Abstract
IRF1 is a transcription factor that participates in interferon signaling. Previous studies of IRF1 binding have utilized in vitro assays. We used ChIP-seq in human monocytes to better define the recognition motif for IRF1. The newly identified 18bp motif (RAAASNGAAAGTGAAASY) is a refinement of the 13bp IRF1 motif commonly used. We utilized the 18bp consensus motif and identified 345 potential target genes. To compare the 18bp motif with the 13bp motif, we compared putative gene targets. Only 56 potential gene targets were defined by both consensus motifs. To compare biological effects of interferon on the 13bp and the 18bp consensus targets, we mined expression data from cells exposed to interferons or transfected with IRF1. In all cases, the 18bp consensus motif was more strongly associated with transcriptional responses than the 13bp motif. Therefore, the new 18bp consensus motif appears to have a greater association with biological activities of IRF1.
Keywords: IRF1, IRF-E, chromatin immunoprecipitation, inflammation, lupus, interferon
1. INTRODUCTION
Interferon regulatory factor (IRF) 1 is the first factor indentified of the IRF family which is comprised of nine members in mammals. IRFs share a helix-turn-helix DNA-binding domain that is characterized by five tryptophan-rich repeats (Escalante et al., 1997). All IRF members are believed to bind similar DNA sites with GAAANN repeats commonly seen as targets (Harada et al., 1989; Naf et al., 1991). The sequence of the 2bp spacer appears to drive some of the specificity of IRF family member binding (Morin et al., 2002). The IRF-1 DNA recognition sequence specifically was initially identified by a polymerase chain reaction (PCR)-assisted method (Tanaka et al., 1993) and analysis of the crystal structure of the DNA-binding domain bound to its cognate DNA sequence element (Fujii et al., 1999). Both single sites of the sequence AANTGAAA have been identified as well as repeats of GAAA with variable spacer nucleotides. At present, it is not clear if IRF1 binding in vivo is reflected by this binding sequence obtained in vitro.
IRF1 was first identified as a transcriptional regulator of type-I interferons (IFNs) and IFN-inducible genes (Miyamoto et al., 1988; Chang et al., 1992; Kimura et al., 1994). Subsequent studies have revealed that IRF1 plays a broad function in a variety of biology process. IRF1 is important for regulating inflammation, cell growth, apoptosis, and oncogenesis (Tamura et al., 1995; Tanaka et al., 1996; Penninger et al., 1997; Taki et al., 1997; Urschel et al., 1997; Ogasawara et al., 1998; Ko et al., 2002) and IRF1 is required for DNA damage-induced growth arrest and apoptosis (Kroger et al., 2001). The IRF1 target genes in this process have not been confirmed, but may include the genes encoding p21/WAF1/CIP1, Caspase-1, Caspase-7, Caspase-8, GAAP-1, and TRAILA (Kim et al., 2004). Recently by performing chromatin immunoprecipitation coupled to a CpG island microarray (ChIP-chip), a set of IRF1-bound genes was identified in breast cancer cells stimulated with γIFN (Frontini et al., 2009). A number of the identified genes were found to be involved in the DNA damage response and DNA repair pathway.
Another central function of IRF1 is the regulation of host defense and the development of various immunologically competent cells. IRF1 can be upregulated by lipopolysaccharide, bacteria, viral infections, and cytokines such as IL-1, IL-12 and type I and type II interferons (Skaar et al., 1998; Taniguchi et al., 2001; Honda and Taniguchi, 2006). Natural killer (NK) cells were dramatically reduced in both the spleen and liver of IRF1 knockout mice and NK cell-medicated cytolytic activity was not observed (Ogasawara et al., 1998; Ohteki et al., 1998). In addition, CD8+ T lymphocytes were diminished in IRF1 knockout mice. IRF1 is also involved in monocyte/macrophage differentiation (Testa et al., 2004) and maturation of dendritic cells (Gabriele et al., 2006). These cells are the effectors and regulators of inflammation and immune responses and are critical in end organ damage in patients with systemic lupus erythematosus (SLE). Our previous studies suggested that IRF1-regulated genes have increased expression in monocytes from patients with SLE (Zhang et al., 2010b). To better define the role of IRF1 in human disease, it became important to identify binding sites in vivo.
Chromatin immunoprecipitation followed by sequencing (ChIP-seq) is a popular strategy to study genome-wide protein-DNA interactions. Next-generation sequencing technology offers higher sensitivity, accuracy and greater coverage and is very useful for analysis of the regulation of gene and epigenetic mechanisms. Here, by employing ChIP-seq, we characterized the IRF1 binding motif in vivo and investigated IRF1 direct target genes in primary monocytes. This study revealed a large number novel IRF1 target genes, which may provide better understanding of IRF1 function in vivo.
2. METHODS
2.1 ChIP-seq and library preparation
Primary human monocytes were purified from healthy people by Ficoll-Paque and adherence. The purity of the preparations was approximately 90% by flow cytometry for CD14 staining. ChIP experiments were carried out as described in our previous paper (23) and utilized the IRF1 antibody from Santa Cruz Biotechnology (H205, Santa Cruz, CA.). The library preparation utilized the SOLiD ChIP-seq kit and was performed according to the manufacturer’s instructions. Emulsion PCR and SOLiD sequencing were performed according to the system standards.
2.2 Analyses of ChIP-seq data
We developed a workflow, CHOPseq, for the processing and analysis of ChIP-seq data. This workflow includes three milestone steps: read alignment, peak calling, and motif discovery (Table 1). Except for read alignment performed using BioScope, all steps were done within R/Bioconductor statistical environment.
Table 1.
Major R Functions Used For Chopseq Workflow
| Task | R Function | R package |
|---|---|---|
| Import reads from BAM file | BamScannerChr | CHOPseq |
| Estimate average fragment length | estimate.mean.fraglen | chipseq |
| Find the best set of alignment scores | MapqCorr | CHOPseq |
| Resize reads | resize | IRanges |
| Convert reads to depth at bases | coverage | IRanges |
| Peak calling | PeakCaller | CHOPseq |
| Count and adjust reads around peaks | PeakStats | CHOPseq |
| Estimate peak FDR | PeakFDR | CHOPseq |
| Motif discovery | GADEM | rGADEM |
| Match PWM to genome | matchPWM | Biostrings |
| Map PWM matches/peaks to genes | findOverlaps | IRanges |
2.2.1 Read Alignment
All 50-base reads were aligned to the GRCh37 version of human genome using BioScope™v1.3 software (Life Technologies Corp.). The alignment used the "classic" setting, allowing up to six mismatches. The alignment results were saved in the BAM format and R was used for further processing. The estimated average length of DNA fragments was 180bp. Accordingly, we extended all reads to 180bp long from their 3' end to cover the regions isolated from sequencing due to protein binding. Each aligned read was assigned a score by BioScope based on the uniqueness and number of mismatches of the alignment. We ran a stepwise procedure to identify a subset of scores that achieved the highest strand-strand correlation of depth.
