Abstract
BACKGROUND:
The antiviral, proapoptotic, antiproliferative gene 2′‐5′ oligoadenylate synthetase (2‐5OAS1) converts adenosine triphosphate into a series of 2′‐5′ oligoadenylates (2‐5A). In turn, 2‐5A activates latent ribonuclease (RNaseL), a candidate hereditary prostate cancer gene. OAS1 polymorphism (reference single nucleotide polymorphism [SNP] 2660 [rs2660]) has been associated with increased susceptibility to infections and various diseases. In general, the low‐enzyme‐activity adenine‐adenine (AA) genotype promotes susceptibility, whereas the high‐enzyme‐activity guanosine‐guanosine (GG) genotype confers protection. In this study, the authors investigated the association of this functional OAS1 polymorphism (rs2660) with prostate cancer.
METHODS:
Sample size and power were calculated using a power calculation software program for case‐control genetic association analyses. Genomic DNA samples from a control group (n = 140) and from a case group of patients with prostate cancer (n = 164) were used for genotyping SNPs rs2660, rs1131454, and rs34137742 in all samples. Statistical analyses were performed using a logistic regression model.
RESULTS:
A significant association was observed between the rs2660 genotype (A/G) and prostate cancer. Genotype AA increased the risk, whereas genotype GG decreased the risk of prostate cancer. The GG genotype was not observed in the African American samples. The AA genotype also increased the risk of prostate cancer with age.
CONCLUSIONS:
The OAS1 SNP rs2660 AA genotype was associated significantly with prostate cancer, whereas the GG genotype protected against prostate cancer. OAS1 rs2660 may be a prostate cancer susceptibility polymorphism, which is a significant observation, especially in a context of the OAS1‐RNaseL pathway. Thus, a functional defect in OAS1 because of the rs2660 SNP not only can attenuate RNaseL function but also can alter cell growth and apoptosis independent of RNaseL. Cancer 2011;. © 2011 American Cancer Society.
Keywords: 2′‐5′, oligoadenylate synthetase 1, prostate, cancer, polymorphism, reference single nucleotide polymorphism 2660
Short abstract
In this study, the authors investigated the association between the functional polymorphism reference single nucleotide polymorphism (rs) number rs2660 in 2′‐5′ oligoadenylate synthetase 1 (OAS1) and prostate cancer. The results suggest that the rs2660 AA genotype is associated significantly with prostate cancer, whereas the GG genotype protects against prostate cancer.
2′‐5′ Oligoadenylate synthetase 1 (2‐5OAS1) is an extensively characterized enzyme induced by interferons (IFNs) that is required for an effective antiviral response.1, 2 2‐5OAS1, in the presence of double‐stranded RNA structures like viral genomes or single‐stranded RNA transcripts that possesses significant double‐stranded character, converts adenosine triphosphate to a series of 2′ to 5′ oligoadenylates (2‐5A).2 One of the primary functions of 2‐5A is to promote dimerization of latent ribonuclease (RNaseL) to form catalytically active RNaseL3, 4 The activated OAS1‐RNaseL system promotes apoptosis,5 attenuates proliferation,6 degrades viral and cellular RNA, and inhibits protein synthesis.1, 7 It also has been reported that OAS affects cellular events independent of RNaseL, such as antiviral activity8, 9 and cell growth and differentiation.10 Evidence also has demonstrated proapoptotic activity of OAS1 isoform 9‐2(p48) (in vitro interaction with B‐cell lymphoma 2 [Bcl‐2] and extra‐large B‐cell lymphoma [Bcl‐xL]).11, 12
Epidemiologic studies, linkage analyses, association‐based studies, and positional cloning/candidate gene approaches have led to the identification of RNaseL as a candidate hereditary prostate cancer gene.13, 14, 15 Experimental studies also have demonstrated that active RNaseL not only degrades viral genome but is also antiproliferative and promotes senescence and apoptosis in prostate cancer cells, thus acting as a true tumor suppressor.16, 17, 18 However, follow‐up studies by several groups have not identified a significant number of RNaseL germline variants among families with hereditary prostate cancer.15, 19, 20 Moreover, significant associations between RNaseL mutations/polymorphisms and sporadic prostate cancer have not been reported.19 These observations led us to believe that alternate mechanisms and pathways leading to the activation/expression of RNaseL may be involved in the prostate cells. It is possible that 1 of these mechanisms/pathways may be at the level of OAS1, which is a rate‐limiting enzyme in RNaseL activation. Thus, a defect in OAS1 may attenuate RNaseL function and also may alter cell growth and apoptosis in an RNaseL‐independent manner.
It is known that single nucleotide polymorphisms (SNPs) modulate OAS1 function at multiple levels, including expression, alternative splicing, and enzyme activity. In total, it is known that approximately 46 polymorphic markers (University of California Santa Cruz genome database) to 57 polymorphic markers (National Center for Biotechnology Information [NCBI] Single Nucleotide Polymorphism database [dbSNP]) exist in the OAS1 gene. Two of these functional, nonsynonymous polymorphic markers (reference SNP [rs] number rs1131454 and rs2660) have been well studied and have demonstrated an association with various diseases, such as susceptibility to hepatitis C virus,21, 22 influenza A virus,23 flavivirus,24 West Nile virus,25 respiratory epithelial cell syncytial virus,26 ocular herpes simplex virus type 1, and severe acute respiratory syndrome (SARS).27 OAS1 polymorphism also increases susceptibility to multiple sclerosis28, 29 and type I diabetes.30, 31 It is noteworthy that an adenine‐to‐guanine (A/G) SNP (rs2660; A→G) near the exon 7 splice‐acceptor site of the OAS1 gene alters 2‐5OAS1 enzyme activity. OAS1 enzyme activity is highest in individuals with the GG genotype, intermediate in those with the AG genotype, and lowest in those with the AA genotype.32
Collectively, these observations led us to investigate the hypothesis that functional polymorphisms, specifically those at rs2660 and rs1131454 in 2‐5OAS1, could be associated with prostate cancer. Our results demonstrate that rs2660, but not rs1131454, is associated with prostate cancer and that a specific rs2660 genotype may confer decreased susceptibility to prostate cancer.
