Abstract
Hepatitis B virus (HBV) can be classified into nine immunological subtypes or eight genotypes. The most prevalent genotypes in Asia are genotypes B and C. HBV is transmitted parenteraly and can produce either asymptomatic or symptomatic disease. Although the consequences of acute hepatitis B can be severe, serious sequelae are associated with chronic infections. HBV seroprevalence ranges from intermediate (2%-7%) to high (≥8%) levels in Asia. Several strategies for the control and prevention of HBV infection have been found to be efficacious. They include vaccination and the administration of HBIG, interferon-a and nucleoside/nucleotide analogues. However, these procedures also apply selective pressures on HBV in infected individuals leading to the generation and accumulation of mutations in the S gene. Most of these mutations occur in the major hydrophilic region (MHR) of the S gene. These mutations create public health concerns as they can be responsible for reactivation of hepatitis B and occult hepatitis B infection. The inability to detect occult infections means that these individuals may become blood donors. This suggests that new strategies for donor evaluation and selection may need to be developed to protect the blood supply.
Keywords: Escape mutants, genotype, hepatitis B virus, review, subtype, surface antigen
Introduction
Hepatitis B virus (HBV) is a member of the Hepadnaviridae family. HBV is transmitted parenteraly and can produce either asymptomatic or symptomatic disease.[1,2] Although the consequences of acute hepatitis B can be severe, the majority of serious sequelae are associated with chronic infections. Worldwide, an estimated 360 million people are chronically infected with HBV.[3] Chronic infection occurs in about 90% of those infected perinataly, 30% in those infected in early childhood under five years of age and 6% when individuals are infected over five years of age.[4–7] Based on data from follow-up studies of individuals infected as infants or young children, about 25% of individuals with chronic hepatitis B die from cirrhosis or liver cancer; the majority remain asymptomatic for decades until the onset of cirrhosis or end-stage liver disease.[8–10] Chronically infected individuals are at increased lifetime risk for cirrhosis and hepatocellular carcinoma and serve as reservoirs for continued HBV transmission.
Estimates of chronic HBV infection are inferred from population sampling for the presence of hepatitis B surface antigen in the population. In Asia HBsAg seroprevalence ranges from intermediate (2%-7%) to high (≥8%) levels. In areas such as southern China, Korea, Melanesia, Micronesia, the Philippines and Polynesia seroprevalence is greater that 10%. Countries like India, Indonesia, Japan and Pakistan have intermediate rates of endemicity.[11–14] However, these rates must be considered to be inaccurate and possibly underestimates as rates of occult HBV infection are not included in these estimates. Prevention and control activities have been shown to reduce seroprevalence rates.[13,14]
Several strategies have been developed to prevent and control HBV infection. Immunization with hepatitis B vaccine with or without administration of hepatitis B immune globulin (HBIG) has proven to be efficacious in the pre-exposure setting.[13] Immunization is also effective in preventing maternal-infant transmission of HBV; however, about 15% of vaccinated infants, who do not develop adequate levels of HBsAg antibodies, may become infected with HBV.[15] In the post-exposure setting, vaccine, HBIG,[13] interferon-α, lamivudine and nucleoside or nucleotide analogues[16] have been used as means of therapy with varying degrees of success. The existence of HBV quasi-species[17] has facilitated the development of mutants with ability to escape antibody detection and antibody neutralization. These escape mutants may lead to reinfection with HBV.
HBV Subtypes
Blumberg et al. first described the Australian antigen, a, in 1965.[18] Further research revealed the immunological heterogeneity of the Australian antigen.[19–22] Two pairs of allelic variations, d/y and w/r , were discovered in 1971 and 1972, respectively.[19,20] The discovery of additional determinants with the appearance of multiple naming conventions[23] occurred at a rapid pace and an international synod was convened in Paris in 1975 to discuss a determinant heterogeneity and reach consensus.[24] At this workshop, four sub-determinants of the a determinant were redefined as w sub-determinants (w1-w4) and the plethora of HBsAg determinants were found to be collapsible into eight subtypes; ayw1, ayw2, ayw3, ayw4, ayr, adw2, adw4 and adr.[24] Following the discovery of the q determinant in 1975,[25] it was determined that adr could be subdivided into q+ and q- categories.[26] The development of monoclonal radioimmunoassays confirmed the existence of these nine subtypes.[27–29] In 2002, Arauz-Ruiz et al.[30] described a tenth subtype, adw3.
The development of DNA sequencing methodologies facilitated the elucidation of the specific amino acids in HBsAg responsible for the reactivity patterns with monoclonal antibodies. Chemical modification of HBsAg revealed the importance of Lys122 for the expression of the d determinant.[31–33] Studies of two blood donors carrying compound subtypes, adyr and adwr, showed that only amino acid positions 122 and 160 are uniquely responsible for the expression of d/y and w/r specificity.[34] These studies found that the d-to-y and w-to-r changes are mediated through a shift from Lys to Arg at positions 122 and 160, respectively.[34] Site-directed mutagenesis later confirmed the specificity of Lys160 for w reactivity[35] and delineated which amino acids at position 127 were responsible for w2-w4 reactivities [Figure 1].[26,36,37] Norder et al.[38] found that Phe134, Ala159 or both, are involved in the expression of w1 reactivity and recent research suggests position 140 may be more important in resolving w1 reactivity than position 134.[39] Additional evidence suggested position 177 is involved in q reactivity of adr specimens[26] and position 178 is involved in q reactivity of adw4 specimens.[38,40] [Figure 1] contains algorithms for determining HBV subtype from the amino acid sequence of the HBsAg.
Figure 1.
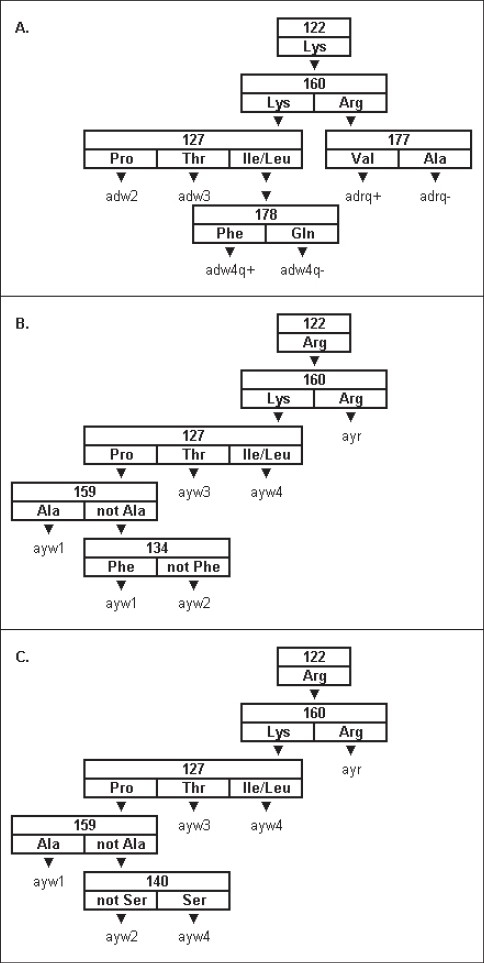
Algorithms for determining HBV subtype from the primary structure of the HBV S gene. A. The decision tree for the ad subtypes. B. The original decision tree for determining the ay subtypes. C. A newly developed decision tree for determining the ay subtypes.[39] Each box in each decision tree contains a number representing an amino acid position in the S gene. The amino acids listed below each number represent the permitted wild type residues at that position. The downward pointing triangles denote paths through each decision tree. The fi nal designation at the end of each possible path is the subtype for that path.