2.2.2 Peak Calling
We used our in-house R function for peak calling. Bases with depth of 10 or higher were identified as initial peak summits. We defined the absolute read enrichment by counting reads on the forward strand within [S−179, S] and the reads on the reverse strand with [S, S+179], where S is the location of the peak summit. To reduce the weight of duplicate reads aligned to the same location, the read count on each strand was adjusted by sqrt(T*U), where sqrt means square root and T and U are the counts of total and uniquely-located reads. The absolute read enrichment at each peak was then calculated as 2*sqrt(F*R) where F and R are the adjusted read counts from two strands. The absolute and relative read enrichment of peaks identified from the IRF1 ChIP sample were plotted in Supplemental Figure 1 as black circles while using the input samples as a reference. To evaluate the false discovery rate (FDR) of the peaks to be selected, we applied the same peak calling and read counting procedure on the DNA input sample while using the ChIP sample as reference. The results were also plotted in Supplemental Figure 1 (red dots), showing that the peaks in the up-right corner were unique to the IRF1 ChIP sample. We used those peaks for the motif discovery analysis (see below).
2.2.3 Motif Discovery
The sequences within the [−50bp, 50bp] region around peak summits were used as seeds for a GADEM analysis. GADEM searched for a consensus motif and reported the matches to this motif in all the seed sequences. If there were multiple matches in one seed, we retained the one with the highest similarity and used the remaining matches to generate a position-weighted matrix (PWM). Furthermore, we searched the matches to this PWM in human genome. Each match was assigned a similarity score. Matches were mapped to genomic regions relative to gene, such as promoter and exon. Gene information was downloaded from the "RefGene" track of UCSC Genome Browser.
2.3 Re-analysis of Transcriptomic Data Sets
Previously processed microarray data series and their annotation information were downloaded from the Gene Expression Omnibus (GEO) database. Through official gene symbols, all measured genes in a data series were split into three groups: 18bp targets, 13bp targets and non-targets. Differential gene expression before and after cytokine treatment was calculated as the log2 ratio of group averages. We called a gene up-/down- regulated by the treatment if the increase/decrease of its expression was among the top 5% of all measured genes. The difference in the group-wise expression change between gene groups was evaluated with Student's t test.
3. RESULTS
3.1 IRF1 consensus motif identification
To identify the IRF1 in vivo binding site, we purified primary human monocytes and performed chromatin immunoprecipitation with a Western-validated IRF1 antibody followed by high throughput sequencing (ChIP-seq). One pair of input samples and IRF1 ChIP-seq samples were used to define the binding motif. We developed a bioinformatic workflow called CHOPseq (Table 1) for the processing and analysis of ChIP-seq data. We filtered the reads aligned to the human genome based on their mapping quality to obtain 40.6 and 36.1 million reads respectively from the input DNA and IRF1 ChIP samples. CHOPseq’s peak calling algorithm was used to identify peak summits. Read counts around the peaks were compared between the IRF1 ChIP samples and the input controls. Peaks of the ChIP sample with both higher total read counts and relative read enrichment were considered as locations of IRF1 binding sites (Supplemental Figure 1). 117 such peaks were selected for further analysis with a false discovery rate (FDR) of 0.1 while 52 of those peaks were located within the 10kb promoter region or 5'-UTR regions. Many of the identified genes were related to immune response, such as AIM2, CD40, IFIT3, and three immunoproteasome genes (Supplemental Table 1).
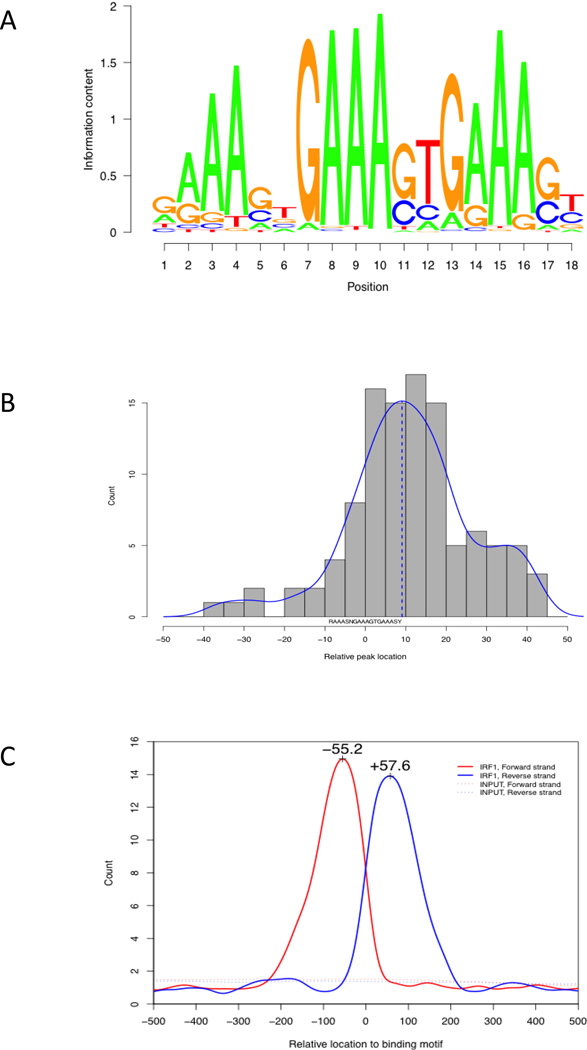
We applied the GADEM method (Li, 2009) for a de novo discovery of the IRF1 binding motif by using 100 bases around 117 peak summits as seed sequences. A single consensus sequence of 18 bases, RAAASNGAAAGTGAAASY, was identified. Matches to this sequence were found at least once within 91% of the seed sequences. A noticeable feature of this motif is the repeated occurrence of the sequence “AAA”, splitting it into three segments (Figure 1a). The bases from position 7 to 18 are similar to the binding motifs of IRF1 (SAAAAGYGAAACC) and IRF2 (GAAAAGYGAAASY) used in the TRANSFAC Matrix Database (Biobase GmbH).
