Abstract
Translocation of transfer RNAs (tRNAs) through the ribosome during protein synthesis involves large-scale structural rearrangements of the ribosome and the ribosome-bound tRNAs that are accompanied by extensive and dynamic remodeling of tRNA-ribosome interactions. The contributions that rearranging individual tRNA-ribosome interactions make to directing tRNA movements during translocation, however, remain largely unknown. To address this question, we have used single-molecule fluorescence resonance energy transfer to characterize the dynamics of ribosomal pre-translocation (PRE) complex analogs carrying either wild-type or systematically mutagenized tRNAs. Our data reveal how specific tRNA-ribosome interactions regulate the rate with which the PRE complex rearranges into a critical, on-pathway translocation intermediate and how these interactions control the stability of the resulting configuration. More interestingly, our results suggest that the conformational flexibility of the tRNA molecule itself plays a crucial role in directing the structural dynamics of the PRE complex during translocation.
INTRODUCTION
During the elongation stage of protein synthesis, ribosome-catalyzed addition of each amino acid to the nascent polypeptide chain is followed by the rapid and unidirectional translocation of the transfer RNA (tRNA)-messenger RNA (mRNA) complex through the ribosome by precisely one codon. Translocation occurs through a multi-step process that requires extensive remodeling of tRNA-ribosome interactions as well as significant structural distortions of the ribosome-bound tRNAs relative to the “ground state” structures of ribosome-free tRNAs1-3. Despite the fundamental importance of translocation to protein synthesis, the mechanism through which the ribosome physically coordinates, regulates, and executes this process remains poorly understood.
Recently, single-molecule fluorescence resonance energy transfer (smFRET) studies of tRNA and ribosome dynamics during translocation have revealed that deacylation of the peptidyl-tRNA bound within the ribosomal P (peptidyl-tRNA binding) site during peptide bond formation enables thermally activated and stochastic structural fluctuations of the resulting pretranslocation (PRE) complexes4-9. These complexes, carrying deacylated tRNA at the P site and peptidyl-tRNA at the A (aminoacyl-tRNA binding) site, oscillate between two major global conformational states. In global state 1 (GS1), the small (30S in Escherichia coli) and large (50S in E. coli) ribosomal subunits are in their non-rotated intersubunit orientation, the tRNAs are positioned in their classical P/P (denoting the 30S P site/50S P site) and A/A configurations, and the ribosomal L1 stalk is in its open conformation; in global state 2 (GS2), the ribosomal subunits are in their rotated intersubunit orientation, the tRNAs are positioned in their hybrid P/E (where E denotes the ribosomal exit, or deacylated tRNA binding, site) and A/P configurations, and the L1 stalk is in its closed conformation5 (Fig. 1a). Biochemical data support the view that GS2 is an authentic on-pathway intermediate in translocation10 and smFRET studies have shown that binding of elongation factor G (EF-G) to the PRE complex dramatically shifts the GS1⇄GS2 dynamic equilibrium towards GS2 as part of the mechanism through which it promotes translocation5-8,11. Moreover, pre-steady state smFRET studies have suggested that the GS1→GS2 transition may limit the rate at which EF-G can productively bind and act on the PRE complex to promote translocation6, findings which have been recently confirmed12.
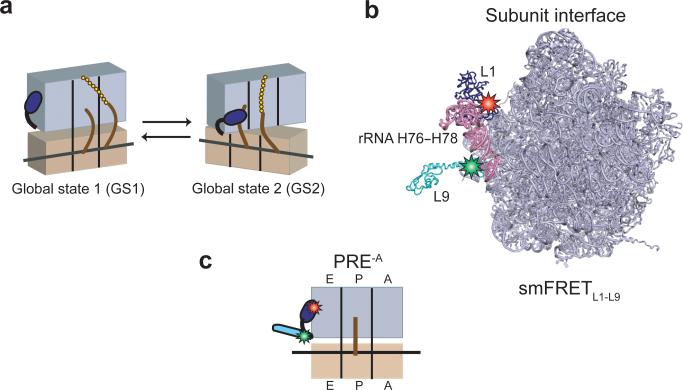
Figure 1. Global states model of the PRE complex, L1-L9 labeling strategy, and PRE-A complexes.
(a) Cartoon representation of the global states model of the PRE complex. The 30S and 50S subunits are shown in tan and lavender, respectively, with the L1 stalk in dark blue. tRNAs are shown as brown curves and the nascent polypeptide as a chain of gold spheres. Upon aa-tRNA selection and peptide bond formation, the PRE complex spontaneously fluctuates between two major global conformational states, which we call global state 1 (GS1) and global state 2 (GS2). (b) Labeling strategy for smFRETL1-L9. The 50S subunit is shown from the perspective of the intersubunit space31 (PDB ID: 2J01). The L1 stalk consists of 23S rRNA helices 76-78 (pink) and r-protein L1 (dark blue). r-protein L9 is shown in cyan. The donor (Cy3) and the acceptor (Cy5) fluorophores are shown as green and red stars on r-proteins L9 and L1, respectively. The image was rendered using PyMol50 (www.pymol.org). (c) Cartoon representation of a PRE-A complex. PRE-A complexes are formed using an L1-L9 labeled 50S subunit and carry a deacylated P-site tRNA.
A key regulator of both the GS1⇄GS2 equilibrium and translocation is the P-site tRNA. Indeed, the ability of the elongating ribosome to actuate the GS1⇄GS2 equilibrium6-8,13-15 and trigger the productive binding of EF-G16,17 that leads to stabilization of GS26-8,11,18,19, ribosome-stimulated GTP hydrolysis16,17, and ultimately translocation17,20, depends critically on the presence of a full-length, deacylated P-site tRNA. In addition to the presence and acylation state of the P-site tRNA, the GS1⇄GS2 equilibrium and translocation are sensitive to the identity of this tRNA, indicating that specific P-site tRNA-ribosome interactions and/or tRNA structural features unique to each tRNA can differentially modulate the GS1⇄GS2 equilibrium and translocation. In the most extensively investigated examples, smFRET studies demonstrated that, relative to PREPhe complexes (where Phe denotes a P-site tRNAPhe), PREfMet complexes exhibit a GS1⇄GS2 equilibrium that is inherently shifted towards GS16-8,10,14,15 and, upon EF-G binding, is shifted towards GS2 through a kinetic mechanism that is distinct from that observed in PREPhe complexes7. Correspondingly, other studies have reported that PREfMet complexes exhibit a slower rate of translocation relative to analogous PRE complexes carrying elongator P-site tRNAs10,20.
At present, however, it is not known which tRNA-ribosome interactions or aspects of tRNA structure give rise to the tRNA-mediated regulation of the GS1⇄GS2 equilibrium and translocation. Here, we begin to close this gap in our understanding of how tRNAs regulate the GS1⇄GS2 equilibrium by using smFRET to compare the kinetic differences between PRE complexes carrying wild-type and strategically mutated tRNAs. We started by conducting a comprehensive characterization of the GS1⇄GS2 equilibrium in the absence and presence of EF-G for PRE complexes carrying an expanded set of wild-type tRNAs at the P site. We demonstrate that, relative to all of the elongator tRNAs examined, tRNAfMet uniquely modulates the GS1⇄GS2 equilibrium and the ability of EF-G to shift this equilibrium towards GS2. Building from these initial experiments, we then examined a series of PRE complexes carrying tRNAfMet mutants, in which sequence elements unique to tRNAfMet were mutated to the corresponding sequences found in elongator tRNAPhe, at the P site. The results of our studies collectively reveal that the kinetics of the GS1⇄GS2 equilibrium and the ability of EF-G to shift this equilibrium towards GS2 are at least partly dictated by the intrinsic conformational flexibility of the tRNA as well as specific tRNA-ribosome interactions. Specifically, our results suggest that the GS1→GS2 transition rate is primarily determined by the intrinsic conformational flexibility of the P-site tRNA itself, while the GS2→GS1 transition rate is largely determined by the minor groove-minor groove interaction between the aminoacyl acceptor stem of the P/E tRNA and H68 of 23S rRNA at the 50S subunit E site. Our proposal that the intrinsic conformational flexibility of the P-site tRNA can modulate the GS1→GS2 transition rate implies that the ease with which the ribosome can distort the tRNA structure is a significant aspect of translocation. This expands the functionally important role of tRNA deformability during translation elongation beyond that already proposed for aminoacyl-tRNA (aa-tRNA) selection21-23. We hypothesize that the differences observed in the dynamics of PRE complexes carrying initiator versus elongator tRNAs in the P site derive from the distinct evolutionary pressures that have been imposed on these tRNAs for optimal performance during the initiation and elongation stages of protein synthesis, respectively.
