Abstract
The nodal gene is expressed on the left side of developing mouse embryos and is implicated in left/right (L-R) axis formation. The transcriptional regulatory regions of nodal have now been investigated by transgenic analysis. A node-specific enhancer was detected in the upstream region (−9.5 to −8.7 kb) of the gene. Intron 1 was also shown to contain a left side-specific enhancer (ASE) that was able to direct transgene expression in the lateral plate mesoderm and prospective floor plate on the left side. A 3.5-kb region of nodal that contained ASE responded to mutations in iv, inv, and lefty-1, all genes that act upstream of nodal. The same 3.5- kb region also directed expression in the epiblast and visceral endoderm at earlier stages of development. Characterization of deletion constructs delineated ASE to a 340-bp region that was both essential and sufficient for asymmetric expression of nodal. Several sequence motifs were found to be conserved between the nodal ASE and the lefty-2 ASE, some of which appeared to be essential for nodal ASE activity. These results suggest that similar transcriptional mechanisms underlie the asymmetric expression of nodal and of lefty-2 as well as the earlier expression of nodal in the epiblast and endoderm.
Keywords: Left/right asymmetry, transcriptional regulation, nodal, lefty
Vertebrates exhibit numerous left/right (L-R) asymmetries such as the positioning of the heart and spleen on the left side of the body. Important insights into the molecular mechanisms that underlie L-R axis formation have been obtained only recently (Fujinaga 1997; King and Brown 1997; Varlet and Robertson 1997; Levin and Mercola 1998; Beddington and Robertson 1999). The entire process by which L-R asymmetry is achieved can be divided into three phases: (1) initial determination of L-R polarity; (2) establishment of such polarity by the asymmetric expression of signaling molecules; and (3) L-R asymmetric morphogenesis induced by these signaling molecules.
The mechanism of initial determination of L-R polarity remains unknown, although several models have been proposed (Brown and Wolpert 1990; Brown et al. 1991; Yost 1991). Several mutations that affect L-R patterning have been identified in mice (Hummel and Chapman 1959; Yokoyama et al. 1993; Heymer et al. 1997; Chen et al. 1998; Melloy et al. 1998; Nonaka et al. 1998) and in human (Gebbia et al. 1997). Two of the more extensively studied of these mutations are iv and inv, the corresponding genes of which encode axonemal dynein and a protein with ankyrin repeats, respectively (Supp et al. 1997; Mochizuki et al. 1998; Morgan et al. 1998). However, the precise functions of the proteins encoded by most of these affected genes (including Lrd, Inv, KIF3A, and HFH4) remain unclear. The initial determination of L-R polarity in mice probably occurs in or near the node, with recent observations suggesting an important role for ciliary movement in the node (Nonaka et al. 1998). In Xenopus, maternal Vg1 is implicated in the initiation of L-R asymmetry (Hyatt et al. 1996).
Three signaling molecules whose asymmetric expression is thought to contribute to the second phase of L-R axis formation have been identified: Nodal, Lefty-1, and Lefty-2, all of which belong to the transforming growth factor-β (TGFβ) superfamily of proteins. The nodal gene is expressed asymmetrically in mice, chick, Xenopus, and zebrafish, suggesting that the encoded protein is a highly conserved component of the pathway of L-R determination (Levin et al. 1995; Collignon et al. 1996; Lowe et al. 1996; Lustig et al. 1996; Sampath et al. 1997). The highly related lefty-1 and lefty-2 genes are also expressed on the left side of developing mouse embryos (Meno et al. 1996 1997). Whereas lefty-1 is expressed predominantly in the prospective floor plate (PFP), lefty-2 is expressed in lateral plate mesoderm (LPM). The results of ectopic expression experiments in chick and Xenopus, as well as of the analysis of knockout mice, support the notion that these genes encode signaling molecules that convey L-R positional information (Levin et al. 1997; Sampath et al. 1997; Meno et al. 1998). Expression of lefty-1, lefty-2, and nodal is affected in iv and inv mutant mouse embryos (Collignon et al. 1996; Lowe et al. 1996; Meno et al. 1996), suggesting that Lefty-1, Lefty-2, and Nodal all act downstream of the proteins encoded by the iv and inv genes.
With regard to the third phase of L-R asymmetry formation, situs-specific morphogenesis in response to L-R asymmetric signals, genetic evidence in mice (Oh and Li 1997) suggests that type II activin receptors (ActRIIA and ActRIIB) serve as functional receptors for Nodal (and also for Lefty-2). Furthermore, recent studies (Logan et al. 1998; Meno et al. 1998; Piedra et al. 1998; Ryan et al. 1998; Yoshioka et al. 1998) have indicated a role for Pitx2, a Bicoid-type homeobox transcription factor, in mediating transmission of signals from Nodal and Lefty-2. In addition, a snail-related gene is expressed on the right side of chick and mouse embryos (Isaac et al. 1997; Sefton et al. 1998) and may be negatively regulated by Nodal.
Although key genes that contribute to the pathway of L-R axis formation have been identified, the precise functions of these genes and how they interact with each other are largely unclear. Several important questions remain to be addressed. For example, it is not known how the initial L-R-specific signal (perhaps generated in the node by Inv and Lrd function) is translated into asymmetric expression of nodal and lefty genes. Elucidation of the mechanisms by which the asymmetric expression of nodal and lefty genes is achieved will require an understanding of the regulation of these genes. Previously, we have analyzed transcriptional regulatory elements of lefty-1 and lefty-2, and have shown that asymmetric expression is conferred on lefty-2 by a left side-specific enhancer [ASE (for asymetric enhancer)], whereas such expression is conferred on lefty-1 by a combination of bilateral enhancers and a right side-specific silencer (Saijoh et al. 1999).
We have now identified the transcriptional regulatory element of nodal that is responsible for the asymmetric expression of this gene. Our results indicate that like lefty-2, nodal is regulated by an ASE that is active exclusively on the left side of the body. Comparison of the nodal and lefty-2 ASE sequences suggests that the asymmetric expression of the two genes is regulated by similar transcriptional mechanisms.
Results
A node-specific enhancer in the upstream region of nodal
At the early somite stage in mice, nodal is expressed in two domains: the LPM and the node (Zhou et al. 1993; Collignon et al. 1996). Expression of nodal in LPM shows a strict L-R specificity, being confined to the left half (Collignon et al. 1996; Lowe et al. 1996). Expression in the node also shows a subtle L-R asymmetry (Collignon et al. 1996). In this study we focus primarily on transcriptional regulatory elements that are required for expression of nodal in the node and left LPM at the early somite stage.
We first measured the transcriptional activity of the 5-kb region upstream of the nodal coding sequence (from −5 kb to −50 bp with respect to the translational initiation codon). Construct 5′-1, in which the 5-kb upstream fragment was linked to a promoterless lacZ gene (Fig. 1A), was injected into the pronuclei of fertilized mouse embryos at the one-cell stage. The embryos were allowed to develop in utero until embryonic day 8.2 (E8.2), when they were recovered and the expression of the lacZ transgene was examined by staining with X-gal (5-bromo-4-chloro-3-indoyl-β-d-galactoside). (Other constructs described below were tested similarly unless otherwise mentioned.) The 5′-1 construct failed to give rise to substantial X-gal staining at E8.2 (Fig. 1B), suggesting that the 5-kb upstream region is not sufficient for nodal expression. The observation that 5′-1 reproducibly yielded low-intensity staining in a few cells of the left LPM (Fig. 1B) suggested the possible existence of a weak cis-regulatory element that contributes to asymmetric expression.
Figure 1.
