Abstract
Data are presented demonstrating that DNA damage leads to specific post-translational modifications of p53 protein. Using two-dimensional peptide mapping of in vivo radiolabeled p53 tryptic phosphopeptides, recombinant truncated p53 protein, and synthetic p53 tryptic peptides, a unique p53 phosphopeptide was identified after exposure of ML-1 cells to ionizing irradiation. This peptide represents the first 24 amino acids of p53 and contains three phosphorylated serine residues. A specific p53 phosphopeptide antibody identified serine-15 as one of the two serines in p53 that becomes phosphorylated following DNA damage induced by either ionizing irradiation (IR) or ultraviolet (UV) irradiation in multiple cell types. IR-induced phosphorylation of p53 does not affect the kinetics of p53 binding to or dissociating from DNA as assessed by electrophoretic mobility-shift assays. However, p53 phosphorylation induced by DNA damage correlates with enhanced transcription of downstream p53 target genes. Low levels of phosphoserine-15 p53 are detectable within 6 hr after IR in AT cells, whereas lymphoblasts from normal individuals exhibit this modification within 1 hr. In contrast, phosphorylation of p53 on serine-15 is similar in normal and AT cells after UV irradiation. Our results indicate that p53 is phosphorylated in response to DNA damage, that this de novo phosphorylation may be involved in the subsequent induction and activation of p53, and that although ATM affects the kinetics of p53 phosphorylation after IR, it is not absolutely required for phosphorylation of p53 on serine-15.
Keywords: p53, phosphorylation, ATM, genotoxic stress
The p53 tumor suppressor protein is a transcription factor that is activated in response to a variety of DNA-damaging agents, including ionizing irradiation (IR) (Kastan et al. 1991), ultraviolet (UV) irradiation (Maltzman and Czyzyk 1984; Nelson and Kastan 1994), hypoxia (Graeber et al. 1994), and ribonucleoside triphosphate depletion (Linke et al. 1996). In response to DNA damage, there is an accumulation of p53 protein through post-transcriptional mechanisms and a subsequent increase in p53 activity (Maltzman and Czyzyk 1984; Kastan et al. 1991, 1992; El-Deiry et al. 1993). Activation of p53 results either in a G1 cell cycle arrest or apoptosis that contributes to suppression of malignant transformation and the maintenance of genomic integrity (Hartwell and Kastan 1994).
The mechanism by which p53 becomes activated after DNA damage is unknown. It has been demonstrated previously that cells with mutant atm (mutated in ataxia telangiectasia) alleles show a defect in the accumulation of p53 protein in response to IR (Kastan et al. 1992; Khanna and Lavin 1993; Canman et al. 1994). The failure of cells derived from ataxia telangiectasia patients to optimally induce p53 in response to IR is also manifested in a failure to induce p53 target genes (Canman et al. 1994; Lavin et al. 1994). Therefore, ATM acts upstream of p53 in the cellular response to IR. It is not known whether ATM directly or indirectly activates p53 in the IR-induced signal transduction pathway.
Post-translational modification involving phosphorylation is one potential mechanism through which the activity of p53 protein may be regulated. Protein kinases, including casein kinase I (Milne et al. 1992), DNA-PK (Lees-Miller et al. 1992), MAP kinase (Milne et al. 1994), and c-Jun kinase (Milne et al. 1995), have been shown to phosphorylate the amino terminus of p53 in vitro. However, the in vivo significance of the amino-terminal p53 phosphorylation sites identified in in vitro studies is unclear. Several observations suggest that phosphorylation within the amino terminus of p53 may have functional consequences in cell lines that overexpress recombinant p53 protein. Simultaneous mutation of serine-9, serine-18, and serine-37 within the transactivation domain of murine p53 (which corresponds to serine-6, serine-15, and serine-33 in human p53) significantly reduced the ability of p53 to suppress transformation of rat embryo fibroblasts transfected with E1A and ras (Mayr et al. 1995). When transfected into a p53 null cell line, this triple mutant p53 construct also displayed decreased transactivation of a CAT reporter plasmid (Mayr et al. 1995). Interestingly, single point mutations of either serine-9, serine-18, or serine-37 in murine p53 had no effect in these assays (Mayr et al. 1995). If aspartic acid, which mimics the charge of a phosphorylated serine, was substituted for serine-9, serine-18, and serine-37 within p53, the phenotype of cells transfected with the recombinant protein resembled that of wild-type p53. These data suggest that phosphorylation of a minimum of two serines within the first 37 amino acids of p53 is important for the function of p53 with regard to transactivation and suppression of transformation. In another study, overexpression of human p53S15A mutant protein in either T98G glioblastoma cells or p53 null Saos-2 cells resulted in partial failure of the mutant protein to inhibit cell cycle progression when compared with cells overexpressing wild-type or p53S37A mutant protein (Fiscella et al. 1993).
In vitro studies have also demonstrated the ability of several kinases to phosphorylate specific residues within the carboxy-terminal regulatory domain of p53 (Ko and Prives 1996). These kinases include cdc2 (Bischoff et al. 1990), casein kinase II (Hall et al. 1996), protein kinase C (Baudier et al. 1992; Takenaka et al. 1995), and the CDK7–cycH–p36 complex (CAK) (Lu et al. 1997). All of these kinases enhanced the in vitro sequence-specific DNA-binding capability of p53 (Hupp et al. 1992; Takenaka et al. 1995; Wang and Prives 1995; Lu et al. 1997). However, the physiologic significance of p53 phosphorylation by these kinases has yet to be determined.
