Abstract
There are still questions about whether macrophage differentiation is predetermined or is induced in response to tissue microenvironments. C2D macrophage cells reside early in the macrophage lineage in vitro, but differentiate to a more mature phenotype after adoptive transfer to the peritoneal cavity (PEC-C2D). Since C2D macrophage cells also traffic to adipose tissue after adoptive transfer, we explored the impact of white adipose tissue (WAT), brown adipose tissue (BAT) and in vitro cultured adipocytes on C2D macrophage cells.
When PEC-C2D macrophage cells were cultured with preadipocytes the cells stretched out and CD11b and Mac-2 expression was lower compared to PEC-C2D macrophage cells placed in vitro alone. In contrast, PEC-C2D cells co-cultured with adipocytes maintained smaller, round morphology and more cells expressed Mac-2 compared to PEC-C2D co-cultured with preadipocytes. After intraperitoneal injection, C2D macrophage cells migrated into both WAT and BAT. A higher percentage of C2D macrophage cells isolated from WAT (WAT-C2D) expressed Ly-6C (33%), CD11b (11%), Mac-2 (11%) and F4/80 (29%) compared to C2D macrophage cells isolated from BAT (BAT-C2D). Overall, BAT-C2D macrophage cells had reduced expression of many cytokine, chemokine and receptor gene transcripts when compared to in vitro grown C2D macrophages, while WAT-C2D macrophage cells and PEC-C2D up-regulated many of these gene transcripts. These data suggest that the C2D macrophage phenotype can change rapidly and distinct phenotypes are induced by different microenvironments.
Keywords: macrophage, plasticity, adipocyte, adipose tissue, trafficking
1. Introduction
Macrophages are found throughout the body and serve as initiators and effectors of the innate immune system [1–6]. Macrophages differentiate from bone marrow hematopoietic stem cells through various stages including, macrophage-colony forming cells to monoblasts, promonocytes and finally into monocytes [7,8]. Monocytes enter the bloodstream, where they circulate before migrating into tissues. There they differentiate into tissue-specific macrophages [9]. Macrophages are a heterogeneous group of cells which have different functions, morphologies and phenotypic properties [7,9]. Heterogeneity is commonly associated with macrophages as a consequence of the functions, organ sites and immune status of the host [9,10]. However, there is controversy about macrophage adaptation to microenvironmental signals in vivo [10–13]. Some think that since subpopulations of macrophages have either proinflammatory (M1) or anti-inflammatory (M2) properties, there are predetermined fates for monocytes and macrophages as opposed to the microenvironmental signaling leading to the macrophage plasticity [10,14].
C2D macrophage cells reside early in the macrophage lineage in vitro, but differentiate to a more mature, phenotype after adoptive transfer to the peritoneal cavity (PEC-C2D) [15]. These macrophage cells differentiate and traffic like primary macrophages and can provide insight into macrophage function [16]. In particular, they can provide evidence about macrophage plasticity in response to different microenvironments.
White adipose tissue (WAT) and brown adipose tissue (BAT) have distinct physiological functions. WAT is an energy storage and endocrine organ [17,18]. In contrast, BAT functions as an energy-dissipating organ through adaptive-thermogenesis [19]. These adipocyte depots display different morphology, cellular characteristics, body localizations and function [19–24].
Previous studies have suggested that macrophage function varies considerably in different fat depots [25]. Some have also suggested that macrophage plasticity is an artifact of in vitro manipulations [10]. Given the controversy about macrophage adaptation to microenvironmental signals in vivo [10–13] and the fact that little is known about BAT-macrophage interactions, we investigated whether macrophage phenotype is predetermined or is adaptable.
2. Materials and methods
2.1 Mouse strains
C57BL/6J (B6) mice were originally obtained from the Jackson Laboratory (Bar Harbor, ME). Male and female, 8–16 week-old mice were bred in the rodent facility of the Division of Biology at Kansas State University and used in these experiments. Mice were fed a normal mouse chow diet (5001, PMI International, St. Louis, MO) and were allowed to feed Ad libitum. All animal experiments were approved by the Institutional Animal Care and Use Committee.
2.2 Antibodies and Reagents
Collagenase (Type II), insulin from bovine pancreas, 3-Isobutyl-1-methylxanthine (IBMX) and dexamethasone were obtained from Sigma-Aldrich Co. (St. Louis, MO). Carboxyfluorescein diacetate, succinmidyl (CFDA-SE) ester was purchased from Molecular probes (Eugene, OR). APC conjugated anti-CD11c, APC conjugated anti-F4/80, APC conjugated anti-CD11b, ALEXA Fluor 647 conjugated anti-Mac2, and their isotype control antibodies were purchased from eBioscience (San Diego, CA). Biotin conjugated anti-Ly-6C (ER-MP20) and its isotype control antibody were from BD Pharmingen (San Jose, CA). APC conjugated Streptavidin was purchased from eBioscience (San Diego, CA).
2.3 Cell lines and cell culture
The C2D macrophage cell line was created as described by our group [26]. These cells were derived from C2D mouse bone marrow and selected in the presence of macrophage colony stimulating factor (M-CSF). These cells have the MHCII−/− and Tlr4Lps-n genotype and are histocompatible with mice of the H-2b haplotype. C2D cells were grown in Dulbecco’s Modified Eagle’s Medium with 4% fetal bovine serum (DMEM4) supplemented with 0.3% Glutamax and 10% Opti-MEM in 150-mm tissue culture plates.
3T3L1 adipocytes were obtained from the American Type Culture Collection (Manassas, VA). Adipocytes were cultured and differentiated as described previously [27]. Briefly, 3T3L1 cell differentiation was induced by culturing cells in DMEM containing 10% FBS (DMEM10), 1 μM dexamethasone, 1.7 μM insulin and 0.5 mM IBMX for 4 days. On the fourth day, the 3T3L1 cells were cultured in DMEM10 with 1.7 μM insulin. On day 8, 3T3L1 cells were maintained in DMEM10. Undifferentiated preadipocytes and adipocytes differentiated for 6–8 days were used in the experiments. 3T3L1 cells (1×106 cells) were directly co-cultured with 1×106 C2D cells grown exclusively in vitro or 1×106 cells adoptively transferred C2D macrophage cells isolated from the peritoneal cavity (PEC-C2D).
Bone marrow derived macrophages (BM-Mo) were differentiated from B6 mouse bone marrow cells isolated from the femora, tibiae, and humeri. Briefly, the bones were recovered and cleaned of all non-osseous tissue. The marrow cavity was flushed with a sterile PBS solution. The red blood cells were lysed by incubating in ammonium chloride lysis buffer (0.15 M NH4Cl, 10 mM KHCO3, and 0.1 mM Na2EDTA, pH 7.3) for 5 min in ice. Cells were centrifuged (300 × g, 5 min) and washed two times with DMEM2. Bone marrow cells were seeded and incubated in M-CSF medium (DMEM10, OPTI-MEM, 0.01 M HEPES, 50 ng/ml gentamycin, 1.5 ng/ml rMCSF-1) for 7 days at 37 °C, 8 % CO2. BM-Mo were indirectly co-cultured with collagenase-digested white adipose tissue (WAT) gonadal fat pads or collagenase- digested BAT perispleen or interscapular fat pads as described below.
2.4 Adoptive transfer of labeled cells
C2D cells were suspended in sterile, pre-warmed (37°C) phosphate buffered saline (PBS; 137 mM NaCl, 10 mM Phosphate, 2.7 mM KCl, pH 7.4) at a concentration of 1.5 × 106 cells per ml, further stained with CFDA-SE according to the manufacturer’s protocol. Briefly, C2D cells were incubated with 22 μM of CFDA-SE solution at 37 °C for 15 minutes. After centrifugation at 370 × g for 10 minutes, cell pellets were suspended in pre-warmed PBS and incubated in 37°C for an additional 20 minutes. Cells were then washed twice in PBS, and suspended at a concentration of 4 × 107 cells per ml in PBS. One and one-half ml of the cell suspension of CFDA-SE labeled C2D or normal C2D cells was injected intraperitoneally (i.p.) per mouse.
2.5 Peritoneal cell extraction and fat tissue isolation
PEC-C2D macrophage cells were obtained from B6 mice by peritoneal lavage 36 hours after intraperitoneal injection of 4 × 107 of C2D macrophage cells labeled with CFDA-SE. The peritoneal exudate red blood cells were lysed as described in section 2.3. One-half of the cells were treated with 1 mg/ml collagenase type II at 37°C with shaking (60 rpm) for 40 minutes. Control or collagenase-treated cells were washed three times with PBS and 3 × 106 cells were plated into 150-mm cell culture plates and incubated in DMEM4 for 16 hours.