MATERIALS AND METHODS
Cell Culture
The prostate cancer cell lines LNCaP, DU145, and PC3 were purchased and cultured according to the supplier's instructions (American Type Culture Collection, Rockville, Md).
Samples
The genotyping protocol and use of human samples for this study were approved by the Clark Atlanta University institutional review board. Genotyping was performed on a total of >300 samples. Cancer samples and control samples constituted approximately 50% each of the total sample set (Table 1). Retrospectively collected buffy coat samples were obtained from the Biospecimen Shared Resource at KU Cancer Center (University of Kansas Medical Center, Kansas City, Kan) and the Cooperative Human Tissue Network (Southern Division; National Cancer Institute, Bethesda, Md) after appropriate protocol review and approval. The purified genomic DNA samples were obtained commercially from BioServe Inc. (Beltsville, Md). All samples were stored at −80 °C until analysis. Deidentified, comprehensive clinical information regarding age, ethnicity, and disease stage was available for all samples. The ethnicity of individuals who provided the samples was as reported by the vendor, and no specific admixture analysis was performed. However, a family history of prostate cancer, prostate‐specific antigen level, and Gleason score were not available for all samples and, thus, were not included in the final statistical analysis.
Table 1.
Demographics of All Samples Used in the Current Study
| Age, y | |||||
|---|---|---|---|---|---|
| Sample Type | Total no. of Samples | Mean±SEM | Median | Range | P |
| Total no. of samples analyzed | 304 | ||||
| Cancer | 164 | 63.707±0.746 | 63.5 | 43‐86 | .273 |
| Normal | 140 | 60.214±1.344 | 64 | 17‐98 | |
| Caucasians | |||||
| Cancer | 84 | 59.750±0.888 | 59.5 | 37‐83 | .736 |
| Normal | 78 | 57.244±1.955 | 61 | 17‐98 | |
| African Americans | |||||
| Cancers | 80 | 67.862±1.028 | 68 | 43‐86 | .073 |
| Normal | 62 | 63.952±1.6 | 67 | 20‐94 | |
Abbreviations: SEM, standard error of the mean.
DNA and RNA Isolation
Genomic DNA was isolated (and stored at −80 °C) from cultured cells or buffy coat using AquaPure total genomic DNA isolation kit (Bio‐Rad, Hercules, Calif). On an average, approximately 30 μg of DNA was routinely isolated from 300 μL of buffy coat. Total RNA from cultured cells was isolated using the Trizol (Invitrogen, Carlsbad, Calif) method. The final RNA pellet was washed with 70% ethanol, air dried, resuspended in diethylpyrocarbonate‐treated H2O at a concentration of 1 mg/mL, and stored at −80C until analysis.
Reverse Transcriptase‐Polymerase Chain Reaction
One microgram of total RNA was reverse transcribed using MMLV‐RT (Promega, Madison, Wis) in a total volume of 20 μL according to the manufacturer's instructions. Reverse transcriptase‐polymerase chain reaction (RT‐PCR) was performed using the gene‐specific primers (NM_001032409) 5′‐TGA CGG TCT ATG CTT GGG AG (forward) and 5′‐CAA GAT GCA CTG GCA TTC AG (reverse) in a 25‐μL reaction using GoTaq Colorless Master Mix (12.5 μL; Promega), 400 picomoles each of the 5′ and 3′ primers, and 2 μL of the reverse‐transcribed RNA.
Genomic PCR
Genomic PCR was carried out in a 25‐μL PCR reaction that consisted of 12.5 μL GoTaq Colorless Master Mix (Promega), 30 ng genomic DNA, and 400 picomoles each of the 5′ and 3′ primers. The PCR was carried out for 35 cycles with annealing temperatures of 62 °C and 52 °C for rs2660 and rs1131454, respectively. The following primers were used (NC_000012.11): for rs2660, the primers were forward 5′‐CGG CTG AAG TCT GTC TGC TG and reverse 5′‐GAC CTG GGT TCT GTC CTG GA, and the nested primer was 5′‐ACA GGC AGA AGA GGA CTG GA; and the primers for rs1131454 were forward 5′‐GGA TCA GGA ATG GAC CTC AA and reverse 5′‐GGA GAA CTC GCC CTC TTT CT.
Western Blot Analysis
Total protein from cultured prostate cancer cells was extracted using M‐per (Pierce, Rockford, Ill) containing appropriate protease inhibitors (Complete Mini; Roche Applied Sciences, Indianapolis, Ind). The total protein concentration was determined with the Bradford method. Twenty micrograms of total protein were separated by 12% sodium dodecyl sulfate‐polyacrylamide gels and blotted onto nitrocellulose membranes (Bio‐Rad). Western blot analysis was performed using protein‐specific antibody for OAS1 (goat antihuman, sc‐49833; Santa Cruz Biotechnology, Santa Cruz, Calif) and actin (rabbit antihuman; Santa Cruz Biotechnology) as primary antibodies, and the results were detected with donkey antigoat (sc‐2020; Santa Cruz Biotechnology)‐horseradish peroxidase (HRP) and antirabbit immunoglobulin‐HRP conjugate (Upstate Biotechnology, Lake Placid, Calif), respectively. HRP was detected by enhanced chemiluminescence (Pierce), and blots were observed using the Fujifilm LAS‐3000 Imager (Fujifilm, Tokyo, Japan).