RESULTS
An intra-ribosomal smFRET signal reports on GS1⇄GS2
Fluctuation of the L1 stalk between open and closed conformations is one of the defining features of the GS1⇄GS2 equilibrium6,7,9 (Fig. 1a). We have previously developed and validated a doubly fluorescently labeled 50S subunit (harboring a Cy5 acceptor fluorophore within ribosomal protein L1 and a Cy3 donor fluorophore within ribosomal protein L9) that yields an smFRET signal that is sensitive to fluctuations of the L1 stalk between open and closed conformations7 (Fig. 1b, Supplementary Methods). Using these doubly labeled 50S subunits, unlabeled 30S subunits, a set of natural, deacylated E. coli tRNAs, and a corresponding set of mRNAs (Supplementary Fig. 1), we non-enzymatically prepared two PRE-AfMet complexes (where -A denotes a PRE complex analog in which the peptidyl-tRNA is absent from the A site) and four PRE-Aelong complexes (Fig. 1c). The two PRE-AfMet complexes carried either one of the two isoacceptors of tRNAfMet, tRNAfMet1 encoded by the metZ gene (PRE-AfMet-1) or tRNAfMet2 encoded by the metY gene (PRE-AfMet-2)24. The four PRE-Aelong complexes carried either tRNAPhe (PRE-APhe), tRNATyr (PRE-ATyr), tRNAGlu (PRE-AGlu), or tRNAVal (PRE-AVal). All six PRE-A complexes were imaged using total internal reflection fluorescence (TIRF) microscopy (see Online Methods and Supplementary Methods for details on sample preparation and TIRF imaging).
Consistent with prior results7, each PRE-A complex exhibits two FRET states centered at FRET efficiencies of 0.56 ± 0.02 and 0.36 ± 0.01, corresponding to the open and closed conformations of the L1 stalk and reporting on GS1 and GS2, respectively. Also as previously reported7, the smFRET versus time trajectories partition into three sub-populations depending on whether they exclusively occupy GS1 (SPGS1), exclusively occupy GS2 (SPGS2), or fluctuate between GS1 and GS2 (SPfluct) prior to photobleaching (Fig. 2). PRE-A complexes carrying different P-site tRNAs exhibited unique population distributions between the open and closed L1 stalk conformations (Fig. 3). Notably, the tRNA-dependent trend we find in the equilibrium constants (Keqs) describing the equilibrium between the open and closed L1 stalk conformations (Table 1) in PRE-AfMet-1, PRE-APhe and PRE-ATyr mirrors the tRNA-dependent trend observed by Cornish et al. in the Keqs describing the equilibrium between the non-rotated and rotated intersubunit orientations in analogous PRE-A complexes8. This observation supports a model in which the open and closed conformations of the L1 stalk are coupled to the non-rotated and rotated intersubunit orientations of the ribosome, respectively, within the GS1 ⇄ GS2 equilibrium5,6 (see Supplementary Discussion).
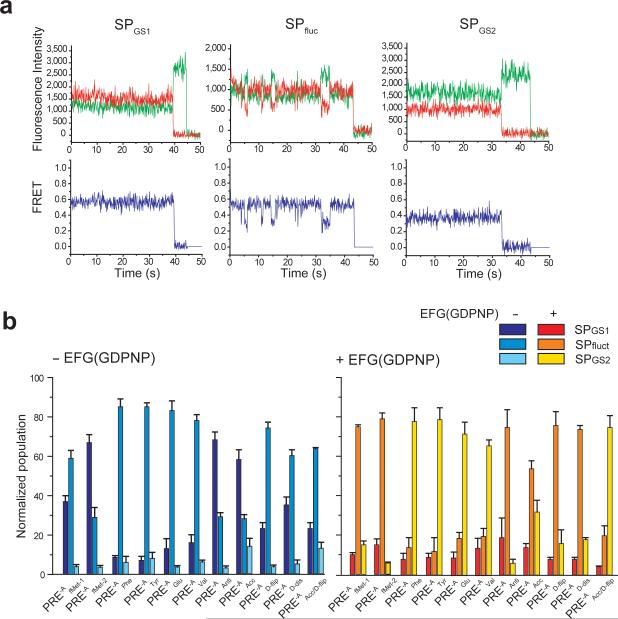
Figure 2. Sample smFRET versus time trajectories and relative occupancies of smFRET trajectory sub-populations.
(a) Sample smFRET versus time trajectories. Three sub-populations of smFRET trajectories were observed. The first sub-population, which exhibits a stable FRET state centered at 0.56 ± 0.02, consists of PRE-A complexes that occupy GS1 and photobleach out of GS1 prior to undergoing a GS1→GS2 transition (SPGS1, left panel); the second sub-population, which exhibits fluctuations between two FRET states centered at 0.56 ± 0.02 and 0.36 ± 0.01, consists of PRE-A complexes that fluctuate between GS1 and GS2 during the observation period (SPfluct, middle panel); and the third sub-population, which exhibits a stable FRET state centered at 0.36 ± 0.01, consists of PRE-A complexes that occupy GS2 and photobleach out of GS2 prior to undergoing a GS2→GS1 transition (SPGS2, right panel). Representative Cy3 and Cy5 emission intensity versus time trajectories are shown in green and red, respectively (top row). The corresponding smFRET versus time trajectories, calculated using E = ICy5 / (ICy3 + ICy5), where E is the FRET efficiency at each time point and ICy3 and ICy5 are the emission intensities of Cy3 and Cy5, respectively, are shown in blue (bottom row). (b) Relative occupancies of the three sub-populations of smFRET trajectories. The percentage of smFRET trajectories occupying SPGS1, SPfluct and SPGS2 for each PRE-A complex in the absence (left panel) and presence (right panel) of EF-G(GDPNP) are shown as bar graphs. Data are the mean ± standard deviation of three independent measurements (see Supplementary Table 1).
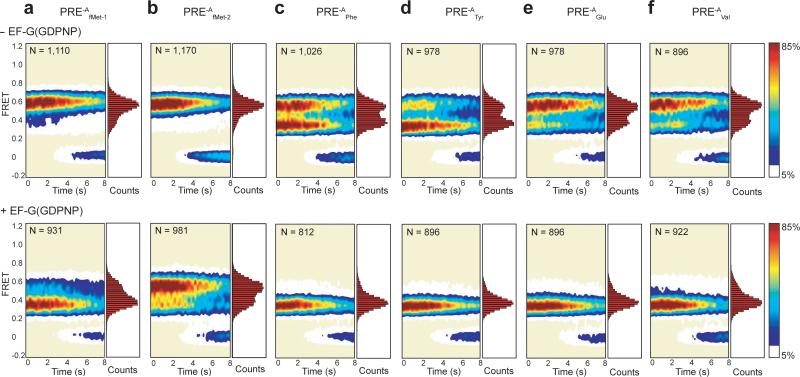
Figure 3. Steady-state smFRET measurements on PRE-A complexes carrying wild-type and elongator tRNAs.
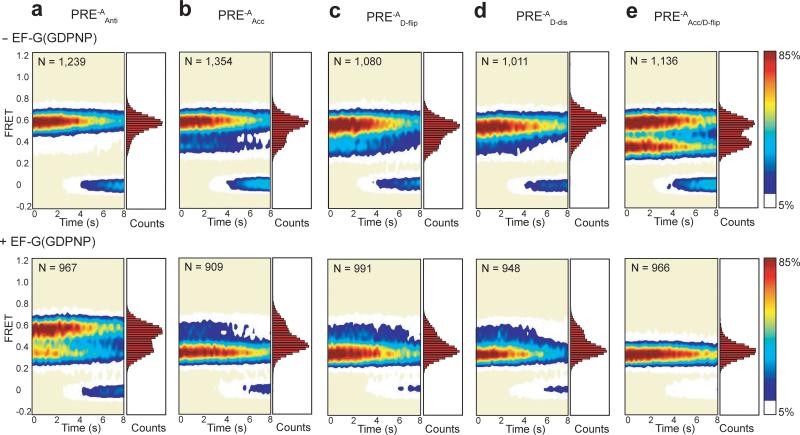
Surface contour plots of the time evolution of population FRET were generated by superimposing the individual smFRET versus time trajectories for each PRE-A complex. Contours are plotted from white (lowest population) to red (highest population) as indicated by the color bar. The number of smFRET trajectories used to construct each contour plot is indicated by “N.” The corresponding one-dimensional FRET histograms plotted along the right-hand y-axis of the surface contour plots were generated using the first 20 time points from all of the FRET trajectories in each dataset. The PRE-A complexes in the absence of EF-G(GDPNP) are shown along the top row, and the corresponding PRE-A complexes in the presence of 2 μM EF-G(GDPNP) are shown along the bottom row. (a) PRE-AfMet-1. (b) PRE-AfMet-2. (c) PRE-APhe. (d) PRE-ATyr. (e) PRE-AGlu. (f) PRE-AVal.
Table 1.