To address whether phosphorylation of p53 may have physiologic significance, we asked whether endogenous p53 is phosphorylated de novo in response to a signal transduction pathway initiated by DNA damage. We demonstrate that in response to DNA damage, p53 is specifically phosphorylated on two serines within the first 24 amino acids of the protein. An antibody raised against a phosphoserine-15 p53 peptide identified serine-15 as one of the two serines that became phosphorylated in response to both IR and UV irradiation. Post-translational modification of p53 in response to IR correlated with both increased levels of p53 protein as well as the ability of p53 to transactivate p21waf1 and mdm2 in vivo. In addition, we present evidence that p53 becomes phosphorylated on serine-15 when cells from ataxia telangiectasia (AT) patients are exposed to either IR or UV irradiation. The implications of these findings with respect to the kinases that are activated in response to DNA damage and the functional significance of p53 phosphorylation are discussed.
Results
p53 is phosphorylated in response to IR in vivo
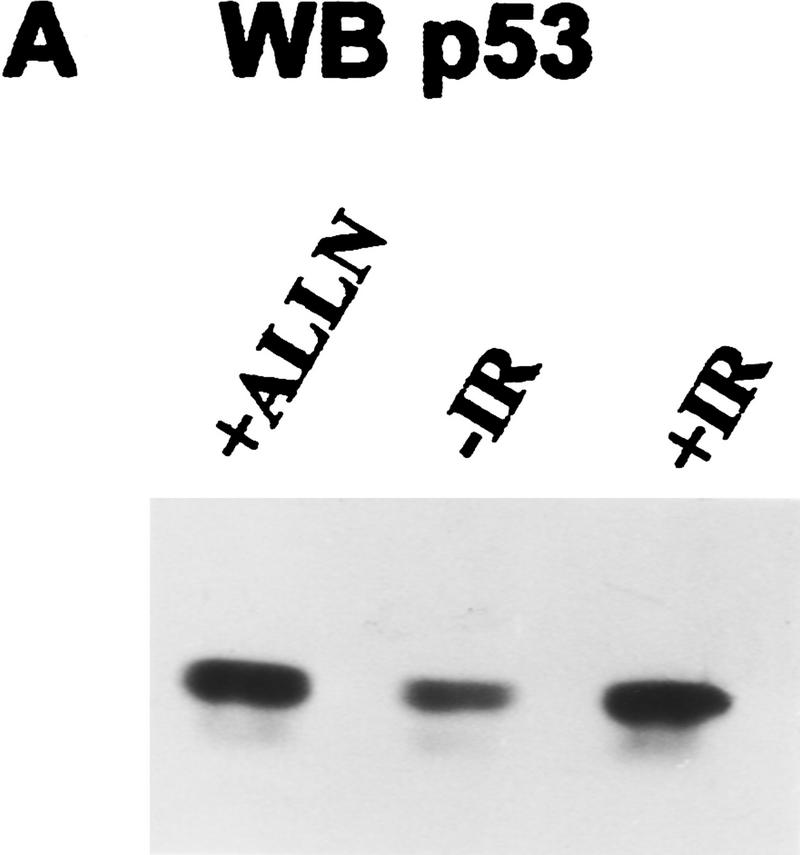
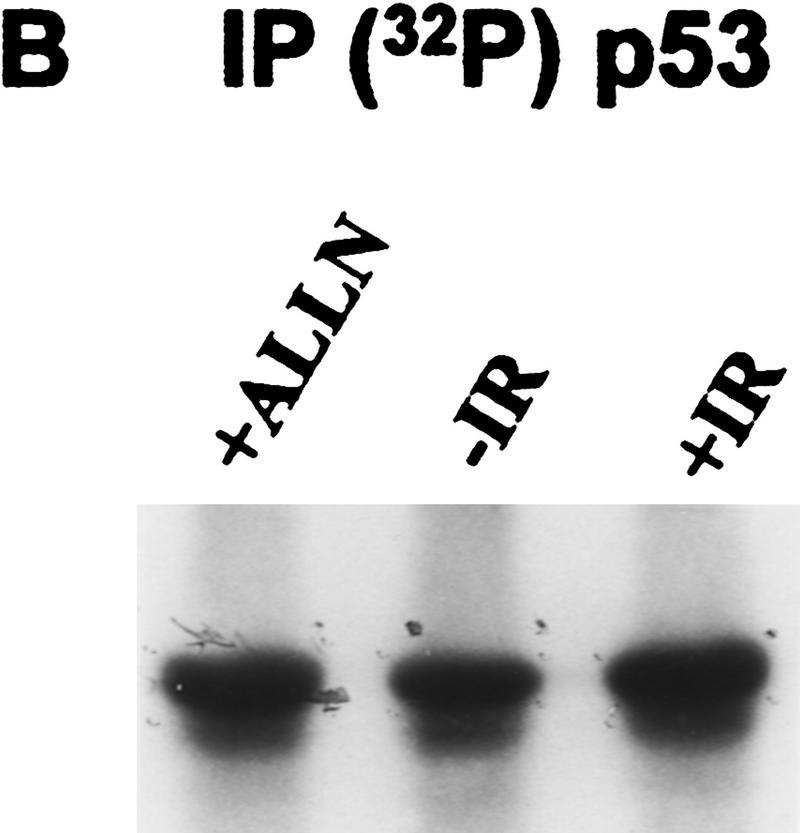
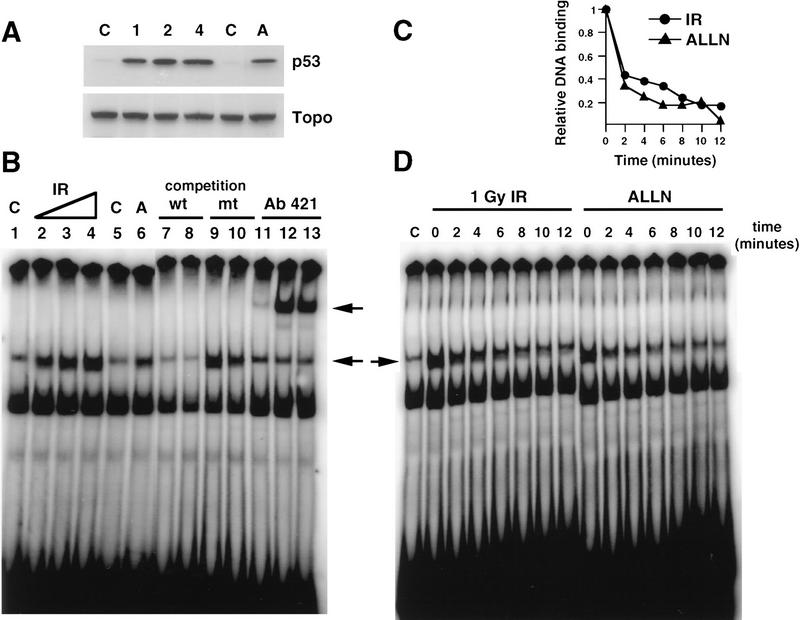
p53 tumor suppressor protein is a transcription factor that is activated in response to a variety of DNA-damaging agents, including IR (Kastan et al. 1992). We investigated whether p53 was phosphorylated in vivo in response to a signal transduction pathway initiated by IR. The myeloid leukemia cell line ML-1 has wild-type p53, and the response to IR has been characterized with respect to p53 protein induction, cell cycle arrest, and induction of p21waf1, mdm2, and gadd45 mRNA (Kastan et al. 1991, 1992; Nelson and Kastan 1994). Initial experiments suggested that labeling periods longer than 30 min would induce DNA damage in the absence of an exogenous source of irradiation (data not shown). Therefore, ML-1 cells were metabolically labeled with 32P for 25 min immediately after 2 Gy of irradiation. p53 was immunoprecipitated from cell lysates and fractionated by 10% SDS-PAGE. After electrophoretic transfer, the radiolabeled protein was isolated from an Immobilon membrane, digested with TPCK–trypsin, and subjected to two-dimensional peptide mapping. To compare equivalent amounts of p53 phosphopeptides in irradiated and unirradiated cells, some cells were treated with the proteosome inhibitor acetyl-Leu-Leu-norleucinal (ALLN). ALLN stabilizes p53 protein levels by inhibiting its degradation by the proteosome (Maki et al. 1996). Under these conditions, Western blot analysis of lysates prepared from ALLN-treated or irradiated cells demonstrated equivalent amounts of p53 protein (Fig. 1A, cf. lanes 1 and 3). Cells treated with either irradiation or ALLN had increased p53 protein when compared with unirradiated cells (Fig. 1A, cf. lane 2 with lanes 1 and 3). Equivalent amounts of 32P-labeled p53 were also immunoprecipitated from cells treated with either ALLN or IR (Fig. 1B, lanes 1,3). The amount of p53 protein obtained in either case was greater than that obtained from the untreated control cells (Fig. 1B, lane 2).
Figure 1.
Immunodetection of p53 protein in ML-1 cells. (A) Western blot analysis of ML-1 lysates. ML-1 cells were untreated (−IR) or treated with either 20 μm of ALLN (+ALLN) or 2 Gy irradiation (+IR). Lysates (50 μg) from each sample were resolved by 10% SDS-PAGE and then electrophoretically transferred to nitrocellulose. p53 was detected by immunoblotting. (B) Immunoprecipitation of p53. 32P-Labeled ML-1 extracts prepared from untreated (−IR), irradiated (+IR), or ALLN-treated (+ALLN) cells were immunoprecipitated with anti-p53 antibodies. Immunoprecipitates were resolved by 10% SDS-PAGE and electrophoretically transferred to a PVDF membrane. Radiolabeled p53 was detected by autoradiography. 32P-Labeled p53 from ALLN- and IR-treated cells had approximately twice as many counts per minute as 32P-labeled p53 from unirradiated cells when the isolated bands were counted in a scintillation counter.
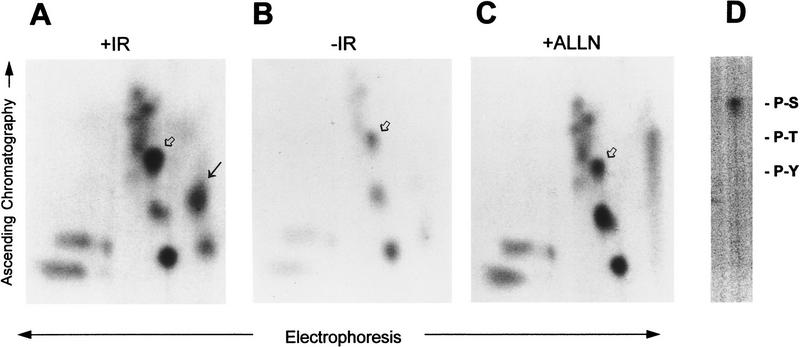
Two-dimensional peptide maps of p53 tryptic phosphopeptides demonstrated clearly the presence of an IR-induced p53 tryptic phosphopeptide (Fig. 2A, black arrow) that was not observed in p53 from unirradiated cells (Fig. 2B) or in p53 from ALLN-treated ML-1 cells (Fig. 2C). A second labeled phosphopeptide seen just below the IR-induced phosphopeptide in Figure 2A was variable in repeated experiments and sometimes was seen in unirradiated samples. The identity of this spot is unknown, and it is not yet clear whether this represents another DNA damage-induced de novo phosphopeptide. The IR-induced p53 phosphopeptide (Fig. 2A, black arrow) was eluted from the thin layer cellulose and subjected to phosphoamino acid analysis. After acid hydrolysis, the phosphorylated amino acid recovered from this peptide was serine (Fig. 2D).
Figure 2.
IR induces phosphorylation of two serines within the first 24 amino acids of p53 in vivo. p53 was immunoprecipitated from 32P-labeled ML-1 cells that were either given 2 Gy irradiation (A), untreated (B), or treated with 20 μm of ALLN (C). Proteins were resolved by 10% SDS-PAGE and electrophoretically transferred to PVDF membrane. Radiolabeled p53 was cut from the membrane and digested with TPCK–trypsin. Radiolabeled peptides were resolved by electrophoresis at pH 3.5 in the first dimension followed by ascending chromatography in the second dimension. The unique, irradiation-induced p53 phosphopeptide (A, black arrow) was eluted from the cellulose and subjected to phosphoamino acid analysis. The position of the unlabeled phosphoamino acid markers are indicated (D). A singly phosphorylated p53 synthetic peptide corresponding to amino acids 1–24 (Ac 1–24; serine-15 P) comigrated with an in vivo peptide in all three maps (A–C, open arrow). The unique, IR-induced phosphopeptide comigrated with a synthetic, triply phosphorylated p53 peptide comprising amino acids 1–24 (Ac 1–24; serine-9 P, serine-15 P, serine-20 P) (A, black arrow).