HBV Genotypes
Originally, genotypes were designated as sequences within a phylogenetic clade having no more than 4% sequence divergence between the members of the clade and more that 8% sequence divergence with extra-clade sequences.[41] To date, eight genotypes, designated, A to H, have been described.[30,37,42] Because of ease of use, genotyping has become more widely accepted as the method for relating phenotype to genetics than serological subtyping. Also, single nucleotide mutations within the a determinant may lead to changes in HBV subtypes; however, this is not the case for genotypes.[43–47] Genotypes B and C are the most prevalent genotypes found in Asia; however, genotypes A and D are the most prevalent genotypes found in India.[11,12]
HBV Subtypes and Subgenotypes
DNA sequencing has led to the elucidation of associations between serologic subtypes and genotypes. Early research indicated an apparent association between genotype/subtype and geographic location.[45] Genotype A (subtypes adw2 and ayw1 ) is most prevalent in North America and northwestern Europe. Genotypes B (subtypes adw2, adw3 and ayw1) and C (subtypes adw2, adw3, ayw3, adr and ayr) are highly prevalent in East Asia. Genotype D (subtypes adw3, ayw2, ayw3 and ayw4) is most prevalent in the Mediterranean and the Middle East. Genotype E (subtype ayw4) has been found in West Africa. Genotypes F (subtypes adw4 and ayw4) and H (subtype adw4) are found in Central and South America. Genotype G (subtype adw2) is found in the United States and Europe.[46,47] Indeed, a study conducted in Sweden concluded that the genotypes of HBV seen in patients at an outpatient clinic correlated more with the country of the patients’ nativity than their current residency in Sweden.[48]
Genotypic sequences, which diverge by less than 4% but form distinct sub-clades within a genotypic clade in a phylogenetic tree, have been designated subgenotypes. Genotype A was found to segregate into two well-defined subgenotypes, Ae in Europe and Aa in Africa and Asia,[49] with a possible third subgenotype found in Cameroon.[50] Genotypes B and C each appear to have about four or five subgenotypes.[46,51–53] One of the B subgenotypes is due to recombination between genotypes B and C.[54] Genotype D segregates into four subgenotypes and genotype F forms two distinct subgenotypes. However, genotypes E, G and H do not appear to have subgenotypes.[46]
There was an indication that subgenotypes might correlate with serological subtypes as was seen with the Ae-adw2 association in northwest Europe and the Aa-ayw1 association in Central Africa.[45] Further research demonstrated this was not always the case.[46,48] Norder et al.[46] found that some subgenotypes were composed of sequences sharing a common serological subtype, while other subgenotypes were composed of sequences from multiple subtypes.
Escape Mutants
As stated above, vaccination with HBsAg has been efficacious in the pre-exposure setting; however, occult infections began to be noted in the late 1980's[55,56] and the first case of a vaccine-induced escape mutant, in a child in southern Italy, who received passive-active post-exposure immunization, was described in 1990.[57] Mutations within the S gene are known to be responsible for occult hepatitis B infections, reactivation of hepatitis B,[58,59] diagnostic assay failure[58,60–63] and reinfection in HBV-infected recipients of orthotopic liver transplantations.[17,64] Occult infections create public health concerns because asymptomatic carriers can be blood donors.[65–67] These mutations are stable and can be transmitted horizontally and vertically.[56,68–71]
HBV replicates to high titers in infected individuals. Because it replicates through an RNA intermediate synthesized by reverse transcriptase, mutant viral genomes[59,72] and quasi-species[71–77] are generated. This results in the production of viral mutants during naturally occurring infections.[78,79] Vaccination and the administration of HBIG and anti-viral drugs like lumavidine exert evolutionary pressures to select mutants.[70,80]
The a determinant is located on the major hydrophilic region (MHR) of the S gene, which is between amino acids 120 and 160. The MHR forms a two-loop structure. There are two alternative models for this double loop structure, which involve two pairs of cysteines forming disulfide bridges at the ends of each loop. The more generally accepted model envisions disulfide bridging between C124 and C137 and between C139 and C147.[58] The alternative model is built on disulfide bridging between C107 and C138 and between C139 and C147.[63]
Research with childhood vaccinations shows that mutations accumulate with higher frequency in vaccinated than unvaccinated children, with more mutations emerging in children vaccinated with plasma-derived vaccine than recombinant vaccine.[80] Vaccinated children generated a preferential accumulation of mutations in the second loop of the MHR, while unvaccinated children generated random mutations.[81] The prevalence of mutations increases over time[80,82] and the frequency of amino acid variation per site increases with age.[73] There is also an accumulation of S gene mutations in HBV related end-stage liver disease.[83]
Mutations accumulate across the MHR [Figure 2]. These mutations can disrupt the antigenicity of HBsAg in a number of ways. One way is by modifying amino acids directly involved in expression of the antigen, such as the amino acid specified by codon 122, which is responsible for the expression of d/y specificity.[84] Alterations to the structure of HBsAg can also disrupt binding of polyclonal antibodies to it. These alterations include mutations to the cysteines involved in the formation of the disulfide bridges responsible for the two-loop structure of the MHR[73,85,86] and modifications to protein hydrophilicity, electric charge and acidity.[59,76,80,81,87] Some mutations involve amino acid insertions into the a determinant[63,78,88,89] or the creation of stop codons.[71,84] Other mutations eliminate the glycosylation site at residue 146[85] or create potential new glycosylation sites in the first loop of the MHR.[90,91] However, not all mutations in the MHR lead to escape mutants.[92,93]
Figure 2.
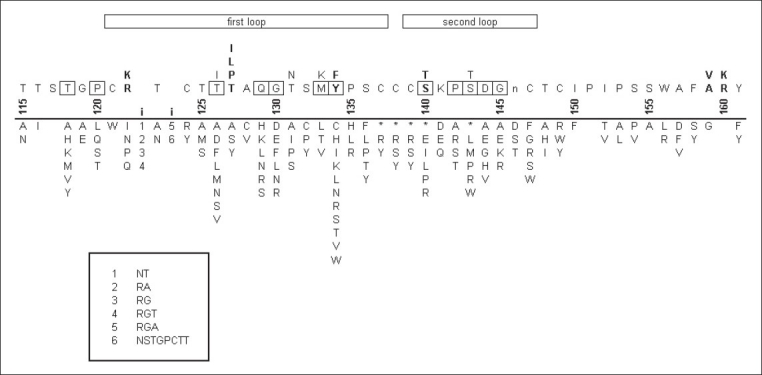
Mutations in the HBsAg a determinant. The numbers above the line represent amino acid positions in HBsAg. The bold i's between the numbers 120 and 125 represent known insertion points in the surface antigen. Above the numbers is the wild type sequence for HBsAg, with multiple letters in a column denoting alternative amino acids seen in various subtypes. Emboldened positions denote putative residues that participate in the a determinant. The lower case n at position 146 is the site of N-linked glycosylation. Boxed positions show the positions at which mutations are most frequently seen. Below the line the columns of letters represent the mutant residues seen at each position. Asterisks denote stop codons. Numbers below the bold i's denote the oligopeptides inserted at these two positions. These oligopeptides are listed in the box at the bottom. The data for this figure come from several sources.[85,86,89,91,93–95,98,108,110–153]
While most research into HBsAg escape mutants has been directed specifically at the MHR, additional research has shown that mutations outside the MHR can also lead to the creation of escape mutants. Mutations near the MHR can alter group-specific antigenicity even though the mutations are not within the MHR.[89,94,95] Monoclonal antibody binding assays have also shown that residues 178-186 are exposed on the surface of the 20 nm particle.[96] This is the region which specifies the q sub-determinant.
Administration of nucleoside and nucleotide analogs, like lamivudine, which inhibit viral polymerase activity, can lead to the generation of mutants in the polymerase gene. Because the polymerase and S genes overlap any change in the polymerase has the potential for causing mutations in the S gene.[93] Although many of these polymerase mutants are in the C domain near the YMDD motif, additional mutations are generated in the B Domain, which overlaps the a determinant.[97,98] Indeed, lamivudine therapy is associated with the generation of escape mutants.[93] Although polymerase mutants exhibit diminished replication activity, some polymerase/a determinant double mutants exhibit increased replication activity in the presence of lamivudine.[99] It has also been found that recombinant HBV vaccine provided limited efficacy when given to post-liver transplant patients who have received prior lamivudine treatment for pre-existing chronic hepatitis B.[100] Besides escape mutants, nucleotide and nucleoside analogs can generate stop codons within the S gene.[97,98]
Ultimately it would be beneficial to be able to correlate escape mutant phenotypes with a specific genetic sequence. Unfortunately, the multitude of mutations does not lend itself to easy categorization. While studies using synthetic peptides and analysis of point mutations allow examinations of individual positions,[101,102] they do not help in investigating the presence of conformational epitopes.[84,88,103] To further confound any analysis of escape mutants, multiple mutations within the MHR lead to generate revertants masking the effects of escape mutants.[92] Nonetheless, it is possible to make some generalizations about HBsAg mutants. Patients with first loop mutants tend to have higher levels of HBV DNA than patients with second loop mutants. G145R, the prototypic HBsAg mutant, is the most stable mutant.[77] In patients with the G145R mutant, a gradual decline in the HBV viral load has been observed.[69] Patients with hepatocellular carcinoma or chronic hepatitis can have a higher frequency of mutations in the class I HLA-A2-restricted CTL epitope between residues 29 to 53 than the a determinant.[104,105] The G145R and M133T mutations have also been implicated in the development of hepatocellular carcinoma.[69,80,106] As stated earlier, there is an accumulation of S gene mutations in HBV related end-stage liver disease.[84]
The Future
HBV escape mutants cause a public health concern through reinfection and occult infection. This is especially true in Asia with its intermediate to high rates of chronic infection.[12,14] The health problems associated with HBV infection are well-documented with the majority of serious sequelae being associated with chronic infections. These consequences are amplified in infected infants and young children.[8–10]
Depending on how blood donors are selected and screened the intermediate to high levels of circulating HBV in Asia increase the probability of contaminated blood entering the blood supply. Although prevention and control strategies have proven to be efficacious, escape mutants are generated in response to these immune pressures.[75,79,106] The large population of infected individuals also creates an environment in which evolutionary processes can lead to the creation of new HBV isolates able to escape detection and enter the blood supply.[33,65,66] Evaluation of donor selection and screening procedures may yield insights into strategies that could potentially safeguard the blood supply. To decrease the levels of post-transfusion hepatitis the feasibility and efficacy of using nucleic acid amplification testing, DNA dot blot hybridisation assays or multiple immunoassays may need to be examined.