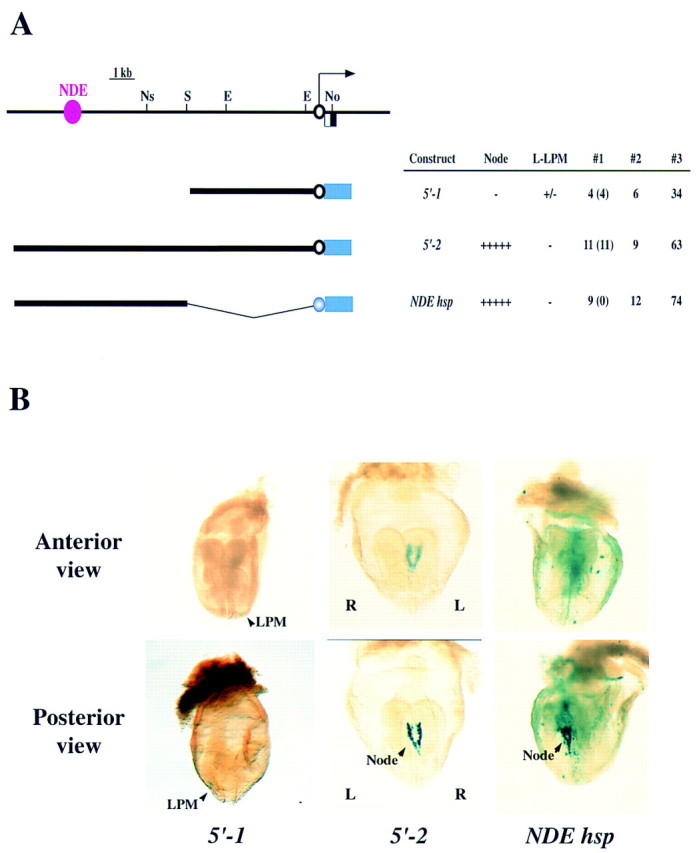
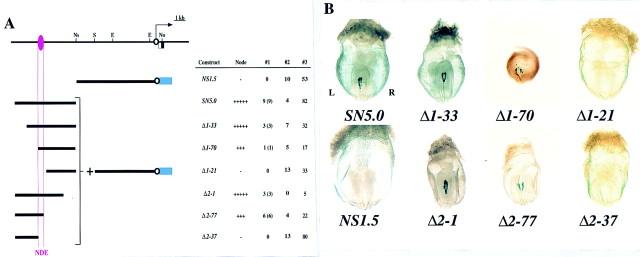
Localization of a NDE in the upstream region of nodal. (A) The structure of the nodal gene, with the arrow indicating the direction of transcription, is shown above those of three lacZ constructs. (E) EcoRI; (No) NotI; (Ns) NsiI; (S) SalI. The blue box indicates the lacZ gene; the open black and closed blue circles indicate the nodal proximal promoter and the hsp68 promoter, respectively. The NDE is shown in pink. In the summary of the expression data in this and subsequent figures, #1 indicates the number of embryos positive for X-gal staining (numbers in parentheses refer to the number of embryos showing the typical staining pattern), #2 indicates the number of embryos that contained the lacZ transgene but were negative for X-gal staining, and #3 indicates the total number of injected embryos that were recovered at E8.2. (L-LPM) Left LPM. (B) X-gal staining of transgenic embryos harboring constructs 5′-1, 5′-2, or NDEhsp. Weak staining in a few cells of the left LPM (arrowhead) was apparent with 5′-1. The 5′-2 construct gave rise to expression in the node, whereas NDEhsp yielded expression in the node and in ectopic sites. The left (L)–right (R) axis is indicated.
Next we examined the effect of construct 5′-2, which contained the upstream region of nodal from −11.5 kb to −50 bp (Fig. 1A). This construct gave rise to X-gal staining in the node (Fig. 1B), suggesting the presence of a node-specific enhancer (NDE) between −11.5 and −5 kb. To examine the specificity of the NDE, we linked this 6.5-kb region (from −11.5 to −5 kb) to a heterologous promoter, that of the hsp68 gene. The resulting construct (NDEhsp) yielded X-gal staining in the node but also in ectopic sites (Fig. 1B), indicating that both NDE and the proximal promoter region of nodal are required for strictly node-specific expression.
To locate the NDE more precisely, we first divided the 6.5-kb region between −11.5 and −5.0 kb into two and attached each portion to the 5-kb proximal promoter region. Construct NS1.5, which contained the 1.5-kb 3′ portion of the upstream region, did not show expression in the node, whereas SN5.0, which contained the 5.0-kb 5′ portion of the upstream region, possessed NDE activity (Fig. 2). Then we prepared two sets of deletion mutants from SN5.0 and examined their activity (Fig. 2) Among the 5′ deletion mutants tested, Δ1–33 and Δ1–70 retained NDE activity, whereas Δ1–21 did not. Of the 3′ deletion mutants, Δ2–1 and Δ2–77 showed X-gal staining in the node, whereas Δ2–37 did not. These results localized the NDE to a 0.8-kb region between −9.5 and −8.7 kb.
Figure 2.
Mapping of the NDE in the upstream region of nodal. (A) Structures and activities of various lacZ constructs. (B) Posterior views of X-gal staining in embryos transgenic for the indicated constructs. X-gal staining in the node was observed with SN5.0, Δ1–33, Δ1–70, Δ2–1, and Δ2–77, but not with NS1.5, Δ1–21, or Δ2–37.
An ASE in intron 1 of nodal
Among five constructs (3′-1 to 3′-5) containing the 5-kb proximal promoter region and various portions of nodal downstream region, construct 3′-1, which contained a 3.5-kb downstream fragment including intron 1, gave rise to marked X-gal staining in the left LPM (Fig. 3), whereas the remaining constructs failed to yield any staining in E8.2 embryos. To confirm the specificity of the 3.5-kb fragment, we linked it to the hsp68 promoter, yielding construct 3′-1hsp. This construct also gave rise to X-gal staining in the left LPM, but the intensity of the staining was much greater than that obtained with 3′-1. Transverse sections confirmed that both 3′-1 and 3′-1hsp induced left-sided expression in LPM (including the splanchnopleure and somatopleure) (Fig. 3B). X-gal staining in the PFP was not apparent with 3′-1, but it was observed with 3′-1hsp. Transverse sections of embryos transgenic for 3′-1hsp indicated that the staining in the PFP was predominantly left-sided (Fig. 3B). These results suggested the presence of an ASE in intron 1 of nodal.
Figure 3.
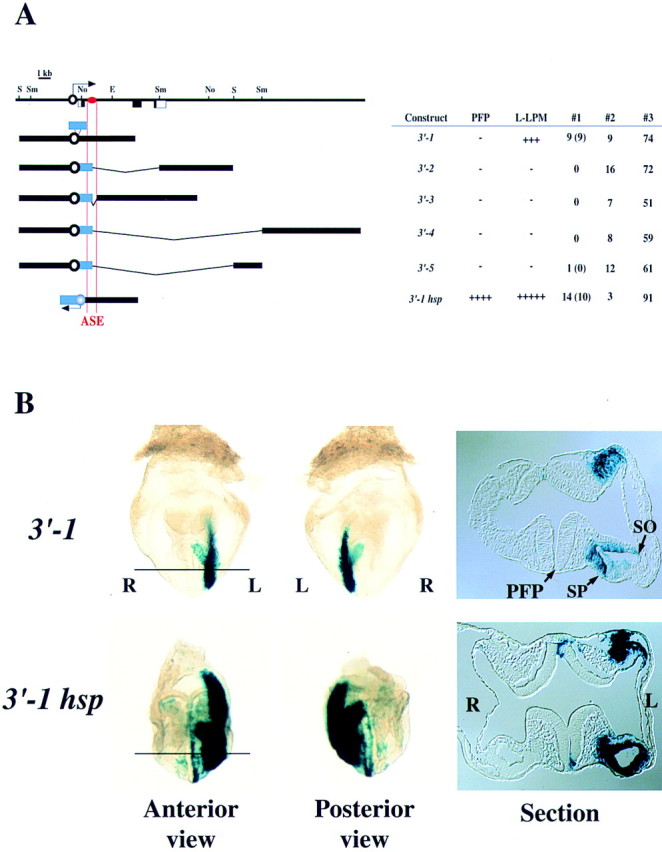
Localization of a left side-specific enhancer (ASE) in intron 1 of nodal. (A) Structures and activities of six constructs containing various portions of nodal downstream plus the proximal promoter region. The approximate location of the ASE is indicated by the red oval. (B) Anterior and posterior views as well as transverse sections of X-gal staining in embryos transgenic for 3′-1 or 3′-1hsp. In both types of transgenic embryos, the somatopleure (so) and splanchnopleure (sp) on the left side are positive for staining. In the embryo harboring 3′-1hsp, left-sided staining was also apparent in the PFP.
