Summary
CopA uses ATP to pump Cu across cell membranes. X-ray crystallography has defined atomic structures of several related P-type ATPases. We have determined a structure of CopA at 10 Å resolution by cryo-electron microscopy of a new crystal form and used computational molecular docking to study the interactions between the N-terminal metal binding domain (NMBD) and other elements of the molecule. We found that the shorter chain lipids used to produce these crystals are associated with movements of the cytoplasmic domains, with a novel dimer interface and with disordering of the NMBD, thus offering evidence for the transience of its interaction with the other cytoplasmic domains. Docking identified a binding site that matched the location of the NMBD in our previous structure by cryo-electron microscopy, allowing a more detailed view of its binding configuration and further support for its role in autoinhibition.
Introduction
Copper is essential to cells as a cofactor for a wide variety of enzymes, such as superoxide dismutase and cytochrome oxidase. If not carefully controlled, however, Cu can be toxic due to its redox potential and its ability to produce free radicals (Rae et al., 1999). As a result, a system of pumps, transporters and metallochaperones have evolved to control the delivery and distribution of Cu. In general, intracellular Cu-scavengers, such as metallothioneins, ensure that there is a vanishingly low concentration of free Cu in the cytoplasm (Rae et al., 1999) and metallochaperones, such as Atox1, CCS and Sco1/2, are used to carry the Cu through the cytoplasm and to deliver it to specific targets (Lutsenko et al., 2007).In mammals, Ctr1 is a secondary transporter on the cell surface that facilitates entry into the cell (Lee et al., 2001) and two transmembrane ATPases, ATP7A and ATP7B, that pump Cu across the endoplasmic reticulum and plasma membrane, respectively (Lutsenko et al., 2007). Mutations in ATP7A gives rise to Menkes disease, resulting from insufficient delivery of Cu in brain and other tissues, whereas mutations to ATP7B are responsible for Wilson’s disease, where Cu overload is responsible for liver and brain dysfunction (Barry et al., 2010; Veldhuis et al., 2009a).
ATP7A, and ATP7B are homolgous to CopA from bacteria and belong to the family of P-type ATPases (Lutsenko and Kaplan, 1995). This family comprises ATP-dependent, transmembrane ion pumps that share a common reaction mechanism, a common and membrane topology domain organization, as well as conserved sequence motifs implicated in ATP hydrolysis and in formation of their hallmark phosphoenzyme intermediate (Kuhlbrandt, 2004; Moller et al., 1996). Phylogenetic analysis of P-type ATPases delineates five subgroups, denoted PI-PV (Axelsen and Palmgren, 1998). CopA, ATP7A, and ATP7B are in the PIB subfamily together with transporters of a diverse array of transition and heavy metal ions, such as Cu+, Cu2+, Zn2+, Cd2+, Co2+, and Pb2+.
PII ATPases are the most thoroughly studied and have become paradigms for the family. They include Ca2+-ATPase and Na+/K+-ATPase, which have ten transmembrane helices (M1-10) and two main cytoplasmic loops inserted between M2/3 and M4/5. The larger cytoplasmic loop (between M4/5) folds into two separate domains, thus composing the nucleotide-binding (N) domain and phosphorylation (P) domain, whereas the smaller loop (between M2/3) composes the so-called actuator (A) domain. Each domain is characterized by sequence motifs related to their individual roles in the reaction cycle: namely DKTGTLT at the phosphorylation site of the P-domain, isolated residues surrounding the ATP binding site in the N-domain, and the conserved TGE in the A-domain (Olesen et al., 2007). The well-characterized reaction cycle alternates between E1 and E2 states, in which transmembrane ion-binding sites are oriented toward the intracellular or extracellular milieu, respectively. Switching between these two states is controlled by phosphorylation of the catalytic aspartate in the conserved DKTGTLT sequence, which instigates coordinated movements of N-, P- and A- and transmembrane domains. By coordinating changes in the affinity and accessibility of the ion-binding sites, ions are transported against a concentration gradient: i.e., E1 binds cytoplasmic ions with high affinity, whereas E2 binds extracellular (or luminal) ions with much lower affinity (Jorgensen and Andersen, 1988).
PIB ATPases such as CopA, although functionally and structurally related to PII ATPases, are distinguished by having only eight transmembrane helices and by bearing one or more N-terminal metal-binding domains (NMBD). These NMBDs are homologous to soluble metallochaperones that carry copper through the cytoplasm (e.g., Atox1). Both NMBDs and metallochaperones bind Cu+ with high affinity via CxxC sequence motifs. Although it has been postulated that NMBDs are mediate transfer of Cu from the metallochaperones to transport sites, there is increasing evidence that these domains are instead involved in autoregulation and, in the case of ATP7A, in targeting the molecule to the basolateral membrane (see reviews by Arguello et al., 2007; Lutsenko et al., 2007). The structural basis for this autoregulation remains to be determined.
X-ray structures have been solved for several intact PII ATPases, including Ca2+-ATPase from sarcoplasmic reticulum (SERCA1) (Toyoshima et al., 2000), Na+-K+-ATPase (Morth et al., 2007) and H+-ATPase (Pedersen et al., 2007). These structures have demonstrated the nature of the E1 and E2 states as well as the role of conformational changes in coordinating the multiple steps of the enzymatic cycle (Olesen et al., 2007). X-ray structures of PIB-type ATPases have so far only involved isolated cytoplasmic domains of CopA from A. fulgidus: specifically, a construct containing the N- and P-domain (Sazinsky et al., 2006b; Tsuda and Toyoshima, 2009) and another construct of the A-domain alone (Sazinsky et al., 2006a). NMR has revealed the binding of ATP to the isolated N-domain (Banci et al., 2010) and has characterized the structural homology between various NMBDs and the soluble metallochaperones (e.g. Banci et al., 2006). Our previous reconstruction of CopA using cryo-electron microscopy (cryo-EM) is the only structure of an intact PIB-type ATPase. Although the resolution was limited (~17 Å), this reconstruction served as a template for building a CopA homology model that defined the location of the NMBD relative to the other cytoplasmic domains and suggested that the NMBD could regulate CopA activity by holding the enzyme in an inactive state in the absence of copper and ATP (Wu et al., 2008). A different conclusion was reached by a more recent study employing chemical cross-linking, which suggests that the NMBD binds to the opposite side of the molecule and represents a static structural element of the cytoplasmic domain (Lubben et al., 2009).
To re-evaluate the location and role of the NMBD, we have determined a higher resolution structure using cryo-EM and helical reconstruction of tubular crystals of CopA from A. fulgidus. This 10-Å resolution structure reveals well-defined cytoplasmic domains that are readily fit with the A-, N-, and P- domains from X-ray crystallography. Although the transmembrane domain is less distinct, the new map leads us to propose a novel location for the extra two N-terminal helices that characterize the PIB subfamily. Surprisingly, the NMBD is disordered in these crystals, presumably due to dramatic changes in the molecular packing of CopA, thus supporting the idea that the NMBD is not a static structural element of the cytoplasmic domain. Based on an all-atom homology model for the cytoplasmic domains of CopA, we used computational methods to dock the NMBD, thus identifying the most favorable site of interaction. The results are consistent with our previous structure from cryo-EM and with the proposed autoinhibitory role for NMBD, which would act by retaining CopA in the E2 conformation.
Results
New crystal form for CopA
One of our primary goals was to obtain a higher resolution structure of CopA in order to understand better the architecture of the various domains that compose the molecule. As in earlier work, we used E. coli to express ΔC-CopA from A. fulgidus which carried the functionally important N-terminal MBD, which is common to all PIB ATPases, but not the C-terminal MBD, which is a unique feature of this particular CopA homolog. Expression levels were quite high, producing a final yield of 6–9 mg of detergent-purified protein from a 6 L expression with a purity >98% (Fig. S1 in Supplemental data).
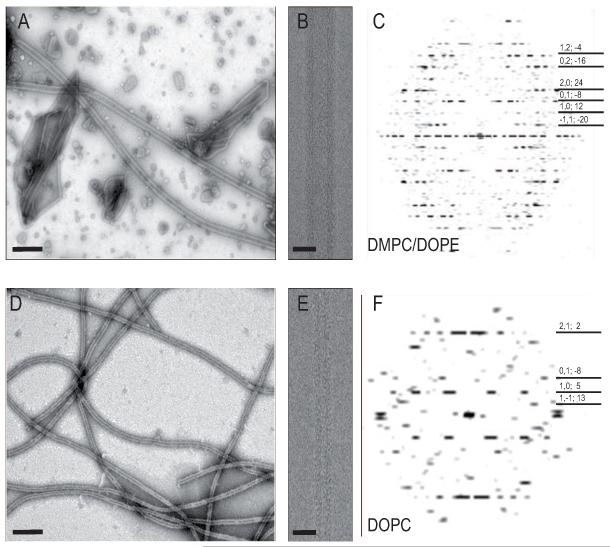
By altering the lipid species and the temperature used for crystallization, we obtained wider tubular crystals with stronger diffraction compared to our previous work. Previously tubular crystals of ΔC-CopA (Wu et al., 2008) were produced at 50°C with a reconstituted membrane of DOPC and in a solution containing the Cu+ chelator BCDS at pH 6.1. These tubes were 35 nm in diameter and the corresponding structure resembled that of SERCA in the E2 enzymatic state, consistent with the absence of Cu+ in the crystallization media. For the current work, the lipid was changed to a mixture of DMPC/DOPE (4:1 weight ratio), which produced substantially wider (60 nm diameter) tubular crystals at 30°C under otherwise identical conditions (Fig. 1). Layer line data from these wider crystals were substantially stronger and were consistent with the presence of several related helical symmetries, characterized by systematic differences in diameter and by the Bessel orders of principal layer lines: (n1,0, n0,1) equal to either (12,-8), (14,-8), or (14,-10). Amongst tubes with (12,-8) symmetry, the variability of unit cell dimensions for the underlying 2D lattice was small (s.d. <0.5 Å and <0.4° for the included angle, Table I), making the Fourier-Bessel approach to helical reconstruction viable (Diaz et al., 2010). Based on two-fold related phase residuals, the final map from an average of 12 tubes was judged to have 10 Å resolution, compared to 17 Å from a similar number of tubes in DOPC (Fig. 2C). The visibility of several individual α-helices in the map (discussed below) is consistent with this estimate of resolution.