Figure 1. IRF1 binding motif discovery.
A) The consensus sequence logo of the 18bp binding motif as a summarization of 107 sites defined by ChIP-seq peaks. B) The relative locations of ChIP-seq peak summits relative to the motif. C) The distribution of the 3' and 5' end bases of sequencing reads around the 107 binding peaks, using the last base of 18bp motif as the reference point. The strand-specific pattern was the result of sequencing from the 3' and 5' ends. The distance between the two peaks corresponds to the size of the IRF1 binding region.
The locations of the peak summits were centered around the last base of the 18bp binding motif (Figure 1b), making it the likely middle point of IRF1 binding. 84% of the peak summits were located within 25bp of this base, suggesting that our peak calling program had high accuracy. The distribution of reads around the binding motif had a distinguishable pattern (Figure 1c). Reads from the forward strand formed the peak at the lower end and those from the reverse strand formed the peak at the higher end. Both peaks are approximately 200 bases in size and topped at 55 to 60 bases from the middle point. This overall pattern suggested that the length of the IRF1 binding region was about 115 bases.
3.2 IRF1 binding sites in human genome
We searched for occurrences of this new binding motif in human genome. No locations were found with a perfect match (100% identity) to the consensus sequence, GAAAGTGAAAGTGAAAGT. The number of hits increased exponentially with a reduction of the percent identity cutoff (Supplemental Figure 2). We selected 28,866 hits that scored 85% or higher for further analysis. The matches were evenly distributed across the human genome with clear enrichment around transcription start sites (Supplemental Figure 3).
We identified 345 unique genes having hits within their 1kb promoter and/or 5'-UTR (trimmed to the first 1kb if longer) regions and used those genes to define potential targets of IRF1 regulation (Supplemental Table 3). This gene list is significantly enriched with genes related to antigen processing and immune response (Table 2).
Table 2.
Gene functional categories significantly enriched in the 345 IRF1 targets
| Term | Count | P Value* | Genes |
|---|---|---|---|
| PIRSF001733: tripartite motif-containing protein | 7 | 1.99E-07 | TRIM5, TRIM34, TRIM50, TRIM26, TRIM21, TRIM74, TRIM73 |
| GO: 0019882~antigen processing and presentation | 11 | 1.21E-06 | HLA-H, PSME1, TAP2, CD209, ERAP1, ERAP2, TAPBPL, PSMB8, TAPBP, PSMB9, B2M |
| IPR007904: APOBEC-like, C-terminal | 5 | 1.45E-05 | APOBEC3B, APOBEC3G, APOBEC3F, APOBEC3C, APOBEC3D |
| GO: 0006955~immune response | 28 | 3.64E-05 | OAS3, TLR3, APOBEC3G, IL15, APOBEC3F, IFI35, B2M, TAPBP, FCGR1C, TAP2, TAP1, ERAP1, ERAP2, DHX58, HLA-H, BST2, IL29, TNFRSF14, PSMB8, PSMB9, IL18BP, APOL1, TNFSF13B, PSEN1, ETS1, CD209, CD274, GBP1 |
| IPR012317: Poly(ADP-ribose) polymerase, catalytic region | 5 | 1.50E-04 | PARP9, PARP14, PARP3, PARP10, PARP4 |
| IPR008405: Apolipoprotein L | 4 | 1.51E-04 | APOL2, APOL1, APOL6, APOL4 |
| Host-virus interaction | 15 | 1.89E-04 | HAVCR1, CCNT1, TNFRSF14, APOBEC3G, SP110, APOBEC3F, PSMB8, APOBEC3C, PSMB9, TRIM5, ISG15, TAP2, CD209, TAP1, RBCK1 |
The p values represent the significance of gene enrichment using all known human genes as background.
To compare the 18bp and 13bp binding motifs, we retrieved the IRF1 binding sites derived from the 13bp motif stored by the "TFBS Conserved" track of UCSC Genome Browser and mapped them to 1kb promoter regions and trimmed the 5'-UTR regions as above to get a larger set of 595 unique genes. This target set is similarly enriched with immune response genes according to DAVID analysis (n=42, p=7.2e-5) although only 56 genes, such as IFIT3 and TAP1, were included in both target sets. IFIT1 and IFIT2 are interferon inducible genes, but have not been previously identified as direct targets of IRF1. They were included in the 18bp target set but not in the 13bp one.
3.3 Expression change of IRF1 targets in response to various treatments
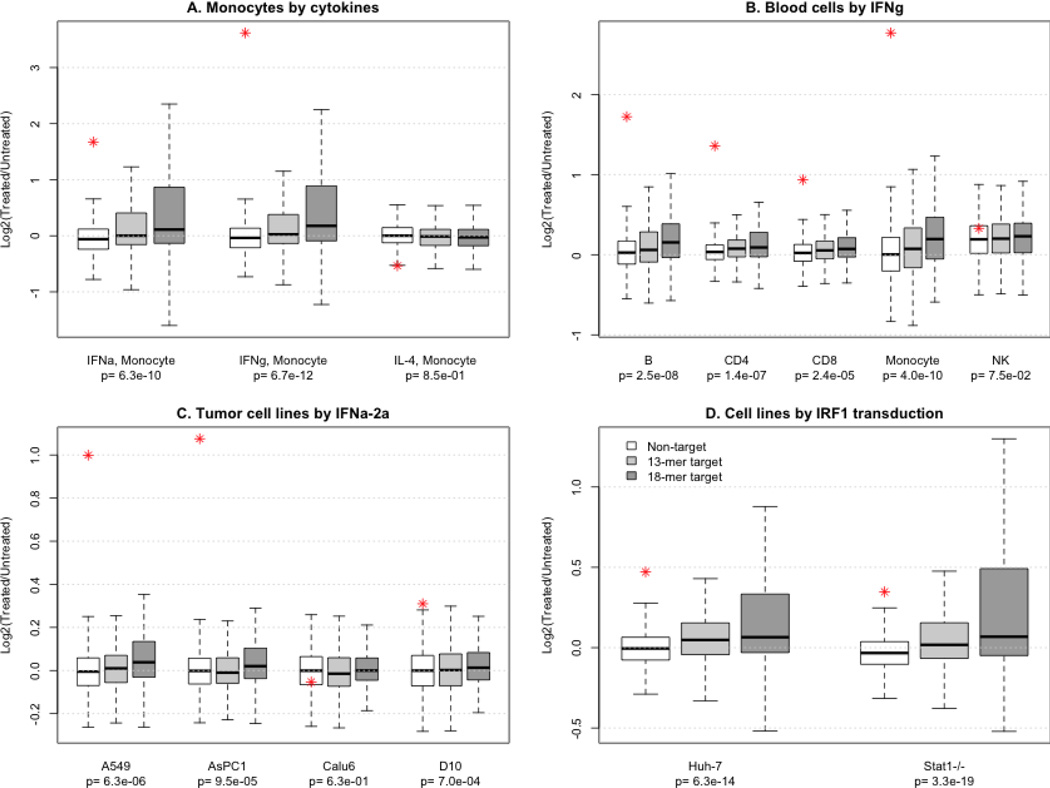
Since IRF1 is a transcription activator induced by interferon, we expected that its target genes would be more likely to be up-regulated by interferon treatment than non-targets. We used our previous microarray study (Zhang et al., 2010a), which measured the transcriptome of monocytes treated with αIFN, γIFN, or IL-4, to compare the expression change of target and non-target genes in response to different cytokine treatments. The expression of IRF1 itself was dramatically increased by both interferons and correspondingly, its targets were significantly up-regulated. IL-4 treatment slightly decreased IRF1 expression and had similar effect on targets and non-targets (Figure 2A). Aside from the overall trend of up-regulation after interferon, the scale of expression change varied among individual target genes. Interestingly, a small number of targets were down-regulated by interferon treatment. Among the genes down-regulated by both interferons, SPHAR is needed for DNA synthesis during S phase (Digweed et al., 1995) and SDC2 encodes a membrane protein enhancing migratory potential of tumor cells (Lee et al., 2009). This observation agrees with previous studies that suggested IRF1 may function as a tumor suppressor and inactivator of cell growth (Romeo et al., 2002).