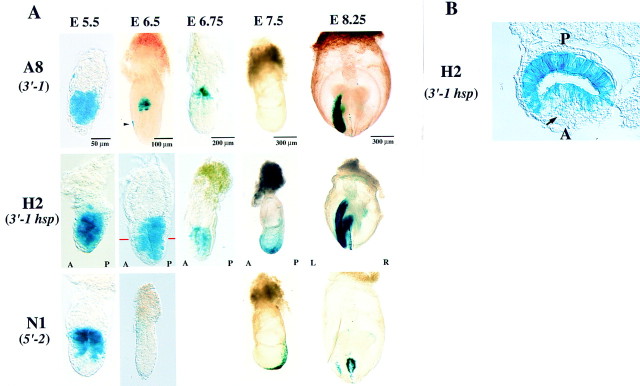
To verify further the presence of a left side-specific enhancer in intron 1, we generated a permanent mouse line (A8) harboring 3′-1 and two such lines (H2 and H13) harboring 3′-1hsp. In all three transgenic lines, X-gal staining was observed in left LPM at the early somite stage (Fig. 4; data not shown). In embryos of lines H2 and H13, left-sided staining was also apparent in the PFP (Fig. 4A). These X-gal staining patterns were indistinguishable from those observed by ‘transient’ transgenic assay (Fig. 3), further confirming the presence of a left side-specific enhancer in intron 1.
Figure 4.
Expression of 3′-1, 3′-1hsp, and 5′-2 in permanent transgenic lines. (A) Permanent transgenic lines harboring 3′-1, 3′-1hsp, or 5′-2 were generated, and the expression of the transgenes was examined at various developmental stages. Lateral views are shown for embryos at E5.5, E6.5, E6.75, and E7.5; posterior views are shown for E8.0 embryos. The arrow indicate the anterior visceral endoderm. A and P indicate the anteroposterior axis. (B) Transverse sections of E6.5 embryos transgenic for 3′-1hsp. In the embryo harboring 3′-1hsp, X-gal staining was apparent in the posterior epiblast and in the anterior visceral endoderm (arrow).
ASE is essential and sufficient for L-R asymmetric expression
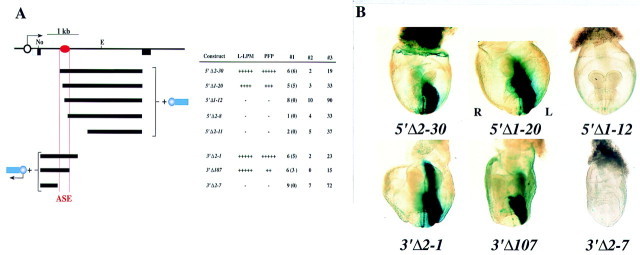
To delineate further the ASE responsible for asymmetric expression of nodal, we prepared two sets of deletion mutants from 3′-1hsp and examined their transcriptional activity in transgenic embryos (Fig. 5). Among the 5′ deletion mutants, left-sided lacZ expression in LPM and PFP was observed with 5′Δ2–30 and 5′Δ1–20, but not with 5′Δ1–12, 5′Δ2–8, or 5′Δ2–11. Of the 3′ deletion mutants, 3′Δ2–1 and 3′Δ107 gave rise to asymmetric expression, whereas 3′Δ2–7 did not. These results localized the ASE to a 340-bp region of intron 1. Among the deletion mutants examined, left PFP was positive for X-gal staining whenever left LPM was positive. Thus, this 340-bp region alone was able to direct asymmetric expression in both the LPM and PFP.
Figure 5.
Mapping of ASE to a 340-bp region of intron 1. (A) Structures and activities of various deletion mutants of 3′-1hsp. The approximate location of ASE is shown by the red oval. (B) X-gal staining in representative embryos. Constructs 5′Δ2–30, 5′Δ1–20, 3′Δ2–1, and 3′Δ107 induced left-sided X-gal staining in LPM and PFP, whereas 5′Δ1–12 and 3′Δ2–7 failed to give rise to X-gal staining.
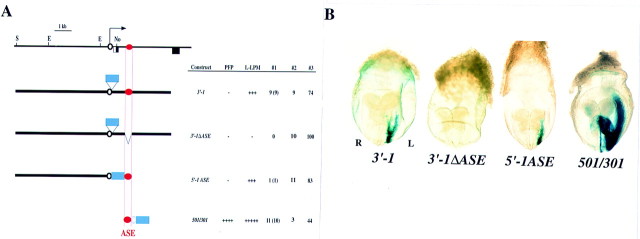
Internal deletion of the 340-bp region containing ASE from construct 3′-1, yielding 3′-1ΔASE, abolished the asymmetric expression of X-gal in LPM (Fig. 6), indicating that ASE is absolutely required for such expression. When the 340-bp region was linked to the 5-kb proximal promoter region of nodal, the resulting construct (5′-1ASE) yielded left-sided X-gal staining in LPM (Fig. 6). Finally, a construct (501/301hsp) in which the 340-bp region was linked to the hsp68 promoter rather than to the nodal promoter, gave rise to X-gal staining in both left LPM and left PFP (Fig. 6). Together, these results suggested that ASE is both essential and sufficient for the asymmetric expression of nodal.
Figure 6.
ASE is essential and sufficient for L-R asymmetric expression of nodal. (A) Structures and activities of the lacZ constructs 3′-1, 3′-1ΔASE, 5′-1ASE, and 501/301hsp. (B) X-gal staining patterns of representative transgenic embryos. Constructs 3′-1, 5′-1ASE, and 501/301hsp yielded left-sided staining in the LPM, whereas 3′-1ΔASE did not. Construct 501/301hsp also gave rise to left-sided staining in PFP.