The IR-induced phosphorylation site observed in the two-dimensional peptide analysis was mapped to within the first 96 amino acids of p53. Comparison of the two-dimensional fingerprint of p53 from irradiated ML-1 cells with the fingerprints of both recombinant full-length p53 and recombinant, truncated p53 protein lacking amino acids 1–96 (wild-type full-length p53 and Δ96 p53 cDNAs; C. Prives, Columbia University, New York, NY) indicated that the de novo phosphopeptide was located within a region of p53 encompassing amino acids 1–96 (data not shown). There are three predicted tryptic fragments within this region. A series of p53 peptides corresponding to two of the three predicted tryptic fragments (amino acids 1–24 and 25–65) within the first 96 amino acids of p53 were chemically synthesized. The IR-induced p53 phosphopeptide was identified by mixing individual synthetic peptides with the endogenous p53 phosphopeptides and then analyzing the mix on a two-dimensional peptide map. Radiolabeled phosphopeptides were detected on the PhosphorImager and compared with the mobility of the synthetic peptides that were detected using ninhydrin. A synthetic, singly phosphorylated p53 peptide corresponding to amino acids 1–24 [Ac (acetylated) 1–24; serine-15 P] comigrated with a radiolabeled p53 tryptic phosphopeptide in unirradiated, irradiated, and ALLN-treated ML-1 cells (Fig. 2A–C, open arrow). The unique, IR-induced p53 phosphopeptide overlapped with a triply phosphorylated peptide comprising amino acids 1–24 (Ac 1–24; serine-9 P, serine-15 P, serine-20 P) (Fig. 2A, black arrow). An in vivo p53 peptide corresponding to amino acids 1–24 with two phosphorylated serines was not observed in any of the two-dimensional maps. Thus, the IR-induced post-translational modification of p53 involves de novo phosphorylation of two serine residues within the first 24 amino acids of p53.
Ionizing irradiation induces phosphorylation of p53 on serine-15
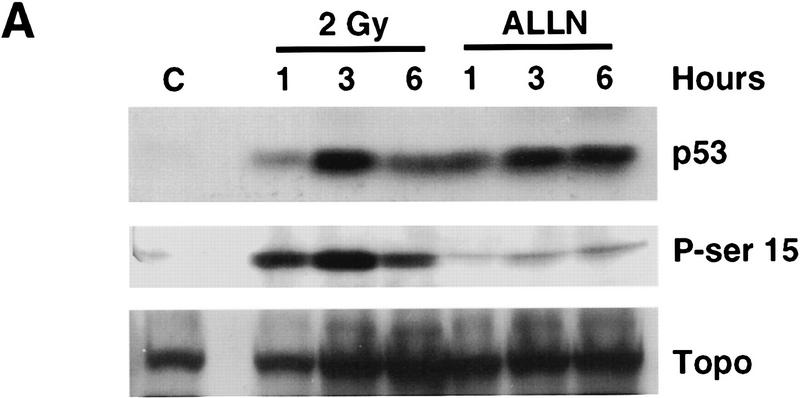
To identify the specific serine residues within amino acids 1–24 of p53 that were phosphorylated in response to IR, an affinity-purified antibody raised against a synthetic p53 serine-15 phosphopeptide (amino acids 9–21, serine-15-PO3) was used in Western blot analysis of lysates from unirradiated, irradiated, or ALLN-treated ML-1 lysates. The anti-phosphoserine-15 p53 peptide antibody used in these studies specifically recognizes p53 only when the protein is phosphorylated on serine-15; there is no immunoreactivity with unphosphorylated p53 (Shieh et al. 1997). ML-1 lysates were characterized with respect to IR-induced increase in p53 protein (Fig. 3A, top panel). Increased levels of p53 protein in ML-1 lysates were observed when cells were harvested 1 hr after 2 Gy irradiation. Levels of p53 protein reached a maximum at 3 hr after IR and then began to decline (Fig. 3A, top panel). Western blot analysis with anti-phosphoserine-15 demonstrated that in response to ionizing radiation, p53 was phosphorylated on serine-15 (Fig. 3A, middle panel). Levels of p53 phosphorylated on serine-15 increased with time after IR, with the greatest amount observed 3 hr after IR. By 6 hr after IR, phosphoserine-15 p53 immunoreactivity had declined. IR induced an increase in levels of phosphoserine-15 p53 (Fig. 3A, middle panel) that correlated with the observed increase in p53 protein (Fig. 3A, top panel). In contrast, there was barely detectable phosphoserine-15 p53 in lysates from unirradiated cells or in cells treated with ALLN (Fig. 3A, middle panel). The lack of phosphoserine-15 p53 immunoreactivity in lysates from ALLN-treated ML-1 cells cannot be attributed to lower total amount of p53 because there was an equivalent amount of p53 protein in lysates from irradiated and ALLN-treated cells (Fig. 3A, top panel). Western blot analysis with anti-topoisomerase was performed as a control for equivalent amounts of protein loaded in each lane (Fig. 3A, bottom panel). Thus, in response to a signal transduction pathway initiated by IR, p53 becomes phosphorylated on serine-15.
Figure 3.
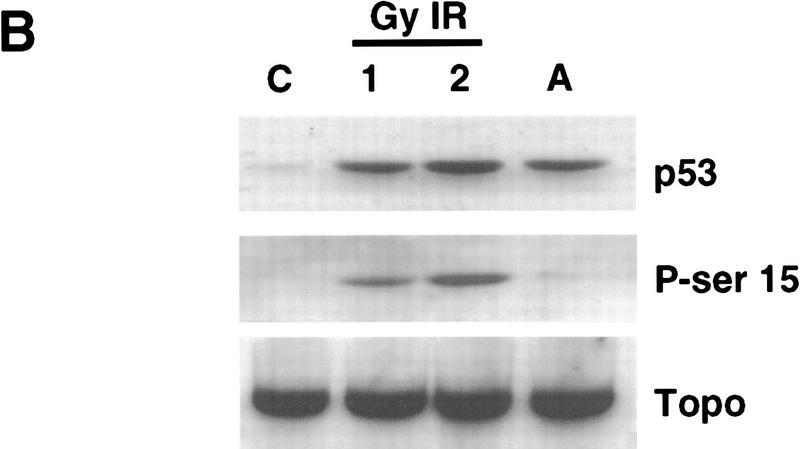
Identification of serine-15 as one of the two sites phosphorylated within p53 in response to IR. (A) Western blot analysis of ML-1 lysates. ML-1 cells were either untreated (control, C) or treated with 2 Gy irradiation or 20 μm of ALLN. Cells were harvested at 1, 3, and 6 hr after treatment. Lysates (50 μg of protein in each lane) were resolved by 10% SDS-PAGE. After electrophoretic transfer, the nitrocellulose was immunoblotted with either anti-p53 ( Ab-6, top), anti-phosphoserine-15 p53 (middle), or anti-topoisomerase (bottom). (B) Western blot analysis of SY5Y lysates. SY5Y cells were either untreated (control, C), treated with 1 or 2 Gy IR, or treated with 20 μm of ALLN. Cells were harvested 3 hr after treatment. After biochemical fractionation, 30 μg of nuclear lysate from each sample was resolved by 10% SDS-PAGE and then electrophoretically transferred to nitrocellose. The nitrocellulose was immunoblotted with either anti-p53 (Ab-6, top), anti-phosphoserine-15 p53 (middle), or anti-topoisomerase (bottom).
IR-induced phosphorylation of p53 on serine-15 was also observed in other cell lines, including a neuroblastoma cell line, SY5Y (Fig. 3B), and several B-lymphoblastoid lines (Fig. 4). Western blot analysis of SY5Y nuclear lysates demonstrated that p53 was phosphorylated on serine-15 in response to IR (Fig. 3B, middle panel). p53 serine-15 phosphorylation was not observed in either unirradiated or ALLN-treated SY5Y cells (Fig. 3B, middle panel) despite an equivalent amount of p53 protein from ALLN-treated cells as compared with irradiated cells (Fig. 3B, top panel).
Figure 4.
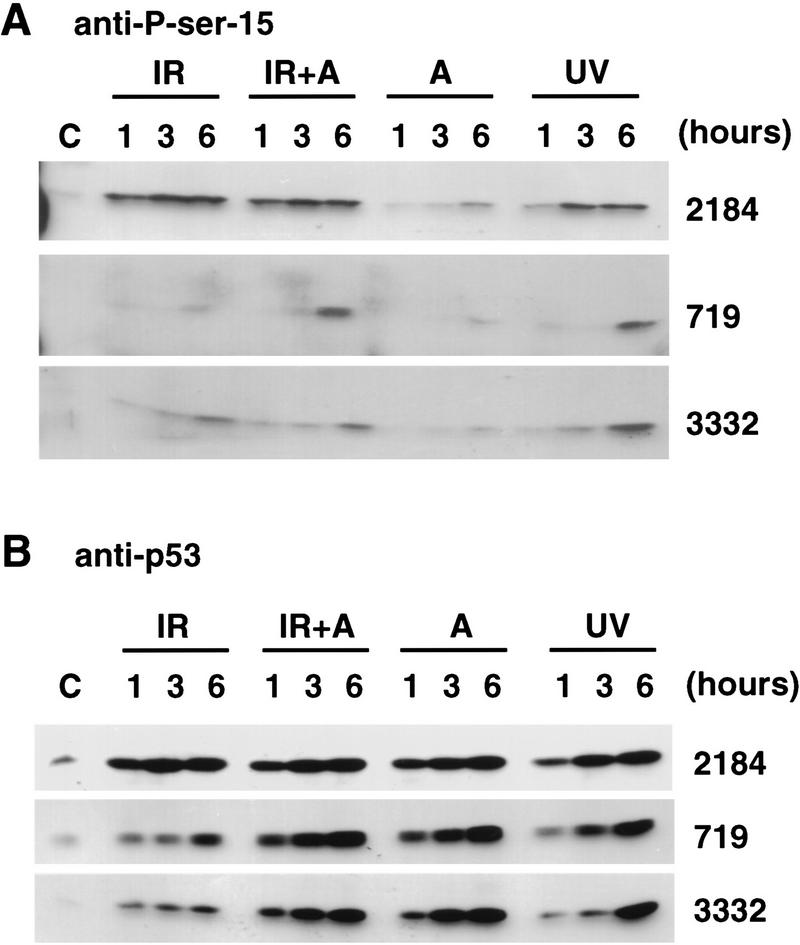
IR and UV irradiation induce phosphorylation of p53 on serine-15 in both normal and AT lymphoblasts. (A) Western blot analysis of normal and AT lymphoblasts with anti-phosphoserine-15 p53 peptide antibody. Normal lymphoblasts (2184) and AT lymphoblasts (719 and 3332) were untreated (control, C) or given either 5 Gy IR (IR), 5 Gy IR + 20 μm of ALLN (IR + A), 20 μm ALLN (A), or 10 J/m2 of UV irradiation (UV). Cells were harvested at 1, 3, and 6 hr after treatment. Lysate (50 μg) from each sample was resolved by 10% SDS-PAGE, followed by electrophoretic transfer to nitrocellulose. The nitrocellulose was immunoblotted with anti-phosphoserine-15 p53 peptide antibody. Phosphoserine-15 p53 was detected by ECL. (B) Western blot analysis of normal and AT lymphoblasts with anti-p53. The identical lysates from the above experiment were immunoblotted with anti-p53 antibody (Ab-6). p53 protein was detected with the ECL reagent.
