Abstract
The ascidian tadpole represents the most simplified chordate body plan. It contains a notochord composed of just 40 cells, but as in vertebrates Brachyury is essential for notochord differentiation. Here, we show that the misexpression of the Brachyury gene (Ci-Bra) of Ciona intestinalis is sufficient to transform endoderm into notochord. Subtractive hybridization screens were conducted to identify potential Brachyury target genes that are induced upon Ci-Bra misexpression. Of 501 independent cDNA clones that were surveyed, 38 were specifically expressed in notochord cells. These potential Ci-Bra downstream genes appear to encode a broad spectrum of divergent proteins associated with notochord formation.
Keywords: Ascidians, notochord, Brachyury, downstream genes, differentiation
Brachyury encodes a sequence-specific activator that contains a T-box DNA-binding domain (Herrmann et al. 1990; Kispert et al. 1995; Conlon et al. 1996). In vertebrates, Brachyury is initially expressed throughout the presumptive mesoderm, and during later stages the expression pattern is gradually restricted to the developing notochord and tailbud. Brachyury notochord differentiation is essential in all vertebrates that have been studied, including mice, frogs, and zebrafish (for review, see Herrmann and Kispert 1994; Smith 1997; Papaioannou and Silver 1998).
Brachyury is expressed exclusively in the notochord precursor cells of two divergent ascidians, Halocynthia roretzi (Yasuo and Satoh 1993) and Ciona intestinalis (Corbo et al. 1997a). The spatial and temporal patterns of the gene expression coincide with the clonal restriction of the notochord lineages. In H. roretzi, notochord formation is induced at the 32-cell stage by signals emanating from the adjacent endoderm (Nakatani and Nishida 1994). Overexpression of the Halocynthia Brachyury gene (As-T) via RNA injection results in notochord formation without a requirement for the inductive event at the 32-cell stage (Yasuo and Satoh 1998). In addition, misexpression of As-T also causes transformation of endoderm and neuronal lineages into notochord (Yasuo and Satoh 1998). These results indicate that the ascidian Brachyury gene is a critical determinant of the notochord. Here we report that the misexpression of the Brachyury gene (Ci-Bra) of C. intestinalis is sufficient to transform endoderm into notochord. Subtractive hybridization screens were conducted to identify potential Brachyury target genes that are induced upon Ci-Bra overexpression. We isolated and characterized 38 different notochord-specific genes that may include potential targets of the ascidian Brachyury.
Results and Discussion
The fork head/HNF-3β gene of C. intestinalis (Ci-fkh) is expressed in the endoderm, endodermal strand, notochord, and ventral ependymal cells of the neural tube (Corbo et al. 1997b). A 2.6-kb genomic DNA fragment from the 5′-flanking region of Ci-fkh is sufficient to direct the expression of a lacZ reporter gene in these tissues after electroporation into one-cell embryos (Fig. 1A). The Ci-Bra gene was misexpressed in ectopic tissues by attaching the Ci-Bra coding sequence to the Ci-fkh promoter region (Fig. 1B). The resulting fusion gene causes extensive transformation of the endoderm into notochord, whereby mutant tailbud embryos contain a large mass of notochord tissue in midtail regions (Fig. 1B).
Figure 1.