SNP Detection
The rs2660 and rs1131454 polymorphisms were detected by sequencing the PCR amplicon spanning the SNPs using the primer pairs described above. An aliquot of the respective PCR reaction was analyzed first on a 1.5% agarose gel to confirm the specificity and quality of the reaction in terms of band size and absence of any background PCR product. Once confirmed, the remaining PCR product was cleaned using ExoSAP‐IT (USB Corporation, Cleveland, Ohio) before sequencing on the AB sequencer (DNA Sequencing Laboratory, Morehouse School of Medicine, Atlanta, Ga). The sequencing was performed using 5′ and 3′ primers for rs1131454 and using nested primers within the rs2660 PCR product. The sequences were scanned using an ABI sequence scanner (Applied Biosystems, Foster City, Calif), and the SNPs were detected manually. The amplicons that had low‐quality reads were resequenced. The SNPs also were detected by using the SNPdetctor software tool developed by Zhang et al33 to ensure that the SNPs were caused by heterozygous allelic variations and not by sequencing artifacts. The sequences aligned with an NCBI reference sequence (NM_001032409) in CLUSTAL W (Conway Institute, University College Dublin, Dublin, Ireland) also were used to validate respective sequence reads.
Sample Size Calculations
The sample size required to achieve statistically significant associations were calculated using the power calculator for case‐control genetic association studies (PGA).34 The parameters used in sample size calculations were as follows: a power of 0.95 (95%), an α level of .05, and a prostate cancer disease prevalence of 250 in 100,000 men (actual for all races, for whites, and for blacks was 156.9, 150.4, and 234.6 per 100,000 men, respectively, according to Surveillance, Epidemiology and End Results data). An rs2660 AA genotype disease frequency of 50% was chosen as the disease allele frequency. On the basis of these variables the PGA estimated a sample size of <100 cases (the actual sample size would have been lower with exact prostate cancer disease prevalence numbers). On the basis of these calculations, we selected a sample size of approximately 1.5 times that estimated by PGA.
Statistical Analysis
Each polymorphism was tested for its association with prostate cancer. Odds ratios (ORs) and 95% confidence intervals (CIs) were calculated for the genotype in association with prostate cancer using logistic regression analysis with adjustment for age. Correlations between genotype distribution and individuals stratified by race and age also were analyzed using the model described above. The SPSS 2007 (version 07.1.19; NCSS, Kaysville, Utah) and SigmaStat (version 3.5; SigmasStat, San Jose, Calif) software packages were used for statistical analyses. The sample size calculations and power were calculated using the “Power for Genetic Association Analyses” (PGA) package in Matlab (available on the National Cancer Institute website; http://dceg.cancer.gov/tools/analysis/pga; accessed on September 19th, 2010).34 After stratifying the data by age group and race, regular logistic regression models were used to calculate the ORs and 95% CIs using the control group as a reference.35 Logistic regression models also were used to assess the interaction between age and genotypes in the case group and to detect differential rs2660 effects at different age intervals. P values < .05 were considered statistically significant.
RESULTS
Structural Organization of OAS1
The OAS1 gene gives rise to 3 alternatively spliced proteins (Isoforms 1, 2, and 3), primarily at the C‐terminal end with various molecular weights, as illustrated in Figure 1A. The rs1131454 SNP in exon C (third exon) is present in all 3 OAS1 isoforms. The rs2660 SNP, however, is present in only OAS1 isoforms 1 and 3. In OAS1 isoform 1, rs2660 is in the 3′‐untranslated region; whereas, in OAS1 isoform 3, it is present toward the C‐terminal end of the protein. Both rs1131454 (in all OAS1 isoforms) and rs2660 (in OAS1 Isoform 3) are nonsynonymous and result in glycine‐to‐serine (G→S) and glycine‐to‐arginine (G→R) amino acid changes, respectively, as outlined in Figure 1A.
Figure 1.
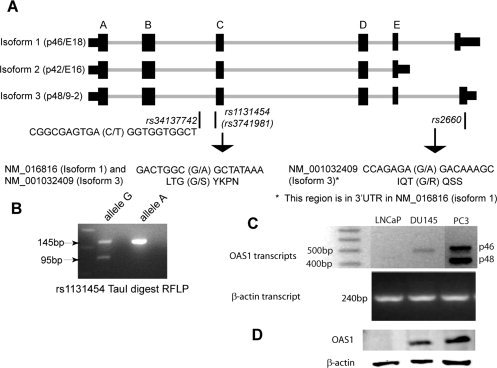
Structure, polymorphisms, and expression of 2′‐5′ oligoadenylate synthetase 1 (OAS1) are illustrated. (A) The structure of the OAS1 gene reveals isoform 1 (p46/E18), Isoform 2 (p42/E16), and Isoform 3 (p48/9‐2). Exons are designated A through E. The last exons in isoform 1 (p46/E18) and Isoform 3 (p48/9‐2) are not labeled. The nonsynonymous single nucleotide polymorphism (SNP) rs1131454 (nucleotide, guanine to adenine [G→A]; protein, glycine to serine [G→S]) appears in all isoforms. The single nucleotide polymorphism rs2660 (G→A) is located in the last exon in Isoform 3 (p48/9‐2), resulting in a glycine‐to‐arginine (G→R) amino acid in the 3′‐untranslated region in isoform 1 (p46/E18). The SNP rs34137742 (C→T) is in the intron between exons B and C. (B) A typical restriction fragment length polymorphism (RFLP) was used to confirm the rs1131454 SNP with Tau1 restriction digest of the amplified genomic polymerase chain reaction (PCR) fragment using the primer pairs described in the text (see Materials and Methods). (C) Reverse transcriptase‐PCR was carried out on messenger RNA isolated from LNCaP, DU145, and PC3 cells using the primer pairs described in the text (see Materials and Methods). These primers were designed to differentially amplify OAS1 Isoforms 1 (p46) and 3 (p48), resulting in approximately 500‐base pair (bp) and 400‐bp bands, respectively. (D) OAS1 protein expression is observed in prostate cancer cell lines with Western blot analysis. Whole cell lysates were used for Western blot analysis with antibody against OAS1 and β‐actin as a loading control. The PCR and Western blot analyses shown here are representative of at least 3 experiments.