Figure 2. Gene expression changes caused by interferon treatment or IRF1 over-expression in various cell types.
The boxes display the overall distribution of differentially expressed gene groups and the stars correspond to the expression change of IRF1 itself. The p values indicate the statistical difference between non-targets, 13bp consensus targets, and targets derived from the 18bp motif. A) Expression data from monocytes treated with three different cytokines were analyzed. The 18bp consensus is more closely associated with genes that respond to IFNs but not IL-4, treatment. B) Expression data from peripheral blood cells treated with interferon gamma were analyzed in the same manner. C) Expression data from tumor cell lines treated with interferon α2a were analyzed in the same manner. Four of the 10 cell lines were plotted. D) Expression data from cell lines transfected with IRF1 were analyzed to define the concordance of gene expression changes with predicted targets.
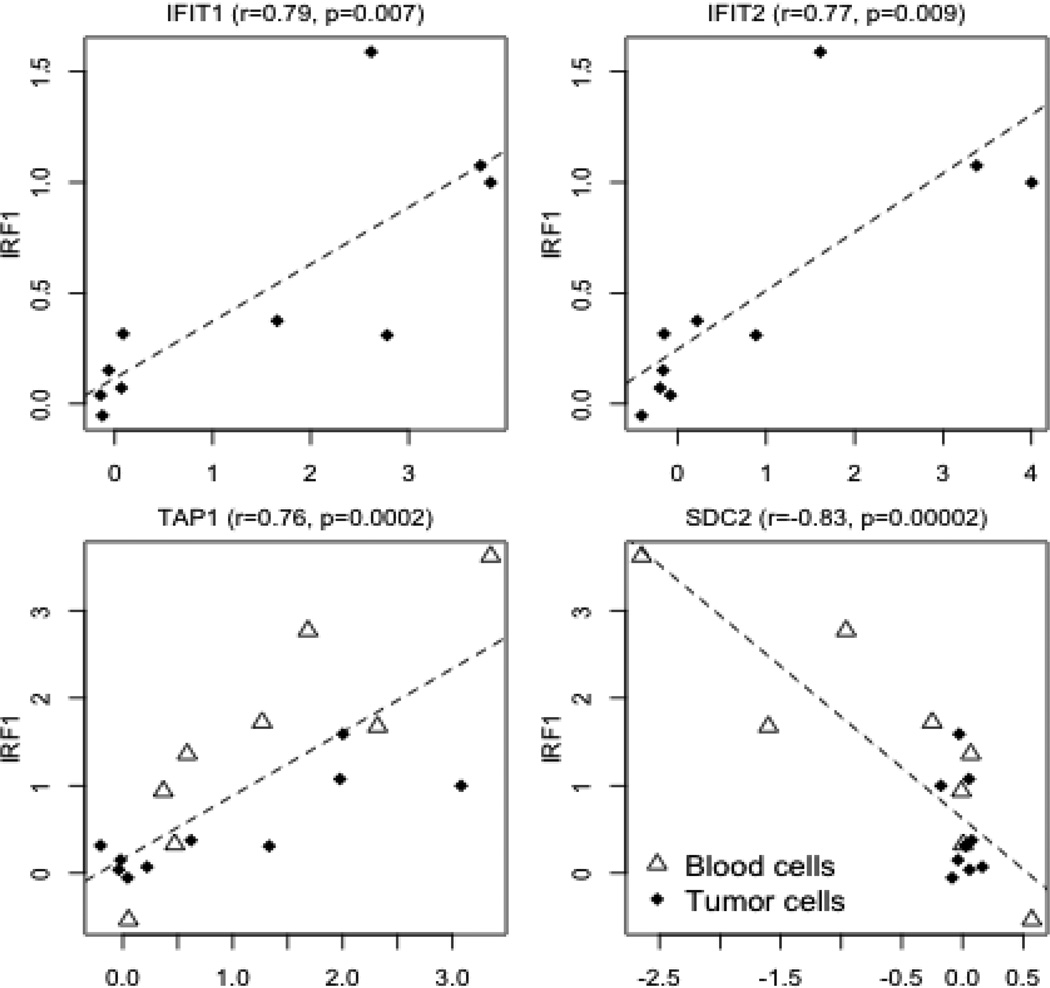
We used additional transcriptomic data stored in the GEO (Gene Expression Omnibus) database to examine the response of IRF1 target genes to interferons in different cell types. In a study of peripheral blood mononuclear cell (PBMC) subpopulations stimulated with interferon gamma (Waddell et al., 2010), up-regulation of IRF1 targets was observed in B cells, CD4+ and CD8 T cells, and monocytes, but not in natural killer cells (Figure 2B). In another study of ten tumor cell lines treated with interferon α2a (Siegrist et al., 2011), the cell lines with over-expression of IRF1 also had an overall up-regulation of target genes (Figure 2C). The expression of both IFIT1 and IFIT2, which were not previously reported as direct targets of IRF1, were significantly up-regulated. The expression of both genes was significantly correlated with the expression of IRF1 in the tumor cell lines after interferon treatment, suggesting that interferons activated these two genes through IRF1 (Figure 3). However, the concurrent expression change of IRF1 and IFIT1 IFIT2 expression was not observed in blood subpopulations, possibly because the regulatory mechanism is more complicated in vivo.