Isolation of adipocytes and CFDA-SE labeled C2D macrophage cells was performed as previously described [15,16]. Adipocytes were isolated from both mouse gonadal fat pads (depots connected to the uterus and ovaries in females and the epididymis and testes in males) and perispleen adipose tissues by collagenase digestion [28,29]. We confirmed BAT origin by quantitating the mRNA of PRDM16 by qRT-PCR [30] and/or UCP-1 [31] in tissues collected from perispleen and interscapular isolates (data not shown). Gonadal fat pads weighed an average of 268 mg while perispleen fat averaged 98 mg. Interscapular fat pads weighed an average of 61 mg. The fat pads were minced and incubated for 10 min in pre-warmed (37°C) Krebs-Ringer phosphate (KRP) buffer (12.5 mM HEPES, 120 mM NaCl, 6 mM KCl, 1.2 mM MgSO4, 1 mM CaCl2, 0.6 mM Na2HPO4, 0.4 mM Na2H2PO4, 2.5 mM D-glucose, and 2 % bovine serum albumin, pH 7.4), thereafter the samples were incubated with Type II collagenase (1mg/ml) for 40 min at 37°C with constant shaking at 60 rpm. The WAT or BAT cells were passed through a 100 μm cell strainer; cells were then centrifuged at 370 × g for 1 minute and washed with Krebs Ringer buffer twice. Additionally, the adipocytes isolated from the paired gonadal fat pads were separated into 2 major fractions. The floating upper layer was primarily white adipocytes and the pelleted fraction was a mixture of stromal-vascular fraction (SVF) cells containing macrophages. Both cell fractions were collected and washed twice with KRP buffer.
WAT and BAT cells were scored for numbers and viability on a hemacytometer using trypan blue exclusion. Viability was 91±0%, 81±3% and 87±0% for PEC, BAT and SVF cells, respectively, after isolation and collagenase treatment. In collagenase-digested samples, we isolated an average of 1.3 × 105 C2D macrophage cells per mouse from gonadal WAT and 8.9 × 104 C2D macrophage cells from perispleen BAT or 3.5 × 104 C2D macrophage cells from interscapular BAT. 1 × 105 cells were pelleted onto a cytospin slide for differential staining. A mixture of white adipocytes (upper layer) and SVF cells was co-incubated at 37°C in DMEM10, for 16 hours at a concentration of 1 × 105 cells/ml in a 150-mm culture dish. The adipocytes remained dispersed in the medium and the SVF cells attached to the 150-mm culture plate. The adipocytes isolated from perispleen adipose were collected from the cell pellets and washed twice with KRP buffer. Cells isolated from perispleen BAT (3 × 106) were cultured at 37°C in DMEM10 in a 150-mm culture plate for 16 hours.
2.6 Flow cytometry analysis of C2D macrophage cells
Cell sorting was based on C2D macrophage cell CFDA-SE fluorescence, with the lowest 10 % of the positive cells not selected. Briefly, cell sorting was performed with either a FACSVantage SE cell sorter (Becton Dickson. Rockville, MD) or a MoFlo XDP Sterile Cell Sorter (Beckman Coulter), using specimen optimization and calibration techniques according to the manufacture’s recommendations. Cells were sorted at a rate of 15,000 cells per second and approximately 1 × 106 viable (trypan blue exclusion), positive cells per group were collected on ice and centrifuged at 350 × g for 5 min at 4°C for PCR Array or qRT-PCR analysis.
We found a loss of cell surface markers following collagenase treatment. For example, Mac-2 was down regulated over 50% after a 40 minute collagenase treatment based on control PEC-C2D (data not shown). However, we were also concerned that this incubation would also influence the cells. Therefore, we also evaluated the changes in TNF gene expression over time after the PEC-C2D cells were cultured in vitro. We found C2D macrophage gene expression was reduced some but was still positive for at least 24 hrs (data not shown). Therefore, we felt a reasonable approach to phenotype the cell surface molecules of the recovered C2D macrophage cells from PEC, BAT and WAT would be to allow the cells to recover in vitro for 16 hours at 37°C in medium. Therefore, control or collagenase-treated C2D macrophage cells isolated from the peritoneal cavity (PEC-C2D), WAT and BAT were resuspended in DMEM4 and incubated for 16 hours prior to labeling cell surface proteins and assessment by flow cytometry.
Cells were transferred to wells of 96-well, round-bottom plates and they were blocked with PBS-goat serum (50:50; 50 μl) at 4 °C for 0.5 hour. Subsequently, macrophage cell surface proteins were identified by direct labeling. Briefly, blocked cells were incubated with the isotype or specific antibody diluted in Hank’s Buffered Salt Solution (HBSS; 0.137 M NaCl, 5.4 mM KCl, 0.25 mM Na2HPO4, 0.44 mM KH2PO4, 1.3 mM CaCl2, 1.0 mM MgSO4, 4.2 mM NaHCO3) for 1 hour in the dark at 4°C. After two washes with HBSS, cells were fixed in 1% formalin. Labeled cell surface proteins were assessed by flow cytometry. We gated on live, CFDA-SE-positive or CFDA-SE negative cells, subsequently we assayed for the presence or absence of the selected cell surface markers.
2.7 Real time quantitative RT-PCR analysis
RNA was obtained by suspending the pelleted cells in 1 ml of TriReagent (Molecular Research Center). The solution was transferred to 2.0 ml Heavy Phase Lock Gel tubes (5 Prime). 200 μl of chloroform was added and the mixture was shaken for 15 seconds. The samples were then centrifuged at 12,000 × g for 10 minutes at 4°C and the aqueous phase was transferred to clean 1.5 ml tubes. 500 μl of isopropanol was added and RNA was precipitated at −20°C for 24 hours. Samples were subsequently centrifuged at 12,000 × g for 10 minutes. The RNA pellet was washed with 1 ml of 70% ethanol and samples were centrifuged at 7.4 × g for 5 minutes. The 70% ethanol was decanted from the pellet; the pellet was allowed to slightly air dry and was resuspended in 50 μl of nuclease-free water. RNA samples were purified and DNAse treated with EZRNA total RNA kit (Omega Bio-Tek, Inc.). One step qRT-PCR was performed using the SuperScript III Platinum SYBR Green kit (Invitrogen; Carlsbad, CA) according to the manufacturer’s protocol. Primers were designed with the PrimerQuest software (IDT; http://www.idtdna.com) using sequence data from NCBI sequence database as following: TNF-αNM_013693) forward 5′-tctcatgcaccaccatcaaggact and reverse 5′-tgaccactctccctttgcagaact; IL-6 NM_031168.1) forward 5′-tctcatgcaccaccatcaaggact and reverse 5′-tgaccactctccctttgcagaact; IL1-βNM_008361) forward 5′-aagggctgcttccaaacctttgac and reverse 5′-atactgcctgcctgaagctcttgt; Arg-1(NM_007482) forward 5′-tggctttaaccttggcttgcttcg and reverse 5′-catgtggcgcattcacagtcactt; Ym-1 (M94584) forward 5′-caccatggccaagctcattcttgt and reverse 5′-tattggcctgtccttagcccaact; Fizz-1(NM_020509.3) forward 5′-actgcctgtgcttactcgttgact and reverse 5′-aaagctgggttctccacctcttca; Prdm16 (BC059838) forward 5′-tcatcccaggagagctgcatcaaa and reverse 5′-atcacaggaacacgctacacggat; Ucp-1 (NM009463.3) forward 5′-ttgagctgctccacagcgcc and reverse 5′-gttgcctgatgcgggcacga; β-actin (NM_007393) forward 5′-tgtgatggtgggaatgggtcagaa and reverse 5′-tgtggtgccagatcttctccatgt. The qRT-PCRs were performed in a Cepheid SmartCycler System (Sunnyvale, CA). Fold increase in transcript expression was calculated: E (gene of interest)Δct target/E (housekeeping)Δct housekeeping where E (efficiency)=10(−1/slope) as was previously described [32].
2.8 PCR array analysis
Expression analysis of 84 cytokines, chemokines and corresponding receptor genes involved in inflammatory responses was performed with the mouse inflammatory cytokines and receptors RT2 profiler™ PCR array system (SuperArray Bioscience Corporation, Frederick, MD). 1.2 μg of total RNA was obtained from CFDA SE labeled C2D cells sorted from the PEC, WAT, and BAT of C57BL/6J mice (n= 2 pooled RNA samples, 4 mice per pooled sample). Genomic DNA was digested with RNAse-free DNAase, followed by first strand cDNA synthesis and then quantitative mRNA analysis according to manufacturer’s protocol. The quantitative real-time PCR array was done on a BioRad iCycler (BioRad Laboratory, Hercules, CA) performed with the RT2 SYBR Green/Fluorescein qPCR Master Mix (SuperArray Bioscience Corporation, Frederick, MD). Expression of mRNA for each gene was normalized to the expression of β-actin and compared to the data obtained with the negative control (RNA from cultured C2D cells) according to the ΔΔCt method [32].