Sensitivity of ASE to iv, inv, and lefty-1 mutations
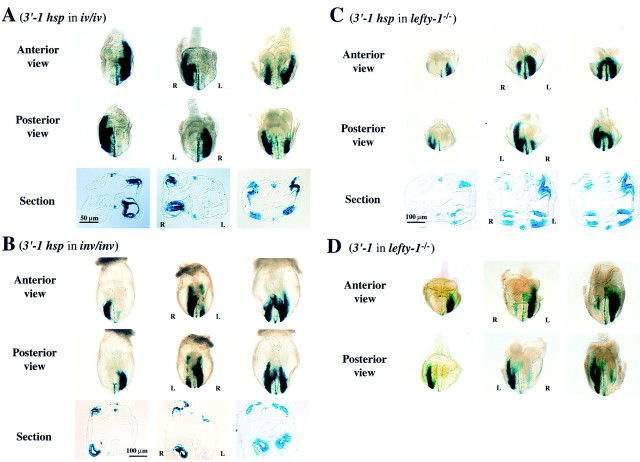
Expression of nodal is affected in situs mutants such as iv/iv, inv/inv, and lefty-1−/− mice; the pattern of expression is randomized (left sided, right sided, or bilateral) in iv/iv mutants, reversed in inv/inv mutants, and bilateral in lefty-1−/− mutants (Collignon et al. 1996; Lowe et al. 1996; Meno et al. 1998). These observations indicate that nodal acts downstream of the iv, inv, and lefty-1 genes. We therefore examined whether expression of nodal–lacZ transgenes was affected by iv, inv, or lefty-1 mutations (Fig. 7). Mice of transgenic lines H2 and H13, which harbor 3′-1hsp, were mated with iv/iv mice, and the lacZ transgene was transferred to the iv/iv background. The iv/iv embryos harboring the transgene showed three different patterns of X-gal staining in LPM (Fig. 7A): left-sided (2/8 embryos), right-sided (2/8 embryos), or bilateral (4/8 embryos). The pattern of X-gal staining in PFP was similarly affected; for example, staining in one PFP was reversed when the staining in LPM was reversed (Fig. 7A).
Figure 7.
Response of the 3′-1hsp construct to iv, inv, and lefty-1 mutations. Expression of the 3′-1hsp transgene was examined in iv/iv (A), inv/inv (B), and lefty-1−/− (C) mutant embryos. Anterior and posterior views, as well as a transverse section, are shown for each transgenic embryo. In iv/iv embryos (A), the staining in the LPM and PFP was either left-sided, right-sided or bilateral. In inv/inv embryos (B), the staining in the LPM and PFP was right-sided (embryo at left) or bilateral (embryos at middle and right). In lefty-1−/− embryos, the staining in the LPM and PFP was initially normal (embryo at left). At later stages (embryos at middle and right), however, the staining appeared on the right LPM. The staining in the PFP, increased and became bilateral (C). In lefty-1−/− embryos harboring 3′-1 transgene (D), X-gal staining in the PFP, which was not detected in the wild-type embryos (Fig. 4A), became apparent. X-gal staining pattern in LPM was similar to that in C. lacZ expression in the PFP always preceded that in the right LPM.
The same transgene was also transferred to the inv/inv background by mating mice of lines H2 and H13 with inv/+ mice (inv/inv homozygotes are lethal). After crossing [inv/+, 3′-1hsp] mice with [inv/+] mice, ∼25% of the lacZ+ embryos, which were inv/inv homozygotes, showed aberrant X-gal staining in the LPM; the staining pattern in such embryos was either right-sided (1/10 embryos), right-side dominant (4/10 embryos), or bilaterally equal (5/10 embryos) (Fig. 7B). Again, the pattern of X-gal staining in PFP was affected similarly to that in LPM. These patterns of X-gal staining observed in the iv/iv and inv/inv mice were almost identical to those of the distribution of nodal mRNA in these mutants (Collignon et al. 1996; Lowe et al. 1996). A minor discrepancy was that, among inv/inv embryos, the frequency of bilateral X-gal staining was somewhat higher than that of bilateral nodal mRNA distribution. The 3.5-kb region of nodal contained in 3′-1hsp, which includes ASE, was thus able to reproduce the response of the intact gene to the iv and inv mutations.
In lefty-1−/− mutant mice, nodal (as well as lefty-2) expression in the LPM becomes bilateral (Meno et al. 1998). We examined whether the 3.5-kb fragment of nodal containing ASE was able to respond to the lefty-1 mutation. The 3′-1hsp transgene was transferred to the lefty-1−/− background by mating mice of lines H2 and H13 with lefty-1 mutant animals. At the two- to four-somite stages, the transgene was expressed on the left side of lefty-1−/− embryos. However, at the four- to eight-somite stages, all lefty-1−/− embryos showed bilateral expression of lacZ in the LPM (Fig. 7C). Thus, lacZ expression in the right LPM was apparent later than that in the left LPM. At the four- to six-somite stages, X-gal staining in the right LPM extended posteriorly (to a greater extent than does lefty-2 expression). In addition, X-gal staining in the PFP was up-regulated and bilateral. These patterns of X-gal staining in lefty-1−/− embryos are consistent with the distribution of nodal mRNA observed previously in these animals (Meno et al. 1998). The lefty-1 mutant mice were also mated with animals of the A8 line containing the 3′-1 transgene. As expected, this transgene was expressed bilaterally in the LPM of embryos with the lefty-1−/− background. Expression of lacZ in the PFP, which was not observed in the wild-type background (Figs. 3 and 4), was apparent in the lefty-1−/− background (Fig. 7D) . These results suggest that the 3.5-kb region of nodal, and most likely ASE, is able to respond not only to the iv and inv mutations but also to the lefty-1−/− mutation.
ASE-directed transgene expression in the epiblast and endoderm of gastrulating embryos
nodal is known to exhibit dynamic expression patterns during development; it is expressed in the epiblast at E5.5 to E6.0, the visceral endoderm at E6.0 to E6.5, and the posterior ectoderm at the early-streak stage (E6.5) (Conlon et al. 1994; Collignon et al. 1996; Varlet et al. 1997). We therefore examined whether ASE conferred similar expression patterns on lacZ transgenes. Embryos were recovered at various developmental stages either from permanent line A8 (harboring the 3′-1 transgene) or from lines H2 and H13 (harboring the 3′-1hsp transgene) and were subjected to X-gal staining (Fig. 4). In embryos containing the 3′-1hsp transgene, X-gal staining was apparent throughout the epiblast at E5.5. At E6.5, lacZ expression was restricted to the posterior epiblast. The visceral endoderm also became transiently positive for X-gal staining at E6.5. The expression of lacZ was restricted to the posterior ectoderm, before appearing in left LPM and left PFP at E8.0. Embryos harboring the 3′-1 transgene showed a similar pattern of X-gal staining in the epiblast (with the exception that the staining was restricted to the proximal epiblast at E6.5). Again, the anterior visceral endoderm was positive for X-gal staining at E6.0 to E6.5. These results suggest that ASE is responsible not only for asymmetric expression of nodal at the early somite stage but also for the expression in the epiblast and visceral endoderm at earlier stages.
We also generated a permanent transgenic line (N1) containing the 5′-2 construct and examined the expression of the transgene in gastrulating embryos. At E5.5, marked X-gal staining was apparent in the proximal epiblast. The expression of lacZ became undetectable at E6.5 (both the ectoderm and the endoderm were negative), but it appeared in the node at E7.5 and was maintained until the early somite stage (E8.0 to E8.2). At E8.0, weak X-gal staining was also detected in the left LPM [consistent with the observation by transient transgenic assay that construct 5′-1 gave rise to faint X-gal staining in left LPM (Fig. 1)]. The observation that lacZ expression was induced in the anterior visceral endoderm by 3′-1 and 3′-1hsp, but not by 5′-2, suggests that nodal expression in the visceral endoderm is regulated by the 3.5-kb intron region, most likely by ASE.
Sequence conservation between lefty-2 ASE and nodal ASE
The lefty-2 and nodal genes exhibit similar expression patterns; both are asymmetrically expressed in the left LPM at the same developmental stage (two- to five-somite pair stage). We recently showed that L-R asymmetric expression of lefty-2 is controlled by a left side-specific enhancer (Saijoh et al. 1999). Our present data suggest that, like that of lefty-2, the expression of nodal is also regulated by such an ASE element. We therefore compared the nucleotide sequence of the nodal ASE (340 bp) with that of the lefty-2 ASE (350 bp). Several blocks of sequences were found to be conserved between the two enhancers (Fig. 8).
