleus, and released in the cytoplasm following GTP hydrolysis7. Recent crystal structures of Ran-GTP complexes of exportins Xpo-1 bound to tRNA8 and Exp-5 bound to pre-miRNA9 have established how the β-karyopherin superfamily use shape, flexibility and electrostatic complementarity to recognize and distinguish between RNA cargos based on their distinct 3'-overhang ends.
By contrast, bulk mRNA export is independent of β-karyopherin-Ran, and dependent on the export factor TAP (also known as NXF1 in metazoans) bound to the protein p1510–13. TAP is a multidomain protein composed of conserved N-terminal RNA recognition (RRM) and leucine-rich repeat (LRR) motifs, a central NTF2-like domain that binds p1514, and a C-terminal ubiquitin associated (UBA)-like fold15. Crystal structures are available of the individual domains in the free state16,17, including a structure of the NTF2-like domain bound to p1514. Both the NTF2-like and UBA-like folds contain binding sites for Phe-Gly-repeats that are prevalent in FG-nucleoporins18, thereby contributing to nuclear pore complex (NPC) association and export of cellular mRNAs19.
Normally, the TAP-p15 heterodimer is only recruited to mature, fully-spliced mRNAs via interactions with export adaptors, Aly/REF and the shuttling SR proteins4. Retroviruses subvert mRNA nuclear export factors to promote the efficient export of unspliced forms of viral RNA, essential for viral replication in the cytoplasm. The TAP-p15 complex is recruited by a cis-acting RNA element embedded in viral pre-mRNAs of simian type D retroviruses, called the constitutive transport element (CTE) RNA11,20,21, and mediates the export of viral unspliced genomic RNA from the nucleus to the cytoplasm11, thereby circumventing their nuclear retention22. A cellular CTE equivalent was also found in the alternatively spliced intron of the Tap gene and shown to facilitate the efficient expression of TAP mRNA containing this intron, thus pointing to a CTE-dependent mechanism to overcome nuclear retention of mRNAs with unspliced introns23.
Given that TAP-p15 recognizes the CTE RNA, we set out to solve the structure of components of this complex, as part of our efforts to define the molecular principles underlying protein-RNA recognition associated with targeting and sequestration of a novel RNA fold for TAP-mediated transport through the nuclear pore. Based on the anticipated structure of TAP bound to CTE RNA, we planned to measure the impact of structure-guided disruption of intermolecular contacts on binding affinities in vitro and on nucleocytoplasmic export activities using intact TAP-p15 complex and full-length CTE RNA at the cellular level.
RESULTS
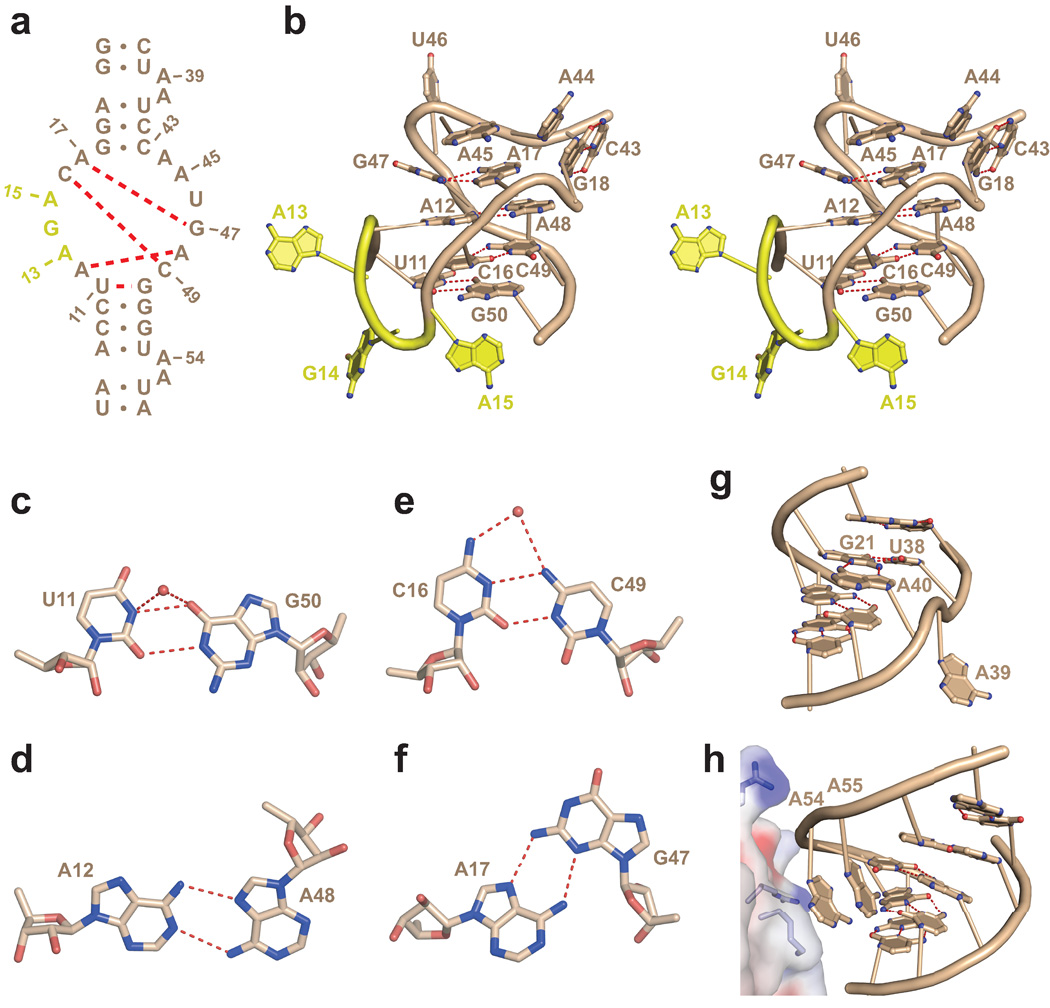
Previous research has established that the N-terminal domain of TAP (TAP-NTD) composed of RRM and LRR domains (Fig. 1a) is sufficient for binding full-length CTE RNA (Fig. 1b)10,12,16,24. The conserved internal loops of the full-length CTE RNA are related by two-fold symmetry, and it has been shown that a single internal loop is sufficient for TAP binding in vitro and in vivo10,24.
Figure 1.
Structure of TAP-NTD bound to the hCTE RNA. (a) Domain architecture of full-length TAP and TAP-NTD construct (96 to 362) consisting of the RRM domain (in green) and the LRR domain (in blue) used for crystallization of the complex. (b) The full-length CTE RNA consisting of two large internal loops whose sequences are related by two-fold symmetry (indicated by thick and thin arrows). (c) Sequence of one symmetrical half of CTE (half CTE, designated hCTE). The 1 to 62 sequence of hCTE corresponds to the half containing the hairpin loop (top half) of the full-length CTE in panel B, with the hairpin loop replaced by a stable GAAA RNA loop (in red) in the construct used for crystallization. The bases colored in red are different from the wild-type CTE sequence and were incorporated to facilitate efficient in vitro transcription (5'-GG), hammerhead ribozyme cleavage site (UC-3') and improve crystal quality (GAAA loop with GC closing pair). The residues are numbered from 1 to 62. (d, e) Two alternate views of the crystal structure of the 1:1 complex of TAP-NTD bound to hCTE. The RRM domain is in green, the LRR domain is in blue, and the RNA is in wheat. Flipped out RNA residues A13, G14 and A15 are in yellow. The N and C termini of TAP-NTD and the 5' and 3' ends of the RNA are labeled.
Structure of TAP-NTD bound to hCTE RNA
We therefore generated a soluble construct of TAP-NTD (amino acids 96 to 362, Fig. 1a) and attempted crystallization trials with both full-length CTE (Fig. 1b) and a 62-nt CTE fragment (Fig. 1c), composed of one symmetrical half (containing a stable GAAA variant of the hairpin loop) of the full-length RNA construct, designated half-CTE (hCTE). We were able to grow crystals of the complex of TAP-NTD with hCTE that diffracted to 2.30 Å resolution, and solved the structure by molecular replacement using the known crystal structure of TAP-NTD in the free state16 and the A-form RNA duplex structures as independent search models (see Online Methods). Similar to previously reported structures of free TAP-NTD16,17, N-terminal amino acids 96–117 were disordered in the crystal structure of the complex with hCTE.