Figure 3. The concurrent expression change of IRF1 and its targets after cytokine treatment.
The scale of the axes corresponds to the log2-ratio of expression levels after versus before treatment. A significant correlation between IFIT1/IFIT2 and IRF1 occurred in tumor cell lines, but not in blood cells. (r: Pearson's correlation coefficient; p: p value obtained from the correlation test)
Also available in GEO were transcriptomic data sets including hepatoma and fibroblast cells transfected with an IRF1-overexpression vector [GSE26817], which made it possible to investigate the exclusive effect of IRF1 over-expression without the involvement of interferons. Target genes had an even stronger response to this direct treatment (Figure 2D). 107 genes in the hepatoma cell line and 122 in the fibroblast cell line (of the 322 measured genes) were upregulated. The response was similar between the two cell lines as 92 genes were up-regulated in both. Therefore, IRF1 over-expression had similar effects in two different cell lines. SDC2 was again down-regulated in both cell lines and SPHAR was down-regulated only in the fibroblast cell line. AMOT, a gene regulates cell migration and tube formation (Troyanovsky et al., 2001), was also down-regulated in both cell lines. In summary, genes with an IRF1 binding motif around their TSS were generally up-regulated with the over-expression of IRF1 and the 18bp targets had a stronger association with IRF1 over-expression than the 13bp targets (Supplemental Table 4). However, individual target genes showed different patterns (Supplemental Table 5). For example, both TAP1 and SDC2 were co-regulated with IRF1 across cell types, but in opposite directions (Figure 3). By correlation analysis, we identified a subset of 56 targets (p<=0.01) that had concurrent changes with IRF1 across different cell types (Supplemental Table 5). Immune response genes were further enriched in this subset (N=12, p=9.2e-6).
3.4 Validation of the IRF1 binding motif/sites in extra samples
To validate the IRF1 binding motif, we performed another run of IRF1 ChIP-seq which included an independent pair of input DNA and IRF1 ChIP-seq samples. Although both samples had substantially lower numbers of aligned reads (20.4 and 20.7 millions respectively), 107 peaks were identified. An almost identical binding motif was obtained from those peaks using the same motif discovery method (Supplemental Figure 4 and Supplemental Table 2), whose consensus sequence was NRAANNGAAASTGAAASY.
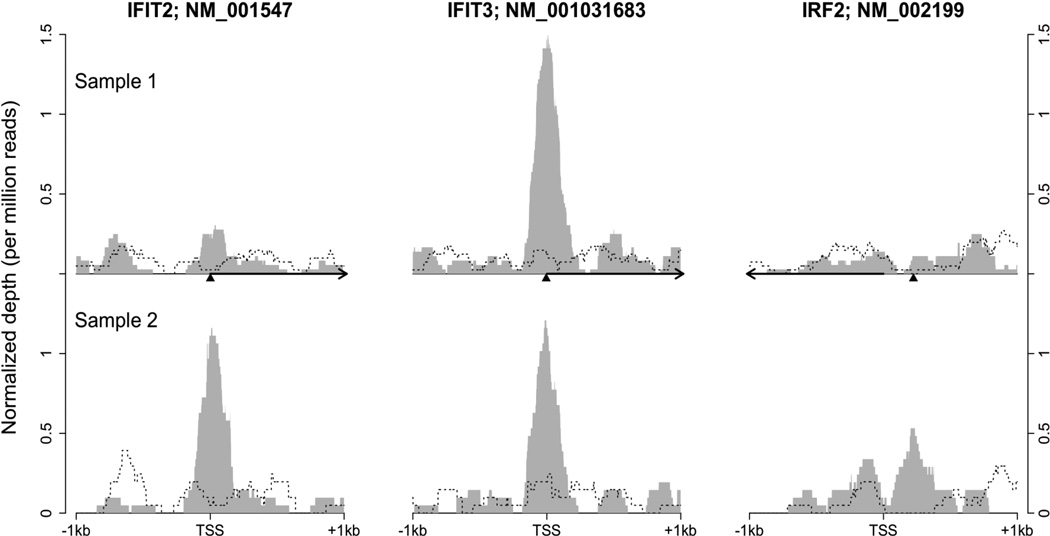
We noted that not all of the peaks identified from the original and the validating samples overlapped (Supplemental Table 6). For example, while there were binding peaks around the TSS of IFIT3 in both samples, only the validating sample had a peak close to the TSS of IFIT2 (Figure 4). Meanwhile, both samples had low or no read coverage at the potential binding site located with the promoter of IRF2, an inhibitor and known target of IRF1. These observations suggest that individual variability of IRF1 binding exists within primary human monocytes. The primary monocytes in this study were analyzed directly ex vivo and probably reflected the individual health status of the donors.
Figure 4. The depth of ChIP-seq signal at three locations of IRF1 binding.
The height represents normalized coverage depth out of one million reads in each sample. The upper half of the figure was based on the original pair of IRF1 ChIP-seq (grey shadow) and DNA input ChIP-seq (dashed line) samples. The lower half was based on the validating pair. Depth information was plotted around the TSS of three IRF1 target genes. Arrows indicate the direction of transcription and the triangles point to the locations of the last base of IRF1 binding motif. NM_001031683 is one of two alternative transcripts of IFIT3.
4. DISCUSSION
The goal of this study was to refine the consensus binding motif for IRF1. As a critical mediator of the inflammatory process and the development of immunologically important cell types, understanding the role of IRF1 has clinical implications for a broad range of diseases. We utilized unstimulated monocytes and were surprised at the large number of binding sites present in unstimulated cells. Whether these truly represent constitutive binding of IRF1 to certain targets or whether the cells experienced some element of activation during the purification process is not known. In a study of IRF1 over-expression in tumor cells, the majority of genes regulated by IRF1 were apoptotic (Tanaka et al., 1994; Yim et al., 1997; Kroger et al., 2001; Kroger et al., 2003; Kim et al., 2004). Nevertheless, many of the best characterized functions of IRF1 relate to host defense (Skaar et al., 1998; Taniguchi et al., 2001; Honda and Taniguchi, 2006). It is possible that IRF1 pre-bound to gene promoters renders them poised for rapid expression (Muse et al., 2007).