Figure 8.
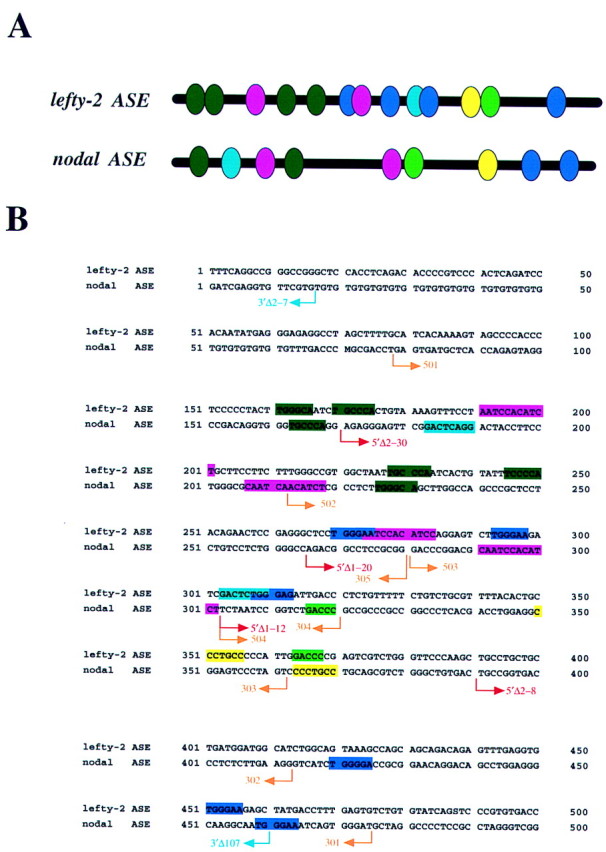
Comparison of the structures of nodal ASE and lefty-2 ASE. (A) Structural organization of nodal ASE (340 bp) and lefty-2 ASE (350 bp). Conserved sequence motifs are indicated by ovals of various colors. (B) Nucleotide sequences of nodal ASE and lefty-2 ASE. The positions of nodal deletion mutants shown in Figs. 5 and 9 are indicated by arrows.
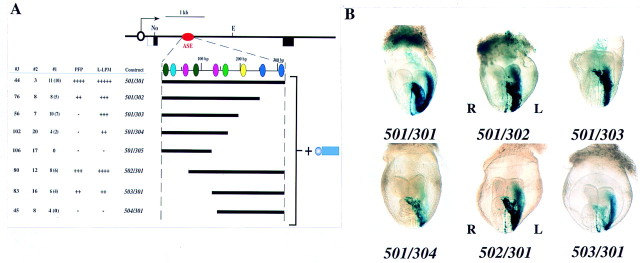
To identify the conserved sequence motifs required for activity of the nodal ASE, we dissected the 340-bp region further. Various deletion mutants were linked to the hsp68 promoter and the expression of the resulting constructs was examined (Fig. 9). Deletion of the 340-bp region from the 3′ end revealed that the 501/302hsp construct was able to induce expression both in the left LPM and the PFP, although the extent of expression was less marked than that apparent with 501/301hsp. The 501/303hsp and 501/304hsp constructs gave rise to expression in the LPM but not in the PFP. Construct 501/305hsp exhibited no ASE activity. Deletion of the 340-bp region from the 5′ end revealed that 502/301hsp and 503/301hsp retained the ability to induce expression in both left LPM and PFP, whereas 504/301hsp had lost ASE activity.
Figure 9.
Dissection of the 340-bp ASE region. (A) Structures and activities of 501/301hsp-based deletion mutants. Each nodal fragment was linked to the hsp68 promoter and lacZ. (B) X-gal staining patterns of representative transgenic embryos.
These results suggest that a 10-bp conserved sequence, CAATCCACAT (shown in pink in Fig. 8), is essential for expression in the left LPM as well as in the PFP. Although two versions of this sequence are present in the 340-bp region, the 3′ version appears more important for asymmetric expression than does the 5′ version. This 10-bp sequence is located between the deletion points of 5′Δ1–20 (which was active) and 5′Δ1–12 (which was inactive) (Figs. 5 and 8). However, additional sequences are also necessary for maximal ASE activity, because activity gradually decreased on deletion of the 340-bp region from either end. These data also suggest that the 3′ region of ASE is necessary for expression in the PFP. In particular, the difference in activity between 501/302hsp and 501/303hsp suggests that a 7-bp conserved sequence, CCCTGCC (shown in yellow in Fig. 8), may be essential for ASE activity in the PFP.
Discussion
Role of ASE in the asymmetric expression of nodal
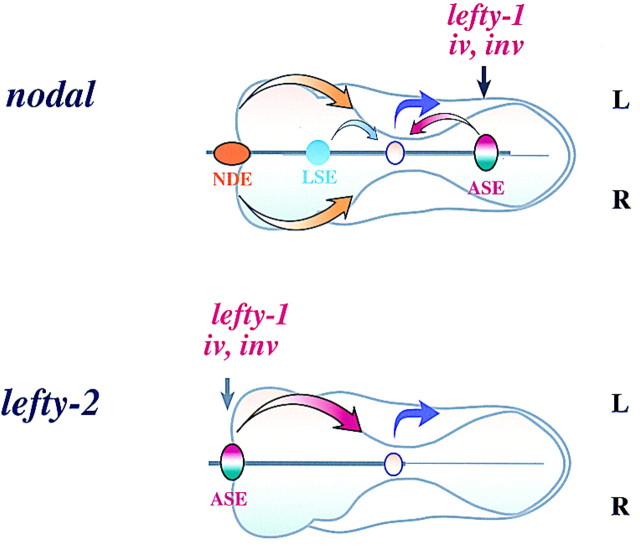
Previously, we have shown that L-R asymmetric expression of lefty-2 is attributable to ASE, whereas that of lefty-1 is mediated by a combination of bilateral enhancers and a right side-specific silencer (Saijoh et al. 1999). We have now shown that expression of nodal, like that of lefty-2, is regulated by a left side-specific enhancer (Fig. 10). Similar results have been obtained by others (Norris and Robertson 1999).
Figure 10.
Models for transcriptional regulation of nodal (top) and of lefty-2 (bottom). The asymmetric expression of both nodal and lefty-2 is determined by similar ASE elements. LSE, a putative weak left side-specific enhancer, is also shown. (See text for further details).
In our transgenic assays, ASE appeared to be both essential and sufficient for the asymmetric expression of nodal. In general, transgenic or transfection assays often overestimate the role of a cis-regulatory element. However, targeted deletion of a 600-bp region spanning the nodal ASE almost abolished the left-sided expression of nodal, confirming the critical role of ASE in the asymmetric expression of this gene (Norris and Robertson 1999).
It is not absolutely clear whether ASE alone is responsible for the asymmetric expression of nodal. Although targeted deletion of ASE in embryonic stem cells largely prevented the asymmetric expression of nodal, weak left-sided expression was still apparent in the left LPM (Norris and Robertson 1999). Furthermore, in our transient transgenic assay, 5′-1, a construct lacking ASE, repeatedly gave rise to faint X-gal staining in a few cells in the left LPM. The permanent line N1, harboring the 5′-2 transgene, also showed similar weak lacZ expression in the left LPM. Thus, although ASE appears to have the major role, an additional enhancer (LSE, for left side-specific enhancer) in the proximal promoter region (between −5 and 0 kb) likely also contributes to the asymmetric expression of nodal. Although ASE and LSE might each act independently as a left side-specific enhancer in our transgenic assays, they may function synergistically to establish asymmetric expression of nodal in vivo.