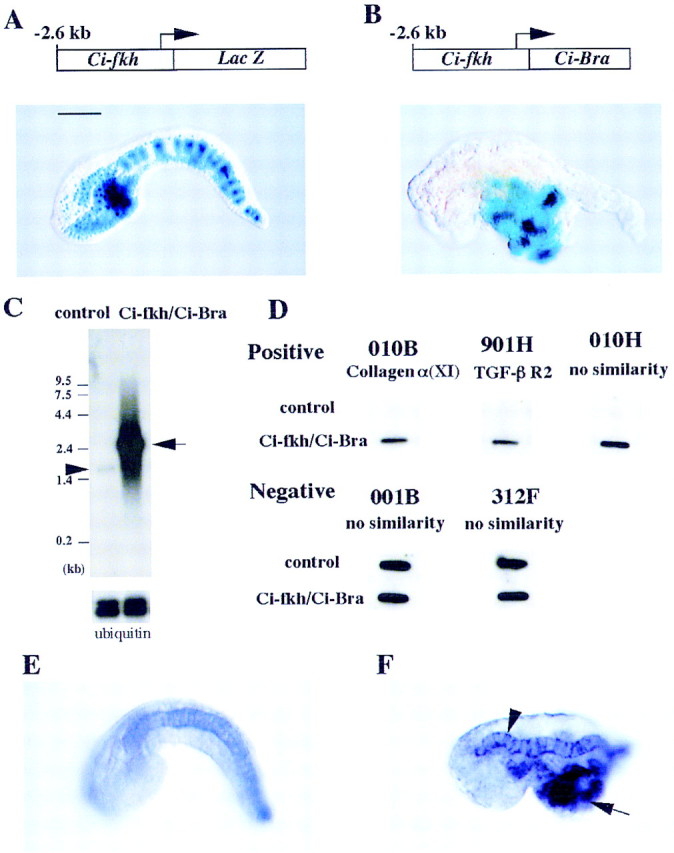
Misexpression of Ci-Bra and the isolation of potential target genes. (A) One-cell embryos were electroporated with a Ci-fkh/lacZ reporter gene and grown to the tailbud stage of development. LacZ staining is detected in the notochord, CNS, endoderm, and endodermal strand. Scale bar, 50 μm. (B) Coelectroporation of the Ci-fkh/lacZ reporter gene and Ci-fkh/Ci-Bra fusion gene results in a severe mutant phenotype, whereby endoderm is transformed into notochord. The lacZ reporter gene is expressed in these transformed cells. (C) Northern assays indicate that the mutant embryos express >100 times more Ci-Bra mRNA than wild-type control embryos. (D) Most of the cDNAs that were isolated upon subtractive hybridization are expressed at higher levels in the mutant embryos as compared with wild-type embryos. The specific dot hybridizations shown here include a cDNA encoding a collagen subunit (α1); another encodes a TGFβ type II receptor cytokine; the other three cDNAs have no obvious sequence similarities with proteins in the various databases. (E,F) One of the cDNAs that was isolated is a member of the ERM family of cytoskeletal proteins (Table 1). In situ hybridization assays reveal localized expression of the gene in the notochord (E). It is overexpressed in mutant embryos that were electroporated with the Ci-fkh/Ci-Bra fusion gene (F). In particular, staining is detected in both the notochord (arrowhead) and transformed endoderm (arrow).
Northern blots were prepared with RNA extracted from wild-type and mutant embryos and subsequently hybridized with a radiolabeled Ci-Bra probe (Fig. 1C). The resulting autoradiographs indicate that the mutant embryos express >100 times more Ci-Bra mRNA as compared with wild type (Fig. 1C). The efficiency of the electroporation method allowed us to obtain large quantities of mutant embryos, thereby facilitating subtractive hybridization reactions using mRNAs extracted from wild-type and mutant embryos. A subtractive cDNA library was prepared that contains mRNAs, which are overexpressed in the mutant embryos relative to the wild-type controls. The library contains 923 cDNA clones. Sequence analysis of ∼500 bp of both 5′ and 3′ regions of the cDNAs suggests that 599 of the clones represent separate genes. As discussed below, these genes appear to encode a broad spectrum of divergent proteins.
Additional experiments were conducted to determine whether the genes that were identified are induced upon ectopic expression of Ci-Bra. The same amounts of RNA prepared from wild-type and mutant embryos were spotted on nitrocellulose and hybridized with radiolabeled probe derived from each of the 599 independent cDNA clones. In most cases, a hybridization signal was observed only with the probe derived from the mutant (three examples shown in Fig. 1D). However, some of the clones exhibited equal levels of hybridization with both probes, indicating that they do not represent genes that are induced upon overexpression of Ci-Bra (two examples in Fig. 1D). These experiments identified 501 of the 599 cDNA clones as potential direct and indirect targets of Ci-Bra.
