Abstract
Calcineurin is a conserved Ca2+/calmodulin-dependent protein phosphatase that plays a critical role in Ca2+ signaling. We describe new components of a calcineurin-mediated response in yeast, the Ca2+-induced transcriptional activation of FKS2, which encodes a β-1,3 glucan synthase. A 24-bp region of the FKS2 promoter was defined as sufficient to confer calcineurin-dependent transcriptional induction on a minimal promoter in response to Ca2+ and was named CDRE (for calcineurin-dependent response element). The product of CRZ1 (YNL027w) was identified as an activator of CDRE-driven transcription. Crz1p contains zinc finger motifs and binds specifically to the CDRE. Genetic analysis revealed that crz1Δ mutant cells exhibit several phenotypes similar to those of calcineurin mutants and that overexpression of CRZ1 in calcineurin mutants suppressed these phenotypes. These results suggest that Crz1p functions downstream of calcineurin to effect multiple calcineurin-dependent responses. Moreover, the calcineurin-dependent transcriptional induction of FKS2 in response to Ca2+, α-factor, and Na+ was found to require CRZ1. In addition, we found that the calcineurin-dependent transcriptional regulation of PMR2 and PMC1 required CRZ1. However, transcription of PMR2 and PMC1 was activated by only a subset of the treatments that activated FKS2 transcription. Thus, in response to multiple signals, calcineurin acts through the Crz1p transcription factor to differentially regulate the expression of several target genes in yeast.
Keywords: S. cerevisiae, calcineurin, calcium signaling, transcriptional activation, cell wall maintenance, ion homeostasis
Changes in intracellular Ca2+ concentration signal a variety of physiological responses in many different cell types (Clapham 1995). The amplitude and duration of dynamic Ca2+ signals contribute to the diversity in signaling of this single ion. One mechanism by which Ca2+ acts is by binding to and activating calmodulin. As an intracellular Ca2+ receptor, calmodulin activates a number of target enzymes such as calmodulin-dependent protein kinases and phosphatases. One of these targets is the serine/threonine-specific protein phosphatase calcineurin that acts as an effector of Ca2+ signaling by regulating the phosphorylation state of proteins (Klee et al. 1988).
Calcineurin activity is critical for many Ca2+-regulated processes, including T-cell activation (Clipstone and Crabtree 1992; O’Keefe et al. 1992) and neutrophil chemotaxis (Hendley et al. 1992; Lawson and Maxfield 1995). The function of calcineurin in different cell types has been assessed in part by examining the effects of FK506 and cyclosporin A, immunosuppressive drugs that specifically inhibit this phosphatase (Liu et al. 1991a). Inhibition of calcineurin by these drugs prevents activation of NFAT, a transcription factor that is necessary for the proliferation of T cells (Clipstone and Crabtree 1992; O’Keefe et al. 1992). Specifically, dephosphorylation of NFAT by calcineurin allows translocation of this transcription factor from the cytoplasm to the nucleus where it induces expression of a number of cytokine genes (Jain et al. 1993; Northrop et al. 1993). In other cell types, calcineurin has been implicated in the control of ion homeostasis. For example, calcineurin regulates the Na+/K+ ATPase in renal tubule cells (Aperia et al. 1992) and the NMDA receptor in neurons (Lieberman and Mody 1994; Tong et al. 1995).
The calcineurin enzyme functions as a heterodimer of catalytic (A) and regulatory (B) subunits that have been highly conserved through evolution. The catalytic subunit contains a carboxy-terminal autoinhibitory domain, and Ca2+–calmodulin binding activates the enzyme by relieving this inhibition (Hubbard and Klee 1989). Truncations of the catalytic subunit that remove the autoinhibitory domain result in a constitutively active enzyme that no longer requires Ca2+ (Hubbard and Klee 1989). In the yeast Saccharomyces cerevisiae, calcineurin catalytic subunits are encoded by CNA1 and CNA2 (Cyert et al. 1991; Liu et al. 1991b), and the regulatory subunit is encoded by CNB1 (Kuno et al. 1991; Cyert and Thorner 1992). The physiological role of calcineurin in yeast has been examined by characterizing cells that lack functional calcineurin, that is, cna1 cna2 mutants, cnb1 mutants, or cells incubated with FK506 or cyclosporin A.
Yeast calcineurin is essential under specific environmental conditions. During prolonged incubation with pheromone, calcineurin is necessary to maintain viability; however, the nature of this requirement is not well understood (Moser et al. 1996; Withee et al. 1997). Calcineurin-deficient cells also grow poorly in the presence of high concentrations of certain ions, including Mn2+, Na+/Li+, and OH− (Nakamura et al. 1993; Mendoza et al. 1994; Farcasanu et al. 1995; Pozos et al. 1996). These ion sensitivities can be explained, at least in part, by altered levels of several ion transporters. Calcineurin is required for transcriptional induction of PMR2, which encodes a Na+ ATPase (Rudolph et al. 1989; Haro et al. 1991), and PMC1 and PMR1, which encode Ca2+ ATPases (Rudolph et al. 1989; Cunningham and Fink 1994, 1996; Mendoza et al. 1994).
Calcineurin also regulates transcription of another gene, FKS2. FKS2 and its homolog, FKS1, encode catalytic subunits of a major, cell wall synthetic enzyme, β-1,3 glucan synthase. fks1 fks2 double mutants are inviable, and cells lacking FKS1 and calcineurin are also inviable because of insufficient expression of FKS2 (Douglas et al. 1994; Eng et al. 1994; Garrett-Engele et al. 1995). In addition, FKS2 mRNA levels increase when cells are incubated with either Ca2+ or mating pheromone, and under both of these conditions, transcriptional activation is completely dependent on calcineurin (Mazur et al. 1995). However, transcriptional regulation of FKS2 is complex and is also regulated by calcineurin-independent mechanisms (Mazur et al. 1995). For example, during growth at elevated temperatures, FKS2 transcription increases, and this induction is the result of the independent and additive effects of both calcineurin and the PKC1-regulated cell integrity pathway (C. Zhao, U. Jung, P. Garrett-Engele, T. Roe, M. Cyert, and D. Levin, in prep.). Furthermore, the expression of FKS2 is induced, independently of calcineurin, by growth on nondextrose carbon sources and at stationary phase (Mazur et al. 1995; C. Zhao, U. Jung, P. Garrett-Engele, T. Roe, M. Cyert, and D. Levin, in prep.). In this study we further characterize the mechanism by which calcineurin activates transcription of FKS2 and identify a transcription factor, Crz1p, that mediates calcineurin-dependent changes in gene expression.
Results
Dissection of the FKS2 promoter identifies a region that is necessary for calcineurin-dependent transcriptional induction
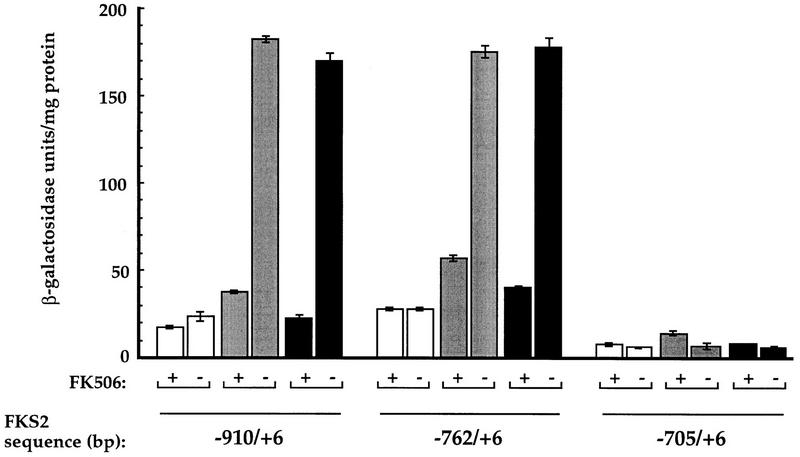
FKS2 mRNA levels increase in response to Ca2+ and mating pheromone (α-factor) in a calcineurin-dependent fashion (Mazur et al. 1995). We previously determined that a lacZ reporter gene containing ∼900 bp of FKS2 promoter sequence displays calcineurin-dependent Ca2+-induced expression, whereas a construct containing ∼700 bp does not (C. Zhao, U. Jung, P. Garrett-Engele, T. Roe, M. Cyert, and D. Levin, in prep.). Here, we have extended that analysis and found that gene fusions containing 762 bp or more of FKS2 promoter sequence exhibited increased expression of β-galactosidase in response to Ca2+ (Fig. 1). We observed a four- to sixfold increase in transcription in response to Ca2+ that was calcineurin dependent because it was eliminated by the addition of FK506, a specific inhibitor of calcineurin (Fig. 1). No such induction was observed with the reporter containing only 705 bp or with the vector lacking any FKS2 regulatory sequences (pLG178; data not shown). We also found that the same constructs that displayed a calcineurin-dependent induction in response to Ca2+ were able to support a three- to fivefold calcineurin-dependent induction in response to α-factor (Fig. 1). Correspondingly, α-factor-induced expression was observed for the 762-bp construct but not the 705-bp construct (Fig. 1). From these results we conclude that the region from −762 bp to −705 bp is necessary for both Ca2+- and α-factor-induced calcineurin-dependent transcription.
Figure 1.
Constructs containing 762 bp or more of FKS2 upstream sequence can support calcineurin-dependent transcriptional induction of expression. Plasmid-based lacZ reporter genes were constructed with varying amounts of FKS2 upstream sequence [−910/+6 (pAMS312), −762/+6 (pAMS317), −705/+6 (pAMS319)]. β-Galactosidase activities are shown for extracts from cells (YPH499) that were either untreated (open bar), treated with 10 μg/ml of α-factor (shaded bar), or treated with 200 mm CaCl2 (solid bar) in the presence or absence of FK506. Each extract was assayed in triplicate, and the s.d. is representative of the error between these samples.