The expression of nodal in the PFP has not been described previously. However, the nodal ASE was able to direct left-sided expression of lacZ in the PFP. This effect was most apparent when the nodal ASE was linked to the hsp68 promoter. For several reasons, we believe that the ASE-induced expression in the PFP is not an artifact of the transgenic assay, but that it rather reflects expression of nodal in the PFP in vivo: (1) X-gal staining in the PFP was repeatedly observed with 3′-1hsp and its deletion mutants in transient as well as permanent transgenic embryos; (2) X-gal staining in the PFP showed an apparent L-R specificity, being observed predominantly in a few cells on the left side, similar to the expression patterns of lefty-1 and lefty-2; and (3) X-gal staining in the PFP, like that in LPM, responded to iv, inv, and lefty-1 mutations. We attempted to detect nodal mRNA in the PFP by whole-mount in situ hybridization; however, although weak signals were detected occasionally in the PFP, we were unable to obtain convincing results (data not shown). It is likely that the extent of nodal expression in the PFP is not sufficient to be detected by this method. A similar reason may also explain why X-gal staining in the PFP was not obvious with 3′-1 (the hsp68 promoter appeared more efficient than did the nodal promoter, given that the intensity of X-gal staining in the left LPM induced by 3′-1hsp was greater than that induced by 3′-1). An alternative possibility is that although ASE possesses an intrinsic ability to induce expression in the PFP, nodal expression is normally repressed in PFP by a negative regulatory element. nodal expression in the PFP may be negatively regulated by Lefty-1 because 3′-1 transgene gave rise to X-gal staining in the absence of lefty-1.
The 3.5-kb intronic fragment containing ASE responded to iv, inv, and lefty-1 mutations. Expression of the 3′-1hsp (and 3′-1) transgene was affected in a manner similar to that of nodal itself, as determined previously by in situ hybridization (Collignon et al. 1996; Lowe et al. 1996; Meno et al. 1998). It is therefore likely that signals derived from iv, inv, and lefty-1 converge within the ASE region of nodal.
Our data suggest that the expression of nodal in the node is regulated by a separate enhancer (NDE) located in the far-upstream region. Although nodal expression in the node exhibits subtle L-R specificity (Collignon et al. 1996), constructs 5′-2 and SN5.0, as well as deletion mutants derived from SN5.0, failed to show L-R specificity of expression in the node. Thus, NDE itself does not appear to possess L-R specificity. Such asymmetric expression of nodal in the node may be achieved by a combination of NDE and an additional cis-regulatory element such as ASE.
Similar transcriptional regulatory mechanisms underlying expression of nodal in the epiblast, endoderm, and left LPM
nodal is known to exhibit very dynamic expression patterns during development; it is expressed throughout the epiblast at E5.5, in the visceral endoderm at E6.0 to E6.5, in the node between E7.5 and E8.0, and in the left LPM at E8.0 (Zhou et al. 1993; Collignon et al. 1996; Lowe et al. 1996; Varlet et al. 1997). Expression of nodal in the visceral endoderm is required for specification of the anterior axis during gastrulation (Varlet et al. 1997). In our transgenic assays, ASE was able to induce expression not only in the left LPM, but also in the epiblast and the visceral endoderm between E5.5 and E6.5. Targeted deletion of a 600-bp region that includes ASE also affected nodal expression in the epiblast, visceral endoderm, and left LPM (Norris and Robertson 1999). Therefore, nodal expression in the epiblast, visceral endoderm, and left LPM appears to be controlled by the same enhancer, ASE, suggesting that similar transcriptional regulatory mechanisms underlie nodal expression in these domains.
The 6.5-kb region including NDE induced expression not only in the node at the early somite stage (E8.0) but also in the proximal epiblast at E5.5 (Fig. 4A) (Norris and Robertson 1999). A similar proximodistal (P-D) graded expression in the epiblast was also observed at E6.5 in a permanent transgenic line harboring 3′-1, consistent with the notion that the epiblast is already specified along with the P-D axis at this early stage (Beddington and Robertson 1999). Thus, the proximal epiblast, which moves posteriorly and later gives rise to the primitive streak, appears to express nodal at a higher level than does the distal epiblast, although previous in situ hybridization analysis failed to detect such a P-D expression gradient. A higher level of nodal expression may be required for the posterior movement of proximal epiblast cells.
Similarities between nodal ASE and lefty-2 ASE
The ASEs of lefty-2 and nodal exhibit several common features: (1) Both enhancers direct left-sided expression not only in the LPM and but also in the PFP; (2) they both respond to iv, inv, and lefty-1 mutations (Fig. 7; Saijoh et al. 1999); and (3) with both enhancers, directed expression in the PFP can be partially separated from that in the LPM. One difference between the two enhancers is that whereas the lefty-2 ASE (350-bp region) can be clearly divided into at least two subdomains (one for the anterior LPM, the other for the posterior LPM), no such subdivision was apparent for nodal ASE (340 bp region). Finally, several conserved sequence motifs are apparent in the nucleotide sequences of the two enhancers. Thus, the asymmetric expression of nodal and of lefty-2 may be regulated by similar transcriptional mechanisms.
Dissection of the 340-bp region of the nodal ASE suggested that most (if not all) of the conserved motifs are required for maximal enhancer activity. However, a 7-bp conserved sequence (CCCTGCC) located in the 3′ portion of this region may be particularly important for expression in the PFP. Moreover, a 10-bp conserved sequence (CAATCCACAT), which exist twice in nodal ASE and lefty-2 ASE, appears essential for expression in both left LPM and PFP. However, database searches (TRANSFAC) failed to identify any previously known transcription factor binding sites that corresponded to these sequences. Clarification of the mechanism by which signals derived from upstream genes, such as the inv and iv genes, induce the asymmetric expression of nodal will require identification of the transcription factors that bind to these conserved sequences.
Materials and methods
lacZ constructs
The nodal gene was isolated from a genomic library derived from 129/Sv mice. The 3′ end of the nodal upstream fragment (SalI–HindIII, 5 kb) is located in the 5′ untranslated region (+80 bp relative to the putative TATA box, −50 bp relative to the translation initiation codon). This upstream fragment (from −5 kb to +80 bp) was linked to the lacZ fragment (HindIII–BamHI, 4 kb) derived from pCH110 (Pharmacia) and was then subcloned in Bluescript or pGEM, yielding construct 5′-1. To generate construct 5′-2, we subcloned a 6.5-kb SalI fragment (−11.5 to −5 kb) into the SalI site of 5′-1. The NDEhsp construct was created by subcloning the blunt-ended 6.5-kb SalI fragment (−11.5 to −5 kb) into the SmaI site of hsp68–lacZpA (Kothary et al. 1989). For generation of constructs 3′-1 to 3′-5, the indicated fragments of nodal (Fig. 3) were subcloned into 5′-1 at the 3′ end of lacZ. For generation of 3′–1hsp, the 3.5-kb intron fragment was subcloned into the SmaI site of hsp68–lacZpA in either orientation. For the deletion mutants derived from 3′-1hsp, deletion proceeded from the SalI site. For all constructs, lacZ fragments free of vector sequences were isolated by gel electrophoresis before microinjection.