2.9 Immunofluorescence and image analysis
Gonadal fat pads and perispleen adipose tissue were washed in Krebs-Ringer phosphate (KRP) buffer fixed in 10% formalin/PBS and were cut into 50-μm-thick slices using a TC-2 tissue sectioner (Sorvall Instruments). Tissue slices were mounted onto glass slides, and differential contrast interference (DIC) images of tissue and CFDA SE-labeled C2D macrophage cells were observed on a model LSM 5 Pascal Zeiss laser scanning confocal microscope. Tissues were visualized with 20X/0.5 and 40X/0.75 Plan Neofluor objectives with DIC. CFDA SE-labeled C2D macrophage cells were visualized using the 488-nm line of an argon ion gas laser (excitation of CFDA SE), an FT 488 primary dichroic beam splitter, a FT 545 secondary dichroic beam slitter, a 505-nm to 530-nm-bandpass filter, a photomultiplier tube, and LSM5 Pa software, version 3.2 SP2. The number of CFDA SE-labeled macrophages per square micrometer of area of adipose tissue was determined using ImageJ v1.37 (NIH). Images were then imported to Adobe Photoshop (Adobe Systems, Inc.) for figure processing.
2.10 Statistical analysis
Flow cytometry data, cell distribution data and qRT-PCR data were presented as the mean ± standard error of mean (SEM) of independent experiments (n=3 samples, 3 mice per sample unless stated otherwise in the Figure legend). Differences in mean were determined using Student’s t test (paired, two-tailed) or were determined using the Mann-Whitney rank-sum test. Differences in cell distribution were assessed using the Chi-Square (χ2) test. All tests were calculated using the StatMost statistical package (Data XIOM, Los Angeles, CA). Differences were considered significantly different when P<0.05. To assess differences in the samples assayed in the PCR Arrays the mean Ct values between macrophage isolates were compared using minimum significant difference (MSD) [33]. Any difference between means greater than or equal to the MSD was considered to be a statistically different, while differences less than the minimum significant difference were considered to be non-significant. An MSD was calculated using the following equation: MSD = 2 × (s_pool) x (√(1/n1+1/n2)), where s pool is the standard deviation pooled across all genes and all groups, n1 and n2 are the numbers of replicates for the two treatments. In this study, n1=n2=2, and the pooled standard deviation was equal to 1.61 and the MSD was equal to 3.22.
3. Results
3.1 Morphological and phenotypic changes of C2D macrophage cells in response to adipocytes in vitro
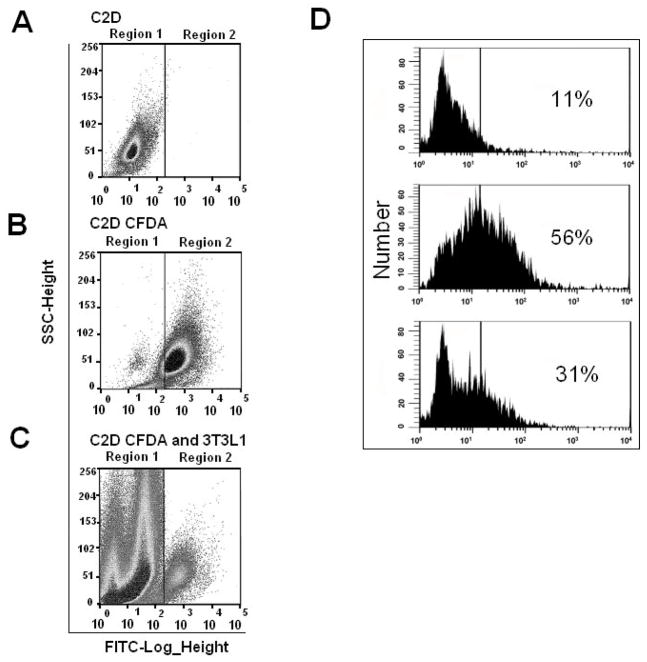
To determine how different tissue environments impact macrophage phenotypes, we investigated macrophage responses to preadipocytes and adipocytes. In our experiments we used the C2D macrophage cell line and 3T3L1 cells before or after, differentiation into adipocytes. We previously established the specific macrophage phenotype expressed by C2D cells after they were injected i.p. (PEC-C2D) or before they respond to other microenvironments [15,16,26]. In order to visualize and identify the C2D macrophages, C2D cells were labeled with CFDA-SE prior to their injection into the animals or their co-culture with (3T3L1) preadipocyte/adipocyte cells in vitro for two days. Subsequently, these cells were recovered and analyzed by fluorescence-activated cell sorting. Cells from peritoneal lavages, BAT and SVF cells from WAT were sorted by FACS analysis as described in Figure 1. C2D cells grown in vitro (Figure 1A, region 1) and C2D CFDA-SE cells (Figure 1B, region 2) were used as negative and positive controls for gating, respectively, and for sorting C2D CFDA-SE positive cells from mixed cell samples such as C2D CFDA-SE macrophages co-cultured with 3T3L1 adipocytes (Figure 1C).
Fig. 1. Gating strategies for cell sorting of CFDA-SE positive macrophage cells and effects of in vitro culture and collagenase treatment on C2D macrophage phenotype.
A) C2D macrophage cells were sorted based on negative expression of CFDA-SE, B) C2D CFDA-SE macrophage cells were sorted based on positive CFDA-SE expression; C) Example of C2D CFDA-SE+ cells that were sorted from a mixed cell sample such as C2D CFDA-SE+ macrophages (region 2) co-cultured with 3T3L1 adipocytes (region 1); D) PEC-C2D macrophages were treated with isotype control antibody (top) or anti Mac-2 antibody (middle and bottom) then assessed by flow cytometry. Cells were treated with PBS (middle) or collagenase (bottom) for 40 minutes before antibody probing.
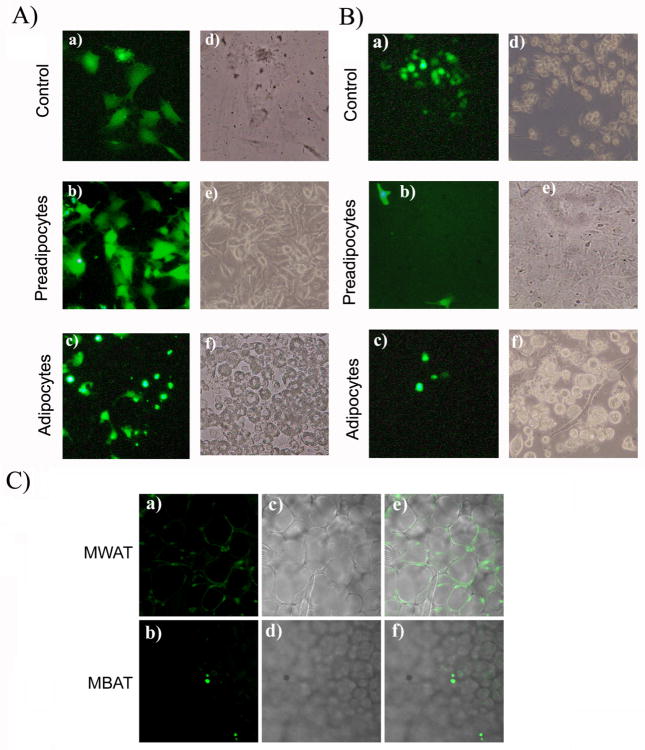
In comparison to the larger stretched morphology of C2D macrophage cells grown in vitro (Figure 2A, panels a and d), the PEC-C2D macrophage cells were round after cell isolation (Figure 2B, panels a and d). When C2D or PEC-C2D macrophage cells were cultured with preadipocytes, the cells stretched out (Figure 2A and 2B, panels b and e). In contrast, when C2D macrophage cells or PEC-C2D cells were co-cultured with differentiated 3T3L1 adipocytes, we found that the cells maintained a mostly smaller, round morphology (Figure 2A and 2B, panels c and f), suggesting that differentiated 3T3L1 adipocytes inhibit normal adherence and stretching of the C2D macrophage cells.