Identification of the calcineurin-dependent response element
To further define the region mediating calcineurin-dependent expression, reporter genes containing heterologous promoters were constructed (see Materials and Methods). No calcineurin-dependent expression was observed with the (60)::cyc1::lacZ construct containing ∼60 bp of FKS2 sequence from the region −762 bp to −705 bp under normal growth conditions or with α-factor addition (data not shown). However, this construct showed an approximately fivefold increase in expression in response to Ca2+ that was calcineurin dependent (Table 1). The 60-bp sequence was subdivided into overlapping 24-bp regions, creating four reporter gene constructs A–D(24)::cyc1::lacZ. Only one 24-bp region, contained in construct A(24)::cyc1::lacZ, was able to support calcineurin-dependent Ca2+-induced expression (4.4 ± 0.9-fold induction; Table 1) that was similar in magnitude to that observed for the larger 60-bp construct. We named this element, which is composed of the sequence CACCAGTCGGTGGCTGTGCGCTTG, the CDRE (calcineurin-dependent response element). Analysis of the CDRE failed to find any matches to consensus binding sites defined previously for yeast transcription factors (Prestridge 1991).
Table 1.
Calcineurin-dependent induction of heterologous reporter genes in response to Ca2+
|
fks2::cyc1::lacZ reportera (bp)
|
FKS2 sequence
|
β-Galactosidase (U/mg protein)b
|
Calcineurin-dependent inductionc (ratio: +Ca2+/+Ca2++FK)
|
|
|---|---|---|---|---|
| +Ca2+
|
+Ca2++FK
|
|||
| 60 | −762 to −705 | 56.4 ± 2.6 | 11.6 ± 0.5 | 4.9 ± 0.6 |
| A (24) | −762 to −738 | 13.6 ± 0.6 | 3.1 ± 0.3 | 4.4 ± 0.9 |
| B (24) | −750 to −726 | 3.1 ± 0 | 3.2 ± 0.1 | 1.0 ± 0 |
| C (24) | −738 to −714 | 3.1 ± 0.2 | 4.5 ± 0.3 | 0.7 ± 0.1 |
| D (24) | −726 to −705 | 1.6 ± 0.3 | 1.6 ± 0.2 | 1.0 ± 0.4 |
The 60-bp, A, B, C, and D reporters are contained on plasmids pAMS327, pAMS342, pAMS344, pAMS455, and pAMS456, respectively.
β-Galactosidase activity per milligram of protein was assayed from cells grown in synthetic medium supplemented as indicated with 200 mm CaCl2 (Ca2+) and 1 μg/ml of FK506 (FK). Data are the averages of extracts assayed in triplicate from one representative experiment, and the standard deviation is representative of the error between these samples.
Calcineurin-dependent induction is presented as the average of the largest possible ratio and the smallest possible ratio, with the error indicating the span between the two numbers.
Multiple copies of the CDRE increase calcineurin-dependent transcriptional activation
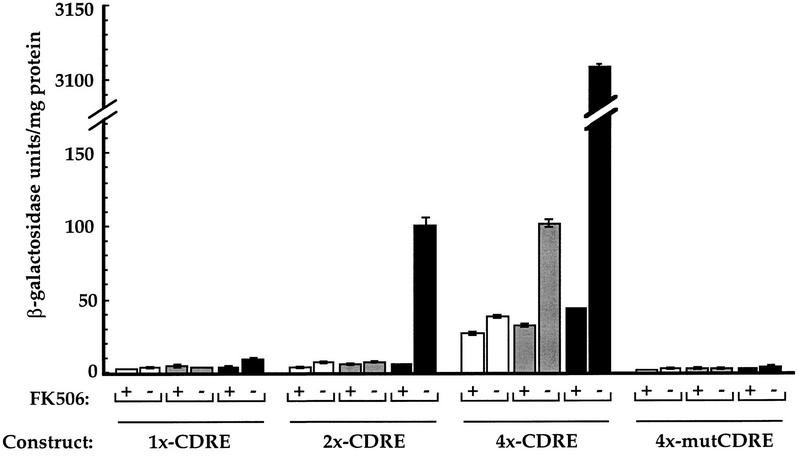
We found that multimerization of the CDRE increased the sensitivity of the reporter gene. In contrast to the 2- to 5-fold calcineurin-dependent Ca2+-induced expression exhibited by one copy of the CDRE, 15-fold and 69-fold inductions were observed in cells carrying heterologous promoter constructs that contained two and four tandem copies of the CDRE, respectively (Fig. 2). Additionally, the most sensitive reporter gene, containing four tandem copies of the CDRE (4×-CDRE::lacZ), supported an approximately threefold increase in expression in response to α-factor that was calcineurin dependent (Fig. 2). No α-factor-induced expression was observed with the other constructs.
Figure 2.
Tandem copies of CDRE increase the response of heterologous promoters to Ca2+ and α-factor. Plasmid-based lacZ reporter genes were constructed containing heterologous promoters with increasing copies of the CDRE or mutCDRE in tandem [1×-CDRE (pAMS342), 2×-CDRE (pAMS363), 4×-CDRE (pAMS366), 4×-mutCDRE (pAMS364)]. β-Galactosidase activities are shown for extracts from cells (YPH499) that were either untreated (open bar), treated with 10 μg/ml of α-factor (shaded bar), or treated with 200 mm CaCl2 (solid bar) in the presence or absence of FK506. Each extract was assayed in triplicate and the s.d. is representative of the error between these samples. (Note discontinuity in y-axis.)
We also constructed a derivative of the CDRE (mutCDRE; see Materials and Methods) that was not able to promote calcineurin-dependent transcriptional regulation. When tandem repeats of mutCDRE were placed upstream of a minimal promoter, no calcineurin-dependent increase in expression was observed with either Ca2+ or α-factor treatment (Fig. 2).
Identification of high-copy plasmids that allow expression of the CDRE reporter gene in the absence of calcineurin
We used the calcineurin mutant strain containing the 4×-CDRE::lacZ reporter (ASY461) to identify gene products that activate CDRE-mediated expression in the absence of calcineurin. We introduced two different multicopy genomic libraries into ASY461 cells and identified plasmids that conferred CDRE-dependent gene expression (see Materials and Methods). Four classes of plasmids were isolated. One class of plasmids contained a copy of CNB1, encoding the calcineurin regulatory subunit. These plasmids complemented the cnb1Δ mutation in the parent strain, thus restoring calcineurin activity and CDRE-driven gene expression. A second class of plasmids contained truncations of CNA2, encoding the calcineurin catalytic subunit. Evidently, enough catalytic activity was supported by truncated Cna2p even in the absence of Cnb1p to test positive in our sensitive reporter system. The third class of plasmids consisted of a single isolate. Specific activation of the reporter by this plasmid was attributable to a single open reading frame (ORF), YMR030w, that is predicted to encode a protein of 43 kD of molecular mass with no significant homology to any other protein. Cells deficient for YMR030w still displayed CDRE-dependent transcriptional activation (data not shown); therefore, this ORF was not studied further. The fourth class of plasmids contained at least part of a previously uncharacterized ORF, YNL027w. YNL027w is predicted to encode a protein of 76 kD of molecular mass containing a polyglutamine tract, which commonly acts as a transcriptional activation domain (Perutz 1994), and two zinc finger domains of the Cys2-His2 type, which mediate protein–nucleic acid interactions in many transcription factors (Evans and Hollenberg 1988; Desjarlais and Berg 1992) (Fig. 3). In addition, YNL027w contains a third putative zinc finger that is less well conserved and only contains one cysteine and one histidine (Fig. 3). We renamed YNL027w CRZ1 for calcineurin-responsive zinc finger protein. This same gene was identified independently by Matheos et al. and named TCN1 (Matheos et al., this issue).
Figure 3.
CRZ1 encodes a putative transcription factor. The predicted amino acid sequence of the transcription factor Crz1p (ORF YNL027w) is shown with the polyglutamine tract at the amino terminus underlined and the three zinc fingers at the carboxyl terminus boxed in gray. Conserved cysteine and histidine residues within each zinc finger are indicated by a mark (▾) above each residue.
CRZ1 encodes an essential component of the CDRE-mediated transcriptional induction
A diploid strain, ASY650, heterozygous for a null allele of CRZ1 (crz1::loxP–kanMX–loxP) was sporulated. All haploid segregants from this diploid were viable, and cells carrying the CRZ1 disruption allele (ASY472) showed no obvious defects in growth or morphology. Thus, CRZ1 is not an essential gene. However, crz1Δ mutant cells were not able to activate the 4×-CDRE::lacZ reporter in the absence (ASY589) or in the presence of calcineurin (ASY587). When incubated with Ca2+, the crz1Δ mutant (ASY587), the cnb1Δ mutant (ASY461), and the cnb1Δ crz1Δ double mutant (ASY589) showed similar low levels of CDRE-mediated expression (4–7 β-galactosidase U/mg of protein) in contrast to the high level of CDRE-mediated expression (183 ± 2 β-galactosidase U/mg of protein) observed with wild-type cells (ASY459) (data not shown). These results indicate that CRZ1 is an essential component of calcineurin-dependent CDRE-mediated gene expression.
Crz1p binds specifically to the CDRE
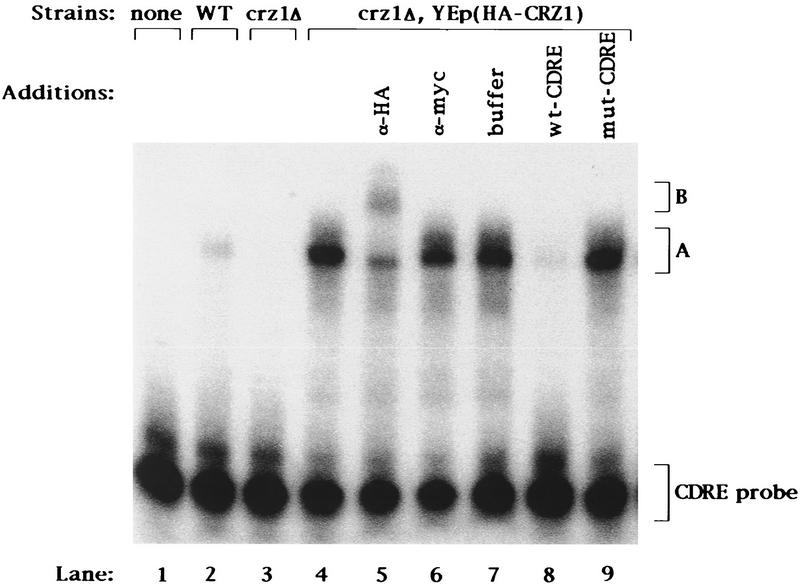
The similarity of Crz1p to other transcription factors (Fig. 3) and the observation that this protein is a required component of calcineurin-dependent CDRE-mediated transcriptional induction suggest that Crz1p may directly bind the CDRE. To test this possibility, we performed gel-shift experiments. Cell extracts were incubated with a 32P-labeled 24-bp double-stranded oligonucleotide corresponding to the CDRE sequence and analyzed by fractionation on nondenaturing polyacrylamide gels. Cell extracts prepared from wild-type cells contained an activity that bound to the oligonucleotide and retarded its migration in the gel, resulting in the formation of a new band (Fig. 4, lane 2, band A). This DNA-binding activity was absent from crz1Δ cells (Fig. 4, lane 3). These results demonstrate that Crz1p is required for assembly of a CDRE-binding activity.