Two alternate views of the structure of the 1:1 complex of TAP-NTD and 62-nt CTE RNA are shown in Fig. 1d,e (structural statistics are listed in Table 1), with the RNA in a stick representation and the protein in a ribbon representation. The same views with the protein in an electrostatic surface representation are shown in Supplementary Fig. 1a,b. Both the protein and RNA adopt L-shaped conformations and are involved in a mutual molecular embrace (Fig. 1d,e).
Table 1.
X-ray data collection and refinement statistics
| Data collection | TAP-NTD/hCTE complex | TAP-NTD |
|---|---|---|
| Space group | P21 | P412121 |
| Cell dimensions | ||
| a, b, c (Å) | 55.4, 117.0, 84.8 | 139.4, 139.4, 207.1 |
| α, β, γ (°) | 90.0, 106.4, 90.0 | 90.0, 90.0, 90.0 |
| Resolution (Å) | 50-2.3 | 50-3.0 |
| Rmerge | 7.4 (23.1)a | 8.8 (51) |
| I / σI | 18.1 (3.1) | 17.9 (1.6) |
| Completeness (%) | 96.6 (92.3) | 99.5 (97.1) |
| Redundancy | 4.8 | 6.9 |
| Refinement | ||
| Resolution (Å) | 20-2.3 | 49-3.0 |
| No. reflections | 42,253 (2,256) | 40,963 (2,055) |
| Rwork / Rfree | 20.3 / 25.6 | 26.0 / 31.0 |
| No. atoms | ||
| Protein | 3971 | 6489 |
| RNA | 2680 | |
| Water | 135 | 8 |
| B-factors | ||
| Protein | 48.4 | 76.7 |
| RNA | 49.4 | |
| Water | 38.2 | 43.5 |
| R.m.s. deviations | ||
| Bond lengths (Å) | 0.007 | 0.010 |
| Bond angles (°) | 1.3 | 1.3 |
Values in parentheses are for highest-resolution shell.
The structures of TAP-NTD in the free and hCTE RNA-bound states are compared following superposition of their LRR domains in Supplementary Fig. 2a. The linker spanning the Ser194 to Leu204 segment of TAP-NTD bound to the hCTE is well ordered and fixes the relative orientations of the RRM and LRR domains on complex formation (Supplementary Fig. 2b).
Structure of the hCTE RNA in complex with TAP
When bound to the TAP protein, the large internal loop of the hCTE RNA zippers up through formation of four mutually stacked non-canonical pairs (schematic in Fig. 2a and stereo view in Fig. 2b), with the pairing alignments of U11•G50, A12•A48, C16•C49 and A17•G47 non-canonical pairs shown in Fig. 2c–f. Three residues A13, G14 and A15 are splayed out (in yellow, in Fig. 1d,e and Fig. 2b) of the zippered up duplex, while three other residues, A44 (stacked on top of G18•C43), A45 (stacked on top of A17•G47) and U46 are also unpaired, and facilitate the formation of the L-shaped scaffold of the bound hCTE RNA.
Figure 2.
Fold of the internal loop and bulged bases in the hCTE-TAP complex. (a) The sequence and numbering of the internal loop and bulged bases of hCTE, with dashed red lines indicating formation of four non-canonical pairs on complex formation. (b) A stereo pair of the structure of the zippered up internal loop of hCTE in the complex. The RNA is in wheat and flipped out A13, G14 and A15 are in yellow. The core of the fold contains four stacked pairs, with junctional U11•G50 and G18-C43 pairs aligned at right angles to each other. (c–f) Pairing alignments of non-canonical pairs within the zippered up internal loop of the complex. The non-canonical pairing alignments are U11•G50 (panel c), A12•A48 (panel d), C16•C49 (panel e) and A17•G47 (panel f). (g, h) Conformation of the A-A bulges in the upper (A39–A40) and lower (A54–A55) stems of the hCTE RNA in the complex. The A39–A40 bulge in the upper stem of the complex is shown in panel g, with A39 flipped out, while A40 is stacked within the duplex and forms an A40•(G21•U38) triple. The A54–A55 bulge in the lower stem of the complex is shown in panel h, with both A54 and A55 flipped out, but mutually stacked on each other.
There are two A-A bulges within the hCTE RNA, with A39–A40 in the upper stem and A54–A55 in the lower stem (Fig. 2a). While A40 inserts into the duplex to form a A40•(G21-U38) triple, A39, A54 and A55 are flipped out of the helix (Fig. 2g,h). For both bulges, base pairs that flank the bulges are collinearly stacked, so that there is no bending or kinking of the stems at either A-A bulge site in the complex.
hCTE RNA recognition by the RRM and LRR domains
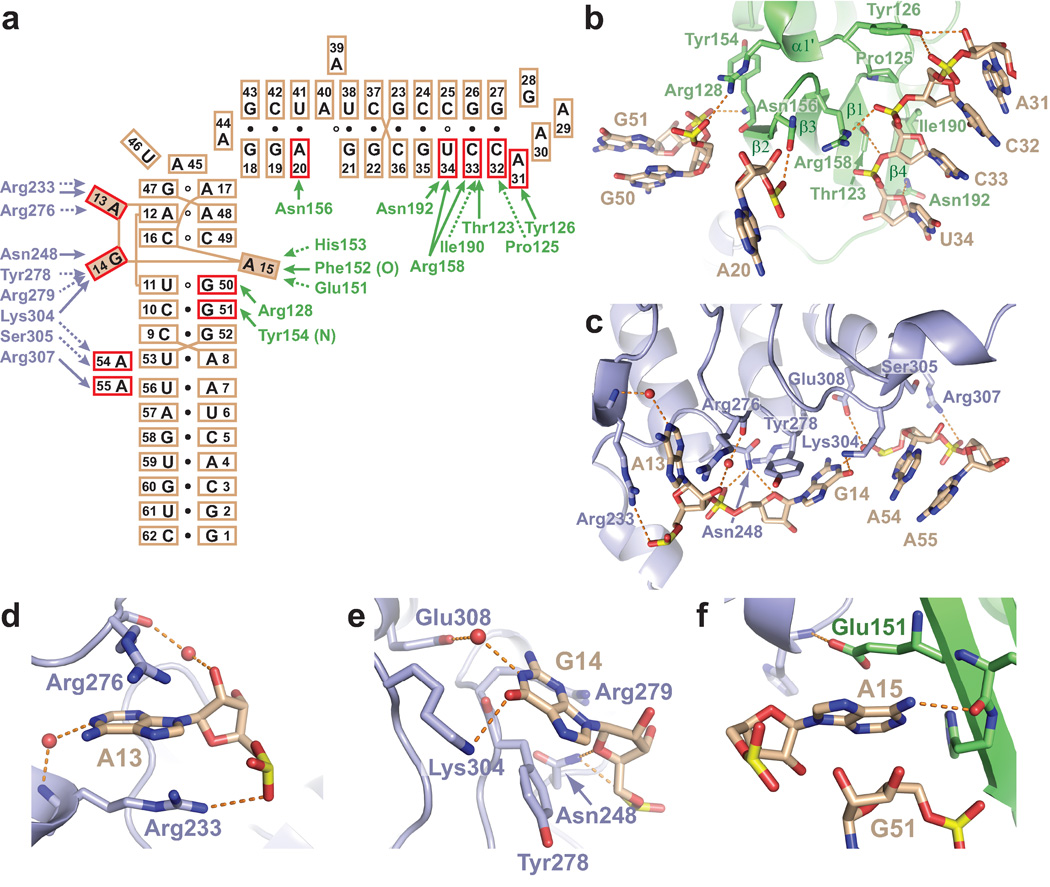
The residues involved in intermolecular protein-RNA contacts in the complex of TAP-NTD bound to hCTE RNA are shown schematically in Fig. 3a, with the basic (Arg and Lys), aromatic (Tyr), and acidic (Glu) amino acid side chains interacting with the L-shaped hCTE scaffold highlighted in a stereo view of the complex in Supplementary Fig. 3a.
Figure 3.