Fig. 2. Change in C2D macrophage cell morphology during co-cultured with adipocytes or pre-adipocytes in vitro and after infiltration into BAT or WAT in vivo.
A) C2D macrophage cells were labeled with CFDA-SE or B) C2D macrophage cells labeled with CFDA-SE and isolated from peritoneal cavity (PEC-C2D) were cultured a) alone or co-cultured with b) 3T3L1 pre-adipocytes or c) adipocytes as described in the Materials and Methods. Panels a, b and c; Cells viewed on the fluorescent microscope (Magnification × 200). Panels d, e and f are phase contrast images of cells in a, b and c. C) WAT-C2D and BAT-C2D were collected from mice two days after adoptive transfer. C2 D macrophages, WAT and BAT were processed as described in Materials and Methods. Panels a and c images from the confocal microscope (× 100). Panels b and d are phase contrast images of the same fields.
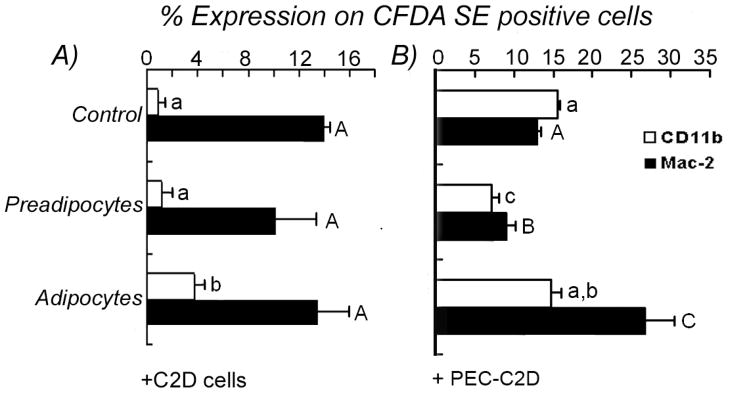
The morphological differences in C2D and PEC-C2D macrophage cells co-cultured with adipocytes were accompanied by changes in cell phenotype defined by cell surface molecules detected using flow cytometry. In vitro, 15% of the C2D macrophage cells expressed Mac-2 but not CD11b; indicative of an immature macrophage phenotype [15]. C2D macrophage cells acquire a more differentiated phenotype after adoptive transfer in vivo with high levels of macrophage-specific molecules CD11b, Mac2, F4/80, cfms and low levels of CD11c, and Gr-1 (Ly6G) [15,16]. We observed no change in the numbers of cells that expressed either of the cell surface markers on C2D macrophage cells co-cultured with pre-adipocytes compared to C2D macrophage cells cultured alone (Figure 3A). However, we observed a statistically significant (P<0.05) increase in the number of C2D macrophages that expressed CD11b when co-cultured with 3T3L1 adipocytes (Figure 3A). For PEC-C2D macrophage cells, their maturation in the peritoneal cavity is accompanied by an increase in the number of cells that express CD11b [15,16]. Significantly more PEC-C2D macrophage cells co-cultured in vitro with adipocytes expressed Mac-2 than PEC-C2D macrophage cells cultured alone or with preadipocytes (Figure 3B, P<0.05). We observed a significant decrease (P<0.05) in the number of PEC-C2D macrophage cells expressing CD11b and Mac-2 when incubated with preadipocytes (Figure 3B).
Fig. 3. Phenotype changes of C2D macrophage cells co-cultured with adipocytes or pre-adipocytes in vitro.
C2D or PEC-C2D cells labeled with CFDA-SE were cultured alone or co-cultured with 3T3L1 adipocytes or pre-adipocytes andthe cell mixtures were immunostained for flow cytometry as described in Materials and Methods. C2D macrophage cells phenotypes were analyzed within CFDA-SE+ population. A) C2D macrophage cells grown in vitro were cultured alone, co-cultured with 3T3L1 adipocytes or with pre-adipocytes. B) PEC-C2D macrophage cells were cultured alone, co-cultured with adipocytes or with pre-adipocytes. The data is presented as the mean ± SEM (n= 3 independently collected samples per treatment group). Different letters indicate a significant difference between control, preadipocytes or adipocytes for CD11b (lower case) or Mac-2 (upper case) cell surface proteins. A P value of < 0.05 was considered significant.
3.2 Assessment of C2D macrophage cells after trafficking into WAT and BAT
We previously found that C2D macrophage cells could be isolated from gonadal WAT [15]. However, macrophage trafficking to BAT has not been well characterized. We used confocal microscopy to visualize and count C2D macrophages in WAT and BAT. We counted an average of 97 C2D macrophages/mm2 in BAT compared to 146 C2D macrophages/mm2 in WAT (P>0.05, T test, n≥9 fields scored per tissue; Figure 2C). We also assessed if C2D macrophage immigration to WAT and BAT induced inflammation. We did differential staining of white blood cells isolated from the adipose tissue 36 hrs after i.p. injection of C2D macrophage cells. This survey revealed distinct cell distributions. The white cell distribution in WAT was 8±1% PMN, 60±2% macrophage/monocyte and 33±1% lymphocytes compared to 3±2% PMN, 79±2% macrophage/monocytes and 18±1% lymphocytes in BAT (P<0.01; χ2 test). We also assessed C2D macrophage cell localization within WAT and BAT. WAT-C2D appeared between adipocytes and some appeared to spread around the adipose cells (Figure 2C, panels a, c, e). In contrast, BAT-C2D appeared only between adipocytes and generally had a round appearance (Figure 2C, panels b, d, f).
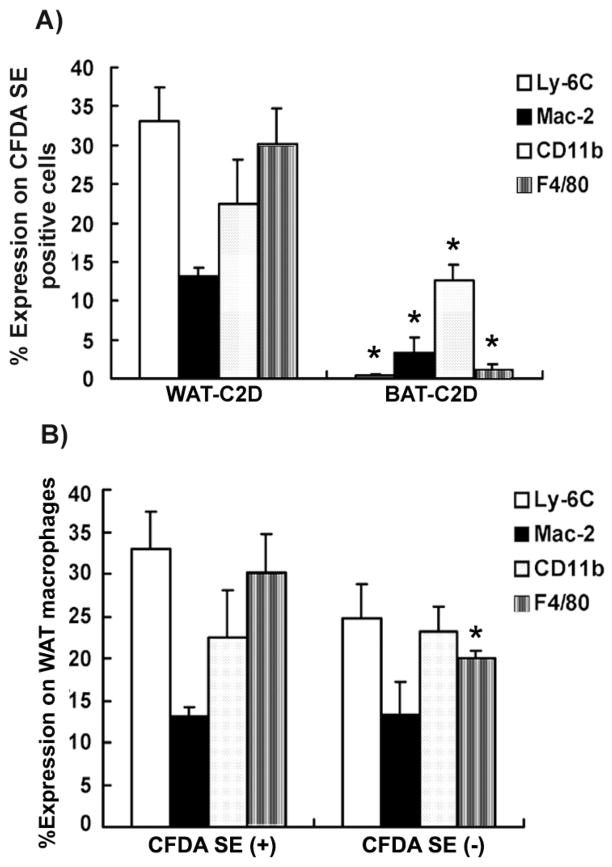
To determine if the C2D macrophage cells isolated from BAT and WAT maintained the same phenotype they expressed in the peritoneum or if they responded to the different tissue environments, we compared BAT-C2D and WAT-C2D for the expression of Ly-6C, Mac-2, CD11b and F4/80. Cells were labeled with CFDA and the CFDA-SE positive cells were assessed (Figure 4A). We observed that 33% of the WAT-C2D expressed Ly-6C, while there were very few Ly6C-positive (<1%) BAT-C2D. A significantly higher number of WAT-C2D macrophage cells expressed (P<0.05) Mac-2 compared to BAT-C2D macrophage cells. We observed over 30% of the WAT-C2D macrophage cells expressed F4/80, but almost no BAT-C2D expressed F4/80 (Figure 4A). Significantly more WAT-C2D cells expressed CD11b than BAT-C2D (Figure 4A). When we compared C2D macrophage phenotype to the phenotype of the recipients’ macrophages, we observed that a higher percentage of WAT-C2D cells expressed F4/80 compared to the recipient’s macrophages (CFDA negative cells; Figure 4B). We detected no significant differences in the % positive cells that expressed Ly-6C, Mac-2 or CD11b when we compared WAT-C2D and recipient macrophages (Figure 4B).
Figure 4. Phenotype changes in C2D macrophage cells isolated from WAT or BAT in vivo after i.p. adoptive transfer.