Each of the 501 clones was used as probes for in situ hybridization assays with whole-mount embryos. A total of 38 clones exhibited notochord-specific patterns of expression (8 examples are presented in Fig. 2). One of the clones, which encodes a Ciona homolog of the ezrin/radixin/moesin (ERM) family of cytoskeletal linker proteins, was used as a probe for hybridization to a mutant embryo that contains the Ci-fkh/Ci-Bra fusion gene. The Ciona ERM gene is expressed ectopically in the mutant (Fig. 1F) as compared with a wild-type control embryo (Fig. 1E). In particular, staining is observed both in the endogenous notochord (Fig. 1F, arrowhead) and the ectopic notochord cells in midtail regions (Fig. 1F, arrow). These results indicate the successful isolation of notochord-specific genes that are induced upon misexpression of Ci-Bra.
Figure 2.
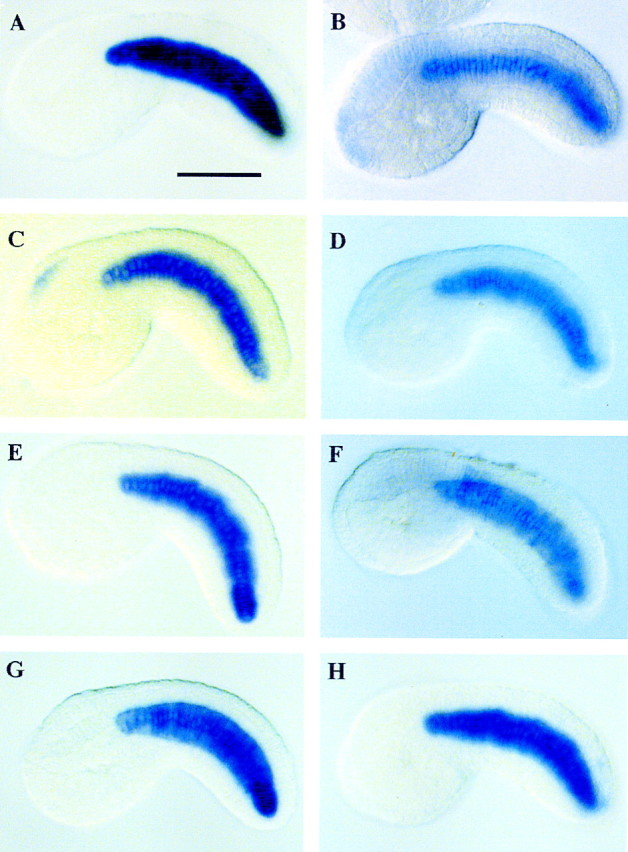
In situ hybridization assays. Digoxigenin-labeled RNA probes were prepared with the cDNA clones isolated via subtractive hybridization. These probes were used for in situ hybridization assays. Results shown were obtained with 8 of the 38 cDNAs that exhibit notochord-specific expression. All of the embryos are at mid-tailbud stage of development. Six of these genes encode proteins that possess homologies to sequences in the databases (see Table 1). For example, the staining pattern shown in A corresponds to a gene that encodes a component of the ECM, a prothrombinase precursor. The genes shown in B and C encode members of the lipoprotein receptor family and sulfate transporter, respectively. (D,E) Genes that encodes putative cytoplasmic proteins; F corresponds to a gene that encodes a nuclear protein (see Table 1); (G,H) genes without sequence similarity. Scale bar, 50 μm.
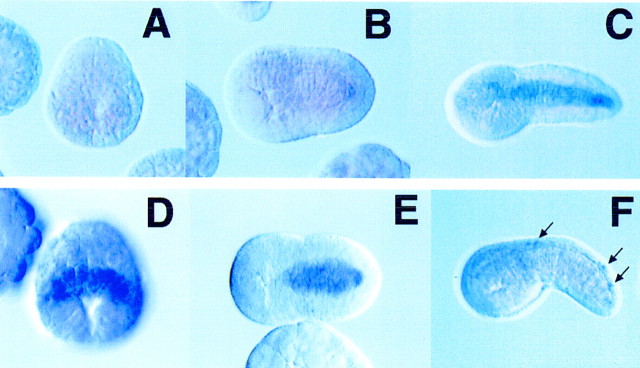
The analysis of staged embryos suggests that at least some of the genes exhibit different temporal patterns of expression (Fig. 3). For example, the ERM gene is first expressed after neurulation (Fig. 3B) and is absent in gastrulating embryos (Fig. 3A). Peak expression is observed just after intercalation of the notochord during tailbud stages of development (Fig. 3C) and persists in 14-hr embryos (data not shown). In contrast, another notochord-specific gene, a Ciona homolog of the leukocyte common antigen related (LAR) family of receptor protein tyrosine phosphatases, is transiently expressed only during early periods of notochord differentiation (Fig. 3D–F). Expression is first detected in gastrulating embryos (Fig. 3D), within a few hours after the first appearance of the Brachyury protein in prospective notochord cells. Staining persists during neurulation (Fig. 3E) but is rapidly lost during tailbud stages (Fig. 3F). After the notochord pattern is lost, staining reappears in neuronal processes (Fig. 3F, arrows), suggesting a role in axonal guidance (Desai et al. 1996; Krueger et al. 1996).