Key interactions between hCTE RNA and the TAP-NTD in the complex. (a) Schematic highlighting intermolecular hydrogen bond and hydrophobic protein-RNA contacts in the complex. hCTE RNA residues involved in base-specific and sugar-phosphate specific recognition are represented as shaded and red lined boxes, respectively. Amino acid residues of RRM (in green) and LRR (in blue) involved in hydrogen-bonding and hydrophobic/stacking interactions are shown by solid and dashed arrows, respectively. (b) Intermolecular contacts between residues of hCTE RNA (A20, A31-U34 and G50–G51) and the RRM domain of TAP-NTD in the complex. (c) Intermolecular contacts between residues of hCTE RNA (A13-G14 and A54–A55) and the LRR domain of TAP-NTD in the complex. (d–f) Intermolecular hydrogen bonds involving flipped out A13, which is inserted between Arg276 and Arg233 (panel d), flipped out G14, which is positioned on the surface patch involving Tyr278, Arg279 and Lys304 (panel e), and flipped out A15, which is inserted between Glu151 and the sugar of G51 (panel f).
The intermolecular contacts between the hCTE RNA and the RRM domain of TAP are shown in Fig. 3b, with an entire RRM perspective shown in Supplementary Fig. 3b. In essence, different segments of the hCTE RNA, spanning the upper stem (A20, A31-U34) and the loop-lower stem junction (G50–G51), form primarily sugar-phosphate backbone contacts with basic and hydroxyl-containing side chains of the RRM domain of TAP-NTD in the complex (Fig. 3a,b and Supplementary Fig. 3b). The intermolecular contacts are either with the edge of the β-sheet (β2 strand) or with side chains projecting from the face of the β-sheet (adjacent β1 and β4 strands), and β1-α1 and β2-β3 loops of the RRM in the complex. In addition, residue A15 is flipped out from the internal loop and forms a base specific-peptide backbone contact with the edge (β2 strand) of the β-sheet in the complex (Fig. 3a, and Supplementary Fig. 3a,b).
The intermolecular contacts between the hCTE RNA and the LRR domain of TAP are shown in Fig. 3c, with an entire LRR perspective shown in Supplementary Fig. 3c. In essence, the A13-G14 segment of the internal loop and the A54–A55 bulge of the lower stem of hCTE RNA form base edge and sugar-phosphate backbone contacts with loop elements projecting from the same face of the LRR domain of TAP in the complex (Fig. 3c and Supplementary Fig. 3c).
Key intermolecular contacts in the CTE-TAP complex
Residues A13, G14 and A15 are flipped out of the internal loop and form key intermolecular contacts with the RRM and LRR domains of TAP in the structure of the complex (Fig. 3a). The adenine ring of A13 interacts with the LRR domain, with the base sandwiched between the side chains of Arg276 and Arg233, and its Watson-Crick edge N1 nitrogen forming a water-mediated hydrogen bond with the amide of Arg233, buttressed by additional intermolecular hydrogen bonds involving its sugar-phosphate backbone (Fig. 3d). The guanine ring of G14 is flipped out and positioned on the surface of the LRR domain that is shaped by the side chains of Tyr278, Arg279 and Lys304 (Fig. 3e). The Watson-Crick edge of G14 forms direct and water-mediated hydrogen bonds to the side chains of Glu308 and Lys304, with its alignment further buttressed by additional intermolecular hydrogen bonds involving its sugar-phosphate backbone. The adenine ring of A15 is flipped out and interacts with the RRM domain, with the base sandwiched between the side chain of Glu151 and the sugar ring of G51, and the NH2-6 proton along its Watson-Crick edge hydrogen-bonded to the backbone carbonyl of Phe152 (Fig. 3f).
The side chains of Arg128 and Arg158 from the RRM domain of TAP form intermolecular contacts with sugar-phosphate backbones of stem segments on either side of the zippered up loop, and contribute to anchoring of the L-shaped RNA scaffold in the complex (Supplementary Fig. 3a and Fig. 3b). The guanidinium group of Arg128 participates in intermolecular hydrogen bond formation with the backbone phosphates of the G50–G51 step, while that of Arg158 forms intermolecular hydrogen bonds with the backbone phosphates of the C32-C33-U34 step (Fig. 3b).
Impact of TAP-CTE mutants on in vitro binding affinity
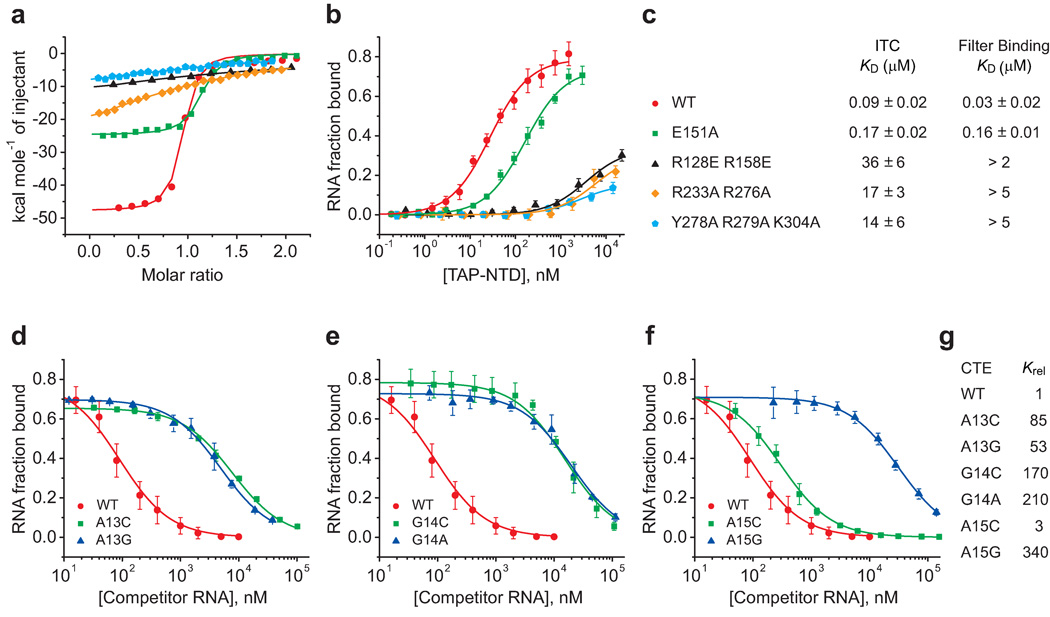
We have investigated the impact of mutations of the amino acids that contact flipped out A13, G14 and A15 nucleotides of the hCTE, and Arg128 and Arg158 of the RRM domain that contact sugar-phosphate backbones of CTE stem segments and contribute to the electrostatic interactions in the complex, on the binding affinities for complex formation between TAP-NTD and CTE. We measured a binding affinity of app. 0.09 µM for binding of wild-type TAP-NTD to hCTE RNA in 200 mM KCl buffer (red curve, Fig. 4a) by isothermal titration calorimetry (ITC). Mutations of both Arg233 and Arg276, that sandwich A13 (Fig. 3d), to Ala, result in a drop in binding affinity of greater than two orders of magnitude (orange curve, Fig. 4a), as does triple mutation of Tyr278, Arg279 and Lys304, that form a platform for G14 (Fig. 3e), to Ala (blue curve, Fig. 4a). A drop in binding affinity is also observed for reverse-charge mutations of both Arg128 and Arg158 on the RRM domain, that interact with the sugar-phosphate backbone of the hCTE (Fig. 3b), to Glu, (black curve, Fig. 4a). By contrast, mutation of Glu151, that is stacked on A15 (Fig. 4f), to Ala (green curve, Fig. 4a), results in a modest 2-fold loss in binding affinity.
Figure 4.
In vitro ITC and direct and competitive filter binding data on TAP-NTD and hCTE RNA mutants. (a, b) Isothermal titration calorimetry (panel a) and nitrocellulose filter (panel b) binding curves for complex formation between TAP-NTD and its mutants with hCTE RNA. Wild-type protein in red circles, dual R233A R276A mutants in orange diamonds, triple Y278A R279A K304A mutants in blue pentagons, single E151A mutant in green squares and dual R128E R158E mutants in black triangles. (c) Summary of the binding constants measured from ITC and filter binding assays. (d–f) Competitive nitrocellulose filter binding assay curves for complex formation between TAP-NTD and hCTE RNA mutants. Wild-type A13 (in red circles) mutated to G13 (in blue triangles) and C13 (in green squares) (panel d), wild-type G14 (in red circles) mutated to A14 (in blue triangles) and C14 (in green squares) (panel e), and wild-type A15 (in red circles) mutated to G15 (in blue triangles) and C15 (in green squares) (panel f). (g) A summary of the binding constants from competitive filter binding assays.