OAS1 Expression in Prostate Cancer Cell Lines
To establish a rationale for investigating the association between OAS1 polymorphism and prostate cancer, first, we investigated the expression of OAS1 in established prostate cancer cell lines. OAS1 expression was observed at both the transcript level and the protein level in DU145 and PC3 cells (Fig. 1C,D). In LNCaP cells, we occasionally observed the OAS1 transcript, but the protein was not detected by Western blot analysis (Fig. 1C,D). The inconsistency at the transcript level may have been caused by alternative splicing, which can result in premature termination of the peptide in LNCaP cells (Fig. 1C,D). The genotypes for each cell line for the SNPs in this study were as follows: rs2660, AG in all 3 cell lines; rs 1131454, GG in LNCaP cells, AG in DU145 cells, and AA in PC3 cells; and rs34137742, cytosine‐cytosine (CC) in all 3 cell lines.
Sample Characteristics
A retrospective case‐control study was performed on 304 samples to analyze the OAS1 rs2660 and rs1131454 polymorphisms. The unrelated, clinically defined cancer samples (N = 164) and control samples (N = 140) comprised roughly 50% each of the total sample population. The mean age (±standard error) of the patients who provided cancer samples was 63.7 ± 0.746 years, and the mean age of individuals who provided normal samples was 60.2 ± 1.3 years, and the groups were age matched (P = .273). The population that was sampled was a mixture of Caucasians and African Americans (Table 1).
Frequency of rs2660 and rs1131454 Polymorphisms
The frequencies of both rs2660 and rs1131454 (previously rs3741981) genotypes in the normal sample set were consistent with those reported for other control populations.22 The rs2660 allele frequency conformed to Hardy‐Weinberg equilibrium in the Caucasian population (chi‐square statistic, 3.546; P = .17). Because there was a lack of samples with the GG genotype (minor allele in rs2660), a marked deviation from Hardy‐Weinberg equilibrium in African Americans was observed. The frequency distributions of rs2660 and rs1131454 in our complete (all samples) and stratified sample sets are listed in Tables 2 and 3, respectively.
Table 2.
Association of the 2′‐5′ Oligoadenylate Synthetase 1 Reference Single Nucleotide Polymorphism rs2660 With Prostate Cancer: Genotype Distribution for All Samples, Caucasian Samples, and African American Samples With Corresponding Odds Ratios, 95% Confidence Intervals, and P Values
| No. (%) | ||||||
|---|---|---|---|---|---|---|
| Genotype | All Samples | Cancer Samples | Normal Samples | OR | 95% CI | P |
| All samples | ||||||
| AA | 205 (67.5) | 125 (76.2) | 80 (57.1) | 2.4 | 1.47‐3.92 | .0006 |
| AG | 75 (24.6) | 37 (22.4) | 38 (27.1) | 0.78 | 0.46‐1.31 | .3533 |
| GG | 24 (7.9) | 2 (1.2) | 22 (15.7) | 0.066 | 0.015‐0.28 | <.0001 |
| Total | 304 (100) | 164 (100) | 140 (100) | |||
| Caucasian samples | ||||||
| AA | 81 (50) | 51 (60.7) | 30 (38.5) | 2.47 | 1.31‐4.65 | .0073 |
| AG | 57 (35.2) | 31 (36.9) | 26 (33.3) | 1.17 | 0.61‐2.23 | .7422 |
| GG | 24 (14.8) | 2 (2.4) | 22 (28.2) | 0.062 | 0.01‐0.27 | <.0001 |
| Total | 162 (100) | 84 (100) | 78 (100) | |||
| African American samples | ||||||
| AA | 124 (87.4) | 74 (92.2) | 50 (80.6) | 2.96 | 1.06‐8.52 | .0434 |
| AG | 18 (12.6) | 6 (7.4) | 12 (19.4) | 0.333 | 0.12‐0.95 | .0423 |
| GG | 0 (0) | 0 (0) | 0 (0) | |||
| Total | 142 (100) | 80 (100) | 62 (100) | |||
Abbreviations: AA, adenine‐adenine genotype; AG, adenine‐guanine genotype; CI, confidence interval; GG, guanine‐guanine genotype; OR, odds ratio.
Table 3.
Association of the 2′‐5′ Oligoadenylate Synthetase 1 Reference Single Nucleotide Polymorphism rs1131454 With Prostate Cancer: Genotype Distribution for All Samples, Caucasian Samples, and African American Samples With Corresponding Odds Ratios, 95% Confidence Intervals, and P Values
| No. (%) | ||||||
|---|---|---|---|---|---|---|
| Genotype | All Samples | Cancer Samples | Normal Samples | OR | 95% CI | P |
| All samples | ||||||
| AA | 56 (18.4) | 24 (14.6) | 32 (22.8) | 0.5786 | 0.322‐1.03 | .0754 |
| AG | 124 (40.7) | 74 (45.1) | 50 (35.7) | 1.4 | 0.931‐2.35 | .102 |
| GG | 124 (40.7) | 66 (40.2) | 58 (41.4) | 0.952 | 0.601‐1.507 | .906 |
| Total | 304 (100) | 164 (100) | 140 (100) | |||
| Caucasian samples | ||||||
| AA | 46 (28.3) | 22 (26.1) | 24 (30.7) | 0.798 | 0.402‐1.582 | .601 |
| AG | 74 (45.6) | 42 (50) | 32 (41) | 1.438 | 0.771‐2.67 | .272 |
| GG | 42 (25.9) | 20 (23.8) | 22 (28.2) | 0.795 | 0.393‐1.608 | .591 |
| Total | 162 (100) | 84 (100) | 78 (100) | |||
| African American samples | ||||||
| AA | 10 (7) | 2 (2.5) | 8 (12.9) | 0.173 | 0.0353‐0.847 | .02 |
| AG | 50 (35.2) | 32 (40) | 18 (29) | 1.63 | 0.802‐3.3 | .215 |
| GG | 82 (57.7) | 46 (57.5) | 36 (58) | 0.977 | 0.499‐1.913 | 1 |
| Total | 142 (100) | 80 (100) | 62 (100) | |||
Abbreviations: AA, adenine‐adenine genotype; AG, adenine‐guanine genotype; CI, confidence interval; GG, guanine‐guanine genotype; OR, odds ratio.