Figure 1.
Images of tubular crystals of ΔC-CopA. (A-C) Tubes grown in a 4:1 mixture of DMPC/DOPE at 30°C. (D-E) Tubes grown in DOPC at 45°C. Except for the difference in lipid composition and temperature, the crystallization conditions were otherwise identical. Low magnification images of negatively stained crystals are shown in A and D, where the scale bars correspond to 200 nm. Higher magnification images of frozen-hydrated tubes used for helical reconstruction are shown in B and E, where the scale bars correspond to 50 nm. Computed Fourier transforms in C and F reveal layer lines characteristic of helical symmetry, which are indexed according to their Miller index and Bessel order on the right margin (h,k;n). The edge of these transforms is at ~20 Å resolution. See also Figure S1.
Table I.
Comparison of unit cell dimensions for tubular crystals of ΔC-CopA
| cytoplasmic domain1 |
membrane2 |
|||
|---|---|---|---|---|
| DMPC/DOPE | DOPC | DMPC/DOPE | DOPC | |
| radius (Å) | 136 | 120 | 200 | 75 |
| a (Å) | 63.9±0.24 | 97.5±1.38 | 82.9±0.29 | 78.7±0.92 |
| b (Å) | 69.8±0.40 | 61.1±0.76 | 78.5±0.42 | 48.7±0.47 |
| γ (°) | 78.6±0.38 | 101.0±0.66 | 99.0±0.40 | 74.4±0.65 |
| area (Å2) | 4370±30.3 | 5851±69.9 | 6427±44.6 | 3656±43.7 |
Unit cell dimensions measured at a radius corresponding to the middle of the A-domain, where major intermolecular contacts occur.
Unit cell dimensions in the middle of the membrane, i.e., half way between the density peaks in the mean radial density profile that correspond to the lipid phosphate headgroups (see Fig. 3).
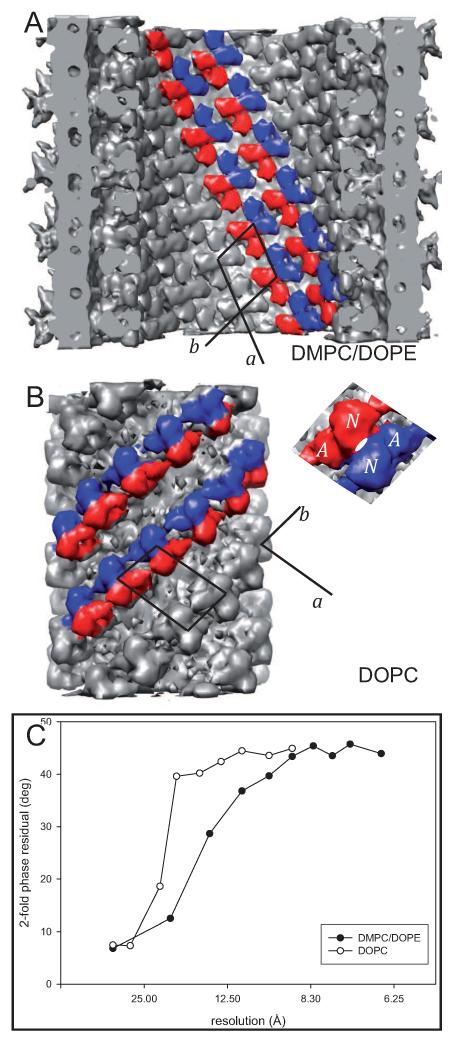
Figure 2.
Packing of ΔC-CopA in the two types of tubular crystals. (A) Cytoplasmic domains of ΔC-CopA face the inside of DMPC/DOPE tubes and are tightly packed with an isotropic set of intermolecular contacts. (B) Cytoplasmic domains face the outside of DOPC tubes and are packed in “dimer ribbons” that run along the b axis of the array. Both types of crystals are composed of ΔC-CopA dimers and two-fold related molecules have been colored red and blue in this figure with the unit cell outlined in black. (C) Two-fold phase statistics for the two helical reconstructions, documenting a resolution of 17 Å and 10 Å for DOPC and DMPC/DOPE crystals, respectively. See also Figure S2.
Packing of ΔC-CopA in tubular crystals
The reconstruction shows that the orientation of ΔC-CopA in DMPC/DOPE tubes is inverted relative to DOPC tubes (Fig. 2 and Fig. S2 in Supplemental materials). Similar to helical crystals of Ca2+-ATPase (Xu et al., 2002) and nicotinic acetylcholine receptor (Toyoshima and Unwin, 1988), ΔC-CopA molecule in DOPC tubes were oriented with their cytoplasmic domains facing the outside of the highly curved bilayer. In these cases, the positive curvature is presumably induced by the larger linear dimension required to accommodate the much larger cytoplasmic domains relative to the transmembrane and luminal domains (Young et al., 1997). Surprisingly, the large cytoplasmic domains of ΔC-CopA face the inside of tubes produced from DMPC/DOPE. This inverted orientation was also observed for tubular crystals of Na+-K+-ATPase grown in native membranes from the duck supraorbital salt gland, though in this case the β subunit added substantial mass to the outside of these tubes (Rice et al., 2001).
The packing of ΔC-CopA molecules is also substantially different in DMPC/DOPE tubes. In both crystal forms, CopA forms dimers that are stabilized by reciprocal interactions between N- and A-domains. In DOPC tubes, the dimers pack into “dimer ribbons” reminiscent of Ca2+-ATPase crystals (Taylor et al., 1986), in which contacts between A- and N-domains form a continuous ribbon of molecules related by two-fold symmetry with another ribbon running in the opposite direction along the b axis of the unit cell (Fig. 2b). In contrast, DMPC/DOPE tubes lack dimer ribbons; instead, the lattice is characterized by a more isotropic set of contacts between neighboring dimers. As will be discussed in more detail below, the geometry of the dimer itself is also quite different, with a 50° angle between the molecules (see Fig. 6). As a result of these differences, the density of cytoplasmic domains in DMPC/DOPE tubes is 33% higher than in DOPC tubes, based on the area of the unit cell at radii corresponding to the middle of these domains (Table I).
Figure 6.
Comparison of the ΔC-CopA dimer seen in the two helical crystals. (A) Dimer from the DMPC/DOPE tubes viewed parallel to the membrane surface. This view illustrates the >30° inclination of the transmembrane domain relative to the membrane normal. (B) The dimer from DMPC/DOPE tubes viewed normal to the membrane surface. (C) Dimer from our previous map from DOPC tubes (pdb code 2VOY, Wu et al., 2008) viewed parallel to the membrane surface showing that in this case the transmembrane domain is closely aligned to the membrane normal. The NMBD, which is visible in these crystals, are shaded in orange for the red monomer and purple for the blue monomer. (D) Dimer from DOPC tubes viewed normal to the membrane surface. Note that B and D are at a smaller scale than A and C; the transparent grey rectangle corresponds to the membrane.
In contrast to the cytoplasmic domains, the transmembrane domains in DMPC/DOPE tubes are less densely packed, with a density at the center of the membrane that is <60% relative to DOPC tubes (Table I). Another way to look at this difference is to compare membrane and cytoplasmic domains within each crystal: the unit cell area within the membrane of DOPC tubes is considerably smaller (64%) than in the region of the cytoplasmic domains, as one would expect given the smaller size of the molecule within the membrane. In contrast, the inverted nature of the DMPC/DOPE tubes provides a much larger area to the membrane domains relative to cytoplasmic domains (150%).
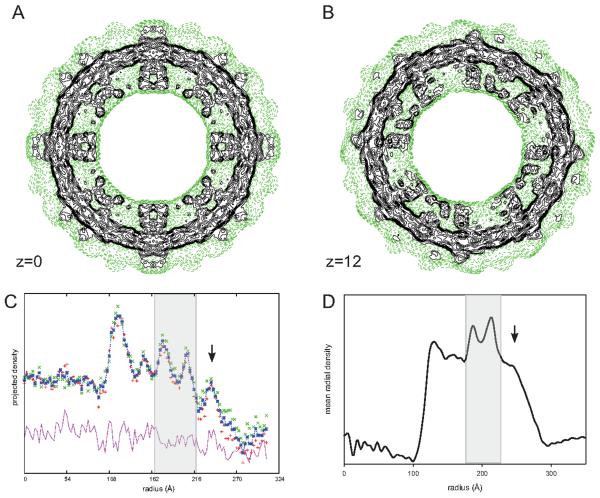
This excess membrane area in DMPC/DOPE tubes may explain the diffuse density seen on the outer surface of the reconstruction (Fig. 3). When images of individual tubes are projected down their cylindrical axes, several discrete peaks are observed in the resulting 1D profile (Fig. 3c). At the innermost radii, these peaks correspond to cytoplasmic domains, followed by two peaks from the phosphate headgroups of the lipid bilayer. Unlike previous tubes of CopA and of Ca2+-ATPase, there is an additional density peak on the outer surface of the membrane in DMPC/DOPE tubes (arrow in Fig. 3c). Helical reconstruction of these tubes produces a considerable amount of low, diffuse density in this region. More organized features with somewhat higher density are visible outside the tubes, but these features lie directly along symmetry axes, where noise builds up during the reconstruction process (e.g., compare sections shown in Figs. 3a and 3b). From these observations, we speculate that there is population of disordered CopA molecules with their cytoplasmic domains facing the outside of the DMPC/DOPE tubes. Indeed, a symmetric distribution of molecules across the membrane is expected from reconstitution and the inverted geometry of the CopA crystals within DMPC/DOPE tubes produces excess membrane area that could readily accommodate extra molecules facing the outside of the tubes. These extra molecules would be unable to participate in regular crystal contacts given the long distance between their cytoplasmic domains. Furthermore, the presence of disordered molecules would explain why, in the helical reconstruction, the protein domains within the membrane have much lower contrast relative to the cytoplasmic domains on the inside of the tube.
Figure 3.