A debate has begun about whether or not the development of vaccines, which protect against mutant strains of HBV, need to be used in conjunction with the wild-type vaccine.[98,107] No matter how this debate is resolved in the future, research has already shown that mutant strain vaccines can be protective against mutant strains.[32,107] If it is decided that mutant strain vaccines would have public health potential, a big part of this discussion will be about which mutants need to be targeted. As far as diagnostics is concerned, there is a concerted effort to understand how mutants affect a determinant antigenicity in order that assays, which can detect mutant strains.[108,109] particularly in blood donors and liver transplant recipients, can be developed. However, the necessity for these assays has been questioned because while one diagnostic assay may fail to detect a particular mutant HBsAg, another assay might detect it, albeit at reduced reactivity.[93,110]
A better understanding of the exact structural and physiochemical changes caused by mutations within the S gene is needed to determine which mutants modulate S gene antigenicity. At present, the best visualization of HBsAg structure is obtained from a 12.5Å resolution cryo-EM reconstruction of a 22-nm noninfectious viral particle.[109] With a higher resolution structure it would be possible to map precisely the resides involved in specifying the a determinant and determine their spatial relationship.[111] This would further allow investigation into the effect of single residue changes in the S gene with respect to changes in HBsAg protein folding and the a determinant structure.[109,110,153]
Source of Support: Nil,
Conflict of Interest: None declared.
Disclaimer: This information is distributed solely for the purpose of pre dissemination peer review under applicable information quality guidelines. It has not been formally disseminated by the Centers for Disease Control and Prevention/ the Agency for Toxic Substances and Disease Registry. It does not represent and should not be construed to represent any agency determination or policy
References
- 1.Krugman S, Overby LR, Mushahwar IK, Ling CM, Frösner GG, Deinhardt F. Viral hepatitis, type B: Studies on natural history and prevention re-examined. N Engl J Med. 1979;300:101–6. doi: 10.1056/NEJM197901183000301. [DOI] [PubMed] [Google Scholar]
- 2.Hoofnagle JH, DiBisceglie AM. Serologic diagnosis of acute and chronic viral hepatitis. Semin Liver Dis. 1991;11:73–83. doi: 10.1055/s-2008-1040426. [DOI] [PubMed] [Google Scholar]
- 3.EASL Jury. EASL International Consensus Conference on Hepatitis B. 13-14 September, 2002: Geneva, Switzerland. Consensus statement (short version) J Hepatol. 2003;38:533–40. doi: 10.1016/s0168-8278(03)00083-7. [DOI] [PubMed] [Google Scholar]
- 4.McMahon BJ, Alward WL, Hall DB, Heyward WL, Bender TR, Francis DP, et al. Acute hepatitis B virus infection: Relation of age to the clinical expression of disease and subsequent development of the carrier state. J Infect Dis. 1985;151:599–603. doi: 10.1093/infdis/151.4.599. [DOI] [PubMed] [Google Scholar]
- 5.Edmunds WJ, Medley GF, Nokes DJ, Hall AJ, Whittle HC. The influence of age on the development of the hepatitis B carrier state. Proc Biol Sci. 1993;253:197–201. doi: 10.1098/rspb.1993.0102. [DOI] [PubMed] [Google Scholar]
- 6.Hyams KC. Risks of chronicity following acute hepatitis B virus infection: A review. Clin Infect Dis. 1995;20:992–1000. doi: 10.1093/clinids/20.4.992. [DOI] [PubMed] [Google Scholar]
- 7.Beasley RP, Hwang LY, Lee GC, Lan CC, Roan CH, Huang FY, et al. Prevention of perinatally transmitted hepatitis B virus infections with hepatitis B immune globulin and hepatitis B vaccine. Lancet. 1983;2:1099–102. doi: 10.1016/s0140-6736(83)90624-4. [DOI] [PubMed] [Google Scholar]
- 8.Beasley RP, Hwang LY, Lin CC, Chin CS. Hepatocellular carcinoma and hepatitis B virus: A prospective study of 22,707 men in Taiwan. Lancet. 1981;2:1129–33. doi: 10.1016/s0140-6736(81)90585-7. [DOI] [PubMed] [Google Scholar]
- 9.Hoofnagle JH, Shafritz DA, Popper H. Chronic type B hepatitis and the “healthy” HBsAg carrier state. Hepatology. 1987;7:758–63. doi: 10.1002/hep.1840070424. [DOI] [PubMed] [Google Scholar]
- 10.McMahon BJ, Alberts SR, Wainwright RB, Bulkow L, Lanier AP. Hepatitis B-related sequelae: Prospective study in 1400 hepatitis B surface antigen-positive Alaska Native carriers. Arch Intern Med. 1990;150:1051–4. doi: 10.1001/archinte.150.5.1051. [DOI] [PubMed] [Google Scholar]
- 11.Acharya SK, Madan K, Dattagupta S, Panda SK. Viral Hepatitis in India. Natl Mad J India. 2006;19:203–17. [PubMed] [Google Scholar]
- 12.Lau GK. Hepatitis B infection in China. Clin Liver Dis. 2001;5:361–79. doi: 10.1016/s1089-3261(05)70170-7. [DOI] [PubMed] [Google Scholar]
- 13.Mast EE, Weinbaum CM, Fiore AE, Alter MJ, Bell BP, Finelli L, et al. A comprehensive immunization strategy to eliminate transmission of hepatitis B virus infection in the United States: Recommendations of the Advisory Committee on Immunization Practices (ACIP) Part II: Immunization of adults. MMWR Recomm Rep. 2006;55:1–33. [PubMed] [Google Scholar]
- 14.Merican I, Guan R, Amarapuka D, Alexander MJ, Chutaputti A, Chien RN, et al. Chronic hepatitis B virus infection in Asian countries. J Gastroenterol Hepatol. 2000;15:1356–61. doi: 10.1046/j.1440-1746.2000.0150121356.x. [DOI] [PubMed] [Google Scholar]
- 15.He C, Nomura F, Itoga S, Isobe K, Nakai T. Prevalence of vaccine-induced escape mutants of hepatitis B virus in the adult population in China: A prospective study in 176 restaurant employees. J Gastroenterol Hepatol. 2001;16:1373–7. doi: 10.1046/j.1440-1746.2001.02654.x. [DOI] [PubMed] [Google Scholar]
- 16.Lok AS. Hepatitis B infection: Pathogenesis and management. J Hepatol. 2000;32:89–97. doi: 10.1016/s0168-8278(00)80418-3. [DOI] [PubMed] [Google Scholar]
- 17.Schätz HM, Sieger E, Jilg W, Nitschko H, Zachoval R. Variability of the hepatitis B surface protein in HBV-infected liver transplant recipients. J Biomed Sci. 1997;4:146–54. doi: 10.1007/BF02255643. [DOI] [PubMed] [Google Scholar]
- 18.Blumberg BS, Alter HJ, Visnich S. A new antigen in leukemia sera. JAMA. 1965;191:541–6. doi: 10.1001/jama.1965.03080070025007. [DOI] [PubMed] [Google Scholar]
- 19.Le Bouvier GL. The heterogeneity of Australian antigen. J Infect Dis. 1971;123:671–5. doi: 10.1093/infdis/123.6.671. [DOI] [PubMed] [Google Scholar]
- 20.Bancroft WH, Mundon FK, Russell PK. Detection of additional antigenic determinants of hepatitis B antigen. J Immunol. 1972;109:842–8. [PubMed] [Google Scholar]
- 21.Levene C, Blumberg BS. Additional specificities of Australian antigen and the possible identifications of hepatitis carriers. Nature. 1969;221:195–6. doi: 10.1038/221195a0. [DOI] [PubMed] [Google Scholar]
- 22.Shokrgozar MA, Shokri F. Subtype specificity of anti-HBs antibodies produced by human B-cell lines isolated from normal individuals vaccinated with recombinant hepatitis B vaccine. Vaccine. 2002;20:2215–20. doi: 10.1016/s0264-410x(02)00116-0. [DOI] [PubMed] [Google Scholar]