This study refined the consensus binding motif for IRF1. It is not known whether other IRF family members can recognize this sequence or whether the expanded sequence drives specificity. IRF family members exhibit some redundancy and therefore it is likely that sequence specificity is not the entire explanation for differential binding (Taniguchi et al., 2001; Decker et al., 2005; Platanias, 2005), however, the variable nucleotide spacers between GAAA repeat motifs clearly contribute to IRF specificity (Morin et al., 2002). Context and relative cell levels of IRF family members may also contribute to the in vivo binding. This study represents an important step in the further understanding of the relative effects of IRF family member interactions with chromatin.
The ChIP-seq approach represents a very direct and efficient strategy to examine in vivo binding for transcription factors, however, it is important to recognize that there could be potential confounders. In our bioinformatic analysis, we did not allow for indels in alignments to the consensus sequence. Therefore, alternative binding sites with bipartite recognition sequences could have been missed. Nevertheless, these data have provided an improved consensus sequence based on in vivo binding.
IRF1 has been studied both as a tumor suppressor and as part of the innate anti-viral response (Pine, 1992; Kashiwaba et al., 1994; Nishizaki et al., 1997; O'Connell et al., 1998; Tirkkonen et al., 1998). We identified IRF1 binding sites upstream of genes with increased acetylation of H4 in monocytes from SLE patients compared to controls (Zhang et al., 2010b). Both cell growth and interferon pathways have been implicated in SLE (Baechler et al., 2003; Bennett et al., 2003; Denny et al., 2006). IRF1 can interact with p300 to acetylate histones and another IRF family member, IRF5, has been implicated in SLE by a genome-wide association study suggesting that this family of transcription factors may be important in SLE (Graham et al., 2006). This study provides an improved consensus motif to facilitate additional mechanistic studies. The new 18bp consensus motif provides better predictive value for interferon-responsive genes than the commonly used 13bp motif and should facilitate IRF1 studies going forward.
Supplementary Material
ACKNOWLEDGEMENTS
This work was supported by NIH grant 1R01AR058547. The authors gratefully acknowledge support from the Nucleic Acid Core Facility, Eric Rappaport, Xiaowu Gai, and Li Song.
Abbreviations
- CD
cluster of differentiation
- ChIP-seq
chromatin immunoprecipitation with next generation sequencing
- IFN
interferon
- IL
interleukin
- IRF1
interferon regulatory factor I
- NK
natural killer cell
- PBMC
peripheral blood mononuclear cells
- SLE
systemic lupus erythematosus
- TSS
transcription start site
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Baechler EC, Batliwalla FM, Karypis G, Gaffney PM, Ortmann WA, Espe KJ, Shark KB, Grande WJ, Hughes KM, Kapur V, Gregersen PK, Behrens TW. Interferon-inducible gene expression signature in peripheral blood cells of patients with severe lupus. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:2610–2615. doi: 10.1073/pnas.0337679100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bennett L, Palucka AK, Arce E, Cantrell V, Borvak J, Banchereau J, Pascual V. Interferon and granulopoiesis signatures in systemic lupus erythematosus blood. Journal of Experimental Medicine. 2003;197:711–723. doi: 10.1084/jem.20021553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang CH, Hammer J, Loh JE, Fodor WL, Flavell RA. The activation of major histocompatibility complex class I genes by interferon regulatory factor-1 (IRF-1) Immunogenetics. 1992;35:378–384. doi: 10.1007/BF00179793. [DOI] [PubMed] [Google Scholar]
- Decker T, Muller M, Stockinger S. The yin and yang of type I interferon activity in bacterial infection. Nat Rev Immunol. 2005;5:675–687. doi: 10.1038/nri1684. [DOI] [PubMed] [Google Scholar]
- Denny MF, Chandaroy P, Killen PD, Caricchio R, Lewis EE, Richardson BC, Lee KD, Gavalchin J, Kaplan MJ. Accelerated macrophage apoptosis induces autoantibody formation and organ damage in systemic lupus erythematosus. J Immunol. 2006;176:2095–2104. doi: 10.4049/jimmunol.176.4.2095. [DOI] [PubMed] [Google Scholar]
- Digweed M, Gunthert U, Schneider R, Seyschab H, Friedl R, Sperling K. Irreversible repression of DNA synthesis in Fanconi anemia cells is alleviated by the product of a novel cyclin-related gene. Molecular and Cellular Biology. 1995;15:305–314. doi: 10.1128/mcb.15.1.305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Escalante CR, Yie J, Thanos D, Aggarwal AK. Expression, purification, and co-crystallization of IRF-I bound to the interferon-beta element PRDI. FEBS Lett. 1997;414:219–220. doi: 10.1016/s0014-5793(97)00996-4. [DOI] [PubMed] [Google Scholar]
- Frontini M, Vijayakumar M, Garvin A, Clarke N. A ChIP-chip approach reveals a novel role for transcription factor IRF1 in the DNA damage response. Nucleic Acids Res. 2009;37:1073–1085. doi: 10.1093/nar/gkn1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujii Y, Shimizu T, Kusumoto M, Kyogoku Y, Taniguchi T, Hakoshima T. Crystal structure of an IRF-DNA complex reveals novel DNA recognition and cooperative binding to a tandem repeat of core sequences. EMBO Journal. 1999;18:5028–5041. doi: 10.1093/emboj/18.18.5028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gabriele L, Fragale A, Borghi P, Sestili P, Stellacci E, Venditti M, Schiavoni G, Sanchez M, Belardelli F, Battistini A. IRF-1 deficiency skews the differentiation of dendritic cells toward plasmacytoid and tolerogenic features. J Leukoc Biol. 2006;80:1500–1511. doi: 10.1189/jlb.0406246. [DOI] [PubMed] [Google Scholar]