Equilibrium constants and transition rates governing the GS1 ⇄ GS2 equilibrium of PRE-A complexesa.
| P-site tRNA | % GS1b | % GS2b | Keqb | kGS1→GS2 (s-1)c | kGS2→GS1 (s-1)c | Normalized kGS1→GS2d | Normalized kGS2→GS1d |
|---|---|---|---|---|---|---|---|
| PRE-A | |||||||
| PRE-AfMet complexes | |||||||
| tRNAfMet1 | 79±1 | 21±1 | 0.27±0.02 | 0.38±0.12 | 1.30±0.28 | 2.1±0.9 | 1.0±0.3 |
| tRNAfMet2 | 88±2 | 12±2 | 0.14±0.02 | 0.18±0.05 | 1.32±0.33 | 1.0±0.4 | 1.0±0.3 |
| PRE-Aelong complexes | |||||||
| tRNAPhe | 50±6 | 50±6 | 1.0±0.2 | 0.50±0.05 | 0.55±0.08 | ||
| tRNATyr | 45±4 | 55±4 | 1.2±0.2 | 0.56±0.04 | 0.43±0.06 | ||
| tRNAGlu | 61±3 | 39±3 | 0.64±0.08 | 0.27±0.05 | 0.59±0.06 | ||
| tRNAVal | 63±1 | 37±1 | 0.58±0.03 | 0.34±0.08 | 0.69±0.18 | ||
| PRE-A complexes with tRNAfMet mutants | |||||||
| tRNAAnti | 89±2 | 11±2 | 0.12±0.02 | 0.18±0.04 | 1.48±0.06 | 1.0±0.4 | 1.1±0.3 |
| tRNAAcc | 73±5 | 27±5 | 0.38±0.11 | 0.06±0.03 | 0.15±0.07 | 0.33±0.18 | 0.12±0.06 |
| tRNAD-flip | 71±2 | 29±2 | 0.40±0.04 | 0.60±0.23 | 1.45±0.30 | 3.4±1.6 | 1.1±0.4 |
| tRNAD-dis | 75±9 | 25±9 | 0.35±0.15 | 0.32±0.10 | 1.53±0.32 | 1.8±0.8 | 1.2±0.4 |
| tRNAAcc/D-flip | 53±2 | 47±2 | 0.88±0.09 | 0.11±0.06 | 0.14±0.07 | 0.61±0.37 | 0.11±0.06 |
| PRE-A + 2 μM EF-G(GDPNP)e | |||||||
| PRE-AfMet complexes | |||||||
| tRNAfMet1 | 33±4 | 67±4 | 2.1±0.4 | 2.37±0.36 | 0.87±0.04 | ||
| tRNAfMet2 | 62±5 | 38±5 | 0.62±0.14 | 0.52±0.17 | 0.84±0.22 | ||
| PRE-A complexes with tRNAfMet mutants | |||||||
| tRNAAnti | 60±1 | 40±1 | 0.67±0.03 | 0.46±0.22 | 0.73±0.23 | ||
| tRNAAcc | 30±8 | 70±8 | 2.4±0.9 | 0.23±0.10 | 0.03±0.10 | ||
| tRNAD-flip | 26±5 | 74±5 | 2.9±0.9 | 1.95±0.20 | 0.58±0.23 | ||
| tRNAD-dis | 30±2 | 70±2 | 2.4±0.2 | 2.17±0.14 | 1.09±0.09 | ||
The mean and standard deviation (mean ± s.d.) of the equilibrium constant and transition rates for each PRE-A complex were calculated from three independent data sets.
Fractional populations of GS1 and GS2 and equilibrium constants were calculated as described in Supplementary Methods.
Transition rates were calculated using dwell-time analysis and corrected for fluorophore photobleaching (see Online Methods and Supplementary Methods).
Transition rates for PRE-A complexes carrying tRNAfMet2 mutants are normalized to PRE-AfMet-2 for comparison.
PRE-AfMet complexes exhibit distinct GS1⇄GS2 dynamics
Relative to all four PRE-Aelong complexes examined, the two PRE-AfMet complexes exhibit a higher occupancy of GS1 (Figs. 2b and 3 and Table 1). These results expand upon previous smFRET7,8 and biochemical10,14 studies, suggesting a general distinction between the dynamics of PRE complexes carrying initiator versus elongator tRNAs. Dwell time analyses of the smFRET data (see Online Methods and Supplementary Methods) reveal the kinetic mechanism underlying this difference, demonstrating that the higher occupancy of GS1 exhibited by the PRE-AfMet-1 complex relative to the PRE-Aelong complexes is driven almost exclusively by a 2-3-fold faster rate of GS2→GS1 transitions (kGS2→GS1), with almost no effect on the rate of GS1→GS2 transitions (kGS1→GS2) (Table 1); for the PRE-AfMet-2 complex, the 2-3-fold faster kGS2→GS1 observed in the PRE-AfMet-1 complex is further augmented by a 30-70% slower kGS1→GS2 relative to the PRE-Aelong complexes (Table 1).
Consistent with previous reports, binding of EF-G to PRE-AfMet-1 and PRE-AfMet-2 complexes in the presence of the non-hydrolyzable GTP analog, guanosine 5’-(β,γ-imido)triphosphate (GDPNP), shifts the GS1⇄GS2 GS2 equilibrium towards GS2 (Fig. 3)6-8,11. This is driven primarily by a 3-6-fold increase in kGS1→GS2 and augmented by a smaller, 30% decrease in kGS2→GS1 (Table 1), thus yielding more frequent fluctuations between GS1 and GS2 relative to PRE-AfMet in the absence of EF-G(GDPNP)7,11 (Fig. 2b). In stark contrast, binding of EF-G(GDPNP) to all four PRE-Aelong complexes results in a strong stabilization of GS2, corresponding to a large reduction in kGS2→GS1, such that fluctuations to GS1, should they occur, are either too rare and/or fast to be observed within our detection limits (Figs. 2b and 3). Confirming previous suggestions from us and others7,8,11, the data we present here unambiguously demonstrate that the presence of tRNAfMet at the P site of PRE complexes distinctly regulates the effect of EF-G binding on the kinetics of GS1→GS2 and GS2→GS1 transitions.
Generation of tRNAfMet mutants
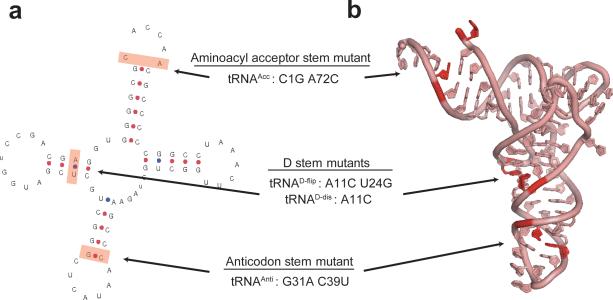
We next sought to determine whether tRNA structural features unique to tRNAfMet are responsible for the ability of P-site tRNAfMet to differentially regulate the GS1⇄GS2 equilibrium. Comparative sequence analysis of bacterial tRNAs reveals the existence of three such structural features25-27: (i) three consecutive GC base pairs located within the anticodon stem of tRNAfMet that are encountered in less than 1% of elongator tRNAs; (ii) a mismatched base pair between nucleotides 1 and 72 of the aminoacyl acceptor stem of tRNAfMet that is a Watson-Crick base pair in elongator tRNAs; and (iii) a purine-pyrimidine base pair between nucleotides 11 and 24 of the D stem of tRNAfMet that is flipped to a pyrimidine-purine base pair in elongator tRNAs. Biochemical and structural studies have revealed that these three unique features of tRNAfMet are specifically recognized by methionyl-tRNA transformylase and translation initiation and elongation factors so as to effectively discriminate tRNAfMet from elongator tRNAs, ensuring proper biosynthesis and selection of fMet-tRNAfMet at the start codon during translation initiation and preventing its incorporation at internal AUG codons during translation elongation26-29. Given that the regions of the P site-bound tRNA which contain these three features all establish extensive interactions with the ribosome that are remodeled during GS1→GS2 and GS2→GS1 transitions30-33, we reasoned that the divergent dynamic behavior observed in PRE-AfMet complexes may originate from one or more of these three unique tRNAfMet structural elements. In order to dissect the contribution that each of these unique structural features of tRNAfMet makes to the GS1⇄GS2 equilibrium, we initially designed three tRNAfMet2 mutants by changing each of its unique features to the corresponding features found in tRNAPhe: (i) a lower anticodon stem G31A C39U mutant (tRNAAnti); (ii), an aminoacyl acceptor stem C1G A72C mutant (tRNAAcc); and (iii) a D stem purine-pyrimidine flip A11C U24G mutant (tRNAD-flip) (Fig. 4, Online Methods and Supplementary Methods).
Figure 4. Design of tRNAfMet2 mutants.
(a) Secondary structure diagram for E. coli tRNAfMet2. The three unique structural features of tRNAfMet that differentiate it from all elongator tRNAs are highlighted in red and the mutations that were designed to convert these three structural features to those found in tRNAPhe are listed. (b) Three-dimensional structure of E. coli tRNAfMet2. The three unique structural features of tRNAfMet are colored as in the secondary structure diagram44 (PDB ID: 3CW6).
Disruption of anticodon stem does not affect GS1⇄GS2
Comparison of Figures 3b and 5a demonstrates that the occupancies of GS1 and GS2 for PRE-AAnti complexes are not significantly altered relative to PRE-AfMet-2 complexes (see also Fig. 2b and Table 1). Moreover, kGS1→GS2 and kGS2→GS1 measured for PRE-AAnti complexes in the absence and presence of EF-G(GDPNP) are within error of those measured for PRE-AfMet-2 complexes under the same conditions (Table 1). These results demonstrate that the unique GS1⇄GS2 dynamics observed in PRE-AfMet complexes do not arise from the highly conserved consecutive GC base pairs found within the anticodon stem of tRNAfMet. More generally, these results suggest that the GS1⇄GS2 equilibrium is relatively insensitive to the identity of the lower anticodon stem base pairs of the tRNA within the P site of the 30S subunit. This observation is consistent with biochemical data13 and with comparative structural analysis of X-ray crystallographic structures and cryogenic electron microscopy (cryo-EM) reconstructions of GS1- and GS2-like ribosomal complexes30-33, all of which indicate that the transition of the P-site tRNA from the P/P to the P/E configuration predominantly remodels interactions between the P-site tRNA and the 50S subunit, leaving interactions between the anticodon stem of the P-site tRNA and the 30S subunit relatively unaltered (Fig. 6a).