Figure 3.
Putative Ci-Bra target genes exhibit distinct temporal patterns of expression. Digoxigenin-labeled RNA probes were hybridized to staged embryos. (A–C) The embryos shown were hybridized with the putative ERM homolog; (D–F) embryos hybridized with a probe that corresponds to a member of the LAR family of receptor tyrosine phosphatases. The ERM gene is expressed only weakly in neurulating embryos (B) and reaches peak expression in early tadpoles (C). There is no staining detected in gastrulating embryos (A). In contrast, the LAR gene exhibits a reciprocal pattern of expression, with staining detected in the prospective notochord cells during gastrulation (D) and neurulation (E). Staining is lost during tailbud stages (F). However, LAR expression might reappear in neuronal processes in older embryos (F, arrows), which is consistent with its role in the axonal guidance of Drosophila motor neurons.
Approximately half (18 of 38) of the notochord-specific genes contain homology to known sequences (Table 1). The deduced proteins represent a broad spectrum of cellular functions, including components of the extracellular matrix (ECM), membrane receptors and adhesion molecules, cytoskeletal proteins, and nuclear proteins. Given the observation that some of the genes exhibit sequential patterns of expression during notochord differentiation (e.g., Fig. 3), it is possible that the encoded proteins comprise a signaling pathway that controls changes in cell adhesion and/or cell shape. Perhaps components of the ECM signal to the notochord through cell surface receptors such as LAR (see Fig. 3D–F). The activation of notochord-specific receptors might lead to the modification or induction of cytoskeletal proteins, such as ERM (see Fig. 3A–C). ERM has been implicated in mediating changes in the cytoskeleton in response to signaling at the cell surface (Bretscher et al. 1997). After intercalation, the notochord is composed of a single row of cells (see Fig. 3). Initially, each cell is columnar in shape but gradually becomes cuboidal. This transition in shape is directly responsible for axial extension and tail morphogenesis. Future studies will determine whether any of the proteins identified in this study are important for regulating intercalation and extension. In addition, it will be important to determine how many of the genes are directly regulated by Ci-Bra and thereby constitute an integrated gene battery.
Table 1.