The binding affinities of the same TAP-NTD mutants for the hCTE RNA were also monitored by nitrocellulose filter-binding assays (see Supplementary Methods) in which the RNA was present in trace quantities, thereby providing an advantage over ITC for accurate determination of tight interactions. The measured binding affinity of 0.03 µM for complex formation with wild-type TAP-NTD drops by greater than two orders of magnitude for the dual R233A R276A mutants, the triple Y278A R279A K304A mutants and the dual reverse charge R128E R158E mutants (Fig. 4b), similar to what was observed by ITC measurements (Fig. 4a). By contrast, a 5-fold drop in binding affinity is observed for the E151A mutant (Fig. 4b). The binding affinities of TAP-NTD mutants measured by ITC and filter-binding assays are summarized in Fig. 4c.
We have also investigated the impact of mutations of the A13, G14 and A15 nucleotides of the hCTE on the binding affinities for complex formation with TAP-NTD by a nitrocellulose filter-binding competition assay. Replacement of A13 by C or G resulted in an affinity drop in the 50 to 85-fold range (Fig. 4d), replacement of G14 by C or A resulted in an affinity drop in the 170 to 210-fold range (Fig. 4e), while replacement of A15 by G resulted in an affinity drop in the 340-fold (Fig. 4f). By contrast, a modest 3-fold drop in affinity was observed following replacement of A15 by C (Fig. 4f). The binding affinities of hCTE mutants measured by competitive filter binding assays are summarized in Fig. 4g.
Impact of TAP-CTE mutants on in vivo export activity
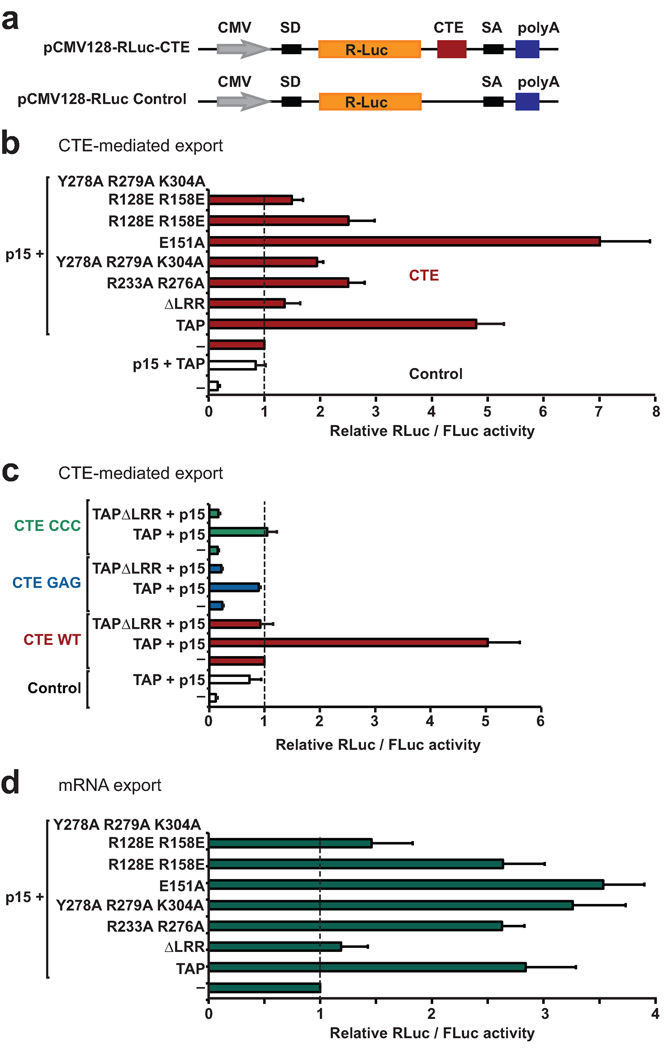
The effect of mutations disrupting the TAP-CTE interaction was tested in vivo using an assay that allows quantification of TAP export activity25. In this assay, human cells are transfected with a reporter plasmid harboring a Renilla luciferase (RLuc) coding sequence within an inefficiently spliced intron, which is therefore retained within the nucleus (Fig. 5a). When the CTE is inserted within the intron, export of the unspliced mRNA is stimulated compared to the control reporter lacking the CTE resulting in increased luciferase activity (Fig. 5b, white versus red bars). Over-expression of TAP-p15 heterodimers bypasses nuclear retention and further stimulates the export of the inefficiently spliced mRNA leading to a further increase in luciferase activity for both the control reporter and the CTE-containing RNA (Fig. 5b).
Figure 5.
In vivo RNA export assay. (a) Schematic representation of the pCMV128-RLuc-CTE reporter and the corresponding control without CTE. SD and SA, splicing donor and acceptor sites. (b) Human cells were transfected with a mixture of three plasmids: one expressing pCMV128-RLuc-CTE reporter or a control reporter lacking the CTE, another expressing Firefly luciferase, and a third expressing TAP (wild type or mutants). A plasmid expressing p15 was included in the transfection mixtures as indicated. Renilla luciferase activity was normalized to that of the Firefly luciferase transfection control and set to 1 in the absence of exogenous TAP. Mean values ± standard deviations are shown. (c) Human cells were transfected with the pCMV128-RLuc-CTE reporter carrying CTE wild-type or mutants. Plasmids expressing TAP and p15 were co-transfected as indicated. A plasmid expressing Firefly luciferase served as a transfection control. Luciferase activity was analyzed as described in panel a. (d) Human cells were transfected with a mixture of three plasmids: one expressing pCMV128-RLuc reporter lacking the CTE, another expressing Firefly luciferase, and a third expressing TAP (wild type or mutants). A plasmid expressing p15 was included in the transfection mixtures. Renilla and Firefly luciferase activities were measured and analyzed as described in panel b.
CTE-mediated mRNA export is reduced by mutations in the TAP protein that disrupt its binding to the CTE RNA, as indicated for the reverse-charge dual mutations R128E and R158E (Fig. 5b). Substitution of three additional residues (Y278A, R279A and K304A) abolishes TAP export activity as efficiently as deletion of the entire LRR domain (ΔLRR). A reduction in mRNA export activity of TAP was also observed for the dual R233A and R276A mutants. By contrast, the E151A mutant, for which there was a modest decrease in binding affinity by in vitro measurements (Fig. 4c), resulted in an increase in transport activity (Fig. 5b). The reason for this increase is unknown and remains to be investigated. Western blot analysis indicated that the expression levels of TAP mutants were comparable to that of the wild-type control (Supplementary Fig. 4). Conversely, CTE mutants within the exposed A13-G14-A15 segment that no longer interact with TAP (replacement of AGA by either GAG or CCC) failed to stimulate export of the unspliced mRNA (Fig. 5c).
To investigate how TAP mutations can affect bulk mRNA export we analyzed the stimulatory effect of TAP-p15 overexpression on the export of the control reporter lacking the CTE. As mentioned above, this mRNA is exported inefficiently but its export can be stimulated by coexpression of TAP-p15 heterodimers (Fig. 5d). Interestingly, with the exception of the mutant lacking the entire LRR or carrying five amino acid substitutions in the LRR and RBD domains, all TAP mutants stimulated the export of the reporter mRNA as efficiently as the wild-type (Fig. 5d). These results suggests different requirements for TAP-mediated export of CTE containing RNAs and mRNAs, in agreement with the idea that TAP is recruited to mRNAs by adaptor proteins. It is important to note that the stimulation of RLuc expression in our assays can be contributed by an increase in export, as well as in translation efficiency, as reported previously26.