Radial distribution of mass in DMPC/DOPC tubes. (A and B). Contour plots of individual sections from the density maps at z=0 and z=12 Å, where green, dashed contours indicate low density and black contours start at a threshold corresponding to the molecular envelope. The boundaries of the bilayer and protein domains inside the tube are clearly delineated. The inside of the tube is devoid of density, but diffuse density is evident on the outside of the membrane. These outer densities are prominent at four-fold and two-fold symmetry axes present in the section at the z=0, but are less prominent at z=12, which does not have these symmetry axes. (C) One-dimensional projection of an individual image along the helical axis. Peaks in this projection correspond to the cytoplasmic domains (at radii of ~120 and 150 Å) and to the phosphate headgroups of the bilayer (at radii of 170 and 210 Å). An additional peak at 230 Å is inconsistent with CopA’s lack of an extracellular domain. (D) Mean radial density distribution from the helical reconstruction of ΔC-CopA. The differing height of the peaks reflects the CTF correction that has been applied to the data. Even considering this correction, the peak at the outer radius has been significantly suppressed during the reconstruction process. The transparent grey rectangle in C and D delineates the membrane. See also Figure S2.
Functional analysis of CopA
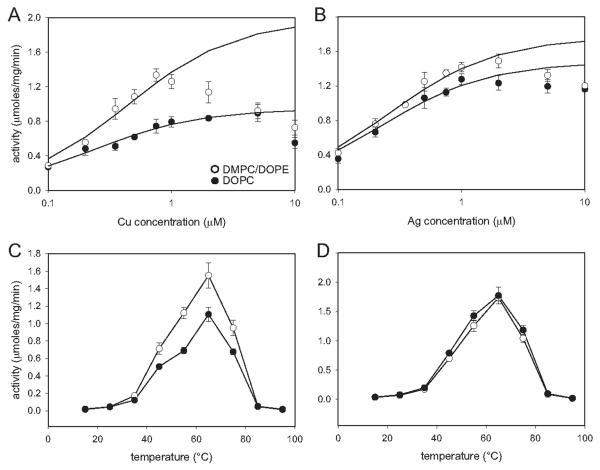
Because of the dramatic effect of bilayer composition on the morphology and molecular packing of the tubular crystals, we used ATPase assays to assess whether lipid had an effect on ΔC-CopA function. In particular, we characterized the dependence of ATPase activity on Cu+ or Ag+ concentrations and on temperature, reasoning that bilayer composition might affect either the mobility or the stability of the enzyme. The results (Fig. 4) indicate that ΔC-CopA has an apparent Kd of 0.44 μM for Cu+ and 0.26 μM for Ag+ in DMPC/DOPE and corresponding Vmax of 1.97 and 1.76 μmol/mg/min. For comparison, affinities in DOPC were 0.23 and 0.22 μM for Cu+ and Ag+ with Vmax of 0.94 and 1.48 μmol/mg/min, respectively. These values are similar to those previously measured by Rice et al. (2006) from detergent-solubilized ΔC-CopA: Kd of 0.11 and 0.2 μM for Cu+ and Ag+, respectively with Vmax of 1.8 and 2.4 μmol/mg/min. In all cases, ATPase activity was partially inhibited at high concentrations of either Cu+ or Ag+, probably reflecting the low affinity ion binding sites involved in ion release to the extracellular side of the membrane. For both lipids, maximal activities were obtained at 65°C, which compares well with published studies of detergent-solubilized CopA by Mandal et al. (2002) and to our own unpublished observations of detergent-solubilized CopA.
Figure 4.
ATPase activity of reconstituted ΔC-CopA. (A,B) Concentration dependence of activity for Cu+ and Ag+ dependent activity. A background level of ~0.3 μmoles/mg/min have been subtracted from these data, which have been fitted with the Michaelis Menton equation for a single binding site. Because there is inhibition at higher ion concentrations, the last 2-3 points of each curve has been omitted for this fit. The resulting values for apparent Kd and Vmax are cited in the text. Temperature dependence of activity for Cu+ (C) and Ag+ (D) shows a peak at ~65°C, which is consistent with previous work with detergent-solubilized CopA. In all plots, the open circles represent ΔC-CopA reconstituted in DMPC/DOPE and filled circles represent ΔC-CopA reconstituted in DOPC.
Atomic model for CopA
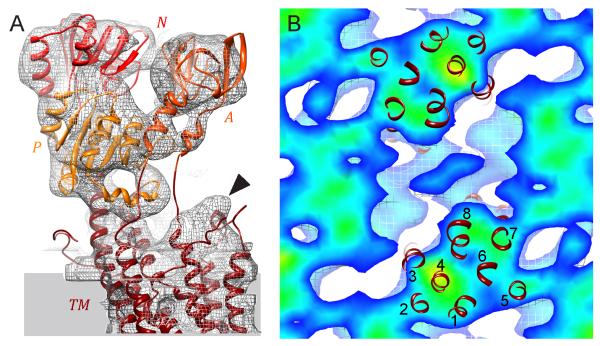
The new density map from DMPC/DOPE tubes was used as a template for building a model of CopA that revealed the juxtaposition of cytoplasmic domains and organization of the transmembrane helices. The first step was to build an all-atom model for the core of CopA using an X-ray crystallographic structure of SERCA1a as a template. For the cytoplasmic domains, X-ray crystal structures for the isolated A-domain of CopA (pdb code 2HC8) (Sazinsky et al., 2006a), and for the isolated pair of N- and P-domains of CopA (pdb code 3A1C and 2B8E) (Sazinsky et al., 2006b; Tsuda and Toyoshima, 2009) were aligned with SERCA1a in the E2 conformation (pdb code 1IWO) (Toyoshima and Nomura, 2002). For the transmembrane helices and their connections to the cytoplasmic domains, the sequence of CopA was aligned with SERCA1a and used to modelled by homology. Two extra transmembrane helices are predicted in the N-terminal region of CopA and, after building α-helices with the corresponding sequences (see methods), they were inserted between M1 and M2 of SERCA1a, as suggested by previous reports (Hatori et al., 2007; Lutsenko and Kaplan, 1995; Wu et al., 2008). The resulting hybrid homology model lacked the N-terminal and C-terminal metal binding domain, which have no counterpart in the SERCA1 template. This hybrid model was used for the docking studies described below.
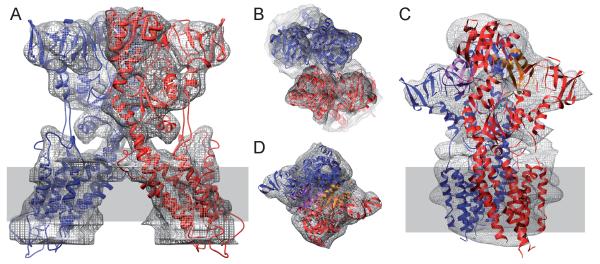
The density map from DMPC/DOPE tubes was used to reposition the individual domains of this ΔC-CopA homology model to produce a fitted model. Specifically, after placing the homology model roughly into the envelope from the helical reconstruction, the loops connecting the domains were broken and the position of each cytoplasmic domain was refined using cross-correlation (using the Fit-in-Map feature of Chimera). The density map accounted well for these domains, with evidence for individual α-helices in several locations (Fig. 5A and Movie S1 in Supplemental data). Surprisingly, the A-, N- and P-domains of CopA accounted for all of the densities in this region of the map, indicating that the NMBD was disordered. In our previous map from DOPC tubes, the NMBD was seen near the dimer interface and its disordering in the current DMPC/DOPE tubes may be due to changes in this interface. Indeed, Fig. 6 shows that there is a ~50° angle between two-fold related molecules in the DMPC/DOPE tubes (defined by axes running from the middle of the transmembrane helices to catalytic aspartate), whereas these molecules were almost parallel in DOPC tubes. There is also a shift in the cytoplasmic domains relative to one another (Fig. 7), which may explain the absence of the NMBD in the map from DMPC/DOPE tubes.
Figure 5.
Fitting an atomic model to the map from DMPC/DOPE tubes. (A) Domains are shown in various shades of red and are labeled accordingly. Isolated density for individual α-helices are visible in A- and P-domains and other α-helices at the periphery of N- and P-domains fit snugly within the isosurface density envelope. The envelope for the transmembrane domain (TM) shows a distinct bulge at top right, consistent with the kinked cytoplasmic end of M1. (B) Section through the transmembrane region of the density map overlaid with transmembrane helices, which are numbered. This is a view from the extracellular side of the membrane. See also Movie S1.
Figure 7.
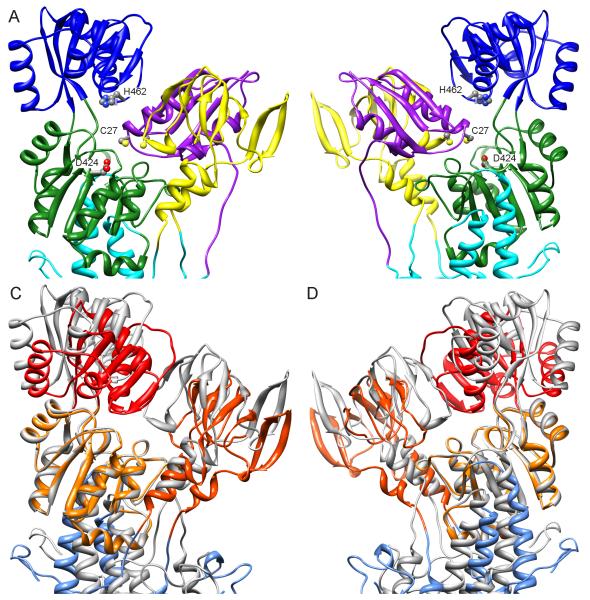
Docking of the NMBD to an all-atom model for ΔC-CopA. (A-B) Two views of the result obtained by the computational docking rotated at 180° about a vertical axis. The NMBD is colored purple, A-domain yellow, N-domain blue, P-domain green and transmembrane helices cyan. Key residues shown are C27 from the GMTCAMC30 Cu-binding loop of the NMBD, H462 from the ERRSEHP463 ATP-binding loop in the N-domain, and the catalytic D424 from the DKTGTLTT431 phosphorylation loop in the P-domain. (C-D) Equivalent views of an overlay between the homology model used for docking (same as in A and B but colored grey and lacking the NMBD) and the atomic model fitted to the density map from DMPC/DOPE tubes (colored shades of red). The P-domains are aligned, revealing relative movements of the N- and A-domains as well as a different angle of the transmembrane domain. See also Figure S3.