Proteins encoded by Brachyury downstream notochord-specific genes in C. intestinalis, deduced from partial 5′ and 3′ sequences of cDNAs
|
cDNA
clones
|
Sequence similarity (BLASTP)
|
Figures
|
|---|---|---|
| I. ECM | ||
| 408h | prothrombinase precursor (fibrinogen-like protein) | 2A |
| 801h | extensin precursor (proline-rich glycoprotein) | |
| 010b | collagen α 1(XI) chain (fragment) | |
| II. Receptor proteins, cell adhesion molecules | ||
| 505a | sulfate adenylyltransferase (SAT) (ATP-sulfurylase) | |
| 103f | low-density lipoprotein receptor-related protein precursor (LRP) | 2B |
| 309h | sulfate transporter | 2C |
| 204d | P-selectin | |
| 108e | LAR–protein tyrosine phosphatase | 3D–F |
| III. Cytoplasmic proteins | ||
| (calcium pathway related) | ||
| 903h | calcium/calmodulin-dependent protein kinase type I | 2D |
| 003b | hspg/agrin | |
| 906g | calmodulin | |
| (Ras pathway related) | ||
| 207f | Ras GTPase-activating-like protein IQGAP1 | |
| 906b | Ras-related protein RAB-2 | |
| (others) | ||
| 406g | cell division control protein 45 | 2E |
| 504g | myosin heavy chain | |
| 002d | ERM | 1E, F; 3A–C |
| IV. Nuclear proteins | ||
| 110g | ATP-citrate (PRO-S-)–lyase (citrate cleavage enzyme) | 2F |
| 403e | testin 2 (TES2) | |
The 38 notochord-specific genes are probably not alone in controlling notochord differentiation. Another 84 cDNAs are expressed in the notochord as well as at least one additional tissue. Among these is the collagen 2A1 gene, which is expressed in both the notochord and neighboring tail muscles (data not shown). Thus, this study has identified 119 genes that are up-regulated in the notochord as a direct or indirect consequence of overexpressing Ci-Bra. Unexpectedly, another 81 cDNA clones exhibit tissue-specific patterns of expression that exclude the notochord. For example, 7 genes are expressed in the cerebral vesicle, 7 in the CNS (cerebral vesicle plus neural tube), 7 in the mesenchyme, 14 in the epidermis, and 16 in the tail muscles. Included among this latter group is a member of the heparin sulfate proteoglycan superfamily (data not shown). The notochord has been implicated in the patterning of the CNS (floor plate; Bronner-Fraser and Fraser 1997), paraxial mesoderm (somites; Bumcrot and McMahon 1995), and derivatives of the endoderm (pancreas; Kim et al. 1997). Perhaps the ectopic expression of Ci-Bra results in the induction of one or more notochord-specific signaling molecules that are secreted from the notochord and influence the development of neighboring tissues.
The present results also provide insights into molecular developmental mechanisms underlying the evolution of the chordate body plan. The origin and evolution of the chordates (urochordates, cephalochordates, and vertebrates) have been debated for more than a century (Berrill 1955; Gee 1996). The ascidian tadpole is thought to represent the most simplified, basic chordate body plan. It contains a dorsal hollow neural tube and prominent notochord (Satoh 1994; Satoh and Jeffery 1995; Di Gregorio and Levine 1998). Our long-range goal is to determine the gene circuits underlying notochord formation in ascidians and use this information to explore the evolutionary origins of the notochord among lower deuterostomes, including echinoderms and hemichordates.
Materials and methods
Ascidian eggs and embryos
C. intestinalis was used in this study. Handling of gametes and embryos was carried out as described (Corbo et al. 1997a).
Electroporation of constructs and Northern blot analysis
Electroporation was carried out as described (Corbo et al. 1997a).
Isolation of poly(A)+ RNA and Northern blot analysis were carried out according to standard procedures. Ubiquitin probe was used as loading control.
Production of subtractive library
Poly(A)+ RNA was isolated from wild-type and mutant embryos at the neurula and early tailbud stages by standard procedures. About 14.3 and 10.1 μg of poly(A)+ RNAs were obtained from wild-type and mutant embryos; 5 μg of each was used for the library construction.
cDNA libraries of wild-type embryos (W-library) and mutant embryos (M-library) were constructed in Uni-ZAP XR using the ZAP-cDNA synthesis kit (Stratagene). From the M-library, single-stranded DNA was isolated as described (Schweinfest et al. 1990) except that VCSM13 helper phage was used instead of R408 helper phage. From the W-library, single-stranded phage was rescued according to the mass excision protocol supplied with the ZAP-cDNA synthesis kit. Single-stranded phage was converted to pBluescript double-stranded phagemid DNA containing a cDNA insert by infection into SoloR cells. The phagemid DNA was prepared with Qiagen tip 100. The phagemid DNA and Ci-Bra plasmid DNA were linearized with XhoI and used as template DNA for in vitro transcription.