- 23.Le Bouvier GL, Williams A. Serotypes of hepatitis B antigen (HBsAg): The problem of “new” determinants, as exemplified by “t”. Am J Med Sci. 1975;270:165–71. doi: 10.1097/00000441-197507000-00023. [DOI] [PubMed] [Google Scholar]
- 24.Couroucé AM, Soulier JP. The a(w) subdeterminants. Bibl Haematol. 1976;42:31–42. doi: 10.1159/000398987. [DOI] [PubMed] [Google Scholar]
- 25.Magnius LO, Kaplan L, Vyas GN, Perkins HA. A new virus-specified determinant of hepatitis B surface antigen. Acta Pathol Microbiol Scand (B) 1975;83:295–7. doi: 10.1111/j.1699-0463.1975.tb00104.x. [DOI] [PubMed] [Google Scholar]
- 26.Couroucé-Pauty AM, Lemaire JM, Roux JF. New hepatitis B surface antigen subtypes inside the ad category. Vox Sang. 1978;35:304–8. doi: 10.1111/j.1423-0410.1978.tb02939.x. [DOI] [PubMed] [Google Scholar]
- 27.Mimms LT, Floreani M, Tyner J, Whitters E, Rosenlof R, Wray L, et al. Discrimination of hepatitis B virus (HBV) subtypes using monoclonal antibodies to the PreS1 and PreS2 domains of the viral envelope. Virology. 1990;176:604–19. doi: 10.1016/0042-6822(90)90031-l. [DOI] [PubMed] [Google Scholar]
- 28.Swenson PD, Riess JT, Krueger LE. Determination of HBsAg subtypes in different high risk populations using monoclonal antibodies. J Virol Methods. 1991;33:27–38. doi: 10.1016/0166-0934(91)90004-j. [DOI] [PubMed] [Google Scholar]
- 29.Wands JR, Wong MA, Shorey J, Brown RD, Marciniak RA, Isselbacher KJ. Hepatitis B viral antigenic structure: Signature analysis by monoclonal radioimmunoassays. Proc Natl Acad Sci USA. 1984;81:2237–41. doi: 10.1073/pnas.81.7.2237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Arauz-Ruiz P, Norder H, Robertson BH, Magnius LO. Genotype H: A new Amerindian genotype of hepatitis B virus revealed in Central America. J Gen Virol. 2002;83:2059–73. doi: 10.1099/0022-1317-83-8-2059. [DOI] [PubMed] [Google Scholar]
- 31.Peterson DL, Paul DA, Lam J, Tribby II, Achord DT. Antigenic structure of hepatitis B surface antigen: Identification of the “d” subtype determinant by chemical modification and use of monoclonal antibodies. J Immunol. 1984;132:920–7. [PubMed] [Google Scholar]
- 32.Okamoto H, Imai M, Shimozaki M, Hoshi Y, Iizuka H, Gotanda T, et al. Nucleotide sequence of a cloned hepatitis B virus genome, subtype ayr: Comparison with genomes of the other three subtypes. J Gen Virol. 1986;67:2305–14. doi: 10.1099/0022-1317-67-11-2305. [DOI] [PubMed] [Google Scholar]
- 33.Ashton-Rickardt PG, Murray K. Mutations that change the immunological subtype of hepatitis B virus surface antigen and distinguish between antigenic and immunogenic determination. J Med Virol. 1989;29:204–14. doi: 10.1002/jmv.1890290311. [DOI] [PubMed] [Google Scholar]
- 34.Okamoto H, Imai M, Tsuda F, Tanaka T, Miyakawa Y, Mayumi M. Point mutation in the S gene of hepatitis B virus for a d/y or w/r subtypic change in two blood donors carrying a surface antigen of compound subtype adyr or adwr. J Virol. 1987;61:3030–4. doi: 10.1128/jvi.61.10.3030-3034.1987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Okamoto H, Omi S, Wang Y, Itoh Y, Tsuda F, Tanaka T, et al. The loss of subtypic determinants in alleles, d/y or w/r, on hepatitis B surface antigen. Mol Immunol. 1989;26:197–205. doi: 10.1016/0161-5890(89)90102-8. [DOI] [PubMed] [Google Scholar]
- 36.Norder H, Hammas B, Lee SD, Bile K, Couroucé AM, Mushahwar IK, et al. Genetic relatedness of hepatitis B viral strains of diverse geographical origin and natural variations in the primary structure of the surface antigen. J Gen Virol. 1993;74:1341–8. doi: 10.1099/0022-1317-74-7-1341. [DOI] [PubMed] [Google Scholar]
- 37.Norder H, Hammas B, Löfdahl S, Couroucé AM, Magnius LO. Comparison of the amino acid sequences of nine different serotypes of hepatitis B surface antigen and genomic classification of the corresponding hepatitis B virus strains. J Gen Virol. 1992;73:1201–8. doi: 10.1099/0022-1317-73-5-1201. [DOI] [PubMed] [Google Scholar]
- 38.Norder H, Couroucé AM, Magnius LO. Molecular basis of hepatitis B virus serotype variations within the four major subtypes. J Gen Virol. 1992;73:3141–5. doi: 10.1099/0022-1317-73-12-3141. [DOI] [PubMed] [Google Scholar]
- 39.Purdy MA, Talekar G, Swenson P, Araujo A, Fields H. A new algorithm for deduction of hepatitis B surface antigen subtype determinants from the amino acid sequence. Intervirology. 2007;50:45–51. doi: 10.1159/000096312. [DOI] [PubMed] [Google Scholar]
- 40.Norder H, Ebert JW, Fields HA, Mushahwar IK, Magnius LO. Complete sequencing of a gibbon hepatitis B virus genome reveals a unique genotype distantly related to the chimpanzee hepatitis B virus. Virology. 1996;218:214–23. doi: 10.1006/viro.1996.0181. [DOI] [PubMed] [Google Scholar]
- 41.Okamoto H, Tsuda F, Sakugawa H, Sastrosoewignjo RI, Imai M, Miyakawa Y, et al. Typing hepatitis B virus by homology in nucleotide sequence: Comparison of surface antigen subtypes. J Gen Virol. 1988;69:2575–83. doi: 10.1099/0022-1317-69-10-2575. [DOI] [PubMed] [Google Scholar]
- 42.Stuyver L, De Gendt S, Van Geyt C, Zoulim F, Fried M, Schinazi RF, et al. A new genotype of hepatitis B virus: Complete genome and phylogenetic relatedness. J Gen Virol. 2000;81:67–74. doi: 10.1099/0022-1317-81-1-67. [DOI] [PubMed] [Google Scholar]
- 43.Araujo A, Yokosawa J, Spelbring J, Tzaneva V, Kamili S, Krawczynski K, et al. Expansion of HBV S gene heterogeneity in chimpanzees after experimental inoculation with HBV sT126N mutant. J Clin Virol. 2006;36:S6. [Google Scholar]
- 44.Norder H, Couroucé AM, Magnius LO. Complete nucleotide sequences of six hepatitis B viral genomes encoding the surface antigen subtypes ayw4, adw4q- and adrq- and their phylogenetic classification. Arch Virol Suppl. 1993;8:189–99. doi: 10.1007/978-3-7091-9312-9_19. [DOI] [PubMed] [Google Scholar]
- 45.Magnius LO, Norder H. Subtypes, genotypes and molecular epidemiology of the hepatitis B virus as reflected by sequence variability of the S-gene. Intervirology. 1995;38:24–34. doi: 10.1159/000150411. [DOI] [PubMed] [Google Scholar]
- 46.Norder H, Couroucé AM, Coursaget P, Echevarria JM, Lee SD, Mushahwar IK, et al. Genetic diversity of hepatitis B virus strains derived worldwide: Genotypes, subgenotypes and HBsAg subtypes. Intervirology. 2004;47:289–309. doi: 10.1159/000080872. [DOI] [PubMed] [Google Scholar]
- 47.Ong HT, Duraisamy G, Kee Peng N, Wen Siang T, Seow HF. Genotyping of hepatitis B virus in Malaysia based on the nucleotide sequence of preS and S genes. Microbes Infect. 2005;7:494–500. doi: 10.1016/j.micinf.2004.12.016. [DOI] [PubMed] [Google Scholar]
- 48.Kidd-Ljunggren K, Myhre E, Bläckberg J. Clinical and serological variation between patients infected with different hepatitis B virus genotypes. J Clin Microbiol. 2004;42:5837–41. doi: 10.1128/JCM.42.12.5837-5841.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sugauchi F, Kumada H, Acharya SA, Shrestha SM, Gamutan MT, Khan M, et al. Epidemiological and sequence differences between two subtypes (Ae and Aa) of hepatitis B virus genotype A. J Gen Virol. 2004;85:811–20. doi: 10.1099/vir.0.79811-0. [DOI] [PubMed] [Google Scholar]