Protein densities were less distinct within the membrane than in the extramembranous regions of the map, though the general outline of the transmembrane domain could be discerned (Fig. 5B). This loss of contrast is common in helical reconstructions and may be exacerbated in this case by the extra, disordered molecules hypothesized to face the outside of the tubes, which would contribute to the background density within the membrane. Despite the poorer definition of membrane domains, we were able to use our all-atom model of ΔC-CopA to interpret the transmembrane densities. Specifically, the six conserved transmembrane helices (M1-M2, M5-M8) fell within strong density. Similar to maps of SERCA from helical crystals (Xu et al., 2002; Young et al., 2001; Zhang et al., 1998), a density was observed at the cytoplasmic surface above M1 (arrowhead in Fig. 5a), which matches the bent extension of this helix seen in atomic structures of SERCA, Na/K-ATPase and H+-ATPase. Additional membrane density was observed surrounding M4, which we fitted with the extra M2 and M3 helices that are characteristic to PIB ATPases. The location of these helices is slightly displaced from our previous model from DOPC tubes, with M2 taking the position of M3 in the previous map and M3 falling between M4 and M8. Given the poor contrast within the transmembrane domains, the precise location of these two extra helices remains tentative.
Computational Docking of the NMBD
Because our previous density map from the thinner DOPC tubes revealed the binding site of the NMBD next to the A-domain, we were disappointed that the current map from DMPC/DOPE tubes did not provide further information at higher resolution. Nevertheless, we hypothesized that the NMBD binding site previously identified next to the A-domain, although perturbed, still represented the most structurally compatible location for docking of the NMBD. To test this hypothesis, we performed in silico docking of the NMBD to the other cytoplasmic domains of CopA models obtained either by homology to the E2 state of SERCA or by fitting to our map from DMPC/DOPE tubes. This procedure uses Monte Carlo sampling to identify favorable docking sites on the molecular surface and ranks them energetically.
For our initial docking, we used the hybrid homology model of ΔNΔC-CopA based on SERCA1 in the E2 conformation as a template, due to its close resemblance to the structure from DOPC tubes in which the NMBD was seen. For a search target we made a homology model of NMBD based on the structure of a CopZ metallochaperone from B. subtilis. This particular template was chosen for its high resolution (1.5 Å) and for its high sequence identity (41%) relative to the CopA NMBD from A. fulgidus. Although the CopZ template structure had Cu+ bound to its CxxC motif, NMR structures of Cu+-free NMBD homologs indicated RMS deviations of only ~1 Å, suggesting that structural changes induced by Cu+ binding are minimal. For docking, the homology models for Cu-free NMBD and CopA were treated as independent, unconstrained molecules free to rotate and translate in all directions in space and the ICM docking routine evaluated numerous alternative binding interfaces, ultimately ranking them according to their predicted binding energy. Table II shows that the lowest energy solution was well separated from the other solutions, making it the clearly preferred location for the NMBD on the molecular surface of CopA. Significantly, this binding pose is consistent with the location of the NMBD in DOPC tubes, despite the lack of physical constraints or any other information related to the EM structure. This docking result places the CxxC Cu-binding loop of the NMBD next to a conserved loop in the N-domain that controls ATP binding and, when bound, the Cu+ ion would lie within 13 Å of the Mg2+ ion at the catalytic site of the P-domain (Fig. 7). Unlike the model previously derived directly from the EM structure, the docking solution packed the two α-helices of the NMBD against the A-domain. Specific interactions at this interface involve the loops between β-strands composing the body of the A-domain (residues 252-256; 270-274; 278-281; 295-298). The surface area buried in this interaction is 1943 Å2, which is divided between the three domains as follows: 1040 Å2 for the A-domain, 507 Å2 for the P-domain and 374 Å2 for the N-domain.
Table II.
Results of computational docking
| rank1 | energy terms |
|||||
|---|---|---|---|---|---|---|
| total2 | vdW H3 | vdW C4 | H-bond5 | electrostatic6 | hydrophobic7 | |
| 1 | −44.9 | −4.71 | −37.83 | −4.71 | −1.10 | −12.32 |
| 2 | −40.6 | −5.76 | −32.46 | −3.68 | −1.31 | −11.48 |
| 3 | −39.4 | −5.76 | −31.49 | −7.75 | −2.43 | −8.80 |
| 4 | −36.2 | −3.55 | −39.56 | −2.65 | 0.42 | −13.89 |
| 5 | −35.9 | −4.56 | −21.10 | −17.63 | −4.53 | −8.71 |
| 6 | −35.6 | −3.30 | −27.97 | −7.93 | −0.51 | −9.68 |
| 7 | −35.3 | 0.07 | −25.46 | −9.20 | −1.05 | −9.59 |
| 8 | −35.3 | −0.30 | −26.78 | −3.53 | −0.55 | −11.70 |
| 9 | −35.2 | −3.05 | −28.23 | −8.29 | −3.38 | −11.30 |
| 10 | −35.2 | −7.25 | −33.14 | −10.48 | −2.26 | −10.74 |
Each docking result was ranked according to its total energy. In our application, the lowest energy solution (rank 1) was well separated from the succeeding solutions, lending confidence to the result.
Total energy of the conformation. These values are reported as kcal/mol by ICM but should only be taken as relative binding energies due to various approximations during the calculation.
van der Waals grid potential using a hydrogen probe.
van der Waals grid potential using a carbon probe.
Hydrogen bonding grid potential.
Electrostatic grid potential.
Hydrophobic grid protential.
For comparison, we also used the fitted model corresponding to the conformation observed in DMPC/DOPE tubes as a template for docking, though in this case the apparent disordering of the NMBD in the corresponding crystals led us to expect a lower binding energy. Indeed, the ICM docking routine determined that the total energy for this pose was almost 17 kcal/mol higher for the DMPC/DOPE tube model compared to the SERCA1 homology model (−28.2 vs. −44.9). These energy units are not absolute, due to inaccuracy in the force field and in the coarseness of the grid on which potential energies were calculated, but are indicative of the relative binding strength. Accordingly, calculations of buried surface area showed a >40% decrease to 1176 Å2. The reason for this reduced binding energy is evident in Fig. 7C & D, where the homology model for CopA is overlaid with the fitted model for CopA from DMPC/DOPE tubes. This reveals a significant shift in the A- and N-domains relative to the P-domain and the loss of binding energy is consistent with an altered molecular surface for NMBD docking due to the disrupted interface between A-, N- and P-domains. Indeed, intermediate values for binding energy were obtained in docking studies with other permutations of the ΔNΔC-CopA model, in which the N-domain was either moved, modified or removed altogether (data not shown). These results illustrate that all three cytoplasmic domains contribute to the NMBD binding site and that the binding was strongest when the conformation resembled the E2 state adopted by SERCA1.
Finally, to complete our model of ΔC-CopA, we constructed a linker between the M1 and the NMBD (Fig. 7). In SERCA, two N-terminal α-helices are attached to the front of the A-domain and are connected to the cytoplasmic end of M1 via a flexible linker. Although the NMBD is associated with the side of the A-domain in our model, the CopA linker sequence is long enough to wrap under the A-domain and to connect to the kinked extension of M1. Thus, our model of ΔC-CopA is consistent with the idea that the physical couplings observed between SERCA transmembrane helices and the cytoplasmic domains are conserved in the PIB subfamily of pumps.
Discussion
Using cryo-EM and image reconstruction, we have produced a new structure of ΔC-CopA from A. fulgidus at 10 Å resolution from a new form of helical crystal grown in membranous tubes composed of shorter chain lipid (DMPC). Although ΔC-CopA forms a dimer as in previous DOPC tubes, there are marked differences in the intermolecular contacts stabilizing this dimer and in the packing of this dimer in the unit cell; furthermore, the transmembrane domain is strongly tilted relative to the thinner DMPC membrane. At 10 Å resolution, X-ray structures of the three isolated cytoplasmic domains could be fitted precisely into the map. However, density for the NMBD is absent, indicating that this domain is disordered. We then used computational molecular docking to explore the site of NMBD binding relative to the other CopA cytoplasmic domains and its conformational determinants. Using a homology model based on the E2 conformation of SERCA1, the lowest energy pose was consistent with the site previously identified in tubular crystals composed of DOPC. Although there were differences with our previous, manually docked model, (i.e., an ~180° rotation of the NMBD relative to the A-domain), this new model is nevertheless consistent with an autoinhibitory interaction between the NMBD and the N- and P-domains in the E2 conformation. Comparison of this CopA homology model with a model fitted to the map from DMPC/DOPE tubes reveals a distinct shift in N- and A-domains, which is predicted to reduce the binding energy of the NMBD by ~17 kcal/mole. This result suggests that the affinity of the NMBD for its preferred site has been lowered to the level of a weak, Brownian association, explaining why it has become disordered in the crystal.
Although our previous model of ΔC-CopA from DOPC tubes is consistent with the E2 enzymatic state of SERCA1, the new structure from DMPC/DOPE tubes reveals a significant conformational change. As shown in Fig. 7, the A-domain has shifted away from the P-domain in the new map, leaving room for the N-domain to swing closer to the catalytic site. Such movement is generally consistent with conformational changes of SERCA1 during its reaction cycle, which depends on an interplay between the A- and N-domains to regulate access to the catalytic aspartate. The overarching idea is that the A-domain interacts with elements of the P-domain in E2 states, thus preventing the approach of the N-domain and its associated ATP. In E1 states, the A-domain swings out of the way to enable transfer of phosphate. Fig. S3 (Supplemental data) compares the position of the A-domain in E2-P and E2 states of SERCA1. In E2-P, the conserved (T/S)GE278 sequence in the A-domain is closest to the catalytic aspartate in the P-domain (DKTGTLT430), making direct interactions with the associated Mg2+ ion. In E2, the (T/S)GE loop has moved back and interacts with the conserved DG(V/I)ND622 loop at the periphery of the P-domain. In our model fitted to DMPC/DOPE tubes, this (T/S)GE loop has moved back even more, though it still appears to interact with residues in the DG(V/I)ND loop. The latter configuration of cytoplasmic domains has not previously been seen in structures of P-type ATPases and could represent a transition from the E2 to the E1 state as the A-domain begins to disengage from the P-domain. The increased angle of the transmembrane domain relative to the P-domain (Fig 7C and D) is consistent with this transition, as it eventually results in a 30°upwards rotation of the P-domain in the E1 state relative to the E2 state (Toyoshima and Nomura, 2002). Normally, the E2 to E1 transition would be initiated by ions binding to their transport sites in the membrane. In the current situation, we speculate that a shearing of the transmembrane domain caused by its oblique angle relative to the membrane might be responsible for inducing this conformation.