Figure 4.
Crz1p binds to the CDRE. Extracts of strains YPH499 containing YEp351 (“WT”), ASY472 containing YEp351 (“crz1Δ”), and ASY472 containing pAMS446 [“crz1Δ, YEp(HA–CRZ1)”] were analyzed by DNA mobility retardation analysis using the CDRE as probe (see Materials and Methods). The plasmid YEp(HA–CRZ1) carries a gene encoding a version of Crz1p with the HA epitope inserted near the amino terminus of the protein. (Lanes 5–7) Equal amounts of α-HA antibody, α-myc antibody, or antibody dilution buffer were added 5 min before gel loading; (lanes 8,9) a 100-fold molar excess of unlabeled CDRE or mut-CDRE oligonucleotide to probe was added before addition of extract.
Further experiments demonstrated that Crz1p binds to the CDRE directly and specifically. First, we created an epitope-tagged version of Crz1p by inserting a DNA segment encoding the hemagglutinin antigen (HA) epitope into CRZ1 near the 5′ end of the coding region (Wilson et al. 1984). The epitope-tagged gene, HA–CRZ1, when expressed from a centromere-based plasmid (pAMS451) fully complemented the CDRE-mediated transcription defect and the ion sensitivities of a crz1Δ mutant strain (see below) indicating that the epitope tag did not impair the function of Crz1p (data not shown). More CDRE-binding activity was observed for cells overexpressing HA–Crz1p than for wild-type cells (Fig. 4, lanes 4 vs. lane 2). This increase in binding activity was attributable to the high level of HA–Crz1p expression and was equivalent to that observed for cells overexpressing native Crz1p (data not shown). To examine the specificity of the observed binding to CDRE, we added an excess of unlabeled CDRE or mutCDRE oligonucleotides to the binding reaction. The CDRE was able to compete for binding (Fig. 4, lane 8), but the mutCDRE had no effect (Fig. 4, lane 9). Thus, the observed Crz1p-dependent binding activity is specific for the intact CDRE sequence. To ascertain whether Crz1p is a component of this CDRE-binding activity, we added anti-HA antibody to the binding reaction containing extract from cells expressing HA-tagged Crz1p. Addition of anti-HA antibodies to this binding reaction resulted in the disappearance of most of band A and the appearance of a new band of slower mobility, band B (Fig. 4, lane 5). This supershift was specific for HA-tagged Crz1p, as no shift was observed for cells containing native Crz1p (data not shown). Addition of anti-myc antibody (Evan et al. 1985), which does not recognize HA-tagged Crz1p, had no effect (Fig. 4, lane 6). These observations establish that Crz1p is a component of the CDRE-specific DNA-binding activity. The remainder of band A that was not supershifted by the addition of anti-HA antibody may represent a distinct protein–CDRE complex. However, as mentioned above, no CDRE-binding activity was detected for crz1Δ cells. Therefore, a more likely explanation is that the epitope present at the amino terminus of the protein is removed from a fraction of HA–Crz1p such that it is not recognized by the antibody but still retains DNA-binding activity.
crz1Δ phenotypes are similar to those of calcineurin mutants
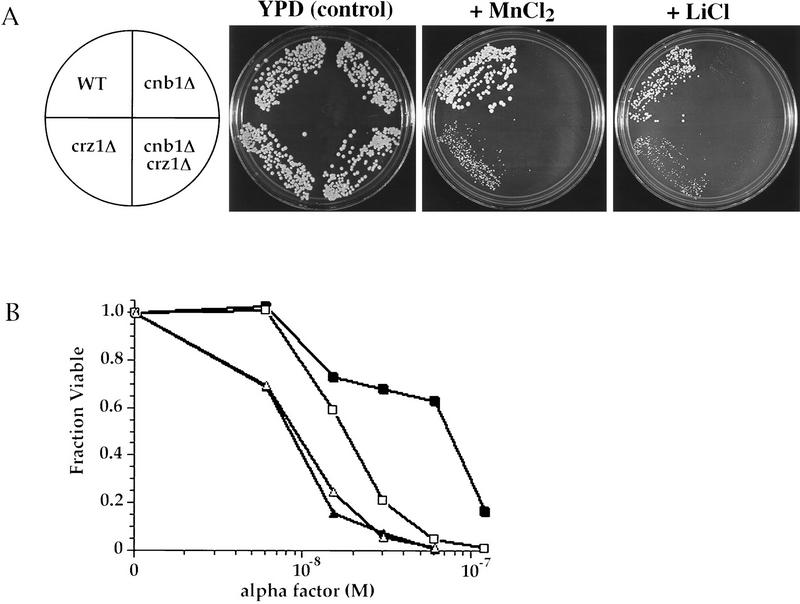
We identified CRZ1 by its capacity to modify one calcineurin mutant phenotype, the defect in CDRE-mediated transcription. To further investigate the relationship between CRZ1 and calcineurin, we analyzed crz1Δ cells for other phenotypes exhibited by calcineurin mutants. Calcineurin mutants are more sensitive to Mn2+, Li+, and high pH than wild-type cells (Nakamura et al. 1993; Mendoza et al. 1994; Farcasanu et al. 1995; Pozos et al. 1996). They also die during prolonged treatment with α-factor (Moser et al. 1996; Withee et al. 1997). The growth of crz1Δ cells was also impaired by Mn2+ and Li+; however, crz1Δ cells were less sensitive to these ions than were calcineurin mutant cells (Fig. 5A). When grown on plates containing lower concentrations of either Mn2+ or Li+, cells lacking both CRZ1 and calcineurin, crz1Δ cnb1Δ (ASY475), displayed the same sensitivity to these treatments as the cnb1Δ single mutant (data not shown). In addition, crz1Δ cells exhibited a survival defect when incubated with α-factor that is less severe than that of cnb1Δ cells (Fig. 5B). Furthermore, as observed for the ion sensitivities described above, crz1Δ cnb1Δ and cnb1Δ cells exhibited equivalent α-factor survival defects (Fig. 5B). These observations, together with the effect of the crz1Δ allele on CDRE reporter activity, indicate that CRZ1 and calcineurin function in the same pathway to regulate both ion homeostasis and CDRE-mediated transcription. However, unlike calcineurin mutant cells, crz1Δ cells do not exhibit sensitivity to high pH (data not shown). In addition, crz1Δ cells exhibit Ca2+-sensitive growth, whereas calcineurin mutant cells are Ca2+ tolerant (data not shown; see Discussion).
Figure 5.
CRZ1 mutants exhibit defects similar to but less severe than those of calcineurin mutants. (A) Saturated overnight cultures were diluted, spread on YPD media containing the specified ions (1.8 mm MnCl2 or 0.16 m LiCl), and grown for 2–5 days at 30°C (Pozos et al. 1996). Strains are YPH499 (“WT”), ASY472 (“crz1Δ”), DD12 (“cnb1Δ”), and ASY475 (“cnb1Δ crz1Δ”). (B) Cells were grown on low pH YPD media with or without the indicated concentrations of α-factor. The fraction viable was determined by comparing growth on plates containing α-factor with growth on plates without α-factor (Moser et al. 1996). Each data point represents an average of three plates. Strains are YPH499 (▪), ASY472 (□), DD12 (▴), and ASY475 (▵).
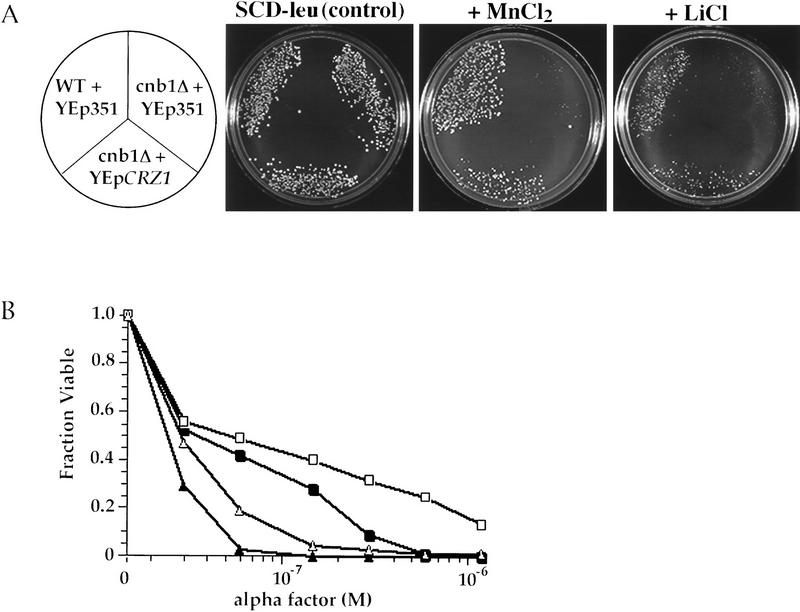
Overproduction of CRZ1 suppresses calcineurin mutant phenotypes
We also characterized the properties of cells overproducing Crz1p. Expression of CRZ1 from a high-copy plasmid YEp(CRZ1) was able to partially suppress the Mn2+ and Li+ sensitivities of calcineurin mutant cells (Fig. 6A). YEp(CRZ1) also increased the viability of calcineurin-deficient cells during prolonged α-factor treatment (Fig. 6B). Furthermore, overproduction of CRZ1 in wild-type cells increased the tolerance of these cells to Mn2+ and Li+ (data not shown) and increased survival during prolonged incubation with pheromone (Fig. 6B).
Figure 6.