C2D macrophage cells were isolated from WAT and BAT and immunostained to detect Ly-6C, Mac-2, CD11b and F4/80 by flow cytometry. A) Surface marker expression was assessed on CFDA-SE-positive cells isolated from WAT or BAT. B) Surface marker expression was assessed on CFDA-SE-positive cells (left) or CFDA-SE-negative cells (right) in the stromal vascular fraction isolated from WAT. The data is represented as mean ± SEM (n= 3–6 independently collected samples per adipose tissue type). Comparisons were done between samples stained for the same surface markers in panels A or B. * indicates a significant difference with a P value of < 0.05.
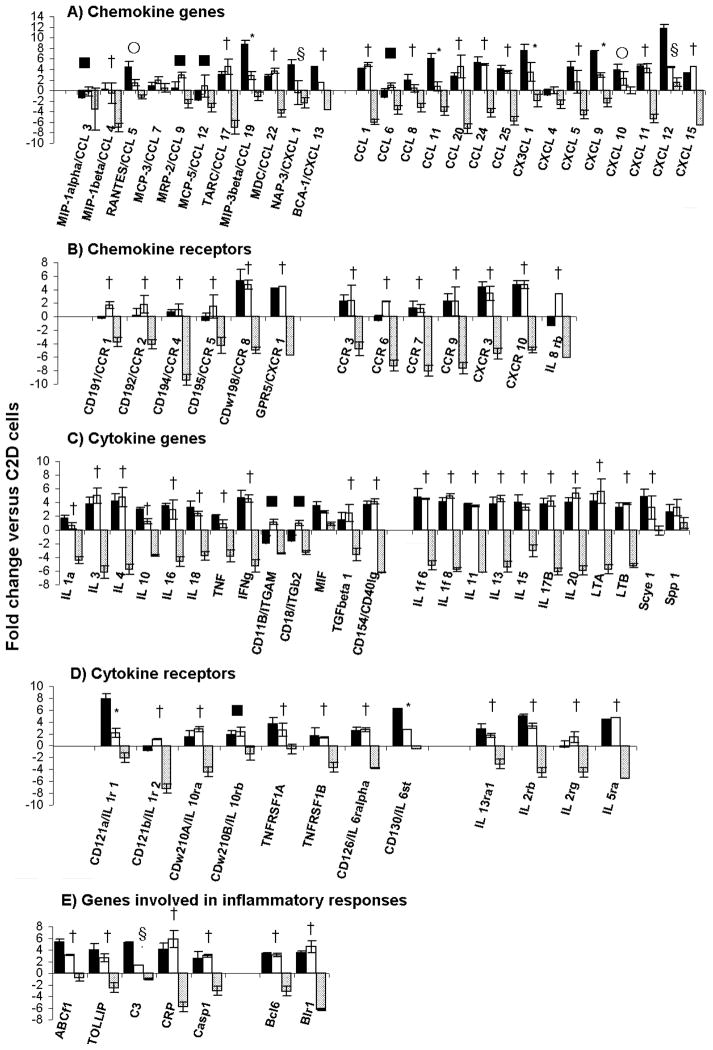
To further characterize the impact of the adipose microenvironment on recently migrated macrophages, we measured transcript levels in C2D macrophages that were isolated and sorted from the peritoneal cavity, WAT or BAT. We measured the expression of an array of inflammatory chemokines and cytokines and their receptor genes by quantitative PCR. As shown in Figure 5, the overall expression of chemokine, cytokine and receptor genes was dramatically down-regulated in BAT-C2D cells relative to the gene expression of C2D cells maintained in vitro, compared to those of PEC-C2D and/or sorted WAT-C2D cells. When WAT-C2D cells were compared to PEC-C2D macrophage cells, transcript levels for several chemokines were lower (Figure 5A). These included MIP-3b/CCL19 (3 vs. 9 fold), NAP-3/CXCL1 (−0.5 vs. 5 fold), CCL11 (1 vs. 6 fold), CXCL5 (2 vs. 4 fold), CXCL9 (3 vs. 7) and CXCL12 (4 vs. 12 fold). MIP-1a/CCL3 and MCP-5/CCL12 were also down-regulated in PEC-C2D compared to C2D macrophages grown in vitro.
Fig. 5. Expression analysis of C2D macrophage cells isolated from WAT or BAT.
C2D macrophage cells were isolated from BAT and WAT by collagenase treatment as described in Materials and Methods. PEC-C2D (black bars), WAT-C2D (white bars) or BAT-C2D (grey bars) were purified by FACS and gene transcripts were quantified by qRT-PCR as described in the Materials and Methods. The data is presented as the mean ± SEM (n=2 independent RNA samples; fat pads from 4 mice per pooled sample). Significant differences found between: * PEC-C2D vs. WAT-C2D, PEC-C2D vs. BAT-C2D and WAT-C2D vs. BAT-C2D; † PEC-C2D vs. BAT-C2D and WAT-C2D vs. BAT-C2D; § PEC-C2D vs. WAT-C2D and PEC-C2D vs. BAT-C2D; ■ WAT-C2D vs. BAT-C2D; ○ PEC-C2D vs. WAT-C2D.
Interestingly we found that two cytokines receptor genes had higher transcript levels in PEC-C2D macrophage cells compared to WAT-C2D macrophage cells; CD121a/IL 1r1 (8 vs. 2 fold) and CD130/IL6st (6 vs. 3 fold). Additionally, C3 (5 vs.1 fold) had higher expression in PEC-C2D cells compared to WAT C2D cells (Figure 5E).
3.3 Macrophages gene expression in response to WAT and BAT in vitro
C2D macrophage cells exhibited distinct phenotypes in response to WAT or BAT adipose environments. Therefore, to confirm that C2D macrophage behavior reflected that of primary macrophages, we indirectly co-cultured BM-Mo in transwell plates (top) with collagenase-digested WAT and BAT (bottom). RNA from the BM-Mo was isolated and the transcript levels of TNF-α, IL-6, IL-1β, Arg-1, Ym-1 and Fizz-1 were assessed using qRT-PCR (Table 1). WAT incubated BM-Mo macrophages had higher TNF-α and IL-6 transcript levels than BAT-BM Mo (Table 1). This would be consistent with the observations seen with C2D macrophages. In contrast, there were no differences in IL-1β or the anti inflammatory genes Arg-1, Ym-1 and Fizz-1 between BM-Mo inoculated in WAT or BAT (Table 1).
Table 1.
Comparison of M1 and M2 gene transcripts in bone marrow derived macrophage cells after 24 h indirect co-culture with digested adipose tissues.
| % gene transcripts compared to bone marrow derived macrophage cells | ||
|---|---|---|
| Transcript | BAT-BM Mo1 | WAT-BM Mo1 |
| TNF-α | 177 ± 39 | 370 ± 46† |
| IL-6 | 285 ± 47 | 564 ± 53† |
| IL-1β | 243,871 ± 154,323 | 170,380 ± 29,734 |
| Arg-1 | 158 ± 105 | 168 ± 45 |
| Ym-1 | 1 ± 1 | 5 ± 3 |
| Fizz-1 | 163 ± 125 | 212 ± 35 |
Bone marrow derived macrophage cells were indirectly co-cultured (plate bottom) with digested fat pads (in transwell insert); WAT-BM Mo bone marrow derived macrophage cells co-cultured with paired gonadal fat pads; BAT-BM Mo bone marrow derived macrophage cells co-cultured with digested perispleen fat.
% gene transcript levels were calculated relative to bone marrow derived macrophages differentiated in vitro as described in the materials and methods.
Number represents average ± standard error of the mean of 3 or 4 independent mouse samples.
indicates statistical difference compared to BAT-BM Mo, P<0.05.
4. Discussion
Adipose tissue contains a heterogeneous array of cells including preadipocytes and adipocytes along with resident and inflammatory macrophages constituting up to 40 percent of the cell population [34]. Additionally, the trafficking of C2D macrophage cells to both WAT and BAT provided a unique opportunity to determine the impact of these distinct adipose environments on recently immigrating macrophages and how different microenvironments affect macrophage plasticity. We used the C2D macrophage cell line to investigate this question. This is a powerful model because the cells are phenotypically defined both in vitro and in vivo and the phenotypic change of the C2D macrophages in response to WAT or BAT paralleled the in vitro response of primary macrophages {Table 1; also see reference [35]}. Therefore, by knowing the characteristics of the C2D macrophages before and after exposure to the different adipose tissues we know exactly what changes are due to their immediate exposure to different microenvironments. Indeed, the finding that preadipocytes allowed C2D macrophage cells to spread regardless of their differentiation state, while white adipocytes inhibited macrophage spreading supports the hypothesis that macrophage phenotype is dependent on the adipose tissue microenvironment.