Factors that may contribute to the unusually high tilt of the membrane domain are (1) the negative curvature of the crystal contacts between cytoplasmic domains, (2) the intermolecular contacts that stabilize the dimer and (3) the thinner bilayer produced by the shorter aliphatic chains of DMPC, which composes the bulk of the bilayer. Indeed, the hydrophobic core thickness of DMPC has been experimentally measured as 23 Å compared to 27 Å for DOPC (Lewis and Engelman, 1983) and this difference would be consistent with a tilt of ~30° in order to accommodate a hydrophobic domain with a fixed thickness (cos−1(23/27)). Given the corresponding difference in the tilt of the CopA molecule in DMPC/DOPE vs.DOPC tubes, it seems plausible that the thinner DMPC bilayer could be the driving force for altering the geometry of the dimer and thus producing the negative curvature of the crystal. Despite the dramatic effect on crystallization, however, the DMPC/DOPE bilayer had only modest effects on ATPase activities. It should be noted that the lipid-to-protein ratio was 6x higher for this assay, where no crystalline arrays were observed to form. Nevertheless, the approximately two-fold increase in Kd and Vmax is comparable to the two-fold changes in these parameters in detergent micelles (Rice et al., 2006) and can therefore be considered to represent the normal range of behavior for CopA in different hydrophobic environments. Furthermore, this range is in line with measurements of other P-type ATPases in lipid and detergent environments (e.g., Cornea and Thomas, 1994; Warren et al., 1974).
From a structural point of view, a shearing of the transmembrane domain would be required to maintain the upper and lower boundaries of the transmembrane helices as the domain was tilted in the bilayer. In other words, one would expect a relative movement of transmembrane helices along their helical axes. Such movements have in fact been described for the M1/M2 helical pair of SERCA, both during the transition from E2 to E1 (Toyoshima and Nomura, 2002) and during Ca2+ occlusion by E1~P (Olesen et al., 2004). In the case of CopA, M2 and M3 are inserted between this M1/M2 pair and placed at the periphery of the transmembrane domain; accordingly, the entire M1-M4 bundle would likely slide relative to M5-M8, thus accommodating the shear force. By analogy with the structural changes during the E2 to E1 transition in SERCA, this movement of M1-M4 would pull the A-domain away from the P-domain, potentially inducing the conformation seen in DMPC/DOPE tubes. A corresponding force on the NMBD, which is directly linked to M1, could also help disengage it from its binding site on the A-domain. According to this hypothesis, M1 would no longer be tethered to the cytoplasmic domain in the E1 state. This implies that the more rigid connection between M4 (M2 in SERCA) and the A-domain may be the important element that drives changes to the transmembrane domain and leads to ion occlusion during the transition to E1~P.
The site for the NMBD identified by in-silico docking is consistent with the corresponding density in our previous map from DOPC tubes (Wu et al., 2008). The specifics of the interface between the NMBD and the A-domain are different in our previous model, presumably reflecting our earlier neglect of binding energy and molecular surface compatibility during manual assembly and fitting. Nevertheless, in both cases, the NMBD binds to the side of the A-domain with additional interactions between the GMTCAMC30 Cu-binding loop and the conserved ERRSEHP463 loop on the N-domain. This ERRSEHP463 loop has been shown in NMR and X-ray crystal structures of isolated domains to be intimately involved in ATP binding (Banci et al., 2010; Dmitriev et al., 2006). In particular, His462 makes critical interactions with the α- and β-phosphates and with the adenine ring of ATP (Sazinsky et al., 2006b; Tsuda and Toyoshima, 2009); its mutation in ATP7A almost completely prevents ATP binding (Morgan et al., 2004) and the H462Q mutation is the most common cause of Wilson’s diseases (Thomas et al., 1995). Interaction between the NMBD and the N-domain has been demonstrated experimentally by Gonzalez-Guerrero et al. (Gonzalez-Guerrero et al., 2009) using pull-down assays. Specifically, they showed that Cu-free NMBD bound to an N/P-domain construct while Cu-loaded NMBD did not. Furthermore, these investigators showed that interaction between the Cu-free NMBD and the N/P-domain construct was disrupted by ADP-Mg2+. Although our docking result indicates that the majority of binding energy for NMBD comes from its interface with the A-domain, the interface between the NMBD and the N- and P-domains still accounts for 891 Å2 of buried surface (out of a total of 1934 Å2), which seems sufficient to explain the pull-down results. It is interesting to note that the altered conformation represented in our DMPC/DOPE crystals results in a loss of 767 Å2 of buried surface (i.e. almost the entire interaction with N- and P-domains). This observation, together with the disordering of the NMBD in these crystals, suggests that the interaction with the A-domain alone is not enough to retain NMDB at its binding site and that its association with the cytoplasmic domains is transient.
This transience is also implied by interaction between the NMBD and metallochaperones. Physiologically relevant metallochaperones have been shown to transfer Cu+ to MBDs of both ATP7A (Walker et al., 2004; Walker et al., 2002) and CopA (Gonzalez-Guerrero and Arguello, 2008) and NMR structures of the complex between the HAH1 metallochaperone and related MBDs of ATP7A indicate that the α-helical face of NMBD interacts with the corresponding face of the metallochaperone during Cu+ transfer (Achila et al., 2006; Banci et al., 2005a; Banci et al., 2005b). According to our model, these α-helices are packed against the A-domain, implying that the NMBD must dissociate prior to interacting with metallochaperones. This dissociation step may be the kinetic barrier to activation that has been revealed by kinetic analysis of the transport cycle (Hatori et al., 2009). An alternative binding site for the NMBD on the opposite side of the A-domain, where it would not have interactions with either the N- or P-domains, has been proposed on the basis of chemical crosslinking (Lubben et al., 2009). This alternative is not compatible with our previous density map or with the results of molecular docking and it is difficult to explain how the NMBD would play an autoinhibitory role from that location.
There is a growing consensus that NMBD’s play a regulatory role for Cu pumps and are not involved in transferring Cu+ to the transport sites within the transmembrane domain (Arguello et al., 2007). This conclusion applies both to the human pumps (ATP7A and ATP7B), which carry six MBD’s on their N-terminus, and CopA from bacteria, which generally have a single N-terminal MBD. In particular, several studies have shown Cu transfer from metallochaperones to the MBD’s (Achila et al., 2006; Banci et al., 2005a; Banci et al., 2005b; Gonzalez-Guerrero and Arguello, 2008; Larin et al., 1999; Ralle et al., 2004; Singleton et al., 2009; Strausak et al., 2003) and from metallochaperones to the transport sites (Gonzalez-Guerrero et al., 2009). However, disruption of the MBDs by either truncation or mutation does not affect transport activity of bacterial pumps (Bal et al., 2001; Fan and Rosen, 2002; Mana-Capelli et al., 2003; Mandal and Arguello, 2003; Mitra and Sharma, 2001; Rice et al., 2006) and disruption of MBD on the human pumps mainly affect enzyme activation and trafficking (Cater et al., 2007; Huster and Lutsenko, 2003; Strausak et al., 1999; Tsivkovskii et al., 2001; Veldhuis et al., 2009b; Walker et al., 2002). An enzymatic study of CopA from T. maritime concluded that the NMBD was responsible for kinetic inhibition of the reaction cycle and that this inhibition was alleviated by its binding of Cu+ (Hatori et al., 2008). Our structure offers a structural explanation for this autoinhibition. Specifically, the NMBD has substantial interactions with both the A- and N-domains in our hybrid model of the E2 conformation. Proteolytic digestion patterns of CopA indicate that these domains undergo large movements during the transition from the E2 state to the E1~P state, similar to SERCA (Hatori et al., 2009) and the binding energy of the NMBD would resist these movements. Cu+ could have a role in disrupting this interaction in three ways: (1) by binding to transport sites and driving a conformational change, (2) by binding directly to the GMTCAMC30 loop at the interface between the NMBD and the N-domain, and (3) by promoting interaction between the NMBD and metallochaperones, which would require dissociation of the NMBD from the A-domain in order to form the corresponding complex.
Experimental Procedures
Protein expression and purification
Plasmids containing the C-terminal truncation of the copA gene (Wu et al., 2008) behind a pBAD promoter were expressed in E.coli (strain LMG1940) using LB broth with 0.1 mg/ml ampicillin at 37°C. After allowing 1L cultures to reach an OD of 1, expression of CopA was induced with 0.02% arabinose for 2 h at 30°C. Cells were then harvested by centrifugation at 10,000g for 10 min and the pellet was suspended in 50 mM Tris-Cl, pH 7.5, 20% glycerol, 50 mM NaCl, 2 mM β-mercaptoethanol, 0.05 mg/ml DNAse I and Complete Protease Inhibitor Cocktail (Hoffmann-La Roche Inc, Nutley, NJ). Cells were broken by French Press at 20,000 psi and, after removing large debris by centrifugation at 10,000g for 20 min, membranes were collected by centrifugation at 150,000g for 1 h and stored at −80°C. Membrane pellets were solubilized in 10 mg/ml n-dodecyl-β-D-maltoside (DDM, Anatrace, Maumee, OH), 25 mM Tris-Cl, pH 7.5, 100 mM NaCl, 20 mM imidazole, 10% glycerol, 2 mM β-mercaptoethanol, protease inhibitor cocktail at a protein concentration of 10 mg/ml. After gentle mixing for 30 min, insoluble material was removed by centrifugation at 150,000g for 30 min and the supernatant was loaded onto a 5 ml HiTrap Chelating HP column (GE Healthcare, Piscataway, NJ), which was charged with 0.1 M NiCl2 and equilibrated with 25 mM Tris-Cl, pH 7.5, 100 mM NaCl, 20 mM imidazole, 10% glycerol, 2 mM β-mercaptoethanol and 1 mg/ml DDM. After washing, purified ΔC-CopA was eluted with 25 mM Tris-Cl, pH 7.5, 100 mM NaCl, 400 mM imidazole, 10% glycerol, 2 mM β-mercaptoethanol and 0.1% DDM. Overnight incubation with thrombin (1 U/mg of protein) was used to remove the polyhistidine-tag. A benzamidine-Sepharose column (Amersham Biosciences) equilibrated with 25 mM Tris-Cl, pH 7.5, 100 mM NaCl, 10% glycerol, 2 mM β-mercaptoethanol and 0.1% DDM was used to remove the thrombin. Finally, purified ΔC-CopA protein was dialyzed against 25 mM Tris-Cl, pH 7.5, 100 mM Na2SO4,10% glycerol, 2 mM DTT and 0.1% DDM. All the steps in this purification were done at 4°C.