Increased expression of CRZ1 suppresses calcineurin mutant phenotypes. (A) Saturated overnight cultures were diluted and spread on synthetic media with or without ions (5 mm MnCl2 or 0.55 m LiCl) and grown for 2–5 days at 30°C (Pozos et al. 1996). Strains are YPH499 containing YEp351 (“WT + YEp351”), DD12 containing YEp351 (“cnb1Δ + YEp351”), and DD12 containing pAMS435 (“cnb1Δ + YEpCRZ1”). (B) Cells were grown on synthetic media with or without the indicated levels of α-factor. The fraction viable was determined by comparing growth on plates containing α-factor with growth on plates without α-factor (Moser et al. 1996). Each data point represents an average of three plates. Strains are YPH499 containing YEp351 (WT + YEp351, ▪), YPH499 containing pAMS435 (WT + YEpCRZ1, □), DD12 containing YEp351 (cnb1Δ + YEp351, ▴), and DD12 containing pAMS435 (cnb1Δ + YEpCRZ1, ▵).
Crz1p functions as a general mediator of calcineurin-dependent transcriptional induction
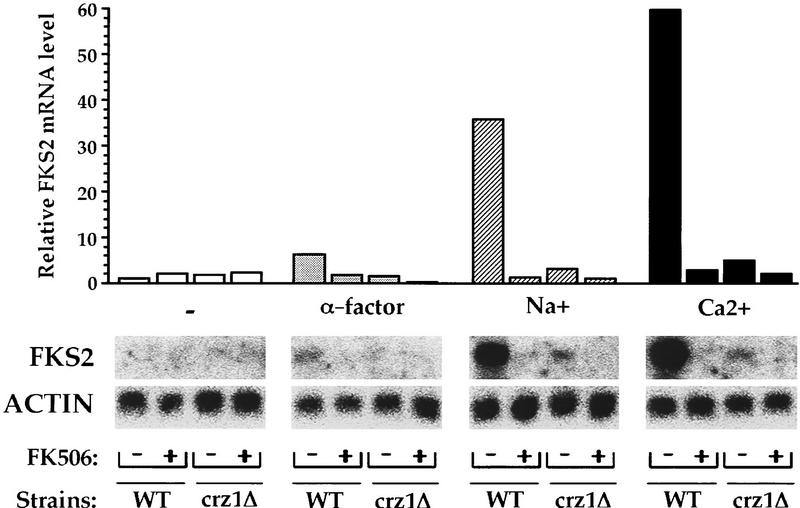
We analyzed the effect of the crz1Δ allele on FKS2 mRNA levels by Northern blot analysis. We found that CRZ1 is necessary for the previously observed calcineurin-dependent increase in FKS2 expression in response to both α-factor and Ca2+ (Fig. 7). These observations confirmed that CRZ1 is required not only for expression of CDRE-driven reporter genes but also for calcineurin-dependent regulation of genomic FKS2. In addition, we observed that FKS2 mRNA levels increase in response to Na+ and that this response was largely calcineurin dependent (Fig. 7). This calcineurin-dependent induction of FKS2 expression in response to Na+ also required CRZ1 (Fig. 7). Thus, CRZ1 is required for the calcineurin-dependent induction of FKS2 in response to α-factor, Na+, or Ca2+ treatment.
Figure 7.
CRZ1 is necessary for calcineurin-dependent transcriptional induction of the FKS2 gene in vivo. YPH499 (“WT”) and ASY472 (“crz1Δ”) cells were incubated for 4 hr in low pH YPD with or without FK506 and inducing treatments. Total RNA isolated from these cells was subjected to Northern blot and hybridized successively to FKS2 and ACT1 probes. FKS2 mRNA levels were normalized to ACT1 mRNA levels and are from one representative experiment with mRNA from cells that were either untreated (open bars) or treated with 10 μg/ml of α-factor (shaded bars), 0.8 m NaCl (hatched bars), or 200 mm CaCl2 (solid bars). The ACT1-normalized FKS2 mRNA level in untreated wild-type cells was arbitrarily designated a value of 1.
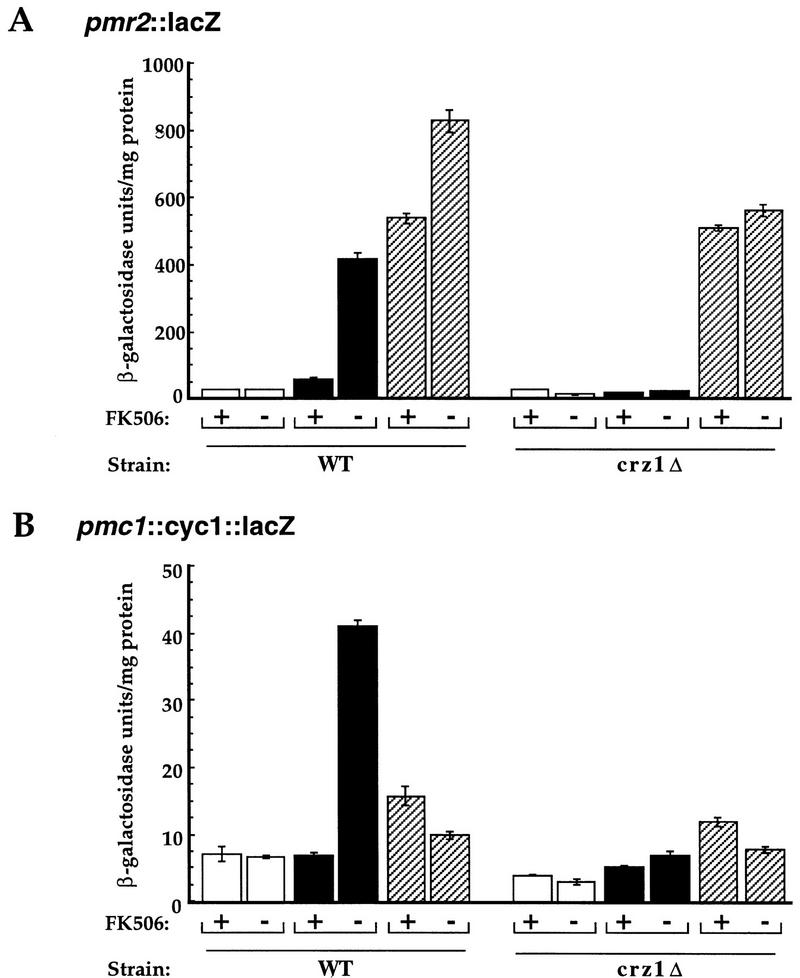
The ion sensitivities of calcineurin mutants are in part attributable to decreased expression of several genes encoding ion transporters (Mendoza et al. 1994; Cunningham and Fink 1996). Because crz1Δ mutant cells similarly exhibit ion sensitivities, we examined whether CRZ1 is required for the calcineurin-dependent transcriptional induction of two genes that encode P-type ATPases, PMR2 and PMC1 (Rudolph et al. 1989; Haro et al. 1991; Cunningham and Fink 1994). CRZ1 is required for the calcineurin-dependent induction of a PMR2 reporter gene (pmr2::lacZ-pFR70; Mendoza et al. 1994) in response to Ca2+ as well as Na+ (Fig. 8A). The crz1Δ mutation had no effect on the Na+-induced expression of pmr2::lacZ that is calcineurin independent. In addition, CRZ1 was required for the calcineurin-dependent expression of a PMC1 reporter gene (pmc1::cyc1::lacZ-pAMS381) in response to Ca2+. No increase in β-galactosidase expression was observed for PMC1 in response to Na+ (Fig. 8B) or with PMR2 or PMC1 reporters in response to α-factor (data not shown).
Figure 8.
CRZ1 is a general mediator of calcineurin-dependent transcriptional regulation. (A) Wild-type cells containing a plasmid-based pmr2::lacZ reporter gene (pFR70; Marquez and Serrano 1996) were grown at 30°C for 4 hr with or without FK506 in either YPD (open bar), synthetic media containing 200 mm CaCl2 (solid bar), or YPD containing 0.8 m NaCl (hatched bar). (B) Wild-type cells containing a plasmid-based pmc1::cyc1::lacZ reporter gene (pAMS381) were grown at 30°C for 4 hr with or without FK506 in either YPD (open bar), low pH YPD containing 200 mm CaCl2 (solid bar), or YPD containing 0.8 m NaCl (hatched bar). β-Galactosidase activities are shown for extracts from cells (YPH499) from one representative experiment. Each extract was assayed in triplicate, and the s.d. is representative of the error between these samples.
Discussion
Identification of a promoter element, the CDRE, and a novel transcription factor, Crz1p, that function downstream of calcineurin to activate transcription in response to Ca2+
Calcineurin is required for the transcriptional induction of FKS2 in response to Ca2+ (Mazur et al. 1995; C. Zhao, U. Jung, P. Garrett-Engele, T. Roe, M. Cyert, and D. Levin, in prep.). We identified a region within the FKS2 upstream sequence from −762 bp to −705 bp that is both necessary and sufficient for this calcineurin-dependent transcriptional induction (see Fig. 1). Furthermore, from within this region, we defined a 24-bp element, the CDRE, that supports calcineurin-dependent transcriptional induction in response to Ca2+ (see Table 1; Fig. 2). We performed a genetic screen and identified a novel transcription factor, Crz1p, that when overexpressed bypasses a requirement for calcineurin inactivation of a CDRE-containing reporter gene. This transcription factor is identical to Tcn1p (Matheos et al., this issue). In addition, we find that Crz1p is required for transcriptional induction of both CDRE-driven reporter genes and genomic FKS2 in response to Ca2+ (see Fig. 7) and physically binds to the 24-bp oligonucleotide comprising the CDRE (see Fig. 4). Thus, CRZ1 encodes the transcription factor that functions downstream of calcineurin to mediate Ca2+-induced transcription of FKS2 through the CDRE.