- 50.Kurbanov F, Tanaka Y, Fujiwara K, Sugauchi F, Mbanya D, Zekeng L, et al. A new subtype (subgenotype) Ac (A3) of hepatitis B virus and recombination between genotypes A and E in Cameroon. J Gen Virol. 2005;86:2047–56. doi: 10.1099/vir.0.80922-0. [DOI] [PubMed] [Google Scholar]
- 51.Huy TT, Ushijima H, Quang VX, Win KM, Luengrojanakul P, Kikuchi K, et al. Genotype C of hepatitis B virus can be classified into at least two subgroups. J Gen Virol. 2004;85:283–92. doi: 10.1099/vir.0.19633-0. [DOI] [PubMed] [Google Scholar]
- 52.Nagasaki F, Niitsuma H, Cervantes JG, Chiba M, Hong S, Ojima T, et al. Analysis of the entire nucleotide sequence of hepatitis B virus genotype B in the Philippines reveals a new subgenotype of genotype B. J Gen Virol. 2006;87:1175–80. doi: 10.1099/vir.0.81525-0. [DOI] [PubMed] [Google Scholar]
- 53.Sugauchi F, Mizokami M, Orito E, Ohno T, Kato H, Suzuki S, et al. A novel variant genotype C of hepatitis B virus identified in isolates from Australian Aborigines: Complete genome sequence and phylogenetic relatedness. J Gen Virol. 2001;82:883–92. doi: 10.1099/0022-1317-82-4-883. [DOI] [PubMed] [Google Scholar]
- 54.Sugauchi F, Orito E, Ichida T, Kato H, Sakugawa H, Kakumu S, et al. Hepatitis B virus of genotype B with or without recombination with genotype C over the precore region plus the core gene. J Virol. 2002;76:5985–92. doi: 10.1128/JVI.76.12.5985-5992.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Coursaget P, Yvonnet B, Bourdil C, Mevelec MN, Adamowicz P, Barrès JL, et al. HBsAg positive reactivity in man not due to hepatitis B virus. Lancet. 1987;2:1354–8. doi: 10.1016/s0140-6736(87)91255-4. [DOI] [PubMed] [Google Scholar]
- 56.Thiers V, Nakajima E, Kremsdorf D, Mack D, Schellekens H, Driss F, et al. Transmission of hepatitis B from hepatitis-B-seronegative subjects. Lancet. 1988;2:1273–6. doi: 10.1016/s0140-6736(88)92891-7. [DOI] [PubMed] [Google Scholar]
- 57.Carman WF, Zanetti AR, Karayiannis P, Waters J, Manzillo G, Tanzi E, et al. Vaccine-induced escape mutant of hepatitis B virus. Lancet. 1990;336:325–9. doi: 10.1016/0140-6736(90)91874-a. [DOI] [PubMed] [Google Scholar]
- 58.Carman WF, Korula J, Wallace L, MacPhee R, Mimms L, Decker R. Fulminant reactivation of hepatitis B due to envelope protein mutant that escaped detection by monoclonal HBsAg ELISA. Lancet. 1995;345:1406–7. doi: 10.1016/s0140-6736(95)92599-6. [DOI] [PubMed] [Google Scholar]
- 59.Kreutz C. Molecular, immunological and clinical properties of mutated hepatitis B viruses. J Cell Mol Med. 2002;6:113–43. doi: 10.1111/j.1582-4934.2002.tb00317.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Harrison TJ, Hopes EA, Oon CJ, Zanetti AR, Zuckerman AJ. Independent emergence of a vaccine-induced escape mutant of hepatitis-B Virus. J Hepatol. 1991;13:S105–7. doi: 10.1016/0168-8278(91)90037-c. [DOI] [PubMed] [Google Scholar]
- 61.Waters JA, Kennedy M, Voet P, Hauser P, Petre J, Carman W, et al. Loss of the common ‘a’ determinant of hepatitis B surface antigen by a vaccine induced escape mutant. J Clin Invest. 1992;90:2543–7. doi: 10.1172/JCI116148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Karthigesu VD, Allison LM, Fortuin M, Mendy M, Whittle HC, Howard CR. A novel hepatitis B virus variant in the sera of immunized children. J Gen Virol. 1994;75:443–8. doi: 10.1099/0022-1317-75-2-443. [DOI] [PubMed] [Google Scholar]
- 63.Weinberger KM, Bauer T, Böhm S, Jilg W. High genetic variability of the group-specific a-determinant of hepatitis B virus surface antigen (HBsAg) and the corresponding fragment of the viral polymerase in chronic virus carriers lacking detectable HBsAg in serum. J Gen Virol. 2000;81:1165–74. doi: 10.1099/0022-1317-81-5-1165. [DOI] [PubMed] [Google Scholar]
- 64.Shields PL, Owsianka A, Carman WF, Boxall E, Hubscher SG, Shaw J, et al. Selection of hepatitis B surface “escape” mutants during passive immune prophylaxis following liver transplantation: Potential impact of genetic changes on polymerase protein function. Gut. 1999;45:306–8. doi: 10.1136/gut.45.2.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jongerius JM, Wester M, Cuypers HT, van Oostendorp WR, Lelie PN, van der Poel CL, et al. New hepatitis B virus mutant form in a blood donor that is undetectable in several hepatitis B surface antigen screening assays. Transfusion. 1998;38:56–9. doi: 10.1046/j.1537-2995.1998.38198141499.x. [DOI] [PubMed] [Google Scholar]
- 66.Levicnik-Stezinar S. Hepatitis B surface antigen escape mutant in a first time blood donor potentially missed by a routine screening assay. Clin Lab. 2004;50:49–51. [PubMed] [Google Scholar]
- 67.Theamboonlers A, Tangkijvanich P, Pramoolsinsap C, Poovorawan Y. Genotypes and subtypes of hepatitis B virus in Thailand. Southeast Asian J Trop Med Public Health. 1998;29:786–91. [PubMed] [Google Scholar]
- 68.Locarnini S. Molecular virology of hepatitis B virus. Semin Liver Dis. 2004;24:3–10. doi: 10.1055/s-2004-828672. [DOI] [PubMed] [Google Scholar]
- 69.Mathet VL, Feld M, Espínola L, Sánchez DO, Ruiz V, Mandó O, et al. Hepatitis B virus S gene mutants in a patient with chronic active hepatitis with circulating Anti-HBs antibodies. J Med Virol. 2003;69:18–26. doi: 10.1002/jmv.10267. [DOI] [PubMed] [Google Scholar]
- 70.Thakur V, Kazim SN, Guptan RC, Hasnain SE, Bartholomeusz A, Malhotra V, et al. Transmission of G145R mutant of HBV to an unrelated contact. J Med Virol. 2005;76:40–6. doi: 10.1002/jmv.20321. [DOI] [PubMed] [Google Scholar]
- 71.Thuy le TT, Ryo H, Van Phung L, Furitsu K, Nomura T. Distribution of genotype/subtype and mutational spectra of the surface gene of hepatitis B virus circulating in Hanoi, Vietnam. J Med Virol. 2005;76:161–9. doi: 10.1002/jmv.20337. [DOI] [PubMed] [Google Scholar]
- 72.Ohishi W, Shirakawa H, Kawakami Y, Kimura S, Kamiyasu M, Tazuma S, et al. Identification of rare polymerase variants of hepatitis B virus using a two-stage PCR with peptide nucleic acid clamping. J Med Virol. 2004;72:558–65. doi: 10.1002/jmv.20026. [DOI] [PubMed] [Google Scholar]
- 73.Wakil SM, Kazim SN, Khan LA, Raisuddin S, Parvez MK, Guptan RC, et al. Prevalence and profile of mutations associated with lamivudine therapy in Indian patients with chronic hepatitis B in the surface and polymerase genes of hepatitis B virus. J Med Virol. 2002;68:311–8. doi: 10.1002/jmv.10205. [DOI] [PubMed] [Google Scholar]
- 74.Torresi J. The virological and clinical significance of mutations in the overlapping envelope and polymerase genes of hepatitis B virus. J Clin Virol. 2002;25:97–106. doi: 10.1016/s1386-6532(02)00049-5. [DOI] [PubMed] [Google Scholar]
- 75.von Weizsäcker F, Pult I, Geiss K, Wirth S, Blum HE. Selective transmission of variant genomes from mother to infant in neonatal fulminant hepatitis B. Hepatology. 1995;21:8–13. doi: 10.1002/hep.1840210103. [DOI] [PubMed] [Google Scholar]
- 76.Liu CJ, Kao JH, Shau WY, Chen PJ, Lai MY, Chen DS. Naturally occurring hepatitis B surface gene variants in chronic hepatitis B virus infection: Correlation with viral serotypes and clinical stages of liver disease. J Med Virol. 2002;68:50–9. doi: 10.1002/jmv.10169. [DOI] [PubMed] [Google Scholar]