After in vitro transcription with T3 RNA polymerase (GIBCO BRL), template DNA was digested with RQ1 DNase (Promega). Fifty micrograms of the synthesized RNA was photobiotinylated with a photobiotin labeling system (GIBCO BRL). Fifty micrograms of biotinylated RNA and 5 μg of Ci-Bra biotinylated RNA were ethanol precipitated together with 5 μg of single-stranded DNA derived from the M-library and 5 μg of poly(A) (Pharmacia); then they were dissolved in 10 μl of hybridization buffer (25 mm HEPES–NaOH at pH 7.5, 0.75 m NaCl, 0.5 mm EDTA, 0.1% SDS). Following incubation at 95°C for 4 min, hybridization was performed at 65°C for 20 hr. After hybridization, the mixture was added to 80 μl of buffer containing 50 mm HEPES–NaOH (pH 7.5), and 437 mm NaCl.
We separated hybrids from single-stranded DNA using streptavidin (GIBCO BRL), as described (Sive and St John 1988). The subtracted single-stranded DNA was ethanol precipitated and dissolved in 34 μl of TE (10 mm Tris-HCl at pH 8.0, 0.1 mm EDTA). The subtraction was performed twice. One-third of the subtracted DNA was converted to double-stranded DNA and was used for transformation of Max Efficiency DH5α competent cells (GIBCO BRL), as described (Schweinfest et al. 1990). Each colony was picked into 96-cell titer plate wells and cultured overnight, and glycerol was stocked with 10 plates (923 clones) at −80°C.
Nucleotide sequencing
Nucleotide sequences were determined for both strands with a Big-Dye Terminator Cycle Sequencing Ready Reaction kit and ABI Prism 377 DNA sequencer (Perkin Elmer).
In situ hybridization
RNA probes were prepared with a DIG RNA labeling Kit (Boehringer Mannheim). Whole-mount in situ hybridization was performed using digoxigenin-labeled antisense probes as described previously (Corbo et al. 1997a). Control embryos hybridized with a sense probe did not show signals above background.
Acknowledgments
This research was supported by a grant from the Human Frontier Science Program (RG212/1997) to N.S. and M.L. This research was supported by a Grant-in-Aid for Specially Promoted Research (no. 07102012) from Monbusho, Japan, to N.S. and a grant from the National Science Foundation (IBN-9514138) to M.L. We thank Yasuo Mitani, Kazuko Hirayama, and Kazuko Hatayama for technical help.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked ‘advertisement’ in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL satoh@ascidian.zool.kyoto-u.ac.jp; FAX 81-75-705-1113.
References
- Berrill NJ. The origin of the vertebrates. Oxford, UK: Oxford University Press; 1955. [Google Scholar]
- Bretscher A, Reczek D, Beeryman M. Ezrin: A protein requiring conformational activation to link microfilaments to the plasma membrane in the assembly of cell surface structures. J Cell Sci. 1997;110:3011–3018. doi: 10.1242/jcs.110.24.3011. [DOI] [PubMed] [Google Scholar]
- Bronner-Fraser M, Fraser SE. Differentiation of the vertebrate neural tube. Curr Opin Cell Biol. 1997;9:885–891. doi: 10.1016/s0955-0674(97)80092-0. [DOI] [PubMed] [Google Scholar]
- Bumcrot DA, McMahon AP. Somite differentiation. Sonic signals somites. Curr Biol. 1995;5:612–614. doi: 10.1016/s0960-9822(95)00123-0. [DOI] [PubMed] [Google Scholar]
- Conlon FL, Sedgwick SG, Weston KM, Smith JC. Inhibition of Xbra transcription activation causes defects in mesodermal patterning and reveals autoregulation of Xbra in dorsal mesoderm. Development. 1996;122:2427–2435. doi: 10.1242/dev.122.8.2427. [DOI] [PubMed] [Google Scholar]
- Corbo JC, Levine M, Zeller RW. Characterization of a notochord-specific enhancer from the Brachyury promoter region of the ascidian, Ciona intestinalis. Development. 1997a;124:589–602. doi: 10.1242/dev.124.3.589. [DOI] [PubMed] [Google Scholar]