Figure 5. Steady-state smFRET measurements on PRE-A complexes carrying tRNAfMet2 mutants.
Data are presented as in Figure 3. (a) PRE-AAnti. (b) PRE-AAcc. (c) PRE-AD-flip. (d) PRE-AD-dis. (e) PRE-AAcc/D-flip.
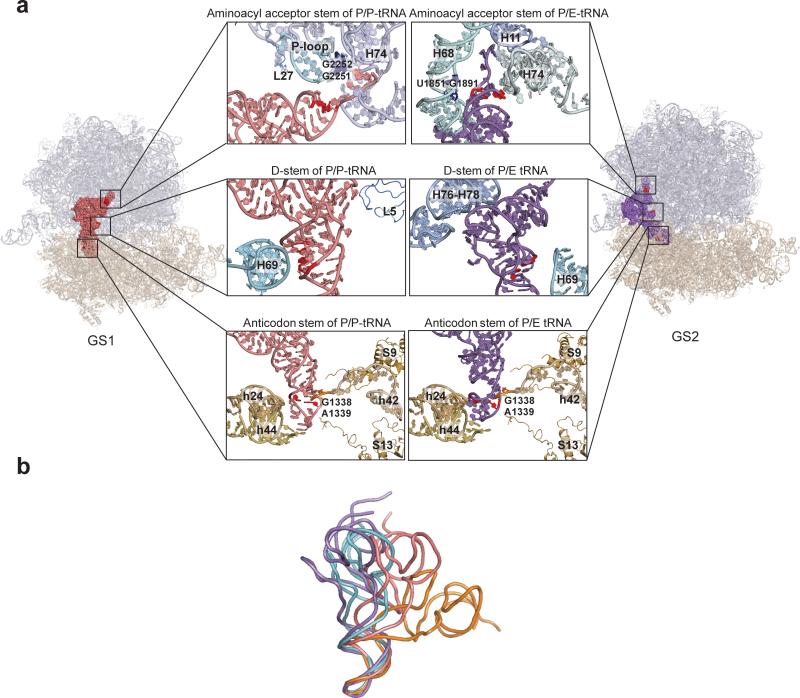
Figure 6. P-site tRNA-ribosome interactions within the GS1 and GS2 state of a PRE complex and comparative structural analysis of ribosome-free and ribosome-bound tRNAs.
(a) P-site tRNA-ribosome interactions within the GS1 and GS2 state of a PRE complex. Quasi-atomic-resolution models for the GS1 (left panel) and GS2 (right panel) states of a PRE complex were generated by real space refinement using rigid body fitting of atomic-resolution structures of the E. coli ribosome (PDB IDs: 2AVY and 2AW4) and a P site-bound tRNA (PDB ID: 2J00) into the electron density obtained from cryo-EM reconstructions of the GS1 and GS2 states of a PRE complex (kindly provided by J. Frank, H. Gao, and X. Aguirrezabala)32. P/P- and P/E-configured tRNAs are shown in pink and purple, respectively. rRNA helices and r-proteins that interact with the aminoacyl acceptor stem (top panels), the D stem (middle panels) and the anticodon stem (bottom panels) of each tRNA are labeled in each figure and the nucleotide positions of the tRNAfMet2 mutations studied in the present work are shown in red. (b) Comparative structural analysis of ribosome-free and ribosome-bound tRNAs. Ribosome-free tRNAfMet(44) (PDB ID: 3CW6), A/T-configured tRNAThr(22) (PDB ID: 2WRN), P/P-configured tRNAfMet(31) (PDB ID: 2J00), and P/E-configured tRNAfMet (quasi-atomic-resolution model generated by molecular dynamics flexible fitting40, kindly provided by K. Schulten and B. Liu) are shown in cyan, orange, pink and purple, respectively. The four tRNAs were superimposed using the anticodon stem loops (nucleotides 31-39 for the alignment of P/P-, P/E-configured tRNA to the ribosome-free tRNA, and nucleotides 32-38 for the alignment of A/T-configured tRNA to the ribosome-free tRNA) with PyMol50 (www.pymol.org).
5’-terminal base pairing in acceptor stem decreases kGS2→GS1
Relative to PRE-AfMet-2 complexes, PRE-AAcc complexes exhibit a GS1⇄GS2 equilibrium that is shifted towards GS2 (Figs. 2b, 3b and 5b and Table 1). Although kGS1→GS2 and kGS2→GS1 are both decreased by conversion of the mismatched C1•A72 base pair within the aminoacyl acceptor stem of tRNAfMet2 to a Watson-Crick GC base pair, the larger, almost one order of magnitude decrease in kGS2→GS1 relative to the smaller ~70% decrease of kGS1→GS2 drives the GS1⇄GS2 equilibrium towards GS2 (Table 1). Biochemical and structural studies have shown that the aminoacyl acceptor stem of a P/P tRNA makes Watson-Crick base pairing interactions with the P loop of 23S rRNA within the 50S subunit P site30,31,34, while the aminoacyl acceptor stem of a P/E tRNA docks into a pocket formed by 23S rRNA H11, H68 and H74 within the 50S subunit E site, making a minor groove-minor groove interaction with H6832,33,35(Fig. 6a). It is therefore possible that replacing the mismatched C1•A72 base pair with a Watson-Crick base pair stabilizes both of these interactions but has a much larger effect on the minor groove-minor groove interaction between the aminoacyl acceptor stem of the tRNA and H68 (see Discussion).
Most importantly, our data identify the interactions of the tRNA aminoacyl acceptor stem with the P loop at the 50S subunit P site and H68 at the 50S E site as important regulators of kGS1→GS2 and kGS2→GS1, respectively. Previously, Dorner et al have shown that PRE complexes carrying a C1G A72C mutant tRNAfMet analogous to tRNAAcc exhibit a faster rate of sparsomycin-promoted translocation relative to PRE complexes carrying P-site tRNAfMet(10) (sparsomycin is a ribosome-targeting antibiotic that has been shown to promote translocation through a mechanism that is closely related to the mechanism of EF-G-promoted translocation36,37). Considered alongside this translocation measurement, the results we present here correlate the greater stability of aminoacyl acceptor stem-H68 interactions in tRNAs carrying a Watson-Crick base pair at positions 1 and 72 with an increased rate of translocation (see Supplementary Discussion).
Altering the D stem or variable loop modulates kGS1→GS2
Similar to PRE-AAcc complexes, PRE-AD-flip complexes exhibit a GS1⇄GS2 equilibrium that is shifted towards GS2 relative to PRE-AfMet-2 complexes (Figs 2b, 3b, and 5c and Table 1). In contrast with PRE-AAcc complexes, however, the shift towards GS2 in PRE-AD-flip complexes primarily arises from an ~3-fold increase in kGS1→GS2, with insignificant effects on kGS2→GS1 (Table 1). In order to further test the role of the A11-U24 base pair in modulating the GS1⇄GS2 equilibrium, we generated an additional tRNAfMet2 A11C mutant, tRNAD-dis, in which we introduced a mismatched C11•U24 base pair (Fig. 4, Supplementary Methods, Supplementary Figs. 2, 3, and 5). Mirroring the results obtained with PRE-AD-flip, the GS1⇄GS2 equilibrium in PRE-AD-dis complexes is shifted towards GS2 relative to that seen in PRE-AfMet-2, and this effect again arises from an ~1.8-fold increase in kGS1→GS2 and an almost negligible effect on kGS2→GS1 (Figs 2b, 3b, 5c and d and Table 1).
Structural studies reveal that the D stem of the P/P tRNA makes a minor groove-minor groove interaction with 23S rRNA H69 within the 50S subunit P site, an interaction which is completely disrupted when the tRNA is repositioned into the P/E configuration within GS230-33 (Fig. 6a). While it is possible that flipping or disrupting the purine-pyrimidine base pair between A11 and U24 destabilizes this minor groove-minor groove interaction, consequently destabilizing GS1 and increasing kGS1→GS2, X-ray structures of GS1-like ribosomal complexes reveal that the interactions between H69 and the D stem of a P/P tRNAfMet or tRNAPhe are almost indistinguishable30,31. Furthermore, deletion of H69 does not have a measurable effect on the yield or rate of translocation, suggesting that this interaction is not required for efficient translocation38. Based on these observations, we posit that the increase in kGS1→GS2 caused by the D stem mutations likely results from the effects that these mutations have on the structural stability of the tRNA itself, rather than from the effects that they might have on the interactions between the D stem and H69 (see Discussion).