Calcineurin is required for the transcriptional induction of PMR2, PMC1, and PMR1 as well as other genes in response to Ca2+. Preliminary analysis using DNA microarray technology indicates that many genes exhibit calcineurin-dependent changes in gene expression that also require CRZ1 (T. Roe, J. DeRisi, A. Stathopoulos, P. Brown, and M. Cyert, in prep.). Here, we show that Crz1p is required for calcineurin-dependent transcription of PMR2 and PMC1 (see Fig. 8). Calcineurin-dependent transcription of PMR1 has also been shown to require Crz1p (Matheos et al., this issue). In addition, we found that crz1Δ cells exhibit sensitivities to Mn2+, Na+/Li+, and to Ca2+ (see Fig. 5A; data not shown). Pmr2p is a Na+ ATPase required for Na+ tolerance (Rudolph et al. 1989; Haro et al. 1991), Pmc1p is a Ca2+ ATPase required for Ca2+ tolerance (Cunningham and Fink 1994), and Pmr1p is a Ca2+ ATPase required for tolerance to Mn2+ (Rudolph et al. 1989; Lapinskas et al. 1995). Thus, it is likely that decreased levels of these ion transporters are responsible for the observed ion sensitivities of crz1Δ cells. We conclude that Crz1p functions downstream of calcineurin to regulate ion homeostasis because cnb1Δ crz1Δ double mutant cells exhibit ion sensitivities similar in extent to those of cnb1Δ cells and overexpression of CRZ1 is sufficient to partially suppress these sensitivities in calcineurin mutants (Fig. 5A and 6A).
Crz1p functions downstream of calcineurin to control transcriptional activation in response to multiple environmental signals
Calcineurin is required not only for transcriptional inductions that result when cells are grown in high levels of Ca2+ but also those that result in response to at least three other environmental signals: Na+, elevated temperature, and mating pheromone (α-factor). Several observations indicate that calcineurin-dependent activation of Crz1p occurs under all of these conditions. First, when cells are incubated in Na+, the expression of at least two genes, FKS2 and PMR2, is increased in a calcineurin-dependent manner (see Fig. 7; Mendoza et al. 1994); we show that Crz1p is required for both of these responses (see Figs. 7 and 8A). Second, elevated temperature induces calcineurin-dependent transcriptional FKS2 (C. Zhao, U. Jung, P. Garrett-Engele, T. Roe, M. Cyert, and D. Levin, in prep.), and preliminary experiments with CDRE reporter genes suggest that Crz1p also mediates this response (A. Stathopoulos, unpubl.). Finally, in response to pheromone, only FKS2 is known to increase in a calcineurin-dependent fashion (Mazur et al. 1995), and we find that this response requires CRZ1 as well (see Fig. 7). Furthermore, like calcineurin, Crz1p is required for cell viability during prolonged treatment with α-factor (see Fig. 5B), and overexpression of CRZ1 can suppress the survival defect of calcineurin mutants during incubation with pheromone (see Fig. 6B). These results indicate that Crz1p-mediated transcriptional regulation contributes to cell viability under these conditions. However, fks2 mutant cells do not exhibit a survival defect when incubated with pheromone (Mazur et al. 1995). Therefore, additional genes, as yet unidentified, must also be transcriptionally regulated by Crz1p and calcineurin in response to α-factor (see Fig. 9).
Figure 9.
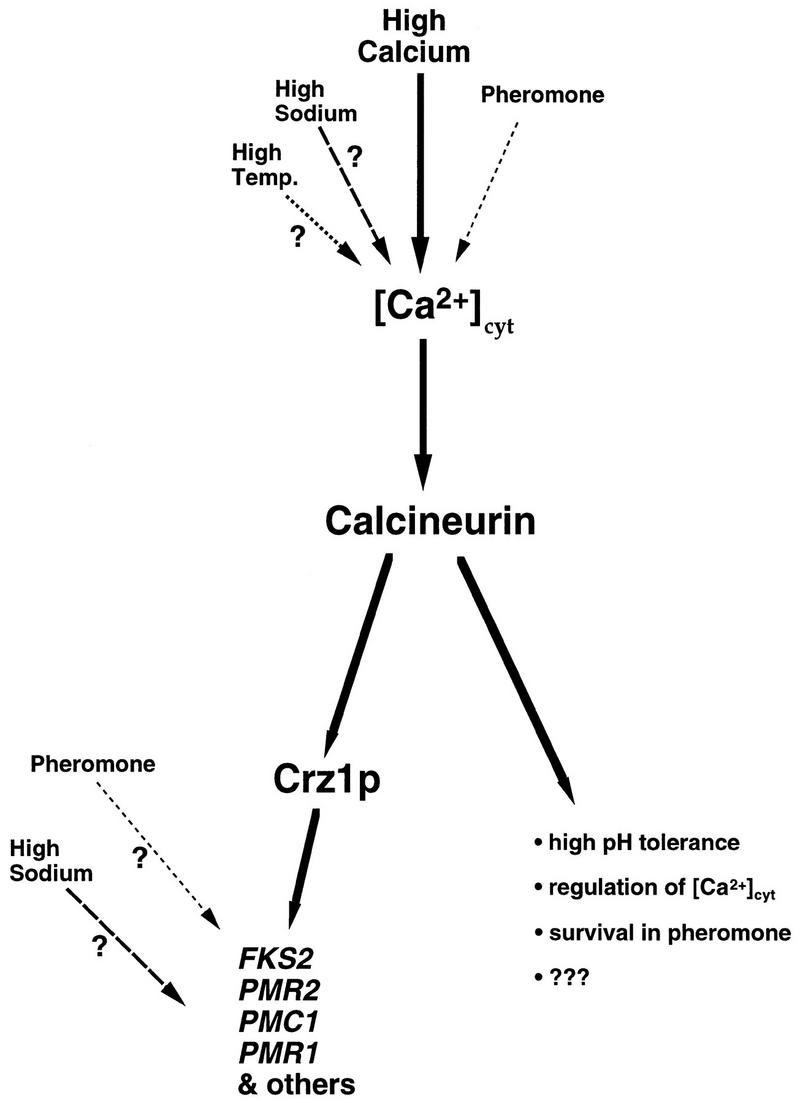
Model describing calcineurin-mediated signal transduction pathways. (See Discussion for description.)
Differential gene expression results from multiple environmental signals and is mediated by both calcineurin and Crz1p
Surprisingly, although calcineurin and Crz1p are both activated in response to a variety of environmental signals (i.e., Ca2+, α-factor, and Na+), the genes transcriptionally regulated by Crz1p are differentially induced by subsets of these signals. FKS2 is induced by Ca2+, α-factor, and Na+, whereas PMR2 is induced only by Ca2+ and Na+, and PMC1 is induced only by Ca2+ (see Figs. 7 and 8). Although calcineurin-dependent regulation of PMR2 and PMC1 requires CRZ1, it is unclear whether Crz1p is directly mediates all these transcriptional inductions by binding to the promoters of these genes. However, even observations with FKS2, which we have shown is directly regulated in a Crz1p/CDRE-dependent manner, suggest that the mechanism of calcineurin- and Crz1p-dependent transcriptional regulation may vary under different environmental conditions. We initially observed that reporters containing the FKS2 promoter sequence exhibit calcineurin-dependent transcriptional induction in response to both Ca2+ and α-factor and that a 57-bp region from −762 bp to −705 bp is necessary for both of these responses. However, whereas only 24 bp of this region, the CDRE, was sufficient to confer Ca2+-induced expression on a reporter gene, even the entire 57 bp was not sufficient to confer α-factor-dependent regulation on a reporter gene (see Table 1; Fig. 2). Although we do observe a small calcineurin-dependent induction of the 4×-CDRE reporter gene when cells are treated with α-factor (see Fig. 2), this response may not reflect true α-factor-dependent signaling but may instead result from the rise in intracellular Ca2+ known to occur during pheromone treatment (Iida et al. 1990). Thus, our observations suggest that though the CDRE responds somewhat to α-factor, the full induction of FKS2 expression in response to α-factor requires additional regulatory sequences. Similarly, with Na+ treatment, although a substantial increase in FKS2 mRNA levels occurs, no induction of CDRE-driven constructs is observed (A. Stathopoulos, unpubl.), suggesting the requirement of additional sequences for this response as well. We conclude that although the CDRE is sufficient for transcriptional induction in response to Ca2+, it may act in conjunction with binding sites for other factors to mediate responses to α-factor and Na+ (see Fig. 9).
How does calcineurin regulate the ability of Crz1p to activate transcription?
The mechanism by which calcineurin regulates the activity of Crz1p remains to be determined. Calcineurin may regulate the binding affinity of Crz1p for the CDRE or the nuclear translocation of this protein. However, we currently favor a different model in which calcineurin regulates the ability of Crz1p to function as a transcriptional activator. Preliminary experiments with a chimeric protein containing Crz1p fused to the Gal4p DNA-binding domain support this model. This fusion protein, which also contains a functional nuclear localization sequence, exhibits calcineurin-dependent transcriptional activation of a reporter gene (UASGAL4) that is induced in a Gal4p-specific manner (A. Stathopoulos, unpubl.). These results suggest that Crz1p contains a calcineurin-dependent transcriptional activation domain. Preliminary two-hybrid experiments failed to detect an interaction between full-length Crz1p and the calcineurin catalytic subunit (A. Stathopoulos, unpubl.). Therefore, more extensive analysis is required to determine whether calcineurin directly dephosphorylates Crz1p or whether instead the calcineurin-dependent regulation of Crz1p is indirect.
A subset of calcineurin-dependent events are mediated by Crz1p
Comparison of the phenotypes exhibited by calcineurin and crz1Δ cells demonstrates that only a subset of calcineurin functions are mediated through Crz1p (see Fig. 9). Calcineurin mutants exhibit a greater degree of sensitivity to Mn2+ and Li+ and a more pronounced α-factor viability defect than crz1Δ cells (see Fig. 5). Thus, calcineurin must also carry out functions that are independent of Crz1p that affect these phenotypes. Similarly, fks1Δ mutants are inviable in the absence of calcineurin activity, whereas these cells are viable though severely growth impaired in the absence of Crz1p (A. Stathopoulos, unpubl.). Furthermore, calcineurin mutants display two phenotypes that are not shared by crz1Δ cells. First, calcineurin mutants are sensitive to high pH (Nakamura et al. 1993), whereas crz1Δ cells are not. Second, calcineurin mutants and cnb1Δ crz1Δ double mutants are Ca2+ tolerant (Tanida et al. 1996; Withee et al. 1997), whereas crz1Δ single mutants are Ca2+ sensitive. An inability to induce PMC1 expression most likely explains the Ca2+ sensitivity exhibited by crz1Δ cells (Fig. 8B). However, in calcineurin mutants despite an inability to induce PMC1 expression, these cells are Ca2+ tolerant because of decreased cytosolic Ca2+ levels (Tanida et al. 1996). Thus, calcineurin also functions independently of Crz1p to inhibit Ca2+ sequestration.