IR and UV irradiation induce phosphorylation of p53 on serine-15 in both normal and AT lymphoblasts
The gene mutated in AT, ATM, acts upstream of p53 in the signal transduction pathway initiated by IR (Kastan et al. 1992; Canman et al. 1994; Lavin et al. 1994). Unlike normal cells, AT cells fail to rapidly induce p53 protein following IR. In contrast, UV irradiation of AT cells induces p53 with kinetics and levels similar to those in normal cells (Khanna and Lavin 1993; Canman et al. 1994). We asked whether cells with defective ATM were able to phosphorylate p53 on serine-15 in response to IR or UV irradiation. Normal B lymphoblasts (2184) and AT lymphoblasts (719 and 3332 cell lines) were treated either with 5 Gy IR, exposed to 10 J/m2 of UV irradiation, or treated with 20 μm of ALLN. In response to IR, p53 was phosphorylated on serine-15 in both normal and AT lymphoblasts. The induction of serine-15 phosphorylation correlated with the induction of total p53 protein. That is, in normal lymphoblasts, the increase in p53 protein was very rapid and occurred within 1 hr after IR (Fig. 4B, top panel); serine-15 phosphorylation also occurred within 1 hr after IR (Fig. 4A, top panel). Maximal levels of phosphoserine-15 p53 were observed 3 hr after IR (Fig. 4A). In contrast, very low levels of p53 induction were observed in both 719 and 3332 AT lymphoblasts even at 6 hr after IR (Fig. 4B, middle and bottom panels). Similarly, low but detectable levels of phosphoserine-15 p53 were also seen in irradiated AT cells (Fig. 4A, middle and bottom panels). Importantly, however, phosphoserine-15 p53 was clearly observed when irradiated AT cells were also treated with ALLN to inhibit the degradation of endogenous p53 (Fig. 4A). p53 was not phosphorylated on serine-15 when cells were either untreated or treated with ALLN alone. As a control of equivalent amounts of protein loaded in each lane, the Western blot was probed with anti-topoisomerase. There were equal amounts of topoisomerase in all samples (data not shown). In conclusion, p53 is phosphorylated on serine-15 in response to IR in normal lymphoblasts and to a much lesser extent in AT lymphoblasts.
In contrast to the delayed and low induction of p53 in AT cells in response to IR, p53 was induced with UV irradiation in both 719 and 3332 AT cell lines and normal lymphoblasts (Fig. 4B). With all three cell lines, p53 was phosphorylated on serine-15 in response to UV irradiation (Fig. 4A). In response to a specific signal induced by UV irradiation, the increased levels of phosphoserine-15 p53 correlated with increased levels of total p53 protein (Fig. 4A,B). Phosphoserine-15 p53 was not observed in either control cells or cells treated with ALLN alone, despite equivalent amounts of p53 protein in the ALLN-treated cells (Fig. 4A,B).
In conclusion, p53 is specifically phosphorylated on serine-15 in response to IR and UV irradiation in both normal and AT lymphoblasts. However, the kinetics and levels of serine-15 phosphorylated p53 in IR-treated AT cells are different from normal lymphoblasts.
IR-induced post-translational modification of p53 correlates with increased transcription of two p53-responsive genes, p21waf1 and mdm2
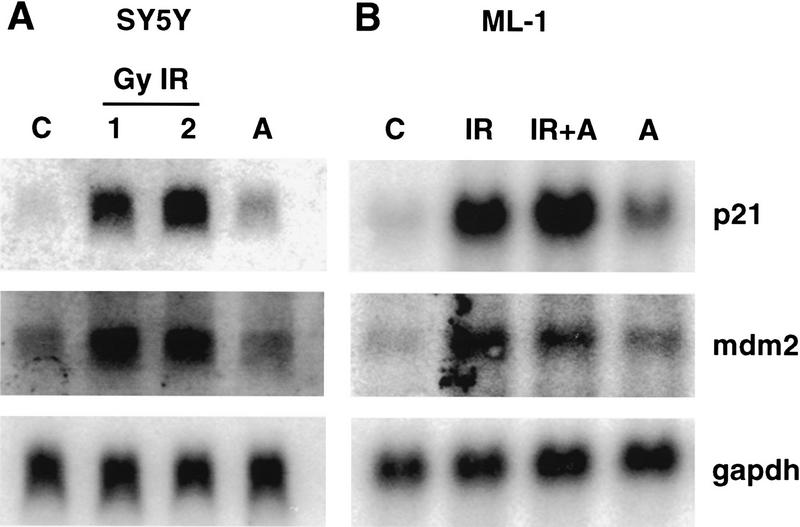
The induction of p53 in response to DNA damage results in the transcriptional activation of p53 target genes, including p21waf1, mdm2, and gadd45 (Kastan et al. 1992; El-Deiry et al. 1993). We asked whether the ability of p53 to transactivate downstream target genes was altered in SY5Y and ML-1 cells that had been treated with ALLN as compared with irradiated cells. Cellular p53 protein levels were equivalent after treatment with IR or ALLN, and both treatments led to greater levels than that observed in unirradiated cells (Fig. 6A, below). However, p53 was post-translationally modified only in irradiated cells (Fig. 2). As seen in Figure 5, there is a dramatic induction in both p21waf1 and mdm2 mRNA in cells that were irradiated (Fig. 5A, top and middle panels). In contrast, p53 did not transactivate these two genes in ALLN-treated cells despite equivalent p53 protein levels to that which was observed in irradiated cells (Fig. 5A, top and middle panels). All lanes had equivalent amounts of RNA as assessed by probing the same Northern blot with GAPDH (Fig. 5A, bottom panel). Northern blot analysis of RNA from unirradiated, irradiated, or ALLN-treated ML-1 cells gave identical results to that observed in SY5Y cells (Fig. 5B). In conclusion, IR-induced post-translational modification of p53 correlated with the ability of p53 to transactivate downstream genes.
Figure 6.
The ability of p53 to bind to or dissociate from a p21waf1 oligonucleotide is not affected by p53 phosphorylation induced by IR. (A) Western blot analysis of SY5Y lysates. Immunoblot analysis with anti-p53 (Ab-6, top) and anti-topoisomerase (bottom) of SY5Y lysates prepared from untreated cells (C), cells treated with 1, 2, or 4 Gy IR, or cells treated with 20 μm ALLN (A). (B) EMSA with SY5Y lysates and a p21waf1 probe. EMSAs of a 32P-labeled p21waf1 oligonucleotide probe were performed with nuclear lysates prepared from either untreated SY5Y cells (lanes 1,5,11), cells given 1 Gy (lane 2), 2 Gy (lanes 3,7,9,12), or 4 Gy (lane 4) irradiation, or cells treated with 20 μm ALLN (lanes 6,8,10,13). Specificity of binding of p53 to the labeled probe was confirmed by competition with either 40-fold excess unlabeled wild-type probe (lanes 7,8) or with excess unlabeled mutant probe (lanes 9,10). Specific binding of p53 to the labeled probe is indicated by the lower arrow. Antibody 421 supershifted this complex (lanes 11–13, upper arrow). (C) Quantitation of rate of p53 dissociation from 32P-labeled p21waf1 oligonucleotide probe. Nuclear lysates were prepared from irradiated (•) or ALLN-treated (▴) SY5Y cells. Binding of p53 to radiolabeled probe was quantitated on a PhosphorImager. (D) Analysis of the dissociation rate of p53 from p21waf1 oligonucleotide probe. SY5Y nuclear lysates prepared from unirradiated (C), irradiated (1Gy IR), or ALLN-treated cells were incubated with radiolabeled p21wafl for 20 min. Excess unlabeled wild-type probe was added. Aliquots of the reaction were applied to a progressively running native gel at various times (0, 2, 4, 6, 8, 10, and 12 min) after the addition of unlabeled wild-type competitor. The gel was dried down and visualized on a PhosphorImager.
Figure 5.