- 77.Hsu HY, Chang MH, Ni YH, Chen HL. Survey of hepatitis B surface variant infection in children 15 years after a nationwide vaccination programme in Taiwan. Gut. 2004;53:1499–503. doi: 10.1136/gut.2003.034223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Yamamoto K, Horikita M, Tsuda F, Itoh K, Akahane Y, Yotsumoto S, et al. Naturally occurring escape mutants of hepatitis B virus with various mutations in the S gene in carriers seropositive for antibody to hepatitis B surface antigen. J Virol. 1994;68:2671–6. doi: 10.1128/jvi.68.4.2671-2676.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Chong-Jin O, Wei Ning C, Shiuan K, Gek Keow L. Identification of hepatitis B surface antigen variants with alterations outside the “a” determinant in immunized Singapore infants. J Infect Dis. 1999;179:259–63. doi: 10.1086/314553. [DOI] [PubMed] [Google Scholar]
- 80.Oon CJ, Chen WN. Current aspects of hepatitis B surface antigen mutants in Singapore. J Viral Hepat. 1998;5:17–23. doi: 10.1046/j.1365-2893.1998.0050s2017.x. [DOI] [PubMed] [Google Scholar]
- 81.Thakur V, Kazim SN, Guptan RC, Malhotra V, Sarin SK. Molecular epidemiology and transmission of hepatitis B virus in close family contacts of HBV-related chronic liver disease patients. J Med Virol. 2003;70:520–8. doi: 10.1002/jmv.10426. [DOI] [PubMed] [Google Scholar]
- 82.Hsu HY, Chang MH, Liaw SH, Ni YH, Chen HL. Changes of hepatitis B surface antigen variants in carrier children before and after universal vaccination in Taiwan. Hepatology. 1999;30:1312–7. doi: 10.1002/hep.510300511. [DOI] [PubMed] [Google Scholar]
- 83.Zhang YY, Nordenfelt E, Hansson BG. Increasing heterogeneity of the ‘a’ determinant of HBsAg found in the presumed late phase of chronic hepatitis B virus infection. Scand J Infect Dis. 1996;28:9–15. doi: 10.3109/00365549609027142. [DOI] [PubMed] [Google Scholar]
- 84.Rodriguez-Frias F, Buti M, Jardi R, Vargas V, Quer J, Cotrina M, et al. Genetic alterations in the S gene of hepatitis B virus in patients with acute hepatitis B, chronic hepatitis B and hepatitis B liver cirrhosis before and after liver transplantation. Liver. 1999;19:177–82. doi: 10.1111/j.1478-3231.1999.tb00032.x. [DOI] [PubMed] [Google Scholar]
- 85.Roznovsky L, Harrison TJ, Fang ZL, Ling R, Lochman I, Orsagova I, et al. Unusual hepatitis B surface antigen variation in a child immunised against hepatitis B. J Med Virol. 2000;61:11–4. doi: 10.1002/(sici)1096-9071(200005)61:1<11::aid-jmv2>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 86.Seddigh-Tonekaboni S, Lim WL, Young B, Hou JL, Waters J, Luo KX, et al. Hepatitis B surface antigen variants in vaccinees, blood donors and an interferon-treated patient. J Viral Hepat. 2001;8:154–8. doi: 10.1046/j.1365-2893.2001.00275.x. [DOI] [PubMed] [Google Scholar]
- 87.Maillard P, Pillot J. At least three epitopes are recognized by the human repertoire in the hepatitis B virus group a antigen inducing protection; possible consequences for seroprevention and serodiagnosis. Res Virol. 1998;149:153–61. doi: 10.1016/s0923-2516(98)80033-2. [DOI] [PubMed] [Google Scholar]
- 88.Hou J, Wang Z, Cheng J, Lin Y, Lau GK, Sun J, et al. Prevalence of naturally occurring surface gene variants of hepatitis B virus in nonimmunized surface antigen-negative Chinese carriers. Hepatology. 2001;34:1027–34. doi: 10.1053/jhep.2001.28708. [DOI] [PubMed] [Google Scholar]
- 89.Kohno H, Inoue T, Tsuda F, Okamoto H, Akahane Y. Mutations in the envelope gene of hepatitis B virus variants co-occurring with antibody to surface antigen in sera from patients with chronic hepatitis B. J Gen Virol. 1996;77:1825–31. doi: 10.1099/0022-1317-77-8-1825. [DOI] [PubMed] [Google Scholar]
- 90.Koyanagi T, Nakamuta M, Sakai H, Sugimoto R, Enjoji M, Koto K, et al. Analysis of HBs antigen negative variant of hepatitis B virus: Unique substitutions, Glu129 to Asp and Gly145 to Ala in the surface antigen gene. Med Sci Monit. 2000;6:1165–9. [PubMed] [Google Scholar]
- 91.Chiou HL, Lee TS, Kuo J, Mau YC, Ho MS. Altered antigenicity of ‘a’ determinant variants of hepatitis B virus. J Gen Virol. 1997;78:2639–45. doi: 10.1099/0022-1317-78-10-2639. [DOI] [PubMed] [Google Scholar]
- 92.Avellón A, Echevarria JM. Frequency of hepatitis B virus ‘a’ determinant variants in unselected Spanish chronic carriers. J Med Virol. 2006;78:24–36. doi: 10.1002/jmv.20516. [DOI] [PubMed] [Google Scholar]
- 93.Kazim SN, Sarin SK, Sharma BC, Khan LA, Hasnain SE. Characterization of naturally occurring and Lamivudine-induced surface gene mutants of hepatitis B virus in patients with chronic hepatitis B in India. Intervirology. 2006;49:152–60. doi: 10.1159/000089376. [DOI] [PubMed] [Google Scholar]
- 94.Kfoury Baz EM, Zheng J, Mazuruk K, Van Le A, Peterson DL. Characterization of a novel hepatitis B virus mutant: Demonstration of mutation-induced hepatitis B virus surface antigen group specific “a” determinant conformation change and its application in diagnostic assays. Transfus Med. 2001;11:355–62. doi: 10.1046/j.1365-3148.2001.00323.x. [DOI] [PubMed] [Google Scholar]
- 95.Miyake Y, Oda T, Li R, Sugiyama K. A comparison of amino acid sequences of hepatitis B virus S gene in 46 children presenting various clinical features for immunoprophylaxis. Tohoku J Exp Med. 1996;180:233–47. doi: 10.1620/tjem.180.233. [DOI] [PubMed] [Google Scholar]
- 96.Paulij WP, de Wit PL, Sünnen CM, van Roosmalen MH, Petersen-van Ettekoven A, Cooreman MP, et al. Localization of a unique hepatitis B virus epitope sheds new light on the structure of hepatitis B virus surface antigen. J Gen Virol. 1999;80:2121–6. doi: 10.1099/0022-1317-80-8-2121. [DOI] [PubMed] [Google Scholar]
- 97.Torresi J, Earnest-Silveira L, Civitico G, Walters TE, Lewin SR, Fyfe J, et al. Restoration of replication phenotype of lamivudine-resistant hepatitis B virus mutants by compensatory changes in the ‘fingers’ subdomain of the viral polymerase selected as a consequence of mutations in the overlapping S gene. Virology. 2002;299:88–99. doi: 10.1006/viro.2002.1448. [DOI] [PubMed] [Google Scholar]
- 98.Torresi J, Earnest-Silveira L, Deliyannis G, Edgtton K, Zhuang H, Locarnini SA, et al. Reduced antigenicity of the hepatitis B virus HBsAg protein arising as a consequence of sequence changes in the overlapping polymerase gene that are selected by lamivudine therapy. Virology. 2002;293:305–13. doi: 10.1006/viro.2001.1246. [DOI] [PubMed] [Google Scholar]
- 99.Bock CT, Tillmann HL, Torresi J, Klempnauer J, Locarnini S, Manns MP, et al. Selection of hepatitis B virus polymerase mutants with enhanced replication by lamivudine treatment after liver transplantation. Gastroenterology. 2002;122:264–73. doi: 10.1053/gast.2002.31015. [DOI] [PubMed] [Google Scholar]
- 100.Lo CM, Liu CL, Chan SC, Lau GK, Fan ST. Failure of hepatitis B vaccination in patients receiving lamivudine prophylaxis after liver transplantation for chronic hepatitis B. J Hepatol. 2005;43:283–7. doi: 10.1016/j.jhep.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 101.Neurath AR, Pride MW, Strick N, Thanavala YM. Toleration of amino acid substitutions within hepatitis B virus envelope protein epitopes established by peptide replacement set analysis. I Region S(139-147) Pept Res. 1990;3:116–22. [PubMed] [Google Scholar]