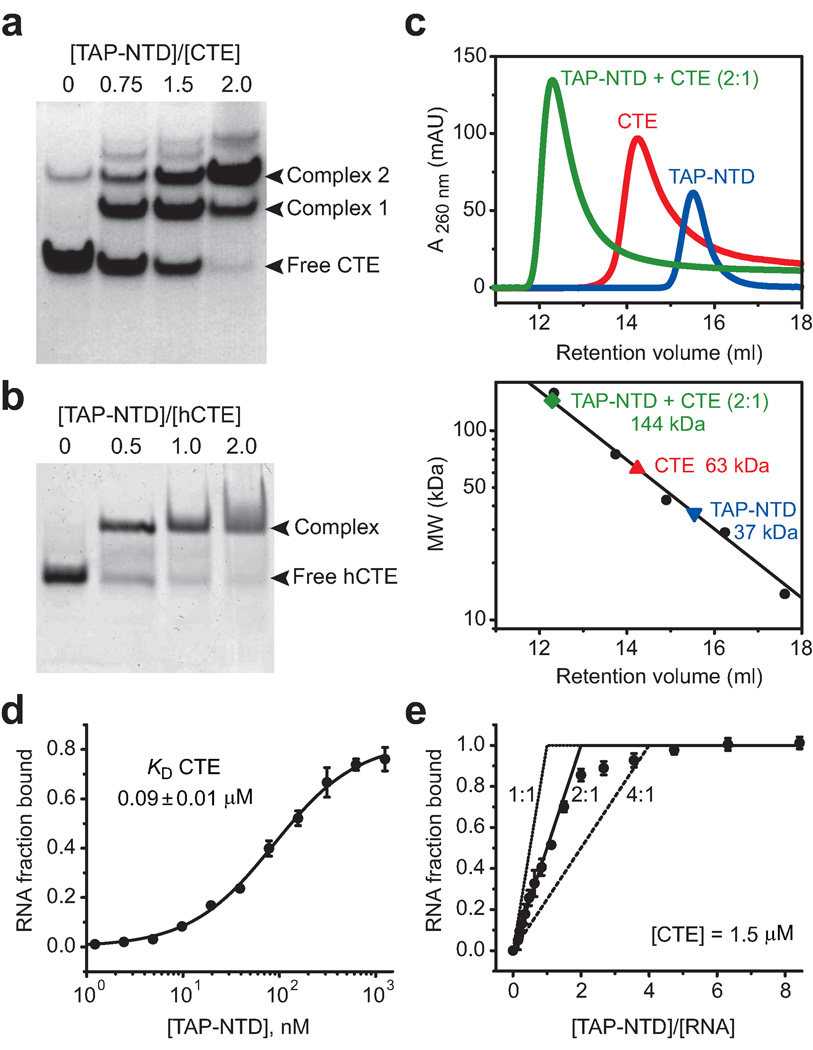
Two TAP molecules can bind one full-length CTE in vitro
TAP has been shown previously to specifically bind full-length CTE in vitro and in vivo, with the symmetry-related CTE internal loops representing two independent binding sites per CTE molecule11,24. We have further explored TAP-NTD binding to a full-length CTE (Fig. 1b) containing both internal loops (158-nt construct) in vitro. TAP-NTD binds CTE with 2:1 stoichiometry (Fig. 6a), while it binds hCTE with 1:1 stoichiometry (Fig. 6b), as determined by electrophoretic mobility shift (gel-shift) assays. The two RNA band shifts observed upon TAP-NTD titration to the CTE (Fig. 6a) most likely represent complexes with one and two molecules of TAP-NTD bound to CTE, respectively. Saturation of binding is reached upon two molar equivalents of TAP-NTD added to the CTE, with the equilibrium shifted towards the higher molecular weight complex. Consistently, the gel-filtration column elution profile of TAP-NTD mixed with CTE at 2:1 protein to RNA ratio (in green), when compared to the elution profiles of the free TAP-NTD (in blue) and CTE (in red), indicate formation of a stable complex (Fig. 6c, top panel; and Supplementary Methods). We next attempted to estimate an apparent molecular weight of the complex and compare it to that of the free components. The observed trend towards higher values estimated for TAP-NTD (37 kDa) and the CTE (63 kDa) compared to their calculated molecular weights (30.5 kDa for TAP-NTD and 51 kDa for CTE) most likely reflects the non-globular shapes of these molecules. An estimated apparent molecular weight of the complex (144 kDa) is consistent with that estimated (137 KDa) for 2:1 TAP-NTD to CTE stoichiometry (Fig. 6c, bottom panel).
Figure 6.
Stoichiometry of TAP-NTD binding to CTE RNA. (a) Electrophoretic mobility gel shift data for binding of TAP-NTD to full-length CTE establishing 2:1 stoichiometry of the complex. The positions of the free CTE and of the TAP-NTD-CTE complexes are indicated on the right. TAP-NTD to CTE molar ratios are listed above the lanes. CTE is fully bound at 2:1 TAP-NTD to CTE molar ratio. (b) Electrophoretic mobility gel shift data for binding of TAP-NTD to hCTE establishing 1:1 stoichiometry of the complex. (c) Gel filtration profiles (upper panel) monitoring the interaction between TAP-NTD (blue) and full-length CTE (red), and a calibration curve for an analytical gel filtration column shown with molecular mass standards (lower panel). The mixture of TAP-NTD and CTE at 2:1 protein to RNA ratio (green) migrates as a single peak corresponding to a higher molecular weight fraction. The elution volumes of TAP-NTD (blue triangle), CTE (red triangle) and TAP-NTD + CTE complex (green diamond) are denoted on the calibration curve. (d) Nitrocellulose filter binding curve for complex between TAP-NTD and full-length CTE. The apparent equilibrium binding constants (KD) measured by non-linear least-squares fit according to equation 1 in the Online Methods section is listed together with ± fitting error. (e) Stoichiometry of TAP-NTD binding to CTE measured by filter binding assay. The total RNA concentration used in the equilibrations is listed and is 16-fold greater then the KD of TAP-NTD for CTE determined by direct titration (panel d). The data are compared to theoretical saturation curves for 1:1, 2:1, and 4:1 protein:RNA stoichiometry. The 2:1 curve most closely approximates the data, establishing that two copies of TAP-NTD interact with a single CTE molecule. The plots in panels d and e represent mean ± standard deviation for two independent measurements.
The binding affinity and stoichiometry of the CTE and TAP-NTD interaction has been further probed based on nitrocellulose filter-binding experiments. The KD of TAP-NTD measured for the full-length CTE is 0.09 µM (Fig. 6d), which is 3-fold weaker than the value measured by the same assay for hCTE. In this regard, KD variations exhibiting 2 to 3-fold differences have been observed when proteins are tested with RNAs of different length27. The stoichiometric binding experiment was conducted under conditions where the RNA concentration of the unlabeled CTE was 16-fold higher than the measured KD. The data were compared to models of saturable protein binding, resulting in a stoichiometric equivalence point of 2 (Fig. 6e and Supplementary Methods). These data clearly establish that two TAP molecules bind to one full-length CTE.
DISCUSSION
Our structure of TAP-NTD bound to hCTE RNA, combined with in vitro binding and in vivo transport assay studies on mutants, provides the first insights into the structural basis of protein-RNA recognition that are critical for nucleocytoplasmic transport of retroviral CTE RNA by the mRNA export factor TAP.
hCTE RNA adopts a L-shaped fold on complex formation
The pair of symmetry-related internal loops with their highly conserved sequences constitute a unique feature of the retroviral CTE RNA (Fig. 1a). There is currently no structure of either CTE or hCTE RNAs in the free state, and based on our current knowledge of RNA architecture28,29, it is more than likely that the internal loop of hCTE RNA interconverts between conformational states in the absence of bound TAP-NTD. By contrast, the internal loop of the hCTE RNA adopts a unique L-shaped fold when bound to the TAP-NTD (Fig. 1d,e). An L-shaped RNA fold has been previously reported for the streptomycin-RNA complex, with the antibiotic bound within a pocket at the elbow of the L-shaped RNA scaffold30, and also for HCV IRES domain II31.
The L-shaped architecture of the bound hCTE RNA is generated by a combination of stacking and looping out interactions on complex formation. The core of the zippered-up internal loop involves a continuous stack of four non-canonical pairs (Fig. 2b), including one each of wobble G•U (Fig. 2c) and water-mediated C•C (Fig. 2e) pairs, together with reversed A•A (Fig. 2d) and sheared G•A (Fig. 2f) pairs (non-canonical pairing alignments are reviewed in32). Towards one end of this zippered-up duplex, the bases of the A13-G14-A15 segment are flipped out and unstacked relative to each other, but interact individually with either the LRR domain (A13 and G14; Fig. 3c–e) or the RRM domain (A15, Fig. 3f), and facilitate continuous stacking of U11•G50 of the internal loop with C10-G51 of the lower stem. At the other end, bases within the A44-A45-U46 are unpaired and aligned orthogonal to each other (i.e. do not participate in intermolecular contacts), and facilitate the orthogonal alignment of the sheared G47•A17 and Watson-Crick G18-C43 pairs (Fig. 2b), thereby contributing to formation of the L-shaped scaffold.