Increased transcription of p21waf1 and mdm2 correlates with IR-induced post-translational modification of p53. (A) Northern blot analysis of SY5Y RNA. Total RNA isolated from SY5Y cells that had been either untreated (control, C) or treated with 1 or 2 Gy IR or treated with 20 μm ALLN (A) was assayed for hybridization to 32P-labeled p21waf1 cDNA (top), 32P-labeled mdm2 cDNA (middle), or 32P-labeled GAPDH cDNA (bottom) coding sequence probes. (B) Northern blot analysis of ML-1 RNA. Total RNA was isolated from ML-1 cells that were either untreated (C) or treated with 5 Gy IR (+IR), 5 Gy IR + 20 μm ALLN (IR + A), or 20 μm ALLN (A). The nitrocellulose was sequentially assayed for hybridization to p21, mdm2, and gadph-labeled cDNA probes (top, middle, and bottom, respectively).
IR-induced phosphorylation of p53 does not affect the ability of p53 to bind to or dissociate from DNA as assessed by electrophoretic mobility shift assays
The above experiments demonstrated that in response to a signal induced by IR, p53 is specifically phosphorylated. Phosphorylation of p53 correlated with both induction of p53 protein and increased transcription of p53-responsive genes. We asked whether the differences in p53 transactivation activity could in part be explained by differential binding of p53 to specific DNA sequences in an electrophoretic mobility shift assay (EMSA). SY5Y nuclear extracts were isolated 3 hr after treatment of cells with 20 μm of ALLN or increasing doses of IR. The extent of p53 binding to an oligonucleotide containing the p21waf1 promoter binding site was compared among the different extracts. IR induced a dose-dependent increase in p53-specific DNA-binding activity (Fig. 6B, lanes 2–4). The increased binding of p53 to the labeled probe was attributable to the increase in p53 protein at higher doses of IR (Fig. 6A). p53 in lysates from ALLN-treated cells was also able to bind to the DNA probe (Fig. 6B, lane 6); the amount of binding to DNA at equilibrium was equivalent to that observed in lysates isolated from cells given 1 Gy IR (Fig. 6B, cf. lane 6 with lane 2). Specificity of DNA binding was verified by competition experiments with either 40-fold excess wild-type (Fig. 6B, lanes 7,8) or mutant oligonucleotide (Fig. 6B, lanes 9,10) and antibody 421 supershift (Fig. 6B, lanes 11–13). In conclusion, there was not a dramatic difference in p53 binding at equilibrium to a p21waf1 oligonucleotide probe when lysates prepared from ALLN- or IR-treated cells were compared.
We then asked whether p53, which had been post-translationally modified in response to IR, had altered stability when bound to DNA. We compared the dissociation rates of p53 from different lysates in an off-rate experiment using p21waf1 oligonucleotide as probe. SY5Y nuclear lysates prepared from control, ALLN, or irradiated cells were incubated with labeled oligonucleotide until equilibrium was achieved. The rate at which p53 dissociated from the radiolabeled probe was measured after the addition of 40-fold excess of wild-type, unlabeled probe (Fig. 6C,D). IR-induced post-translational modification of p53 did not appear to alter the binding stability of p53 to the p21waf1 oligonucleotide. There was no significant difference in dissociation rate of p53 to probe when lysates from IR- or ALLN-treated cells were compared (Fig. 6C,D).
Discussion
p53 is a critical mediator of cellular responses to genotoxic stress. Following DNA damage, there is a post-transcriptional induction of p53 protein and subsequent activation of p53. Ultimately, activation of p53 leads to either G1 arrest or cell death. The mechanism by which p53 becomes activated is unknown. Phosphorylation is one potential mechanism by which cells might regulate the activity of p53 after genotoxic stress. In vivo experiments have demonstrated clearly that p53 is a phosphoprotein (Ko and Prives 1996). In vitro studies have identified multiple sites of phosphorylation within the amino and carboxyl termini within p53. Although several kinases have been shown to phosphorylate specific sites within p53 in vitro, it has not yet been demonstrated that phosphorylation at these sites is important for the function and regulation of p53 in vivo. Interestingly, Tegtmeyer and colleagues have demonstrated that simultaneous mutation of several serines within the amino terminus of p53 has a biologic effect, namely, a reduction in the ability of the recombinant, overexpressed p53 to suppress transformation as well as a decrease in the ability of p53 to transactivate a reporter construct (Mayr et al. 1995). In addition, Fiscella et al. (1993) reported that mutation of serine-15 results in partial failure of p53 to inhibit cell cycle progression.
Interest in the biological significance of phosphorylation of p53 at the carboxyl terminus results from the observations from several laboratories that the carboxyl terminus of p53 acts to regulate negatively the sequence-specific DNA-binding ability of p53 (Hupp et al. 1992; Gottlieb and Oren 1995; Ko and Prives 1996). Phosphorylation at the carboxyl terminus of p53 may be important in this regulation. Several protein kinases have been demonstrated to phosphorylate p53 within this domain. In all cases, phosphorylation within the carboxyl terminus of p53 enhanced the ability of p53 to bind DNA in vitro. However, as is the case for phosphorylation sites within the amino terminus of p53, de novo phosphorylation of p53 at the carboxyl terminus in response to a signal transduction pathway in vivo has not been demonstrated.
In this study we demonstrated that endogenous p53 becomes phosphorylated within the transactivation domain of the protein in response to a signal transduction pathway initiated by DNA damage. We identified the site of de novo phosphorylation as within amino acids 1–24 of p53. In addition, we showed that post-translational modification of serine-15 within p53 occurs in response to both UV irradation and IR. In response to DNA damage, phosphorylation of p53 on serine-15 occurs in several different cell types, including cells derived from AT patients. These data also demonstrate that the signal transduction pathway activated following IR leads to phosphorylation of an additional serine residue within amino acids 1–24 of p53 (serine-6, serine-9, or serine-20).
Several kinases have been implicated in signal transduction pathways initiated by IR and UV irradiation. These kinases are potential candidates that may be involved, directly or indirectly, in the in vivo post-translational modification of p53 that we have characterized. First, ATM protein, encoded by the gene mutated in AT, may activate p53 in response to DNA damage. ATM is a member of a family of proteins involved in cell cycle control and DNA repair and recombination (Zakian 1995; Shiloh and Rotman 1996; Morgan and Kastan 1997). Although it has not been demonstrated formally that ATM is a protein kinase, a domain within ATM is homologous to phosphatidylinositol-3-kinase (Savitsky et al. 1995). ATM has been shown to act upstream of p53 in the signal transduction pathway initiated by IR as cells with defective ATM poorly induce p53 protein following IR (Canman et al. 1994; Xu and Baltimore 1996). Given these observations, it is possible that ATM phosphorylates p53 in response to IR. However, the data presented here suggest a more complex scenario as IR induced p53 serine-15 phosphorylation in cells that lack ATM. In addition, UV irradiation of AT cells also results in phosphorylation of p53 on serine-15. Therefore, in response to specific types of DNA damage, a particular signal transduction pathway is initiated, and ATM is not absolutely required for p53 phosphorylation on serine-15. It is possible, however, that ATM participates in signaling to the kinase that phosphorylates p53 after IR; a defect in ATM would contribute to the altered induction of p53 phosphorylation in AT cells. It remains to be determined whether ATM can phosphorylate p53 at an alternative site, namely, the additional serine within amino acids 1–24 that is also phosphorylated in response to IR.
It is also possible that other ATM family members, such as DNA-PK, can phosphorylate p53 in response to DNA damage. DNA-PK is a serine/threonine kinase that is activated by double-strand breaks in DNA and is involved in DNA double-strand-break repair and V(D)J recombination (Danska et al. 1996). DNA-PK has been shown to phosphorylate human p53 on serine-15 and serine-37 in vitro (Lees-Miller et al. 1992). However, cells from scid mice, which have defective DNA-PK, are still able to induce p53 and undergo G1 arrest in response to IR, making this kinase an unlikely candidate (Bogue et al. 1996; Guidos et al. 1996; Huang et al. 1996; Nacht et al. 1996; Rathmell et al. 1997). Clearly, additional experiments are necessary to determine whether ATM or related proteins directly phosphorylate p53 in vivo in response to IR.
A second candidate kinase that may phosphorylate p53 in response to IR is casein kinase I (CKI). In vitro experiments have demonstrated that CKI phosphorylates murine p53 at multiple sites within the amino terminus. In addition, CKI from Drosophila melanogaster becomes activated when Drosophila embryos are exposed to low doses of IR (Santos et al. 1996). It is possible that IR activates CKI, which then phosphorylates p53 within the amino terminus.
Third, UV irradiation-induced DNA damage may activate a kinase that is able to phosphorylate p53 on serine-15 or other sites within the amino terminus. This kinase could be a different kinase than the one activated by IR. Candidate kinases that may be involved in UV irradiation-induced p53 phosphorylation are the Jun kinases, which have been shown to be activated by UV irradiation (Derijard et al. 1994; Rosette and Karin 1996). In vitro experiments have demonstrated that c-Jun kinase can phosphorylate both a murine p53 recombinant peptide on serine-34 (Milne et al. 1995) and recombinant GST–p53 protein (Adler et al. 1997). It will be important to determine in vivo whether Jun kinases directly phosphorylate p53 on serine-15 or other serines within p53 in response to UV irradiation.