Further evidence that perturbations to the structural stability of the P-site tRNA might modulate the GS1⇄GS2 equilibrium comes from a comparison of PRE-AfMet-1 and PRE-AfMet-2 complexes (Figs. 3a and b). Similar to PRE-AD-flip and PRE-AD-dis complexes, the GS1⇄GS2 equilibrium within PRE-AfMet-1 complexes is shifted towards GS2 relative to that in PRE-AfMet-2 complexes, through a kinetic mechanism involving an ~2-fold increase in kGS1→GS2 and no detectable change in kGS2→GS1 (Table 1). The only difference between tRNAfMet1 and tRNAfMet2 is a change at nucleotide position 46 within the variable loop from a 7-methylguanosine (7mG) in tRNAfMet1 to an adenosine (A) in tRNAfMet224,27. In this case, however, structures of GS1- and GS2-like ribosomal complexes demonstrate that the variable loop of tRNAfMet does not directly contact the ribosome when tRNAfMet is in either the P/P or P/E configuration31,32, strongly suggesting that slight differences in the structural stability of P site-bound tRNAfMet1 versus tRNAfMet2 are responsible for the observed increase in kGS1→GS2. The notion that the relatively subtle differences in the sequences within the D stems and variable loops of tRNAfMet2, tRNAD-flip, tRNAD-dis, and tRNAfMet1 lead to differences in the structural stabilities of these tRNAs is strengthened by their distinct migrations on a native gel (Supplementary Fig. 2 and Supplementary Discussion).
tRNAfMet double-mutant exhibits elongator-like behavior
Despite their ability to shift the GS1⇄GS2 equilibrium towards GS2, both PRE-AAcc and PRE-AD-flip complexes demonstrate rapid fluctuations between GS1 and GS2 in the presence of EF-G(GDPNP) that are characteristic of PRE-AfMet complexes (Figs. 3a and b, 5b and c, and Table 1). This suggests that neither replacing the C1•A72 mismatched base pair with a Watson-Crick base pair within the aminoacyl acceptor stem nor flipping the purine-pyrimidine base pair between A11 and U24 to a pyrimidine-purine base pair within the D stem of tRNAfMet can individually enable EF-G to modulate the GS1⇄GS2 equilibrium of the corresponding PRE complexes in the same manner as it does in the PRE-Aelong complexes which we have examined. This therefore prompted us to design a tRNAfMet2 double-mutant that combines the aminoacyl acceptor stem and D stem mutations (tRNAAcc/D-flip). We find that PRE-AAcc/D-flip complexes exhibit kinetic effects on the GS1⇄GS2 equilibrium that are roughly additive of those observed for PRE-AAcc and PRE-AD-flip complexes. Prior to the addition of EF-G(GDPNP), the net effect of the double mutation is a large, ~6-fold shift in the Keq governing the GS1⇄GS2 equilibrium towards GS2 that is predominantly driven by an order of magnitude decrease in kGS2→GS1 and a slight, ~40% decrease in kGS1→GS2 relative to the corresponding parameters for the PRE-AfMet-2 complex (Figs. 3b and 5e, and Table 1). It is interesting to note that, although Keq for the PRE-AAcc/D-flip complex now falls within the range of Keqs for the PRE-Aelong complexes, kGS1→GS2 and kGS2→GS1 for the PRE-AAcc/D-flip complex are both ~50-80% slower than those for the PRE-Aelong complexes investigated in this study. This indicates that, although we have uncovered two structural elements of tRNAfMet that influence its unique GS1⇄GS2 dynamics, it is likely that additional, unidentified structural features of tRNAfMet also collaboratively play a role in this regulation. Nevertheless, in contrast to the PRE-AAcc and PRE-AD-flip complexes, the PRE-AAcc/D-flip complex is highly stabilized in GS2 in the presence of EF-G(GDPNP), exhibiting thermodynamic and kinetic behavior that is indistinguishable from that of the PRE-Aelong complexes we have examined (Figs 2b, 3 and 5e). This result implies that the structural stability of the P-site tRNA as well as the interactions that this tRNA makes with the ribosome can effectively regulate the ability of EF-G to stabilize GS2 and, consequently suggests that these same features may affect the efficiency with which EF-G catalyzes translocation.
DISCUSSION
Interactions of P/E tRNA with H68 affect stability of GS2
The results we present here demonstrate that PRE-AfMet complexes exhibit GS1⇄GS2 dynamics in the absence and presence of EF-G(GDPNP) that differ significantly from those observed in PRE-Aelong complexes. Perhaps most importantly, our findings reveal how specific tRNA-ribosome interactions and tRNA structural features modulate kGS1→GS2 and kGS2→GS1 in order to drive these dynamic differences. The effects of altering individual tRNAfMet structural features on PRE-A complex dynamics can be interpreted in terms of the ability of each alteration to stabilize or destabilize GS1 and/or GS2. In this regard, structural interpretations based on the available X-ray and cryo-EM structures of GS1- and GS2-like ribosomes30-33 are especially enlightening.
The largest effect we have observed is an order of magnitude decrease in kGS2→GS1 caused by replacing the mismatched C1•A72 base pair with a Watson-Crick base pair within the aminoacyl acceptor stem of tRNAfMet. We speculate that introducing this Watson-Crick base pair likely stabilizes GS2 by stabilizing the minor groove-minor groove interaction between H68 and nucleotides 70 and 71 in the tRNA aminoacyl acceptor stem30-33,35. Such an interpretation suggests that this minor groove-minor groove interaction is exquisitely sensitive to the detailed helical geometry of the aminoacyl acceptor stem. Highlighting the functional importance of this interaction, biochemical studies have shown that PRE complexes carrying a P-site tRNA in which the 2’ hydroxyl at nucleotide 71 has been modified to disrupt its minor groove-minor groove interaction with H68 exhibit at least a 90% reduction in the rate of EF-G-promoted translocation39. In addition to this minor groove-minor groove interaction, a recent molecular dynamics simulation comparing the interactions that H68 makes with the aminoacyl acceptor stem of either P/E tRNAfMet or tRNAPhe suggests that the universally conserved 23S rRNA U1851•G1891 wobble base pair within H68 can be disrupted such that U1851 can flip out of H68 and establish a wobble base pairing interaction with G70 of tRNAPhe. Interestingly, this interaction is not observed in an analogous simulation using a P/E tRNAfMet, thus providing an additional rationale for the enhanced ability of tRNAPhe to stabilize GS2 relative to that of tRNAfMet(40).
Flexibility of P/P tRNA modulates stability of GS1
tRNAD-flip, tRNAD-dis, and tRNAfMet1, differ from tRNAfMet2 at a single base pair within the D stem (tRNAD-flip and tRNAD-dis) or at a single nucleotide within the variable loop (tRNAfMet1). As discussed in the Results, the effect of these differences on kGS1→GS2 for the corresponding PRE-A complexes likely originates from differences in the structural stabilities of the tRNAs themselves. The characteristic L-shaped tertiary structure of tRNA is determined and stabilized by coaxial stacking of the aminoacyl acceptor and T stems, coaxial stacking of the D and anticodon stems, and a network of base pairing and base stacking interactions between the T, D, and variable loops41. Even from the earliest structural studies of tRNA, the delicate nature of the network of tertiary interactions that stabilizes its L-shaped structure was noted, and the possibility that the structurally-determined, intrinsic conformational flexibility of the tRNA might be functionally important during protein synthesis was proposed41,42.
Perhaps the best studied mechanistic step during translation in which the conformational flexibility of tRNA features prominently is the elongation factor Tu (EF-Tu)-catalyzed aa-tRNA selection step of the elongation cycle. During this process, the aa-tRNA adopts a functionally critical intermediate conformation, termed the A/T configuration, which requires a dramatic distortion of the aa-tRNA centered at the junction between the anticodon and D stems21-23 (Fig. 6b). Similarly, P/P tRNAs exhibit a pronounced distortion that is centered at the very same junction2,21,22,30,31. More specifically, the D stem of the P/P tRNA is partially unwound relative to its anticodon stem and the tRNA is kinked at a hinge formed by the G26-A44 base pair at the junction between the anticodon and D stems such that it is positioned towards the 50S subunit and slightly towards the A site2,30,31 (Fig. 6b). It is therefore conceivable that the stability of the distorted conformation adopted by a particular P/P tRNA makes a significant contribution to the stability of GS1 and, consequently, to the kGS1→GS2 exhibited by the corresponding PRE-A complex.