Bulge bases are a common feature of higher order RNAs and confer structural diversity to RNA scaffolds33,34. We observe stable base pair formation of flanking pairs and coaxial stacking of stems flanking both A39–A40 (Fig. 2g) and A54–A55 (Fig. 2h) bulge sites.
Novel features of RRM-RNA interactions
The RRM domain of TAP-NTD utilizes a non-canonical RNA binding interface in its complex with the hCTE RNA (Supplementary Fig. 3b). In most RRM-RNA complexes, the single-stranded RNA traverses and makes base-specific intermolecular contacts with side chains projecting from the central β-strands of the β-sheet of the RRM (reviewed in35,36,37), as observed in the structure of the ternary complex of spliceosomal U2B” bound to a fragment of U2 small nuclear RNA38 (Supplementary Fig. 5a). Rather, the intermolecular interactions in the TAP-hCTE RNA complex are between the sugar-phosphate backbone of multiple double-stranded segments (G50-G51G, A20 and A31-U34), as well as a single flipped out base (A15), and the periphery of the β-sheet (Supplementary Fig. 3b). A15, G50–G51 segments contact one edge (β2 strand) of the β-sheet (Supplementary Fig. 3b and Fig. 3b,f) in a manner similar to that observed in the complex of nascent RNA transcripts containing UUU-3’ termini bound to the RRM domain of the La protein39 (Supplementary Fig. 5b). Intermolecular interactions are also observed between the side chains projecting from loop elements of the RRM and the upper hCTE stem segments (A20 and A31-U34) (Supplementary Fig. 3b and Fig. 3b).
Novel features of LRR-RNA interactions
The intermolecular contacts involving the LRR domain are restricted solely to the internal loop (A13-G14) and the lower bulge (A54–A55) RNA residues that target side chains projecting from loop elements on the same face of the domain in the complex (Figs. 3a,c). The intermolecular contacts involve both specific bases (A13-G14) and sugar-phosphate backbone (A13-G14 and A54–A55), with the emphasis on intermolecular contacts at the single-stranded RNA level. There are similarities with the LRR-RNA interface in the structure of the complex of Toll-like receptor 3 (TLR3) with dsRNA40 (Supplementary Fig. 5c), except that in the TLR3 complex, loop elements projecting from the same face of the LRR domain form intermolecular contacts with the sugar-phosphate backbone at the double-strand RNA level.
In vitro and in vivo disruption of TAP-CTE interactions
Our in vitro studies on TAP-NTD bound to hCTE RNA by ITC (Fig. 4a,c) and direct (Fig. 4b,c) and competitive filter-binding assays (Fig. 4d–g) have focused on mutants of flipped out residues A13, G14 and A15 of the RNA, the amino acids that contact these flipped out nucleotides, and Arg128 and Arg158 of the RRM domain, that participate in intermolecular protein-RNA contacts in the complex (Fig. 3 and Supplementary Fig. 3). We observe a dramatic reduction in binding affinity following disruption of contacts involving A13 and G14 independent of whether the mutation is in the RNA or protein residues involved in intermolecular recognition (Fig. 4). The intermolecular contacts involving A13 (Fig. 3d) and G14 (Fig. 3e) include a combination of base stacking, Watson-Crick edge hydrogen-bonding and sugar-phosphate backbone recognition, and appear to be critical for complex formation. Similarly, intermolecular contacts involving Arg128 and Arg158 (Fig. 3b) are important for molecular recognition, since reverse charge mutations result in a dramatic reduction in binding affinity (Fig. 4a–c).
Flipped out residue A15 exhibits a more nuanced response to mutations, with replacement by G15 impacting dramatically on binding affinity, while replacement by C15 has a minimal effect on binding affinity (Fig. 4f). Both adenine and cytosine have amino groups at similar positions and hence the observed intermolecular hydrogen bond involving the amino group of A15 and the backbone carbonyl (Fig. 3f) is most likely retained (perhaps through a bridging water molecule) on replacement by cytosine.
It is noteworthy that the in vivo export studies on TAP-p15 complex bound to full-length CTE RNA establish that the RNA export activity of TAP is either reduced or eliminated by the same protein (Fig. 5b) and RNA (Fig. 5c) mutations investigated in the in vitro binding studies. Therefore, our studies establish that the intermolecular contacts identified in the crystal structure of the minimalist complex of TAP-NTD and hCTE RNA (Fig. 3) are also critical for nucleocytoplasmic export activity at the cellular level mediated by full-length TAP and CTE. Interactions observed in the crystal structure finally explain the effects of alanine mutations of RRM Arg128, Lys129 and Lys132, and LRR Arg233, Arg276, Tyr278 and Lys304 on CTE-dependent nuclear export and CTE-binding in vivo that have been previously identified by analysis of 38 TAP surface mutants17. While five of these side chains (Arg128, Arg233, Arg276, Tyr278 and Lys304) directly contact CTE (Fig. 3), the remaining two (Lys132 and Lys129) participate in water-mediated hydrogen-bonding and electrostatic interactions with lower and upper RNA stems, respectively.
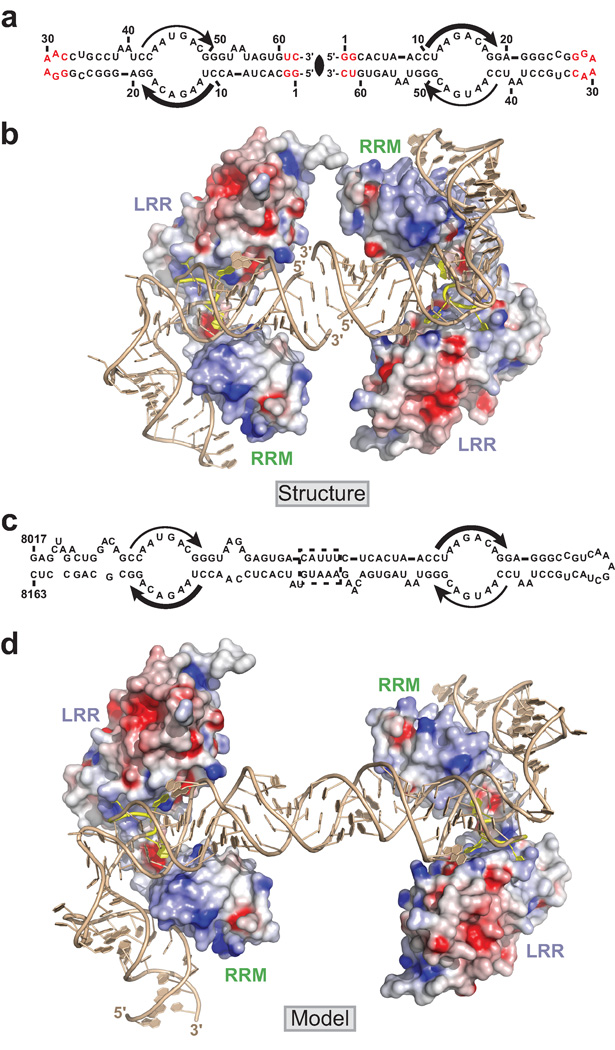
Model of two TAP-NTD’s binding to full-length CTE RNA
The conserved internal loops of full-length CTE RNA are related by two-fold symmetry (Fig. 1b). Similarly, the two hCTE RNA molecules in the asymmetric unit of the crystal of the complex are related by two-fold non-crystallographic symmetry as they stack with 5' to 3' directionality of their blunt ends (Fig. 7a). The packing arrangement of the two molecules of the complex in the asymmetric unit is shown in Fig. 7b, with the RNA in a ribbon and stick representation and the protein in an electrostatic surface representation. In essence, the observed packing arrangement provides insights into how individual TAP-NTD domains could target each of the two internal loop sites in full-length CTE RNA. There are five additional base pairs in the center of the full-length CTE RNA (dashed box region in Fig. 7c) together with two adjacent CAA and UA bulges compared to the end-to-end stacked hCTE RNAs in the structure of the complex (Fig. 7a). In addition, the lower half of CTE differs from the upper half by the presence of a 6-nt asymmetric AAGA/CA internal loop (Fig. 7c) compared to a 2-nt AA-bulge (A54–A55) of the hCTE in the structure of the complex (Fig. 7a), and hence binding to full-length CTE will most likely require rotation and translation of one TAP-NTD molecule of the complex relative to the other. We have generated a model of TAP-NTD bound to each internal loop of full-length CTE to form a 2:1 complex as shown in Fig. 7d (details of modeling of the various bulges based on RNA structures available in the data base are outlined in Supplementary Methods), in which the bound NTD-CTE molecules are separated by roughly one turn of helix, when compared with the packing alignments of the two 1:1 complexes of TAP-NTD bound to hCTE in the asymmetric unit in the crystal structure of the complex (Fig. 7b).