We have demonstrated that although inhibition of p53 degradation by ALLN can elevate p53 concentration to levels equivalent to those seen in cells that have been irradiated, stabilized p53 from ALLN-treated cells was unable to transactivate several p53 target genes in vivo. This was not attributable to the inability of p53 to bind DNA. EMSAs demonstrated that p53 in lysates prepared from either ALLN- or IR-treated SY5Y cells was capable of binding to and dissociating from DNA to the same extent. We have also shown that p53 is phosphorylated in response to DNA damage, whereas ALLN treatment raises p53 protein levels but does not induce p53 phosphorylation. Therefore, de novo phosphorylation of p53 within its transactivation domain, rather than an increase in p53 levels per se, correlated with an increase in the ability of p53 to transactivate downstream genes in response to IR. There are several possible mechanisms that may explain how IR-induced phosphorylation of the amino terminus of p53 affects the ability of p53 to transactivate its target genes. First, IR-induced phosphorylation of p53 at these sites might affect the ability of p53 to interact with other protein components of the transcriptional machinery that are required for transactivation of p53 target genes. For example, the TATA box-binding protein (TBP), the subunit of the basal transcription factor TFIID, binds to the amino terminus of p53 (Farmer et al. 1996). In addition, several TBP-associated factors (TAFs) also bind to the amino terminus of p53, including dTAFII40 and dTAFII60 (and the corresponding human homologs TAFII32 and TAFII70) (Thut et al. 1995). Optimal interaction of these transcriptional activators with p53 might require post-translational modification within the amino terminus of p53.
Second, IR-induced phosphorylation of p53 might affect its interaction with Mdm2. The direct interaction of Mdm2 with p53 results in inhibition of p53 transcriptional activity (Momand et al. 1992; Zauberman et al. 1993). It has been demonstrated recently that the mechanism for this inhibition involves binding Mdm2 to p53 and targeting p53 for rapid degradation (Haupt et al. 1997; Kubbutat et al. 1997). Binding of Mdm2 to p53 has been mapped to the amino terminus of p53 (Chen et al. 1993; Picksley et al. 1994). IR-induced phosphorylation of p53 may therefore affect this interaction by interfering with the ability of Mdm2 to bind p53 (Shieh et al. 1997). Third, IR-induced phosphorylation of p53 might result in a conformational change in p53 that affects directly the ability of p53 to transactivate downstream genes. Fourth, the increased transcription of p21waf1 and mdm2 in response to IR but not observed with ALLN-treated cells could be attributable to IR-induced modification and/or alteration in activity of proteins other than p53.
Activation of p53 is a critical component in the cellular response to DNA damage. It will be important to determine if the specific phosphorylation of p53 described here affects whether a cell undergoes a G1 checkpoint arrest versus apoptosis in response to irradiation. In addition, it will be important to delineate whether de novo p53 phosphorylation also occurs when cells are grown either under hypoxic conditions or under conditions where ribonucleotide pools are altered. Finally, identification of the kinases that phosphorylate p53 in vivo will further elucidate the signal transduction pathway initiated by cells in response to DNA damage.
Materials and methods
Cell lines
Epstein–Barr virus (EBV)-immortalized normal human B-lymphoblast line 2184 and the EBV-immortalized human AT homozygote lymphoblast lines 1526, 719, and 3332 were obtained from the Human Genetic Mutant Cell Repository (Camden, NJ). The human SH-SY5Y neuroblastoma cell line was generously provided by G. Brodeur (The Children’s Hospital of Philadelphia, PA). The human myeloid leukemia cell line, ML-1, has been described previously (Kastan et al. 1991). All cell lines were grown in RPMI medium supplemented with 10% fetal bovine serum, gentamicin, and glutamine.
Induction of DNA damage
Cells were irradiated with a 137Cs source and received 2 Gy IR or treated with UV irradiation at 2 J/m2 per sec as described previously (Canman et al. 1994; Nelson and Kastan 1994). The proteosome inhibitor ALLN (Sigma) was added to cells at a final concentration of 20 μm. Cells that received both irradiation and ALLN were first irradiated and then immediately given ALLN. Cells were harvested at indicated times after DNA damage or ALLN treatment.
Metabolic labeling and immunoprecipitation of p53
Exponentially growing ML-1 cells were washed with phosphate-free RPMI supplemented with 10% dialyzed fetal bovine serum and further incubated in the same medium for 30 min before irradiation. The cells were irradiated at 2 Gy and then [32P]orthophosphate (NEN) was added to the cells in phosphate-free medium (0.6 mCi/ml). The cells were labeled for 25 min at 37°C and then harvested by washing once in ice-cold PBS. Cells were lysed at 4°C in ice-cold lysis buffer (250 mm NaCl, 1% NP-40, 20 mm Tris-HCl at pH 7.4, 1 mm EDTA, 2 μg/ml of aprotinin, 2 μg/ml of leupeptin, 1 μg/ml of pepstatin, 2.5 μg/ml of antipain, 1 μg/ml of chymostatin, 1 mm Na3VO4, 10 mm NaF, 1 mm sodium molybdate, and 0.5 mm PMSF). Lysates were clarified at 12,000g for 15 min at 4°C. Ten micrograms of affinity-purified mouse monoclonal anti-p53 antibodies (Ab-2 and Ab-6, Calbiochem) were added to the lysates. As a control for nonspecific binding, an immunoprecipitation reaction was also done with an irrelevant primary mouse monoclonal antibody of the same isotype as the anti-p53 antibodies. Immunoprecipitation reactions were incubated on ice at 4°C for 2 hr. Protein G Plus/Protein A Agarose (Calbiochem) was added, and lysates were further incubated at 4°C for 1 hr, with rocking. The beads were collected by centrifugation and washed three times with ice-cold wash buffer (150 mm NaCl, 1% NP-40, 20 mm Tris-HCl at pH 7.4, 1 mm EDTA, and the same protease inhibitors as in the lysis buffer). The beads were boiled in SDS sample buffer, and immunoprecipitated p53 was electrophoresed on a 10% SDS–polyacrylamide gel. Protein was electrophoretically transferred to Immobilon-P membrane [polyvinylidene difluoride (PVDF) membrane, Millipore]. Radiolabeled p53 protein was visualized by autoradiography, and the appropriate band was cut out and prepared for tryptic digest as described below.
Two-dimensional phosphopeptide mapping and phosphoamino acid analysis
After transfer to a PVDF membrane, the radiolabeled p53 band was cut from the membrane and digested with TPCK–trypsin (Sigma) according to the methods described by Boyle et al. (1991). Peptides were resolved in two dimensions on a TLC cellulose sheet (EM Sciences) by electrophoresis in the first dimension followed by ascending chromatography in the second dimension. 32P-Labeled tryptic peptides were visualized on a PhosphorImager (Molecular Dynamics). 32P-Labeled phosphopeptides were recovered from the thin-layer cellulose plate and hydrolyzed in 6 n hydrochloric acid for 60 min at 110°C, and the phosphoamino acid content of the hydrolyzed peptides was determined by the method of Boyle et al. (1991). [32P]-Phosphoamino acids were detected with a PhosphorImager. Phosphoamino acid standards were visualized by spraying the plate with 1% (wt/vol) ninhydrin in acetone.
Synthesis of phosphorylated p53 peptides
Peptides were synthesized by the solid-phase method with Fmoc chemistry using an Applied Biosystems 430A peptide synthesizer (Foster City, CA). Phosphoserine residues were incorporated as Fmoc-Ser[PO(OBzl)OH]-OH (Novabiochem, San Diego, CA) (Wakamiya et al. 1994). Cleavage of the peptide from the resin and removal of the side-chain protecting groups were performed using reagent K (TFA/phenol/thioanisole/H2O/EDT = 82.5:5:5:5:2.5) for 3 hr at room temperature (King et al. 1990). The peptides were purified by HPLC on a pH-stable Vydac C-8 column (Hesperia, CA) with 0.2% hexafluoroaceton-NH4OH (pH 7.0)/acetonitrile. The mass of the peptides was confirmed by electrospray ionization mass spectrometry on a Finnigan MAT SSQ 7000 (Finnigan MAT, San Jose, CA).
Immunoblot analysis
Cells were harvested, solubilized in Laemmli sample buffer, fractionated by SDS/10% PAGE, and electrophoretically transferred to nitrocellulose. Affinity-purified antibodies to p53 (Ab-6, Calbiochem) and topoisomerase (Topogen) were used for immunochemical detection as described (Canman et al. 1994). The phosphoserine-15 p53 peptide antibodies were raised in rabbits against a chemically synthesized p53 phosphoserine-15 peptide (amino acids 9–21) as described previously (Kitagawa et al. 1996). For immunodetection of serine-15-phosphorylated p53 protein, lysates were fractionated by SDS/10% PAGE and electrophoretically transferred to nitrocellulose. Nonspecific binding was blocked by incubating the membrane in PBST (PBS with 0.05% Tween -20 and 1% nonfat dried milk). The membrane was incubated with the primary antibody, which had been preincubated previously with 1 μg/ml of unphosphorylated p53 peptide, amino acids 9–21, washed several times with PBST, followed by incubation with a HRP-conjugated goat anti-rabbit secondary antibody (Pierce). After several washes, p53 protein specifically phosphorylated on serine-15 was detected using the enhanced chemiluminescence (ECL) reagent (Amersham).
RNA analysis
Total RNA was isolated from cells with guanidine isothiocyanate–phenol–chloroform, fractionated by electrophoresis on a formaldehyde–agarose gel, and transferred to nitrocellulose. RNA was assayed for hybridization to 32P-labeled human cDNA probes comprising either the entire p21waf1, mdm2, or gadph coding sequences as described previously (Canman et al. 1995).