Association of OAS1 Genotype With Prostate Cancer
rs2660
The combined cancer and control groups revealed that AA was a major genotype (67.5%), whereas GG was a minor genotype (7.9%) (Table 2). The AA genotype was over represented in cancer samples (76% vs 57% in control samples; ratio, 3:2), whereas distribution of the AG genotype essentially was similar in the cancer group (22.4%) and the normal group (27.1%) (Table 2). Contrary to the AA genotype distribution, the GG genotype was over represented in the normal group (15.7%) compared with the cancer group (1.2%) (Table 2). Sample stratification demonstrated that rs2660 genotype distribution was associated with race: The AA genotype was over represented in the African American group (87.4% vs 49% in the Caucasian group), whereas the AG genotype (13% vs 36% in the Caucasian group) and the GG genotype (0% vs 15% in the Caucasian group) were over represented in Caucasian group. The genotype distribution suggested that the G allele was significantly under‐represented in African Americans (6.5%), whereas the A allele was over represented in African Americans (93.5%).
Comprehensive statistical analysis revealed that individuals in the combined sample with the AA genotype were 2.4 times more likely to develop prostate cancer than those with other genotypes (OR, 2.4; P = .0006) (Table 2). A logistic regression analysis also suggested a 61% increased risk of prostate cancer with the AA genotype than with other genotypes (Fig. 2A). This result was consistent with both the African American group (OR, 3.0; P = .042) (Table 2) and the Caucasian group (OR, 2.473; P = .0073) (Table 2), suggesting that, irrespective of race, the AA genotype has a stronger association with prostate cancer. Conversely, the GG genotype reduced the incidence of prostate cancer by 15‐fold (1/0.06; OR, 0.06; P < .0001) (Table 2) compared with the AG and GG genotypes. Converting this OR into probability terms suggested that the presence of the GG genotype reduced the incidence of prostate cancer by 92% (using other genotypes as base and for the same age group). The AG genotype was not identified as a risk factor when it was analyzed in the context of all samples (OR, 0.775; P = .3533) and within Caucasian population (OR, 1.17; P = .7422).
Figure 2.
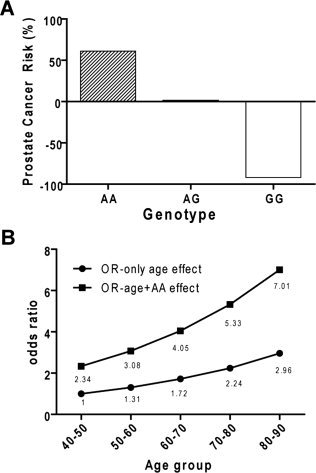
(A) The probability of a particular genotype and prostate cancer was calculated from a logistic regression model (P[case|genotype]). Each genotype was used as a dependent variable to calculate its association with prostate cancer in a logistic regression model. The graph shows the probability from this model expressed as a percentage. On the basis of this model, the adenine‐adenine (AA) genotype increases the risk of prostate cancer by 61% compared with other genotypes, the adenine‐guanine (GA) genotype is neutral to the risk of prostate cancer (ie, it neither increases nor decreases the risk of prostate cancer), and the GG genotype decreases the risk of prostate cancer by 92%. (B) This graph illustrates the effect of age and reference single nucleotide polymorphism 2660 (rs2660) genotype AA on prostate cancer. Prostate cancer incidence increases with age, as indicated by an increased odds ratio (OR) (OR‐only age effect). Genotype AA has an additive effect on prostate cancer incidence (OR‐age + AA effect).
We also used statistical analyses to examine the effect of age on the incidence of prostate cancer. Data stratification by age demonstrated that, after age 50 years, the incidence of prostate cancer was 1.22‐fold higher than it was in the previous decade, but the likelihood jumped to 3.60‐fold compared with the previous decade for the AA genotype. Therefore, with increasing age, the AA genotype significantly increased the incidence of prostate cancer compared with the effect of age alone on prostate cancer (Fig. 2B).
rs1131454 and rs34137742
No significant association between rs1131454 (A→G) was observed with prostate cancer (Table 3). The rs34137742 genotype was observed while analyzing the sequence from rs1131454 genotyping (C→T; 209 bp upstream of rs1131454 in intron) (Table 4). Statistical analysis failed to demonstrate any significant association of rs34137742 with prostate cancer (Table 4). However, both rs1131454 and rs34137742 did conform to Hardy‐Weinberg equilibrium, and no ethnic disparity was observed in their genotype distribution.
Table 4.
Association of the 2′‐5′ Oligoadenylate Synthetase 1 Reference Single Nucleotide Polymorphism rs34137742 With Prostate Cancer: Genotype Distribution for All Samples, Caucasian Samples, and African American Samples With Corresponding Odds Ratios, 95% Confidence Intervals, and P Values
| No. (%) | ||||||
|---|---|---|---|---|---|---|
| Genotype | All Samples | Cancer Samples | Normal Samples | OR | 95% CI | P |
| All samples | ||||||
| CC | 202 (66.4) | 104 (63.4) | 98 (70) | 0.742 | 0.459‐1.202 | .272 |
| CT | 86 (28.2) | 52 (31.7) | 34 (24.2) | 1.447 | 0.871‐2.4 | .162 |
| TT | 16 (5.2) | 8 (4.8) | 8 (5.7) | 0.846 | 0.309‐2.317 | .8004 |
| Total | 304 (100) | 164 (100) | 140 (100) | |||
| Caucasian samples | ||||||
| CC | 144 (88.8) | 72 (85.7) | 72 (92.3) | 0.5 | 0.1777‐1.405 | .216 |
| CT | 16 (9.8) | 10 (11.9) | 6 (7.6) | 1.622 | 0.560‐4.69 | .436 |
| TT | 2 (1.2) | 2 (2.3) | 0 (0) | 4.758 | 0.224 ‐100.7 | .496 |
| Total | 162 (100) | 84 1(00) | 78 (100) | |||
| African American samples | ||||||
| CC | 58 (40.8) | 32 (40) | 26 (41.9) | 0.923 | 0.470‐1.812 | .864 |
| CT | 70 (49.2) | 42 (52.5) | 28 (45.1) | 1.342 | 0.689‐2.611 | .402 |
| TT | 14 (9.8) | 6 (7.5) | 8 (12.9) | 0.547 | 0.179‐1.670 | .395 |
| Total | 142 (100) | 80 (100) | 62 (100) | |||
Abbreviations: CC, cytosine‐cytosine genotype; CI, confidence interval; CT, cytosine‐thymine genotype; OR, odds ratio; TT, thymine‐thymine genotype.