Figure 7.
Packing of two molecules of complex (TAP-NTD bound to hCTE) in the crystallographic asymmetric unit and a model of the 2:1 complex of TAP-NTD bound to full-length CTE. (a) The alignment of two hCTEs in the crystallographic asymmetric unit of the complex. The bases colored in red reflect differences from the wild-type CTE sequence. (b) The orientation of two molecules of complex (TAP-NTD bound to hCTE RNA) in the crystallographic asymmetric unit. (c) The full-length CTE RNA consisting of two large internal loops whose sequences are related by two-fold symmetry (indicated by thick and thin arrows). (d) Model of the complex of full-length CTE with two TAP-NTD molecules bound to two CTE internal loops generated based on the structure of the two molecules of complex (TAP-NTD bound to hCTE) in the crystallographic asymmetric unit. The missing parts of the full-length CTE were modeled using idealized A-form RNA duplexes and RNA structural elements from previously determined structures (see Supplementary Methods for details) followed by rounds of idealization of geometric parameters with REFMAC and Coot programs.
It is apparent that one face of hCTE RNA interacts with the TAP-NTD, leaving the other face exposed to solvent, in the structure of the complex (Fig. 1d,e and Fig. 7b,d). It is conceivable that the C-terminal domain of TAP and/or p15, that forms a complex with TAP, could assist in fully encapsulating the CTE RNA, thereby facilitating efficient nucleocytoplasmic transport.
Implications for mechanism of TAP-mediated mRNA export
Although it has been shown that TAP readily crosslinks to mRNA in vivo, suggestive of a direct stable interaction during export, TAP binds RNA weakly in vitro, with a measured KD of app. 70 µM for 15-mer RNA41. Weak non-specific RNA binding to TAP might be one factor that restricts cellular mRNAs from export before splicing. TAP is recruited to fully-spliced mRNAs through multiple adaptors, such as REF and SR proteins whose own recruitment is regulated by RNA-processing events reviewed in42,43. Both REF and SR proteins bind the N-terminal region of TAP through Gly-Arg rich sequences adjacent to their RRMs44, with a contribution of the RRM of REF/SR to stable binding and efficient recruitment of mRNA for export41. Since the nonspecific RNA-binding site on TAP has been mapped to an Arg-rich motif (61–118 aa), a region that overlaps with minimal REF/SR protein-binding region (1–202 aa)44, it is possible that the RRM domain of TAP might be involved in interaction with REF and SR proteins. We have shown that the RRM binds CTE RNA via a portion of its β-sheet face and edge, together with loop segments, leaving its α-helical face available for further interactions with protein or RNA. However, it remains to be investigated whether TAP RRM could also act as a protein-protein interaction module. Similarly, the precise role of the LRR domain of TAP in cellular mRNA export remains to be elucidated at this time. We have shown that loop elements projecting from the same face of the LRR domain interact with flipped out bases of the conserved internal loop and an AA bulge of the CTE RNA. This interaction leaves the opposite concave β-sheet surface of the LRR available for interaction in a manner similar to that observed between U2A’ (LRR fold) and U2B" (RRM fold) in the structure of the ternary complex of these proteins with RNA38. Further structural studies aimed at elucidation of ternary TAP-RNA complexes with adaptor proteins should clarify this issue.
Summary
Our combined structural, in vitro binding and in vivo transport studies define the principles of molecular recognition by which a cis-acting CTE of retroviruses subvert the messenger RNA export factor TAP, thereby facilitating nucleocytoplasmic export of viral genomic RNA.
METHODS
Methods and any associated references are available in the online version of the paper at http://www.nature.com/nsmb/.
Supplementary Material
Acknowledgements
This research was supported by funds from the NIH (CA049982) to D.J.P. and the Max-Planck Society to E.I. X-ray data was collected at the X-29 beamline at the National Synchrotron Light Source at the Brookhaven National Laboratory and we are grateful to the staff for their assistance.
Footnotes
Accession codes. The atomic coordinates and structure factors for the free TAP-NTD and TAP-NTD/hCTE complex have been deposited in the protein data bank under accession codes 3RW7 and 3RW6, respectively.
AUTHOR CONTRIBUTIONS
Constructs design, protein and RNA preparation and purification, crystallization of complex and its structure determination and in vitro binding assays were undertaken by M.T. with the assistance of N.W.K. under the supervision of D.J.P. The in vivo transport assays were performed by L.W. under the supervision of E.I. The paper was written by M.T., D.J.P. and E.I. with input from the other authors.
REFERENCES
- 1.Chook YM, Blobel G. Karyopherins and nuclear import. Curr Opin Struct Biol. 2001;11:703–715. doi: 10.1016/s0959-440x(01)00264-0. [DOI] [PubMed] [Google Scholar]
- 2.Conti E, Izaurralde E. Nucleocytoplasmic transport enters the atomic age. Curr Opin Cell Biol. 2001;13:310–319. doi: 10.1016/s0955-0674(00)00213-1. [DOI] [PubMed] [Google Scholar]
- 3.Conti E, Muller CW, Stewart M. Karyopherin flexibility in nucleocytoplasmic transport. Curr Opin Struct Biol. 2006;16:237–244. doi: 10.1016/j.sbi.2006.03.010. [DOI] [PubMed] [Google Scholar]
- 4.Cook A, Bono F, Jinek M, Conti E. Structural biology of nucleocytoplasmic transport. Annu Rev Biochem. 2007;76:647–671. doi: 10.1146/annurev.biochem.76.052705.161529. [DOI] [PubMed] [Google Scholar]
- 5.Kohler A, Hurt E. Exporting RNA from the nucleus to the cytoplasm. Nat Rev Mol Cell Biol. 2007;8:761–773. doi: 10.1038/nrm2255. [DOI] [PubMed] [Google Scholar]
- 6.Stewart M. Molecular mechanism of the nuclear protein import cycle. Nat Rev Mol Cell Biol. 2007;8:195–208. doi: 10.1038/nrm2114. [DOI] [PubMed] [Google Scholar]
- 7.Cook AG, Conti E. Nuclear export complexes in the frame. Curr Opin Struct Biol. 2010;20:247–252. doi: 10.1016/j.sbi.2010.01.012. [DOI] [PubMed] [Google Scholar]
- 8.Cook AG, Fukuhara N, Jinek M, Conti E. Structures of the tRNA export factor in the nuclear and cytosolic states. Nature. 2009;461:60–65. doi: 10.1038/nature08394. [DOI] [PubMed] [Google Scholar]
- 9.Okada C, et al. A high-resolution structure of the pre-microRNA nuclear export machinery. Science. 2009;326:1275–1279. doi: 10.1126/science.1178705. [DOI] [PubMed] [Google Scholar]
- 10.Braun IC, Rohrbach E, Schmitt C, Izaurralde E. TAP binds to the constitutive transport element (CTE) through a novel RNA-binding motif that is sufficient to promote CTE-dependent RNA export from the nucleus. EMBO J. 1999;18:1953–1965. doi: 10.1093/emboj/18.7.1953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gruter P, et al. TAP, the human homolog of Mex67p, mediates CTE-dependent RNA export from the nucleus. Mol Cell. 1998;1:649–659. doi: 10.1016/s1097-2765(00)80065-9. [DOI] [PubMed] [Google Scholar]