WAT has been well characterized. There are differences in CD68+ macrophages between visceral and subcutaneous WAT [36]. “Obese” WAT has increased proinflammatory cytokine transcripts [37,38] and secreted cytokines such as TNF [39], angiotensinogen, PAI-1, PGAR/FIAF, IL-6, leptin, and resistin [40–42]. In particular, isolated adipocytes secrete TNF-alpha, IL-6, IL-8, IL-1Ra, IL-10, leptin, adiponectin, resistin [41] and visfatin [40,43] and various populations of CD14+ CD31+ adipose tissue macrophages (ATMs) [40] or MGL1+ ATMs have increased IL10, Arg1, and Pgc1b transcript levels [44] or secrete MCP-1, MIP-1α and IL-8 [43]. Nos2 and IL1b transcripts also go up in MGL1− CCR2+ macrophage populations around necrotic adipocytes [44]. One explanation for the differences may be the origins of the tissues. Brown adipose cells may be more closely related to muscle cells than white adipose cells [45,46]. Macrophages in normal muscle are angiogenic or anti-inflammatory [47]. In addition, BAT and WAT express and secrete different autocrine, paracrine and endocrine signals. WAT has been recognized as an endocrine organ. It produces and secretes a plethoric collection of adipokines [48–50]. Among them for example, adipsin, leptin and adiponectin are highly expressed in WAT, whereas their production in BAT is associated only with thermogenically inactive BAT cells [51]. In contrast, BAT has been reported to express other cellular mediators, such as basic fibroblast growth factor [52] and prostaglandins E2 and F2α [53]. BAT cells also produce T4 thyroxine deiodinase type II [54] and nitric oxide synthase enzymes (eNOS and iNOS) [55] which enables it to produce T3 and NO, respectively. Uncoupling protein, unique to BAT, also exhibits chloride channel properties [56]. Chloride channels can regulate NADPH oxidase membrane depolarization [57] and can regulate phagocyte cell function [58]. Therefore, it is possible that BAT can regulate the C2D macrophage cell phenotype because of UCP’s unique ability to regulate oxidative metabolism. While it is not clear at present what specific molecular mechanisms are responsible for the distinct phenotypic differences between WAT-C2D and BAT-C2D, we have shown that there are significant interactions between adipocytes and macrophages that is mediated by cytokines and cell-cell contact that affects the differentiation and function of both macrophages and adipocytes [35]. This current study extends the macrophage interaction to include brown adipocytes by showing that C2D macrophages traffic to the BAT and they acquire a phenotype unique to that tissue. The data support the hypothesis that macrophage plasticity is dependent upon environmental signals and is not predetermined as some have suggested [10].
It is possible that the adoptive transfer technique or procedures used in recovering macrophages for our study may have impacted the results. First, adoptive transfer could have induced a peritonitis or inflammation in the adipose tissue. We do not believe this to be the case. Although, we detected some neutrophils (28% PMNs in PEC, 3% in BAT and 10% in WAT), the lack of an acute inflammatory response where one would expect a large PMN inflammation (>80% PMN) [59] and extensive macrophage activation [60] suggests that we did not induce a peritonitis or abnormal inflammation in these tissues. We found that the recipient host macrophages that were isolated from the SVF had a similar cell surface phenotype to the WAT-C2D cells. These data suggest that the C2D macrophages were acquiring a “resident macrophage phenotype” as opposed to a proinflammatory phenotype which would be expected of recently immigrating macrophages in obese mice [18,43]. This hypothesis is supported by the fact that we saw inconsistent evidence of a proinflammatory phenotype in WAT-C2D because both M1 (e.g. TNF-α) and M2 (IL-10) [61,62] markers were up regulated. The differential counts of the cells in WAT and BAT also did not reflect an inflammatory milieu. The CD11b expression on WAT-C2D macrophages would also be reflective of cells which are undergoing normal cell trafficking [63].
Ruan et al. found that the isolation of adipocytes with collagenase for 2 hours led to the activation of the adipocytes when they were assayed in vitro [64]. Our macrophages were isolated with a 40 minute collagenase treatment and that exposure could have affected them and we cannot rule out this possibility. However, the adipocytes were separated from the macrophages quickly and the expression of some of the genes of interest (e.g. TNF and TNFR) take several hours to upregulate [64]. In addition, if collagenase had a general activating action on the C2D macrophage cells [65], we probably would not have seen the general down regulation of C2D macrophage cell gene transcripts in BAT unless BAT had a suppressive environment.
Lastly, we were concerned that the 16 h incubation that we included before we assessed surface marker expression could have affected the macrophage phenotype [66]. We see changes in C2D macrophages when they are reintroduced to in vitro culture. However, two observations suggest that the cell surface expressions we report are an accurate sampling of the C2D macrophage phenotype. 1) The changes induced in vivo were still evident after an additional 16 hours of in vitro culture and 2) the differential expression of surface markers such as CD11b in BAT and WAT paralleled the general changes in transcript level in those same tissues. The RNA used for those analyses was not subject to the 16 h. incubations.
In summary, the WAT microenvironment altered C2D macrophage cells differently than BAT. The changes in WAT were dependent upon the differentiation of both the macrophages and the adipocytes. In addition, WAT caused C2D macrophage cells to upregulate many genes and molecules compared to when they were isolated from BAT. To our knowledge, this is the first study to directly compare the macrophages that have recently trafficked to different adipose tissues in the absence of complicating chronic diseases or altered genetic states. The evidence that infiltrating macrophages begin to display unique tissue-specific phenotypes in normal mice reaffirms the adaptive nature of macrophages to their environment. Determining the properties of adipose tissue that make BAT and WAT so different may give us clues on how to regulate macrophages to prevent disease.
Highlights.
C2D macrophage phenotype change rapidly in response to different microenvironments.
Macrophage phenotype responses are dependent on the differentiated stages of the macrophages and adipocytes.
Brown adipose tissue has distinct impact on macrophages compared to white adipose tissue.
Acknowledgments
We thank Ms. Tammy Koopman for her assistance with flow cytometry, Dr. Dan Boyle for his help with confocal microscopy, and Dr. Kurt Zhang for his help with statistical analysis. We thank Ms. Alison Luce-Fedrow for her laboratory assistance and Ms. Lea Dib for her input and review of the manuscript. This project has been supported by American Heart Association grant 0950036G, NIH grants AI55052, AI052206, AI088070, RR16475, RR17686, RR17708, NASA grants NAG2-1274 and NNX08BA91G, European Commission (Grant HEALTH-F4-2008-223450), funding from Diabetes UK and the Wellcome Trust, the Kansas Agriculture Experiment station and the Terry C. Johnson Center for Basic Cancer Research. This is Kansas Agriculture Experiment Station publication 09-132-J.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Meltzer M, Occhionero M, Ruco L. Macrophage activation for tumor cytotoxicity: Regulatory mechanisms for induction and control of cytotoxic activity. Fed Proc. 1982;41:2198–2205. [PubMed] [Google Scholar]