EMSAs
Approximately 20–30 million cells were lysed on ice in buffer containing 20 mm HEPES (pH 7.6), 25% glycerol, 1.5 mm MgCl2, 10 mm NaCl, 0.2 mm EDTA, 2 mm DTT, 0.1% Triton X-100, 1 mm PMSF, 2 μg/ml of pepstatin A, 20 μg/ml of leupeptin, 10 μg/ml of aprotinin, 1 mm sodium orthovanadate, and 1 mm NaF. Nuclei were pelleted, extracted for 30 min in the above buffer that had been adjusted to 400 mm NaCl, and then cleared by centrifugation at 13,000g for 15 min. For DNA-binding assays, 30 μg of nuclear extract was incubated at room temperature with 1 ng of 32P-end-labeled oligonucleotide encoding the p21WAF1 promoter binding site for p53 (5′-ATCAATTCTCGAGGAACATGTCCCAACATGTTGCTCGAGGAT-3′) (El-Deiry et al. 1993) in 30 μl of buffer containing 20 mm Tris-HCl (pH 7.6), 5% glycerol, 40 mm KCl, 1 mm EDTA, 2 mm MgCl2, 2 mm DTT, 30 μg of BSA, and 1 μg of sheared salmon sperm DNA. Competition for p53 binding activity was carried out in the presence of either a 40-fold excess of unlabeled wild-type or mutated (5′-ATCAATTCTCGAGGAAACGTTCCCAAACGTTTGCTCGAGGAT-3′) p21waf1 oligonucleotide probe. DNA–protein complexes were separated by electrophoresis through a 4% native polyacrylamide gel, dried, and then visualized on a PhosphorImager. For off-rate analysis, extracts were incubated with 32P-end-labeled p21WAF1 probe until equilibrium. Fortyfold excess of unlabeled wild-type probe was added, and aliquots were applied to a progressively running 4% acrylamide gel.
Acknowledgments
We thank Dr. Stephen V. Desiderio and Ann Marie Egloff for their valuable advice and help on two-dimensional peptide mapping and phosphoamino acid analysis. We thank Dr. Carol Prives for helpful discussions and sharing unpublished information. We thank Ella Freulich and Dr. Sheau-Yann Shieh for the recombinant full-length p53 and Δ96 p53 construct in addition to metabolic labeling of full-length p53 and Δ96 p53 infected Sf9 cells. We thank Beverly Plunkett for her excellent technical help. We thank Dr. Robert F. Siliciano for his helpful comments regarding the manuscript. We also would like to thank all the members of the Kastan laboratory for many helpful discussions. This work was supported by grants from the National Institutes of Health (ES05777 and CA71387). M.B.K. is the Steven Birnbaum Scholar of the Leukemia Society of America. C.E.C. is supported by a training grant from the American Society of Hematology scholar fellowship award.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL jsilicia@welchlink.welch.jhu.edu; FAX (410) 614-4095.
References
- Adler V, Pincus MR, Minamoto T, Fuchs SY, Bluth MJ, Brandt-Rauf PW, Friedman FK, Robinson RC, Chen JM, Wang XW, Harris CC, Ronai Z. Conformation-dependent phosphorylation of p53. Proc Natl Acad Sci. 1997;94:1686–1691. doi: 10.1073/pnas.94.5.1686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudier J, Delphin C, Grunwald D, Khochbin S, Lawrence JJ. Characterization of the tumor suppressor protein p53 as a protein kinase C substrate and a S100b-binding protein. Proc Natl Acad Sci. 1992;89:11627–11631. doi: 10.1073/pnas.89.23.11627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bischoff JR, Friedman PN, Marshak DR, Prives C, Beach D. Human p53 is phosphorylated by p60-cdc2 and cyclin B-cdc2. Proc Natl Acad Sci. 1990;87:4766–4770. doi: 10.1073/pnas.87.12.4766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogue MA, Zhu C, Aguilar-Cordova E, Donehower LA, Roth DB. p53 is required for both radiation-induced differentiation and rescue of V(D)J rearrangement in scid mouse thymocytes. Genes & Dev. 1996;10:553–565. doi: 10.1101/gad.10.5.553. [DOI] [PubMed] [Google Scholar]
- Boyle WJ, van der Geer P, Hunter T. Phosphopeptide mapping and phosphoamino acid analysis by two-dimensional separation on thin-layer cellulose plates. Methods Enzymol. 1991;201:110–149. doi: 10.1016/0076-6879(91)01013-r. [DOI] [PubMed] [Google Scholar]
- Canman CE, Wolff AC, Chen C, Fornace AJ, Kastan MB. The p53-dependent G1 cell cycle checkpoint pathway and ataxia-telangiectasia. Cancer Res. 1994;54:5054–5058. [PubMed] [Google Scholar]
- Canman CE, Gilmer T, Coutts S, Kastan MB. Growth factor modulation of p53-mediated growth arrest versus apoptosis. Genes & Dev. 1995;9:600–611. doi: 10.1101/gad.9.5.600. [DOI] [PubMed] [Google Scholar]
- Chen J, Marechal V, Levine AJ. Mapping of the p53 and mdm-2 interaction domains. Mol Cell Biol. 1993;13:4107–4114. doi: 10.1128/mcb.13.7.4107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Danska JS, Holland DP, Mariathasan S, Williams KM, Guidos CJ. Biochemical and genetic defects in the DNA-dependent protein kinase in murine scid lymphocytes. Mol Cell Biol. 1996;16:5507–5517. doi: 10.1128/mcb.16.10.5507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derijard B, Hibi M, Wu IH, Barrett T, Su B, Deng T, Karin M, Davis RJ. JNK1: A protein kinase stimulated by UV light and Ha-Ras that binds and phosphorylates the c-Jun activation domain. Cell. 1994;76:1025–1037. doi: 10.1016/0092-8674(94)90380-8. [DOI] [PubMed] [Google Scholar]
- El-Deiry WS, Tokino T, Velculescu VE, Levy DB, Parsons R, Trent JM, Lin D, Mercer WE, Kinzler KW, Vogelstein B. WAF1, a potential mediator of p53 tumor suppression. Cell. 1993;75:817–825. doi: 10.1016/0092-8674(93)90500-p. [DOI] [PubMed] [Google Scholar]
- Farmer G, Colgan J, Nakatani Y, Manley JL, Prives C. Functional interaction between p53, the TATA-binding protein (TBP), and TBP-associated factors in vivo. Mol Cell Biol. 1996;16:4295–4304. doi: 10.1128/mcb.16.8.4295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiscella M, Ullrich SJ, Zambrano N, Shields MT, Lin D, Lees-Miller SP, Anderson CW, Mercer WE, Appella E. Mutation of the serine-15 phosphorylation site of human p53 reduces the ability of p53 to inhibit cell cycle progression. Oncogene. 1993;8:1519–1528. [PubMed] [Google Scholar]
- Gottlieb TM, Oren M. p53 in growth control and neoplasia. Biochim Biophys Acta. 1995;1287:77–102. doi: 10.1016/0304-419x(95)00019-c. [DOI] [PubMed] [Google Scholar]
- Graeber TG, Peterson JF, Tsai M, Monica K, Fornace Jr AJ, Giaccia AJ. Hypoxia induces accumulation of p53 protein, but activation of a G1-phase checkpoint by low-oxygen conditions is independent of p53 status. Mol Cell Biol. 1994;14:6264–6277. doi: 10.1128/mcb.14.9.6264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guidos CJ, Williams CJ, Grandal I, Knowles G, Huang MTF. V(D)J recombination activates a p53-dependent DNA damage checkpoint in scid lymphocyte precursors. Genes & Dev. 1996;10:2038–2054. doi: 10.1101/gad.10.16.2038. [DOI] [PubMed] [Google Scholar]
- Hall SR, Campbell LE, Meek DW. Phosphorylation of p53 at the casein kinase II site selectively regulates p53-dependent transcriptional repression but not transactivation. Nucleic Acids Res. 1996;24:1119–1126. doi: 10.1093/nar/24.6.1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartwell LH, Kastan MB. Cell cycle control and cancer. Science. 1994;266:1821–1828. doi: 10.1126/science.7997877. [DOI] [PubMed] [Google Scholar]
- Haupt Y, Maya R, Kazaz A, Oren M. Mdm2 promotes the rapid degradation of p53. Nature. 1997;387:296–299. doi: 10.1038/387296a0. [DOI] [PubMed] [Google Scholar]
- Huang L, Clarkin KC, Wahl GM. p53-dependent cell cycle arrests are preserved in DNA-activated protein kinase-deficient mouse fibroblasts. Cancer Res. 1996;56:2940–2944. [PubMed] [Google Scholar]
- Hupp TR, Meek DW, Midgley CA, Lane DP. Regulation of the specific DNA binding function of p53. Cell. 1992;71:875–886. doi: 10.1016/0092-8674(92)90562-q. [DOI] [PubMed] [Google Scholar]
- Kastan MB, Onyekwere O, Sidransky D, Vogelstein B, Craig RW. Participation of p53 protein in the cellular response to DNA damage. Cancer Res. 1991;51:6304–6311. [PubMed] [Google Scholar]