Viewed through this lens, the specific interactions that define and stabilize the tertiary structure of a particular tRNA would govern its conformational flexibility and influence the stability of the distorted conformation it adopts within the P/P configuration. For example, the identity of nucleotide 46 in tRNAfMet (7mG46 in tRNAfMet1 and A46 in tRNAfMet2) might affect the conformational flexibility of tRNAfMet via the highly conserved base triple interaction between nucleotide 46 and the C13-G22 base pair within the D stem of tRNAfMet(41,43,44). Interestingly, while this base triple is observed in ribosome-free tRNAfMet(41,43,44), it is apparently disrupted when tRNAfMet adopts the distorted P/P configuration30,31. Because the A46•C13-G22 base triple is weaker than the 7mG46•C13-G22 base triple43-45, it is likely that disrupting this tertiary interaction in tRNAfMet2 is less energetically costly than disrupting it in tRNAfMet1. Thus, we expect tRNAfMet2 to be energetically more stable than tRNAfMet1 when it adopts the distorted P/P configuration within GS1, providing a molecular basis for the observed higher stability of GS1 in PRE-AfMet-2, and hence its slower kGS1→GS2, relative to PRE-AfMet-1. Likewise, perturbations to the D stem of tRNAfMet (as in tRNAD-flip and tRNAD-dis) might alter the conformational flexibility of the tRNA by directly affecting the structural integrity of the D stem. Nevertheless, it is difficult to predict the effect that a particular sequence alteration will have on the conformational flexibility of a tRNA based solely on the X-ray crystal structure of that tRNA; this is primarily due to the difficulty of assessing the conformational entropy of a biomolecule using its X-ray crystal structure46. For example, the observation that weakening the A11-U24 base pair via the A11C mutation in tRNAD-dis has a smaller effect on kGS1→GS2 than strengthening it via the A11C U24G mutations in tRNAD-flip suggests a complex interplay between the tertiary structure and conformational flexibility of a particular tRNA and the stability of its corresponding PRE-A complex in GS1; additional X-ray structures, smFRET studies, and computational simulations will likely be necessary to fully understand this interplay.
In addition to its role in modulating the stability of GS1, it is also possible that the intrinsic conformational flexibility of the tRNA may directly influence the transition from its P/P to its P/E configuration (see Supplementary Discussion). Regardless, based on our data and the discussion presented here, we would predict that variations in the structure of tRNA within or proximal to the junction between the anticodon and D stems would generally influence kGS1→GS2. Future smFRET experiments to evaluate the effect of systematic mutations within this region of a single tRNA species should allow testing of this hypothesis and a more thorough mapping of the relationship between the stability of this junction and kGS1→GS2.
As this article was completed, we became aware of an X-ray crystal structure of a GS2-like ribosomal complex carrying a full-length deacylated P/E tRNAPhe(47). This new X-ray crystal structure provides near-atomic-resolution views of the ribosome-tRNA interactions and tRNA distortion that were originally identified at lower resolution through cryo-EM studies of GS2-like PRE complexes32,33, and confirms the structural interpretations reported above.
tRNA-mediated PRE complex dynamics may regulate elongation
Collectively, our results demonstrate that the P-site tRNA is a key regulator of PRE complex dynamics. Interestingly, each PRE-A complex that we have investigated exhibits unique GS1⇄GS2 dynamics (Table 1). Based on the discussion above, we expect that the particular structural features of each tRNA species will differentially regulate the GS1⇄GS2 equilibrium. Considered together with data suggesting that the GS1→GS2 transition may be rate-limiting for EF-G-promoted translocation6,12 and that PRE complexes which preferentially occupy GS2 are more efficiently translocated by EF-G10,20, it is possible that incorporation of specific tRNAs at particular codons of an mRNA may be used to regulate the rate of translation elongation at those codons. In this view, it is plausible that tRNA-mediated control of the GS1⇄GS2 equilibrium would allow selective attenuation of EF-G-promoted translocation and serve as a point of translational regulation (see Supplementary Discussion). Consistent with this possibility, the observed lower occupancy of GS2 in PREfMet complexes relative to PREelong complexes provides a mechanistic rationale for the recent observation by Puglisi and co-workers that EF-G-promoted translocation of the PREfMet complex during the first round of translation elongation is slower than EF-G-promoted translocation during subsequent rounds of translation elongation48,49.
Distinct dynamics may reflect unique selective pressures
Based on our data, we hypothesize that the unique dynamics exhibited by PREfMet complexes relative to the PREelong complexes we studied may generally arise from the different biochemical functions of tRNAfMet and elongator tRNAs and the distinct selective pressures under which these two types of tRNAs have evolved. The presence of a Watson-Crick or wobble base pair versus a mismatched base pair between nucleotides 1 and 72 of the aminoacyl acceptor stem is the primary feature through which EF-Tu discriminates elongator tRNAs from tRNAfMet during translation elongation27. Our results demonstrate that this sequence and structural feature primarily modulates the stability of GS2. In addition, elongator tRNAs undergo distortions at the junction between the anticodon and D stems as the incoming aa-tRNA passes through the A/T configuration during aa-tRNA selection, and as the newly formed peptidyl-tRNA is positioned into the P/P configuration after translocation from the A site into the P site. Thus, the conformational flexibility of each elongator tRNA has likely been optimized for, among other things, high fidelity aa-tRNA selection and translocation of peptidyl-tRNA from the A site to the P site. In contrast, tRNAfMet does not undergo aa-tRNA selection into the A site nor is it loaded into the P site through a translocation event from the A site. Instead, tRNAfMet, with only a single amino acid attached to its aminoacyl acceptor end, binds directly to the P site of the 30S subunit as part of the formation of the 30S initiation complex and adopts the P/P configuration as the 50S subunit joins to the 30S initiation complex during translation initiation27. Thus, in contrast to elongator tRNAs, the intrinsic conformational flexibility of tRNAfMet has been optimized for proper positioning within the 30S initiation complex, participation in the mechanism of 50S subunit joining, and maintenance of the P/P configuration even in the absence of a bone fide polypeptide at its aminoacyl acceptor end. In summary, the unique selective pressures under which tRNAfMet has evolved relative to elongator tRNAs have generated unique sequence and structural elements in tRNAfMet that translate into the unique GS1⇄GS2 dynamics and translocation properties that are observed in ribosomal complexes carrying this P-site tRNA.
Supplementary Material
ACKNOWLEDGEMENTS
We would like to thank Uttam RajBhandary for providing us with the plasmids and strains necessary to generate, overexpress, and purify all tRNAfMet mutants reported here; Joachim Frank, Haixiao Gao, and Xabier Aguirrezabala for providing us with structural models for GS1 and GS2; Klaus Schulten and Bo Liu for providing us with a quasi-atomic-resolution model of a hybrid P/E configured tRNA; Jamie Cate for providing us with an early preprint of ref. 47; Daniel MacDougall, Wei Ning, Somdeb Mitra, Corey Perez, and J. Cate for valuable discussions and comments on the manuscript; and C. Perez for managing the Gonzalez laboratory. This work was supported by grants to R.L.G. from the Burroughs Wellcome Fund (CABS 1004856), the National Science Foundation (MCB 0644262), and the National Institute of General Medical Sciences (GM 084288). A.C.R. was supported, in part, by the Amgen Scholars Program at Columbia University.
ONLINE METHODS
mRNA preparation
All mRNAs used in the present study were derived from a previously described variant of an mRNA encoding the first 20 amino acids of gene product 32 from T4 bacteriophage51. Five mRNAs were designed such that their first codons code for fMet (AUG), Phe (UUC), Tyr (UAC), Glu (GAA), or Val (GUU). All five mRNAs are otherwise identical and contain no other codons coding for the anticodons of tRNAfMet, tRNAPhe, tRNATyr, tRNAGlu, or tRNAVal in any reading frame (Supplementary Fig. 1). All mRNAs were in vitro transcribed from linearized plasmid DNA templates using T7 RNA polymerase following a previously described protocol52-55. A 3’-biotinylated DNA oligonucleotide (TGTGTAAGTTTTAGGTTGATTTG-Biotin; Integrated DNA Technologies) complementary to the 5’ end of the mRNAs was then hybridized to these mRNAs to enable surface immobilization as previously described54,55. mRNA transcripts hybridized to a 3’-biotinylated DNA oligonucleotide are hereafter referred to as “biotin-mRNAs”.
tRNA mutagenesis and purification
The pUC13.trnfM plasmid (a kind gift from Prof. Uttam RajBhandary, MIT) carrying the E. coli metY gene, which encodes tRNAfMet isoacceptor 2, tRNAfMet2 (24), was mutated to generate tRNAfMet1 and all tRNAfMet2 mutants. tRNAfMet2 mutants included: tRNAAnti (G31A C39U), in which the G31-C39 base pair in the anticodon stem of tRNAfMet2 is changed to the A31-U39 base pair found in tRNAPhe; tRNAAcc (C1G A72C), in which the mismatched C1•A72 base pair in the aminoacyl acceptor stem of tRNAfMet2 is changed to the G1-C72 Watson-Crick base pair found in tRNAPhe; tRNAD-flip (A11C U24G), in which the purine-pyrimidine A11-U24 base pair within the D stem of tRNAfMet2 is flipped to the pyrimidine-purine C11-G24 base pair found in tRNAPhe; tRNAD-dis (A11C), in which the A11-U24 base pair within the D stem is disrupted by changing A11 to C11; and tRNAAcc/D-flip (C1G A11C U24G A72C), in which the mutations generated in tRNAAcc and tRNAD-flip are combined. All tRNAs were expressed in E. coli strain B105, which lacks the metY gene and therefore endogenous tRNAfMet2, and were purified using a previously published protocol45,56,57 with slight modifications. Briefly, tRNAfMet1 and tRNAfMet2 (or tRNAfMet2 mutants) were separated from each other as well as from elongator tRNAs and all other cellular RNA species by native polyacrylamide gel electrophoresis (PAGE) on a 15 % (w/v) gel (Supplementary Fig. 2). tRNA bands were identified by UV shadowing at 254 nm wavelength, cut from the gel, and eluted from the gel slices using RNA Elution Buffer (10 mM Tris hydrochloride (pH25 °C = 7.5), 1 mM ethylenediamine tetraaceticacid, and 10 mM sodium chloride). The PAGE-purified tRNAs were further purified on a Phenyl 5PW TSK-Gel hydrophobic interaction chromatography (HIC) column (Tosoh Bioscience) using a gradient from HIC Buffer A (1.7 M ammonium sulfate and 10 mM ammonium acetate, pH=6.3) to HIC Buffer B (10 % (v/v) methanol and 10 mM ammonium acetate, pH=6.3)) (Supplementary Fig. 3).