- Corbo JC, Erives A, Di Gregorio A, Chang A, Levine M. Dorsoventral patterning of the vertebrate neural tube is conserved in a protochordate. Development. 1997b;124:2335–2344. doi: 10.1242/dev.124.12.2335. [DOI] [PubMed] [Google Scholar]
- Desai CJ, Gindhart JG, Jr, Goldstein LSB, Zinn K. Receptor tyrosine phosphatases are required for motor axon guidance in the Drosophilaembryo. Cell. 1996;84:599–609. doi: 10.1016/s0092-8674(00)81035-1. [DOI] [PubMed] [Google Scholar]
- Di Gregorio A, Levine M. Ascidian embryogenesis and the origins of the chordate body plan. Curr Opin Genet Dev. 1998;8:457–463. doi: 10.1016/s0959-437x(98)80118-4. [DOI] [PubMed] [Google Scholar]
- Gee H. Before the backbone. Views on the origin of the vertebrates. London, UK: Chapman & Hall; 1996. [Google Scholar]
- Herrmann BG, Kispert A. The Tgenes in embryogenesis. Trends Genet. 1994;10:280–286. doi: 10.1016/0168-9525(90)90011-t. [DOI] [PubMed] [Google Scholar]
- Herrmann BG, Labeit S, Poustka A, King TR, Lehrach H. Cloning of the Tgene required in mesoderm formation in the mouse. Nature. 1990;343:617–622. doi: 10.1038/343617a0. [DOI] [PubMed] [Google Scholar]
- Kim SK, Hebrok M, Melton DA. Notochord to endoderm signaling is required for pancreas development. Development. 1997;124:4243–4252. doi: 10.1242/dev.124.21.4243. [DOI] [PubMed] [Google Scholar]
- Kispert A, Koschorz B, Herrmann BG. The T protein encoded by Brachyuryis a tissue-specific transcription factor. EMBO J. 1995;14:4763–4772. doi: 10.1002/j.1460-2075.1995.tb00158.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krueger NX, Van Vactor D, Wan HI, Gelbart WM, Goodman CS, Saito H. The transmembrane tyrosine phosphatase DLAR controls motor axon guidance in Drosophila. Cell. 1996;84:611–622. doi: 10.1016/s0092-8674(00)81036-3. [DOI] [PubMed] [Google Scholar]
- Nakatani Y, Nishida H. Induction of notochord during ascidian embryogenesis. Dev Biol. 1994;166:289–299. doi: 10.1006/dbio.1994.1315. [DOI] [PubMed] [Google Scholar]
- Papaioannou VE, Silver LM. The T-box gene family. BioEssays. 1998;20:9–19. doi: 10.1002/(SICI)1521-1878(199801)20:1<9::AID-BIES4>3.0.CO;2-Q. [DOI] [PubMed] [Google Scholar]
- Satoh N. Developmental biology of ascidians. New York, NY: Cambridge University Press; 1994. [Google Scholar]
- Satoh N, Jeffery WR. Chasing tails in ascidians: Developmental insights into the origin and evolution of chordates. Trends Genet. 1995;11:354–359. doi: 10.1016/s0168-9525(00)89106-4. [DOI] [PubMed] [Google Scholar]
- Schweinfest CW, Henderson KW, Gu JR, Kottaridis SD, Besbeas S, Panotopoulou E, Papas TS. Subtraction hybridization cDNA libraries from colon carcinoma and hepatic cancer. Genet Anal Tech Appl. 1990;7:64–70. doi: 10.1016/0735-0651(90)90042-e. [DOI] [PubMed] [Google Scholar]
- Sive HL, St John T. A simple subtractive hybridization technique employing photoactivatable biotin and phenol extraction. Nucleic Acids Res. 1988;16:10937. doi: 10.1093/nar/16.22.10937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith J. Brachyuryand the T-box genes. Curr Opin Genet Dev. 1997;7:474–480. doi: 10.1016/s0959-437x(97)80073-1. [DOI] [PubMed] [Google Scholar]
- Yasuo H, Satoh N. Function of vertebrate Tgene. Nature. 1993;364:582–583. doi: 10.1038/364582b0. [DOI] [PubMed] [Google Scholar]
- ————— Conservation of the developmental role of Brachyury in notochord formation in a urochordate, the ascidian Halocynthia roretzi. Dev Biol. 1998;200:158–170. doi: 10.1006/dbio.1998.8958. [DOI] [PubMed] [Google Scholar]