We have demonstrated that Crz1p is a component of one or more calcineurin-dependent pathways controlling transcriptional activation and that calcineurin carries out additional functions that are not mediated by Crz1p. The identification and characterization of the CDRE and Crz1p should facilitate the identification of additional components of these and other calcineurin-dependent signal transduction pathways. This analysis should more clearly define the roles of the multifunctional calcineurin phosphatase in yeast and other eukaryotic cells.
Materials and methods
Media and general methods
Yeast media and culture conditions were those recommended (Sherman et al. 1986) except that nutritional supplements in synthetic media were added at twice the specified level and 3.5 grams of ammonium chloride per liter was substituted for ammonium sulfate as indicated. In addition, low pH YP media was adjusted to pH 5.0 by adding succinate (10 gram/liter). Where indicated, α-factor (Star Biochemicals) or the chloride salt of certain ions were added to media at the specified level. A stock solution of FK506 (Fugisawa, Inc.; 20 mg/ml in 90% ethanol–10% Tween 20) was also added to liquid media to a final concentration of 1 μg/ml where noted.
All procedures involving recombinant DNA in S. cerevisiae and Escherichia coli were performed using standard techniques (Ausubel et al. 1987). DNA was introduced into yeast cells by lithium acetate transformation (Ausubel et al. 1987) and into bacteria by electroporation (Ausubel et al. 1987). Double-stranded DNA templates used for sequencing were prepared according to the manufacturer’s instructions (Promega Wizard Miniprep). The Sequenase version of phage T7 DNA polymerase and nonradioactive nucleotides were obtained from U.S. Biochemical Corporation. [α-35S]dATP was obtained from Amersham. Sequencing reactions were conducted according to the manufacturer’s instructions.
Plasmids
A series of fks2::lacZ reporter genes was constructed from cyc1::lacZ reporters pLGΔ178 and pLGΔ312 (Guarente and Mason 1983). The fks2::lacZ reporter plasmids pAMS312, pAMS317, and pAMS319 were constructed by removing a region of CYC1 sequence from pLGΔ178 with XhoI and BamHI and inserting PCR products flanked by XhoI and BamHI sites containing various segments of the FKS2 gene upstream sequence (−910/+6, −762/+6, and −705/+6 bp, respectively). pAMS327 was constructed by removing the CYC1 upstream activating sequence (UAS) from pLGΔ312 with SmaI and XhoI and inserting a PCR product flanked by blunt and SalI sites containing the FKS2 sequence from −762 bp to −705 bp. Plasmids pAMS342, pAMS344, pAMS455, and pAMS456 were constructed by removing the UAS sequence from pLGΔ312 with SmaI and XhoI and inserting annealed complementary oligonucleotides flanked by blunt and SalI sites that contained the FKS2 sequence from −762 bp to −738 bp, −750 bp to −726 bp, −738 bp to −714 bp, and −726 bp to −705 bp, respectively. Plasmids pAMS363 and pAMS366 contain two and four tandem repeats, respectively, of the CDRE flanked by XhoI and SalI sites (XS-CDRE: 5′-TCGACACCAGTCGGTGGCTGTGCGCTTGC-3′ and 3′-GTGGTCAGCCACCGACACGCGAACGAGCT-5′). Two and four tandem repeats of the CDRE were constructed by iterative insertion of the CDRE into the XhoI site of pBluescript (Stratagene) creating plasmids pAMS346 and pAMS347, respectively. A XhoI–SalI fragment containing the tandem repeats plus pBluescript polylinker sequence was inserted into the XhoI site of pLGΔ178. Plasmid pAMS364 was constucted exactly as pAMS366 except four tandem copies of mutCDRE (XS–mutCDRE: 5-TCGACTCCTGTGGGACCGTGAGCCCTAGC-3′ and 3′-GAGGACACCCTGGCACTCGGGATCGAGCT-5′), constructed by iterative insertion into the XhoI site of pBluescript to create pAMS350, were used.
Another series of CDRE reporter genes was constructed from the gal1::lacZ integrating reporter pJL638 (Li and Herskowitz 1993). Plasmids pAMS367 and pAMS369 were created by inserting BamHI–BglII fragments of a CDRE tetramer from pAMS341 and a mutCDRE tetramer from pAMS354, respectively, into the BglII sites of pJL638. Plasmids pAMS341 and pAMS354 were created by iterative insertion of a double-stranded oligonucleotide flanked by BamHI and BglII sites containing the CDRE sequence or mutCDRE sequence, respectively, into the BamHI and BglII sites of LITMUS 28 (NEB).
Plasmid pAMS433 containing CRZ1 flanked by BamHI and SalI sites was isolated by gap repair in yeast strain DD12 of EcoRI-digested plasmid pAMS417. PCR fragments (575 and 600 bp) were amplified from the 5′ and 3′ regions of CRZ1 using genomic yeast DNA as a template and were flanked by BamHI and EcoRI or EcoRI and SalI sites, respectively. Both PCR products were inserted into BamHI- and SalI-digested pRS316 (Sikorski and Hieter 1989) to create plasmid pAMS417. A 3.2-kb BamHI–SalI fragment from pAMS433, containing CRZ1, was inserted into the BamHI and SalI sites of YEp351 to create pAMS435. A 130-bp PCR fragment containing a triple-HA epitope tag flanked by XbaI sites was amplified from plasmid pMPY-3XHA (Schneider et al. 1995) and inserted into the unique SpeI site of pAMS435, placing the epitope between the twelfth and thirteenth codons of Crz1p, creating pAMS446. BamHI–SalI (3.2-kb) fragments from pAMS435 and pAMS437 were inserted into pRS315 (Sikorski and Heiter 1989) creating pAMS452 and pAMS450, respectively.
A pmc1::cyc1::lacZ reporter construct, pAMS381, was created by first removing the UAS sequence from pLGΔ312 with SmaI and XhoI and then inserting a StuI–XhoI PCR fragment containing the PMC1 sequence from −568 to −207 relative to the initiation codon.
Yeast strains
The yeast strains used in this study are listed in Table 2 and were constructed through transformation or isogenic crosses by using standard techniques (Sherman et al. 1986). Each reporter gene construct (pAMS367, pAMS369, pJL638) was integrated at the URA3 locus as described (Li and Herskowitz 1993) into both strains YPH499 and DD12 to create reporter strains ASY459, ASY460, ASY461, ASY462, ASY465, and ASY466. The crz1::loxP-kanMX-loxP disruption that removes only the CRZ1 coding region was created by homologous recombination in yeast of a PCR-amplified fragment that was transformed into yeast cells (Güldener et al. 1996). This PCR-amplified fragment was generated with primers that recognize the loxP-kanMX-loxP cassette in plasmid pUG6 (Güldener et al. 1996) and also contain 40 bp of homology to the DNA flanking the CRZ1 coding sequence. Correct integration of the crz1::loxP-kanMX-loxP disruption fragment in ASY650, ASY472, ASY475, ASY587, and ASY589 was confirmed by PCR.
Table 2.
Yeast strains used in this study
| Strain
|
Relevant genotype
|
Reference
|
|---|---|---|
| YPH499 | MATa ura3-52 lys2-801 ade2-101 trp-Δ63 his3-Δ200 leu2-Δ1 | (Sikorski and Hieter 1989) |
| YPH500 | MATα ura3-52 lys2-801 ade2-101 trp-Δ63 his3-Δ200 leu2-Δ1 | (Sikorski and Hieter 1989) |
| YPH501 | MATa/α YPH499 × YPH500 | (Sikorski and Hieter 1989) |
| MCY3 | same as YPH501 except CNB1/cnb1::LEU2 | (Cyert and Thorner 1992) |
| DD12 | same as YPH499, except cnb1::hisG | (Cyert and Thorner 1992) |
| ASY459 | same as YPH499, except ura3-52::URA3–4×–CDRE::gal1::lacZ | this study |
| ASY460 | same as YPH499, except ura3-52::URA3–4×–mutant CDRE::gal1::lacZ | this study |
| ASY465 | same as YPH499, except ura3-52::URA3–gal1::lacZ | this study |
| ASY461 | same as DD12, except ura3-52::URA3–4×–CDRE::gal1::lacZ | this study |
| ASY462 | same as DD12, except ura3-52::URA3–4×–mutant CDRE::gal1::lacZ | this study |
| ASY466 | same as DD12, except ura3-52::URA3–gal1::lacZ | this study |
| ASY650 | same as MCY3, except CRZ1/crz1::loxP–kanMX–loxP | this study |
| ASY472 | same as YPH499, except crz1::loxP–kanMX–loxP | this study |
| ASY475 | same as DD12, except crz1::loxP–kanMX–loxP | this study |
| ASY587 | same as ASY459, except crz1::loxP–kanMX–loxP | this study |
| ASY589 | same as ASY461, except crz1::loxP–kanMX–loxP | this study |
β-Galactosidase assays
Quantitative assay
Exponentially growing cells (OD600 of 0.8–1.2) were incubated at 21°C in synthetic media containing ammonium chloride for 6 hr unless otherwise noted. When indicated, salts, α-factor and/or FK506 were added to the media at the start of this incubation. Cells were then harvested, and the cell pellet was frozen at −20°C. Cell extracts were prepared essentially as described in Withee et al. (1997). Protein concentration of the extracts was determined using the Bio-Rad Bradford assay kit, with dilutions of bovine serum albumin used to generate the standard curve. The β-galactosidase activity was determined at room temperature using the substrate ONPG (O-nitrophenyl-β-d-galactopyranoside, Sigma) as described (Miller 1972) and are given in units of nanomoles ONPG converted per minute per milligram of protein.
Qualitative assay
Colonies were scored for β-galactosidase activity essentially as described (Hannon et al. 1993) with minor modifications. Supported nitrocellulose filters (Schleicher & Schuell) were used instead of Whatman 50 filters. Positive colonies showed blue color in 15 min to 16 hr (overnight), whereas negatives remained white even after an overnight incubation.