ATPase activity assays
For measuring ATPase activity, proteoliposomes were made with a 2.5:1 lipid-to-protein weight ratio using either 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC) or a 4:1 weight ratio of 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC) /1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE). To make these proteoliposomes, ΔC-CopA (solubilized in DDM) and the relevant lipid were dialyzed against 50 mM Tris-SO4, pH 7.5, 50 mM Na2SO4, 50 mM K2SO4, 10 mM MgSO4 and 2 mM β-mercaptoethanol for 5 days at 4°C. As described in our previous work on CopA (Rice et al., 2006), the malachite green assay (Kimura et al., 1996) was used to quantitate phosphate production in 50 mM Tris-SO4, 200 mM Na2SO4, 3 mM MgSO4, 5 mM glutathione, 2.4 mM ATP, 10% glycerol, 0.025% NaN . To test dependence on either Cu+3 or Ag+, assays were performed at 70°C; to test temperature dependence, assays were performed at either 2 μM Cu+ or 1 μM Ag+. In order to prevent buildup of transport ions inside the vesicles, a small, sub-solubilizing amount of detergent (0.1 mg/ml DDM) was added to make the vesicles leaky. All non-reducing buffers were pretreated with 100 mg/ml Chelex (BioRad Laboratories, Hercules, CA) for at least 1 h. Aliquots of ΔC-CopA proteoliposomes at 0.02 and 0.04 mg/ml were prepared and stored on ice, where the enzyme is completely inactive. Using a programmable PCR thermocycler (Eppendorf North America, Hauppauge, NY), duplicate 45-μl aliquots of the reaction mixture were heated to 70°C for 3, 5, 7, and 9 min then cooled to 4°C to stop the reaction. Phosphate was then quantified in 96-well microtiter plates using a development time of 3 min; absorbance was read at 630 nm. In order to maintain a pH of 6.3 at a range of different temperatures, the following buffers were adjusted to the corresponding pHs at room temperature: MES-SO4 pH 6.2 for 15°C, MES-SO4 pH 6.3 for 25°C, MES-SO4 pH 6.4 for 35°C, MES-SO4 pH 6.5 for 45°C, MES-SO4 pH 6.6 for 55°C, MOPS-SO4 pH 6.9 for 65°C, Tris-SO4 pH 7.7 for 75°C, Tris-SO4 pH 8 for 85°C and Tris-SO4 pH 8.3 for 95°C. Like previous work, we observed a background of 0.3-0.35 μmol/mg/min in the absence of added Cu+ or Ag+; this background activity was abolished by the Cu chelator bathocuproinedisulphonic acid (BCDS), suggesting that it arises from trace Cu+ in the solutions. After subtracting the background activity, the data was fitted with the Michaelis–Menten binding model to determine the apparent dissociation constant (Kd).
Crystallization and Cryo-electron Microscopy
ΔC-CopA tubular crystals were grown with a 4:1 mixture of DMPC/DOPE at a protein concentration of 1 mg/ml and at a lipid-to-protein weight ratio of 0.4. Dialysis was carried out for 5 d in 50μl dialysis buttons (Hampton Research, Aliso Viejo, CA) at 30°C against 500 ml of 50 mM MES, pH 6.1, 25 mM Na2SO4, 25 mM K2SO4, 200 μM BCDS, 10 mM MgSO4 and 2 mM β-mercaptoethanol. Stock solutions of lipid for both crystallization and for ATPase assays were made in dodecyl octaethylene glycol ether (C12E8) at 1mg lipid per 2 mg detergent.
For cryo-EM, a suspension of crystals was deposited on a holey carbon grid and rapidly frozen in liquid ethane. Samples were imaged at −175°C with a CT3500 cryoholder (Gatan Inc, Pleasanton, CA) at 50,000x nominal magnification with a CM200 FEG electron microscope (FEI Company, Hillsboro OR) operating at 200 kV. Electron micrographs were recorded on Kodak SO-163 film and, after screening by optical diffraction, suitable images were digitized at 14 μm intervals with a Zeiss SCAI microdensitometer (Intergraph Corp., Huntsville, AL).
Helical reconstruction
Tubular crystals with a diameter of ~600Å, displaying symmetric layer lines by optical diffraction, and with defocus values between 1.5 and 2.5 μm were selected for helical processing using Fourier-Bessel methods (Diaz et al., 2010). Layer lines were catalogued with a consistent set of Miller indices, which reflect the underlying 2D lattice within the membrane. To define the helical symmetry, Bessel orders were assigned to all of these layer lines (Fig. 1). Initial assignments were based on phase relationships across the meridian, the radial location of the diffraction maximum and the self-consistency of assignments across the transform (e.g. the construction of a plausible n,l plot). To verify the Bessel order assignments, several alternative indexing schemes were tested, using the EMIP helical reconstruction software (http://cryoem.nysbc.org/EmIP.html) to compare phase residuals for the determination of out-of-plane tilt (Ω). For highly tilted tubes (Ω~5°), this phase residual was definitive in identifying the correct indexing. Near/far phase residuals and two-fold phase residuals were also consistently lower with the correct indexing. In the end, EMIP was used to apply the unbending methods of Beroukhim and Unwin (1997) to 12 tubes. Correction for the contrast transfer function was based on an amplitude contrast of 7% (Toyoshima et al., 1993) and the resolution of the final maps were truncated at 9 Å resolution, where the two-fold related phase residual approached 43° (random data produces a 45° residual).
Modeling and Docking
All modeling and docking procedures were carried out using the ICM protein docking software (v3.6-1f, Abagyan and Totrov, 1994; Cardozo et al., 1995) as previously described (Allen and Stokes, 2011). Briefly, the sequences of CopA and SERCA1a were aligned by Needleman and Wunsch global alignment using zero end gap penalties (Abagyan and Batalov, 1997). A 3D model of CopA (omitting N- and C-terminal metal binding domains) was then constructed by threading its sequence onto the X-ray crystallographic structure for SERCA1a in the E2 conformation (pdb code 1IWO) using the alignment as a guide. For this homology model, the geometries of conserved residues were fixed, whereas the non-conserved residues were set to their standard geometries and a local minimization was performed to relieve any clashes. Structural databases were consulted for probable configurations of the non-conserved loops or, in the event no reasonable solution was found, the loop was modeled freely to its lowest energy conformation (Cardozo et al., 1995). Existing X-ray crystal structures of CopA cytoplasmic domains (the isolated P/N-domain pair - 3A1C - and the isolated A-domain - 2HC8) were then aligned with the initial homology model and this model was adjusted to exactly match the X-ray structures in overlapping regions. Thus, the resulting hybrid homology model of CopA inherits X-ray crystallographic information from both the SERCA1a E2 conformation and the isolated structures of the CopA cytoplasmic domains. A homology model for the NMBD of CopA based on the X-ray crystallographic structure of the metallochaperone, CopZ (pdb code 2QIF) was built in ICM using the same approach as above. Finally we used Coot (Emsley et al., 2010) to make minor adjustments or refinements to the model, Phyre (Bennett-Lovsey et al., 2008) to build the extra M2 and M3 helices of CopA, and then several rounds of energy minimization were performed in ICM. Finally, Chimera (Pettersen et al., 2004) was used to visualize results and to make figures.
After building these atomic models, the NMBD was docked to the hybrid model of CopA (N-, P- and A- domains) using ICM protein-protein docking (Fernandez-Recio et al., 2005). ICM first predicted surface patches on the cytoplasmic domains that were likely to produce energetically favorable protein interactions using the Optimal Docking Areas method (ODA). Next, ICM computed electrostatic, van der Waals, hydrogen bond, solvation, and hydrophobic potentials on a coarse grid over these surface regions. Interaction energies were determined for a Monte Carlo sampling of NMBD poses at each of the surface patches. The 400 most favorable results were then subjected to side chain energy minimization and ranked according to their overall binding energy. Buried surface area was determined in CNS (Brunger, 2007) with an 1.4 Å probe radius.
Supplementary Material
Research Highlights.
Dramatic changes in crystal morphology of CopA were obtained with short-chain lipids
Movements of cytoplasmic domains suggest transition between E2 and E1 conformations
Disordering of the NMBD illustrates its transient interaction with other domains
Best docking result matches previous structure, supporting model of autoinhibition
Acknowledgements
Coordinates for both models have been deposited in the Protein Data Bank (accession code pending) and the map has been deposited in the Electron Microscopy Databank (EMDB 5271). The authors gratefully acknowledge the facilities at the New York Structural Biology Center and support from NIH grants R01 GM56960 and U54 GM094598 to D.L.S. as well as NIH grant DP2 OD004631 to T.C.