- 2.Ruco L, Meltzer M. Macrophage activation for tumor cytotoxicity: Induction of tumoricidal macrophages by supernatants of PPD-stimulated Bacillus Calmette-Guerin-immune spleen cell cultures. J Immunol. 1977;119:889–896. [PubMed] [Google Scholar]
- 3.Wiktor-Jedrzejczak W, Dzwigala B, Szperl M, Maruszynski M, Urbanowska E, Szwech P. Colony-stimulation factor 1-dependent resident macrophages play a regulatory role in fighting Escherichia coli fecal peritonitis. Infect Immun. 1996;64:1577–1581. doi: 10.1128/iai.64.5.1577-1581.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Held TK, Weihua X, Yuan L, Kalvakolanu DV, Cross AS. Gamma interferon augments macrophage activation by lipopolysaccharide by two distinct mechanisms, at the signal transduction level and via an autocrine mechanism involving tumor necrosis factor alpha and interleukin-1. Infect Immun. 1999;67:206–12. doi: 10.1128/iai.67.1.206-212.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dorger M, Munzing S, Allmeling AM, Messmer K, Krombach F. Phenotypic and functional differences between rat alveolar, pleural, and peritoneal macrophages. Exp Lung Res. 2001;27:65–76. doi: 10.1080/019021401459770. [DOI] [PubMed] [Google Scholar]
- 6.Cailhier JF, Partolina M, Vuthoori S, Wu S, Ko K, Watson S, Savill J, Hughes J, Lang RA. Conditional macrophage ablation demonstrates that resident macrophages initiate acute peritoneal inflammation. J Immunol. 2005;174:2336–42. doi: 10.4049/jimmunol.174.4.2336. [DOI] [PubMed] [Google Scholar]
- 7.Takahashi K, Naito M, Takeya M. Development and heterogeneity of macrophages and their related cells through their differentiation pathways. Pathol Int. 1996;46:473–85. doi: 10.1111/j.1440-1827.1996.tb03641.x. [DOI] [PubMed] [Google Scholar]
- 8.Naito M, Umeda S, Yamamoto T, Moriyama H, Umezu H, Hasegawa G, Usuda H, Shultz L, Takahashi K. Development, differentiation, and phenotypic heterogeneity of murine tissue macrophages. J Leukoc Biol. 1996;59:133–138. doi: 10.1002/jlb.59.2.133. [DOI] [PubMed] [Google Scholar]
- 9.Gordon S, Taylor PR. Monocyte and macrophage heterogeneity. Nat Rev Immunol. 2005;5:953–64. doi: 10.1038/nri1733. [DOI] [PubMed] [Google Scholar]
- 10.Geissmann F, Manz MG, Jung S, Sieweke MH, Merad M, Ley K. Development of monocytes, macrophages, and dendritic cells. Science. 2010;327:656–61. doi: 10.1126/science.1178331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rutherford MS, Witsell A, Schook LB. Mechanisms generating functionally heterogeneous macrophages: chaos revisited. J Leukoc Biol. 1993;53:602–18. doi: 10.1002/jlb.53.5.602. [DOI] [PubMed] [Google Scholar]
- 12.Laskin DL, Weinberger B, Laskin JD. Functional heterogeneity in liver and lung macrophages. J Leukoc Biol. 2001;70:163–70. [PubMed] [Google Scholar]
- 13.Stout RD, Suttles J. Functional plasticity of macrophages: reversible adaptation to changing microenvironments. J Leukoc Biol. 2004;76:509–13. doi: 10.1189/jlb.0504272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Geissmann F, Jung S, Littman DR. Blood monocytes consist of two principal subsets with distinct migratory properties. Immunity. 2003;19:71–82. doi: 10.1016/s1074-7613(03)00174-2. [DOI] [PubMed] [Google Scholar]
- 15.Potts BE, Hart ML, Snyder LL, Boyle D, Mosier DA, Chapes SK. Differentiation of C2D macrophage cells after adoptive transfer. Clin Vaccine Immunol. 2008;15:243–252. doi: 10.1128/CVI.00328-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Potts BE, Chapes SK. Functions of C2D macrophage cells after adoptive transfer. J Leukoc Biol. 2008;83:602–609. doi: 10.1189/jlb.0607365. [DOI] [PubMed] [Google Scholar]
- 17.Fain JN, Madan AK, Hiler ML, Cheema P, Bahouth SW. Comparison of the release of adipokines by adipose tissue, adipose tissue matrix, and adipocytes from visceral and subcutaneous abdominal adipose tissues of obese humans. Endocrinology. 2004;145:2273–82. doi: 10.1210/en.2003-1336. [DOI] [PubMed] [Google Scholar]
- 18.Fantuzzi G. Adipose tissue, adipokines, and inflammation. J Allergy Clin Immunol. 2005;115:911–9. doi: 10.1016/j.jaci.2005.02.023. quiz 920. [DOI] [PubMed] [Google Scholar]
- 19.Klaus S. Functional differentiation of white and brown adipocytes. Bioessays. 1997;19:215–23. doi: 10.1002/bies.950190307. [DOI] [PubMed] [Google Scholar]
- 20.Adachi K, Miki M, Tamai H, Tokuda M, Mino M. Adipose tissues and vitamin E. J Nutr Sci Vitaminol (Tokyo) 1990;36:327–37. doi: 10.3177/jnsv.36.4-supplementi_327. [DOI] [PubMed] [Google Scholar]
- 21.Himms-Hagen J. Brown adipose tissue thermogenesis: interdisciplinary studies. Faseb J. 1990;4:2890–8. [PubMed] [Google Scholar]
- 22.Nedergaard J, Bengtsson T, Cannon B. Unexpected evidence for active brown adipose tissue in adult humans. Am J Physiol Endocrinol Metab. 2007;293:E444–52. doi: 10.1152/ajpendo.00691.2006. [DOI] [PubMed] [Google Scholar]
- 23.Virtanen KA, Lidell ME, Orava J, Heglind M, Westergren R, Niemi T, Taittonen M, Laine J, Savisto NJ, Enerback S, Nuutila P. Functional brown adipose tissue in healthy adults. N Engl J Med. 2009;360:1518–25. doi: 10.1056/NEJMoa0808949. [DOI] [PubMed] [Google Scholar]
- 24.Lee P, Greenfield JR, Ho KK, Fulham MJ. A critical appraisal of prevalence and metabolic significance of brown adipose tissue in adult humans. Am J Physiol Endocrinol Metab. 2010 doi: 10.1152/ajpendo.00298.2010. [DOI] [PubMed] [Google Scholar]
- 25.Villena JA, Cousin B, Penicaud L, Casteilla L. Adipose tissues display differential phagocytic and microbicidal activities depending on their localization. Int J Obes Relat Metab Disord. 2001;25:1275–80. doi: 10.1038/sj.ijo.0801680. [DOI] [PubMed] [Google Scholar]
- 26.Beharka AA, Armstrong JW, Chapes SK. Macrophage cell lines derived from major histocompatibility complex II-negative mice. In Vitro Cell Dev Biol. 1998;34:499–507. doi: 10.1007/s11626-998-0085-y. [DOI] [PubMed] [Google Scholar]
- 27.Xie L, Boyle D, Sanford D, Scherer PE, Pessin JE, Mora S. Intracellular trafficking and secretion of adiponectin is dependent on GGA-coated vesicles. J Biol Chem. 2006;281:7253–9. doi: 10.1074/jbc.M511313200. [DOI] [PubMed] [Google Scholar]
- 28.Rodbell M. Localization of Lipoprotein Lipase in Fat Cells of Rat Adipose Tissue. J Biol Chem. 1964;239:753–5. [PubMed] [Google Scholar]
- 29.Rodbell M. Metabolism of Isolated Fat Cells. I. Effects of Hormones on Glucose Metabolism and Lipolysis. J Biol Chem. 1964;239:375–80. [PubMed] [Google Scholar]
- 30.Seale P, Kajimura S, Yang W, Chin S, Rohas LM, Uldry M, Tavernier G, Langin D, Spiegelman BM. Transcriptional control of brown fat determination by PRDM16. Cell Metab. 2007;6:38–54. doi: 10.1016/j.cmet.2007.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Murholm M, Dixen K, Qvortrup K, Hansen LH, Amri EZ, Madsen L, Barbatelli G, Quistorff B, Hansen JB. Dynamic regulation of genes involved in mitochondrial DNA replication and transcription during mouse brown fat cell differentiation and recruitment. PLoS One. 2009;4:e8458. doi: 10.1371/journal.pone.0008458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pfaffl MW. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001;29:e45. doi: 10.1093/nar/29.9.e45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Phillips BM, Hunt JW, Anderson BS, Puckett HM, Fairey R, Wilson CJ, Tjeerdema R. Statistical significance of sediment toxicity test results: threshold values derived by the detectable significance approach. Environ Toxicol Chem. 2001;20:371–3. [PubMed] [Google Scholar]