Genetic screen to identify activators of CDRE-dependent expression
Under standard growth conditions, wild-type cells (ASY459) expressed enough β-galactosidase from the 4×-CDRE::lacZ reporter to make colonies blue when incubated with its chromogenic substrate, X-gal. This expression was calcineurin dependent, because calcineurin mutant cells (ASY461) incubated under these same conditions remained white. We confirmed that the lacZ reporter was functional because plasmids containing CNB1, thus complementing the cnb1Δ mutation, allowed for ASY461 to activate the reporter and turn blue. No β-galactosidase production was observed for strains containing the 4×-mutCDRE::lacZ reporter (ASY460 and ASY462).
Two high-copy genomic libraries, 2J351 (Hill et al. 1986; Engebrecht et al. 1990) and Y2HL (James et al. 1996), were screened for plasmids that allowed ASY461 to form blue colonies. Plasmids were harvested from blue colonies and then amplified in E. coli TOP10. These plasmids were then used to retransform the original strain (ASY461), a strain containing the mutCDRE reporter (ASY462), and a strain containing a lacZ reporter with no upstream activating sequence (ASY466). Because strains ASY462 and ASY466 were not able to support calcineurin-dependent transcriptional induction, they served as negative controls to eliminate positives that induced expression independently of an intact CDRE. Only plasmids were further characterized that specifically induced expression in strain ASY461 but not in strains ASY462 or ASY466 and therefore conferred CDRE-dependent gene expression in the absence of calcineurin. The ends of the genomic inserts were sequenced, and this sequence was used to scan the yeast genome to identify all ORFs contained within the insert.
DNA mobility retardation assays
The synthetic double-stranded XS-CDRE oligonucleotide (described above) was used as a probe in all binding experiments. XS-CDRE was end-labeled and passed over a G25 Sephadex column (Sigma) to separate out unincorporated nucleotides. XS-mutCDRE (also described above) contains multiple sequence changes and was shown to abolish transcriptional induction activity in vivo.
Cells were grown at 21°C in synthetic media and harvested when cultures had reached logarithmic growth (OD600 of 0.8–1.2). Protein extracts were prepared as described (Cox and Walter 1996). Binding reactions (10 μl) included ∼0.1 ng (3000 cpm) of radiolabeled CDRE oligonucleotide probe, 0.7 μg of poly[d(I-C)] (Pharmacia), and 1× binding buffer (20 mm HEPES at pH 7.9, 50 mm KCl, 2.5 mm EDTA, 5% glycerol, 0.5 mm dithiothreitol, 1 mm phenylmethanesulfonyl fluoride). Ten micrograms of crude extract (in 2 μl) was added last, and binding was allowed to proceed at room temperature for 10 min. Where appropriate, addition of 9 μl of antibody [either affinity-purified anti-HA from Boehringer Mannheim or anti-myc ascities fluid diluted 1:25 in antibody buffer (10 mm KPO4 and 70 mm NaCl)] occurred 5 min before gel loading to a final binding reaction volume of 20 μl, 1× in binding buffer. For competition assays, a 100-fold molar excess as compared with probe of unlabeled oligonucleotide (10 μg in 0.5 μl) was added before addition of the extract. The reactions were loaded on a 4% polyacrylamide gel containing 5% glycerol and electrophoresed at 100 V for 1 hr in 1× TBE at room temperature. Gels were dried and exposed to phosphor screens, and images were scanned using a Bio-Rad Molecular Imager System GS-363.
α-Factor viability assay
The viability of different strains during prolonged exposure to α-factor was assayed by determining their plating efficiency on solid growth media containing various amounts of synthetic α-factor (Star Biochemicals). This assay was performed essentially as described (Moser et al. 1996; see LD50 determination for α-factor).
Northern analysis
Northern analysis of total RNA prepared from yeast cells grown to exponential phase in YPD was performed according to established protocols (Ausubel et al. 1987). DNA fragments used for synthesizing probes to FKS2 and ACT1 were made as described (Mazur et al. 1995). Radioactive probes were synthesized using the Amersham rediprime kit and [α-32P]dCTP (Amersham) according to the manufacturer’s instructions. Washed filters were exposed to phosphor screens, images were scanned on a Bio-Rad Molecular Imager System GS-363, and the band intensities were quantified using the Molecular Analyst program.
Acknowledgments
We give special thanks to Tai Roe for providing pAMS312 and Daphna Axelrod for providing pAMS317. We also thank Phil Garrett-Engele, Joachim Li, Francisco Rubio, Philip James, Johannes Hegemann, and Scott Erdman for providing other invaluable reagents. In addition, we wish to thank Mohamad Moghadam and Bob Fisher for help with the gel-shift experiments and Kyle Cunningham for sharing results before publication. We are grateful to members of the Cyert laboratory for helpful discussions and support and to Tai Roe, Kirstie Saltsman, James Withee, Tim Stearns, Pamela Carroll, and Tamara Pozos for critical reading of the manuscript. M.S.C. is supported by a biomedical scholar award from the Lucille P. Markey Charitable Trust, National Science Foundation Young Investigator award MCB-9357017, funds from the Procter and Gamble Company, and National Institutes of Health (NIH) research grant GM48729. A.M.S. is supported by NIH training grant 5T32GM07276-22.
The publication costs of this article were defrayed in part by payment of page charges. This article must therefore be hereby marked “advertisement” in accordance with 18 USC section 1734 solely to indicate this fact.
Footnotes
E-MAIL mcyert@leland.stanford.edu; FAX (650) 725-8309.
References
- Aperia A, Ibarra F, Svensson L-B, Klee C, Greengard P. Calcineurin mediates alpha-adrenergic stimulation of Na+, K+-ATPase activity in renal tubule cells. Proc Natl Acad Sci. 1992;89:7394–7397. doi: 10.1073/pnas.89.16.7394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K, editors. Current protocols in molecular biology. 1 and 2. New York, NY: John Wiley and Sons; 1987. [Google Scholar]
- Clapham DE. Calcium signaling. Cell. 1995;80:259–268. doi: 10.1016/0092-8674(95)90408-5. [DOI] [PubMed] [Google Scholar]
- Clipstone NA, Crabtree GR. Identification of calcineurin as a key signalling enzyme in T-lymphocyte activation. Nature. 1992;357:695–697. doi: 10.1038/357695a0. [DOI] [PubMed] [Google Scholar]
- Cox JS, Walter P. A novel mechanism for regulating activity of a transcription factor that controls the unfolded protein response. Cell. 1996;87:391–404. doi: 10.1016/s0092-8674(00)81360-4. [DOI] [PubMed] [Google Scholar]
- Cunningham KW, Fink GR. Calcineurin-dependent growth control in Saccharomyces cerevisiae mutants lacking PMC1, a homolog of plasma membrane Ca2+ ATPases. J Cell Biol. 1994;124:351–363. doi: 10.1083/jcb.124.3.351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- ————— Calcineurin inhibits VCX1-dependent H+/Ca2+ exchange and induces Ca2+ ATPases in Saccharomyces cerevisiae. Mol Cell Biol. 1996;16:2226–2237. doi: 10.1128/mcb.16.5.2226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cyert MS, Thorner J. Regulatory subunit (CNB1 gene product) of yeast Ca2+/calmodulin-dependent phosphoprotein phosphatases is required for adaption to pheromone. Mol Cell Biol. 1992;12:3460–3469. doi: 10.1128/mcb.12.8.3460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cyert MS, Kunisawa R, Kaim D, Thorner J. Yeast has homologs (CNA1 and CNA2 gene products) of mammalian calcineurin, a calmodulin-regulated phosphoprotein phosphatase. Proc Natl Acad Sci. 1991;88:7376–7380. doi: 10.1073/pnas.88.16.7376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desjarlais JR, Berg JM. Toward rules relating zinc finger protein sequences and DNA binding site preferences. Proc Natl Acad Sci. 1992;89:7345–7349. doi: 10.1073/pnas.89.16.7345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douglas CM, Foor F, Marrinan JA, Morin N, Nielsen JB, Dahl AM, Mazur P, Baginsky W, Li W, el-Sherbeini M, Clemas JA, Mandela SM, Frommer BR, Kurtz MB. The Saccharomyces cerevisiae FKS1 (ETG1) gene encodes an integral membrane protein which is a subunit of 1,3-β-D-glucan synthase. Proc Natl Acad Sci. 1994;91:12907–12911. doi: 10.1073/pnas.91.26.12907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng W-K, Faucette L, McLaughlin MM, Cafferkey R, Koltin Y, Morris RA, Young PR, Johnson RK, Livi GP. The yeast FKS1 gene encodes a novel membrane protein, mutations in which confer FK506 and cyclosporin A hypersensitivity and calcineurin-dependent growth. Gene. 1994;151:61–71. doi: 10.1016/0378-1119(94)90633-5. [DOI] [PubMed] [Google Scholar]
- Engebrecht J, Hirsch H, Roeder GS. Meiotic gene conversion and crossing over: Their relationship to each other and to other chromosome synapsis and segregation. Cell. 1990;62:927–937. doi: 10.1016/0092-8674(90)90267-i. [DOI] [PubMed] [Google Scholar]
- Evan GI, Lewis GK, Ramsay G, Bishop JM. Isolation of monoclonal antibodies specific for human c-myc proto-oncogene product. Mol Cell Biol. 1985;5:3610–3616. doi: 10.1128/mcb.5.12.3610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Evan RM, Hollenberg SM. Zinc fingers: Gilt by association. Cell. 1988;52:1–3. doi: 10.1016/0092-8674(88)90522-3. [DOI] [PubMed] [Google Scholar]