Abbreviations
- DMPC
1,2-dimyristoyl-sn-glycero-3-phosphocholine
- DOPE
1,2-dioleoyl-sn-glycero-3-phosphoethanolamine
- DOPC
(1,2-dioleoyl-sn-glycero-3-phosphocholine
- NMBD
N-terminal metal binding domain
- DDM
n-dodecyl-β-D-maltoside
- EM
electron microscopy
- BCDS
bathocuproinedisulphonic acid
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abagyan R, Totrov M. Biased probability Monte Carlo conformational searches and electrostatic calculations for peptides and proteins. J. Mol. Biol. 1994;235:983–1002. doi: 10.1006/jmbi.1994.1052. [DOI] [PubMed] [Google Scholar]
- Abagyan RA, Batalov S. Do aligned sequences share the same fold? J. Mol. Biol. 1997;273:355–368. doi: 10.1006/jmbi.1997.1287. [DOI] [PubMed] [Google Scholar]
- Achila D, Banci L, Bertini I, Bunce J, Ciofi-Baffoni S, Huffman DL. Structure of human Wilson protein domains 5 and 6 and their interplay with domain 4 and the copper chaperone HAH1 in copper uptake. Proc. Nat. Acad. Sci. 2006;103:5729–5734. doi: 10.1073/pnas.0504472103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen GS, Stokes DL. Modeling, docking, fitting of atomic structures to 3D maps from cryo-electron microscopy. Methods Mol Biol. 2011 doi: 10.1007/978-1-62703-176-9_13. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arguello JM, Eren E, Gonzalez-Guerrero M. The structure and function of heavy metal transport P1B-ATPases. Biometals. 2007;20:233–248. doi: 10.1007/s10534-006-9055-6. [DOI] [PubMed] [Google Scholar]
- Axelsen KB, Palmgren MG. Evolution of substrate specificities in the P-type ATPase superfamily. J. Mol. Evol. 1998;46:84–101. doi: 10.1007/pl00006286. [DOI] [PubMed] [Google Scholar]
- Bal N, Mintz E, Guillain F, Catty P. A possible regulatory role for the metal-binding domain of CadA, the Listeria monocytogenes Cd2+-ATPase. FEBS Lett. 2001;506:249–252. doi: 10.1016/s0014-5793(01)02927-1. [DOI] [PubMed] [Google Scholar]
- Banci L, Bertini I, Cantini F, Chasapis CT, Hadjiliadis N, Rosato A. A NMR study of the interaction of a three-domain construct of ATP7A with copper(I) and copper(I)-HAH1: the interplay of domains. J. Biol. Chem. 2005a;280:38259–38263. doi: 10.1074/jbc.M506219200. [DOI] [PubMed] [Google Scholar]
- Banci L, Bertini I, Cantini F, DellaMalva N, Herrmann T, Rosato A, Wuthrich K. Solution structure and intermolecular interactions of the third metal-binding domain of ATP7A, the Menkes disease protein. J. Biol. Chem. 2006;281:29141–29147. doi: 10.1074/jbc.M603176200. [DOI] [PubMed] [Google Scholar]
- Banci L, Bertini I, Cantini F, Inagaki S, Migliardi M, Rosato A. The binding mode of ATP revealed by the solution structure of the N-domain of human ATP7A. J. Biol. Chem. 2010;285:2537–2544. doi: 10.1074/jbc.M109.054262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banci L, Bertini I, Ciofi-Baffoni S, Chasapis CT, Hadjiliadis N, Rosato A. An NMR study of the interaction between the human copper(I) chaperone and the second and fifth metal-binding domains of the Menkes protein. FEBS J. 2005b;272:865–871. doi: 10.1111/j.1742-4658.2004.04526.x. [DOI] [PubMed] [Google Scholar]
- Barry AN, Shinde U, Lutsenko S. Structural organization of human Cutransporting ATPases: learning from building blocks. J Biol Inorg Chem. 2010;15:47–59. doi: 10.1007/s00775-009-0595-4. [DOI] [PubMed] [Google Scholar]
- Bennett-Lovsey RM, Herbert AD, Sternberg MJ, Kelley LA. Exploring the extremes of sequence/structure space with ensemble fold recognition in the program Phyre. Proteins. 2008;70:611–625. doi: 10.1002/prot.21688. [DOI] [PubMed] [Google Scholar]
- Beroukhim R, Unwin N. Distortion correction of tubular crystals: Improvements in the acetylcholine receptor structure. Ultramicroscopy. 1997;70:57–81. doi: 10.1016/s0304-3991(97)00070-3. [DOI] [PubMed] [Google Scholar]
- Brunger AT. Version 1.2 of the Crystallography and NMR system. Nat Protoc. 2007;2:2728–2733. doi: 10.1038/nprot.2007.406. [DOI] [PubMed] [Google Scholar]
- Cardozo T, Totrov M, Abagyan R. Homology modeling by the ICM method. Proteins. 1995;23:403–414. doi: 10.1002/prot.340230314. [DOI] [PubMed] [Google Scholar]
- Cater MA, La Fontaine S, Mercer JF. Copper binding to the N-terminal metal-binding sites or the CPC motif is not essential for copper-induced trafficking of the human Wilson protein (ATP7B) Biochem J. 2007;401:143–153. doi: 10.1042/BJ20061055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cornea RL, Thomas DD. Effects of membrane thickness on the molecular dynamics and enzymatic activity of reconstituted Ca-ATPase. Biochem. 1994;33:2912–2920. doi: 10.1021/bi00176a022. [DOI] [PubMed] [Google Scholar]
- Diaz R, Rice WJ, Stokes DL. Fourier-Bessel reconstruction of helical assemblies. Methods Enzymol. 2010;482:131–165. doi: 10.1016/S0076-6879(10)82005-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dmitriev O, Tsivkovskii R, Abildgaard F, Morgan CT, Markley JL, Lutsenko S. Solution structure of the N-domain of Wilson disease protein: distinct nucleotide-binding environment and effects of disease mutations. Proc. Nat. Acad. Sci. 2006;103:5302–5307. doi: 10.1073/pnas.0507416103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Lohkamp B, Scott W, Cowtan K. Features and development of Coot. Acta Crystallographica Section D - Biological Crystallography. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan B, Rosen BP. Biochemical characterization of CopA, the Escherichia coli Cu(I)-translocating P-type ATPase. J. Biol. Chem. 2002;277:46987–46992. doi: 10.1074/jbc.M208490200. [DOI] [PubMed] [Google Scholar]
- Fernandez-Recio J, Totrov M, Skorodumov C, Abagyan R. Optimal docking area: a new method for predicting protein-protein interaction sites. Proteins. 2005;58:134–143. doi: 10.1002/prot.20285. [DOI] [PubMed] [Google Scholar]
- Gonzalez-Guerrero M, Arguello JM. Mechanism of Cu+-transporting ATPases: soluble Cu+ chaperones directly transfer Cu+ to transmembrane transport sites. Proc Natl Acad Sci U S A. 2008;105:5992–5997. doi: 10.1073/pnas.0711446105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez-Guerrero M, Hong D, Arguello JM. Chaperone-mediated Cu+ delivery to Cu+ transport ATPases: requirement of nucleotide binding. J. Biol. Chem. 2009;284:20804–20811. doi: 10.1074/jbc.M109.016329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatori Y, Hirata A, Toyoshima C, Lewis D, Pilankatta R, Inesi G. Intermediate phosphorylation reactions in the mechanism of ATP utilization by the copper ATPase (CopA) of Thermotoga maritima. J. Biol. Chem. 2008;283:22541–22549. doi: 10.1074/jbc.M802735200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatori Y, Lewis D, Toyoshima C, Inesi G. Reaction cycle of Thermotoga maritima copper ATPase and conformational characterization of catalytically deficient mutants. Biochem. 2009;48:4871–4880. doi: 10.1021/bi900338n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hatori Y, Majima E, Tsuda T, Toyoshima C. Domain organization and movements in heavy metal ion pumps: papain digestion of CopA, a Cu+-transporting ATPase. J. Biol. Chem. 2007;282:25213–25221. doi: 10.1074/jbc.M703520200. [DOI] [PubMed] [Google Scholar]
- Huster D, Lutsenko S. The distinct roles of the N-terminal copper-binding sites in regulation of catalytic activity of the Wilson’s disease protein. J. Biol. Chem. 2003;278:32212–32218. doi: 10.1074/jbc.M305408200. [DOI] [PubMed] [Google Scholar]
- Jorgensen PL, Andersen JP. Structural basis for E1-E2 conformational transitions in Na,K-pump and Ca-pump proteins. J. Membr. Biol. 1988;103:95–120. doi: 10.1007/BF01870942. [DOI] [PubMed] [Google Scholar]
- Kimura Y, Kurzydlowski K, Tada M, MacLennan DH. Phospholamban regulates the Ca2+-ATPase through intramembrane interactions. J. Biol. Chem. 1996;271:21726–21731. doi: 10.1074/jbc.271.36.21726. [DOI] [PubMed] [Google Scholar]
- Kuhlbrandt W. Biology, structure and mechanism of P-type ATPases. Nat. Rev. Mol. Cell Biol. 2004;5:282–295. doi: 10.1038/nrm1354. [DOI] [PubMed] [Google Scholar]
- Larin D, Mekios C, Das K, Ross B, Yang AS, Gilliam TC. Characterization of the interaction between the Wilson and Menkes disease proteins and the cytoplasmic copper chaperone, HAH1p. J. Biol. Chem. 1999;274:28497–28504. doi: 10.1074/jbc.274.40.28497. [DOI] [PubMed] [Google Scholar]
- Lee J, Prohaska JR, Thiele DJ. Essential role for mammalian copper transporter Ctr1 in copper homeostasis and embryonic development. Proc Natl Acad Sci U S A. 2001;98:6842–6847. doi: 10.1073/pnas.111058698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis BA, Engelman DM. Lipid bilayer thickness varies linearly with acyl chain length in fluid phosphatidylcholine vesicles. J. Mol. Biol. 1983;166:211–217. doi: 10.1016/s0022-2836(83)80007-2. [DOI] [PubMed] [Google Scholar]
- Lubben M, Portmann R, Kock G, Stoll R, Young MM, Solioz M. Structural model of the CopA copper ATPase of Enterococcus hirae based on chemical cross-linking. Biometals. 2009;22:363–375. doi: 10.1007/s10534-008-9173-4. [DOI] [PubMed] [Google Scholar]
- Lutsenko S, Barnes NL, Bartee MY, Dmitriev OY. Function and regulation of human copper-transporting ATPases. Physiol. Rev. 2007;87:1011–1046. doi: 10.1152/physrev.00004.2006. [DOI] [PubMed] [Google Scholar]