- Kastan MB, Zhan Q, El-Deiry WS, Carrier F, Jacks T, Walsh WV, Plunkett BS, Vogelstein B, Fornace Jr AJ. A mammalian cell cycle checkpoint pathway utilizing p53 and GADD45 is defective in ataxia-telangiectasia. Cell. 1992;71:587–597. doi: 10.1016/0092-8674(92)90593-2. [DOI] [PubMed] [Google Scholar]
- Khanna KK, Lavin MF. Ionizing radiation and UV induction of p53 protein by different pathways in ataxia-telangiectasia cells. Oncogene. 1993;8:3307–3312. [PubMed] [Google Scholar]
- King DS, Fields CG, Fields GB. A cleavage method which minimizes side reactions following Fmoc solid phase peptide synthesis. Int J Peptide Protein Res. 1990;36:255–266. doi: 10.1111/j.1399-3011.1990.tb00976.x. [DOI] [PubMed] [Google Scholar]
- Kitagawa M, Higashi H, Jung HK, Suzuki-Takahashi I, Ikeda M, Tamai K, Kato J, Segawa K, Yoshida E, Nishimura S, Taya Y. The consensus motif for phosphorylation by cyclin D1-Cdk4 is different from that for phosphorylation by cyclin A/E-Cdk2. EMBO J. 1996;15:7060–7069. [PMC free article] [PubMed] [Google Scholar]
- Ko LJ, Prives C. p53: Puzzle and paradigm. Genes & Dev. 1996;10:1054–1072. doi: 10.1101/gad.10.9.1054. [DOI] [PubMed] [Google Scholar]
- Kubbutat MHG, Jones SN, Vousden KH. Regulation of p53 stability by Mdm2. Nature. 1997;387:299–303. doi: 10.1038/387299a0. [DOI] [PubMed] [Google Scholar]
- Lavin MF, Khanna KK, Beamish H, Teale B, Hobson K, Watters D. Defect in radiation signal transduction in ataxia-telangiectasia. Int J Radiat Biol. 1994;66:S151–S156. [PubMed] [Google Scholar]
- Lees-Miller SP, Sakaguchi K, Ullrich SJ, Appella E, Anderson CW. Human DNA-activated protein kinase phosphorylates serines 15 and 37 in the amino-terminal transactivation domain of human p53. Mol Cell Biol. 1992;12:5041–5049. doi: 10.1128/mcb.12.11.5041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Linke SP, Clarkin KC, DiLeonardo A, Tsou A, Wahl GM. A reversible, p53-dependent G0/G1 cell cycle arrest induced by ribonucleotide depletion in the absence of detectable DNA damage. Genes & Dev. 1996;10:934–947. doi: 10.1101/gad.10.8.934. [DOI] [PubMed] [Google Scholar]
- Lu H, Fisher RP, Bailey P, Levine AJ. The CDK7-cycH-p36 complex of transcription factor IIH phosphorylates p53, enhancing its sequence-specific DNA binding activity in vitro. Mol Cell Biol. 1997;17:5923–5934. doi: 10.1128/mcb.17.10.5923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maki CG, Huibregtse JM, Howley PM. In vivo ubiquitination and proteasome-mediated degradation of p53. Cancer Res. 1996;56:2649–2654. [PubMed] [Google Scholar]
- Maltzman W, Czyzyk L. UV irradiation stimulates levels of p53 cellular tumor antigen in nontransformed mouse cells. Mol Cell Biol. 1984;4:1689–1694. doi: 10.1128/mcb.4.9.1689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayr GA, Reed M, Wang P, Wang Y, Schwedes JF, Tegtmeyer P. Serine phosphorylation in the NH2 terminus of p53 facilitates transactivation. Cancer Res. 1995;55:2410–2417. [PubMed] [Google Scholar]
- Milne DM, Palmer RH, Campbell DG, Meek DW. Phosphorylation of the p53 tumour-suppressor protein at three N-terminal sites by a novel casein kinase I-like enzyme. Oncogene. 1992;7:1361–1369. [PubMed] [Google Scholar]
- Milne DM, Campbell DG, Caudwell FB, Meek DW. Phosphorylation of the tumor suppressor protein p53 by mitogen-activated protein kinases. J Biol Chem. 1994;269:9253–9260. [PubMed] [Google Scholar]
- Milne DM, Campbell LE, Campbell DG, Meek DW. p53 is phosphorylated in vitro and in vivo by an ultraviolet radiation-induced protein kinase characteristic of the c-Jun kinase, JNK1. J Biol Chem. 1995;270:5511–5518. doi: 10.1074/jbc.270.10.5511. [DOI] [PubMed] [Google Scholar]
- Momand J, Zambetti GP, Olson DC, George DL, Levine AJ. The mdm-2 oncogene product forms a complex with the p53 protein and inhibits p53-mediated transactivation. Cell. 1992;69:1237–1245. doi: 10.1016/0092-8674(92)90644-r. [DOI] [PubMed] [Google Scholar]
- Morgan SE, Kastan MB. p53 and ATM: Cell cycle, cell death and cancer. Adv Cancer Res. 1997;71:1–25. doi: 10.1016/s0065-230x(08)60095-0. [DOI] [PubMed] [Google Scholar]
- Nacht M, Strasser A, Chan YR, Harris AW, Schlissel M, Bronson RT, Jacks T. Mutations in the p53 and SCID genes cooperate in tumorigenesis. Genes & Dev. 1996;10:2055–2066. doi: 10.1101/gad.10.16.2055. [DOI] [PubMed] [Google Scholar]
- Nelson WG, Kastan MB. DNA strand breaks: The DNA template alterations that trigger p53-dependent DNA damage response pathways. Mol Cell Biol. 1994;14:1815–1823. doi: 10.1128/mcb.14.3.1815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Picksley SM, Vojtesek B, Sparks A, Lane DP. Immunochemical analysis of the interaction of p53 with MDM2; fine mapping of the MDM2 binding site on p53 using synthetic peptides. Oncogene. 1994;9:2523–2529. [PubMed] [Google Scholar]
- Rathmell WK, Kaufmann WK, Hurt JC, Byrd LL, Chu G. DNA-dependent protein kinase is not required for accumulation of p53 or cell cycle arrest after DNA damage. Cancer Res. 1997;57:68–74. [PubMed] [Google Scholar]
- Rosette C, Karin M. Ultraviolet light and osmotic stress: Activation of the JNK cascade through multiple growth factor and cytokine receptors. Science. 1996;274:1194–1197. doi: 10.1126/science.274.5290.1194. [DOI] [PubMed] [Google Scholar]
- Santos JA, Logarinho E, Tapia C, Allende CC, Allende JE, Sunkel CE. The casein kinase 1α gene of Drosophila melanogaster is developmentally regulated and the kinase activity of the protein induced by DNA damage. J Cell Sci. 1996;109:1847–1856. doi: 10.1242/jcs.109.7.1847. [DOI] [PubMed] [Google Scholar]
- Savitsky K, Bar-Shira A, Gilad S, Rotman G, Ziv Y, Vanagaite L, Tagle DA, Smith S, Uziel T, Sfez S, Ashkenazi M, Pecker I, Frydman M, Harnik R, Patanjali SR, Simmons A, Clines GA, Sartiel A, Gatti RA, Chessa L, Sanal O, Lavin MF, Jaspers NGJ, Taylor AMR, Arlett CF, Miki T, Weissman SM, Lovett M, Collins FS, Shiloh Y. A single ataxia telangiectasia gene with a product similar to PI-3 kinase. Science. 1995;268:1749–1753. doi: 10.1126/science.7792600. [DOI] [PubMed] [Google Scholar]
- Shieh SY, Ikeda M, Taya Y, Prives C. DNA damage-induced phosphorylation of p53 alleviates inhibition by MDM2. Cell. 1997;91:325–334. doi: 10.1016/s0092-8674(00)80416-x. [DOI] [PubMed] [Google Scholar]
- Shiloh Y, Rotman G. Ataxia-telangiectasia and the ATM gene—linking neurodegeneration, immunodeficiency, and cancer to cell cycle checkpoints. J Clin Immunol. 1996;16:254–260. doi: 10.1007/BF01541389. [DOI] [PubMed] [Google Scholar]
- Takenaka I, Morin F, Seizinger BR, Kley N. Regulation of the sequence-specific DNA binding function of p53 by protein kinase C and protein phosphatases. J Biol Chem. 1995;270:5405–5411. doi: 10.1074/jbc.270.10.5405. [DOI] [PubMed] [Google Scholar]
- Thut CJ, Chen J-L, Klemm R, Tjian R. p53 transcriptional activation mediated by coactivators TAFII40 and TAFII60. Science. 1995;267:100–104. doi: 10.1126/science.7809597. [DOI] [PubMed] [Google Scholar]
- Wakamiya, T., K. Saruta, J. Yasuoka, and S. Kusumoto. 1994. An efficient procedure for solid-phase synthesis of phosphopeptides by the Fmoc strategy. Chem. Lett. 1099–1102.
- Wang Y, Prives C. Increased and altered DNA binding of human p53 by S and G2/M but not G1 cyclin-dependent kinases. Nature. 1995;376:88–91. doi: 10.1038/376088a0. [DOI] [PubMed] [Google Scholar]
- Xu Y, Baltimore D. Dual roles of ATM in the cellular response to radiation and in cell growth control. Genes & Dev. 1996;10:2401–2410. doi: 10.1101/gad.10.19.2401. [DOI] [PubMed] [Google Scholar]
- Zakian VA. ATM-related genes: What do they tell us about functions of the human gene? Cell. 1995;82:685–687. doi: 10.1016/0092-8674(95)90463-8. [DOI] [PubMed] [Google Scholar]
- Zauberman A, Barak Y, Ragimov N, Levy N, Oren M. Sequence-specific DNA binding by p53: Identification of target sites and lack of binding to p53-MDM2 complexes. EMBO J. 1993;12:2799–2808. doi: 10.1002/j.1460-2075.1993.tb05941.x. [DOI] [PMC free article] [PubMed] [Google Scholar]