Assembly and purification of PRE-A complexes
PRE-A complexes for smFRET experiments were assembled using 30S subunits purified from wild-type E. coli strain BW25113 and L1- and L9-labeled 50S subunits derived from a variant of strain BW25113 as previously described 7,55. A mixture of 30 pmol of biotin-mRNA, 20 pmol of deacylated tRNA, and 15 pmol of 30S subunits, in a total reaction volume of 30 μL of Ribosome Assembly Buffer (50 mM Tris hydrochloride (pH25 °C = 7.5), 70 mM ammonium chloride, 30 mM potassium chloride, 6 mM 2-mercaptoethanol, and 7 mM magnesium chloride), was incubated for 10 min at 37 °C. 10 pmol of L1- and L9-labeled 50S subunits were then added to the reaction followed by an additional incubation for 20 min at 37 °C. The reaction was then diluted to 100 μL with Tris-polymix buffer (50 mM Tris acetate (pH25 °C = 7.0), 100 mM potassium chloride, 5 mM ammonium acetate, 0.5 mM calcium acetate, 0.1 mM ethylenediamine tetraacetic acid, 10 mM 2-mercaptoethanol, 5 mM putrescine dihydrochloride and 1 mM spermidine, free base) containing 26 mM magnesium acetate in order to bring the final concentration of magnesium ions to 20 mM. The resulting PRE-A complexes were then purified by 10-40% (w/v) sucrose density gradient ultracentrifugation4,6.
smFRET experiments and data analysis
smFRET experiments were performed in Trispolymix buffer containing 15mM magnesium acetate and supplemented with an oxygen-scavenging system (300 μg mL-1 glucose oxidase, 40 μg mL-1 catalase and 1% (w/v) β-D-glucose)4,7,58 and a triplet-state quencher cocktail (1 mM 1,3,5,7-cyclooctatetraene (Aldrich) and 1 mM 3-nitrobenzyl alcohol (Fluka))59. smFRET versus time trajectories were recorded using a laboratory-built, prism-based TIRF microscope with a 532 nm diode-pumped solid-state laser as an excitation source and an electron-multiplying charge-coupled device camera operating at a time resolution of 10 frames s-1, unless otherwise specified, as a detector6. Each smFRET trajectory was idealized by hidden Markov modeling using the vbFRET software package (http://vbfret.sourceforge.net)60. With the exception of PRE-AfMet-2, PRE-AAnti and PRE-AAcc, dwell times spent in GS1 prior to transitioning to GS2 and in GS2 prior to transitioning to GS1 were extracted from the idealized smFRET trajectories and the lifetimes of GS1 and GS2 were determined from exponential fits to the corresponding one-dimensional dwell-time histograms. kGS1→GS2 and kGS2→GS1 were calculated by taking the inverse of the lifetimes of GS1 and GS2, respectively, and correcting for the rate of photobleaching from each state6,7. For the smFRET trajectories recorded for PRE-AfMet-2, PRE-AAnti and PRE-AAcc, which exhibited extended dwell times in GS1, a dwell-time analysis slightly modified from that described above was used. For these PRE-A complexes, the slow kGS1→GS2 was calculated by the following procedure: (i) assuming a two-state GS1⇄GS2 equilibrium model; (ii) calculating the corresponding equilibrium constant (Keq) from the ratio of GS1 and GS2 occupancies (Keq = (occupancy of GS2)/(occupancy of GS1)); (iii) calculating the lifetime of GS2, and the corresponding kGS2→GS1, from a standard dwell-time analysis as described above; and (iv) setting kGS1→GS2 = Keq × kGS2→GS1. Detailed descriptions and references for all materials and methods can be found in the Supplementary Information.
References
- 1.Frank J, Gao H, Sengupta J, Gao N, Taylor DJ. The process of mRNA-tRNA translocation. Proc Natl Acad Sci U S A. 2007;104:19671–8. doi: 10.1073/pnas.0708517104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Korostelev A, Noller HF. The ribosome in focus: new structures bring new insights. Trends Biochem Sci. 2007;32:434–41. doi: 10.1016/j.tibs.2007.08.002. [DOI] [PubMed] [Google Scholar]
- 3.Shoji S, Walker SE, Fredrick K. Ribosomal Translocation: One Step Closer to the Molecular Mechanism. ACS Chem Biol. 2009 doi: 10.1021/cb8002946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blanchard SC, Kim HD, Gonzalez RL, Jr., Puglisi JD, Chu S. tRNA dynamics on the ribosome during translation. Proc Natl Acad Sci U S A. 2004;101:12893–8. doi: 10.1073/pnas.0403884101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Frank J, Gonzalez RL., Jr. Structure and dynamics of a processive brownian motor: the translating ribosome. Annu Rev Biochem. 2010;79:381–412. doi: 10.1146/annurev-biochem-060408-173330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Fei J, Kosuri P, MacDougall DD, Gonzalez RL., Jr. Coupling of ribosomal L1 stalk and tRNA dynamics during translation elongation. Mol Cell. 2008;30:348–59. doi: 10.1016/j.molcel.2008.03.012. [DOI] [PubMed] [Google Scholar]
- 7.Fei J, et al. Allosteric collaboration between elongation factor G and the ribosomal L1 stalk directs tRNA movements during translation. Proc Natl Acad Sci U S A. 2009;106:15702–7. doi: 10.1073/pnas.0908077106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cornish PV, Ermolenko DN, Noller HF, Ha T. Spontaneous intersubunit rotation in single ribosomes. Mol Cell. 2008;30:578–88. doi: 10.1016/j.molcel.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Cornish PV, et al. Following movement of the L1 stalk between three functional states in single ribosomes. Proc Natl Acad Sci U S A. 2009;106:2571–6. doi: 10.1073/pnas.0813180106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dorner S, Brunelle JL, Sharma D, Green R. The hybrid state of tRNA binding is an authentic translation elongation intermediate. Nat Struct Mol Biol. 2006;13:234–41. doi: 10.1038/nsmb1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Munro JB, Altman RB, Tung CS, Sanbonmatsu KY, Blanchard SC. A fast dynamic mode of the EF-G-bound ribosome. EMBO J. 2010;29:770–81. doi: 10.1038/emboj.2009.384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Munro JB, Wasserman MR, Altman RB, Wang L, Blanchard SC. Correlated conformational events in EF-G and the ribosome regulate translocation. Nat Struct Mol Biol. 2010;7:7. doi: 10.1038/nsmb.1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moazed D, Noller HF. Intermediate states in the movement of transfer RNA in the ribosome. Nature. 1989;342:142–8. doi: 10.1038/342142a0. [DOI] [PubMed] [Google Scholar]
- 14.Ermolenko DN, et al. Observation of intersubunit movement of the ribosome in solution using FRET. J Mol Biol. 2007;370:530–40. doi: 10.1016/j.jmb.2007.04.042. [DOI] [PubMed] [Google Scholar]
- 15.Munro JB, et al. Spontaneous formation of the unlocked state of the ribosome is a multistep process. Proc Natl Acad Sci U S A. 2009;107:709–14. doi: 10.1073/pnas.0908597107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zavialov AV, Ehrenberg M. Peptidyl-tRNA regulates the GTPase activity of translation factors. Cell. 2003;114:113–22. doi: 10.1016/s0092-8674(03)00478-1. [DOI] [PubMed] [Google Scholar]
- 17.Modolell J, Cabrer B, Vaquez D. The interaction of elongation factor G with N-acetylphenylalanyl transfer RNA-ribosome complexes. Proc Natl Acad Sci U S A. 1973;70:3561–5. doi: 10.1073/pnas.70.12.3561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Connell SR, et al. Structural basis for interaction of the ribosome with the switch regions of GTP-bound elongation factors. Mol Cell. 2007;25:751–64. doi: 10.1016/j.molcel.2007.01.027. [DOI] [PubMed] [Google Scholar]
- 19.Valle M, et al. Locking and unlocking of ribosomal motions. Cell. 2003;114:123–34. doi: 10.1016/s0092-8674(03)00476-8. [DOI] [PubMed] [Google Scholar]
- 20.Studer SM, Feinberg JS, Joseph S. Rapid kinetic analysis of EF-G-dependent mRNA translocation in the ribosome. Journal of Molecular Biology. 2003;327:369–81. doi: 10.1016/s0022-2836(03)00146-3. [DOI] [PubMed] [Google Scholar]