- Lutsenko S, Kaplan JH. Organization of P-type ATPases: significance of structural diversity. Biochem. 1995;34:15607–15613. doi: 10.1021/bi00048a001. [DOI] [PubMed] [Google Scholar]
- Mana-Capelli S, Mandal AK, Arguello JM. Archaeoglobus fulgidus CopB is a thermophilic Cu2+-ATPase: functional role of its histidine-rich-N-terminal metal binding domain. J. Biol. Chem. 2003;278:40534–40541. doi: 10.1074/jbc.M306907200. [DOI] [PubMed] [Google Scholar]
- Mandal AK, Arguello JM. Functional roles of metal binding domains of the Archaeoglobus fulgidus Cu+-ATPase CopA. Biochem. 2003;42:11040–11047. doi: 10.1021/bi034806y. [DOI] [PubMed] [Google Scholar]
- Mandal AK, Cheung WD, Arguello JM. Characterization of a thermophilic P-type Ag+/Cu+-ATPase from the extremophile Archaeoglobus fulgidus. J. Biol. Chem. 2002;277:7201–7208. doi: 10.1074/jbc.M109964200. [DOI] [PubMed] [Google Scholar]
- Mitra B, Sharma R. The cysteine-rich amino-terminal domain of ZntA, a Pb(II)/Zn(II)/Cd(II)-translocating ATPase from Escherichia coli, is not essential for its function. Biochem. 2001;40:7694–7699. doi: 10.1021/bi010576g. [DOI] [PubMed] [Google Scholar]
- Moller JV, Juul B, le Maire M. Structural organization, ion transport, and energy transduction of P-type ATPases. Biophys. Biochim. Acta. 1996;1286:1–51. doi: 10.1016/0304-4157(95)00017-8. [DOI] [PubMed] [Google Scholar]
- Morgan CT, Tsivkovskii R, Kosinsky YA, Efremov RG, Lutsenko S. The distinct functional properties of the nucleotide-binding domain of ATP7B, the human copper-transporting ATPase: analysis of the Wilson disease mutations E1064A, H1069Q, R1151H, and C1104F. J. Biol. Chem. 2004;279:36363–36371. doi: 10.1074/jbc.M404553200. [DOI] [PubMed] [Google Scholar]
- Morth JP, Pedersen BP, Toustrup-Jensen MS, Sorensen TL, Petersen J, Andersen JP, Vilsen B, Nissen P. Crystal structure of the sodium-potassium pump. Nature. 2007;450:1043–1049. doi: 10.1038/nature06419. [DOI] [PubMed] [Google Scholar]
- Olesen C, Picard M, Winther AM, Gyrup C, Morth JP, Oxvig C, Moller JV, Nissen P. The structural basis of calcium transport by the calcium pump. Nature. 2007;450:1036–1042. doi: 10.1038/nature06418. [DOI] [PubMed] [Google Scholar]
- Olesen C, Sorensen TL, Nielsen RC, Moller JV, Nissen P. Dephosphorylation of the calcium pump coupled to counterion occlusion. Science. 2004;306:2251–2255. doi: 10.1126/science.1106289. [DOI] [PubMed] [Google Scholar]
- Pedersen BP, Buch-Pedersen MJ, Morth JP, Palmgren MG, Nissen P. Crystal structure of the plasma membrane proton pump. Nature. 2007;450:1111–1114. doi: 10.1038/nature06417. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Rae TD, Schmidt PJ, Pufahl RA, Culotta VC, O’Halloran TV. Undetectable intracellular free copper: the requirement of a copper chaperone for superoxide dismutase. Science. 1999;284:805–808. doi: 10.1126/science.284.5415.805. [DOI] [PubMed] [Google Scholar]
- Ralle M, Lutsenko S, Blackburn NJ. Copper transfer to the N-terminal domain of the Wilson disease protein (ATP7B): X-ray absorption spectroscopy of reconstituted and chaperone-loaded metal binding domains and their interaction with exogenous ligands. J Inorg Biochem. 2004;98:765–774. doi: 10.1016/j.jinorgbio.2004.02.009. [DOI] [PubMed] [Google Scholar]
- Rice WJ, Kovalishin A, Stokes DL. Role of metal-binding domains of the copper pump from Archaeoglobus fulgidus. Biochem. Biophys. Res. Commun. 2006;348:124–131. doi: 10.1016/j.bbrc.2006.07.012. [DOI] [PubMed] [Google Scholar]
- Rice WJ, Young HS, Martin DW, Sachs JR, Stokes DL. Structure of Na+,K+-ATPase at 11 Å resolution: comparison with Ca2+-ATPase in E1 and E2 states. Biophys. J. 2001;80:2187–2197. doi: 10.1016/S0006-3495(01)76191-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sazinsky MH, Agarwal S, Arguello JM, Rosenzweig AC. Structure of the actuator domain from the Archaeoglobus fulgidus Cu+-ATPase. Biochem. 2006a;45:9949–9955. doi: 10.1021/bi0610045. [DOI] [PubMed] [Google Scholar]
- Sazinsky MH, Mandal AK, Arguello JM, Rosenzweig AC. Structure of the ATP binding domain from the Archaeoglobus fulgidus Cu+-ATPase. J. Biol. Chem. 2006b;281:11161–11166. doi: 10.1074/jbc.M510708200. [DOI] [PubMed] [Google Scholar]
- Singleton C, Hearnshaw S, Zhou L, Le Brun NE, Hemmings AM. Mechanistic insights into Cu(I) cluster transfer between the chaperone CopZ and its cognate Cu(I)-transporting P-type ATPase, CopA. Biochem J. 2009;424:347–356. doi: 10.1042/BJ20091079. [DOI] [PubMed] [Google Scholar]
- Strausak D, Howie MK, Firth SD, Schlicksupp A, Pipkorn R, Multhaup G, Mercer JF. Kinetic analysis of the interaction of the copper chaperone Atox1 with the metal binding sites of the Menkes protein. J. Biol. Chem. 2003;278:20821–20827. doi: 10.1074/jbc.M212437200. [DOI] [PubMed] [Google Scholar]
- Strausak D, La Fontaine S, Hill J, Firth SD, Lockhart PJ, Mercer JF. The role of GMXCXXC metal binding sites in the copper-induced redistribution of the Menkes protein. J. Biol. Chem. 1999;274:11170–11177. doi: 10.1074/jbc.274.16.11170. [DOI] [PubMed] [Google Scholar]
- Taylor KA, Dux L, Martonosi A. Three-dimensional reconstruction of negatively stained crystals of the Ca++-ATPase from muscle sarcoplasmic reticulum. J. Mol. Biol. 1986;187:417–427. doi: 10.1016/0022-2836(86)90442-0. [DOI] [PubMed] [Google Scholar]
- Thomas GR, Forbes JR, Roberts EA, Walshe JM, Cox DW. The Wilson disease gene: spectrum of mutations and their consequences. Nat Genet. 1995;9:210–217. doi: 10.1038/ng0295-210. [DOI] [PubMed] [Google Scholar]
- Toyoshima C, Nakasako M, Nomura H, Ogawa H. Crystal structure of the calcium pump of sarcoplasmic reticulum at 2.6 Å resolution. Nature. 2000;405:647–655. doi: 10.1038/35015017. [DOI] [PubMed] [Google Scholar]
- Toyoshima C, Nomura H. Structural changes in the calcium pump accompanying the dissociation of calcium. Nature. 2002;418:605–611. doi: 10.1038/nature00944. [DOI] [PubMed] [Google Scholar]
- Toyoshima C, Unwin N. Ion channel of acetylcholine receptor reconstructed from images of postsynaptic membranes. Nature. 1988;336:247–250. doi: 10.1038/336247a0. [DOI] [PubMed] [Google Scholar]
- Toyoshima C, Yonekura K, Sasabe H. Contrast transfer for frozen-hydrated specimens: II. Amplitude contrast at very low frequencies. Ultramicroscopy. 1993;48:165–176. [Google Scholar]
- Tsivkovskii R, MacArthur BC, Lutsenko S. The Lys1010-Lys1325 fragment of the Wilson’s disease protein binds nucleotides and interacts with the N-terminal domain of this protein in a copper-dependent manner. J. Biol. Chem. 2001;276:2234–2242. doi: 10.1074/jbc.M003238200. [DOI] [PubMed] [Google Scholar]
- Tsuda T, Toyoshima C. Nucleotide recognition by CopA, a Cu+-transporting P-type ATPase. EMBO J. 2009;28:1782–1791. doi: 10.1038/emboj.2009.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veldhuis NA, Gaeth AP, Pearson RB, Gabriel K, Camakaris J. The multi-layered regulation of copper translocating P-type ATPases. Biometals. 2009a;22:177–190. doi: 10.1007/s10534-008-9183-2. [DOI] [PubMed] [Google Scholar]
- Veldhuis NA, Valova VA, Gaeth AP, Palstra N, Hannan KM, Michell BJ, Kelly LE, Jennings I, Kemp BE, Pearson RB, et al. Phosphorylation regulates copper-responsive trafficking of the Menkes copper transporting P-type ATPase. Int J Biochem Cell Biol. 2009b;41:2403–2412. doi: 10.1016/j.biocel.2009.06.008. [DOI] [PubMed] [Google Scholar]
- Walker JM, Huster D, Ralle M, Morgan CT, Blackburn NJ, Lutsenko S. The N-terminal metal-binding site 2 of the Wilson’s Disease Protein plays a key role in the transfer of copper from Atox1. J. Biol. Chem. 2004;279:15376–15384. doi: 10.1074/jbc.M400053200. [DOI] [PubMed] [Google Scholar]
- Walker JM, Tsivkovskii R, Lutsenko S. Metallochaperone Atox1 transfers copper to the NH2-terminal domain of the Wilson’s disease protein and regulates its catalytic activity. J. Biol. Chem. 2002;277:27953–27959. doi: 10.1074/jbc.M203845200. [DOI] [PubMed] [Google Scholar]
- Warren GB, Toon PA, Birdsall NJM, Lee AG, Metcalfe JC. Reconstitution of a calcium pump using defined membrane components. Proc. Natl. Acad. Sci. 1974;71:622–626. doi: 10.1073/pnas.71.3.622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu CC, Rice WJ, Stokes DL. Structure of a copper pump suggests a regulatory role for its metal-binding domain. Structure. 2008;16:976–985. doi: 10.1016/j.str.2008.02.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu C, Rice WJ, He W, Stokes DL. A structural model for the catalytic cycle of Ca2+-ATPase. J. Mol. Biol. 2002;316:201–211. doi: 10.1006/jmbi.2001.5330. [DOI] [PubMed] [Google Scholar]
- Young H, Xu C, Zhang P, Stokes D. Locating the thapsigargin binding site on Ca2+-ATPase by cryoelectron microscopy. J. Mol. Biol. 2001;308:231–240. doi: 10.1006/jmbi.2001.4558. [DOI] [PubMed] [Google Scholar]
- Young HS, Rigaud J-L, Lacapere J-J, Reddy LG, Stokes DL. How to make tubular crystals by reconstitution of detergent-solubilized Ca2+-ATPase. Biophys. J. 1997;72:2545–2558. doi: 10.1016/S0006-3495(97)78898-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang P, Toyoshima C, Yonekura K, Green NM, Stokes DL. Structure of the calcium pump from sarcoplasmic reticulum at 8 Å resolution. Nature. 1998;392:835–839. doi: 10.1038/33959. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.