- 12.Izaurralde E. Friedrich Miescher Prize awardee lecture review. A conserved family of nuclear export receptors mediates the exit of messenger RNA to the cytoplasm. Cell Mol Life Sci. 2001;58:1105–1112. doi: 10.1007/PL00000924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Segref A, et al. Mex67p, a novel factor for nuclear mRNA export, binds to both poly(A)+ RNA and nuclear pores. EMBO J. 1997;16:3256–3271. doi: 10.1093/emboj/16.11.3256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fribourg S, Braun IC, Izaurralde E, Conti E. Structural basis for the recognition of a nucleoporin FG repeat by the NTF2-like domain of the TAP/p15 mRNA nuclear export factor. Mol Cell. 2001;8:645–656. doi: 10.1016/s1097-2765(01)00348-3. [DOI] [PubMed] [Google Scholar]
- 15.Herold A, et al. TAP (NXF1) belongs to a multigene family of putative RNA export factors with a conserved modular architecture. Mol Cell Biol. 2000;20:8996–9008. doi: 10.1128/mcb.20.23.8996-9008.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Liker E, Fernandez E, Izaurralde E, Conti E. The structure of the mRNA export factor TAP reveals a cis arrangement of a non-canonical RNP domain and an LRR domain. EMBO J. 2000;19:5587–5598. doi: 10.1093/emboj/19.21.5587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ho DN, Coburn GA, Kang Y, Cullen BR, Georgiadis MM. The crystal structure and mutational analysis of a novel RNA-binding domain found in the human Tap nuclear mRNA export factor. Proc Natl Acad Sci U S A. 2002;99:1888–1893. doi: 10.1073/pnas.042698599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Grant RP, Neuhaus D, Stewart M. Structural basis for the interaction between the Tap/NXF1 UBA domain and FG nucleoporins at 1A resolution. J Mol Biol. 2003;326:849–858. doi: 10.1016/s0022-2836(02)01474-2. [DOI] [PubMed] [Google Scholar]
- 19.Bachi A, et al. The C-terminal domain of TAP interacts with the nuclear pore complex and promotes export of specific CTE-bearing RNA substrates. RNA. 2000;6:136–158. doi: 10.1017/s1355838200991994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ernst RK, Bray M, Rekosh D, Hammarskjold ML. A structured retroviral RNA element that mediates nucleocytoplasmic export of intron-containing RNA. Mol Cell Biol. 1997;17:135–144. doi: 10.1128/mcb.17.1.135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tabernero C, Zolotukhin AS, Valentin A, Pavlakis GN, Felber BK. The posttranscriptional control element of the simian retrovirus type 1 forms an extensive RNA secondary structure necessary for its function. J Virol. 1996;70:5998–6011. doi: 10.1128/jvi.70.9.5998-6011.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cullen BR. Viral RNAs: lessons from the enemy. Cell. 2009;136:592–597. doi: 10.1016/j.cell.2009.01.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li Y, et al. An intron with a constitutive transport element is retained in a Tap messenger RNA. Nature. 2006;443:234–237. doi: 10.1038/nature05107. [DOI] [PubMed] [Google Scholar]
- 24.Kang Y, Cullen BR. The human Tap protein is a nuclear mRNA export factor that contains novel RNA-binding and nucleocytoplasmic transport sequences. Genes Dev. 1999;13:1126–1139. doi: 10.1101/gad.13.9.1126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Braun IC, Herold A, Rode M, Conti E, Izaurralde E. Overexpression of TAP/p15 heterodimers bypasses nuclear retention and stimulates nuclear mRNA export. J Biol Chem. 2001;276:20536–20543. doi: 10.1074/jbc.M100400200. [DOI] [PubMed] [Google Scholar]
- 26.Jin L, Guzik BW, Bor YC, Rekosh D, Hammarskjold ML. Tap and NXT promote translation of unspliced mRNA. Genes Dev. 2003;17:3075–3086. doi: 10.1101/gad.1155703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Serganov A, Polonskaia A, Ehresmann B, Ehresmann C, Patel DJ. Ribosomal protein S15 represses its own translation via adaptation of an rRNA-like fold within its mRNA. EMBO J. 2003;22:1898–1908. doi: 10.1093/emboj/cdg170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Batey RT, Rambo RP, Doudna JA. Tertiary Motifs in RNA Structure and Folding. Angew Chem Int Ed Engl. 1999;38:2326–2343. doi: 10.1002/(sici)1521-3773(19990816)38:16<2326::aid-anie2326>3.0.co;2-3. [DOI] [PubMed] [Google Scholar]
- 29.Hermann T, Patel DJ. Adaptive recognition by nucleic acid aptamers. Science. 2000;287:820–825. doi: 10.1126/science.287.5454.820. [DOI] [PubMed] [Google Scholar]
- 30.Tereshko V, Skripkin E, Patel DJ. Encapsulating streptomycin within a small 40-mer RNA. Chem Biol. 2003;10:175–187. doi: 10.1016/s1074-5521(03)00024-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dibrov SM, Johnston-Cox H, Weng YH, Hermann T. Functional architecture of HCV IRES domain II stabilized by divalent metal ions in the crystal and in solution. Angew Chem Int Ed Engl. 2007;46:226–229. doi: 10.1002/anie.200603807. [DOI] [PubMed] [Google Scholar]
- 32.Leontis NB, Westhof E. Geometric nomenclature and classification of RNA base pairs. RNA. 2001;7:499–512. doi: 10.1017/s1355838201002515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hermann T, Patel DJ. RNA bulges as architectural and recognition motifs. Structure. 2000;8:R47–R54. doi: 10.1016/s0969-2126(00)00110-6. [DOI] [PubMed] [Google Scholar]
- 34.Jiang F, et al. Anchoring an extended HTLV-1 Rex peptide within an RNA major groove containing junctional base triples. Structure. 1999;7:1461–1472. doi: 10.1016/s0969-2126(00)88337-9. [DOI] [PubMed] [Google Scholar]
- 35.Clery A, Blatter M, Allain FH. RNA recognition motifs: boring? Not quite. Curr Opin Struct Biol. 2008;18:290–298. doi: 10.1016/j.sbi.2008.04.002. [DOI] [PubMed] [Google Scholar]
- 36.Lunde BM, Moore C, Varani G. RNA-binding proteins: modular design for efficient function. Nat Rev Mol Cell Biol. 2007;8:479–490. doi: 10.1038/nrm2178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Serganov A, Patel DJ. Towards deciphering the principles underlying an mRNA recognition code. Curr Opin Struct Biol. 2008;18:120–129. doi: 10.1016/j.sbi.2007.12.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Price SR, Evans PR, Nagai K. Crystal structure of the spliceosomal U2B"-U2A' protein complex bound to a fragment of U2 small nuclear RNA. Nature. 1998;394:645–650. doi: 10.1038/29234. [DOI] [PubMed] [Google Scholar]
- 39.Teplova M, et al. Structural basis for recognition and sequestration of UUU(OH) 3' temini of nascent RNA polymerase III transcripts by La, a rheumatic disease autoantigen. Mol Cell. 2006;21:75–85. doi: 10.1016/j.molcel.2005.10.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu L, et al. Structural basis of toll-like receptor 3 signaling with double-stranded RNA. Science. 2008;320:379–381. doi: 10.1126/science.1155406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hautbergue GM, Hung ML, Golovanov AP, Lian LY, Wilson SA. Mutually exclusive interactions drive handover of mRNA from export adaptors to TAP. Proc Natl Acad Sci U S A. 2008;105:5154–5159. doi: 10.1073/pnas.0709167105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Stutz F, Izaurralde E. The interplay of nuclear mRNP assembly, mRNA surveillance and export. Trends Cell Biol. 2003;13:319–327. doi: 10.1016/s0962-8924(03)00106-5. [DOI] [PubMed] [Google Scholar]
- 43.Walsh MJ, Hautbergue GM, Wilson SA. Structure and function of mRNA export adaptors. Biochem Soc Trans. 2010;38:232–236. doi: 10.1042/BST0380232. [DOI] [PubMed] [Google Scholar]
- 44.Huang Y, Gattoni R, Stevenin J, Steitz JA. SR splicing factors serve as adapter proteins for TAP-dependent mRNA export. Mol Cell. 2003;11:837–843. doi: 10.1016/s1097-2765(03)00089-3. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.