- 102.Pride MW, Thanavala YM, Strick N, Houghten RA, Neurath AR. Toleration of amino acid substitutions within hepatitis B virus envelope protein epitopes established by peptide replacement set analysis. II Region S(122-136) Pept Res. 1992;5:217–26. [PubMed] [Google Scholar]
- 103.Stirk HJ, Thornton JM, Howard CR. A topological model for hepatitis B surface antigen. Invetvirology. 1992;33:148–58. doi: 10.1159/000150244. [DOI] [PubMed] [Google Scholar]
- 104.Srey CT, Ijaz S, Tedder RS, Monchy D. Characterization of hepatitis B surface antigen strains circulating in the Kingdom of Cambodia. J Viral Hepat. 2006;13:62–6. doi: 10.1111/j.1365-2893.2005.00656.x. [DOI] [PubMed] [Google Scholar]
- 105.Chen WN, Oon CJ. Mutation “hot spot” in HLA class I-restricted T cell epitope on hepatitis B surface antigen in chronic carriers and hepatocellular carcinoma. Biochem Biophys Res Commun. 1999;262:757–61. doi: 10.1006/bbrc.1999.1267. [DOI] [PubMed] [Google Scholar]
- 106.Hsu HY, Chang MH, Ni YH, Lin HH, Wang SM, Chen DS. Surface gene mutants of hepatitis B virus in infants who develop acute chronic infections despite immunoprophylaxis. Hepatology. 1997;26:786–91. doi: 10.1002/hep.510260336. [DOI] [PubMed] [Google Scholar]
- 107.Hu H, Peng XM, Huang YS, Gu L, Xie QF, Gao ZL. Yeast expression and DNA immunization of hepatitis B virus S gene with second-loop deletion of alpha determinant region. World J Gastroenterol. 2004;10:2989–93. doi: 10.3748/wjg.v10.i20.2989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Ijaz S, Ferns RB, Tedder RS. A ‘first loop’ linear epitope accessible on native hepatitis B surface antigen that persists in the face of ‘second loop’ immune escape. J Gen Virol. 2003;84:269–75. doi: 10.1099/vir.0.18667-0. [DOI] [PubMed] [Google Scholar]
- 109.Gilbert RJ, Beales L, Blond D, Simon MN, Lin BY, Chisari FV, et al. Hepatitis B small surface antigen particles are octahedral. Proc Natl Acad Sci USA. 2005;102:14783–8. doi: 10.1073/pnas.0505062102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Ijaz S, Torre F, Tedder RS, Williams R, Naoumov NV. Novel immunoassay for the detection of hepatitis B surface ‘escape’ mutants and its application in liver transplant recipients. J Med Virol. 2001;63:210–6. doi: 10.1002/1096-9071(200103)63:3<210::aid-jmv1002>3.0.co;2-c. [DOI] [PubMed] [Google Scholar]
- 111.Alexopoulou A, Baltayiannis G, Jammeh S, Waters J, Dourakis SP, Karayiannis P. Hepatitis B surface antigen variant with multiple mutations in the a determinant in an agammaglobulinemic patient. J Clin Microbiol. 2004;42:2861–5. doi: 10.1128/JCM.42.6.2861-2865.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Allain JP. Occult hepatitis B virus infection: Implications in transfusion. Vox Sang. 2004;86:83–91. doi: 10.1111/j.0042-9007.2004.00406.x. [DOI] [PubMed] [Google Scholar]
- 113.Bahn A, Gerner P, Martiné U, Bortolotti F, Wirth S. Detection of different viral strains of hepatitis B virus in chronically infected children after seroconversion from HBsAg to anti-HBs indicating viral persistence. J Hepatol. 1997;27:973–8. doi: 10.1016/s0168-8278(97)80139-0. [DOI] [PubMed] [Google Scholar]
- 114.Banerjee K, Guptan RC, Bisht R, Sarin SK, Khandekar P. Identification of a novel surface mutant of hepatitis B virus in a seronegative chronic liver disease patient. Virus Res. 1999;65:103–9. doi: 10.1016/s0168-1702(99)00106-9. [DOI] [PubMed] [Google Scholar]
- 115.Basuni AA, Butterworth L, Cooksley G, Locarnini S, Carman WF. Prevalence of HBsAg mutants and impact of hepatitis B infant immunization in four Pacific Island countries. Vaccine. 2004;22:2791–9. doi: 10.1016/j.vaccine.2004.01.046. [DOI] [PubMed] [Google Scholar]
- 116.Carman WF, Trautwein C, van Deursen FJ, Colman K, Dornan E, McIntyre G, et al. Hepatitis B virus envelope variation after transplantation with and without hepatitis B immune globulin prophylaxis. Hepatology. 1996;24:489–93. doi: 10.1002/hep.510240304. [DOI] [PubMed] [Google Scholar]
- 117.Chan HL, Tsang SW, Leung NW, Tse CH, Hui Y, Tam JS, et al. Occult HBV infection in cryptogenic liver cirrhosis in an area with high prevalence of HBV infection. Am J Gastroenterol. 2002;97:1211–5. doi: 10.1111/j.1572-0241.2002.05706.x. [DOI] [PubMed] [Google Scholar]
- 118.Chen HB, Fang DX, Li FQ, Jing HY, Tan WG, Li SQ. A novel hepatitis B virus mutant with A-to-G at nt551 in the surface antigen gene. World J Gastroenterol. 2003;9:304–8. doi: 10.3748/wjg.v9.i2.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Chen WN, Oon CJ, Moh MC. Detection of hepatitis B virus surface antigen mutants in paraffin-embedded hepatocellular carcinoma tissues. Virus Genes. 2000;20:263–7. doi: 10.1023/a:1008100930584. [DOI] [PubMed] [Google Scholar]
- 120.Cooreman MP, Leroux-Roels G, Paulij WP. Vaccine- and hepatitis B immune globulin-induced escape mutations of hepatitis B virus surface antigen. J Biomed Sci. 2001;8:237–247. doi: 10.1007/BF02256597. [DOI] [PubMed] [Google Scholar]
- 121.Fang DX, Gan RB, Li ZP, Duan SC. A Hepatitis B Virus Variant with an Ile to Ser Mutation at aa126 of HBsAg. Sheng Wu Hua Xue Yu Sheng Wu Wu Li Xue Bao (Shanghai) 1996;28:429–33. [PubMed] [Google Scholar]
- 122.Ghany MG, Ayola B, Villamil FG, Gish RG, Rojter S, Vierling JM, et al. Hepatitis B virus S mutants in liver transplant recipients who were reinfected despite hepatitis B immune globulin prophylaxis. Hepatology. 1998;27:213–22. doi: 10.1002/hep.510270133. [DOI] [PubMed] [Google Scholar]
- 123.Hawkins AE, Gilson RJ, Gilbert N, Wreghitt TG, Gray JJ, Ahlers-de Boer I, et al. Hepatitis B virus surface mutations associated with infection after liver transplantation. J Hepatol. 1996;24:8–14. doi: 10.1016/s0168-8278(96)80179-6. [DOI] [PubMed] [Google Scholar]
- 124.He JW, Lu Q, Zhu QR, Duan SC, Wen YM. Mutations in the ‘a’ determinant of hepatitis B surface antigen among Chinese infants receiving active postexposure hepatitis B immunization. Vaccine. 1998;16:170–3. doi: 10.1016/s0264-410x(97)00182-5. [DOI] [PubMed] [Google Scholar]
- 125.Hino K, Okuda M, Hashimoto O, Ishiko H, Okazaki M, Fujii K, et al. Glycine-to-arginine substitution at codon 145 of HBsAg in two infants born to hepatitis B e antigen-positive carrier. Dig Dis Sci. 1995;40:566–70. doi: 10.1007/BF02064370. [DOI] [PubMed] [Google Scholar]
- 126.Hou J, Karayiannis P, Waters J, Luo K, Liang C, Thomas HC. A unique insertion in the S gene of surface antigen-negative hepatitis B virus Chinese carriers. Hepatology. 1995;21:273–8. doi: 10.1016/0270-9139(95)90080-2. [DOI] [PubMed] [Google Scholar]
- 127.Huang X, Lu D, Ji G, Sun Y, Ma L, Chen Z, et al. Hepatitis B virus (HBV) vaccine-induced escape mutants of HBV S gene among children from Qidong area, China. Virus Res. 2004;99:63–8. doi: 10.1016/j.virusres.2003.10.007. [DOI] [PubMed] [Google Scholar]
- 128.Karthigesu VD, Allison LM, Ferguson M, Howard CR. A hepatitis B virus variant found in the sera of immunised children induces a conformational change in the HBsAg “a” determinant. J Med Virol. 1999;58:346–52. doi: 10.1002/(sici)1096-9071(199908)58:4<346::aid-jmv5>3.0.co;2-7. [DOI] [PubMed] [Google Scholar]