Transient transgenic assay
Transient transgenic assays were performed essentially as described previously (Saijoh et al. 1999). Transgenic embryos were generated by injection of lacZ constructs (4 ng/1 μl) into pronuclei of fertilized eggs obtained from crosses between (C57BL/6 × C3H) F1 mice (Hogan et al. 1994; Sasaki and Hogan 1996). The injected embryos were transferred into pseudopregnant recipients and allowed to develop in utero until E8.2. The E8.2 embryos were examined for the presence of the transgene (PCR) and for lacZ expression (by X-gal staining) according to standard protocols. The amount of β-galactosidase activity was estimated from the extent of X-gal staining after incubation in the staining buffer for 2, 8, and 24 hr. When a given construct was active, most of X-gal-positive embryos showed the staining in restricted sites without ectopic lacZ expression, whereas some embryos had variable ectopic expression as well. This was the case for all of the active constructs used in this study except for NDEhsp, which always gave rise to X-gal staining in ectopic sites as well as in the node. For each construct, a representative embryo showing a typical staining pattern is presented in the figures. Primers used to amplify the lacZ sequence were 5′-CTCAAACTGGCAGATGCACGGT-3′ and 5′-CGTTGCACCACAGATGAAACGC-3′.
Permanent transgenic lines
For some lacZ constructs, the transgenic embryos were allowed to develop to term and permanent transgenic lines were established. The presence of the transgene was examined by Southern blot analysis with the lacZ fragment as a probe. The lacZ transgenes were introduced into the background of iv/iv or inv/+ by first mating transgenic mice with iv/iv or inv/+ mice, respectively. The genotype of the resulting offspring was determined by PCR or Southern blot analysis. [iv/iv, lacZ] or [inv/+, lacZ] mice thus obtained were mated, and transgene expression was examined at E8.2.The iv alleles were genotyped by PCR with primers 5′-GCCAGCAATGAACGAGTGGCCCTCAAACCT-3′ and 5′-AGCTCTGGAAACCGTGGCTGGTGTGGCTGT-3′, followed by TaqI digestion (the wild-type allele is sensitive to TaqI, yielding 50-bp fragments; the mutant allele is resistant to TaqI, yielding a 100-bp fragment) (Supp et al. 1997).
Acknowledgments
We thank Dominic Norris and Elizabeth Robertson for communicating their unpublished data prior to submission; Chikara Meno for lefty-1 mutant mice; Kazuko Miyama, Tomoe Tanabe, and Yasuko Kasakawa for excellent technical and secretarial assistance; Atsusi Kuroiwa for mouse nodal cDNA; Alexandra Joyner and Janet Rossant for hsp68–lacZpA; Takahiko Yokoyama and Paul Overbeek for inv+/− mice; and Martina Brueckner for information on iv genotyping. This work was supported by CREST JST, and by Grants-in-Aid for Scientific Research from the Ministry of Education, Science, Sports and Culture of Japan (nos. 10780461 and 08408033).
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked ‘advertisement’ in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL hamada@imcb.osaka-u.ac.jp; FAX 81-6-878-9846.
References
- Beddington R, Robertson EJ. Axis development and early asymmetry in mammals. Cell. 1999;96:195–209. doi: 10.1016/s0092-8674(00)80560-7. [DOI] [PubMed] [Google Scholar]
- Brown NA, Wolpert L. The development of handedness in left/right asymmetry. Development. 1990;109:1–9. doi: 10.1242/dev.109.1.1. [DOI] [PubMed] [Google Scholar]
- Brown NA, McCarthy A, Wolpert L. Development of handed body asymmetry in mammals. Ciba Found Symp. 1991;162:182–201. doi: 10.1002/9780470514160.ch11. [DOI] [PubMed] [Google Scholar]
- Chen J, Knowles HJ, Hebert JL, Hackett B. Mutation of the mouse hepatocyte nuclear factor/forkhead homologue 4 gene results in an absence of cilia and random left-right asymmetry. J Clin Invest. 1998;102:1077–1082. doi: 10.1172/JCI4786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collignon J, Varlet I, Robertson EJ. Relationship between asymmetric nodal expression and the direction of embryonic turning. Nature. 1996;381:155–158. doi: 10.1038/381155a0. [DOI] [PubMed] [Google Scholar]
- Conlon FL, Lyons KM, Takaesu N, Barth KS, Kispert AK, Herrmann B, Robertson EJ. A primary requirement of nodalin the formation and maintenance of the primitive streak in mouse. Development. 1994;120:1919–1928. doi: 10.1242/dev.120.7.1919. [DOI] [PubMed] [Google Scholar]
- Fujinaga M. Development of sidedness of asymmetric body structures in vertebrates. Int J Dev Biol. 1997;41:153–186. [PubMed] [Google Scholar]
- Gebbia M, Ferrero GB, Pilia G, Bassi MT, Aylsworth AS, Penman-Splitt M, Bird L, Bamforth JS, Burn J, Schlessinger D, Nelson D, Caskey B. X-Linked situs abnormalities result from mutation in ZIC3. Nat Genet. 1997;17:305–308. doi: 10.1038/ng1197-305. [DOI] [PubMed] [Google Scholar]
- Heymer J, Kuehn M, Ruther U. The expression pattern of nodal and leftyin the mouse mutant Ft suggests a function in the establishment of handedness. Mech Dev. 1997;66:5–11. doi: 10.1016/s0925-4773(97)00084-1. [DOI] [PubMed] [Google Scholar]
- Hogan B, Beddington R, Constantini F, Lacy E. Manipulating the mouse embryo: A laboratory manual. 2nd ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1994. [Google Scholar]
- Hummel KP, Chapman DB. Visceral inversion and associated anomalies in the mouse. J Hered. 1959;50:9–13. [Google Scholar]
- Hyatt BA, Lohr JL, Yost HJ. Initiation of vertebrate left-right asymmetry by maternal Vg1. Nature. 1996;384:62–65. doi: 10.1038/384062a0. [DOI] [PubMed] [Google Scholar]
- Isaac A, Sargent MG, Cooke J. Control of vertebrate left-right asymmetry by a Snail-related zinc finger gene. Science. 1997;275:1301–1304. doi: 10.1126/science.275.5304.1301. [DOI] [PubMed] [Google Scholar]
- King T, Brown NA. Embryonic asymmetry: Left TGFβ at the right time. Curr Biol. 1997;7:212–215. doi: 10.1016/s0960-9822(06)00103-5. [DOI] [PubMed] [Google Scholar]
- Kothary R, Clapoff S, Darling S, Perry MD, Moran LA, Rossant J. Inducible expression of an hsp68–lacZhybrid gene in transgenic mice. Development. 1989;105:707–714. doi: 10.1242/dev.105.4.707. [DOI] [PubMed] [Google Scholar]
- Levin M, Mercola M. The compulsion of chirality: Toward an understanding of left-right asymmetry. Genes & Dev. 1998;12:763–769. doi: 10.1101/gad.12.6.763. [DOI] [PubMed] [Google Scholar]
- Levin M, Johnson RL, Stern CD, Kuehn MR, Tabin C. Molecular pathway determining left-right asymmetry in chick embryogenesis. Cell. 1995;82:803–814. doi: 10.1016/0092-8674(95)90477-8. [DOI] [PubMed] [Google Scholar]
- Levin M, Pagan S, Roberts DJ, Cooke J, Kuehn MR, Tabin C. Left/right patterning signals and the independent regulation of different aspects of situsin the chick embryo. Dev Biol. 1997;189:57–67. doi: 10.1006/dbio.1997.8662. [DOI] [PubMed] [Google Scholar]