- 34.Neels JG, Olefsky JM. Inflamed fat: what starts the fire? J Clin Invest. 2006;116:33–5. doi: 10.1172/JCI27280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Xie L, Ortega MT, Mora S, Chapes SK. Interactive changes between macrophages and adipocytes. Clin Vaccine Immunol. 2010;17:651–9. doi: 10.1128/CVI.00494-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bouloumie A, Curat CA, Sengenes C, Lolmede K, Miranville A, Busse R. Role of macrophage tissue infiltration in metabolic diseases. Curr Opin Clin Nutr Metab Care. 2005;8:347–54. doi: 10.1097/01.mco.0000172571.41149.52. [DOI] [PubMed] [Google Scholar]
- 37.Hotamisligil G, Shargill N, Spiegelman B. Adipose expression of tumor necrosis factor-alpha: Direct role in obesity-linked insulin resistance. Science. 1993;259:87–91. doi: 10.1126/science.7678183. [DOI] [PubMed] [Google Scholar]
- 38.Rotter V, Nagaev I, Smith U. Interleukin-6 (IL-6) induces insulin resistance in 3T3-L1 adipocytes and is, like IL-8 and tumor necrosis factor-alpha, overexpressed in human fat cells from insulin-resistant subjects. J Biol Chem. 2003;278:45777–84. doi: 10.1074/jbc.M301977200. [DOI] [PubMed] [Google Scholar]
- 39.Cancello R, Tordjman J, Poitou C, Guilhem G, Bouillot JL, Hugol D, Coussieu C, Basdevant A, Bar Hen A, Bedossa P, Guerre-Millo M, Clement K. Increased infiltration of macrophages in omental adipose tissue is associated with marked hepatic lesions in morbid human obesity. Diabetes. 2006;55:1554–61. doi: 10.2337/db06-0133. [DOI] [PubMed] [Google Scholar]
- 40.Fantuzzi G. Adipose tissue, adipokines, and inflammation. J Allergy Clin Immunol. 2005;115:911–9. doi: 10.1016/j.jaci.2005.02.023. [DOI] [PubMed] [Google Scholar]
- 41.Guerre-Millo M. Adipose tissue hormones. J Endocrinol Invest. 2002;25:855–61. doi: 10.1007/BF03344048. [DOI] [PubMed] [Google Scholar]
- 42.Ruan H, Lodish HF. Insulin resistance in adipose tissue: direct and indirect effects of tumor necrosis factor-alpha. Cytokine Growth Factor Rev. 2003;14:447–55. doi: 10.1016/s1359-6101(03)00052-2. [DOI] [PubMed] [Google Scholar]
- 43.Lumeng CN, DelProposto JB, Westcott DJ, Saltiel AR. Phenotypic switching of adipose tissue macrophages with obesity is generated by spatiotemporal differences in macrophage subtypes. Diabetes. 2008;57:3239–46. doi: 10.2337/db08-0872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Guzik TJ, Mangalat D, Korbut R. Adipocytokines - novel link between inflammation and vascular function? J Physiol Pharmacol. 2006;57:505–28. [PubMed] [Google Scholar]
- 45.Tseng YH, Kokkotou E, Schulz TJ, Huang TL, Winnay JN, Taniguchi CM, Tran TT, Suzuki R, Espinoza DO, Yamamoto Y, Ahrens MJ, Dudley AT, Norris AW, Kulkarni RN, Kahn CR. New role of bone morphogenetic protein 7 in brown adipogenesis and energy expenditure. Nature. 2008;454:1000–4. doi: 10.1038/nature07221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Seale P, Bjork B, Yang W, Kajimura S, Chin S, Kuang S, Scime A, Devarakonda S, Conroe HM, Erdjument-Bromage H, Tempst P, Rudnicki MA, Beier DR, Spiegelman BM. PRDM16 controls a brown fat/skeletal muscle switch. Nature. 2008;454:961–7. doi: 10.1038/nature07182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Arnold L, Henry A, Poron F, Baba-Amer Y, van Rooijen N, Plonquet A, Gherardi RK, Chazaud B. Inflammatory monocytes recruited after skeletal muscle injury switch into antiinflammatory macrophages to support myogenesis. J Exp Med. 2007;204:1057–1069. doi: 10.1084/jem.20070075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ahima RS, Flier JS. Adipose tissue as an endocrine organ. Trends Endocrinol Metab. 2000;11:327–32. doi: 10.1016/s1043-2760(00)00301-5. [DOI] [PubMed] [Google Scholar]
- 49.Maury E, Brichard SM. Adipokine dysregulation, adipose tissue inflammation and metabolic syndrome. Mol Cell Endocrinol. 2009 doi: 10.1016/j.mce.2009.07.031. [DOI] [PubMed] [Google Scholar]
- 50.Wozniak SE, Gee LL, Wachtel MS, Frezza EE. Adipose tissue: the new endocrine organ? A review article. Dig Dis Sci. 2009;54:1847–56. doi: 10.1007/s10620-008-0585-3. [DOI] [PubMed] [Google Scholar]
- 51.Cannon B, Nedergaard J. Brown adipose tissue: function and physiological significance. Physiol Rev. 2004;84:277–359. doi: 10.1152/physrev.00015.2003. [DOI] [PubMed] [Google Scholar]
- 52.Asano A, Kimura K, Saito M. Cold-induced mRNA expression of angiogenic factors in rat brown adipose tissue. J Vet Med Sci. 1999;61:403–9. doi: 10.1292/jvms.61.403. [DOI] [PubMed] [Google Scholar]
- 53.Portet R, de Marco F, Zizine L, Bertin R, Senault C. Perinatal variations of prostaglandins E2 and F alpha levels in brown adipose tissue of the rat; effects of ambient temperature. Biochimie. 1980;62:715–8. doi: 10.1016/s0300-9084(80)80030-7. [DOI] [PubMed] [Google Scholar]
- 54.Silva JE, Larsen PR. Adrenergic activation of triiodothyronine production in brown adipose tissue. Nature. 1983;305:712–3. doi: 10.1038/305712a0. [DOI] [PubMed] [Google Scholar]
- 55.Kikuchi-Utsumi K, Gao B, Ohinata H, Hashimoto M, Yamamoto N, Kuroshima A. Enhanced gene expression of endothelial nitric oxide synthase in brown adipose tissue during cold exposure. Am J Physiol Regul Integr Comp Physiol. 2002;282:R623–6. doi: 10.1152/ajpregu.00310.2001. [DOI] [PubMed] [Google Scholar]
- 56.Huang SG, Klingenberg M. Chloride channel properties of the uncoupling protein from brown adipose tissue mitochondria: a patch-clamp study. Biochemistry. 1996;35:16806–14. doi: 10.1021/bi960989v. [DOI] [PubMed] [Google Scholar]
- 57.Ahluwalia J. Chloride channels activated by swell can regulate the NADPH oxidase generated membrane depolarisation in activated human neutrophils. Biochem Biophys Res Commun. 2008;365:328–33. doi: 10.1016/j.bbrc.2007.10.176. [DOI] [PubMed] [Google Scholar]
- 58.Moreland JG, Davis AP, Bailey G, Nauseef WM, Lamb FS. Anion channels, including ClC-3, are required for normal neutrophil oxidative function, phagocytosis, and transendothelial migration. J Biol Chem. 2006;281:12277–88. doi: 10.1074/jbc.M511030200. [DOI] [PubMed] [Google Scholar]
- 59.Chapes SK, Haskill S. Evidence for granulocyte-mediated macrophage activation after C. parvum immunization. Cell Immunol. 1983;75:367–377. doi: 10.1016/0008-8749(83)90334-9. [DOI] [PubMed] [Google Scholar]
- 60.Knudsen E, Iversen PO, Van Rooijen N, Benestad HB. Macrophage-dependent regulation of neutrophil mobilization and chemotaxis during development of sterile peritonitis in the rat. Eur J Haematol. 2002;69:284–96. doi: 10.1034/j.1600-0609.2002.02657.x. [DOI] [PubMed] [Google Scholar]
- 61.Munder M, Eichmann K, Modolell M. Alternative metabolic states in murine macrophages reflected by the nitric oxide synthase/arginase balance: competitive regulation by CD4+ T cells correlates with Th1/Th2 phenotype. J Immunol. 1998;160:5347–54. [PubMed] [Google Scholar]
- 62.Edwards JP, Zhang X, Frauwirth KA, Mosser DM. Biochemical and functional characterization of three activated macrophage populations. 2006:1298–1307. doi: 10.1189/jlb.0406249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Broxmeyer HE, Cooper S, Hangoc G, Gao JL, Murphy PM. Dominant myelopoietic effector functions mediated by chemokine receptor CCR1. J Exp Med. 1999;189:1987–92. doi: 10.1084/jem.189.12.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Ruan H, Zarnowski MJ, Cushman SW, Lodish HF. Standard Isolation of Primary Adipose Cells from Mouse Epididymal Fat Pads Induces Inflammatory Mediators and Down-regulates Adipocyte Genes. J Biol Chem. 2003;278:47585–47593. doi: 10.1074/jbc.M305257200. [DOI] [PubMed] [Google Scholar]
- 65.Moore C, Hutson JC. Physiological relevance of tumor necrosis factor in mediating macrophage-Leydig cell interactions. Endocrinology. 1994;134:63–69. doi: 10.1210/endo.134.1.8275970. [DOI] [PubMed] [Google Scholar]
- 66.Watkins SK, Egilmez NK, Suttles J, Stout RD. IL-12 Rapidly Alters the Functional Profile of Tumor-Associated and Tumor-Infiltrating Macrophages In Vitro and In Vivo. J Immunol. 2007;178:1357–1362. doi: 10.4049/jimmunol.178.3.1357. [DOI] [PubMed] [Google Scholar]