- Farcasanu IC, Hirata D, Tsuchiya E, Nishiyama F, Miyakawa T. Protein phosphatase 2B of Saccharomyces cerevisiae is required for tolerance to manganese in blocking the entry of ions into the cell. Eur J Biochem. 1995;232:712–717. [PubMed] [Google Scholar]
- Garrett-Engele P, Moilanen B, Cyert MS. Calcineurin, the Ca2+/calmodulin-dependent protein phosphatase, is essential in yeast mutants with cell integrity defects and in mutants that lack a functional vascuolar H+-ATPase. Mol Cell Biol. 1995;15:4103–4114. doi: 10.1128/mcb.15.8.4103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guarente L, Mason T. Heme regulates transcription of the CYC1 gene of S. cerevisiae via an upstream activation site. Cell. 1983;32:1279–1286. doi: 10.1016/0092-8674(83)90309-4. [DOI] [PubMed] [Google Scholar]
- Güldener U, Heck S, Fiedler T, Beinhauer J, Hegemann JH. A new efficient gene disruption cassette for repeated use in budding yeast. Nucleic Acids Res. 1996;24:2519–2524. doi: 10.1093/nar/24.13.2519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hannon GJ, Demetrick D, Beach D. Isolation of the Rb-related p130 through its interaction with CDK2 and cyclins. Genes & Dev. 1993;7:2378–2391. doi: 10.1101/gad.7.12a.2378. [DOI] [PubMed] [Google Scholar]
- Haro R, Graciadebles B, Rodriguez-Navarro A. A novel P-type APTase from yeast involved in sodium transport. FEBS Lett. 1991;291:189–191. doi: 10.1016/0014-5793(91)81280-l. [DOI] [PubMed] [Google Scholar]
- Hendey B, Klee CB, Maxfield FR. Inhibition of neutrophil chemokinesis on vitronectin by inhibitors of calcineurin. Science. 1992;258:296–299. doi: 10.1126/science.1384129. [DOI] [PubMed] [Google Scholar]
- Hill JE, Myers AM, Koerner JJ, Tzagaloff A. Yeast/E. coli shuttle vectors with multiple unique restriction sites. Yeast. 1986;2:163–167. doi: 10.1002/yea.320020304. [DOI] [PubMed] [Google Scholar]
- Hubbard MJ, Klee CB. Functional domain structure of calcineurin A: Mapping by limited proteolysis. Biochemistry. 1989;28:1868–1874. doi: 10.1021/bi00430a066. [DOI] [PubMed] [Google Scholar]
- Iida H, Yagawa Y, Anraku Y. Essential role for induced Ca2+ influx followed by [Ca2+]i rise in maintaining viability of yeast cells late in the mating pheromone response pathway. J Biol Chem. 1990;265:13391–13399. [PubMed] [Google Scholar]
- Jain J, McCaffrey PG, Miner Z, Kerppola TK, Lambert JN, Verdine GL, Curran T, Rao A. The T-cell transcription factor NFATp is a substrate for calcineurin and interects with Fos and Jun. Nature. 1993;365:352–355. doi: 10.1038/365352a0. [DOI] [PubMed] [Google Scholar]
- James P, Halladay J, Craig EA. Genomic libraries and a host strain designed for highly efficient two-hybrid selection in yeast. Genetics. 1996;144:1425–1436. doi: 10.1093/genetics/144.4.1425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klee CB, Draetta GF, Hubbard MJ. Calcineurin. Adv Enzym. 1988;61:149–200. doi: 10.1002/9780470123072.ch4. [DOI] [PubMed] [Google Scholar]
- Kuno T, Tanaka H, Mukai J, Chang C, Hiraga K, Miyakawa T, Tanaka C. cDNA cloning of a calcineurin B homolog in Saccharomyces cerevisiae. Biochem Biophys Res Commun. 1991;180:1159–1163. doi: 10.1016/s0006-291x(05)81188-x. [DOI] [PubMed] [Google Scholar]
- Lapinskas PJ, Cunningham KW, Liu XF, Fink GR, Culotta VC. Mutations in PMR1 suppress oxidative damage in yeast cells lacking superoxide dismutase. Mol Cell Biol. 1995;15:1382–1388. doi: 10.1128/mcb.15.3.1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawson MA, Maxfield FR. Ca2+- and calcineurin-dependent recycling of an integrin to the front of migrating neutrophils. Nature. 1995;377:75–79. doi: 10.1038/377075a0. [DOI] [PubMed] [Google Scholar]
- Li JJ, Herskowitz I. Isolation of ORC6, a component of the yeast origin recognition complex by a one-hybrid system. Science. 1993;262:1870–1874. doi: 10.1126/science.8266075. [DOI] [PubMed] [Google Scholar]
- Lieberman DN, Mody I. Regulation of NMDA channel function by endogenous Ca2+-dependent phosphatase. Nature. 1994;369:235–239. doi: 10.1038/369235a0. [DOI] [PubMed] [Google Scholar]
- Liu J, Farmer JJD, Lane WL, Friedman J, Weissman I, Schreiber SL. Calcineurin is a common target of cyclophilin-CsA and FKBP-FK506 complexes. Cell. 1991a;66:807–815. doi: 10.1016/0092-8674(91)90124-h. [DOI] [PubMed] [Google Scholar]
- Liu Y, Ishii S, Tokai M, Tsutsumi H, Ohke O, Akada R, Tanaka K, Tsuchiya E, Fukui S, Miyakawa T. The Saccharomyces cerevisiae genes (CMP1 and CMP2) encoding calmodulin-binding proteins homologous to the catalytic subunit of mammalian protein phosphatase 2B. Mol & Gen Genet. 1991b;227:52–59. doi: 10.1007/BF00260706. [DOI] [PubMed] [Google Scholar]
- Marquez JA, Serrano R. Multiple transduction pathways regulate the sodium-extrusion gene PMR2/ENA1 during salt stress in yeast. FEBS Lett. 1996;382:89–92. doi: 10.1016/0014-5793(96)00157-3. [DOI] [PubMed] [Google Scholar]
- Matheos, D.P., T.J. Kingbury, U.S. Ahsan, and K.W. Cunningham. 1997. Tcn1p/Crz1p, a calcineurin-dependent transcription factor that differentially regulates gene expression in Saccharomyces cerevisiae. Genes & Dev. (this issue). [DOI] [PMC free article] [PubMed]
- Mazur P, Morin N, Baginsky W, El-Sherbeini M, Clemas JA, Nielsen JB, Foor F. Differential expression and function of two homologous subunits of yeast 1,3-β-D-glucan synthase. Mol Cell Biol. 1995;15:5671–5681. doi: 10.1128/mcb.15.10.5671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mendoza I, Rubio F, Rodriguez-Navarro A, Pardo JM. The protein phosphatase calcineurin is essential for NaCl tolerance of Saccharomyces cerevisiae. J Biol Chem. 1994;269:8792–8796. [PubMed] [Google Scholar]
- Miller J. Experiments in molecular genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1972. [Google Scholar]
- Moser MJ, Geiser JR, Davis TN. Ca2+-calmodulin promotes survival of pheromone-induced growth arrest by activation of calcineurin and Ca2+-calmodulin-dependent protein kinase. Mol Cell Biol. 1996;16:4824–4831. doi: 10.1128/mcb.16.9.4824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura T, Liu Y, Hirata D, Namba H, Harada S, Hirokawa T, Miyakawa T. Protein phosphatase type 2B (calcineurin)-mediated, FK506-sensitive regulation of intracellular ions in yeast is an important determinant for adaptation to high salt stress conditions. EMBO J. 1993;12:4063–4071. doi: 10.1002/j.1460-2075.1993.tb06090.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Northrop JP, Ullman KS, Crabtree GR. Characterization of the nuclear and cytoplasmic components of the lymphoid-specific nuclear factor of activated T cells (NF-AT) complex. J Biol Chem. 1993;268:2917–2923. [PubMed] [Google Scholar]
- O’Keefe SJ, Tamura J, Kincaid RL, Tocci MJ, O’Neill EA. FK506- and CsA-sensitive activation of the interleukin-2 promoter by calcineurin. Nature. 1992;357:692–694. doi: 10.1038/357692a0. [DOI] [PubMed] [Google Scholar]
- Perutz M. Polar zippers: Their role in human disease. Prot Sci. 1994;3:1629–1637. doi: 10.1002/pro.5560031002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pozos TC, Sekler I, Cyert MS. The product of HUM1, a novel yeast gene, is required for vascuolar Ca2+/H+ exchange and is related to mammalian Na+/Ca2+ exchangers. Mol Cell Biol. 1996;16:3730–3741. doi: 10.1128/mcb.16.7.3730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prestridge DS. SIGNAL SCAN: A computer program that scans DNA sequences for eukaryotic transcriptional elements. Comp Appl Biosci. 1991;7:203–206. doi: 10.1093/bioinformatics/7.2.203. [DOI] [PubMed] [Google Scholar]
- Rudolph HK, Antebi A, Fink GR, Buckley CM, Dorman TE, LeVitre J, Davidow LS, Mao JI, Moir DT. The yeast secretory pathway is perturbed by mutations in PMR1, a member of a Ca2+ ATPase family. Cell. 1989;58:133–145. doi: 10.1016/0092-8674(89)90410-8. [DOI] [PubMed] [Google Scholar]
- Schneider BL, Seufert W, Steiner B, Yang QH, Futcher AB. Use of polymerase chain reaction epitope tagging for protein tagging in Saccharomyces cerevisiae. Yeast. 1995;11:1265–1274. doi: 10.1002/yea.320111306. [DOI] [PubMed] [Google Scholar]
- Sherman F, Fink GR, Hicks JB. Methods in yeast genetics. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory; 1986. [Google Scholar]
- Sikorski RS, Heiter P. A system of shuttle vectors and yeast host strains designed for efficient manipulation of DNA in Saccharomyces cerevisiae. Genetics. 1989;122:19–27. doi: 10.1093/genetics/122.1.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanida I, Takita Y, Hasegawa A, Ohya Y, Anraku Y. Yeast Cls2p/Csg2p localized on the endoplasmic reticulum membrane regulates a non-exchangeable intracellular Ca2+ pool cooperatively with calcineurin. FEBS Lett. 1996;379:38–42. doi: 10.1016/0014-5793(95)01478-0. [DOI] [PubMed] [Google Scholar]
- Tong G, Shepherd D, Jahr CE. Synaptic desensitization of NMDA receptors by calcineurin. Science. 1995;267:1510–1512. doi: 10.1126/science.7878472. [DOI] [PubMed] [Google Scholar]
- Wilson IA, Niman HL, Houghten RA, Cherenson AR, Connolly ML, Lerner RA. The structure of an antigenic determinant in a protein. Cell. 1984;37:767–778. doi: 10.1016/0092-8674(84)90412-4. [DOI] [PubMed] [Google Scholar]
- Withee JL, Mulholland J, Jeng R, Cyert MS. An essential role of the yeast pheromone-induced Ca2+ siganl is to activate calcineurin. Mol Biol Cell. 1997;8:263–277. doi: 10.1091/mbc.8.2.263. [DOI] [PMC free article] [PubMed] [Google Scholar]