- 21.Villa E, et al. Ribosome-induced changes in elongation factor Tu conformation control GTP hydrolysis. Proc Natl Acad Sci U S A. 2009;106:1063–8. doi: 10.1073/pnas.0811370106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schmeing TM, et al. The crystal structure of the ribosome bound to EF-Tu and aminoacyl-tRNA. Science. 2009;326:688–94. doi: 10.1126/science.1179700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Voorhees RM, Schmeing TM, Kelley AC, Ramakrishnan V. The mechanism for activation of GTP hydrolysis on the ribosome. Science. 2010;330:835–8. doi: 10.1126/science.1194460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ishii S, Kuroki K, Imamoto F. tRNAMetf2 gene in the leader region of the nusA operon in Escherichia coli. Proc Natl Acad Sci U S A. 1984;81:409–13. doi: 10.1073/pnas.81.2.409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Varshney U, Lee CP, RajBhandary UL. From elongator tRNA to initiator tRNA. Proc Natl Acad Sci U S A. 1993;90:2305–9. doi: 10.1073/pnas.90.6.2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.RajBhandary UL. Initiator transfer RNAs. J Bacteriol. 1994;176:547–52. doi: 10.1128/jb.176.3.547-552.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Laursen BS, Sorensen HP, Mortensen KK, Sperling-Petersen HU. Initiation of protein synthesis in bacteria. Microbiol Mol Biol Rev. 2005;69:101–23. doi: 10.1128/MMBR.69.1.101-123.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mayer C, Stortchevoi A, Kohrer C, Varshney U, RajBhandary UL. Initiator tRNA and its role in initiation of protein synthesis. Cold Spring Harb Symp Quant Biol. 2001;66:195–206. doi: 10.1101/sqb.2001.66.195. [DOI] [PubMed] [Google Scholar]
- 29.Kolitz SE, Lorsch JR. Eukaryotic initiator tRNA: finely tuned and ready for action. FEBS Lett. 2010;584:396–404. doi: 10.1016/j.febslet.2009.11.047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Korostelev A, Trakhanov S, Laurberg M, Noller HF. Crystal structure of a 70S ribosome-tRNA complex reveals functional interactions and rearrangements. Cell. 2006;126:1065–77. doi: 10.1016/j.cell.2006.08.032. [DOI] [PubMed] [Google Scholar]
- 31.Selmer M, et al. Structure of the 70S ribosome complexed with mRNA and tRNA. Science. 2006;313:1935–42. doi: 10.1126/science.1131127. [DOI] [PubMed] [Google Scholar]
- 32.Agirrezabala X, et al. Visualization of the hybrid state of tRNA binding promoted by spontaneous ratcheting of the ribosome. Mol Cell. 2008;32:190–7. doi: 10.1016/j.molcel.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Julian P, et al. Structure of ratcheted ribosomes with tRNAs in hybrid states. Proc Natl Acad Sci U S A. 2008;105:16924–7. doi: 10.1073/pnas.0809587105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Samaha RR, Green R, Noller HF. A base pair between tRNA and 23S rRNA in the peptidyl transferase centre of the ribosome. Nature. 1995;377:309–14. doi: 10.1038/377309a0. [DOI] [PubMed] [Google Scholar]
- 35.Yusupov MM, et al. Crystal structure of the ribosome at 5.5 A resolution. Science. 2001;292:883–96. doi: 10.1126/science.1060089. [DOI] [PubMed] [Google Scholar]
- 36.Fredrick K, Noller HF. Catalysis of ribosomal translocation by sparsomycin. Science. 2003;300:1159–62. doi: 10.1126/science.1084571. [DOI] [PubMed] [Google Scholar]
- 37.Southworth DR, Green R. Ribosomal translocation: sparsomycin pushes the button. Curr Biol. 2003;13:R652–4. doi: 10.1016/s0960-9822(03)00574-8. [DOI] [PubMed] [Google Scholar]
- 38.Ali IK, Lancaster L, Feinberg J, Joseph S, Noller HF. Deletion of a conserved, central ribosomal intersubunit RNA bridge. Mol Cell. 2006;23:865–74. doi: 10.1016/j.molcel.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 39.Feinberg JS, Joseph S. Identification of molecular interactions between P-site tRNA and the ribosome essential for translocation. Proc Natl Acad Sci U S A. 2001;98:11120–5. doi: 10.1073/pnas.211184098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Trabuco LG, et al. The Role of L1 Stalk-tRNA Interaction in the Ribosome Elongation Cycle. J Mol Biol. 2010;402:741–60. doi: 10.1016/j.jmb.2010.07.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rich A, RajBhandary UL. Transfer RNA: molecular structure, sequence, and properties. Annu Rev Biochem. 1976;45:805–60. doi: 10.1146/annurev.bi.45.070176.004105. [DOI] [PubMed] [Google Scholar]
- 42.Alexander RW, Eargle J, Luthey-Schulten Z. Experimental and computational determination of tRNA dynamics. FEBS Lett. 2010;584:376–86. doi: 10.1016/j.febslet.2009.11.061. [DOI] [PubMed] [Google Scholar]
- 43.Daniel WE, Jr., Cohn M. Changes in tertiary structure accompanying a single base change in transfer RNA. Proton magnetic resonance and aminoacylation studies of Escherichia coli tRNAMet f1 and tRNAMet f3 and their spin-labeled (s4U8) derivatives. Biochemistry. 1976;15:3917–24. doi: 10.1021/bi00663a003. [DOI] [PubMed] [Google Scholar]
- 44.Barraud P, Schmitt E, Mechulam Y, Dardel F, Tisne C. A unique conformation of the anticodon stem-loop is associated with the capacity of tRNAfMet to initiate protein synthesis. Nucleic Acids Res. 2008;36:4894–901. doi: 10.1093/nar/gkn462. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mandal N, RajBhandary UL. Escherichia coli B lacks one of the two initiator tRNA species present in E. coli K-12. J Bacteriol. 1992;174:7827–30. doi: 10.1128/jb.174.23.7827-7830.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Uhlenbeck OC. RNA biophysics has come of age. Biopolymers. 2009;91:811–4. doi: 10.1002/bip.21269. [DOI] [PubMed] [Google Scholar]
- 47.Dunkle JA, et al. Structures of the bacterial ribosome in classical and hybrid states of tRNA binding. Science. 2011;332:981–4. doi: 10.1126/science.1202692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Uemura S, et al. Real-time tRNA transit on single translating ribosomes at codon resolution. Nature. 2010;464:1012–7. doi: 10.1038/nature08925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Aitken CE, Puglisi JD. Following the intersubunit conformation of the ribosome during translation in real time. Nat Struct Mol Biol. 2010;17:793–800. doi: 10.1038/nsmb.1828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Schrodinger LLC. The PyMOL Molecular Graphics System, Version 1.3r1. 2010 [Google Scholar]
- 51.Alberts BM, Frey L. T4 bacteriophage gene 32: a structural protein in the replication and recombination of DNA. Nature. 1970;227:1313–8. doi: 10.1038/2271313a0. [DOI] [PubMed] [Google Scholar]
- 52.Milligan JF, Groebe DR, Witherell GW, Uhlenbeck OC. Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res. 1987;15:8783–98. doi: 10.1093/nar/15.21.8783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wyatt JR, Chastain M, Puglisi JD. Synthesis and purification of large amounts of RNA oligonucleotides. Biotechniques. 1991;11:764–9. [PubMed] [Google Scholar]
- 54.Sternberg SH, Fei J, Prywes N, McGrath KA, Gonzalez RL., Jr. Translation factors direct intrinsic ribosome dynamics during translation termination and ribosome recycling. Nat Struct Mol Biol. 2009;16:861–8. doi: 10.1038/nsmb.1622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Fei J, et al. A highly-purified, fluorescently-labeled in vitro translation system for single-molecule studies of protein synthesis. Methods in Enzymology. 2010;472:221–59. doi: 10.1016/S0076-6879(10)72008-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee CP, Mandal N, Dyson MR, RajBhandary UL. The discriminator base influences tRNA structure at the end of the acceptor stem and possibly its interaction with proteins. Proc Natl Acad Sci U S A. 1993;90:7149–52. doi: 10.1073/pnas.90.15.7149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Mandal N, Mangroo D, Dalluge JJ, McCloskey JA, Rajbhandary UL. Role of the three consecutive G:C base pairs conserved in the anticodon stem of initiator tRNAs in initiation of protein synthesis in Escherichia coli. RNA. 1996;2:473–82. [PMC free article] [PubMed] [Google Scholar]
- 58.Stone MD, et al. Stepwise protein-mediated RNA folding directs assembly of telomerase ribonucleoprotein. Nature. 2007;446:458–61. doi: 10.1038/nature05600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gonzalez RL, Jr., Chu S, Puglisi JD. Thiostrepton inhibition of tRNA delivery to the ribosome. RNA. 2007;13:2091–7. doi: 10.1261/rna.499407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bronson JE, Fei J, Hofman JM, Gonzalez RL, Jr., Wiggins CH. Learning rates and states from biophysical time series: a Bayesian approach to model selection and single-molecule FRET data. Biophys J. 2009;97:3196–205. doi: 10.1016/j.bpj.2009.09.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.