- Graham RR, Kozyrev SV, Baechler EC, Reddy MV, Plenge RM, Bauer JW, Ortmann WA, Koeuth T, Gonzalez Escribano MF, Argentine, Spanish Collaborative, G. Pons-Estel B, Petri M, Daly M, Gregersen PK, Martin J, Altshuler D, Behrens TW, Alarcon-Riquelme ME. A common haplotype of interferon regulatory factor 5 (IRF5) regulates splicing and expression and is associated with increased risk of systemic lupus erythematosus. Nat Genet. 2006;38:550–555. doi: 10.1038/ng1782. [DOI] [PubMed] [Google Scholar]
- Harada H, Fujita T, Miyamoto M, Kimura Y, Maruyama M, Furia A, Miyata T, Taniguchi T. Structurally similar but functionally distinct factors, IRF-1 and IRF-2, bind to the same regulatory elements of IFN and IFN-inducible genes. Cell. 1989;58:729–739. doi: 10.1016/0092-8674(89)90107-4. [DOI] [PubMed] [Google Scholar]
- Honda K, Taniguchi T. IRFs: master regulators of signalling by Toll-like receptors and cytosolic pattern-recognition receptors. Nat Rev Immunol. 2006;6:644–658. doi: 10.1038/nri1900. [DOI] [PubMed] [Google Scholar]
- Kashiwaba M, Tamura G, Ishida M. Aberrations of the APC gene in primary breast carcinoma. J Cancer Res Clin Oncol. 1994;120:727–731. doi: 10.1007/BF01194271. [DOI] [PubMed] [Google Scholar]
- Kim PK, Armstrong M, Liu Y, Yan P, Bucher B, Zuckerbraun BS, Gambotto A, Billiar TR, Yim JH. IRF-1 expression induces apoptosis and inhibits tumor growth in mouse mammary cancer cells in vitro and in vivo. Oncogene. 2004;23:1125–1135. doi: 10.1038/sj.onc.1207023. [DOI] [PubMed] [Google Scholar]
- Kimura T, Nakayama K, Penninger J, Kitagawa M, Harada H, Matsuyama T, Tanaka N, Kamijo R, Vilcek J, Mak TW, et al. Involvement of the IRF-1 transcription factor in antiviral responses to interferons. Science. 1994;264:1921–1924. doi: 10.1126/science.8009222. [DOI] [PubMed] [Google Scholar]
- Ko J, Gendron-Fitzpatrick A, Splitter GA. Susceptibility of IFN regulatory factor-1 and IFN consensus sequence binding protein-deficient mice to brucellosis. Journal of Immunology. 2002;168:2433–2440. doi: 10.4049/jimmunol.168.5.2433. [DOI] [PubMed] [Google Scholar]
- Kroger A, Dallugge A, Kirchhoff S, Hauser H. IRF-1 reverts the transformed phenotype of oncogenically transformed cells in vitro and in vivo. Oncogene. 2003;22:1045–1056. doi: 10.1038/sj.onc.1206260. [DOI] [PubMed] [Google Scholar]
- Kroger A, Ortmann D, Krohne TU, Mohr L, Blum HE, Hauser H, Geissler M. Growth suppression of the hepatocellular carcinoma cell line Hepa1-6 by an activatable interferon regulatory factor-1 in mice. Cancer Res. 2001;61:2609–2617. [PubMed] [Google Scholar]
- Lee JH, Park H, Chung H, Choi S, Kim Y, Yoo H, Kim TY, Hann HJ, Seong I, Kim J, Kang KG, Han IO, Oh ES. Syndecan-2 regulates the migratory potential of melanoma cells. Journal of Biological CHemistry. 2009;284:27167–27175. doi: 10.1074/jbc.M109.034678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L. GADEM: a genetic algorithm guided formation of spaced dyads coupled with an EM algorithm for motif discovery. J Comput Biol. 2009;16:317–329. doi: 10.1089/cmb.2008.16TT. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto M, Fujita T, Kimura Y, Maruyama M, Harada H, Sudo Y, Miyata T, Taniguchi T. Regulated expression of a gene encoding a nuclear factor, IRF-1, that specifically binds to IFN-beta gene regulatory elements. Cell. 1988;54:903–913. doi: 10.1016/s0092-8674(88)91307-4. [DOI] [PubMed] [Google Scholar]
- Morin P, Braganca J, Bandu MT, Lin R, Hiscott J, Doly J, Civas A. Preferential binding sites for interferon regulatory factors 3 and 7 involved in interferon-A gene transcription. Journal of Molecular Biology. 2002;316:1009–1022. doi: 10.1006/jmbi.2001.5401. [DOI] [PubMed] [Google Scholar]
- Muse GW, Gilchrist DA, Nechaev S, Shah R, Parker JS, Grissom SF, Zeitlinger J, Adelman K. RNA polymerase is poised for activation across the genome. Nat Genet. 2007;39:1507–1511. doi: 10.1038/ng.2007.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naf D, Hardin SE, Weissmann C. Multimerization of AAGTGA and GAAAGT generates sequences that mediate virus inducibility by mimicking an interferon promoter element. Proc Natl Acad Sci U S A. 1991;88:1369–1373. doi: 10.1073/pnas.88.4.1369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishizaki T, Chew K, Chu L, Isola J, Kallioniemi A, Weidner N, Waldman FM. Genetic alterations in lobular breast cancer by comparative genomic hybridization. Int J Cancer. 1997;74:513–517. doi: 10.1002/(sici)1097-0215(19971021)74:5<513::aid-ijc6>3.0.co;2-6. [DOI] [PubMed] [Google Scholar]
- O'Connell P, Pekkel V, Fuqua SA, Osborne CK, Clark GM, Allred DC. Analysis of loss of heterozygosity in 399 premalignant breast lesions at 15 genetic loci. J Natl Cancer Inst. 1998;90:697–703. doi: 10.1093/jnci/90.9.697. [DOI] [PubMed] [Google Scholar]
- Ogasawara K, Hida S, Azimi N, Tagaya Y, Sato T, Yokochi-Fukuda T, Waldmann TA, Taniguchi T, Taki S. Requirement for IRF-1 in the microenvironment supporting development of natural killer cells. Nature. 1998;391:700–703. doi: 10.1038/35636. [DOI] [PubMed] [Google Scholar]