DISCUSSION
In this study, we report for the first time an association between OAS1 polymorphism and prostate cancer. OAS1 activity is a rate‐limiting enzyme in determining RNaseL antiviral and tumor suppressor activities as an important part of the total constitutive activity of OAS enzymes.36 Recent studies have demonstrated that OAS1 also can promote apoptosis and acts as an antiviral agent independent of RNaseL through paracrine mechanisms.9 Thus, OAS1 is emerging as a multifunction protein that is involved not only in innate immune response but also as a potent tumor suppressor.
For this study, we selected 2 functional OAS1 polymorphic markers (rs2660 and rs1131454) that are associated with a disease/viral infection. In general, the rs2660 AA genotype promotes disease susceptibility, whereas the GG genotype confers protection (SARS‐associated coronavirus infection,37 multiple sclerosis28, 29), possibly because the AA, AG, and GG genotypes result in the lowest, intermediate, and highest OAS1 enzyme activity, respectively.32 Consistent with these observations, the genotype AA appears to be a risk factor for prostate cancer (61% increased risk), whereas the higher activity GG genotype protects against prostate cancer (92% lower risk). The A allele also is a significant risk factor with age in prostate cancer: In our logistic regression model, age was a risk factor for prostate cancer, as expected. However, the AA genotype further increased the odds of prostate cancer.
The absence of an rs2660 homozygous GG genotype in the African American population is essentially consistent with population‐wide genotype distribution reported in the NCBI dbSNP database. An analysis of the NCBI dbSNP database revealed that the GG genotype is absent in African American and Sub‐Saharan African populations: the GG genotype is absent in ss24415263 African American samples (NCBI AFD_AFR_panel; n = 46) and in ss38908277 Sub‐Saharan African samples (NCBI HapMap‐YRI; n = 120). The frequency of the G allele in the ss38908277 African American set is only 0.09% (NCBI AoD_African_American; n = 90) compared with a frequency of 9.7% (exclusively from AG genotype) in our normal African American sample set (Table 2). Thus, at least 3 independent studies performed on 3 different platforms (PERLEGEN [ss24415263], ABI XPLORE [ss38908277], and our sequencing‐based analysis) on 2 diverse populations have demonstrated the lack of a GG genotype in individuals of African ethnicity (Sub‐Saharan and African American). These results suggest that OAS1 polymorphisms and perhaps spliced variants may be determined genetically in African Americans. Surprisingly, the GG genotype was observed in the Caucasian sample set, and its frequency was heavily biased in favor of normal Caucasians (28.2% in normal samples vs 2.4% in cancer samples; OR, 0.062; P < .0001). In contrast, the AA genotype associated with prostate cancer is frequent in African Americans compared with Caucasians. For the sake of brevity, we propose that the higher incidence of prostate cancer in African Americans may be caused by absence of the GG genotype; however, a larger cohort and specific biologic endpoint measurements, such as the effect of this genotype on expression, enzyme activity, apoptosis, proliferation, and other yet unknown biologic activities, are needed to support this statement.
The rs2660 genotype is in linkage disequilibrium with rs10774671.32 The G and A alleles in rs10774671 govern splicing to the last OAS1 exon (exon 7), which harbors rs2660. The GG genotype (in rs10774671) retains the splice site, resulting in high enzyme activity; whereas the AA genotype disrupts the splice site, leading to low enzyme activity.32 Because of altered splicing, the 3 genotypes reportedly generate different OAS1 isoforms: p46 (GG), p46 and p48 (AG), and p48 (AA).38 The p46 and p48 isoforms have identical exons 1 through 6 but divergent exon 7. As expected, Two transcripts corresponding to p46 (approximately 500 bp) and p48 (approximately 400 bp) isoforms were observed in the PC3 and DU145 prostate cancer cell lines, which harbor the AG genotype (rs2660), using primer pairs spanning exons 3 through 7. Surprisingly, LNCaP cells, which also have the heterozygous AG genotype, did not express any of the OAS1 spliced variants, suggesting a splicing event 5′ from exon 3 or between exons 3 and 7. In addition to rs2660, rs10774671 also is in linkage disequilibrium with rs1051042 and rs3177979 (all on exon 7).32 It will be interesting to investigate whether the proposed linkage disequilibrium exists between this block in prostate cancer cells and in our sample set.
The rs1131454 SNP (previously rs3741981) in the evolutionary conserved exon 3 also is nonsynonymous (G→A allele results in G→S amino acid change) and is close to the double‐stranded RNA binding domain of all OAS1 isoforms. In a genetic study of type 1 diabetes, it was suggested that this polymorphism is functional, possibly because of congenital rubella virus infection.39 In our dataset, no statistically significant association was observed between rs1131454 genotype racial distribution and prostate cancer.