- 129.Kidd-Ljunggren K, Ekdahl K, Oberg M, Kurathong S, Lolekha S. Hepatitis B virus strains in Thailand: Genomic variants in chronic carriers. J Med Virol. 1995;47:454–61. doi: 10.1002/jmv.1890470427. [DOI] [PubMed] [Google Scholar]
- 130.Kim KH, Lee KH, Chang HY, Ahn SH, Tong S, Yoon YJ, et al. Evolution of hepatitis B virus sequence from a liver transplant recipient with rapid breakthrough despite hepatitis B immune globulin prophylaxis and lamivudine therapy. J Med Virol. 2003;71:367–75. doi: 10.1002/jmv.10503. [DOI] [PubMed] [Google Scholar]
- 131.Komatsu H, Fujisawa T, Sogo T, Isozaki A, Inui A, Sekine I, et al. Acute self-limiting hepatitis B after immunoprophylaxis failure in an infant. J Med Virol. 2002;66:28–33. doi: 10.1002/jmv.2107. [DOI] [PubMed] [Google Scholar]
- 132.Lada O, Benhamou Y, Poynard T, Thibault V. Coexistence of hepatitis B surface antigen (HBs Ag) and anti-HBs antibodies in chronic hepatitis B virus carriers: influence of “a” determinant variants. J Virol. 2006;80:2968–75. doi: 10.1128/JVI.80.6.2968-2975.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Lee KM, Kim YS, Ko YY, Yoo BM, Lee KJ, Kim JH, et al. Emergence of vaccine-induced escape mutant of hepatitis B virus with multiple surface gene mutations in a Korean child. J Korean Med Sci. 2001;16:359–62. doi: 10.3346/jkms.2001.16.3.359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Lee PI, Chang LY, Lee CY, Huang LM, Chang MH. Detection of hepatitis B surface gene mutation in carrier children with or without immunoprophylaxis at birth. J Infect Dis. 1997;176:427–30. doi: 10.1086/514060. [DOI] [PubMed] [Google Scholar]
- 135.Lee SY, Choi MS, Lee D, Lee JH, Koh KC, Paik SW, et al. Overlapping gene mutations of hepatitis B virus in a chronic hepatitis B patient with hepatitis B surface antigen loss during lamivudine therapy. J Korean Med Sci. 2005;20:433–7. doi: 10.3346/jkms.2005.20.3.433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Louisirirotchanakul S, Kanoksinsombat C, Theamboonlert A, Puthavatana P, Wasi C, Poovorawan Y. Mutation of the “a” determinant of HBsAg with discordant HBsAg diagnostic kits. Viral Immunol. 2004;17:440–4. doi: 10.1089/vim.2004.17.440. [DOI] [PubMed] [Google Scholar]
- 137.Matsumoto T, Nakata K, Hamasaki K, Daikokoku M, Nakao K, Yamashita Y, et al. Efficacy of immunization of high-risk infants against hepatitis B virus evaluated by polymerase chain reaction. J Med Virol. 1997;53:255–60. doi: 10.1002/(sici)1096-9071(199711)53:3<255::aid-jmv13>3.0.co;2-g. [DOI] [PubMed] [Google Scholar]
- 138.Mele A, Tancredi F, Romanò L, Giuseppone A, Colucci M, Sangiuolo A, et al. Effectiveness of hepatitis B vaccination in babies born to hepatitis B surface antigen-positive mothers in Italy. J Infect Dis. 2001;184:905–8. doi: 10.1086/323396. [DOI] [PubMed] [Google Scholar]
- 139.Ngui SL, O’Connell S, Eglin RP, Heptonstall J, Teo CG. Low detection rate and maternal provenance of hepatitis B virus S gene mutants in cases of failed postnatal immunoprophylaxis in England and Wales. J Infect Dis. 1997;176:1360–5. doi: 10.1086/514133. [DOI] [PubMed] [Google Scholar]
- 140.Ogura Y, Kurosaki M, Asahina Y, Enomoto N, Marumo F, Sato C. Prevalence and significance of naturally occurring mutations in the surface and polymerase genes of hepatitis B virus. J Infect Dis. 1999;180:1444–51. doi: 10.1086/315094. [DOI] [PubMed] [Google Scholar]
- 141.Oon CJ, Lim GK, Ye Z, Goh KT, Tan KL, Yo SL, et al. Molecular epidemiology of hepatitis B virus vaccine variants in Singapore. Vaccine. 1995;13:699–702. doi: 10.1016/0264-410x(94)00080-7. [DOI] [PubMed] [Google Scholar]
- 142.Oon CJ, Chen WN, Goh KT, Mesenas S, Ng HS, Chiang G, et al. Molecular characterization of hepatitis B virus surface antigen mutants in Singapore patients with hepatocellular carcinoma and hepatitis B virus carriers negative for HBsAg but positive for anti-HBs and anti-HBc. J Gastroenterol Hepatol. 2002;17:S491–6. doi: 10.1046/j.1440-1746.17.s4.16.x. [DOI] [PubMed] [Google Scholar]
- 143.Poovorawan Y, Theamboonlers A, Chongsrisawat V, Sanpavat S. Molecular analysis of the a determinant of HBsAg in children of HBeAg-positive mothers upon failure of postexposure prophylaxis. Int J Infect Dis. 1998;2:216–20. doi: 10.1016/s1201-9712(98)90056-x. [DOI] [PubMed] [Google Scholar]
- 144.Schories M, Peters T, Rasenack J. Isolation, characterization and biological significance of hepatitis B virus mutants from serum of a patient with immunologically negative HBV infection. J Hepatol. 2000;33:799–811. doi: 10.1016/s0168-8278(00)80313-x. [DOI] [PubMed] [Google Scholar]
- 145.Singh H, Aggarwal R, Singh RL, Naik SR, Naik S. Frequency of infection by hepatitis B virus and its surface mutants in a northern Indian population. Indian J Gastroenterol. 2003;22:132–7. [PubMed] [Google Scholar]
- 146.Song BC, Kim SH, Kim H, Ying YH, Kim HJ, Kim YJ, et al. Prevalence of naturally occurring surface antigen variants of hepatitis B virus in Korean patients infected chronically. J Med Virol. 2005;76:194–202. doi: 10.1002/jmv.20354. [DOI] [PubMed] [Google Scholar]
- 147.Tai PC, Banik D, Lin GI, Pai S, Pai K, Lin MH, et al. Novel and frequent mutations of hepatitis B virus coincide with a major histocompatibility complex class I-restricted T-cell epitope of the surface antigen. J Virol. 1997;71:4852–6. doi: 10.1128/jvi.71.6.4852-4856.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Theamboonlers A, Chongsrisawat V, Jantaradsamee P, Poovorawan Y. Variants within the «a» determinant of HBs gene in children and adolescents with and without hepatitis B vaccination as part of Thailand's Expanded Program on Immunization (EPI) Tohoku J Exp Med. 2001;193:197–205. doi: 10.1620/tjem.193.197. [DOI] [PubMed] [Google Scholar]
- 149.Wallace LA, Echevarria JE, Echevarria JM, Carman WF. Molecular characterization of envelope antigenic variants of hepatitis B virus from Spain. J Infect Dis. 1994;170:1300–3. doi: 10.1093/infdis/170.5.1300. [DOI] [PubMed] [Google Scholar]
- 150.Wang YM, Ng WC, Lo SK. Detection of pre-S/S gene mutants in chronic hepatitis B carriers with concurrent hepatitis B surface antibody and hepatitis B surface antigen. J Gastroenterol. 1999;34:600–6. doi: 10.1007/s005350050379. [DOI] [PubMed] [Google Scholar]
- 151.Wang Z, Zhang J, Yang H, Li X, Wen S, Guo Y, et al. Quantitative analysis of HBV DNA level and HBeAg titer in hepatitis B surface antigen positive mothers and their babies: HBeAg passage through the placenta and the rate of decay in babies. J Med Virol. 2003;71:360–6. doi: 10.1002/jmv.10493. [DOI] [PubMed] [Google Scholar]
- 152.Wu L, He JW, Yao X, Li HM, Wen YM. A novel hepatitis B virus variant S 129 (Gln->Leu): Lack of correlation between antigenicity and immunogenicity. J Med Virol. 1999;59:424–30. [PubMed] [Google Scholar]
- 153.Yoshida EM, Ramji A, Erb SR, Davis JE, Steinbrecher UP, Sherlock CH, et al. De novo acute hepatitis B infection in a previously vaccinated liver transplant recipient due to a strain of HBV with a Met 133 Thr mutation in the “a” determinant. Liver. 2000;20:411–4. doi: 10.1034/j.1600-0676.2000.020005411.x. [DOI] [PubMed] [Google Scholar]