- Ohteki T, Yoshida H, Matsuyama T, Duncan GS, Mak TW, Ohashi PS. The transcription factor interferon regulatory factor 1 (IRF-1) is important during the maturation of natural killer 1.1+ T cell receptor-alpha/beta+ (NK1+ T) cells, natural killer cells, and intestinal intraepithelial T cells. Journal of Experimental Medicine. 1998;187:967–972. doi: 10.1084/jem.187.6.967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Penninger JM, Sirard C, Mittrucker HW, Chidgey A, Kozieradzki I, Nghiem M, Hakem A, Kimura T, Timms E, Boyd R, Taniguchi T, Matsuyama T, Mak TW. The interferon regulatory transcription factor IRF-1 controls positive and negative selection of CD8+ thymocytes. Immunity. 1997;7:243–254. doi: 10.1016/s1074-7613(00)80527-0. [DOI] [PubMed] [Google Scholar]
- Pine R. Constitutive expression of an ISGF2/IRF1 transgene leads to interferon-independent activation of interferon-inducible genes and resistance to virus infection. J Virol. 1992;66:4470–4478. doi: 10.1128/jvi.66.7.4470-4478.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Platanias LC. Mechanisms of type-I- and type-II-interferon-mediated signalling. Nat Rev Immunol. 2005;5:375–386. doi: 10.1038/nri1604. [DOI] [PubMed] [Google Scholar]
- Romeo G, Fiorucci G, Chiantore MV, Percario ZA, Vannucchi S, Affabris E. IRF-1 as a negative regulator of cell proliferation. Journal of Interferon and Cytokine Research. 2002;22:39–47. doi: 10.1089/107999002753452647. [DOI] [PubMed] [Google Scholar]
- Siegrist F, Ebeling M, Certa U. The small interferon-induced transmembrane genes and proteins. Journal of Interferon and Cytokine Research. 2011;31:183–197. doi: 10.1089/jir.2010.0112. [DOI] [PubMed] [Google Scholar]
- Skaar TC, Prasad SC, Sharareh S, Lippman ME, Brunner N, Clarke R. Two-dimensional gel electrophoresis analyses identify nucleophosmin as an estrogen regulated protein associated with acquired estrogen-independence in human breast cancer cells. J Steroid Biochem Mol Biol. 1998;67:391–402. doi: 10.1016/s0960-0760(98)00142-3. [DOI] [PubMed] [Google Scholar]
- Taki S, Sato T, Ogasawara K, Fukuda T, Sato M, Hida S, Suzuki G, Mitsuyama M, Shin EH, Kojima S, Taniguchi T, Asano Y. Multistage regulation of Th1-type immune responses by the transcription factor IRF-1. Immunity. 1997;6:673–679. doi: 10.1016/s1074-7613(00)80443-4. [DOI] [PubMed] [Google Scholar]
- Tamura T, Ishihara M, Lamphier MS, Tanaka N, Oishi I, Aizawa S, Matsuyama T, Mak TW, Taki S, Taniguchi T. An IRF-1-dependent pathway of DNA damage-induced apoptosis in mitogen-activated T lymphocytes. Nature. 1995;376:596–599. doi: 10.1038/376596a0. [DOI] [PubMed] [Google Scholar]
- Tanaka N, Ishihara M, Kitagawa M, Harada H, Kimura T, Matsuyama T, Lamphier MS, Aizawa S, Mak TW, Taniguchi T. Cellular commitment to oncogene-induced transformation or apoptosis is dependent on the transcription factor IRF-1. Cell. 1994;77:829–839. doi: 10.1016/0092-8674(94)90132-5. [DOI] [PubMed] [Google Scholar]
- Tanaka N, Ishihara M, Lamphier MS, Nozawa H, Matsuyama T, Mak TW, Aizawa S, Tokino T, Oren M, Taniguchi T. Cooperation of the tumour suppressors IRF-1 and p53 in response to DNA damage. Nature. 1996;382:816–818. doi: 10.1038/382816a0. [DOI] [PubMed] [Google Scholar]
- Tanaka N, Kawakami T, Taniguchi T. Recognition DNA sequences of interferon regulatory factor 1 (IRF-1) and IRF-2, regulators of cell growth and the interferon system. Molecular and Cellular Biology. 1993;13:4531–4538. doi: 10.1128/mcb.13.8.4531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi T, Ogasawara K, Takaoka A, Tanaka N. IRF family of transcription factors as regulators of host defense. Annu Rev Immunol. 2001;19:623–655. doi: 10.1146/annurev.immunol.19.1.623. [DOI] [PubMed] [Google Scholar]
- Testa U, Stellacci E, Pelosi E, Sestili P, Venditti M, Orsatti R, Fragale A, Petrucci E, Pasquini L, Belardelli F, Gabriele L, Battistini A. Impaired myelopoiesis in mice devoid of interferon regulatory factor 1. Leukemia. 2004;18:1864–1871. doi: 10.1038/sj.leu.2403472. [DOI] [PubMed] [Google Scholar]
- Tirkkonen M, Tanner M, Karhu R, Kallioniemi A, Isola J, Kallioniemi OP. Molecular cytogenetics of primary breast cancer by CGH. Genes Chromosomes Cancer. 1998;21:177–184. [PubMed] [Google Scholar]
- Troyanovsky B, Levchenko T, Mansson G, Matvijenko O, Holmgren L. Angiomotin: an angiostatin binding protein that regulates endothelial cell migration and tube formation. J Cell Biol. 2001;152:1247–1254. doi: 10.1083/jcb.152.6.1247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urschel JD, Bertsch DJ, Takita H. Thoracic packing for postoperative hemorrhage. J Cardiovasc Surg (Torino) 1997;38:673–675. [PubMed] [Google Scholar]
- Waddell SJ, Popper SJ, Rubins KH, Griffiths MJ, Brown PO, Levin M, Relman DA. Dissecting interferon-induced transcriptional programs in human peripheral blood cells. PLoS ONE. 2010;5:e9753. doi: 10.1371/journal.pone.0009753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yim JH, Wu SJ, Casey MJ, Norton JA, Doherty GM. IFN regulatory factor-1 gene transfer into an aggressive, nonimmunogenic sarcoma suppresses the malignant phenotype and enhances immunogenicity in syngeneic mice. Journal of Immunology. 1997;158:1284–1292. [PubMed] [Google Scholar]
- Zhang Z, Maurer K, Perin JC, Song L, Sullivan KE. Cytokine-induced monocyte characteristics in SLE. J Biomed Biotechnol. 2010a;2010:507475. doi: 10.1155/2010/507475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z, Song L, Maurer K, Petri MA, Sullivan KE. Global H4 acetylation analysis by ChIP-chip in systemic lupus erythematosus monocytes. Genes Immun. 2010b;11:124–133. doi: 10.1038/gene.2009.66. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.