Collectively, the association studies and data stratification presented in this study suggest that rs2660 is specifically associated with prostate cancer with significant racial disparity and age. Prostate cancer is caused by multiple genes possibly interacting in complex biologic pathways and influenced by environmental factors. The OAS1‐RNaseL pathway fits this general hypothesis: Irrespective of RNaseL mutations/expression, low or lack of OAS1 activity will attenuate RNaseL action and subsequently host innate immune response and tumor suppression. Thus, polymorphisms in OAS1 like rs2660 that alter its activity32 not only can modulate the effects of higher risk susceptibility genes (RNaseL) responsible for hereditary prostate cancer but also may promote sporadic prostate cancer independently.
FUNDING SOURCES
This work was supported by National Institutes of Health (NIH)/National Center on Minority Health and Health Disparities Grant P20MD002285‐01. The DNA Sequencing Laboratory and the Research Core at Morehouse School of Medicine (Atlanta, Ga) were supported through the Research Centers at Minority Institutions (RCMI) Program (NIH/National Center for Research Resources [NCCR]/RCMI Grant G12‐RR03034). Support for core facilities and shared/additional resources were funded in part by NIH/NCRR/RCMI Grant G12‐RR03062.
CONFLICT OF INTEREST DISCLOSURES
The authors made no financial disclosures.
We thank Ms. Qi Yang, DNA Sequencing Laboratory, Morehouse School of Medicine, Atlanta, Ga, and the Research Cores at Morehouse School of Medicine.
REFERENCES
- 1. Hovanessian AG, Justesen J. The human 2′‐5′ oligoadenylate synthetase family: unique interferon‐inducible enzymes catalyzing 2′‐5′ instead of 3′‐5′ phosphodiester bond formation. Biochimie. 2007; 89: 779‐788. [DOI] [PubMed] [Google Scholar]
- 2. Hovnanian A, Rebouillat D, Mattei MG, et al. The human 2′,5′‐oligoadenylate synthetase locus is composed of 3 distinct genes clustered on chromosome 12q24.2 encoding the 100‐, 69‐, and 40‐kDa forms. Genomics. 1998; 52: 267‐277. [DOI] [PubMed] [Google Scholar]
- 3. Zhou A, Hassel BA, Silverman RH. Expression cloning of 2–5A‐dependent RNAase: a uniquely regulated mediator of interferon action. Cell. 1993; 72: 753‐765. [DOI] [PubMed] [Google Scholar]
- 4. Naik S, Paranjape JM, Silverman RH. RNase L dimerization in a mammalian 2‐hybrid system in response to 2′,5′‐oligoadenylates. Nucleic Acids Res. 1998; 26: 1522‐1527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Domingo‐Gil E, Esteban M. Role of mitochondria in apoptosis induced by the 2–5A system and mechanisms involved. Apoptosis. 2006; 11: 725‐738. [DOI] [PubMed] [Google Scholar]
- 6. Yaffe A, Schwarz Y, Hacohen D, Kinar Y, Nir U, Salzberg S. Inhibition of 2–5A synthetase expression by antisense RNA interferes with interferon‐mediated antiviral and antiproliferative effects and induces anchorage‐independent cell growth. Cell Growth Differ. 1996; 7: 969‐978. [PubMed] [Google Scholar]
- 7. Nilsen TW, Maroney PA, Baglioni C. Double‐stranded RNA causes synthesis of 2′,5′‐oligo(A) and degradation of messenger RNA in interferon‐treated cells. J Biol Chem. 1981; 256: 7806‐7811. [PubMed] [Google Scholar]
- 8. Kajaste‐Rudnitski A, Mashimo T, Frenkiel MP, Guenet JL, Lucas M, Despres P. The 2′,5′‐oligoadenylate synthetase 1b is a potent inhibitor of West Nile virus replication inside infected cells. J Biol Chem. 2006; 281: 4624‐4637. [DOI] [PubMed] [Google Scholar]
- 9. Kristiansen H, Scherer CA, McVean M, et al. Extracellular 2′‐5′ oligoadenylate synthetase stimulates RNase L‐independent antiviral activity: a novel mechanism of virus‐induced innate immunity. J Virol. 2010; 84: 11898‐11904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Salzberg S, Hyman T, Turm H, et al. Ectopic expression of 2–5A synthetase in myeloid cells induces growth arrest and facilitates the appearance of a myeloid differentiation marker. Cancer Res. 1997; 57: 2732‐2740. [PubMed] [Google Scholar]
- 11. Ghosh A, Sarkar SN, Rowe TM, Sen GC. A specific isozyme of 2′‐5′ oligoadenylate synthetase is a dual function proapoptotic protein of the Bcl‐2 family. J Biol Chem. 2001; 276: 25447‐25455. [DOI] [PubMed] [Google Scholar]
- 12. Gomos JB, Rowe TM, Sarkar SN, Kessler SP, Sen GC. The proapoptotic 9‐2 isozyme of 2‐5 (A) synthetase cannot substitute for the sperm functions of the proapoptotic protein, Bax. J Interferon Cytokine Res. 2002; 22: 199‐206. [DOI] [PubMed] [Google Scholar]
- 13. Carpten J, Nupponen N, Isaacs S, et al. Germline mutations in the ribonuclease L gene in families showing linkage with HPC1. Nat Genet. 2002; 30: 181‐184. [DOI] [PubMed] [Google Scholar]
- 14. Rennert H, Zeigler‐Johnson CM, Addya K, et al. Association of susceptibility alleles in ELAC2/HPC2, RNASEL/HPC1, and MSR1 with prostate cancer severity in European American and African American men. Cancer Epidemiol Biomarkers Prev. 2005; 14: 949‐957. [DOI] [PubMed] [Google Scholar]
- 15. Wiklund F, Jonsson BA, Brookes AJ, et al. Genetic analysis of the RNasaL gene in hereditary, familial, and sporadic prostate cancer. Clin Cancer Res. 2004; 10: 7150‐7156. [DOI] [PubMed] [Google Scholar]