- Logan M, Pagan-Westphal SM, Smith DM, Paganessi L, Tabin C. The transcription factor Pitx2 mediates situs-specific morphogenesis in response to left-right asymmetric signals. Cell. 1998;94:307–318. doi: 10.1016/s0092-8674(00)81474-9. [DOI] [PubMed] [Google Scholar]
- Lowe L, Supp DM, Sampath K, Yokoyama T, Wright CVE, Potter SS, Overbeek P, Kuehn MR. Conserved left-right asymmetry of nodal expression and alteration in murine situs inversus. Nature. 1996;381:158–161. doi: 10.1038/381158a0. [DOI] [PubMed] [Google Scholar]
- Lustig KD, Kroll K, Sun E, Ramos R, Elmenford H, Kirschner MW. A Xenopus nodal-related gene that acts in synergy with noggin to induce complete secondary axis and notochord formation. Development. 1996;122:3275–3282. doi: 10.1242/dev.122.10.3275. [DOI] [PubMed] [Google Scholar]
- Melloy PG, Ewart JL, Cohen MF, Desmond ME, Kuehn MR, Lo CW. No turning, a mouse mutation causing left-right and axial patterning defects. Dev Biol. 1998;193:77–89. doi: 10.1006/dbio.1997.8787. [DOI] [PubMed] [Google Scholar]
- Meno C, Saijoh Y, Fujii H, Ikeda M, Yokoyama T, Yokoyama M, Toyoda Y, Hamada H. Left-right asymmetric expression of the TGFβ-family member leftyin mouse embryos. Nature. 1996;381:151–155. doi: 10.1038/381151a0. [DOI] [PubMed] [Google Scholar]
- Meno C, Itoh Y, Saijoh Y, Matsuda Y, Tashiro K, Kuhara S, Hamada H. Two closely-related left-right asymmetrically expressed genes, lefty-1 and lefty-2: Their distinct expression domains, chromosomal linkage and direct neuralizing activity in Xenopusembryos. Genes Cells. 1997;2:513–524. doi: 10.1046/j.1365-2443.1997.1400338.x. [DOI] [PubMed] [Google Scholar]
- Meno C, Shimono A, Saijoh Y, Yashiro K, Oishi S, Mochida K, Noji S, Kondoh H, Hamada H. lefty-1 is required for left-right determination as a regulator of lefty-2and nodal. Cell. 1998;94:287–298. doi: 10.1016/s0092-8674(00)81472-5. [DOI] [PubMed] [Google Scholar]
- Mochizuki T, Saijoh Y, Tsuchiya K, Shirayoshi Y, Takai S, Taya C, Yonekawa H, Yamada K, Nihei H, Nakatsuji N, Overbeek P, Hamada H, Yokoyama T. Cloning of inv, a gene that controls left/right asymmetry and kidney development. Nature. 1998;395:177–181. doi: 10.1038/26006. [DOI] [PubMed] [Google Scholar]
- Morgan D, Turnpenny L, Goodship J, Dai W, Majumder K, Mattews L, Gardner A, Schuster G, Vien L, Harrison W, Elder FFB, Pennman-Splitt M, Overbeek P, Strachan T. Inversin, a novel gene in the vertebrate left-right axis pathway, is partially deleted in the inv mouse. Nat Genet. 1998;20:149–156. doi: 10.1038/2450. [DOI] [PubMed] [Google Scholar]
- Nonaka S, Tanaka Y, Okada Y, Takeda S, Harada A, Kanai Y, Kido M, Hirokawa N. Randomization of left-right asymmetry due to loss of nodal cilia generating leftward flow of extraembryonic fluid in mice lacking KIF3B motor protein. Cell. 1998;95:829–837. doi: 10.1016/s0092-8674(00)81705-5. [DOI] [PubMed] [Google Scholar]
- Norris, D.P. and E.J. Robertson. 1999. Node-specific and asymmetric nodal expression patterns are controlled by two distinct cis-acting regulatory elements. Genes & Dev. (this issue). [DOI] [PMC free article] [PubMed]
- Oh SP, Li E. The signaling pathway mediated by the type IIB activin receptor controls axial patterning and lateral asymmetry in the mouse. Genes & Dev. 1997;11:1812–1826. doi: 10.1101/gad.11.14.1812. [DOI] [PubMed] [Google Scholar]
- Piedra ME, Icardo JM, Albajar M, Rodriguez-Rey JC, Ros MA. Pitx2 participates in the late phase of the pathway controlling left-right asymmetry. Cell. 1998;94:319–324. doi: 10.1016/s0092-8674(00)81475-0. [DOI] [PubMed] [Google Scholar]
- Ryan A K, Blumberg B, Rodoriguez-Estaban C, Yonei-Tamura S, Tamura K, Tsukui T, de la Pena J, Sabbagh W, Greenwald J, Choe S, Norris D, Robertson EJ, Evans RM, Rosenfeld MG, Izpisua-Belmonte JC. Pitx2 determines left-right asymmetry of internal organs in vertebrates. Nature. 1998;394:545–551. doi: 10.1038/29004. [DOI] [PubMed] [Google Scholar]
- Saijoh Y, Adachi H, Hirao A, Mochida K, Ohishi S, Hamada H. Distinct transcriptional regulatory mechanisms underlie left-right asymmetric expression of lefty-1 and lefty-2. Genes & Dev. 1999;13:259–269. doi: 10.1101/gad.13.3.259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sampath K, Cheng AMS, Frisch A, Wright CVE. Functional differences among nodal- related genes in left-right axis determination. Development. 1997;124:3293–3302. doi: 10.1242/dev.124.17.3293. [DOI] [PubMed] [Google Scholar]
- Sasaki H, Hogan BLM. Enhancer analysis of the mouse HNF3β gene: Regulatory elements for node/notochord and floor plate are independent and consist of multiple sub-elements. Genes Cells. 1996;1:59–72. doi: 10.1046/j.1365-2443.1996.04004.x. [DOI] [PubMed] [Google Scholar]
- Sefton M, Sanchez S, Nieto MA. Conserved and divergent roles for members of the snail family of transcription factors in the chick and mouse embryos. Development. 1998;125:3111–3121. doi: 10.1242/dev.125.16.3111. [DOI] [PubMed] [Google Scholar]
- Supp DM, Witte DP, Potter SS, Brueckner M. Mutation of an axonemal dynein affects left-right asymmetry in inversus viscerummice. Nature. 1997;389:963–966. doi: 10.1038/40140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varlet I, Robertson EJ. Left-right asymmetry in vertebrates. Curr Opin Genet Dev. 1997;6:519–523. doi: 10.1016/s0959-437x(97)80080-9. [DOI] [PubMed] [Google Scholar]
- Varlet I, Collignon J, Robertson EJ. Nodal expression in the primitive endoderm is required for the specification of the anterior axis during mouse gastrulation. Development. 1997;124:1033–1044. doi: 10.1242/dev.124.5.1033. [DOI] [PubMed] [Google Scholar]
- Yokoyama T, Copeland NG, Jenkins NA, Montgomery CA, Elder FFB, Overbeek P. Reversal of left-right asymmetry: A situs inversusmutation. Science. 1993;260:679–682. doi: 10.1126/science.8480178. [DOI] [PubMed] [Google Scholar]
- Yoshioka H, Meno C, Koshiba K, Sugihara M, Itoh H, Ishimaru Y, Inoue T, Ohuchi H, Semina EV, Murray JC, Hamada H, Noji S. Pitx2, a bicoid type homeobox gene, is involved in a Lefty-signaling pathway in determination of left-right asymmetry. Cell. 1998;94:299–305. doi: 10.1016/s0092-8674(00)81473-7. [DOI] [PubMed] [Google Scholar]
- Yost HJ. Development of the left-right axis in amphibians. Ciba Found Symp. 1991;162:165–176. doi: 10.1002/9780470514160.ch10. [DOI] [PubMed] [Google Scholar]
- Zhou X, Sasaki H, Lowe L, Hogan BL, Kuehn MR. Nodal is a novel TGFβ-like gene expressed in the mouse node during gastrulation. Nature. 1993;361:543–547. doi: 10.1038/361543a0. [DOI] [PubMed] [Google Scholar]