- 16. Xiang Y, Wang Z, Murakami J, et al. Effects of RNase L mutations associated with prostate cancer on apoptosis induced by 2′,5′‐oligoadenylates. Cancer Res. 2003; 63: 6795‐6801. [PubMed] [Google Scholar]
- 17. Andersen JB, Li XL, Judge CS, Zhou A, Jha BK, Shelby S, et al. Role of 2–5A‐dependent RNase‐L in senescence and longevity. Oncogene. 2007; 26: 3081‐3088. [DOI] [PubMed] [Google Scholar]
- 18. Silverman RH. Implications for RNase L in prostate cancer biology. Biochemistry. 2003; 42: 1805‐1812. [DOI] [PubMed] [Google Scholar]
- 19. Wang L, McDonnell SK, Elkins DA, et al. Analysis of the RNASEL gene in familial and sporadic prostate cancer. Am J Hum Genet. 2002; 71: 116‐123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Noonan‐Wheeler FC, Wu W, Roehl KA, et al. Association of hereditary prostate cancer gene polymorphic variants with sporadic aggressive prostate carcinoma. Prostate. 2006; 66: 49‐56. [DOI] [PubMed] [Google Scholar]
- 21. Knapp S, Yee LJ, Frodsham AJ, et al. Polymorphisms in interferon‐induced genes and the outcome of hepatitis C virus infection: roles of MxA, OAS‐1 and PKR. Genes Immun. 2003; 4: 411‐419. [DOI] [PubMed] [Google Scholar]
- 22. Li CZ, Kato N, Chang JH, et al. Polymorphism of OAS‐1 determines liver fibrosis progression in hepatitis C by reduced ability to inhibit viral replication. Liver Int. 2009; 29: 1413‐1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Min JY, Krug RM. The primary function of RNA binding by the influenza A virus NS1 protein in infected cells: inhibiting the 2′‐5′ oligo(A)synthetase/RNase L pathway. Proc Natl Acad Sci U S A. 2006; 103: 7100‐7105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Urosevic N. Is flavivirus resistance interferon type I‐independent? Immunol Cell Biol. 2003; 81: 224‐229. [DOI] [PubMed] [Google Scholar]
- 25. Yakub I, Lillibridge KM, Moran A, et al. Single nucleotide polymorphisms in genes for 2′‐5′‐oligoadenylate synthetase and RNase L in patients hospitalized with West Nile virus infection. J Infect Dis. 2005; 192: 1741‐1748. [DOI] [PubMed] [Google Scholar]
- 26. Behera AK, Kumar M, Lockey RF, Mohapatra SS. 2′‐5′ Oligoadenylate synthetase plays a critical role in interferon‐gamma inhibition of respiratory syncytial virus infection of human epithelial cells. J Biol Chem. 2002; 277: 25601‐25608. [DOI] [PubMed] [Google Scholar]
- 27. Hamano E, Hijikata M, Itoyama S, et al. Polymorphisms of interferon‐inducible genes OAS‐1 and MxA associated with SARS in the Vietnamese population. Biochem Biophys Res Commun. 2005; 329: 1234‐1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Fedetz M, Matesanz F, Caro‐Maldonado A, et al. OAS1 gene haplotype confers susceptibility to multiple sclerosis. Tissue Antigens. 2006; 68: 446‐449. [DOI] [PubMed] [Google Scholar]
- 29. O'Brien M, Lonergan R, Costelloe L, et al. OAS1: a multiple sclerosis susceptibility gene that influences disease severity. Neurology. 2010; 75: 411‐418. [DOI] [PubMed] [Google Scholar]
- 30. Bonnevie‐Nielsen V, Martensen PM, Justesen J, et al. The antiviral 2′,5′‐oligoadenylate synthetase is persistently activated in type 1 diabetes. Clin Immunol. 2000; 96: 11‐18. [DOI] [PubMed] [Google Scholar]
- 31. Field LL, Bonnevie‐Nielsen V, Pociot F, Lu S, Nielsen TB, Beck‐Nielsen H. OAS1 splice site polymorphism controlling antiviral enzyme activity influences susceptibility to type 1 diabetes. Diabetes. 2005; 54: 1588‐1591. [DOI] [PubMed] [Google Scholar]
- 32. Bonnevie‐Nielsen V, Field LL, Lu S, et al. Variation in antiviral 2′,5′‐oligoadenylate synthetase (2′5′AS) enzyme activity is controlled by a single‐nucleotide polymorphism at a splice‐acceptor site in the OAS1 gene. Am J Hum Genet. 2005; 76: 623‐633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Zhang J, Wheeler DA, Yakub I, et al. SNPdetector: a software tool for sensitive and accurate SNP detection [serial online]. PLoS Comput Biol. 2005; 1: e53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Menashe I, Rosenberg PS, Chen BE. PGA: power calculator for case‐control genetic association analyses. BMC Genet. 2008; 9: 36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Fears TR, Brown CC. Logistic regression methods for retrospective case‐control studies using complex sampling procedures. Biometrics. 1986; 42: 955‐960. [PubMed] [Google Scholar]
- 36. Mullan PB, Hosey AM, Buckley NE, et al. The 2,5 oligoadenylate synthetase/RNaseL pathway is a novel effector of BRCA1‐ and interferon‐gamma‐mediated apoptosis. Oncogene. 2005; 24: 5492‐5501. [DOI] [PubMed] [Google Scholar]
- 37. He WW, Sciavolino PJ, Wing J, et al. A novel human prostate‐specific, androgen‐regulated homeobox gene (NKX3.1) that maps to 8p21, a region frequently deleted in prostate cancer. Genomics. 1997; 43: 69‐77. [DOI] [PubMed] [Google Scholar]
- 38. Lim JK, Lisco A, McDermott DH, et al. Genetic variation in OAS1 is a risk factor for initial infection with West Nile virus in man [serial online]. PLoS Pathog. 2009; 5: e1000321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Tessier MC, Qu HQ, Frechette R, et al. Type 1 diabetes and the OAS gene cluster: association with splicing polymorphism or haplotype? J Med Genet. 2006; 43: 129‐132. [DOI] [PMC free article] [PubMed] [Google Scholar]


